Abstract
B cells from patients with common variable immunodeficiency (CVID) who are heterozygous for transmembrane activator and CAML interactor (TACI) mutation C104R, which abolishes ligand binding, fail to produce Igs in response to TACI ligand. It is not known whether this is due to haploinsufficiency or dominant interference. Using in vitro transfection assays, here we demonstrate that C104R and the corresponding murine TACI mutant, C76R, which also does not bind ligand, dominantly interfere with TACI signaling. This effect was dependent on preassociation of the mutants with WT TACI in the absence of ligand. The mutants did not interfere with ligand binding by WT TACI, suggesting that they may act by disrupting ligand-induced receptor rearrangement and signaling. This work demonstrates that TACI preassembles as an oligomeric complex prior to ligand binding and provides a mechanistic insight into how the heterozygous C104R TACI mutation can potentially lead to CVID.
Introduction
Common variable immunodeficiency (CVID) is the most common primary immunodeficiency disease that comes to medical attention. CVID is characterized by markedly reduced serum levels of IgG, IgA, and often IgM (1). In addition to having low levels of Igs, patients with CVID fail to produce specific antibodies after vaccination or exposure to antigens (2). CVID patients suffer from recurrent sinopulmonary, ear, and gastrointestinal infections and have increased risk of autoimmune and lymphoproliferative diseases (1). Most cases of CVID are sporadic, but about 10% are familial with a predominantly autosomal dominant inheritance (3). It has been recently found that TNFRSF13B, which encodes transmembrane activator and calcium-modulating ligand (CAML) interactor (TACI), is mutated in a small subgroup (approximately 5%–10%) of patients with CVID (4, 5). Most of these patients are heterozygous with respect to TACI mutation.
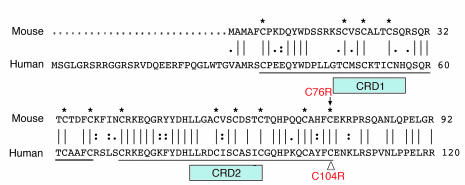
TACI is a cell-surface receptor that belongs to the TNF receptor (TNFR) superfamily and is mainly expressed on B cells (6). Structurally, TACI is a type III transmembrane protein. The extracellular (EC) domain contains 2 cysteine-rich domains (CRDs), which are hallmarks of the TNFR superfamily (6). Each CRD contains 6 highly conserved cysteines that participate in the formation of 3 intrachain disulfide bonds (Figure 1). Both human TACI (hTACI) and murine TACI (mTACI) have 2 CRD domains that are conserved between the 2 species (Figure 1). The TNF family members BAFF (B cell–activating factor) and APRIL (a proliferation-inducing ligand) are ligands for TACI. Recently, TACI was found to bind to proteoglycans including syndecan-2 (7, 8). Functionally, TACI has been shown to be important in Ig class switching, Ig production, and regulation of B cell homeostasis (9–12). TACI-deficient mice have low levels of serum IgA and IgM and impaired antibody response to the type II T-independent antigens pneumococcal vaccine polyvalent (Pneumovax) and TNP-Ficoll (11). In addition, as they age, these mice develop fatal disorders characterized by autoimmune nephritis and lymphoproliferation, leading to lymphoma in 15% of the mice (10).
Figure 1. Aligned aa sequences of human and murine TACI.
Cysteine residues that make up the TNFR motif have an asterisk above them; the CRD1 and CRD2 domains are underlined. The vertical lines indicate identical residues and the dots, similar residues. The open triangle denotes the human C104R mutation found in CVID and its corresponding mutation C76R in mTACI.
One of the most common TACI mutations in patients with CVID is C104R, which is located in the EC domain of the receptor (13). The mutated C104 residue is located in the second CRD of TACI (Figure 1) and forms a disulfide bond with C93. This residue is highly conserved together with its surrounding sequence in all species examined (our unpublished observations). It has been demonstrated that the C104R mutation abolishes binding of BAFF and APRIL to TACI (4, 5). Most CVID patients with a C104R mutation are heterozygous. Furthermore, we have reported that B cells from such patients fail to produce Igs in response to APRIL (4). Two explanations have been proposed to explain the deleterious effect of the heterozygous C104R mutation on B cell function. One invokes haploinsufficiency because one normal copy of TACI may not be sufficient for proper function. However, mice with a single copy of the TACI gene have a normal phenotype (11), strongly arguing that TACI haploinsufficiency may not cause B cell dysfunction. Alternatively, the C104R mutant may act as a dominant negative and interfere with the function of the normal allele.
Given the variable age of onset of CVID, which extends into the seventh decade of life, and the genetic heterogeneity of the human population, we concluded that the causality of TACI missense mutations is best established in a mouse model where mutations in mTACI that correspond to hTACI mutations found in CVID patients are introduced into a uniform genetic background. Prior to the generation of such a model, it was important to confirm that the mouse mutants behave similarly to the human mutants. We have taken advantage of the fact that the C104 residue in hTACI and its surrounding sequence are highly conserved in the mouse to test whether mutation of the corresponding residue C76 in mTACI abolishes ligand binding. We show that the C76R mutation abolishes the capacity of mTACI to bind ligand. More importantly, both the human and mouse mutants preassociate with WT TACI in a ligand-independent manner and act as dominant negative. Coexpression of the mutant with WT TACI did not interfere with ligand binding, suggesting that the mutant may disrupt ligand-induced receptor rearrangement and signaling.
Results
TACI monomers engage in ligand-independent homotypic interaction in transfected 293T cells.
