Abstract
Tsc22d3 coding for glucocorticoid-induced leucine zipper (GILZ) was initially identified as a dexamethasone-responsive gene involved in the control of T lymphocyte activation and apoptosis. However, the physiological role of this molecule and its function in the biological activity of glucocorticoids (GCs) has not been clarified. Here, we demonstrate that GILZ interacts directly with Ras in vitro and in vivo as shown by GILZ and Ras coimmunoprecipitation and colocalization upon PMA activation in primary mouse spleen T lymphocytes and thymus cells. The analysis of GILZ mutants showed that they bound Ras through the tuberous sclerosis complex box (TSC) and, depending on the Ras activation level, formed a trimeric complex with Ras and Raf, which we previously identified as a GILZ binder. As a consequence of these interactions, GILZ diminished the activation of Ras and Raf downstream targets including ERK1/2, AKT/PKB serine/threonine kinase, and retinoblastoma (Rb) phosphorylation and cyclin D1 expression, leading to inhibition of Ras- and Raf-dependent cell proliferation and Ras-induced NIH-3T3 transformation. GILZ silencing resulted in an increase in concanavalin A–induced T cell proliferation and, most notably, inhibition of dexamethasone antiproliferative effects. Together, these findings indicate that GILZ serves as a negative regulator of Ras- and Raf-induced proliferation and is an important mediator of the antiproliferative effect of GCs.
Introduction
Glucocorticoids (GCs) are widely used as antiinflammatory and immunosuppressive agents to control several chronic and acute diseases. Their therapeutic activity is due to the enhanced intensity of the physiological effects of endogenous steroids (1, 2). GCs function via the GC receptor (GR), which regulates transcription of several target genes and also mediates GC effects indirectly via a negative or positive regulation of transcription factors and other signaling proteins (3, 4).
Most GC-related antiinflammatory and immunosuppressive effects are linked to GC modulation of cell proliferation, via various mechanisms such as inhibition of cyclin D3 and c-myc, and increased p27Kip1 expression (5–7). Another intriguing and as yet not completely understood molecular mechanism is GC interference with the MAPK signaling pathway (8). By cycling between active GTP-bound and inactive GDP-bound states, small G proteins, members of the Ras superfamily, control the MAPK pathways (9). Once activated, Ras interacts with and activates several effectors, which in turn trigger a multitude of signaling pathways (10). Raf and PI3K, the best characterized Ras effectors, activate MEK/ERK kinase and AKT/PKB serine/threonine kinase (AKT) cascades, respectively (11). Both cooperate in regulating cyclin D1 expression and normal and neoplastic cell proliferation (12–18).
Ras signal transmission efficiency is upregulated and downregulated by several proteins that enhance or disrupt physical interactions between pathway components (19, 20). Raf kinase activity, for example, is modulated by many binding partners, which function as adaptors and/or connectors with enhancer functions. These include the kinase suppressor of Ras (KSR) (21–23); BCL-2 binding athanogene-1 (24); soc-2 (suppressor of clear) homolog (C. elegans), a leucine-rich protein that acts as a scaffold to enhance signal transduction of Ras-MAPK by facilitating the interaction between Ras and Raf (25); cofactors such as 14-3-3 (26); chaperones such as Hsp90 and Hsp70 (27); activators or inhibitors such as impedes mitogenic signal propagation (IMP) (28); and Raf kinase inhibitor protein (RKIP) (29). RKIP interacts with Raf, MEK, and ERK and suppresses Raf-induced transformation (29). Such control mechanisms serve to maintain normal cell growth rate and function. In fact, aberrant Ras activation has been found in many human cancers, and Raf and PI3K activation underlies oncogenic transformation of mammalian cells (30).
The GC-MAPK cross-talk occurs at many points of the signaling cascade, where other signals are also integrated. GR is purified with 14-3-3 and Raf-1 proteins (31). Dexamethasone (DEX), a synthetic GC, suppresses ERK1/2 activation in several cellular systems (32). Moreover, GR modulates PI3K activity (33) and represses cyclin D1 by targeting the TCF/β-catenin complex (34). DEX upregulates gene 33, which blunts EGF-induced activation of Ras, ERK, JNK, AKT (35), and docking protein 1 (DOK1), the inhibitory adaptor protein that suppresses MAPK pathway downstream activation (36).
Tsc22d3 coding for glucocorticoid-induced leucine zipper (GILZ), one of the GC-induced genes (37, 38), binds and inhibits Raf-1 with consequent inhibition of its downstream pathway (39). Similarly, GILZ overexpression inhibits ERK phosphorylation in epithelial kidney cells (40), thus confirming that GILZ is involved in controlling the MAPK pathway and suggesting that GILZ induction is one of the mechanisms by which steroid hormones modulate MAPK pathway activity.
In the present study, we show that GILZ interacts directly with Ras to inhibit Ras-induced cell proliferation and transformation. This effect is associated with decreased ERK, AKT, and retinoblastoma (Rb) phosphorylation and decrease in cyclin D1 expression. These results identify GILZ as a Ras signal suppressor that may contribute to the effects of GCs on cell growth.
Results
GILZ interacts with Ras.
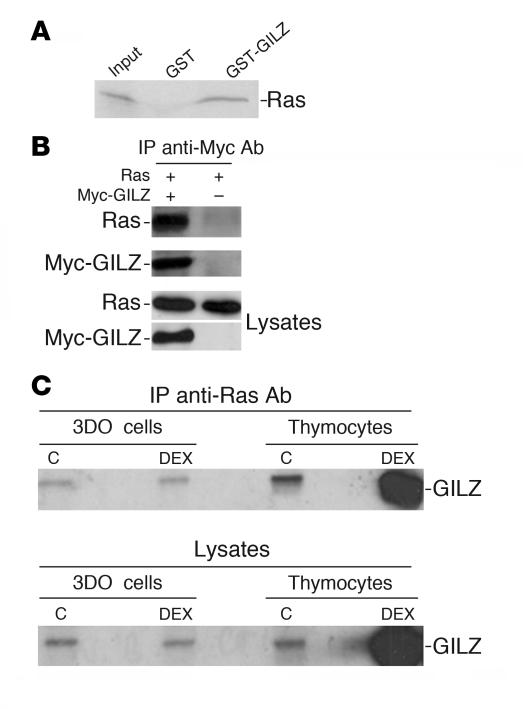
We previously demonstrated that GILZ binds to Raf-1 and inhibits ERK phosphorylation, c-Fos expression, and AP-1 transcription activity (39). In the present study, we investigated whether GILZ interacts with Ras. First, an N-terminal glutathione-S-transferase–GILZ (GST-GILZ) fusion protein was prepared and used in GST pulldown experiments using in vitro translated Ras. The GST-GILZ fusion protein bound directly to in vitro translated Ras (Figure 1A). The GILZ-Ras interaction was analyzed in vivo in COS-7 cells cotransfected with myc-tagged GILZ and a vector expressing wild-type H-Ras. Anti-myc Ab immunoprecipitates containing immunoreactive Ras were obtained from cells cotransfected with myc-GILZ and Ras but not from cells transfected with Ras alone (Figure 1B).
Figure 1. GILZ interacts with Ras.
(A) The 35S-labeled in vitro transcription and translation product of full-length wild-type H-Ras was incubated with GST or GST-GILZ immobilized on glutathione-sepharose beads. The proteins bound to the resin were eluted, resolved by SDS-PAGE, and visualized by autoradiography. Input indicates 10% volume of the 35S-labeled product used in the pulldown assay. (B) COS-7 cells were cotransfected with pUSEamp-Ras wild-type and Myc-GILZ vectors. Immunoprecipitation was performed with anti-myc Ab, and immunoreactive proteins were revealed with anti-Ras or anti-myc Ab. Whole-cell lysates were loaded to control plasmid expression. (C) 3DO cells and mouse thymocytes were treated for 3 hours with DEX. Cell lysates were immunoprecipitated with anti-Ras Ab and revealed with anti-GILZ Ab. Whole-cell lysates were loaded to control GILZ regulation by DEX. C, control.
Because GILZ modulates T cell function (41), we evaluated whether endogenous GILZ binds Ras in thymocytes and 3DO T cells. We treated cells with DEX for 3 hours to rapidly upregulate GILZ expression in thymocytes (37). The anti-Ras Ab coimmunoprecipitated GILZ in all groups (Figure 1C). DEX-treated thymocytes expressed (Figure 1C, lower panel) and coimmunoprecipitated (Figure 1C, upper panel) more GILZ compared with untreated control. DEX treatment did not change Ras expression (not shown). These results indicate that GILZ binds Ras under physiological conditions in T cells.
GILZ interacts mainly with activated Ras.
