Abstract
Anthocyanin concentration is an important determinant of the colour of many fruits. In apple (Malus × domestica), centuries of breeding have produced numerous varieties in which levels of anthocyanin pigment vary widely and change in response to environmental and developmental stimuli. The apple fruit cortex is usually colourless, although germplasm does exist where the cortex is highly pigmented due to the accumulation of either anthocyanins or carotenoids. From studies in a diverse array of plant species, it is apparent that anthocyanin biosynthesis is controlled at the level of transcription. Here we report the transcript levels of the anthocyanin biosynthetic genes in a red-fleshed apple compared with a white-fleshed cultivar. We also describe an apple MYB transcription factor, MdMYB10, that is similar in sequence to known anthocyanin regulators in other species. We further show that this transcription factor can induce anthocyanin accumulation in both heterologous and homologous systems, generating pigmented patches in transient assays in tobacco leaves and highly pigmented apple plants following stable transformation with constitutively expressed MdMYB10. Efficient induction of anthocyanin biosynthesis in transient assays by MdMYB10 was dependent on the co-expression of two distinct bHLH proteins from apple, MdbHLH3 and MdbHLH33. The strong correlation between the expression of MdMYB10 and apple anthocyanin levels during fruit development suggests that this transcription factor is responsible for controlling anthocyanin biosynthesis in apple fruit; in the red-fleshed cultivar and in the skin of other varieties, there is an induction of MdMYB10 expression concurrent with colour formation during development. Characterization of MdMYB10 has implications for the development of new varieties through classical breeding or a biotechnological approach.
Keywords: anthocyanin, Malus × domestica, MdMYB10, regulation, transcription factor
Introduction
The accumulation of anthocyanin pigments in apple fruit is an important determinant of fruit quality. Usually restricted to the skin of apples, these pigments provide essential cultivar differentiation for consumers and are implicated in the health attributes of apple fruit (Boyer and Liu, 2004). Anthocyanins belong to the diverse group of ubiquitous secondary metabolites collectively known as flavonoids. In plants, flavonoids are believed to have a variety of functions, including defence and protection against light stress, and the pigmented anthocyanin compounds play an important physiological role as attractants in plant/animal interactions (Harborne and Grayer, 1994; Koes et al., 1994).
Differences in colour attributable to anthocyanins may be due to a number of factors including the number of hydroxyl groups on the B-ring, the sugars and acyl side groups (Harborne, 1967), the environment of the vacuole including its pH or the accumulation of specific metal ions (Brouillard, 1988), or cellular ultrastructure (Noda et al., 1994). One of the most common anthocyanin pigments is cyanidin, which, in the form of cyanidin 3-O-galactoside, is the pigment primarily responsible for red colouration in apple skin (Lancaster, 1992; Tsao et al., 2003). The enzymes operating in this biosynthetic pathway in apple have been well characterized (Honda et al., 2002; Kim et al., 2003).
In apple, pigment biosynthesis may be induced by light, particularly UV, and various stress treatments including cold (Dong et al., 1998). There is also evidence that the anthocyanin biosynthetic enzymes are coordinately induced during development of apple fruit (Lister and Lancaster, 1996). This suggests that expression of the genes encoding the biosynthetic enzymes is coordinately regulated by one or a few regulatory proteins.
There are numerous reports of the regulation of genes in the anthocyanin pathway by transcription factors (TFs), and collectively these have established that the components of the regulatory complex controlling anthocyanin biosynthesis are conserved in all higher plants (Holton and Cornish, 1995). MYB TFs have been shown to play an important role in transcriptional regulation of anthocyanins (reviewed in Martin and Paz-Ares, 1997). Plant MYBs have been implicated in controlling pathways as diverse as secondary metabolism (including the anthocyanin pathway), development, signal transduction and disease resistance (Jin and Martin, 1999). They are characterized by a structurally conserved DNA-binding domain consisting of single or multiple imperfect repeats; those associated with the anthocyanin pathway are of the two-repeat (R2R3) class. Regulation also appears to be specific to discrete subsets of structural genes, acting either early or late in the anthocyanin biosynthetic pathway (Davies and Schwinn, 2003). In the leaves of perilla (Perilla frutescens), coordinated transcriptional regulation controls virtually all steps of anthocyanin biosynthesis from the genes encoding chalcone synthase (CHS) to the final genes encoding anthocyanin transporter proteins (Saito and Yamazaki, 2002). In contrast, in grape (Vitis vinifera), specific regulation by MybA is restricted to the very late steps in anthocyanin production, including UDP-glucose:flavonoid 3-O-glycosyltransferase (UFGT), but not the earlier biosynthetic steps (Kobayashi et al., 2002).
There are 126 R2R3 MYB TFs in Arabidopsis thaliana L. Heynh, which can be divided on the basis of their sequence into 24 subgroups (Stracke et al., 2001). The Production of Anthocyanin Pigment 1(PAP1) MYB gene of Arabidopsis (Borevitz et al., 2000) falls into subgroup 10 (when the phylogeny of Stracke et al., 2001 is used), and its protein demonstrates a high degree of amino acid conservation with other known MYB regulators of anthocyanin production. When PAP1 was over-expressed in transgenic Arabidopsis, this led to up-regulation of a number of genes in the anthocyanin biosynthesis pathway from phenylalanine ammonia-lyase (PAL) to chalione synthase (CHS) and dihydroflavonol 4-reductase (DFR) (Borevitz et al., 2000; Tohge et al., 2005).
The R2R3 MYB factors that regulate anthocyanin biosynthesis have been shown to interact closely with basic helix-loop-helix (bHLH) TFs (Mol et al., 1996; Winkel-Shirley, 2001). Examples include the maize ZmC1 MYB and ZmB bHLH, the petunia AN2 MYB and AN1/JAF13 bHLHs, and the Antirrhinum Rosea1, Rosea2 and Venosa MYBs and the Mutabilis and Delila bHLHs (Goff et al., 1992; Goodrich et al., 1992; Mol et al., 1998; Schwinn et al., 2006). Over-expressing lines of PAP1 have elevated transcript levels of the TT8 gene (Genbank accession NM_117050) that encodes a bHLH protein involved in regulating condensed tannin and anthocyanin biosynthesis (Nesi et al., 2000; Tohge et al., 2005). The dependency of these specific MYB-related proteins on a bHLH co-regulator to activate transcription is linked to the presence of a small number of amino acid residues (a signature motif) in the highly conserved R2R3 binding domain of the MYB proteins, as revealed by comparison between the structure of the maize P protein (which activates in a bHLH-independent manner) and the bHLH-dependent maize C1 protein (Grotewold et al., 2000). Substitution of just six amino acids from the R2R3 domain of MYB C1 into the corresponding positions in MYB P resulted in a mutant with bHLH-dependent behaviour, similar to C1. More recently, it was suggested that this might be a key mechanism that allows MYBs to discriminate between target genes (Hernandez et al., 2004).
An additional level of anthocyanin regulation involves a separate class of proteins, containing WD40 domains, which form complexes with MYB and bHLH proteins (Baudry et al., 2004, and reviewed in Ramsay and Glover, 2005). Examples include an11 in petunia (de Vetten et al., 1997) and TTG1 in Arabidopsis (Walker et al., 1999). However, a ubiquitous role for the WD repeat in the transcriptional complex regulating anthocyanin biosynthesis has yet to be demonstrated. Indeed, in maize, the WD repeat protein appears to function in regulating anthocyanin biosynthesis only in kernels. It may be that some bHLH proteins are more dependent on the participation of WD repeat proteins in the regulatory complex than others.