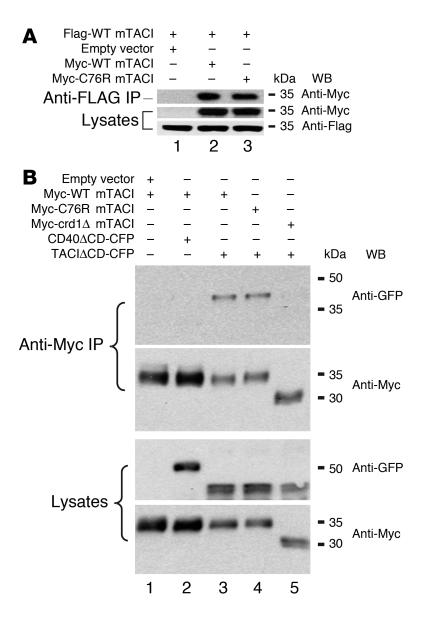
We first sought to determine whether TACI molecules transfected into 293T cells associate in the absence of ligand. 293T cells were cotransfected with vectors that encode Myc-tagged WT mTACI (Myc-WT mTACI) and Flag-tagged WT mTACI (Flag-WT mTACI). Figure 2A shows that Myc-WT mTACI was coprecipitated with Flag-WT mTACI (Figure 2A, lane 2). This interaction was not dependent on the presence of the cytoplasmic domain (CD), as it was still retained when the CD of TACI was replaced by cyan fluorescent protein (CFP) to generate TACIΔCD-CFP, as Myc-WT mTACI was able to coimmunoprecipitate TACIΔCD-CFP (Figure 2B, lane 3). However, deletion of CRD1, encoded by aa 1–40 (Figure 1), to generate TACIΔCRD1, abrogated association with TACIΔCD-CFP (Figure 2B, lane 5). The homotypic TACI interaction detected in 293T cells was specific, as another TNFR family receptor, CD40, did not interact with TACI under the same conditions (Figure 2B, lane 2). These results suggest that TACI monomers engage in a ligand-independent homotypic interaction that is dependent on the CRD1 domain located at the N terminus of the receptor.
Figure 2. Self association of mTACI monomers in transfectants.
(A and B) 293T cells were cotransfected with the indicated plasmids. IP was performed 48 hours later, followed by SDS-PAGE and Western blotting (WB). Lysates were probed for expression of tagged TACI. Results are representative of 3 experiments. The anti-GFP mAb used, JL-8, recognizes the CFP variant of GFP.
TACI assembles at least into trimers in ER microsomal membranes.
The above data indicate that TACI monomers interact but do not define the stoichiometry of the interaction. To examine how many TACI monomers preassemble prior to ligand binding, we used sequential nondenaturing IP (snIP), a technique that has been used to elucidate the molecular mechanisms for the assembly of the T cell receptor–CD3 complex and NK cell receptors (14–16). snIP allows the isolation of intact radiolabeled receptor complexes that have been assembled in vitro using ER microsomes purified from lymphocytes. Specialized affinity tags are used that permit the elution of assembled complexes under nondenaturing conditions after the IP such that the eluate can be used for subsequent IPs. The 12-aa protein C–derived peptide (PC) tag allows elution from the calcium-dependent anti-PC mAb by EDTA, and the 47-aa streptavidin-binding (SA-binding) peptide (SBP) tag allows elution from SA with free biotin.
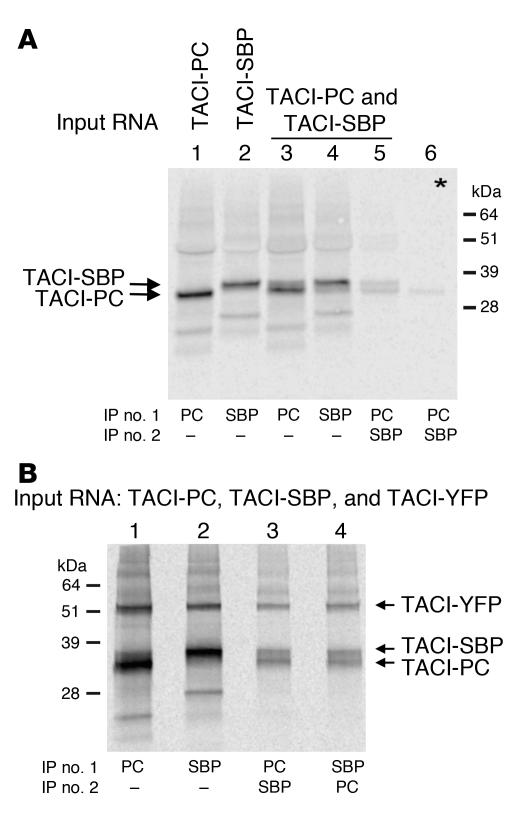
To our knowledge, the snIP technique has never been used to study assembly of receptors from the TNFR superfamily. To determine whether snIP is suitable for studying the composition of TACI oligomers, we first cloned TACI into PC- and SBP-tagged vectors. Equal amounts of mRNA encoding the tagged proteins were used in an in vitro translation system in the presence of 35S-labeled aa and ER microsomes. Figure 3A shows that individually translated PC-tagged and SBP-tagged TACIs were successfully precipitated from ER membranes (lanes 1 and 2). When PC-tagged and SBP-tagged TACIs were cotranslated together, anti-PC beads captured predominantly PC with a minor amount of SBP (lane 3), and the converse was true for SA beads (lane 4). More importantly, snIP, using first anti-PC followed by EDTA elution and SA bead precipitation of the elutant, resulted in the recovery of both PC-tagged and SBP-tagged TACI (lane 5). When PC-tagged and SBP-tagged TACIs were assembled separately and then mixed together, no TACI-SBP was coprecipitated with TACI-PC (lane 6), ruling out nonspecific association of the tagged proteins. The same results were obtained when the SDS-PAGE was run under nonreducing conditions (data not shown), excluding the possibility that covalent complexes were being immunoprecipitated by this technique. Thus, these results suggest that noncovalent, ligand-independent preassociation of TACI monomers can occur during assembly in ER microsomes.
Figure 3. snIP analysis of TACI receptor assembly in ER microsomes.
(A and B) Assembly of differentially tagged TACI monomers was examined in an in vitro translation system using ER microsomes. Membranes were solubilized with digitonin, and complexes were isolated by snIP using the protein C (PC) and SA-binding protein (SBP) affinity tags on the C terminus of the TACI chains. SBP-tagged complexes were captured by SA beads and PC tagged complexes were captured by anti-PC mAb. Asterisk (lane 6) represents mixing control in which TACI-SBP and TACI-PC monomers were translated separately and mixed prior to solubilization and IP.
