Abstract
Cre is widely used for DNA tailoring and, in combination with recombineering techniques, to modify BAC/PAC sequences for generating transgenic animals. However, mammalian genomes contain recombinase recognition sites (cryptic loxP sites) that can promote illegitimate DNA recombination and damage when cells express the Cre recombinase gene. We have created a new bioinformatic tool, FuzznucComparator, which searches for cryptic loxP sites and we have applied it to the analysis of the whole mouse genome. We found that cryptic loxP sites occur frequently and are homogeneously distributed in the genome. Given the mammalian nature of BAC/PAC genomic inserts, we hypothesised that the presence of cryptic loxP sites may affect the ability to grow and modify BAC and PAC clones in E. coli expressing Cre recombinase. We have observed a defect in bacterial growth when some BACs and PACs were transformed into EL350, a DH10B-derived bacterial strain that expresses Cre recombinase under the control of an arabinose-inducible promoter. In this study, we have demonstrated that Cre recombinase expression is leaky in un-induced EL350 cells and that some BAC/PAC sequences contain cryptic loxP sites, which are active and mediate the introduction of single-strand nicks in BAC/PAC genomic inserts.
INTRODUCTION
BAC and PAC clones have become the preferred tool for generating transgenic animals, since they accommodate large genomic DNA fragments, are well characterized, stable and easy to propagate and purify. In recent years, efficient and reliable methods have been developed to modify their sequence in E. coli. These techniques are generally termed recombineering (homologous recombination-mediated genetic engineering) and they are based on homologous recombination between a linear double-stranded DNA cassette or synthetic oligonucleotides and a circular DNA molecule (plasmid, BACs or PACs) (1,2). A variety of methods and strains have been described for homologous recombination, but the most widely used methods derive the homologous recombination machinery from the bacteriophage λ-encoded Red system (3). The DY380 strain of E. coli is derived from DH10B and encodes a defective λ-prophage, in which the recombination apparatus is expressed under the control of the temperature-sensitive λ repressor (cI857) (3,4). Two further strains have been derived from DY380 in which the tetracycline resistance gene, (encoded on the bacterial genome) is substituted by either Flp (strain EL250) or Cre (strain EL350) genes (5). The Flp or Cre genes are expressed under the control of the arabinose-inducible AraC-PBAD promoter to allow further manipulation of introduced BAC/PACs. Cre DNA recombinase catalyses the recombination between two 34-bp loxP elements. The outcome (excision or inversion) depends on the relative orientation of the 2 loxP elements (6). The loxP element contains a core spacer sequence of 8-bp flanked by two palindromic sequences each of 13-bp to which Cre binds (7). Up to 5 mismatches from the consensus in each of the two palindromic sequences can be tolerated by Cre, without significantly reducing DNA binding (8,9). The core sequence is the cleavage site; its asymmetry defines the direction of the loxP site and homology in this region between two loxP sites is required for efficient recombination (10). When recombination occurs between a mutant loxP site, which bears a deletion in the spacer region, and a wild-type loxP site, the introduction of single- and double-strand breaks in the DNA may occur (11). On a statistical basis, the 34-bp consensus loxP site is not expected to be present in mammalian genomes, but the presence of functional recombinase recognition sites, that diverge significantly from the native loxP site, has been identified in both the human and mouse genomes (12). These cryptic (or pseudo) loxP sites, can support Cre-mediated recombination at high efficiency when cryptic loxP sites with the same spacer region are involved in the recombination (12). Studies have shown that Cre expression in mammalian cells causes recombination events between cryptic loxP sites resulting in gross chromosomal rearrangements in spermatids (13) and, in cultured mammalian cells, growth inhibition accompanied by DNA damage (14). Furthermore, recombination can occur even when the spacer regions display a relatively high level of non-homology (15). Moreover, it is generally acknowledged that some large DNAs grow slowly in Cre-containing E. coli and it has been reported that modified strains of E. coli are not ideal for receiving PACs and BACs and some BAC clones cannot be transformed into such strains for reasons that are not fully understood (16).
Our hypothesis is that cryptic loxP sites, encoded in the mammalian genomic DNA insert, may act as substrates for Cre recombinase, expression of which may be leaky in some bacterial strains.