Although studies into the activation and repression of anthocyanin biosynthesis in apple fruit have shown developmental and environmental control, to date this has not been characterized at the level of transcriptional regulation. The control of anthocyanin accumulation in apple is a key question in understanding and manipulating the colour of the fruit. Identification of the factors that exert this control will provide tools for moderating the extent and distribution of anthocyanin-derived pigmentation in fruit tissue. In this study, we compared two genotypes, the white-fleshed commercial cultivar Malus × domestica‘Sciros’ (Pacific RoseTM) and the red-fleshed cultivar Malus × domestica‘Red Field’ Open Pollinated (‘Red Field’), which is highly pigmented throughout all tissues including fruit (skin and cortex) and leaves. We were able to isolate the cDNA of a subgroup 10 MYB, MdMYB10, from the highly pigmented red-fleshed cultivar. We compared expression of the major anthocyanin biosynthetic genes, and our candidate TF, in the different apple varieties by quantitative RT-PCR (qPCR). From our apple EST database (Newcomb et al., 2006), several candidate bHLH partners were also isolated. We further show that this transcription factor is functionally active in driving anthocyanin accumulation in both heterologous and homologous systems, generating pigmented tobacco leaf patches using transient transformation methods and highly pigmented anthocyanin-rich apple plants using stable transformation techniques. In expression analysis studies, the behaviour of MdMYB10 suggests that it is responsible for red apple fruit colour.
Results
The RNA expression profile of anthocyanin biosynthetic enzymes suggests coordinated regulation in apple fruit
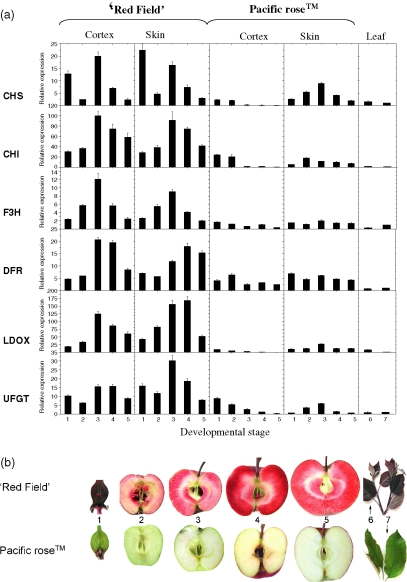
The levels of transcripts encoding six anthocyanin biosynthetic genes were compared between white-fleshed Pacific RoseTM and red-fleshed ‘Red Field’ apples by qPCR. Striking differences were observed. For all the genes assayed, transcript levels in ‘Red Field’ were significantly higher during all stages of fruit development than the levels found in Pacific RoseTM (Figure 1a). Transcript abundance in ‘Red Field’ was enhanced throughout fruit development in both skin and cortex, with the highest transcript levels at stage 3 (102 days after full bloom, DAFB). This correlated with the degree of pigmentation observed during tissue sampling, with the most intense pigmentation being observed early in development (40 DAFB) and then again in mid-summer (102 DAFB). These levels were subsequently sustained through to fruit maturation in late summer (Figure 1b). In Pacific RoseTM cortex tissue, there were comparatively low transcript levels for all the anthocyanin biosynthesis genes assayed, with a general decline in expression during fruit development. Moderate activity was observed in the skin of Pacific RoseTM, with a peak of expression midway through development (102 DAFB), concomitant with the onset of enhanced levels of pigmentation during fruit maturation.
Figure 1.
Expression profiling of apple anthocyanin genes.
(a) Data from qPCR analysis of the apple anthocyanin biosynthetic genes in the cortex, skin and leaf of the apple cultivars ‘Red Field’ and Pacific RoseTM. CHS, chalcone synthase; CHI, chalcone isomerase; F3H, flavanone 3β-hydroxylase; DFR, dihydroflavonol 4-reductase (denoted as DFR1 in the text); LDOX, leucoanthocyanidin dioxygenase; UFGT, uridine diphosphate (UDP)-glucose:flavonoid 3-O-glycosyltransferase. Samples referred to on the x axis: (1) 40 days after full bloom (DAFB), (2) 67 DAFB, (3) 102 DAFB, (4) 130 DAFB, (5) 146 DAFB, (6) ‘Red Field’ leaf and (7) Pacific RoseTM leaf. Error bars are SE for three replicate reactions.
(b) Fruit development series and leaves of ‘Red Field’ and Pacific RoseTM. Numbering from 1 to 7 as for (a).
Expression of all genes was detectable in the leaves of both cultivars but was relatively low, with little difference between ‘Red Field’ and Pacific RoseTM. As multiple copies of these biosynthetic genes exist in apple, it is possible that different family members or alleles may be differentially expressed in such diverse tissues as fruit and leaf. To test this, two family members of DFR were identified, denoted as DFR1 (Genbank accession AF117268) (shown in Figure 1) and DFR2 (Genbank accessionDT040651), and were assayed with gene-specific primers. Transcript levels of DFR1 were shown to be up-regulated in pigmented fruit tissue but not to any great extent in leaf (as shown in Figure 1). In contrast, DFR2 showed little or no activity in fruit but was highly elevated in the leaves of ‘Red Field’ (Figure S1).
Cloning of transcription factor candidates from pigmented apple flesh
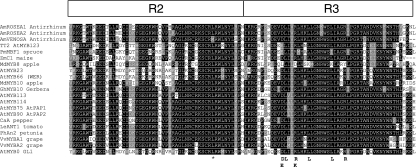
Previous studies, in other species, have shown that subgroup 10 MYBs are key determinants of plant pigmentation. Within publicly available apple EST databases (199 900 nucleotide sequences in April 2006); there were no MYB TFs showing high homology in BLAST searches to Arabidopsis PAP1 or subgroup 10 MYBs from other species. The qPCR results for gene transcripts encoding the enzymes of the anthocyanin biosynthetic pathway were analysed to determine the most suitable tissue/time point for relevant MYB transcription factor isolation. We chose tissue from the cortex of ‘Red Field’ that showed the highest expression levels for the anthocyanin biosynthesis genes, and then applied a degenerate PCR approach to gene isolation. Overlapping sequence alignments of cDNAs cloned after PCR showed that the best candidate, MdMYB10, when translated into protein, shared a high degree of homology with other MYB TFs at the R2R3 domain, and, in particular, with anthocyanin regulators from other species (Figure 2).
Figure 2.
Protein sequence alignment of the R2R3 DNA-binding domains of MdMYB10 and other known anthocyanin MYB regulators from other species. Arrows indicate specific residues that contribute to a motif implicated in bHLH co-factor interaction in Arabidopsis (Zimmermann et al., 2004). These same residues are evident in MdMYB10, suggesting a similar protein–protein interaction. The asterisk represents a two-nucleotide change that results in a leucine to a glutamine amino acid change in some alleles. Identical residues are shown in black, conserved residues in dark grey, and similar residues in light grey. MdMYB8 and AtGL1 are included as examples of MYBs that are not involved in anthocyanin regulation. The accession number of these proteins, or translated products, in the GenBank database are as follows: AmROSEA1, ABB83826; AmROSEA2, ABB83827; AmVENOSA, ABB83828; MdMYB10, DQ267896; AtPAP1, CAB09230; AtPAP2, NP176813; AtMYB113, NM105308; AtMYB114, NM105309; AtMYB66, NM121479; VvMYBA1, AB242302; VvMYBA2, AB097924; Ca A, CAE75745; PhAN2, AAF66727; LeANT1, AAQ55181; GhMYB10, CAD87010; PmMBF1, AAA82943; ZmC1, AAK81903; AtTT2, Q9FJA2; MdMYB8, DQ267899; AtGL1, AAC97387.
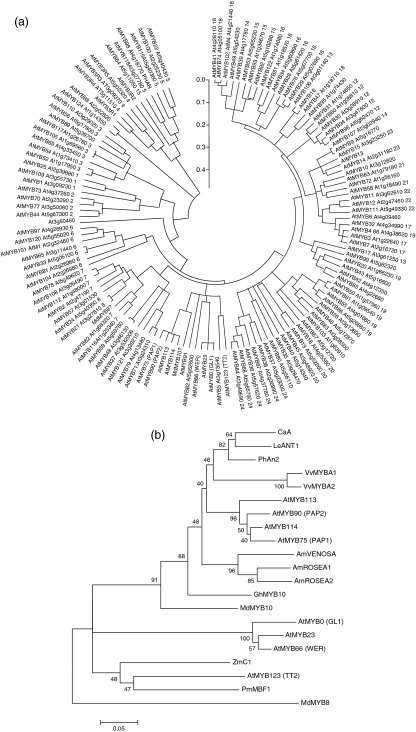
MdMYB10 is closely related to the Arabidopsis subgroup 10 MYBs, including PAP1, with 77% amino acid identity over the R2R3 DNA-binding domain and 58% identity to the entire protein. For Arabidopsis PAP2, these amino acid percentage identities are 75% and 57%, respectively, whilst for other species figures for overall identity are as follows: petunia AN2 60%, tomato ANT1 57%, maize C1 58% and maize P 26%. A bootstrapped circular phylogenic tree, generated using MEGA version 3.1 (Kumar et al., 2004), shows MdMYB10 in the same clade as Arabidopsis PAP1, PAP2, AtMYB113 and AtMYB114 (Figure 3a). MdMYB10 clusters with known MYBs involved in anthocyanin regulation from a diverse range of species (Figure 3b). Sequences of MdMYB10 from three cultivars, ‘Red Field’ (GenBank accession number DQ267896), Pacific RoseTM (DQ267897) and Granny Smith (DQ267989), were analysed. Within the coding region there were only minor nucleotide differences, which produced no changes in the predicted amino acids encoded. The only exception is a two-base change resulting in a leucine to glutamine amino acid change in helix 3 of the R2 domain (see marked residue in Figure 2). Both versions appear in the ‘Red Field’ and Pacific RoseTM cultivars.