We generated a third TACI monomer tagged with yellow fluorescent protein (TACI-YFP) to examine whether preassembled TACI trimers can be detected in ER microsomes. Assembly reactions were set up with TACI-PC, TACI-SBP, and TACI-YFP. Single IP with anti-PC captured predominantly PC-tagged TACI along with SBP- and YFP-tagged TACI (Figure 3B, line 1), while single IP with SA beads captured predominantly SBP-tagged TACI along with PC- and YFP-tagged TACI (Figure 3B, line 2). More importantly, 2-step snIP that targeted first the PC and then the SBP epitope (PC→SBP) and vice versa (SBP→PC) coprecipitated all 3 tagged molecules, including TACI-YFP (Figure 3B, lanes 3 and 4). These results demonstrate that TACI monomer can assemble at least into trimers in the absence of ligand and that this assembly can take place in ER microsomes.
TACI assembles independently of ligand in living cells.
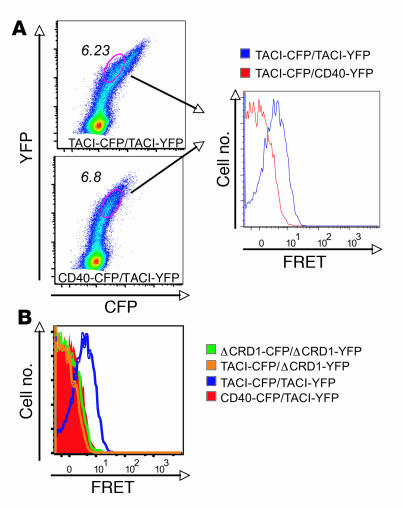
To confirm TACI homotypic interaction in living cells, we used a flow-cytometric approach to analyze fluorescence resonance energy transfer (FRET) between TACI monomers in which the CD was replaced with CFP or YFP. Energy transfer was detected between TACIΔCD-CFP and TACIΔCD-YFP in the absence of ligand, supporting the notion of a preassembled receptor complex (Figure 4A). More importantly, there was no FRET signal between WT TACI and TACIΔCRD1 and no detectable FRET with CD40 control (Figure 4B). Thus, mTACI molecules are in close proximity in living cells, and this is dependent on the CRD1 domain.
Figure 4. TACI self-associates through CRD1 in living cells.
(A) Flow cytometric analysis of 293T cells cotransfected with CFP- or YFP-tagged mTACI and CFP- or YFP-tagged CD40 control. The dual-expressing cells were electronically gated, and more than 20,000 events were acquired into the FRET gate and used for subsequent analysis. (B) Overlay of FRET plots derived as in A for interactions between indicated TNFR molecules.
Endogenous TACI exists as an oligomeric complex on the surface of B cells.
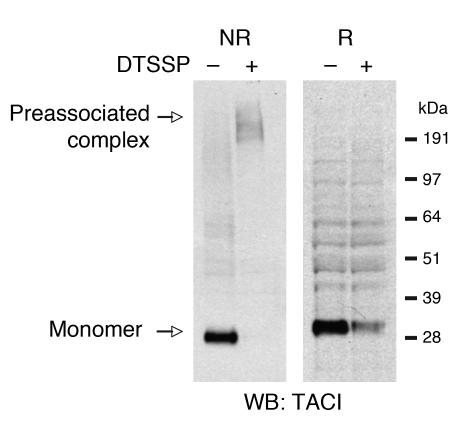
We next examined whether endogenous TACI forms trimers or higher-order oligomers on the cell surface in the absence of ligand. We treated A20 murine B cells with membrane-impermeant, thiol-cleavable chemical cross-linker 3,3′-dithiobis[sulfosuccinimidyl-propionate] (DTSSP) to covalently cross-link cell-surface molecules that are closely associated, with less than 12 υ distance from each other. Detergent-solubilized lysates were run on a gel under nonreducing conditions or under reducing conditions (100 mM DTT) and probed for TACI. In the absence of DTSSP, under nonreducing conditions a strong band that corresponds to monomeric TACI was visualized (Figure 5). In the presence of DTSSP, under nonreducing conditions monomeric TACI was no longer detected; instead, a high-molecular-weight complex that exhibited a molecular size greater than 191 kDa became evident. The presence of DTT in the same sample resulted in the disappearance of the higher-molecular-weight complex and appearance of TACI monomer. The decreased recovery of monomeric TACI under reducing conditions in DTSSP cross-linked sample seen in Figure 5 was due to decreased affinity of the monoclonal anti-TACI to modified TACI monomer, as no difference in band intensity was observed when polyclonal antibody was used for Western blotting (data not shown). These data show that native TACI monomers assemble as oligomers on the surface of B cells.
Figure 5. Preassociation of endogenous mTACI in B cells.
Mouse lymphoma A20 B cells were treated as indicated. Lysates were run on SDS-PAGE under nonreducing (NR) or reducing (R) conditions and probed with anti-TACI mAb. The ladder visible in the reduced samples represents nonspecific bands and is visible, albeit less strongly, under nonreducing conditions without DTSSP.
The murine C76R mutant fails to bind ligand but preassociates with WT TACI.