Here, we describe the use of a bioinformatics tool to identify such cryptic sites and experiments to test these predictions carried out on BAC/PACs.
MATERIAL AND METHODS
In silico identification of cryptic loxP sites
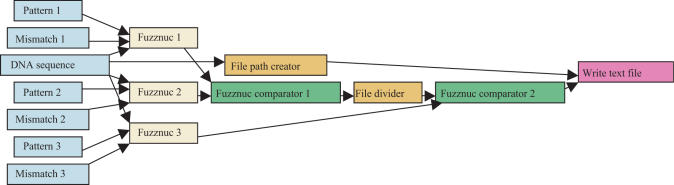
To automate the identification of cryptic loxP sites within a given DNA sequence a bioinformatics workflow was created using Taverna [TAVERNA] (17). Taverna provides a workbench application that enables the construction and enactment of workflows within a graphical environment. A schematic representation of the workflow is illustrated in Figure 1. This workflow takes a DNA sequence and searches for matches to three different patterns using Fuzznuc, which is freely available as part of the European Molecular Biology Open Software Suite (EMBOSS) [EMBOSS] (18) and is accessed via a Soaplab [SOAPLAB] web service (19).
Figure 1.
Schematic representation of the workflow created to automate the identification of cryptic loxP sites.
Pattern 1 is described by the sequence ATAACTTCGTATA (N)8 TATACGAAGTTAT (12) and it selects for high homology in the 13-bp palindromic regions of the wild-type loxP. This pattern is augmented by a mismatch parameter that allows up to 10 mismatches to be tolerated (8,9). Pattern 2 is described by the sequence ATNAC(N)2CNTATA (N)8 TATANG(N)2GTNAT. It selects for conservation of those base pairs in the loxP site believed to be contact points for the Cre enzyme (underlined bases in the sequence ATAACTTCGTATA ATGTATGC TATACGAAGTTAT). This pattern is augmented by a mismatch parameter that allows up to 5 mismatches to be tolerated. We hypothesized that the mismatches allowed in this search could be tolerated by the Cre recombinase. Pattern 3 is described by the sequence (N)9TATA (N)8 TATA(N)9. It ensures that the TATA motif surrounding the core 8-bp spacer region is present. This pattern is augmented by a mismatch parameter that ensures no mismatches are tolerated. The fulfilment of these three criteria defines a primary cryptic loxP site and provides a wider classification then previously proposed (12).
A web service called FuzznucComparator was developed that compares the output from two Fuzznuc processes and outputs only those sequences present in both. When the result of the comparison contains more than one sequence, the FuzznucComparator tool performs 2 pairwise alignments of the core 8-bp spacer regions. The first alignment is calculated using the sequences in their given orientation. The second complements one sequence prior to making the alignment. The output file format consists of the result of the pairwise comparison (if any) followed by those sequences present in both input files in fuzznuc's seqtable format.
To isolate those sequences that match all three patterns two comparisons are required. First, a FuzznucComparator process is used to isolate those sequences that match patterns 1 and 2. A fileDivider process splits the output content and outputs only the fuzznuc seqtable section. Second, a FuzznucComparator process compares the output from the fileDivider process with those sequences that match pattern 3. The final step in the workflow is to write those sequences that match all three patterns to file. The Scufl workflow can be downloaded from http://www.bioinf.mvm.ed.ac.uk/projects/workflows/loxp. This workflow can be opened and enacted within the Taverna workbench. A web page interface to this workflow is also freely available at: http://wilkie226.dmed.ed.ac.uk:8080/loxpFinder.
The default mismatch values enable discovery of primary cryptic loxP sites. The Taverna workbench and the web page interface enable users to edit the workflow and thus change the number of mismatches tolerated for each pattern. By relaxing the number of allowed mismatches the workflow can find secondary cryptic loxP sites. Additional information regarding how to use these resources is available in the online manual.
Plasmid, BAC/PAC vectors, E. coli strains and growth conditions
pROSA26/Tet is a pBluescript-modified plasmid containing a tetracycline resistance gene sub-cloned between two loxP consensus sequences (pROSA26 unpublished, personal gift from Yuri Kotelevtsev, University of Edinburgh. Original source Igor Samokhvalov, RIKEN Center for Developmental Biology, Kobe Japan). This plasmid contains no AraC-PBAD promoter elements.