Figure 3.
Phylogenetic relationships between Arabidopsis MYB transcription factors and anthocyanin-related MYBs of other species.
(a) MdMYB10 clusters next to PAP1 (AtMYB75), within the anthocyanin MYB regulator subgroup 10. Subgroup numbers are those described by Stracke et al. (2001) and are shown as a suffix after most MYB descriptors. Arabidopsis genes are identified by Arabidopsis unique identifiers. (b) Phylogeny of MdMYB10 and MYB genes from other species involved in the regulation of anthocyanin biosynthesis. Full-length sequences were aligned using Clustal W (opening = 15, extension = 0.3) in Vector NTI 9.0. Conserved motifs were extracted and re-aligned as above. Phylogenetic and molecular evolutionary analyses were conducted using MEGA version 3.1 (Kumar et al., 2004) using a minimum evolution phylogeny test and 1000 bootstrap replicates. Accession numbers for genes from other species are given in Figure 2.
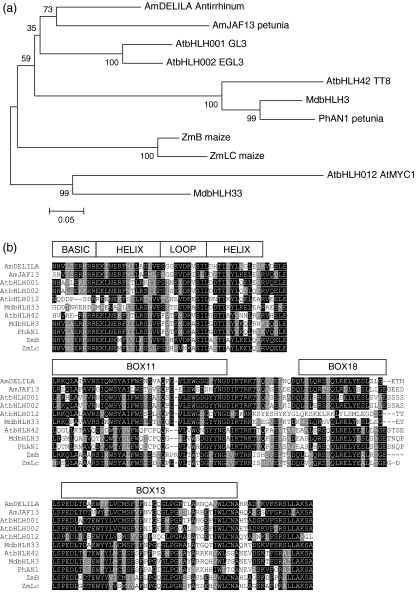
MdMYB10 has the amino acid residues that specify interaction with bHLHs (arrows on Figure 2) (Grotewold et al., 2000; Zimmermann et al., 2004). Candidates for bHLH co-factors were therefore selected from the HortResearch EST database (Newcomb et al., 2006). In the large phylogenetic family that constitutes the bHLH type TF, there is a smaller clade termed IIIf (Heim et al., 2003) that has been shown to be involved in the regulation of anthocyanin biosynthesis. Two apple ESTs encoded bHLH TFs that clustered within this clade (Figure 4a). These ESTs were sequenced and given the identifiers MdbHLH3 (GenBank accession number CN934367), a putative homologue of the Arabidopsis TT8 gene, and MdbHLH33 (GenBank accession number DQ266451), a putative homologue of Delila (from Antirrhinum, Goodrich et al., 1992). N-terminal domains identified as important for MYB interaction (Heim et al., 2003) were present in both MdbHLH3 and MdbHLH33 (Figure 4c).
Figure 4.
MdbHLH3 and MdbHLH33 share homology at the bHLH motif with anthocyanin bHLH regulators from other species.
(a) Unrooted phylogenic tree showing that MdbHLH3 clusters in the same group as AtTT8 whilst MdbHLH33 clusters in the same group as AtMYC1. Full-length sequences were aligned using Clustal W (opening = 15, extension = 0.3) in Vector NTI 9.0. Conserved motifs were extracted and re-aligned as above. Phylogenetic and molecular evolutionary analyses were conducted using MEGA version 3.1 (Kumar et al., 2004) using a minimum evolution phylogeny test and 1000 bootstrap replicates.
(b) Protein sequence alignment, with the structure of the bHLH binding domain indicated. The accession numbers of these proteins, or translated products, in the GenBank database are as follows: AmDELILA, AAA32663; ATMYC1, BAA11933; EGL1, Q9CAD0; G13, NP680372; PhJAF13, AAC39455; MdbHLH3, CN934367; MdbHLH33, DQ266451; PhAN1, AAG25928; AtTT8, CAC14865; ZmB, CAA40544; ZmLC, AAA33504.
(c) N-terminus of the same bHLH subset showing regions (boxes 11, 18 and 13) conserved within the bHLH IIIf clade (according to Heim et al., 2003). Identical residues are shown in black, conserved residues in dark grey, and similar residues in light grey.
Transient luminescent assays of apple transcription factor activity
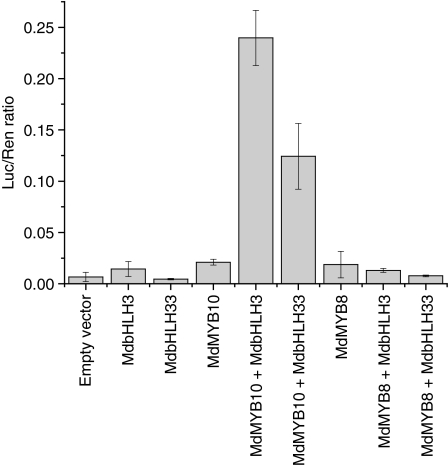
The dual luciferase system has been demonstrated to provide a rapid method of transient gene expression analysis (Hellens et al., 2005; Matsuo et al., 2001). It requires no selectable marker and results can be quantified by a simple enzymatic assay. We used Nicotiana tabacum to test the interaction of our candidate TFs with an Arabidopsis anthocyanin biosynthesis gene promoter AtDFR (TT3, GenBank accession numberNM_123645). This is known to be regulated by Arabidopsis PAP1 and PAP2 MYB TFs (Tohge et al., 2005; Zimmermann et al., 2004). To investigate the interaction between MYB and bHLH TFs, co-transformations were performed with bHLH class putative regulators from apple: MdbHLH3 and MdbHLH33. The results from the promoter assay indicated a significant increase in activity when MdMYB10 was co-transformed with an apple bHLH (Figure 5). The highest activity was observed when MdMYB10 was co-infiltrated with MdbHLH3, although MdbHLH33 also activated MdMYB10. These results reflect previous work in a transient protoplast transfection system in which an Arabidopsis DFR promoter:GUS fusion was activated by PAP1 only in the presence of a bHLH (Zimmermann et al., 2004). The apple MYB MdMYB8 (which falls in subgroup 8, Figure 3) was selected to probe the specificity of MdMYB10 against the DFR promoter: MdMYB8 activity was low under all conditions and showed no activation by either bHLH (Figure 5).
Figure 5.
Interaction of MdMYB10 and apple bHLH TFs in transient tobacco transformation assays affects the activity of the DFR gene promoter.
The dual luciferase assay shows promoter activity expressed as a ratio of DFR promoter luciferase (LUC) to 35S Renilla (REN), where an increase in activity equates to an increase in LUC relative to REN. Error bars are the SE for six replicate reactions.
Transient colour assays of apple transcription factor activity
We have established a simple method to reveal anthocyanin pigment accumulation in Nicotiana tabacum via Agrobacterium infiltration. Subgroup 10 MYBs and anthocyanin-related bHLHs were syringe-infiltrated into the underside of expanding tobacco leaves. Pigmentation was evident at infiltration points as early as 4 days post-infiltration for MdMYB10 when co-infiltrated with an apple bHLH (Figure 6a). The degree of pigmentation increased over the experimental period of up to 10 days.