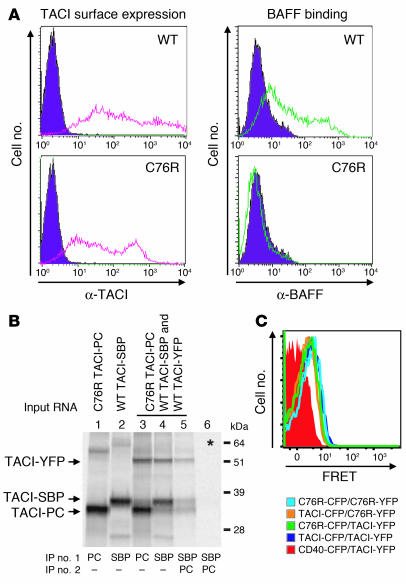
Having established that TACI receptor monomers preassociate in a ligand-independent manner, we next determined whether the C76R mTACI mutant can preassociate with WT TACI. First we established that the C76R mTACI mutant behaves similarly to the C104R hTACI mutant, in that it is expressed on the cell surface but fails to bind the ligand. Figure 6A shows that the C76R mTACI mutant is expressed on the cell surface. However, like its hTACI C104R counterpart, it was unable to bind BAFF. There was consistently a slight difference in the intensity of expression of C76R TACI compared with WT TACI, possibly because the missense mutation alters the epitope recognized by the Ab. However, this difference was small and cannot account for the complete inability of the mutant to bind BAFF. When 293T cells were cotransfected with Myc-tagged C76R mTACI and Flag-tagged WT mTACI, the Myc-tagged mutant coprecipitated with Flag-WT mTACI (Figure 2A, lane 3). This interaction was independent of the presence of the CD, as Myc-tagged C76R mTACI coprecipitated with CFP-tagged TACIΔCD (Figure 2B, lane 4). The ligand-independent preassociation of the C76R mTACI mutant with WT TACI was confirmed with the in vitro translation and assembly system using PC-tagged C76R mutant and SBP- and YFP-tagged WT TACI. Assembly reactions were set up with C76R TACI-PC, WT TACI-SBP, and WT TACI-YFP. Single IP with anti-PC captured predominantly PC-tagged C76R TACI with a minor amount of SBP- and YFP-tagged WT TACI (Figure 6B, lane 3), while single IP with SA beads captured predominantly SBP-tagged WT TACI with a minor amount of C76R TACI-PC and WT TACI-YFP (Figure 6B, lane 4). More importantly, 2-step snIP that targeted the SBP and PC epitopes (SBP→PC) brought down all 3 tagged molecules (Figure 6B, lane 5). This preassociation occurred in the ER and was specific, as no complex was observed in mixing control, in which WT TACI-YFP, WT TACI-SBP, and C76R TACI-PC were translated in separate reactions, the contents of which were combined before solubilization and IP (Figure 6B, lane 6). These results demonstrate that WT TACI and C76R TACI monomers can assemble at least into trimers in the absence of ligand in ER microsomes. Association of the C76R mutant and WT TACI was further demonstrated in living cells by FRET analysis (Figure 6C).
Figure 6. C76R mTACI associates with WT TACI.
(A) Surface expression and recombinant human BAFF (rhBAFF) binding of WT and mutant mTACI in transfected 293T cells. (B) snIP analysis of the association of WT and mutant mTACI in ER microsomes. Asterisk (lane 6) represents mixing control in which TACI-SBP, TACI-PC, and TACI-YFP monomers were translated separately and mixed prior to solubilization and IP. (C) FRET analysis of the association of WT and mutant mTACI in 293T cells.
The murine C76R mutant inhibits signaling by WT TACI.
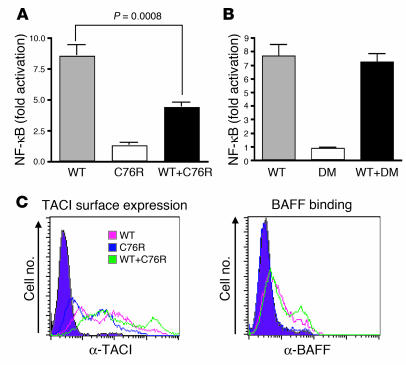
It has been previously reported that engagement of TACI by its ligands BAFF and APRIL leads to activation of NF-κB in transfected 293T cells (6, 10, 17, 18). To understand the functional significance of ligand-independent preassociation of the C76R mutant and WT TACI, a quantitative luciferase assay was used to measure signaling via TACI in 293T cells transfected with WT TACI, C76R mutant, or equivalent amounts of both. Simultaneous expression of both mutant and WT TACI on the surface of cotransfected cells was confirmed using differentially tagged constructs (data not shown). Figure 7A shows that ligation of WT TACI with BAFF caused NF-κB activation. As expected, the C76R mutant, which fails to bind BAFF, was unable to activate NF-κB. NF-κB activation was significantly reduced (P < 0.001) in 293T cells cotransfected with equal amounts of WT and mutant TACI. These results demonstrate that the C76R mutant dominantly interferes with signaling by WT TACI.
Figure 7. C76R mTACI dominantly interferes with signaling, but not ligand binding, by WT mTACI.
(A) 293T cells were transfected with 0.1 μg WT mTACI alone, 0.1 μg C76R mTACI, or 0.1 μg of each along with 0.1 μg of NF-κB–luc reporter plasmid and 10 ng control pRL-TK plasmid. Equal amounts of DNA were used for each transfection by using empty vector DNA when needed. After 4 hours, recombinant mBAFF (0.1 μg/ml) was added, and reporter gene activity was determined 20 hours later. NF-κB luciferase induction was normalized to Renilla luciferase intensity and total protein of each sample. Fold induction was calculated by dividing the normalized NF-κB luciferase result of BAFF-treated samples by the NF-κB luciferase result of untreated samples. n = 7. (B) The same assay as described in A was performed, with C76R mTACI that lacked the CRD1 domain (DM) substituting for C76R mTACI. Values represent the fold increase by BAFF stimulation compared with unstimulated sample and are the mean ± SEM from 6 independent experiments. (C) TACI surface expression and rhBAFF binding by 293T cells transfected with 1 μg of WT mTACI, 1 μg of C76R mTACI alone, or 1 μg of each.
To ascertain that the dominant-negative effect of the mutant is dependent on its ability to preassociate with WT TACI, we examined the effect of CRD1-deleted C76R mTACI double mutant (DM) on WT TACI signaling. Simultaneous expression of the DM and WT TACI on the surface of cotransfected cells was demonstrated using differentially tagged constructs (data not shown). Figure 7B shows that the DM had no effect on signaling by WT TACI. These data suggest that the ability of the C76R mutant to act as a dominant negative is dependent on its ability to preassociate with WT TACI.
Preassociation of C76R with WT TACI could interfere with ligand binding by the oligomeric complex. Alternatively, C76R may not interfere with ligand binding but could still interfere with signaling if all subunits of the receptor oligomer need to bind ligand for signaling to proceed. To test whether C76R TACI mutant interferes with ligand binding to WT TACI, we examined the binding of Flag-tagged BAFF to 293T cells transfected with equal amounts of WT and C76R mutant TACI. Figure 7C shows that the presence of C76R mutant did not decrease ligand binding to WT TACI. No interference by the mutant with ligand binding to WT TACI was observed either under limiting concentrations of BAFF or when the mutant was used at a concentration 4-fold that of WT TACI (data not shown). These data indicate that the C76R mutant does not interfere with ligand binding by WT TACI.
C104R mutant hTACI preassembles with WT hTACI.