PAC111L11 (20) (kindly provided by Craig A. Jones, Buffalo, New York) maps on human chromosome 1 and spans the Ren gene locus. The vector backbone (pCYPAC2) contains a single consensus loxP site and encodes kanamycin resistance. BACN10 was isolated by the screening of a mouse genomic library (129/Ola mouse strain) (Invitrogen Corporation, Formally Research Genetics) using a renin gene probe. It maps on mouse chromosome 1 and spans the Ren1c and Ren1d gene locus. The vector backbone (pBeloBAC11) contains a single loxP site and encodes chloramphenicol resistance. ASBAC (kindly provided by Keith Parker Dallas, Texas) also comes from a pBeloBAC11 library, and maps to chromosome 15, spanning the cyp11b1 and cyp11b2 genes. [BAC ends coordinates refer to RP23-23009 clone sequence T7: 205723 bp, SP6: 82689 bp]. The DY380, EL250 and EL350 strains of E. coli have been described (3), and were kindly provided by Neal Copeland (Mouse Cancer Genetics Program, National Cancer Institute-Frederik).
EL350 and EL250 differ from DY380 in that they encode the Cre and Flp recombinase genes respectively under the control of AraC-PBAD promoter and they are not tetracycline resistant (5).
One hundred nanograms of PAC or BAC DNA were transformed into EL350 and DY380 bacterial strains by electroporation (Easyjet Plus, Equibio; 1.75 kV, 200 ohms, 2 μF). Following selection on kanamycin/chloramphenicol plates, a 10−6 dilution of cells was plated on LB agar plates containing either 0.2% arabinose or 0.2% glucose and incubated overnight at 32°C.
In vivo assay for Cre activity in EL350
EL350 cells were transformed with 40 ng of pROSA26/TET plasmid DNA by electroporation and plated on LB agar containing 50 μg/ml ampicillin or 25 μg/ml tetracycline or both antibiotics. After overnight growth at 32°C, colonies were counted and the ratio of tetracycline-resistant colonies to ampicillin-resistant colonies calculated as a measure of the Cre gene expression in EL350 in the un-induced state. This measure was then compared to the Cre activity after induction of the PBAD-promoter with 0.1% arabinose, according to a protocol adapted from Lee et al. (2001) (5) which is available at http://recombineering.ncifcrf.gov/Protocol.asp. Two cell dilutions (10−4 and 10−6) were plated on Amp, Amp/Tet and Tet plates.
In vitro assay for BAC/PAC DNA nicking
One microgram of BAC/PAC DNA extracted from DY380 cells was incubated with 1 or 2 μl of Cre enzyme (1000 U/ml, New England Biolabs) overnight. After the incubation, each reaction was phenol-chloroform extracted and ethanol precipitated. An aliquot of DNA was then digested with EcoRV or HindIII restriction enzymes (Promega). DNA, re-suspended in alkaline gel loading buffer (50 mM NaOH, 1 mM EDTA, 2.5% Ficoll and 0.025% bromoCresol green), was loaded on a denaturing gel as previously described (21). Following electrophoresis, the gel was soaked in 0.25 N HCl for 6–7 min, rinsed and neutralised in 0.5 N NaOH, 1.5 M NaCl for 30 min, blotted onto a positively charged membrane and hybridized according to standard procedures with specific radiolabelled probes, generated using the Ready-To-Go™ DNA Labelling beads (Amersham Pharmacia Biotech). Probes P1 (for the detection of cryptic loxP site 1) P2 and P3 (to detect cryptic loxP site 2) were generated by PCR from the PAC111L11 clone, using primers c-loxP1F 5′-CTCAGACACTTTGGTGGGTC-3′ and c-loxP1R 5′-GACTTTCAGTATGGCTGCCTAAC-3′ for probe 1 (P1); c-loxP2F 5′-CAGGAGTTAGAGACCAGC-3′ and c-loxP2R 5′-GCTATCTCGGCTCACTG-3′ for probe 2 (P2) and c-loxP3F 5′-GAAGGGCTGAGGTTAGGCAG-3′ and c-loxP3R 5′-GAACACCTACTGAGCTTGAG-3′ for probe 3 (P3).