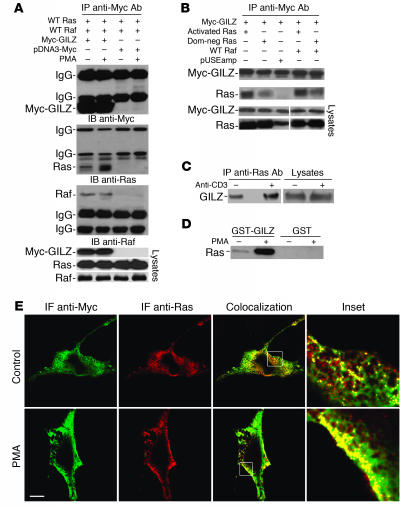
Because several mammalian proteins associate directly with Ras in a GTP-dependent manner (25), we determined whether GILZ binds Ras-GTP with higher affinity than Ras-GDP and investigated whether Raf plays a role in GILZ-Ras binding. COS-7 cells were transfected with myc-GILZ, wild-type H-Ras, and Raf-1 and were treated with PMA. Anti-myc Ab–immunoprecipitated myc-GILZ and coimmunoprecipitated Ras and Raf. (Figure 2A). PMA activation increased GILZ-Ras binding but did not affect GILZ-Raf binding (Figure 2A). COS-7 cells were transfected with myc-GILZ along with either constitutively activated Ras or dominant-negative Ras. Anti-myc Ab coimmunoprecipitated more Ras when it was constitutively activated than when it was dominant negative (Figure 2B). The concomitant presence of Raf did not affect this trend (Figure 2B). Furthermore, GILZ binding to Ras was poorly influenced by the state of Raf activation (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI30724DS1). To test even further the hypothesis that GILZ binds mainly to active Ras, mouse spleen T lymphocytes were activated for 30 minutes with anti-CD3 Ab (42). Cell lysates were immunoprecipitated with anti-Ras and revealed by Western blot with anti-GILZ Ab. Ras bound more GILZ protein in the activated group than in the control group (Figure 2C). Furthermore, upon PMA treatment, lysates of 3DO T cells, absorbed by GST-GILZ fusion protein, showed strongly increased amounts of Ras bound to GILZ (Figure 2D). Confocal fluorescent microscopy revealed extensive iuxta-membrane colocalization between Ras and GILZ when GILZ-Ras–cotransfected COS-7 cells were treated with PMA (Figure 2E). These results suggest that upon activation, Ras increases its affinity for GILZ.
Figure 2. GILZ interacts mainly with Ras in the active form.
(A) COS-7 cells were cotransfected with the indicated vectors. After 48 hours, the cells were treated with PMA (100 ng/ml) for 20 minutes. Total cell lysates were subjected to immunoprecipitation with anti-myc Ab and analyzed by immunoblot for the indicated Abs. Whole-cell lysates were loaded to control plasmid expression. (B) COS-7 cells were cotransfected with the indicated vectors, immunoprecipitated, and analyzed by Western blot as described in A. Dom-neg Ras, dominant-negative Ras. (C) Splenic T lymphocytes were activated by cross-linked anti-CD3 Ab for 30 minutes. Cell lysates were immunoprecipitated by anti-Ras Ab and analyzed by immunoblot for anti-GILZ Ab. (D) 3DO cells were treated with PMA (100 ng/ml) for 20 minutes, and pulldown assays were performed by incubating total lysates with GST-GILZ fusion protein or GST alone for 18 hours at 4°C. Immunoblot was analyzed by anti-Ras Ab. (E) Confocal analysis of Ras (red) and GILZ (green) localization in myc-GILZ/pUSEamp-Ras wild-type–cotransfected COS-7 cells treated with PMA for 20 minutes. PMA treatment produced iuxta-membrane accumulation of Ras and an increase in the colocalization with GILZ in the same areas (inset). IF, immunofluorescence. Scale bar, 10 μm. Original magnification, ×5 (top inset); ×6 (bottom inset).
Finally, we determined whether GTP-Ras competes with GDP-Ras for binding to GILZ. Lysates from COS-7 cells, transfected with wild-type Xpress-Ras (Xp-Ras; Xpress is an epitope engineered into pcDNA3.1C vectors), were loaded with GDP and used with GST-GILZ fusion protein as bait in a binding assay. The GST-GILZ-Ras-GDP complex was then incubated with COS-7 cell lysates loaded with GTP, and the complex was analyzed by Western blot. Adding GTP-Ras resulted in an increased amount of Ras binding GILZ (Supplemental Figure 2). As expected, the same results were obtained when a fusion protein corresponding to the human Ras-binding domain of Raf-1 (GST-Raf-RBD) was used to evaluate Ras activation (Supplemental Figure 2). Moreover, no binding was detected with GST protein alone (Supplemental Figure 2).
Different GILZ domains interact with Ras and Raf.
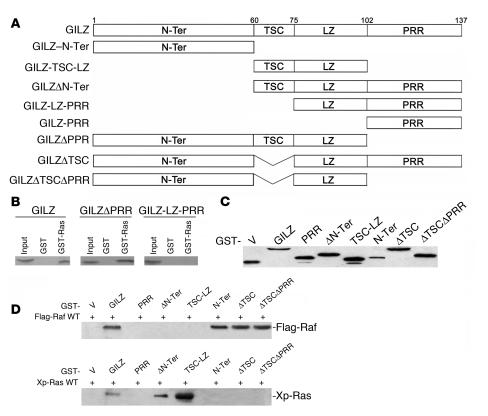
Using deleted GILZ mutants, we demonstrated that GILZ loses its ability to interact with Raf when 1–75 aa at the N-terminal domain are deleted (39). To identify the GILZ domain that binds Ras, we performed in vitro GST-pulldown experiments using GST-Ras as a bait with in vitro translated GILZ or deleted forms of GILZ (Figure 3A) lacking either the C-terminal proline-rich region (PRR), GILZΔPRR (1–101 aa), or the whole N-terminal domain, GILZ–leucine zipper–PRR (GILZ-LZ-PRR) (76–137 aa). Ras bound full-length GILZ (1–137 aa) and GILZΔPRR (1–101 aa) but not GILZ-LZ-PRR (76–137 aa) (Figure 3B), indicating that the same GILZ domain (1–75 aa) interacts with Raf and Ras.
Figure 3. Mapping the Ras-binding domains of GILZ.
(A) Schematic of GILZ deletion constructs. N-Ter, N-terminal. (B) GST-Ras fusion protein was incubated for 18 hours with 35S-labeled in vitro–transcribed and –translated proteins, GILZ, or GILZ mutants. (C) GST-GILZ or GST-GILZ mutants (in pEBG) were transfected in COS-7, and lysates were analyzed by Western blot using anti-GST Ab to control plasmid expression. (D) COS-7 cells were cotransfected with GST-GILZ or GST-GILZ mutants, as described in C, together with either Flag-Raf (top) or Xp-Ras (bottom). GST-GILZ and GST-GILZ mutants were purified by glutathione-sepharose resin, and the copurified Raf and Ras were detected by anti-Flag and anti-Xp Abs, respectively. V, empty vector.
To further address this issue, we constructed a series of mutants (Figure 3A) and expressed them in COS-7 as GST-tagged proteins along with either wild-type Xp-Ras or Flag-Raf. The expression level of transfected GILZ deletion mutants was assessed by immunoblotting using the anti-GST Ab (Figure 3C). Lysates expressing GST-GILZ deletion mutants and Raf were immunoprecipitated with Glutathione Sepharose 4B. Raf was detected in full-length GILZ (1–137 aa) and GILZ–N-terminal (GILZ–N-Ter) (1–60 aa) immunocomplexes. None of the other GILZ mutants, including GILZ–tuberous sclerosis complex box–LZ (GILZ-TSC-LZ) (61–100 aa), GILZΔN-Ter (61–137 aa), or GILZ-PRR (103–137 aa) bound Raf, suggesting that the N-terminal GILZ domain (1–60 aa) is necessary and sufficient for GILZ-Raf binding (Figure 3D).
Lysates expressing GILZ deletion mutants and wild-type Xp-Ras were immunoprecipitated with Glutathione Sepharose 4B, and the immunocomplexes revealed with anti-Xp Ab. Surprisingly, GILZ–N-Ter (1–60 aa) did not bind Ras; GILZΔN-Ter (61–137 aa) bound Ras to the same extent as full-length GILZ. There was no binding between GILZ-PRR (103–137 aa) and Ras, and GILZ-TSC-LZ (61–100 aa) immunoprecipitated more Ras than full-length GILZ. Therefore, although deletion of the whole GILZ N-terminal domain (GILZ-LZ-PRR) abrogated both GILZ-Ras and GILZ-Raf binding, the motif underlying each interaction appears different. In the GILZ-Ras interaction, unlike the GILZ-Raf, the TSC plays a major role. When TSC was deleted in GILZΔTSC (Δ60-75aa), no binding with Ras was detected, although it was still evident with Raf. As GILZ-TSC-LZ (61–100 aa) immunoprecipitated more than GILZΔN-Ter (61–137 aa), it is possible that other regions, including the PRR, played a negative role. Deletion of PRR and TSC (GILZΔTSCΔPRR) did not clarify this point because deleting only TSC inhibits GILZ-Ras binding (Figure 3D). These results support the view that different GILZ domains interact with Ras and Raf.