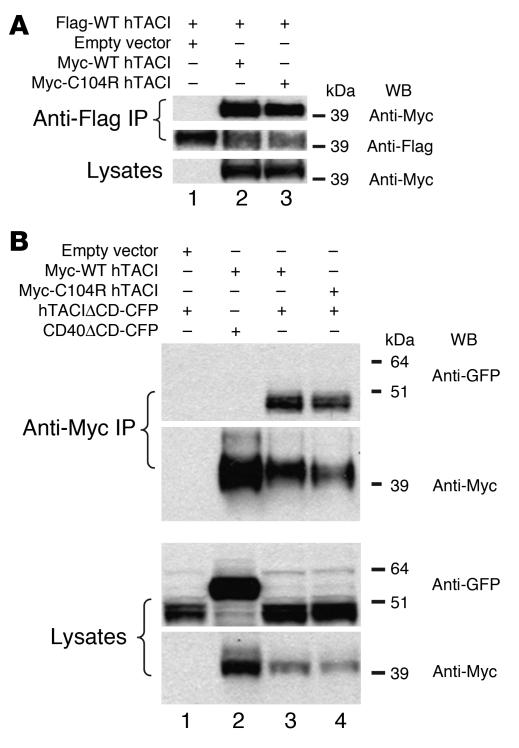
We examined the association of WT hTACI and C104R hTACI mutant in 293T cells. 293T cells were cotransfected with full-length Flag-WT hTACI in combination with Myc-WT hTACI or Myc-C104R hTACI. As shown in Figure 8, hTACI monomers were found to homotypically associate (Figure 8A, lane 2). More importantly, C104R hTACI was able to associate with WT hTACI (Figure 8A, lane 3). This homotypic interaction was not dependent on the intracellular (IC) domain, as it was still retained when the IC domain of WT hTACI was replaced by CFP to generate hTACIΔCD-CFP. Figure 8B shows that WT hTACIΔCD-CFP associated with Myc-WT hTACI (lane 3) as well as with Myc-C104R hTACI (lane 4). These associations were specific because there was no association of CD40ΔCD-CFP with Myc-WT hTACI (Figure 8B, lane 2).
Figure 8. C104R hTACI associates with WT hTACI.
(A and B) 293T cells were cotransfected with the indicated hTACI plasmids. IP was performed 48 hours later followed by SDS-PAGE and Western blotting. Lysates were probed for expression of tagged TACI. Results are representative of 3 experiments.
The C104R mutant inhibits signaling by WT hTACI.
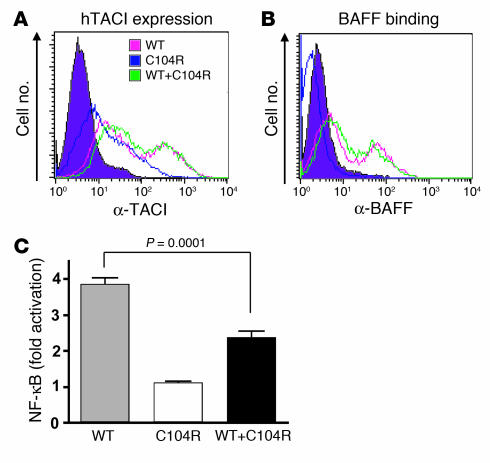
Previously we have demonstrated that B cells from 3 CVID patients heterozygous for the C104R hTACI mutation failed to secrete IgG and IgA in response to APRIL, indicating that TACI-mediated signaling was impaired (4). One can argue that this could possibly be due to the ability of the mutant to interfere with WT hTACI surface expression. However, simultaneous expression of mutant and WT hTACI on the surface of cotransfected cells was confirmed using differentially tagged constructs (Figure 9). C104R did not interfere with the expression of WT hTACI, and more importantly, it did not interfere with ligand binding by WT hTACI in cotransfected cells (Figure 10, A and B). To get a better understanding of the molecular mechanism by which C104R could be interfering with WT hTACI function, we used a quantitative luciferase assay to measure signaling via TACI in 293T cells transfected with WT hTACI, C104R mutant, or equivalent amounts of both. Figure 10C shows that ligation of WT hTACI with BAFF caused NF-κB activation, while the C104R mutant, which fails to bind BAFF, was unable to activate NF-κB. TACI signaling was significantly reduced in 293T cells cotransfected with equal amounts of WT and mutant hTACI. These results demonstrate that the C104R mutant dominantly interferes with signaling by WT hTACI.
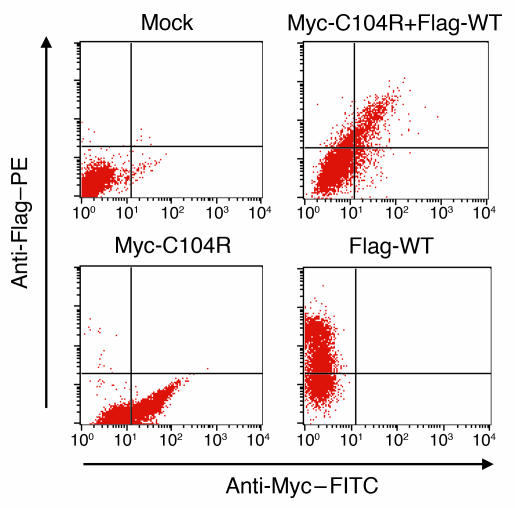
Figure 9. C104R mutant does not interfere with surface expression of WT hTACI.
293T cells were cotransfected with the indicated hTACI plasmids, and surface expression of Flag-tagged WT hTACI and Myc-tagged C104R hTACI was analyzed 24 hours after transfection using anti-Myc–FITC and anti-Flag–PE antibodies. Mock, untransfected.
Figure 10. C104R hTACI dominantly interferes with signaling, but not ligand binding, by WT hTACI.
hTACI surface expression (A) and rhBAFF binding (B) by 293T cells transfected with 1 μg of WT hTACI, 1 μg of C104R hTACI alone, or 1 μg of each together. (C) 293T cells were transfected with 10 ng WT mTACI alone, 10 ng C104R mTACI alone, or 10 ng of each together along with 0.5 mg of NF-κB–luc reporter plasmid and 10 ng control pRL-null plasmid. Equal amounts of DNA were used for each transfection by using empty vector DNA when needed. After 4 hours, recombinant hBAFF (0.1 μg/ml) was added, and reporter gene activity was determined 20 hours later. NF-κB luciferase induction was normalized to Renilla luciferase intensity. Values represent the fold increase by BAFF stimulation compared with unstimulated sample and are the mean ± SEM from 6 independent experiments.