RESULTS
Mouse genome-wide distribution of cryptic loxP sites
In order to assess the distribution and frequency of cryptic loxP sites in the genome, a new bioinformatics tool (FuzznucComparator) was developed to perform a mouse genome-wide search (see materials and methods). Two stringencies were applied. Primary cryptic loxP sites were identified as sequences conforming to three patterns, which together define a primary cryptic loxP site (homology in the 13-bp palindromic sequences of the loxP consensus sequence; conservation of base pairs in the loxP site believed to be contact points for the Cre enzyme and presence of the four bases (TATA) flanking the core sequence). Figure 1 shows the workflow for the search of cryptic loxP sites. FuzznucComparator also defined secondary cryptic loxP sites, using less stringent criteria where the mismatch allowance for the three patterns is increased arbitrarily by the operator. In our hypothesis, some of these secondary sites have the potential to bind Cre and mediate DNA damage, if they are located near primary cryptic loxP sites.
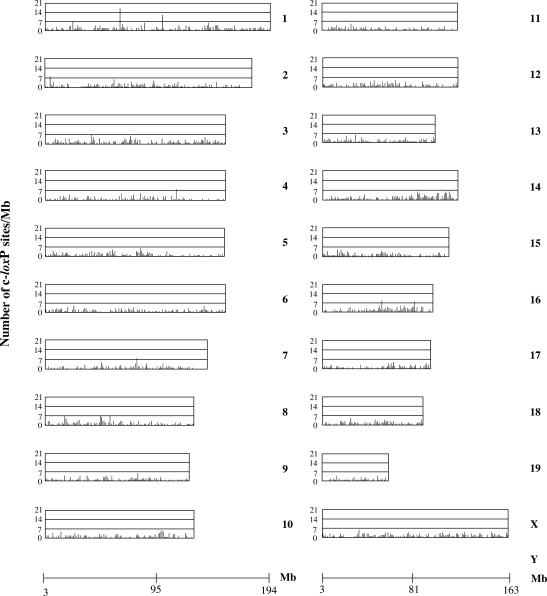
The NCBI m34 mouse assembly (http://www.ncbi.nlm.nih.gov/genome/seq/NCBIContigInfo.html) was split into one-megabase regions and submitted to the Fuzznuc and FuzznucComparator search for the three patterns defining a primary cryptic loxP site. The output of the search is represented in Figure 2. The overall frequency of primary cryptic loxP sites in the mouse genome is 1.2 per megabase. Some chromosomes show a more tightly clustered distribution of cryptic loxP sites than others (Figure 2, Chr 3, 13, 14, 15, 16, 18 and X). A few chromosomes (Figure 2, Chr 1, 2, 4, 7, 16) present with hot spots of cryptic loxP sites, generally 8 or 9 in a Mb DNA window, but up to 17 in the case of chromosome 1. Chromosome Y has no primary cryptic loxP sites.
Figure 2.
Mouse genome-wide search for cryptic loxP sites. Each graph shows the distribution and the number of cryptic loxP sites in 1 Mb regions along the 21 mouse chromosomes. The length of each graph is proportional to the corresponding chromosomal length. A megabase scale is present at the bottom of each graph.
About 10% of the spacer regions in primary cryptic loxP sites are not unique, and occur more than once.