GILZ forms a trimeric complex with Ras and Raf.
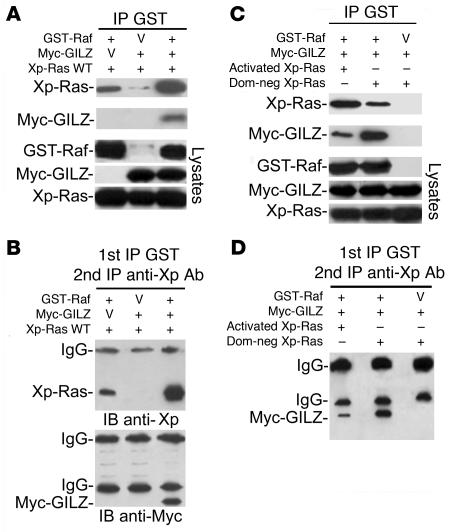
We characterized the formation of a Ras-Raf-GILZ complex in vivo by sequential coimmunoprecipitation experiments. GST-Raf, myc-GILZ, and wild-type Xp-Ras were cotransfected in COS-7 cells. GST-Raf and associated proteins were purified, eluted with glutathione-sepharose resin, and analyzed by Western blot with anti-myc and anti-Xp Abs. GILZ coexpression significantly increased the amount of Ras copurified with GST-Raf (Figure 4A), suggesting that GILZ is able to bridge this interaction. The purified GST complex underwent a second immunoprecipitation with anti-Xp Ab and Western blot with anti-Xp Ab to detect Xp-Ras and anti-myc Ab to detect myc-GILZ. Myc-GILZ was present after sequential immunoprecipitation, and again in the presence of GILZ the amount of Ras present in the complex was increased (Figure 4B). Therefore, we studied the Ras-Raf-GILZ complex as a function of the Ras activation state. For this purpose, we transfected COS-7 with Raf, GILZ, and either dominant-negative Ras or activated Ras. As expected, in the presence of dominant-negative Ras, GST-Raf bound Ras poorly but bound GILZ strongly. In the presence of activated Ras, the behavior was opposite — Raf bound Ras strongly but bound GILZ poorly (Figure 4C). After sequential immunoprecipitation, the amount of GILZ in the trimeric complex was higher in the presence of dominant-negative Ras than in the presence of activated Ras (Figure 4D). These data provide strong evidence that, depending on the Ras activation state, Ras, Raf, and GILZ can form a ternary complex in vivo.
Figure 4. GILZ forms a ternary complex with Ras and Raf.
(A) GST-Raf and wild-type Xp-Ras were transfected in COS-7 cells with or without myc-GILZ. GST-Raf was purified by adsorption to glutathione-sepharose beads, and copurified proteins were revealed with anti-Xp and anti-myc Abs using Western blot. Lysates were loaded to control expression levels. (B) The overexpressed glutathione-purified proteins were subjected to immunoprecipitation with anti-Xp Ab, followed by Western blot with anti-Xp and anti-myc antibodies. (C) GST-Raf and myc-GILZ were transfected in COS-7 cells together with either activated or dominant-negative Xp-Ras. Cell lysates, purified by glutathione-sepharose resin, were eluted by glutathione and analyzed by Western blot with the indicated Abs. (D) The glutathione-purified proteins described in C were immunoprecipitated with anti-Xp Ab and revealed by Western blot with anti-myc Ab.
GILZ inhibits Ras-activated signaling.
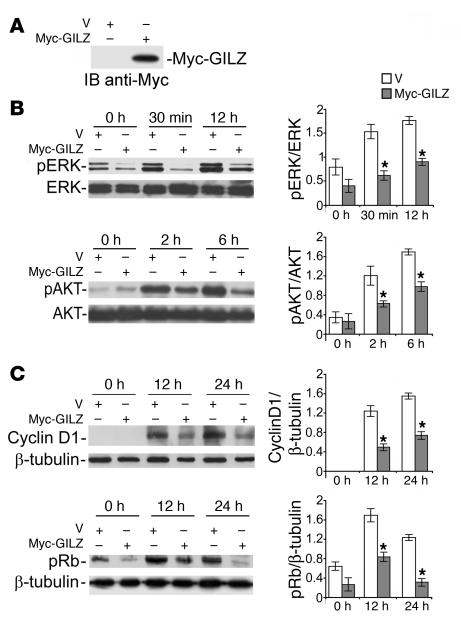
Ras downstream signals involve AKT and ERK pathways that cooperate to promote cyclin D1 expression and Rb phosphorylation (17, 18). We investigated whether the interaction of GILZ with Ras and Raf affects the activity of downstream Ras-dependent signaling. We first evaluated the effect of GILZ expression on ERK and AKT activation levels and cyclin D1 expression. For this purpose, we transfected NIH-3T3 cells with myc-GILZ or control vector. After 12 days of selection with antibiotics and 24 hours of starvation in low-serum medium, cell lysates were evaluated for GILZ expression (Figure 5A) and cells recultured in FCS-containing medium. ERK and AKT phosphorylation was increased after serum stimulation in both control and GILZ-expressing cells (Figure 5B). GILZ-transfected cells displayed significantly lower ERK and AKT phosphorylation levels than control cells (P < 0.05; Figure 5B), suggesting that GILZ can inhibit both the ERK and AKT pathways. Moreover, the inhibition of ERK and AKT pathways was correlated with decreased cyclin D1 expression and Rb phosphorylation (P < 0.05; Figure 5C). Together, these results indicate that there is an inhibition of downstream Ras-dependent signaling following GILZ overexpression.
Figure 5. GILZ inhibits Ras signaling.
NIH-3T3 cells, transfected with myc-GILZ or control vector, were cultured for 12 days with G418, starved for 24 hours (1% FCS), and then stimulated with 10% FCS for the indicated times. Total cell lysates were subjected to Western blot with the indicated Abs. Results of quantitative densitometry analysis are shown at right. (A) GILZ expression evaluated 12 days after transfection. (B) Phosphorylated ERK (pERK) and AKT. (C) Cyclin D1 expression and phosphorylated Rb. After stripping, membranes were reprobed with anti-ERK1/2 or anti-AKT or with anti–β-tubulin Ab. Three independent lysates for each condition were immunoblotted and the data were quantified and expressed as the relative ratio of phosphoprotein to total protein or β-tubulin (mean ± SD). *P < 0.05.
GILZ expression decreases Ras- and Raf-induced cell proliferation.
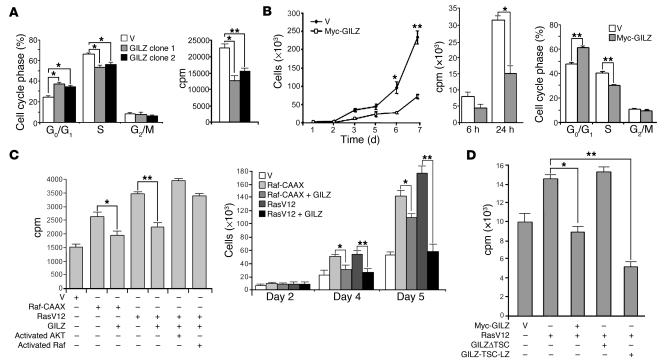
Given the association of GILZ with 2 molecules that affect cell proliferation, such as Ras and Raf, and the effect on downstream signals, we investigated whether GILZ plays a role in controlling cell proliferation. First, we evaluated the proliferation of stable 3DO T cell clones overexpressing GILZ, which we used in our previous studies, after starvation for 48 hours (37, 41). GILZ induced a significant increase in the G0/G1 phase of the cell cycle (P < 0.05; Figure 6A) and a decrease in thymidine uptake (P < 0.05 and P < 0.01; Figure 6A).
Figure 6. GILZ impairs cell proliferation rate.
(A) Empty vector– or GILZ-transfected 3DO stable clones (clones 1 and 2) were starved for 48 hours and then cultured for 24 hours in the presence of 10% FCS. Proliferation was evaluated by PI staining and cell cycle analysis (left) and [3H]-thymidine uptake assays (right). (B) NIH-3T3 cells were transfected, selected, and controlled for vector expression by Western blot as described in Figure 5, then starved for 24 hours (1% FCS). Growth curves (left) and [3H]-thymidine uptake (center) were quantified following FCS stimulation at the indicated times. Cell cycle analysis of PI-labeled cells (right) was performed 24 hours after FCS stimulation. (C) Inhibitory effect of GILZ on NIH-3T3 cell proliferation, transiently transfected with Raf-CAAX or RasV12 and reversion of GILZ-mediated inhibition by activated AKT or activated Raf. [3H]-thymidine uptake (left) and cell number (right) are shown. (D) Cell proliferation of NIH-3T3 cells transfected with RasV12 and full-length GILZ or GILZ mutant lacking the TSC or expressing only the TSC and LZ regions, as evaluated by [3H]-thymidine uptake. Data represent the average of 3 independent experiments; triplicate samples were counted at each time point. *P < 0.05; **P < 0.01.