Discussion
Our data clearly demonstrate that endogenous mTACI exists as a preassociated oligomeric complex and that a missense C76R mutation that abolishes ligand binding functions as a dominant negative.
Our work provides new insight into the structure and function of TACI receptor by showing that it preassociates into an oligomeric complex prior to ligand binding. Homotypic interaction of TACI molecules was demonstrated by coimmunoprecipitation of differentially tagged TACI molecules (Figure 2) and was confirmed in living cells by FRET (Figure 4). We used a novel snIP method to explore the stoichiometry of the TACI oligomeric complex. Using 3 differentially tagged TACI molecules, we demonstrated that a minimum of 3 TACI monomers preassociate in the absence of ligand. However, with only 3 differentially tagged monomers, we could not determine, using this method, whether TACI can preassociate into an oligomer of a higher order than a trimer. The results of chemical cross-linking studies on A20 B cells, which express endogenous TACI, suggest that the TACI receptor exist on the cell surface as a preassociated higher-order oligomer (Figure 5), although the presence of other molecules in this complex cannot be ruled out. Thus, TACI joins the TNFR family members Fas, TNFR1 (p60), and TNFR2 (p80) as a receptor with the ability to preassociate independently of ligand (19, 20).
Recently published data demonstrate that BAFF and APRIL can form higher-order oligomers under physiologic conditions. BAFF has been reported to assemble into a virus-like cage complex of 60 monomers (21–23). It has been suggested that the increased multivalency of the 60-mer affects the quality of the signal transduced rather than the avidity of binding (23). APRIL has been shown to bind to heparan sulfate proteoglycans and oligomerize (24). This oligomerization was shown to be essential for proper signaling (25). Given these observations and our data, it is logical to posit that TACI exists on the cell surface in a preassociated oligomeric state ready to efficiently bind its oligomeric ligands in order to transduce a rapid and strong signal. Higher-order clustering with simultaneous engagement of oligomeric receptors may be required to transduce a signal above a certain threshold, and the inclusion of non–ligand-binding subunits in preassociated oligomers may inhibit this process.
The demonstration that WT TACI monomers preassociate prior to ligand binding is critical to understanding the molecular mechanisms by which heterozygous mutations in TACI that affect ligand binding can cause B cell dysfunction. The prototype of such a mutation is C104R, one of the most common TACI mutations associated with CVID. According to the haploinsufficiency explanation, a single copy of WT TACI is insufficient to transduce a signal. The alternative explanation is that C104R TACI acts as a dominant-negative mutant that interferes with signaling. Our demonstration that the C104R hTACI mutant, and the C76R mTACI mutant that corresponds to it, associates with WT TACI and functions as a dominant negative favors the second hypothesis. Preassociation of the C76R mutant with WT mTACI was demonstrated by several techniques that included coimmunoprecipitation, snIP, and FRET (Figures 2 and 6). Coexpression of the mutant with WT mTACI significantly inhibited ligand-induced activation of NF-κB (Figure 7A). This inhibition was strictly dependent on the assembly of the mutant with WT mTACI, as a C76R mutant that lacked the CRD1 domain and failed to associate with WT mTACI also failed to inhibit signaling by WT mTACI (Figure 7B).
These results are strikingly similar to the findings with EC Fas mutations cloned from patients with autosomal dominant autoimmune lymphoproliferative syndrome (ALPS). Those Fas mutations also failed to bind FasL and preassociated with wild-type receptors, impairing FasL-mediated apoptosis (19). One possible explanation for the dominant effect of the C76R mutant is that inclusion of the mutant in the preassociated TACI oligomeric complex would interfere with ligand binding to the complex. This explanation was ruled out because coexpression of the mutant with WT mTACI had no detectable effect on ligand binding (Figure 7C). This result is consistent with the previously reported observation that B cells from CVID patients heterozygous for the C104R hTACI mutations show normal APRIL binding (5). These data raise the possibility that the mutant may interfere with signaling by disrupting ligand-induced conformational changes in the receptor that allow proper recruitment of downstream signaling molecules. Further biophysical and structural studies are necessary to prove this hypothesis.
The present work provides new insight into the structure of the mTACI receptor by demonstrating that it preassociates as an oligomer prior to ligand binding. We have also shown that the hTACI mutant C104R, which is associated with CVID, preassociates with WT hTACI (Figure 8A). As with mTACI, this preassociation is independent of the IC domain (Figure 8B). More importantly, the hTACI mutant C104R can interfere with signaling by WT hTACI without interfering with the surface expression of WT hTACI or with its ability to bind ligand (Figures 9 and 10). Thus, ligand-independent preassociation of C104R mutant with WT hTACI provides a potential explanation for how the heterozygous C104R mutation may disrupt TACI signaling in B cells, illustrated by the impaired ability of B cells from CVID patients heterozygous for C104R to produce IgG and IgA in response to APRIL (4). Final proof that this B cell dysfunction results in CVID awaits the analysis of mice carrying the C76R mutation.
Methods
Antibodies and reagents.
SA, and anti-Myc coupled to agarose beads and anti-Flag antibody were purchased from Sigma-Aldrich. Anti-myc (9B11) antibody was purchased from Cell Signaling Technology. Calcium-dependent anti-PC (mouse mAb HPC4) was purchased from Roche. Anti-GFP (Living Colors A.v. Monoclonal Antibody JL-8) was purchased from Clontech. Digitonin was purchased from Biosynth International. mTACI surface expression was analyzed using rat monoclonal anti-TACI–PE antibody (R&D Systems), and hTACI surface expression was analyzed using goat polyclonal anti-TACI antibody (R&D Systems) as described previously (4).
cDNA constructs and in vitro transcription.
mTACI sequence was amplified from mouse B cells by RT-PCR. hTACI sequence was amplified from human PBMCs by RT-PCR. We obtained blood by venipuncture from a healthy adult control after obtaining informed consent. The study protocol was approved by the Ethics Committee/Internal Review Board of Children’s Hospital, Boston, Massachusetts, USA. PBMCs were isolated from the blood and used to obtain mRNA. BALB/c wild-type mouse spleen was used to purify B cells and obtain mRNA. The procedure performed on the mouse was reviewed and approved by the Animal Care and Use Committee of the Children’s Hospital Boston. The sequences were cloned into pCMVTag3A (Myc tag), pCMVTag2A (Flag tag), pcDNA6, and modified pSP64 vector with PC and SBP tags described in ref. 14. For FRET, TACI was cloned into modified YFP and CFP vectors, which have mutations introduced into CFP and YFP that inhibit their inherent tendency to form hetero- or homodimers as described in ref. 26 (gift from C. Carman, Harvard Medical Center and Beth Israel Deaconess Medical Center, Boston, Massachusetts, USA). Mutation was introduced by QuikChange Mutagenesis Kit (Stratagene). In vitro transcription was performed from cDNA constructs using T7 RiboMAX Large-Scale RNA Production Kit and m7G cap analog (Promega).