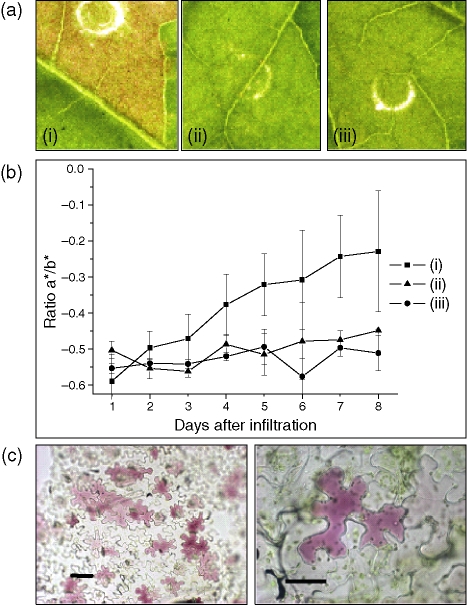
Figure 6.
Development of colour due to transient transformation of Nicotiana tabacum leaves.
(a) Digital images of infiltration sites 8 days after transformation with: (i) MdMYB10 + MdbHLH3, (ii) MdMYB10 alone, and (iii) empty vector control.
(b) Colour measurement by Minolta chromameter, shown as a*/b* ratio. A shift towards positive indicates a colour change from green towards red; (i) to (iii) as above. Error bars are the SE for six replicate reactions.
(c) Microscope images showing anthocyanin accumulation in tobacco epidermal cells infiltrated with MdMYB10 + MdbHLH3 at magnifications of 20× (left) and 40× (right). Scale bars = 50 μm.
Pigmentation was also observed with infiltration of MdMYB10 alone (Figure 6a), but not when either apple bHLH or MdMYB8 alone were infiltrated (data not shown). The results demonstrate the efficacy of this assay as a useful visual reporter system to study the regulation of the pigmentation processes.
Colour intensity and hue were quantified by measurement with a Minolta chromometer, using the L*a*b* system. This confirmed the visible transition from green to red. The data are shown as a ratio of a*/b* (Figure 6b), where the change from negative towards positive indicates a shift from green to red. It should be noted that there was variability in pigmentation between replicates of a given treatment, particularly between leaves of different ages (Figure 6b).
To verify cellular build-up of anthocyanin compounds, microscopic images were obtained from epidermal peels, removed from the abaxial surface of the leaf, 1 week after co-inoculation with MdMYB10 and MdbHLH3 (Figure 6c). This illustrated the transformation of individual cells with the candidate genes and the accumulation of anthocyanin pigments within the vacuoles of epidermal cells.
HPLC analysis of anthocyanins in tobacco and apple
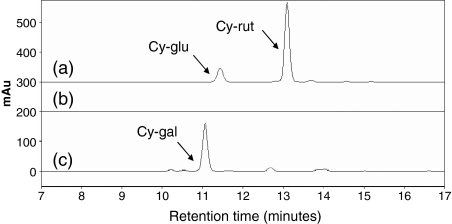
To confirm the identity of the anthocyanins synthesized after transformation of tobacco with selected TFs, samples of leaf tissue were removed and extracted, and the soluble anthocyanins analysed by HPLC. When MdMYB10 and MdbHLH3 were co-transformed in tobacco leaves, two major peaks were observed (Figure 7a), representing cyanidin-3-glucoside (LC-MS molecular ion [M+] with a mass of 449 Da; major fragment of 287 Da corresponding to the M+ of cyanidin with a ‘neutral loss’ of 162 Da corresponding to a hexose sugar residue) and cyanidin-3-0-rutinoside (LC-MS M+ at 595 Da, with major fragments of 449 Da [cyanidin hexoside], 287 Da [cyanidin] and neutral losses of 146 Da [deoxyhexose sugar; probably rhamnose residue] and a 162 Da hexose sugar residue). No observable anthocyanin peaks were found in the extracts of tobacco leaf transformed with Agrobacterium carrying empty vector control alone (Figure 7b). To compare with compounds naturally occurring in apple, anthocyanins from the skin of apple (Pacific RoseTM, mature fruit) were also extracted. HPLC confirmed the predominance of cyanidin-3-galactoside (LC-MS as for glucoside above) in apple skin, with traces of cyanidin glucoside and three pentosides (Figure 7c) as previously described (Tsao et al., 2003).
Figure 7.
Anthocyanin profiles of N. tabacum and apple.
HPLC traces at 520 nm of (a) MdMYB10 + MdbHLH3, (b) tobacco leaf infiltrated with empty vector control, and (c) Pacific RoseTM apple fruit skin. Peaks identified are as follows: cy-glu, cyanidin-3-glucoside; cy-rut, cyanidin-3-O-rutinoside; cy-gal, cyanidin-3-galactoside.
An HPLC comparison (not shown) of apple flesh phenolic extracts showed that the phenolic profile at 280 nm of ‘Red Field’ was almost identical to that of Pacific RoseTM, with the exception that anthocyanins were present only in the flesh of the former. No significant change in levels of phenolic acids, flavonoids, chalcones or procyanidins was observed.
Generation of 35S-MdMYB10 transgenic apple plants
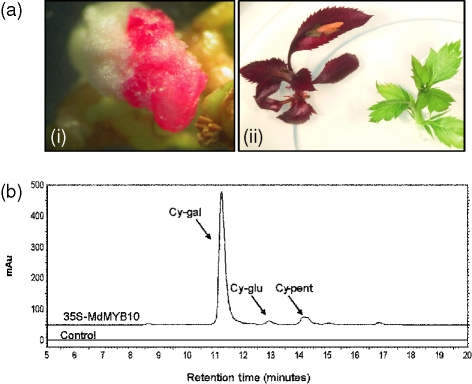
To test the function of MdMYB10 in a homologous system, apple explants (cultivar ‘Royal Gala’) were transformed with 35S-MdMYB10 and a kanamycin selectable marker. Regenerated callus of kanamycin-positive transformants was highly pigmented (Figure 8a). Shoots regenerated from this callus maintained colour, whilst control plants transformed with an empty vector consistently showed no pigmentation at either the callus or regenerated shoot stages. qPCR analysis confirmed a massive elevation of MdMYB10 transcript level but no apparent elevation in either MdbHLH3 or MdbHLH33 transcript levels (Figure S2). The suggestion that different gene family members of biosynthetic enzymes are leaf- or fruit-specific was confirmed in these transgenic plants. MdDFR1 showed a slight increase in transcript levels in the 35S-MdMYB10 transgenic lines compared to the control plants, and MdDFR2 showed even higher levels (Figure S1). HPLC analysis of MdMYB10 transgenic apple leaves showed a predominant peak of cyanidin-3-galactoside (Figure 8b) and confirmed the presence of smaller amounts of cyanidin glucoside and three derivatives of cyanidin pentoside.
Figure 8.
Over-expression of MdMYB10 in apple elevates anthocyanin production.
(a) Pigmented callus (i) and apple plants (ii) transformed with 35S-MdMYB10 (left) and empty vector control plant (right).
(b) Anthocyanin profiles of extracts of 35S-MdMYB10 apple leaf (top line) and empty vector control (bottom line). Peaks identified from HPLC traces at 520 nm: cy-gal, cyanidin-3-galactoside; with minor traces of cy-glu, cyanidin-3-glucoside and cy-pent, cyanidin-3-pentoside.