To further assess whether sustained GILZ expression could affect the rate of cell growth, we analyzed the proliferation of the GILZ-transfected NIH-3T3 cells used in the experiment described in the Figure 5 legend. As shown in Figure 6B, GILZ overexpression induced a decrease in cell proliferation rate evaluated by growth curves (P < 0.01; day 7) and by thymidine uptake assays (P < 0.05). Consistent with these results, GILZ overexpression induced a significant increase (P < 0.01) of cells in the G0/G1 phase and a decrease (P < 0.01) in the S phase 24 hours after FCS stimulation (Figure 6B). No difference in apoptosis level was detected (data not shown).
We also evaluated the GILZ effect in cells transfected with oncogenic Ras (RasV12) or Raf-CAAX. GILZ overexpression diminished thymidine uptake and proliferation rates (Figure 6C) of NIH-3T3 cells transfected with either RasV12 (P < 0.01) or Raf-CAAX (P < 0.05), suggesting that the GILZ effect on cell proliferation occurred downstream from Ras and either downstream or parallel to Raf. The effect on Ras was more evident. We also investigated whether constitutively active Raf or AKT could reverse the GILZ-induced effect on cell proliferation. For this purpose, NIH-3T3 cells were transfected with RasV12 and GILZ along with either constitutively activated AKT or Raf, and proliferation was evaluated by thymidine uptake assays. The results indicate that either activated Raf or activated AKT was sufficient to revert GILZ inhibition of RasV12-induced cell proliferation (Figure 6C), suggesting that GILZ targeted both pathways downstream from Ras. Together, these results indicate that GILZ inhibits Ras- and Raf-dependent cell proliferation and that these effects are due to the inhibition of both ERK- and AKT-dependent downstream Ras pathways.
Finally, we examined whether the binding with Ras was necessary for GILZ inhibitory function on Ras-driven proliferation. For this purpose, we transfected NIH-3T3 cells with RasV12 along with full-length GILZ; GILZΔTSC, which does not bind Ras; or GILZ-TSC-LZ, which binds Ras strongly, and performed thymidine uptake assays. Full-length GILZ and GILZ-TSC-LZ significantly inhibited thymidine uptake (P < 0.05 and P < 0.01, respectively), while GILZΔTSC did not (Figure 6D), suggesting that the inhibitory function of GILZ requires binding to Ras.
GILZ inhibits Ras-mediated cell transformation and tumorigenesis.
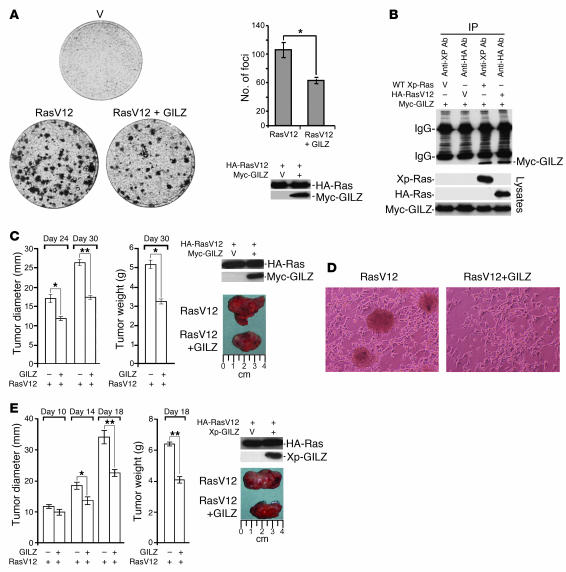
The Raf-MEK-ERK and PI3K-AKT pathways are involved in Ras-induced cell transformation (43–45). Since GILZ decreased cell proliferation and ERK and AKT phosphorylation, we reasoned that it might also have a negative role in cell transformation. We therefore examined the potential GILZ contribution to Ras-transforming activity. We expressed GILZ together with the oncogenic RasV12 in NIH-3T3 cells, evaluated the expression by Western blot (Figure 7A), and performed focus formation assays. GILZ significantly inhibited the number of RasV12-induced transformation foci (P < 0.05; Figure 7A), suggesting that GILZ inhibition of cell transformation occurred through interference of GILZ with Ras signaling. Immunoprecipitation experiments to detect the binding of GILZ to RasV12 showed that GILZ bound oncogenic Ras at the same level as wild-type Ras (Figure 7B).
Figure 7. GILZ reduces Ras-driven transformation and tumorigenicity.
(A) NIH-3T3 cells were transiently transfected with HA-tagged RasV12 together with either control or myc-GILZ vector. After 14 days, the dishes were stained with 0.1% crystal violet. Quantification of the foci in each dish and the expression of transfected vectors are shown at right. The data represent an average from 3 dishes in 3 independent experiments each. (B) Lysates of NIH-3T3 cells transiently cotransfected with Myc-GILZ and either HA-RasV12 or wild-type Xp-Ras were immunoprecipitated with anti-HA and anti-Xp Abs, respectively. Immunoreactive proteins were revealed with anti-myc Ab. Whole-cell lysates were loaded to control plasmid expression. (C) NIH-3T3 cells were transiently transfected as described above and injected subcutaneously into SCID mice (5 per group). Tumor growth was scored at the indicated times as tumor diameter and tumor weight (left). Plasmid expression was evaluated 2 days after transfection by Western blot (right). (D) NIH-3T3 cells transfected with HA-RasV12 with or without Xp-GILZ were double-selected with antibiotics, and plates were photographed 2 weeks after transfection, at which time the cells were used to assess expression levels by Western blot with the Abs indicated in E. (E) The cells described in D were subcutaneously injected into SCID mice, and tumor growth was scored at the indicated times. After excision, representative tumors were photographed by a digital camera. *P < 0.05; **P < 0.01.
Raf-induced cell transformation was also inhibited by GILZ, although this inhibition was less evident due to the low transforming activity of Raf (data not shown). We next evaluated whether GILZ reduces the tumorigenic potential of NIH-3T3 transfected with oncogenic RasV12. For this purpose, we injected NIH-3T3 cells expressing RasV12 or RasV12+GILZ (Figure 7C) subcutaneously in immunocompromised SCID mice and followed the tumor growth up to 30 days. In mice injected with RasV12-transfected NIH-3T3 cells, large tumors were readily observed, whereas in mice injected with RasV12-GILZ–transfected NIH-3T3 cells, tumors appeared later and were smaller. Tumor weight was scored 30 days after injection and mice inoculated with cells transfected with both RasV12 and GILZ had tumors with significantly lower weight than those injected with RasV12-transfected cells (P < 0.05; Figure 7C). After excision, representative tumors were photographed by digital camera (Figure 7C). To evaluate the effect of sustained GILZ expression, we next generated RasV12 or RasV12-GILZ stable-transfected cell lines. The morphological changes such as transformation foci and irregular growth pattern associated with NIH-3T3 transformation were less in RasV12-GILZ-transfected cells compared with RasV12-transfected cells (Figure 7D). When stable-transfected cells, analyzed by Western blot for protein expression (Figure 7E), were injected in SCID mice, although the tumor incidence was similar in RasV12 and RasV12-GILZ-injected mice (data not shown), this last group had smaller tumors (P < 0.01 for tumor diameter and weight on day 18; Figure 7E). These data indicate that GILZ interferes with Ras-induced transformation of NIH-3T3 cells.
GILZ regulates concanavalin A–induced T cell growth and is required for the antiproliferative effect of DEX.
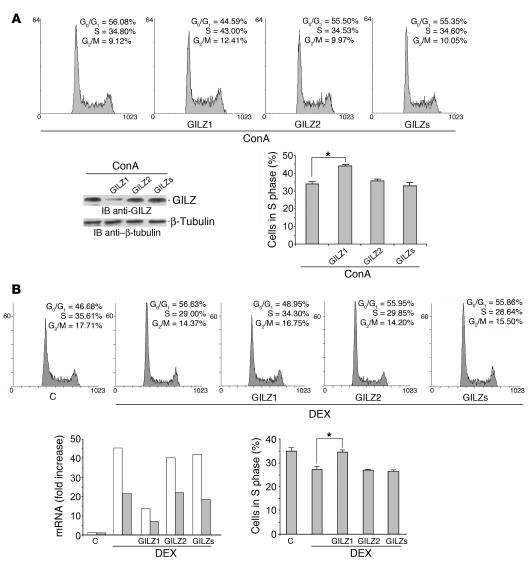
We evaluated the role of GILZ in controlling proliferation of primary T lymphocytes. For this purpose, splenic T lymphocytes transfected with GILZ-specific siRNA (GILZ1, GILZ2) or scrambled GILZ siRNA (GILZs) were stimulated with concanavalin A (ConA) and the cell cycle evaluated by propidium iodide (PI) assay. GILZ silencing was evaluated by Western blot (Figure 8A). The results indicated that weakening GILZ expression decreased the percentage of cells in the G0/G1 phase and increased the percentage of cells in the S phase of the cell cycle (P < 0.05; Figure 8A). These results confirm the physiological role of GILZ in cell proliferation mechanisms.