In vitro translation and assembly.
The in vitro translation and assembly reactions followed by IP and analysis were performed as previously described (14, 15). All in vitro translation and assembly reactions were performed at 30°C. Each 25-μl reaction contained 17.5 μl nuclease-treated rabbit reticulocyte lysate (Promega), 0.5 μl amino acid mixture minus methionine (Promega), 0.5 μl SUPERase-In RNase inhibitor (Ambion), 1–2 μl 35S-labeled methionine (Amersham), equimolar amounts of each RNA (300 ng), and 2.0 μl ER microsome from murine hybridoma (IVD12) as previously described (14). Reactions were performed with an initial translation period of 30 minutes under reducing conditions, followed by a 2-hour assembly period after addition of oxidized glutathione to 4 mM. Reaction volumes were 25–100 μl as required for optimal signal with multistep snIP procedures.
IP and electrophoretic analysis.
Translation assembly reactions were stopped by dilution with 1 ml of ice-cold PBS with 10 mM iodoacetamide, and microsomes were pelleted and rinsed. Pellets were lysed in 400 μl of IP buffer (PBS plus 0.5% digitonin, 10 mM iodoacetamide, 0.1% BSA with 1 mM CaCl when anti–protein C mAb was used) and rotated for 2 hours at 4°C. Lysates were precleared for 1 hour with Tris/BSA-blocked protein G beads, and primary captures were performed overnight at 4°C. IPs were washed twice with 500 μl of IP buffer. Nondenaturing elution of SA-captured complexes was performed by incubation with 400 μl of IP buffer with 100 μM free biotin for 1 hour at 4°C, and eluted complexes were incubated with PC antibody and protein G beads for 2 hours at 4°C and washed as described above. Nondenaturing elution of PC-captured complexes was performed by incubation with 400 μl of IP buffer with EDTA for 1 hour at 4°C.
Cross-linking experiment.
A20 B cell lymphoma cells were washed and resuspended in PBS. Cells were then treated with 1 mM solution of the cross-linker DTSSP (Pierce) for 30 minutes at room temperature, and the reaction was quenched with 50 mM Tris-Cl (pH 7.5) for 20 minutes on ice. The cells were lysed in 1% Triton, 10 mM HEPES, 150 mM NaCl, 1 mM EDTA, 100 mM PMSF, and protease inhibitors. Equal amounts of lysates were subjected to electrophoresis under nonreducing (without DTT) or reducing (with 100 mM DTT) conditions and were analyzed for TACI complexes with monoclonal rat anti-TACI or polyclonal goat anti-TACI antibodies (R&D Systems).
293T coimmunoprecipitation.
293T cells were transfected using FuGENE 6 (Roche). Forty-eight hours after transfection, cells were harvested and lysed in IP buffer: 0.5% NP-40 or 1% Triton, 50 mM HEPES, 10 mM iodoacetamide, 250 mM NaCl, 5 mM EDTA, 100 mM PMSF, and complete protease inhibitor cocktail (Sigma-Aldrich). After preclearing with protein G–agarose beads (Upstate), proteins were immunoprecipitated with either anti-Myc or anti-Flag agarose beads (Sigma-Aldrich). Immune complexes were washed 3–5 times with lysis buffer. Proteins were electrophoresed on Tris/glycine gels, transferred to nitrocellulose membranes, and blotted with the indicated antibodies.
FRET.
TACI receptors with C-terminal in-frame fusions (at position 156 in place of the IC domain) to monomeric CFP and YFP were generated by standard PCR cloning techniques. After sequence and protein expression of these constructs were confirmed, they were transfected into 293T cells, and flow cytometric detection of FRET was performed as previously described (19, 27) using a CyAn cytofluorimeter (Dako) and 408/488 nm dual laser excitation. Data was analyzed by FlowJo software (version 8.3; Tree Star) and fluorescence plotted on biexponential axes.
Statistics.
Significance of experimental values was analyzed by using the 2-tailed Student’s t test.
Acknowledgments
The authors thank Chris Carman for the modified CFP and YFP vectors and members of the Geha laboratory for useful discussions. L. Garibyan was funded by the Medical Scientist Training Program grant to Harvard Medical School. This work was supported by NIH grants P01-AI-031541 and T32-AI-007512 to R.S. Geha and R01 AI054520 to K.W. Wucherpfennig.