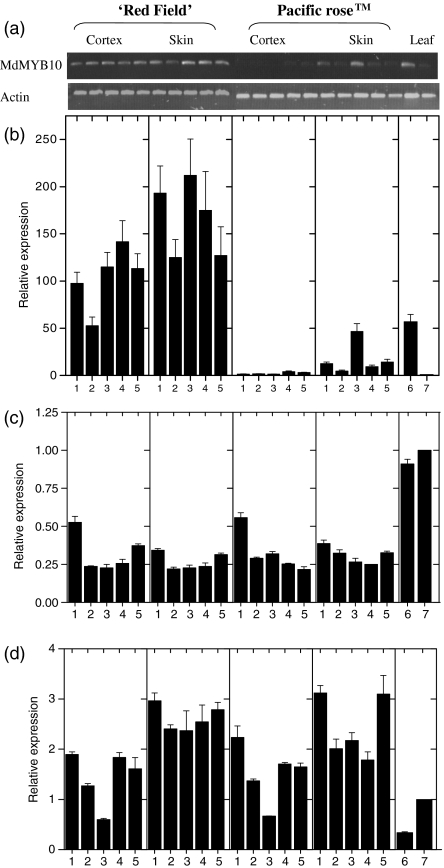
Quantitative PCR expression analysis of MdMYB10 in apple fruit
qPCR analysis of the expression of TF gene transcripts in the developmental series from both red- and white-fleshed varieties of apple revealed massive increases in the relative transcript levels of MdMYB10 in the fruit tissues of ‘Red Field’ compared with Pacific RoseTM (Figure 9). In Pacific RoseTM cortex, transcript levels were barely detectable, whilst in Pacific RoseTM skin, transcript was detectable only by mid-season (Figure 9), and the level of transcript correlated with changes in the transcript levels of the biosynthetic genes, particularly at the 102 DAFB time point. The link between MdMYB10 expression and UFGT expression was particularly strong (Figure 1a). Expression levels of MdMYB10 in ‘Red Field’ generally followed the transcript patterns of the assayed genes encoding enzymes, with highly elevated levels throughout fruit tissues, particularly at 40 DAFB and then again at 102, 130 and 146 DAFB (Figure 9) and in leaf tissue. To confirm specificity, qPCR amplicons were sequenced and found to encode MdMYB10.
Figure 9.
Expression analysis of the transcripts of MdMYB10, MdbHLH3 and MdbHLH33 during apple fruit development and in leaves.
(a) RT-PCR analysis of MdMYB10 (220 bp) and actin (131 bp) in ‘Red Field’ (cortex, skin and leaf) and Pacific RoseTM (cortex, skin and leaf), and corresponding qPCR data for MdMYB10 (b), MdbHLH3 (c) and MdbHLH33 (d). Gel lane and x axis numbering as follows: (1) 40 DAFB, (2) 67 DAFB, (3) 102 DAFB, (4) 130 DAFB, (5) 146 DAFB, (6) ‘Red Field’ leaf and (7) Pacific RoseTM leaf. Error bars shown in qPCR data are of the SE for three replicate reactions.
Transcript levels of MdbHLH3 and MdbHLH33 were also assayed (Figure 9c,d) but did not appear to follow the pattern displayed for the biosynthetic genes, or for MdMYB10, with more consistent levels of expression throughout the developmental series and in both cultivars. Again, amplicons were sequenced to verify authenticity. Transcript levels of MdMYB8 (GenBank accession number DQ267899), an apple MYB unrelated to the anthocyanin subgroup 10 (Figure 3), were also analysed and no correlative patterns with the transcript levels of the anthocyanin biosynthetic genes were observed (data not shown).
Discussion
Apple anthocyanin biosynthesis is coordinately regulated
Modern domesticated cultivars of apple show differential anthocyanin accumulation during development. Specific anthocyanin pigments accumulate in floral tissues and during fruit maturation in apple skin. Environmental factors also cause increases in the expression of genes encoding anthocyanin biosynthetic enzymes, as demonstrated by their responsiveness to light (Kim et al., 2003). It is possible that white-fleshed apples have lost the ability to produce pigmentation throughout the fruit due to a mutation in one or more biosynthetic gene(s), or in the regulatory genes as has been observed for white grapes (Boss et al., 1996). In terms of the genes encoding the biosynthetic enzymes, we detected some transcript in both cortex and skin of white-fleshed Pacific RoseTM, but at low levels (Figure 1a). A previous study in apple showed that all the late anthocyanin biosynthetic genes are present in the genome of a green cultivar but that transcripts were barely detectable by RNA gel-blot analysis (Kim et al., 2003). Our data differ from these results in that we were able to detect expression of at least six genes, including the late biosynthetic genes, in non-pigmented apple cortex. This is probably due to the enhanced sensitivity of qPCR over RNA gel-blot analysis. In grape, it has been shown that the only difference in expression of the biosynthetic genes in red grape skin versus white grape skin was the loss of expression of UFGT in the latter (Boss et al., 1996).
The anthocyanin genes in ‘Red Field’ followed a pattern whereby the expression levels of the biosynthetic gene transcripts peaked at 102 and 130 DAFB. There were large increases in all expressed genes in the red-fleshed compared to the white-fleshed cultivar. Only CHS differed significantly from this general pattern, with the highest expression seen early in the development of ‘Red Field’ fruit skin (40 DAFB), although expression did rise again at 102 DAFB. It is likely that at least some of the anthocyanin biosynthetic genes are differentially expressed according to tissue type. This is demonstrated by the transcript levels of the two versions of DFR measured: MdDFR1 showed a large variation in differently pigmented fruit tissues but not in differently pigmented leaf tissue, whilst MdDFR2 demonstrated the reverse.
Transcription factors controlling apple colour
Sequence analysis of the MdMYB10-encoded protein showed that it shares homology with anthocyanin-regulating MYB TFs from other species (Figure 2). Its probable orthology to PAP1 is supported by its position in the same clade of the MYB TF family (Figure 3a). Further analysis of MdMYB10 cDNAs from two other apple cultivars showed a high degree of sequence conservation between such strikingly different varieties as ‘Granny Smith’ (white cortex and predominantly green skin) and the highly pigmented ‘Red Field’. In grape, a retrotransposon-induced insertion into the gene encoding the grape MYB-related VvmybA1 TF has resulted in a lack of anthocyanin production, generating white grape cultivars (Kobayashi et al., 2004). The similarity between the sequences of MdMYB10 in these three apple cultivars suggests that the dramatic difference in anthocyanin accumulation in apple is not due to alterations in the coding sequence of this regulator. However, the difference in anthocyanin accumulation appears to be due to differences in the expression levels and pattern of MdMYB10 in the two varieties.
Expression analysis of MdMYB10 showed a pattern very similar to that observed for the genes encoding the biosynthetic enzymes in ‘Red Field’ apples. A large increase in expression of MdMYB10 in the fruit of this variety suggests that it is likely to be the regulator of the apple anthocyanin pathway. This association is further strengthened by expression analysis of MdMYB10 in Pacific RoseTM; it was virtually undetectable in leaf and cortex but was expressed in the pigmented skin, particularly at 102 DAFB, when colour was developing. We did not detect evidence for increased transcript levels for the two bHLH genes assayed, suggesting that, in apple, neither of these genes is positively regulated by MdMYB10. This is in contrast to the results for over-expressing PAP1 in Arabidopsis (Tohge et al., 2005), where PAP1 appears to up-regulate its bHLH interactor. The proposed model for TTG1-dependent (WD40-containing) transcriptional networks (Zhang et al., 2003) predicts increasing specificity moving down the hierarchy from WD40 through the bHLHs and down to the MYBs. Mutations in TTG1 (encoding a WD40-containing protein) affect both anthocyanin levels and epidermal cell fate (Walker et al., 1999). The MYB component of this WD40–bHLH–MYB complex appears to provide the most anthocyanin-specific effect. Our data suggest that, in apple, the MYB component, MdMYB10, is the limiting feature for pigment accumulation.
Evidence for MYB:bHLH interaction
A signature motif for the interaction between MYB and bHLH proteins, based on specific residues within the R2R3 DNA-binding domain of the MYB partner, has been proposed for maize and Arabidopsis and confirmed experimentally (Grotewold et al., 2000; Zimmermann et al., 2004). MdMYB10 shares a similar residue pattern to the bHLH-dependent maize C1 protein (unlike the bHLH-independent maize MYB-related P protein). We identified the interaction signature motif in MdMYB10, and confirmed, using transient assays, that the protein interacts with bHLH proteins to activate transcription of DFR.
The results from transient assays in tobacco showed interaction between MdMYB10 and two selected apple bHLH candidates (Figures 5 and 6). Whilst MdbHLH3, when co-infiltrated with MdMYB10, does appear to have a greater effect than MdbHLH33 on the activity of the DFR promoter (Figure 5), further work is required to confirm whether either of these associations is significant in apple tissue. One possible reason for a background level of anthocyanin accumulation with MdMYB10 alone may be because there is an endogenous tobacco bHLH protein working within this transient assay. Using this assay, transient expression demonstrated that MdMYB10 is effective at driving anthocyanin accumulation (Figure 6). In the presence of an apple co-activator, this accumulation was intensified and the colour appeared several days earlier, demonstrating partial dependence of MdMYB10 activity on the presence of an apple bHLH protein in this heterologous system.