Figure 8. GILZ controls T cell proliferation and is responsible for GC-mediated anti-proliferative activity.
(A) Splenic T lymphocytes were transfected with GILZ1, GILZ2, or GILZs siRNAs and stimulated with ConA (1 μg/ml) for 3 days. Shown are the cell cycle, evaluated by PI assay (DNA content is shown on x axis and number of nuclei is shown on y axis), and GILZ expression, evaluated by Western blot, of a representative experiment. (B) 3DO cells were transfected with GILZ1, GILZ2, or GILZs siRNAs 1 hour before and 3 hours after DEX treatment (100 nM) (DNA content is shown on x axis and number of nuclei is shown on y axis). Shown are GILZ mRNA levels by real-time PCR 6 and 18 hours (white and gray bars, respectively) after silencing and cell cycle analysis by PI assay 18 hours after silencing, from a representative experiment. Histograms in A and B represent the average percent of cells in S phase from 3 independent experiments. *P < 0.05.
It has been shown that GCs inhibit cell proliferation (5). The effect of GILZ on cell proliferation prompted us to examine directly whether GILZ could be responsible for DEX-induced antiproliferative effects. We treated 3DO T cells with GILZ-specific siRNA prior to DEX treatment. According to previous data (37), GILZ was upregulated in 3DO T cells after 6 hours of DEX treatment and then declined over time (Figure 8B). Treatment with GILZ1 but not GILZ2 or GILZs siRNAs resulted in inhibition of transcript expression at 6 and 18 hours after transfection (Figure 8B). As expected, DEX treatment significantly reduced the number of cells in S phase, while GILZ silencing completely blocked the antiproliferative effect of DEX (Figure 8B; upper panel shows a representative cell cycle experiment; lower panel the statistically significant increase in number of cells in S phase upon GILZ1 transfection, P < 0.05).
Although 3DO treatment with DEX for 18 hours induced apoptosis in 9%–12% of cells, GILZ silencing did not result in a difference in apoptosis level (data not shown). Together, these results indicate that GILZ has a physiological role in T cell proliferation and is required to achieve the full antiproliferative effects of GCs.
Discussion
The results described in this study indicate that GILZ binds Ras, inhibits downstream Ras-dependent signals, functions as a physiological brake on cell proliferation, and is required for the antiproliferative activity of GCs.
We have previously shown that GILZ binds to Raf and downmodulates ERK activation (39). Here, we show that GILZ binds not only Raf but also Ras and that this binding depends on different molecular domains. In fact, while the first 60 aa of GILZ are responsible for binding to Raf at the N-terminal domain, binding to Ras is mediated via the TSC, which by itself is necessary and sufficient for binding. In addition, we found that Ras, Raf, and GILZ interact to form a ternary complex, the formation of which depends on the activation state of Ras. This suggests that GILZ is particularly important in activated cells. However, the formation of the trimeric complex does not preclude the formation of dimeric GILZ-Ras and GILZ-Raf complexes from occurring. As Ras activation levels regulate Ras-GILZ and Ras-Raf binding, it is reasonable to propose that a trimeric conformation may coexist with dimeric Ras-GILZ and Raf-GILZ conformations. When Ras is not activated, GILZ mainly binds Raf. However, the activation of Ras leads to both a greater affinity of GILZ for Ras and an increase in the affinity of Raf for Ras that is greater than the affinity of Raf for GILZ. In addition, more GILZ was found in the trimeric conformation when Ras was less activated, which suggests that, for trimeric conformation, Raf-GILZ binding is more important than Ras-GILZ binding. Importantly, GILZ interactions with Ras and Raf are physiological, as they are detectable in primary spleen T lymphocytes and thymocytes and their presence suggests that GILZ has a regulatory role on Ras activity.
As a consequence of binding to Ras and Raf, GILZ inhibits downstream signals such as ERK, AKT, and Rb phosphorylation as well as cyclin D1 expression, which are important mediators of cell proliferation. In fact, we found that stable GILZ transfection of NIH-3T3 cells induces a significant decrease in cell number and [3H]-thymidine uptake as well as an enrichment of the G0/G1 phase of the cell cycle, which correlates with inhibition of ERK, AKT, and Rb phosphorylation and cyclin D1 expression. In particular, GILZ binding to Ras is necessary for GILZ antiproliferative activity, as GILZΔTSC did not bind to Ras and did not inhibit Ras-triggered cell proliferation, while GILZ-TSC-LZ, which binds Ras, inhibited Ras-driven proliferation. This suggests that, by binding Ras, GILZ blocks the propagation of signaling to Raf and AKT, which are both involved in mediating Ras-triggered cell proliferation (12, 46). In fact, the inhibitory effect of GILZ can be overcome by either activated AKT or activated Raf.
Interestingly, GILZ inhibited Ras- and Raf-induced cell proliferation, thus suggesting that the antiproliferative effect may be mediated by trimeric Ras-Raf-GILZ or dimeric Ras-GILZ and Raf-GILZ conformations. Raf-ERK and PI3K-AKT are considered the 2 major pathways modulating cyclin D1 transcriptional activation (17, 18), wherein ERK, but not AKT, is considered important for Raf-regulated cell signaling and both ERK and AKT are considered important for Ras-regulated cell signaling (11). Because GILZ inhibits AKT and ERK activation and affects Ras- and Raf-dependent cell proliferation, it is conceivable that GILZ may target Ras-dependent AKT and ERK activation as well as Raf-dependent ERK activation. Thus, GILZ can bind directly to Raf and inhibit ERK but not AKT, and this is sufficient to inhibit the cell proliferation. On the other hand, the binding of GILZ to Ras, by an allosteric effect that has a negative impact on the activation of ERK and AKT (generally thought to be indispensable for mediating the transforming potential of Ras [ref. 45]), inhibits both pathways, which cooperate in inducing cell proliferation (11, 12).
A notable result here is that Ras-GILZ interaction affects cell transformation. The effect of GILZ on the Ras-induced transformation of NIH-3T3, together with the signaling regulation, supports previous reports that Ras transformation requires multiple downstream pathways including ERK (43) and AKT (47). The inhibitory effect of GILZ on Ras-mediated transformation suggests that GILZ could contribute to modulation of Ras oncogenic activity and warrants further investigation as to the possible structural and/or functional alteration of GILZ in human tumors.
The antiproliferative action of GILZ suggests that it plays a physiological role in the control of cell growth and that its upregulation may be a mechanism of pharmacological importance. Endogenous GCs, the end-effectors of the hypothalamus-pituitary-adrenal axis, play an important role in regulating the homeostasis of the immune system (48). Moreover, GCs are used as pharmacological agents due to their immunosuppressive and antiinflammatory properties. Their activity is due to their complex action on innate and adaptive immunity reflecting GC capacity to inhibit T lymphocyte activation (49, 50). Previous studies demonstrate that GCs inhibit T cell proliferation (5–7). In particular, GCs have been shown to block antigen and mitogen-induced T lymphocyte proliferation, and various molecular mechanisms have been suggested as being responsible for this effect (5). Our results show that GILZ inhibition by siRNA results in increased cell proliferation consequent to ConA stimulation, which suggests that GILZ plays a role in regulating T cell activation and growth. Most notable, silencing GILZ in the 3DO T cell line completely inhibited the DEX-induced antiproliferative effect. These results indicate that GILZ has a physiological role in cell proliferation machinery and is required for the pharmacological action of GCs.
Methods
Cells, cell lines, and animals.
The COS-7 and the NIH-3T3 cell lines were maintained in culture in DMEM supplemented with 10% FCS and antibiotics streptomycin and penicillin. A spontaneously dividing CD3+CD4+ subline of the Ova-specific hybridoma T cell line, 3DO, was used for the experiments. Cells were maintained in logarithmic growth in RPMI 1640 supplemented with FCS, 20 mM HEPES, and antibiotics. Thymus and spleen cells were obtained from 4- to 6-week-old C3H/HeN mice (Charles River Laboratories). Spleen T cells were separated by negative selection using Pan T Cell Isolation Kit (Miltenyi Biotec), according to the manufacturer’s instructions, and stimulated for 30 minutes with plastic-bound anti-CD3 Ab (BD Biosciences — Pharmingen) as previously described (41). SCID mice, 8–10 weeks old, were from Charles River Laboratories. Animal care was in compliance with regulations in Italy (Decreto Ministeriale 116192), Europe (Official Journal of European Commission L 358/1 12/18/1986), and USA (Animal Welfare Assurance A5594-01, Department of Health and Human Services, Washington, DC, USA). Animal care and use was approved by the Ethics Committee of the University of Perugia.
Plasmid construction.