In silico identification of cryptic loxP in PAC/BAC sequences
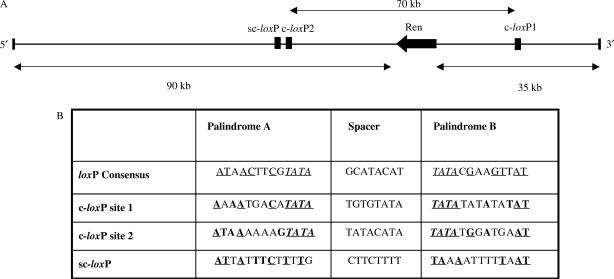
Three BAC and PAC clones (BACN10, ASBAC and PAC111L11) were scanned for cryptic loxP sites (in addition to the consensus loxP site in the backbone of their respective vectors) to see whether their presence caused instability in Cre-expressing host cells. Both PAC111L11 and BACN10, which span the human and mouse renin locus respectively, show the presence of primary cryptic loxP sites that match all three patterns. The search returned two potential cryptic loxP sequences (c-loxP1 and c-loxP2) in PAC111L11 with 5 out of 8 bp matching in the spacer region (62.5% identity) (Table 1). A search for secondary cryptic loxP sites within 6 kb of each primary site revealed one hit (sc-loxP), located 4.3 kb upstream of c-loxP2 in PAC111L11. The sc-loxP has twelve mismatches to the consensus loxP in the 13-bp palindromic arms, 12 out of 18 conserved Cre contact points and 3 mismatches in the TATA sequences flanking the spacer region. When aligned with the complementary strand of the c-loxP2 site, 4 out of 8 bp match in the spacer region.
Table 1.
Number of primary cryptic loxP sites in three BAC/PAC molecules and their characteristics
| PAC/BAC name | Putative primary cryptic loxP sites (search pattern1 and 2) | Pairs of primary c-loxP sites with 6/8 matches in the spacer region | Pairs of primary c-loxP sites with 5/8 matches in the spacer region | Pairs that have conservation in the 4 bases flanking the spacer region |
|---|---|---|---|---|
| PAC111L11(H) | 2 | 0 | 1 | 1 |
| ASBAC (M) | 5 | 0 | 1 | 0 |
| BACN10 (M) | 4 | 1 | 1 | 2 |
BACN10 is predicted to contain four primary cryptic loxP sites. Two of these are identical at 6 out of 8 bp in the spacer region and the other two match at 5; both pairs of sites show conservation in the four bases flanking the spacer region (Table 1). A search for secondary cryptic loxP sites in the regions surrounding the primary sites returned one hit with the same characteristics outlined for the sc-loxP site in PAC111L11.
ASBAC contains 5 sequences that match the first two criteria for primary cryptic loxP sites but, when considered in pairs, none show conservation in the four bases flanking the core region (Table 1). Since not all three criteria for cryptic loxP definition are satisfied, these sites are not predicted to be functional.
Differential growth of PAC/BAC-transformed EL350 E. coli strain compared to DY380 and EL250
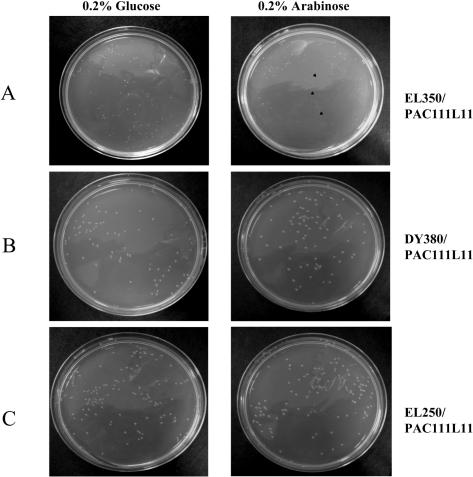
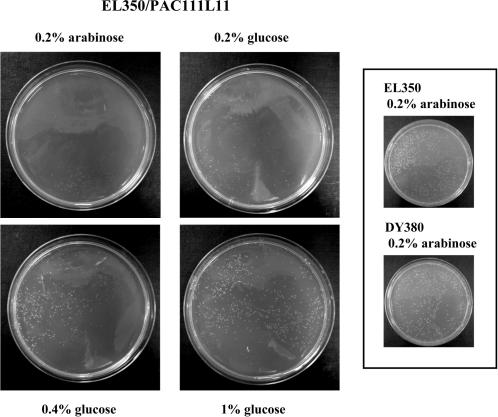
To test the hypothesis that cryptic loxP sites mediate DNA damage in E. coli strains expressing Cre recombinase, PAC111L11 (depicted in Figure 3) was transformed into DY380 (expressing neither Cre nor Flp), EL250 (expressing Flp) and EL350 (expressing Cre) E. coli (3). Transformed cells were grown in the presence of arabinose (to induce the AraC-PBAD promoter) or glucose (to ensure catabolite repression of the promoter) (22–24). Whilst the growth of PAC111L11-transformed DY380 and EL250 cells was similar on plates containing glucose or arabinose (Figure 4B and C), the growth of PAC111L11-transformed EL350 cells differed on arabinose, with a similar number of colonies of much smaller size (Figure 4A). Statistically significant reduced colony diameter is observed on arabinose plates when comparing PAC111L1-transformed EL350 (colony diameter 0.51 ± 0.02 mm) cells to PAC111L11-transformed DY380 (colony diameter 1.69 ± 0.02 mm on arabinose, p < 0.001) or EL250 (colony diameter 1.44 ± 0.02 mm on arabinose, p < 0.01) cells. In addition, the EL350-PAC111L11 colonies formed in the presence of glucose were appreciably smaller than those formed by DY380 or EL250 transformants. The effect of glucose on PAC111L11-transformed EL350 colony size is concentration-dependent with 1% glucose resulting in the largest colonies (Figure 5). Un-transformed DY380 and EL350 cells grow with the same efficiency in 0.2% arabinose-containing agar, despite the presence of one primary cryptic loxP site in the E. coli bacterial genome (Figure 5, inset).