Tobacco foliar anthocyanin accumulation
We have used transient transformation of tobacco leaves as an assay for the anthocyanin pathway. HPLC data and microscopy show significant accumulation of anthocyanin compounds, driven by MdMYB10 and a bHLH, confirming the functional presence of anthocyanin biosynthetic enzymes in tobacco leaves and the cellular machinery required to transport anthocyanins to the vacuole. Transient expression of MdMYB10 in tobacco results in major anthocyanin peaks representing cyanidin-3-galactoside and cyanidin-3-0-rutinoside. Accumulation of an alternative compound to that normally observed in apple is a probable consequence of differences in the genes encoding the biosynthetic enzymes available in the two species. However, this apple TF-driven enhancement ably demonstrates the proposed conservation of MYB regulators between the species.
Anthocyanin accumulation in 35S-MdMYB10 apple and the ‘Red Field’ red-fleshed cultivar
Over-expression of MdMYB10 in apple generated a strong phenotype, with highly pigmented plants (Figure 8) due to enhanced levels of anthocyanin. No anthocyanin was detected in control plants by HPLC analysis in contrast to the considerable level of anthocyanins in the MdMYB10 plants. The effect of MdMYB10 over-expression on the pigmentation of fruit cortex will be apparent when these plantlets have fruited. In naturally occurring red-fleshed apples, high levels of MdMYB10 expression appear to be the cause of this startling phenotype (Figure 9). Further, genetic mapping of an MdMYB10-derived molecular marker indicates that this locus co-segregates with the red-fleshed apple phenotype (D. Chagné, HortResearch, Auckland, New Zealand, personal communication).
Conclusion
The combination of bioinformatic, expression and functional data suggests that MdMYB10 is the apple orthologue of known anthocyanin regulators from other species, such as Arabidopsis PAP1. Over-expression of MdMYB10 in apple is reminiscent of PAP1 transgenic Arabidopsis plants (Borevitz et al., 2000; Xie et al., 2006). Isolation of this gene provides a crucial step in understanding the molecular regulation of anthocyanin gene transcription in apple fruit. The high levels of expression of MdMYB10 in the cortex of the red apple variety ‘Red Field’ suggest that it is differences in the activity of this gene that determine the colour differences between the cortex tissues in these varieties. These differences in activity appear to result from differences in expression of the MdMYB10 gene, as the coding sequences of this gene are virtually identical in the two varieties. Once again, a MYB transcription factor appears to be the primary determinant of differences in colour between cultivated plant varieties, as has been reported for potato, tomato, pepper (De Jong et al., 2004) and grape (Kobayashi et al., 2004), and in various species of Antirrhinum (Schwinn et al., 2006). These results confirm the previous findings that it is the MYB component from the proposed regulatory complex of MYB–bHLH–WD40 that is primarily (and specifically) responsible for the level of anthocyanin accumulation. Our elucidation of the primary molecular control of colour in apple provides important information for the breeding of apples with coloured flesh.
Experimental procedures
Real-time qPCR expression analysis
Apple fruit were collected at six time points during the season from spring through to late summer (2003–2004): stage 1, 7 days after full bloom (DAFB); stage 2, 40 DAFB; stage 3, 67 DAFB; stage 4, 102 DAFB; stage 5, 130 DAFB; stage 6, 146 DAFB, from trees at the HortResearch orchard (Nelson, New Zealand). RNA was isolated (by a method adapted from that described by Chang et al., 1993) from the fruit (six fruit from the same tree, skin and cortex separately) and the leaves of two genotypes: the white-fleshed commercial cultivar Malus × domestica‘Sciros’ (Pacific RoseTM, derived from a cross between ‘Gala’ and ‘Splendour’), and an open-pollinated red-fleshed seedling of Malus × domestica‘Red Field’ (a cross between ‘Wolf River’ and Malus × domestica niedzwetzkyana; Brooks and Olmo, 1972). For the first developmental fruit time point (7 DAFB), successful excision of skin from cortex was not possible, so data from this sample have been excluded. Following DNase treatment, first-strand cDNA synthesis was carried out in triplicate for each sample (to minimize variation in RNA template levels) using oligo(dT) according to the manufacturer's instructions (Transcriptor; Roche Diagnostics, Mannheim, Germany). Samples were then pooled.
Genes encoding apple anthocyanin pathway enzymes and regulators were identified by homology in the HortResearch EST database, and, where putative gene family members existed, candidates were selected when they appeared in fruit library tissues. Gene-specific primers corresponding to these genes were designed using Vector NTI version 9.0.0 (http://www.invitrogen.com) to a stringent set of criteria, enabling application of universal reaction conditions. To check reaction specificity, RT-PCR reactions were carried out according to the manufacturer's instructions (Platinum Taq, Invitrogen, Carlsbad, CA, USA), with a thermal profile as follows: pre-incubation at 95°C for 5 min followed by 35 cycles of 95°C (30 sec), 60°C (30 sec) and 72°C (30 sec), with a final extension at 72°C for 5 min. The sequences of each primer pair and the relevant accession numbers are shown in Table S1.
qPCR DNA amplification and analysis was carried out using the LightCycler System (Roche LightCycler 1.5; Roche Diagnostics). All reactions were performed using the LightCycler FastStart SYBR Green Master Mix (Roche Diagnostics) according to the procedure described by the manufacturer. Reactions were performed in triplicate using 2 μl 5 × Master Mix, 0.5 μm each primer, 1 μl diluted cDNA and nuclease-free water (Roche Diagnostics) to a final volume of 10 μl. A negative water control was included in each run. Fluorescence was measured at the end of each annealing step. Amplification was followed by a melting curve analysis with continual fluorescence data acquisition during the 65–95°C melt. The raw data were analysed with the LightCycler software, version 4, and expression was normalized to Malus × domestica actin (MdActin, accession number CN938023) to minimize variation in cDNA template levels, with the Pacific RoseTM leaf sample acting as calibrator with a nominal value of 1. MdActin was selected for normalization due to its consistent transcript level throughout fruit tissues and leaf, with crossing threshold (Ct) values changing by < 2. For each gene, a standard curve was generated using a cDNA serial dilution, and the resultant PCR efficiency calculations (ranging between 1.839 and 1.945) were imported into relative expression data analysis. To ensure that the transcripts of single genes had been amplified, qPCR amplicons were sequenced and confirmed as the expected plant DNA sequences. Error bars shown in qPCR data are technical replicates, representing the means ± SE of three replicate qPCR reactions.
Isolation of MdMYB10
PCR was performed using cDNA from the cortex sample of ‘Red Field’ (102 DAFB) using degenerate primers (with a 32-fold degeneracy) designed to the consensus DNA sequence of the R2R3 DNA-binding domain based on the sequence of anthocyanin regulators in diverse species. Numerous cDNAs encoding R2R3 MYB domains were obtained. Results from sequencing data revealed one cDNA with high identity to anthocyanin regulators, and the full-length sequence was obtained using 5′ RACE (GeneRacer, Invitrogen). The complete sequence for the MdMYB10 cDNA was compiled from overlapping fragments (Genbank accession number DQ267896). To compare the transcript from ‘Red Field’, full-length cDNAs were subsequently isolated from Malus × domestica Pacific RoseTM (DQ267897) and ‘Granny Smith’ (DQ267898). cDNA of MdMYB8 (DQ267899), an apple MYB bearing little sequence homology to known anthocyanin regulators, was isolated from the HortResearch EST collection to act as a negative control.
Dual luciferase assay of transiently transformed tobacco leaves
The promoter sequence for Arabidopsis DFR was inserted into the cloning site of pGreen 0800-LUC (Figure S3) (Hellens et al., 2005) and modified to introduce an NcoI site at the 3′ end of the sequence, allowing the promoter to be cloned as a transcriptional fusion with the firefly luciferase gene (LUC). Thus, TFs that bind the promoter and increase the rate of transcription could be identified as an increase in luminescence activity. Arabidopsis DFR (TT3, AT5g42800) was isolated from genomic Arabidopsis DNA. In the same construct, a luciferase gene from Renilla (REN), under the control of a 35S promoter, provided an estimate of the extent of transient expression. Activity is expressed as a ratio of LUC to REN activity such that where interaction between a TF (with or without bHLH) and the promoter occurred, a significant increase in LUC activity relative to REN would be observed.