A full-length cDNA encoding GILZ was inserted into pcDNA3.1/Myc-His for myc (myc-GILZ) or pcDNA6/His for Xp (Xp-GILZ) epitope tagging (Invitrogen) as previously described (39). Deletion and substitution mutants of GILZ were generated by PCR, using pCR3.1-GILZ as a template and mutant synthetic oligonucleotides as primers, and subcloned in expression vectors. GST-tagged vectors, containing GILZ or its mutants, were generated by subcloning into BamHI and NotI sites of the mammalian expressing vector pEBG (kindly provided by W. Kolch, Beatson Institute for Cancer Research, Glasgow, United Kingdom). pUSEamp containing human wild-type or activated Raf-1 (substituting aspartic acid for tyrosine at residue 340), human wild-type and activated H-Ras (substituting leucine for glutamine at position 61), and dominant-negative H-Ras (substituting asparagine for serine at position 17) were from Upstate. Wild-type, activated, and dominant-negative Xp-Ras was obtained by subcloning them into pcDNA6/His. HA-tagged H-RasV12 was kindly provided by Kun-Liang Guan (University of Michigan Medical School, Ann Arbor, Michigan, USA); pEF-m/Raf-CAAX was provided by Richard Marais (Institute of Cancer Research, London, United Kingdom); and pEBG-Raf-1 and Flag-Raf-1 were provided by W. Kolch (Beatson Institute for Cancer Research).
Cell transfection.
Transfected 3DO clones were prepared as previously described (37). NIH-3T3 cells were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions, with Myc-GILZ, H-Ras (wild-type or V12), and/or Raf (wild-type, constitutively active, or CAAX). In some experiments, 48 hours after transfection, the cells were selected by G418 (700 μg/ml; Invitrogen) for 12 days before starting the experiments. For RasV12-GILZ stable-transfected cell line, NIH-3T3 were first transfected with H-RasV12, selected for 7 days with G418, and then transfected with Xp-GILZ and selected for a further 7 days with blasticidin (1 μg/ml; Sigma-Aldrich). For growth curves, 10,000 or 5,000 cells were plated in 12-well plates. At each time point, duplicate wells were trypsinized and the cells were counted in a Beckman Coulter counter. COS-7 cells were cotransfected using DEAE-dextran as previously described (39). In some experiments, transfected cells were serum starved (1% FCS) and treated for 20 minutes with PMA (100 ng/ml; Sigma-Aldrich).
Coimmunoprecipitation and Western blot.
Forty-eight hours after transfection, whole-cell extracts were prepared and immunoprecipitations performed in IP buffer (41). Antigen-Ab complexes were precipitated with protein A or G bound to Sepharose beads (Sigma-Aldrich).
Extracted or immunoprecipitated proteins were separated by SDS-PAGE and analyzed by Western blotting. Primary Abs included polyclonal rabbit antiserum recognizing GILZ (37), anti-mouse phospho-ERK1/2 and anti-mouse ERK1/2, anti–phospho-Rb (Cell Signaling Technology), anti–Raf-1, anti–pan Ras, anti–cyclin D1 (Santa Cruz Biotechnology Inc.), and Abs specific for Xp and Myc epitopes (Invitrogen) or GST (EMD Biosciences). Secondary Abs included horseradish peroxidase-labeled (Pierce Biotechnology) and anti–β-tubulin mAb (Sigma-Aldrich) were used as controls. The bands were analyzed using an 1D Image Analysis software (version 3.5.0; Eastman Kodak).
For analysis of complex formation, wild-type Ras or activated or dominant-negative Xp-tagged Ras was cotransfected along with myc-GILZ and either GST-Raf or pEBG empty vector. Cell extracts were incubated with glutathione-sepharose beads (Amersham Biosciences) in buffer containing 25 mM HEPES (pH 7.5), 150 mM NaCl, 1% Igepal CO-630 (Sigma-Aldrich), 10 mM MgCl2, 1 mM EDTA, 2% glycerol, and 10 μg/ml leupeptin and aprotinin. GST fusion and associated proteins were eluted with 5 mM glutathione and subjected to immunoprecipitation with the indicated anti-tag Abs.
The antigen-Ab complexes were revealed by enhanced chemiluminescence according to the manufacturer’s instructions (SuperSignal; Pierce Biotechnology). Immunoprecipitation experiments were only considered to be valid once they had repeated at least 3 times with similar results and were controlled with the appropriate pre-immune serum (data not shown).
Transfection efficiency, assessed by intracellular triple staining of transfected proteins and FACS analysis, was 63% ± 5.6%. Triple intracellular staining was done using the BD Biosciences Cytofix/Cytoperm fixation/permeabilization kit and a combination of FITC-conjugated anti-myc, anti-Xp and biotin-conjugated anti-GST antibodies, followed by CY5-conjugated streptavidin and PE-conjugated anti-mouse Ab (BD Biosciences). FACS analyses included the use of isotype control Abs and were performed using Expo 32 ADC Analysis 1.1C (Beckman Coulter).
In vitro protein-binding assay.
Extracts were made from 3DO cells or thymocytes untreated or treated with DEX (100 nM) or PMA (100 ng/ml) as previously described (40). GST-GILZ and GST-Ras fusion proteins were prepared as previously described (37, 39). Pulldown assays were performed by incubating the fusion proteins, loaded on glutathione-sepharose beads, with cellular lysates in binding buffer (39), for 18 hours at 4° C. The beads were washed extensively, resuspended in sample buffer, and analyzed by SDS-PAGE and Western blotting with the indicated Abs.
Wild-type H-Ras was translated in vitro as previously described (40) with [35S]Methionine (Amersham Biosciences). In vitro–translated proteins were diluted with the binding buffer (25 mM HEPES, pH 7.5, 10% glycerol, 50 mM NaCl, 0.05% NP-40, 1 mM DTT) and precleared with glutathione-sepharose beads for 45 minutes at 4°C. GST, GST-GILZ, or GST-Ras, bound to glutathione-sepharose beads, was incubated with in vitro–translated proteins for 18 hours at 4°C. The beads were subsequently washed 5 times with binding buffer. Bound proteins were analyzed by SDS-PAGE and revealed by autoradiography.
Cell cycle analysis.
Cell cycle profiles were analyzed by flow cytometry to determine DNA content of cell nuclei stained with PI (37). Briefly, cells were collected by centrifugation, washed in PBS, and fixed for 30 minutes at 4°C in 70% ethanol. DNA was stained by incubating the cells in PBS containing PI (50 μg/ml) and RNase A (50 μg/ml) (Sigma-Aldrich), and fluorescence was measured and analyzed using a Becton Dickinson FACScan and the Cell Fit software.
[3H]-thymidine uptake.
Twenty-four hours after transfection, NIH-3T3 cells were grown in low-serum–containing medium for 24 hours and then trypsinized. A total of 10,000; 5,000; or 2,500 cells was seeded in triplicate in 96-well plates and cultured for the indicated times (Figure 6B). Empty-vector– or GILZ-transfected 3DO stable clones (1 × 105/well) were starved for 48 hours and then cultured for 24 hours in the presence of serum. In all experiments, [3H]-thymidine (2 μCi/well; Amersham Biosciences) was added 6 hours before harvesting with a multiple suction-filtration apparatus (Mash II; Skatron Instruments) fitted on a fiberglass filter. Radioactivity was counted in a β-counter (Packard).
Focus-forming and transformation assays.
For focus assays, NIH-3T3 cells were transfected with H-RasV12 or pEF-m/Raf-CAAX, along with myc-GILZ expression vectors in 6-well plates. Cells were split 3 days following transfection. One set was used to assess whether the transfected cDNAs were successfully expressed, while the 2 remaining sets were used for focus assays. The growth medium was replaced every 3 days. Foci visualized 2 weeks after transfection by crystal violet staining were counted.
SCID mice were inoculated subcutaneously with pools of NIH-3T3 cells (2 × 106 per injection) transfected with H-RasV12 with or without myc-GILZ expression vectors. Vector control cells were injected into the opposite flank. Equal numbers of cells were injected, and Western blotting with anti-tag Ab was performed to ensure that equal levels of the proteins were expressed. At the times indicated (Figure 7, C and E), tumor growth was scored and animals were killed because of the tumor burden in compliance with animal welfare regulations. GILZ tumor thickness was evaluated using a digital micrometer (Mitutoyo).
Indirect immunofluorescence and microscope analysis.