Footnotes
Nonstandard abbreviations used: APRIL, a proliferation-inducing ligand; BAFF, B cell–activating factor; CAML, calcium-modulating ligand; CD, cytoplasmic domain; CFP, cyan fluorescent protein; CRD, cysteine-rich domain; CVID, common variable immunodeficiency; DM, double mutant; DTSSP, 3,3′-dithiobis[sulfosuccinimidyl-propionate]; EC, extracellular; FRET, fluorescence resonance energy transfer; IC, intracellular; PC, protein C–derived peptide; SA, streptavidin; SBP, SA-binding peptide; snIP, sequential nondenaturing IP; TACI, transmembrane activator and CAML interactor; TNFR, TNF receptor; YFP, yellow fluorescent protein.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 117:1550–1557 (2007). doi:10.1172/JCI31023
References
- 1.Cunningham-Rundles C., Bodian C. Common variable immunodeficiency: clinical and immunological features of 248 patients. Clin. Immunol. 1999;92:34–48. doi: 10.1006/clim.1999.4725. [DOI] [PubMed] [Google Scholar]
- 2.Kokron C.M., et al. Clinical and laboratory aspects of common variable immunodeficiency. An. Acad. Bras. Cienc. 2004;76:707–726. doi: 10.1590/s0001-37652004000400007. [DOI] [PubMed] [Google Scholar]
- 3.Schroeder H.W., Jr., Schroeder H.W., 3rd, Sheikh S.M. The complex genetics of common variable immunodeficiency. J. Investig. Med. 2004;52:90–103. doi: 10.1136/jim-52-02-17. [DOI] [PubMed] [Google Scholar]
- 4.Castigli E., et al. TACI is mutant in common variable immunodeficiency and IgA deficiency. Nat. Genet. 2005;37:829–834. doi: 10.1038/ng1601. [DOI] [PubMed] [Google Scholar]
- 5.Salzer U., et al. Mutations in TNFRSF13B encoding TACI are associated with common variable immunodeficiency in humans. Nat. Genet. 2005;37:820–828. doi: 10.1038/ng1600. [DOI] [PubMed] [Google Scholar]
- 6.von Bulow G.U., Bram R.J. NF-AT activation induced by a CAML-interacting member of the tumor necrosis factor receptor superfamily. Science. 1997;278:138–141. doi: 10.1126/science.278.5335.138. [DOI] [PubMed] [Google Scholar]
- 7.Mackay F., Schneider P., Rennert P., Browning J. BAFF and APRIL: a tutorial on B cell survival. Annu. Rev. Immunol. 2003;21:231–264. doi: 10.1146/annurev.immunol.21.120601.141152. [DOI] [PubMed] [Google Scholar]
- 8.Bischof D., et al. Selective activation of TACI by syndecan-2. Blood. 2006;107:3235–3242. doi: 10.1182/blood-2005-01-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castigli E., et al. TACI and BAFF-R mediate isotype switching in B cells. J. Exp. Med. 2005;201:35–39. doi: 10.1084/jem.20032000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seshasayee D., et al. Loss of TACI causes fatal lymphoproliferation and autoimmunity, establishing TACI as an inhibitory BLyS receptor. Immunity. 2003;18:279–288. doi: 10.1016/s1074-7613(03)00025-6. [DOI] [PubMed] [Google Scholar]
- 11.von Bulow G.U., van Deursen J.M., Bram R.J. Regulation of the T-independent humoral response by TACI. Immunity. 2001;14:573–582. doi: 10.1016/s1074-7613(01)00130-3. [DOI] [PubMed] [Google Scholar]
- 12.Yan M., et al. Activation and accumulation of B cells in TACI-deficient mice. Nat. Immunol. 2001;2:638–643. doi: 10.1038/89790. [DOI] [PubMed] [Google Scholar]
- 13.Hymowitz S.G., et al. Structures of APRIL-receptor complexes: like BCMA, TACI employs only a single cysteine-rich domain for high affinity ligand binding. J. Biol. Chem. 2005;280:7218–7227. doi: 10.1074/jbc.M411714200. [DOI] [PubMed] [Google Scholar]
- 14.Call M.E., Pyrdol J., Wiedmann M., Wucherpfennig K.W. The organizing principle in the formation of the T cell receptor-CD3 complex. Cell. 2002;111:967–979. doi: 10.1016/s0092-8674(02)01194-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng J., Garrity D., Call M.E., Moffett H., Wucherpfennig K.W. Convergence on a distinctive assembly mechanism by unrelated families of activating immune receptors. Immunity. 2005;22:427–438. doi: 10.1016/j.immuni.2005.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garrity D., Call M.E., Feng J., Wucherpfennig K.W. The activating NKG2D receptor assembles in the membrane with two signaling dimers into a hexameric structure. Proc. Natl. Acad. Sci. U. S. A. 2005;102:7641–7646. doi: 10.1073/pnas.0502439102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marsters S.A., et al. Interaction of the TNF homologues BLyS and APRIL with the TNF receptor homologues BCMA and TACI. Curr. Biol. 2000;10:785–788. doi: 10.1016/s0960-9822(00)00566-2. [DOI] [PubMed] [Google Scholar]
- 18.Yan M., et al. Identification of a receptor for BLyS demonstrates a crucial role in humoral immunity. Nat. Immunol. 2000;1:37–41. doi: 10.1038/76889. [DOI] [PubMed] [Google Scholar]
- 19.Siegel R.M., et al. Fas preassociation required for apoptosis signaling and dominant inhibition by pathogenic mutations. Science. 2000;288:2354–2357. doi: 10.1126/science.288.5475.2354. [DOI] [PubMed] [Google Scholar]
- 20.Chan F.K., et al. A domain in TNF receptors that mediates ligand-independent receptor assembly and signaling. Science. 2000;288:2351–2354. doi: 10.1126/science.288.5475.2351. [DOI] [PubMed] [Google Scholar]
- 21.Kim H.M., et al. Crystal structure of the BAFF-BAFF-R complex and its implications for receptor activation. Nat. Struct. Biol. 2003;10:342–348. doi: 10.1038/nsb925. [DOI] [PubMed] [Google Scholar]
- 22.Cachero T.G., et al. Formation of virus-like clusters is an intrinsic property of the tumor necrosis factor family member BAFF (B cell activating factor). Biochemistry. 2006;45:2006–2013. doi: 10.1021/bi051685o. [DOI] [PubMed] [Google Scholar]
- 23.Liu Y., et al. Crystal structure of sTALL-1 reveals a virus-like assembly of TNF family ligands. Cell. 2002;108:383–394. doi: 10.1016/s0092-8674(02)00631-1. [DOI] [PubMed] [Google Scholar]
- 24.Hendriks J., et al. Heparan sulfate proteoglycan binding promotes APRIL-induced tumor cell proliferation. Cell Death Differ. 2005;12:637–648. doi: 10.1038/sj.cdd.4401647. [DOI] [PubMed] [Google Scholar]
- 25.Ingold K., et al. Identification of proteoglycans as the APRIL-specific binding partners. J. Exp. Med. 2005;201:1375–1383. doi: 10.1084/jem.20042309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zacharias D.A., Violin J.D., Newton A.C., Tsien R.Y. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 27.Siegel R.M., et al. Measurement of molecular interactions in living cells by fluorescence resonance energy transfer between variants of the green fluorescent protein. Sci. STKE. 2000;2000:PL1.. doi: 10.1126/stke.2000.38.pl1. [DOI] [PubMed] [Google Scholar]