The promoter–LUC fusion in pGreenII 0800-LUC was used in transient transformation by mixing 100 μl of Agrobacterium strain GV3101 (MP90) transformed with the reporter cassette with 450 μl each of two other Agrobacterium cultures transformed with cassettes containing a MYB TF gene fused to the 35S promoter and a bHLH TF gene in either pART27 (Gleave, 1992) or pGreenII 62-SK binary vectors (Figure S4) (Hellens et al., 2000).
Nicotiana tabacum‘Samsun’ plants were grown under glasshouse conditions in full potting mix, using natural light with daylight extension to 16 h, until at least six leaves were available for infiltration with Agrobacterium. Plants were maintained in the glasshouse for the duration of the experiment. Agrobacterium was cultured on Lennox agar (Invitrogen) supplemented with selection antibiotics and incubated at 28°C. A 10 μl loop of confluent bacterium was re-suspended in 10 ml of infiltration buffer (10 mm MgCl2, 0.5 μm acetosyringone) to an OD600 of 0.2, and incubated at room temperature without shaking for 2 h before infiltration. Infiltrations were performed according to the methods described by Voinnet et al. (2003). Approximately 150 μl of this Agrobacterium mixture was infiltrated at six points into a young leaf of N. tabacum, and transient expression was assayed 3 days after inoculation.
Firefly luciferase and renilla luciferase were assayed using the dual luciferase assay reagents (Promega, Madison, WI, USA). Three days after inoculation, 2 cm leaf discs (six technical replicates from each plant) were removed and ground in 500 μl of passive lysis buffer (PLB), then 10 μl of a 1/100 dilution of this crude extract was assayed in 40 μl of luciferase assay buffer, and the chemiluminescence measured. Stop and GlowTM buffer (40 μl; Promega) was then added and a second chemiluminescence measurement taken. Absolute relative luminescence units (RLU) were measured in a Turner 20/20 luminometer (Turner BioSystems, Sunnyvale, CA, USA), with a 5 sec delay and 15 sec integrated measurement.
Induction of anthocyanins by transient transformation of tobacco
Nicotiana tabacum plants were grown as described above and maintained in the glasshouse for the duration of the experiment. Agrobacterium cultures were incubated as for the dual luciferase assay, and separate strains containing the MYB TF gene and the bHLH TF gene fused to the 35S promoter in the pART27 binary vector were mixed (500 μl each) and infiltrated into the abaxial leaf surface as for the luciferase assay. Six separate infiltrations were performed into N. tabacum leaves (two plants per treatment) and changes in colour were measured daily using a Minolta CR-300 chromometer (calibrated to D65 light) (Konica Minolta, Mahwah, NJ, USA) by the L*a*b* system (CIE, 1986). Infiltrations comprising MdMYB10 together with an apple bHLH resulted in visible pigmentation after 4 days. The level of pigmentation increased throughout the experimental period; digital photographs and microscope images were taken 8 days after infiltration. To control for leaf-to-leaf variability, at least two leaves were infiltrated, and each leaf included positive controls (Agrobacterium cultures carrying MdMYB10 + MdbHLH3) and negative controls (Agrobacterium with empty vector).
Phylogeny
Apple EST sequences were trimmed of vector, adapter and low-quality sequence regions and uploaded to Vector NTI version 9.0.0 (http://www.invitrogen.com). Full-length sequences were aligned using Vector NTI Clustal W (opening = 15, extension = 0.3). Phylogenetic and molecular evolutionary analyses were conducted using MEGA version 3.1 (Kumar et al., 2004) using a minimum evolution phylogeny test and 1000 bootstrap replicates. Full MSF format files are available (Figure S6).
Identification and quantification of anthocyanins
Tobacco or apple samples were freeze-dried and coarsely ground before resuspension in 5 ml methanol and 0.1% HCl, extraction at room temperature for 2 h, and centrifugation at 3000 g. Aliquots of 1 ml were dried down to completion in a Labconco Centrivap Concentrator (Labconco, Kansas City, MO, USA). Samples were resuspended in 20% methanol (250 μl). Anthocyanins were identified by LC-MS analysis as described previously (Montefiori et al., 2005). Identification was based on both masses (M+) of molecular ions and characteristic fragments, and comparison of retention times and fragmentation with authentic standards of cyanidin-3-O-glucoside and cyanidin-3-O-galactoside. Anthocyanins and other phenolic compounds were quantified by HPLC as described previously (Stevenson et al., 2006). Quantification was achieved by reference to standards of anthocyanins and other phenolic compounds, using LC-MS data to confirm identification of peaks.
Transformation of apple
The binary vector pSAK277-MdMYB10 (Figure S5) containing the MdMYB10 cDNA driven by the 35S promoter was transferred into Agrobacterium tumefaciens strain GV3101 by the freeze–thaw method. Transgenic Malus domestica‘Royal Gala’ plants were generated by Agrobacterium-mediated transformation of leaf pieces, using a method previously reported (Yao et al., 1995).
Supplementary Material
The following supplementary material is available for this article online:
Expression data for DFR family members.
TF expression data in transgenic apple.
pGreen II 0800-LUC vector map.
pGreen II 62 SK binary vector map.
pSak 277 MDMYBIO binary vector map.
Protein sequence alignments.
Primers for qPCR
This material is available as part of the online article from http://www.blackwell-synergy.com
Acknowledgments
We thank Kevin Davies (Crop and Food Research, Palmerston North, New Zealand) for helpful discussions and advice, and Bernd Weisshaar (Universitat Bielefeld, Bielefeld, Germany) for providing sequence information on the bHLH family. We are also grateful to Janine Cooney (HortResearch, Ruakura, New Zealand) for LC-MS data, Richard Volz and Nnadozie Oraguzie (HortResearch, Hawke's Bay, New Zealand) for providing invaluable advice and sample fruit, Julie Nichols for maintaining the glasshouse plants, Karen Bolitho for Agrobacterium transformations, and Sakuntala Karunairetnam for vector construction of the EST-derived genes. This work was supported by a grant (contract number PREV0401) from the New Zealand Foundation for Research, Science and Technology and PREVAR Limited, New Zealand.
[Note added in proof: a similar gene, MdMYB1 (Dq886414-DQ886416), has been isolated from apple and shown to regulate anthocyanin production in apple skin (Takos, A.M., Jaffe, F.W., Jacob, S.R., Bogs, J., Robinson, S.P. and Walker, A.R. (2006) Light induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol.142, 1216–1232.]