Myc-GILZ/pUSEamp-Ras–cotransfected COS-7 cells, treated or untreated for 20 minutes with PMA, were adhered directly onto glass slides. Cells were permeabilized by incubation in methanol for 20 minutes at –20°C. After washing 3 times in PBS and blocking in PBS containing 3% BSA and 1% glycine (blocking buffer), the cells were incubated for 1 hour at room temperature with anti-myc and anti-Ras Abs. Cells were then incubated for 1 hour with secondary Abs (anti-mouse FITC-conjugated and anti-rabbit Texas Red–conjugated Abs) in blocking buffer containing 1 ng/ml DAPI, then washed, and the slides were mounted with a coverglass. The slides were analyzed by Zeiss LSM 510 confocal microscope (Zeiss) using the 488-nm and 543-nm laser lines for excitation. Green and red fluorescence emissions were collected with the single-track technique (51). The 3D for LSM 510 software (version 3.5; Zeiss) was used for confocal regulations and image acquisition. The images were then transferred to an Silicon Graphics Octane workstation for further processing. Colocalization analysis was performed using the Imaris software (version 4.5.2; Bitplane AG).
siRNA preparation, transfection, and real-time PCR.
GILZ-specific double-stranded–nucleotide siRNA (GILZ1, sense 5′-CAAUUUCUCCACCUCCUUCUUtt-3′ and antisense 5′-AAGAAGGAGGUGGAGAAAUUGtt-3′; GILZ2, sense 5′-CAAGAUUGAGCAGGCCAUGtt-3′ and antisense 5′-CAUGGCCUGCUCAAUCUUGtt-3′) and scrambled siRNA (GILZs, sense 5′-GUACCGGACGAGUUAGAACtt-3′ and antisense 5′-GUUCUAACUCGUCCGGUACtt-3′) were synthesized and annealed by the manufacturer (MWG-Biotech). siRNA transfection was performed as previously described (52). siRNA nucleotides (6.7 μg/ml) were added to splenic T cells 1 hour before and 24 hours and 48 hours after ConA treatment (1 μg/ml) and to 3DO cells 1 hour before and 3 hours after DEX treatment (100 nM).
For real-time PCR, the sense primer for GILZ was 5′-GGTGGCCCTAGACAACAAGA-3′, and the antisense primer was 5′-tcttctcaagcagctcacga-3′. For GAPDH the sense primer was 5′-GCCTTCCGTGTTCCTACCC-3′ and the antisense primer was 5′-CAGTGGGCCCTCAGATGC-3′. Polymerase chain reaction was done in the Chromo 4 (Bio-Rad) using DyNAmo HS SYBR Green qPCR Kit (Finnzymes; Celbio). Gene expression was quantified relative to GAPDH expression. The relative expression value for the control samples were defined as 1, and all other values were plotted in comparison.
Statistics.
Each experiment was performed at least 3 times. Representative experiments are shown. Because the distribution of the data was not normal, nonparametric tests (Kruskal-Wallis) were adopted for statistical evaluation. P values less than 0.05 were considered to be significant.
Supplementary Material
Acknowledgments
We are grateful to W. Kolch for providing pEBG-Raf-1 and Flag-Raf-1 expression plasmids, to K.L. Guan for providing H-RasV12, and to R. Marais for providing pEF-m/RAF-CAAX vectors. This study was supported by the Associazione Italiana per la Ricerca sul Cancro (AIRC), Milan, Italy.
Footnotes
Nonstandard abbreviations used: AKT, AKT/PKB serine/threonine kinase; ConA, concanavalin A; DEX, dexamethasone; GC, glucocorticoid; GILZ, glucocorticoid-induced leucine zipper; GILZs, scrambled GILZ; GR, GC receptor; GST, glutathione-S-transferase; LZ, leucine zipper; PI, propidium iodide; PRR, proline-rich region; Rb, retinoblastoma; TSC, tuberous sclerosis complex box; Xp, Xpress.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 117:1605–1615 (2007). doi:10.1172/JCI30724
References
- 1.Payne D.N., Adcock I.M. Molecular mechanisms of corticosteroid actions. Paediatr. Respir. Rev. 2001;2:145–150. doi: 10.1053/prrv.2000.0122. [DOI] [PubMed] [Google Scholar]
- 2.De Bosscher K., Vanden Berghe W., Haegeman G. Mechanisms of anti-inflammatory action and of immunosuppression by glucocorticoids: negative interference of activated glucocorticoid receptor with transcription factors. J. Neuroimmunol. 2000;109:16–22. doi: 10.1016/s0165-5728(00)00297-6. [DOI] [PubMed] [Google Scholar]
- 3.Adcock I.M. Glucocorticoid-regulated transcription factors. Pulm. Pharmacol. Ther. 2001;14:211–219. doi: 10.1006/pupt.2001.0283. [DOI] [PubMed] [Google Scholar]
- 4.Wikstrom A.C. Glucocorticoid action and novel mechanisms of steroid resistance: role of glucocorticoid receptor-interacting proteins for glucocorticoid responsiveness. J. Endocrinol. 2003;178:331–337. doi: 10.1677/joe.0.1780331. [DOI] [PubMed] [Google Scholar]
- 5.Almawi W.Y., Abou Jaoude M.M., Li X.C. Transcriptional and post-transcriptional mechanisms of glucocorticoid antiproliferative effects. Hematol. Oncol. 2002;20:17–32. doi: 10.1002/hon.684. [DOI] [PubMed] [Google Scholar]
- 6.Ausserlechner M.J., Obexer P., Bock G., Geley S., Kofler R. Cyclin D3 and c-MYC control glucocorticoid-induced cell cycle arrest but not apoptosis in lymphoblastic leukemia cells. Cell Death Differ. 2004;11:165–174. doi: 10.1038/sj.cdd.4401328. [DOI] [PubMed] [Google Scholar]
- 7.Rhee K., Bresnahan W., Hirai A., Hirai M., Thompson E.A. c-Myc and cyclin D3 (CcnD3) genes are independent targets for glucocorticoid inhibition of lymphoid cell proliferation. Cancer Res. 1995;55:4188–4195. [PubMed] [Google Scholar]
- 8.Clark A.R., Lasa M. Crosstalk between glucocorticoids and mitogen-activated protein kinase signalling pathways. Curr. Opin. Pharmacol. 2003;3:404–411. doi: 10.1016/s1471-4892(03)00073-0. [DOI] [PubMed] [Google Scholar]
- 9.Malumbres M., Barbacid M. RAS oncogenes: the first 30 years. Nat. Rev. Cancer. 2003;3:459–465. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- 10.Vojtek A.B., Der C.J. Increasing complexity of the Ras signaling pathway. J. Biol. Chem. 1998;273:19925–19928. doi: 10.1074/jbc.273.32.19925. [DOI] [PubMed] [Google Scholar]
- 11.Katz M.E., McCormick F. Signal transduction from multiple Ras effectors. Curr. Opin. Genet. Dev. 1997;7:75–79. doi: 10.1016/s0959-437x(97)80112-8. [DOI] [PubMed] [Google Scholar]
- 12.Mirza A.M., et al. Cooperative regulation of the cell division cycle by the protein kinases RAF and AKT. Mol. Cell. Biol. 2004;24:10868–10881. doi: 10.1128/MCB.24.24.10868-10881.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Downward J. Cell cycle: routine role for Ras. Curr. Biol. 1997;7:R258–R260. doi: 10.1016/s0960-9822(06)00116-3. [DOI] [PubMed] [Google Scholar]
- 14.Gille H., Downward J. Multiple ras effector pathways contribute to G(1) cell cycle progression. J. Biol. Chem. 1999;274:22033–22040. doi: 10.1074/jbc.274.31.22033. [DOI] [PubMed] [Google Scholar]
- 15.Kerkhoff E., Rapp U.R. Cell cycle targets of Ras/Raf signalling. Oncogene. 1998;17:1457–1462. doi: 10.1038/sj.onc.1202185. [DOI] [PubMed] [Google Scholar]
- 16.Stacey D.W. Cyclin D1 serves as a cell cycle regulatory switch in actively proliferating cells. Curr. Opin. Cell Biol. 2003;15:158–163. doi: 10.1016/s0955-0674(03)00008-5. [DOI] [PubMed] [Google Scholar]
- 17.Lavoie J.N., L’Allemain G., Brunet A., Muller R., Pouyssegur J. Cyclin D1 expression is regulated positively by the p42/p44MAPK and negatively by the p38/HOGMAPK pathway. J. Biol. Chem. 1996;271:20608–20616. doi: 10.1074/jbc.271.34.20608. [DOI] [PubMed] [Google Scholar]
- 18.Muise-Helmericks R.C., et al. Cyclin D expression is controlled post-transcriptionally via a phosphatidylinositol 3-kinase/Akt-dependent pathway. J. Biol. Chem. 1998;273:29864–29872. doi: 10.1074/jbc.273.45.29864. [DOI] [PubMed] [Google Scholar]
- 19.Morrison D.K., Davis R.J. Regulation of MAP kinase signaling modules by scaffold proteins in mammals. Annu. Rev. Cell Dev. Biol. 2003;19:91–118. doi: 10.1146/annurev.cellbio.19.111401.091942. [DOI] [PubMed] [Google Scholar]
- 20.Kolch W. Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nat. Rev. Mol. Cell Biol. 2005;6:827–837. doi: 10.1038/nrm1743. [DOI] [PubMed] [Google Scholar]
- 21.Stewart S., et al. Kinase suppressor of Ras forms a multiprotein signaling complex and modulates MEK localization. Mol. Cell. Biol. 1999;19:5523–5534. doi: 10.1128/mcb.19.8.5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kortum R.L., Lewis R.E. The molecular scaffold KSR1 regulates the proliferative and oncogenic potential of cells. Mol. Cell. Biol. 2004;24:4407–4416. doi: 10.1128/MCB.24.10.4407-4416.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roy F., Laberge G., Douziech M., Ferland-McCollough D., Therrien M. KSR is a scaffold required for activation of the ERK/MAPK module. Genes Dev. 2002;16:427–438. doi: 10.1101/gad.962902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang H.G., Takayama S., Rapp U.R., Reed J.C. Bcl-2 interacting protein, BAG-1, binds to and activates the kinase Raf-1. Proc. Natl. Acad. Sci. U. S. A. 1996;93:7063–7068. doi: 10.1073/pnas.93.14.7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li W., Han M., Guan K.L. The leucine-rich repeat protein SUR-8 enhances MAP kinase activation and forms a complex with Ras and Raf. Genes Dev. 2000;14:895–900. [PMC free article] [PubMed] [Google Scholar]
- 26.Irie K., et al. Stimulatory effects of yeast and mammalian 14-3-3 proteins on the Raf protein kinase. Science. 1994;265:1716–1719. doi: 10.1126/science.8085159. [DOI] [PubMed] [Google Scholar]
- 27.Silverstein A.M., Grammatikakis N., Cochran B.H., Chinkers M., Pratt W.B. p50(cdc37) binds directly to the catalytic domain of Raf as well as to a site on hsp90 that is topologically adjacent to the tetratricopeptide repeat binding site. J. Biol. Chem. 1998;273:20090–20095. doi: 10.1074/jbc.273.32.20090. [DOI] [PubMed] [Google Scholar]
- 28.Matheny S.A., et al. Ras regulates assembly of mitogenic signalling complexes through the effector protein IMP. Nature. 2004;427:256–260. doi: 10.1038/nature02237. [DOI] [PubMed] [Google Scholar]
- 29.Yeung K., et al. Suppression of Raf-1 kinase activity and MAP kinase signalling by RKIP. Nature. 1999;401:173–177. doi: 10.1038/43686. [DOI] [PubMed] [Google Scholar]
- 30.Giehl K. Oncogenic Ras in tumour progression and metastasis. Biol. Chem. 2005;386:193–205. doi: 10.1515/BC.2005.025. [DOI] [PubMed] [Google Scholar]
- 31.Widen C., Zilliacus J., Gustafsson J.A., Wikstrom A.C. Glucocorticoid receptor interaction with 14-3-3 and Raf-1, a proposed mechanism for cross-talk of two signal transduction pathways. J. Biol. Chem. 2000;275:39296–39301. doi: 10.1074/jbc.M006943200. [DOI] [PubMed] [Google Scholar]
- 32.Kassel O., et al. Glucocorticoids inhibit MAP kinase via increased expression and decreased degradation of MKP-1. EMBO J. 2001;20:7108–7116. doi: 10.1093/emboj/20.24.7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leis H., et al. Glucocorticoid receptor counteracts tumorigenic activity of Akt in skin through interference with the phosphatidylinositol 3-kinase signaling pathway. Mol. Endocrinol. 2004;18:303–311. doi: 10.1210/me.2003-0350. [DOI] [PubMed] [Google Scholar]
- 34.Takayama S., Rogatsky I., Schwarcz L.E., Darimont B.D. The glucocorticoid receptor represses cyclin D1 by targeting the Tcf-beta-catenin complex. J. Biol. Chem. 2006;281:17856–17863. doi: 10.1074/jbc.M602290200. [DOI] [PubMed] [Google Scholar]
- 35.Xu D., Makkinje A., Kyriakis J.M. Gene 33 is an endogenous inhibitor of epidermal growth factor (EGF) receptor signaling and mediates dexamethasone-induced suppression of EGF function. J. Biol. Chem. 2005;280:2924–2933. doi: 10.1074/jbc.M408907200. [DOI] [PubMed] [Google Scholar]
- 36.Hiragun T., Peng Z., Beaven M.A. Dexamethasone up-regulates the inhibitory adaptor protein Dok-1 and suppresses downstream activation of the mitogen-activated protein kinase pathway in antigen-stimulated RBL-2H3 mast cells. Mol. Pharmacol. 2005;67:598–603. doi: 10.1124/mol.104.008607. [DOI] [PubMed] [Google Scholar]
- 37.D’Adamio F., et al. A new dexamethasone-induced gene of the leucine zipper family protects T lymphocytes from TCR/CD3-activated cell death. Immunity. 1997;7:803–812. doi: 10.1016/s1074-7613(00)80398-2. [DOI] [PubMed] [Google Scholar]
- 38.Cannarile L., et al. Cloning, chromosomal assignment and tissue distribution of human GILZ, a glucocorticoid hormone-induced gene. Cell Death Differ. 2001;8:201–203. doi: 10.1038/sj.cdd.4400798. [DOI] [PubMed] [Google Scholar]
- 39.Ayroldi E., et al. Glucocorticoid-induced leucine zipper inhibits the Raf-extracellular signal-regulated kinase pathway by binding to Raf-1. Mol. Cell. Biol. 2002;22:7929–7941. doi: 10.1128/MCB.22.22.7929-7941.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soundararajan R., Zhang T.T., Wang J., Vandewalle A., Pearce D. A novel role for glucocorticoid-induced leucine zipper protein in epithelial sodium channel-mediated sodium transport. . J. Biol. Chem. 2005;280:39970–39981. doi: 10.1074/jbc.M508658200. [DOI] [PubMed] [Google Scholar]
- 41.Ayroldi E., et al. Modulation of T-cell activation by the glucocorticoid-induced leucine zipper factor via inhibition of nuclear factor kappaB. Blood. 2001;98:743–753. doi: 10.1182/blood.v98.3.743. [DOI] [PubMed] [Google Scholar]
- 42.Franklin R.A., et al. Ligation of the T cell receptor complex results in activation of the Ras/Raf-1/MEK/MAPK cascade in human T lymphocytes. J. Clin. Invest. 1994;93:2134–2140. doi: 10.1172/JCI117209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cowley S., Paterson H., Kemp P., Marshall C.J. Activation of MAP kinase kinase is necessary and sufficient for PC12 differentiation and for transformation of NIH 3T3 cells. Cell. 1994;77:841–852. doi: 10.1016/0092-8674(94)90133-3. [DOI] [PubMed] [Google Scholar]
- 44.Kato-Stankiewicz J., et al. Inhibitors of Ras/Raf-1 interaction identified by two-hybrid screening revert Ras-dependent transformation phenotypes in human cancer cells. Proc. Natl. Acad. Sci. U. S. A. 2002;99:14398–14403. doi: 10.1073/pnas.222222699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li W., Zhu T., Guan K.L. Transformation potential of Ras isoforms correlates with activation of phosphatidylinositol 3-kinase but not ERK. J. Biol. Chem. 2004;279:37398–37406. doi: 10.1074/jbc.M405730200. [DOI] [PubMed] [Google Scholar]
- 46.Roovers K., Assoian R.K. Integrating the MAP kinase signal into the G1 phase cell cycle machinery. Bioessays. 2000;22:818–826. doi: 10.1002/1521-1878(200009)22:9<818::AID-BIES7>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 47.Sheng H., Shao J., DuBois R.N. Akt/PKB activity is required for Ha-Ras-mediated transformation of intestinal epithelial cells. J. Biol. Chem. 2001;276:14498–14504. doi: 10.1074/jbc.M010093200. [DOI] [PubMed] [Google Scholar]
- 48.Webster J.I., Tonelli L., Sternberg E.M. Neuroendocrine regulation of immunity. Annu. Rev. Immunol. 2002;20:125–163. doi: 10.1146/annurev.immunol.20.082401.104914. [DOI] [PubMed] [Google Scholar]
- 49.Rook G.A. Glucocorticoids and immune function. Baillieres Best Pract. Res. Clin. Endocrinol. Metab. 1999;13:567–581. doi: 10.1053/beem.1999.0044. [DOI] [PubMed] [Google Scholar]
- 50.Ashwell J.D., Lu F.W., Vacchio M.S. Glucocorticoids in T cell development and function. Annu. Rev. Immunol. 2000;18:309–345. doi: 10.1146/annurev.immunol.18.1.309. [DOI] [PubMed] [Google Scholar]
- 51. Hibbs, A.R., MacDonal, G., and Garsha, K. 2006. Practical confocal microscopy. In Handbook of biological confocal microscopy. 3rd edition. J.B. Pawley, editor. Springer Science and Business Media LLC. New York, New York, USA. 650–671. [Google Scholar]
- 52.Cannarile L., et al. 2006Increased GILZ expression in transgenic mice up-regulates Th-2 lymphokines. Blood. 1071039 – 1047. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.