References
- Baudry A. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 2004;39:366–380. doi: 10.1111/j.1365-313X.2004.02138.x. [DOI] [PubMed] [Google Scholar]
- Borevitz JO. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell. 2000;12:2383–2394. doi: 10.1105/tpc.12.12.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boss PK. Expression of anthocyanin biosynthesis pathway genes in red and white grapes. Plant Mol. Biol. 1996;32:565–569. doi: 10.1007/BF00019111. [DOI] [PubMed] [Google Scholar]
- Boyer J. Apple phytochemicals and their health benefits. Nutr. J. 2004;3:5. doi: 10.1186/1475-2891-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks RM. Register of New Fruit and Nut Varieties. Berkeley, California: University of California Press; 1972. [Google Scholar]
- Brouillard R. Flavonoids and flower colour. In: Harborne JB, editor. The Flavonoids: Advances in Research Since 1980. London: Chapman & Hall; 1988. pp. 525–538. [Google Scholar]
- Chang S. A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 1993;11:113–116. [Google Scholar]
- CIE . Colorimetry. 2. Vienna: Central Bureau of the Commission Internationale de L'Eclairage (CIE Publication Number 15.2); 1986. [Google Scholar]
- Davies KM. Transcriptional regulation of secondary metabolism. Funct. Plant. Biol. 2003;30:913–925. doi: 10.1071/FP03062. [DOI] [PubMed] [Google Scholar]
- De Jong WS. Candidate gene analysis of anthocyanin pigmentation loci in the Solanaceae. Theor. Appl. Genet. 2004;108:423–432. doi: 10.1007/s00122-003-1455-1. [DOI] [PubMed] [Google Scholar]
- Dong YH. Postharvest stimulation of skin colour in Royal Gala apple. J. Am. Soc. Hort. Sci. 1998;120:95–100. [Google Scholar]
- Gleave A. A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 1992;20:1203–1207. doi: 10.1007/BF00028910. [DOI] [PubMed] [Google Scholar]
- Goff SA. Functional analysis of the transcriptional activator encoded by the maize B gene: evidence for a direct functional interaction between two classes of regulatory proteins. Genes Dev. 1992;6:864–875. doi: 10.1101/gad.6.5.864. [DOI] [PubMed] [Google Scholar]
- Goodrich J. A common gene regulates pigmentation pattern in diverse plant species. Cell. 1992;68:955–964. doi: 10.1016/0092-8674(92)90038-e. [DOI] [PubMed] [Google Scholar]
- Grotewold E. Identification of the residues in the Myb domain of maize C1 that specify the interaction with the bHLH cofactor R. Proc. Natl Acad. Sci. USA. 2000;97:13579–13584. doi: 10.1073/pnas.250379897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harborne JB. Comparative Biochemistry of the Flavonoids. London: Academic Press; 1967. [Google Scholar]
- Harborne JB. Flavonoids and insects. In: Harborne JB, editor. The Flavonoids: Advances in Research Since 1986. London: Chapman & Hall; 1994. pp. 589–618. [Google Scholar]
- Heim MA. The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 2003;20:735–747. doi: 10.1093/molbev/msg088. [DOI] [PubMed] [Google Scholar]
- Hellens RP. pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 2000;42:819–832. doi: 10.1023/a:1006496308160. [DOI] [PubMed] [Google Scholar]
- Hellens RP. Transient plant expression vectors for functional genomics, quantification of promoter activity and RNA silencing. Plant Methods. 2005;1:13. doi: 10.1186/1746-4811-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez JM. Different mechanisms participate in the R-dependent activity of the R2R3 MYB transcription factor C1. J. Biol. Chem. 2004;279:48205–48213. doi: 10.1074/jbc.M407845200. [DOI] [PubMed] [Google Scholar]
- Holton TA. Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell. 1995;7:1071–1083. doi: 10.1105/tpc.7.7.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda C. Anthocyanin biosynthetic genes are coordinately expressed during red coloration in apple skin. Plant Physiol. Biochem. 2002;40:955–962. [Google Scholar]
- Jin H. Multifunctionality and diversity within the plant MYB-gene family. Plant Mol. Biol. 1999;41:577–585. doi: 10.1023/a:1006319732410. [DOI] [PubMed] [Google Scholar]
- Kim S-H. Molecular cloning and analysis of anthocyanin biosynthesis genes preferentially expressed in apple skin. Plant Sci. 2003;165:403–413. [Google Scholar]
- Kobayashi S. Myb-related genes of the Kyoho grape (Vitis labruscana) regulate anthocyanin biosynthesis. Planta. 2002;215:924–933. doi: 10.1007/s00425-002-0830-5. [DOI] [PubMed] [Google Scholar]
- Kobayashi S. Retrotransposon-induced mutations in grape skin colour. Science. 2004;304:982. doi: 10.1126/science.1095011. [DOI] [PubMed] [Google Scholar]
- Koes RE. The flavonoid biosynthetic pathway in plants: function and evolution. BioEssays. 1994;16:123–132. [Google Scholar]
- Kumar S. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Lancaster J. Regulation of skin color in apples. Crit. Rev. Plant. Sci. 1992;10:487–502. [Google Scholar]
- Lister CE. Developmental changes in enzymes of flavonoid biosynthesis in the skins of red and green apple cultivars. J. Sci. Food Agric. 1996;71:313–320. [Google Scholar]
- Martin C. MYB transcription factors in plants. Trends Genet. 1997;13:67–73. doi: 10.1016/s0168-9525(96)10049-4. [DOI] [PubMed] [Google Scholar]
- Matsuo N. Dual luciferase assay for monitoring transient gene expression in higher plants. Plant Biotechnol. 2001;18:71–75. [Google Scholar]
- Mol JJ. Signal perception, transduction, and gene expression involved in anthocyanin biosynthesis. Crit. Rev. Plant. Sci. 1996;15:525–557. [Google Scholar]
- Mol J. How genes paint flowers and seeds. Trends Plant. Sci. 1998;3:212–217. [Google Scholar]
- Montefiori M. Pigments in the fruit of red-fleshed kiwifruit (Actinidia chinensis and Actinidia deliciosa) J. Agric. Food Chem. 2005;53:9526–9530. doi: 10.1021/jf051629u. [DOI] [PubMed] [Google Scholar]
- Nesi N. The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell. 2000;12:1863–1878. doi: 10.1105/tpc.12.10.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomb RD, et al. Analyses of expressed sequence tags from apple (Malus × domestica) Plant Physiol. 2006;141:147–167. doi: 10.1104/pp.105.076208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda K-I. Flower colour intensity depends on specialized cell shape controlled by a Myb-related transcription factor. Nature. 1994;369:661–664. doi: 10.1038/369661a0. [DOI] [PubMed] [Google Scholar]
- Ramsay NA. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 2005;10:63–70. doi: 10.1016/j.tplants.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Saito K. Biochemistry and molecular biology of the late-stage of biosynthesis of anthocyanin: lessons from Perilla frutescens as a model plant. New Phytol. 2002;155:9–23. doi: 10.1046/j.1469-8137.2002.00440.x. [DOI] [PubMed] [Google Scholar]
- Schwinn KE. A small family of MYB-regulatory genes controls floral pigmentation intensity and patterning in the genus Antirrhinum. Plant Cell. 2006;18:831–851. doi: 10.1105/tpc.105.039255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson DE. Direct acylation of flavonoid glycosides with phenolic acids catalysed by Candida antarctica lipase B (Novozym 435®) Enzyme Microb. Technol. 2006;39:1236–1241. [Google Scholar]
- Stracke R. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001;4:447–456. doi: 10.1016/s1369-5266(00)00199-0. [DOI] [PubMed] [Google Scholar]
- Tohge T, et al. Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 2005;42:218–235. doi: 10.1111/j.1365-313X.2005.02371.x. [DOI] [PubMed] [Google Scholar]
- Tsao R. Polyphenolic profiles in eight apple cultivars using high-performance liquid chromatography (HPLC) J. Agric. Food Chem. 2003;51:6347–6353. doi: 10.1021/jf0346298. [DOI] [PubMed] [Google Scholar]
- de Vetten N. The an11 locus controlling flower pigmentation in petunia encodes a novel WD-repeat protein conserved in yeast, plants, and animals. Genes Dev. 1997;11:1422–1434. [Google Scholar]
- Voinnet O. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 2003;33:949–956. doi: 10.1046/j.1365-313x.2003.01676.x. [DOI] [PubMed] [Google Scholar]
- Walker AR. The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell. 1999;11:1337–1350. doi: 10.1105/tpc.11.7.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B. Flavonoid biosynthesis. A colourful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001;126:485–493. doi: 10.1104/pp.126.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D-Y. Metabolic engineering of proanthocyanidins through co-expression of anthocyanidin reductase and the PAP1 MYB transcription factor. Plant J. 2006;45:895–907. doi: 10.1111/j.1365-313X.2006.02655.x. [DOI] [PubMed] [Google Scholar]
- Yao J-L. Regeneration of transgenic plants from the commercial apple cultivar Royal Gala. Plant Cell Rep. 1995;14:407–412. doi: 10.1007/BF00234044. [DOI] [PubMed] [Google Scholar]
- Zhang F. A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development. 2003;130:4859–4869. doi: 10.1242/dev.00681. [DOI] [PubMed] [Google Scholar]
- Zimmermann IM. Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 2004;40:22–34. doi: 10.1111/j.1365-313X.2004.02183.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression data for DFR family members.
TF expression data in transgenic apple.
pGreen II 0800-LUC vector map.
pGreen II 62 SK binary vector map.
pSak 277 MDMYBIO binary vector map.
Protein sequence alignments.
Primers for qPCR