Figure 3.
(A) Positions of the cryptic loxP sites on the PAC111L11 insert. (B) Sequence comparison between the loxP consensus site and PAC111L11 cryptic loxP sites (primary c-loxP and secondary sc-loxP). Underlined nucleotides — Cre contact points on loxP consensus sequence; bold — conserved nucleotides.
Figure 4.
Growth of PAC111L11-transformed EL350 (A), DY380 (B) and EL250 (C) cells on LB agar supplemented with either 0.2% glucose or 0.2% arabinose. Arrows indicate very small and barely detectable colonies formed on arabinose-containing agar by EL350/PAC111L11.
Figure 5.
Growth of PAC111L11-transformed EL350 on LB agar supplemented with increasing concentrations of glucose. Inset: Un-transformed EL350 and DY380 on agar with 0.2% arabinose.
EL350 cells transformed with BACN10 also gave rise to very small colonies that failed to grow or had delayed growth on arabinose (data not shown), whereas EL350 cells transformed with ASBAC showed no difference in growth rate.
In vivo assay for Cre activity in EL350
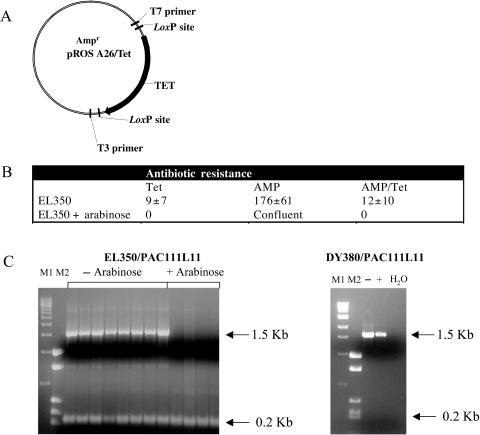
Low levels of expression from the PBAD promoter can occur in the absence of the inducer arabinose if the expression at maximum induction is very high. Leaky Cre expression from the AraC-PBAD promoter has been described for multi-copy plasmids (25). However, in EL350 cells the AraC-PBAD-Cre gene is present in a single copy. To test if Cre was produced from the un-induced AraC-PBAD promoter in EL350, a functional test was performed, using a plasmid vector (pROSA26/Tet) encoding a tetracycline resistance gene between two loxP consensus sequences (Figure 6A). The plasmid pROSA26/Tet also confers ampicillin resistance to the host. If recombination occurs between the loxP sequences, the Tet resistance gene is excised and the E. coli strain can only grow under Amp selection. This was the case when Cre expression in EL350 cells was induced with arabinose. Two cell dilutions (10−4 and 10−6) were plated on LB agar plates containing either 50 μg/ml ampicillin or 25 μg/ml tetracycline or both. Although many colonies were obtained on ampicillin, no colonies were observed on Amp/Tet or Tet plates (Figure 6B), even after 24 h. Following transformation with pROSA26/Tet, Tet resistant colonies were obtained in the absence of arabinose (when the AraC-PBAD promoter is not induced in EL350 cells), however, the number of colonies was reduced approximately twenty-fold (Figure 6B). These data show that about 95% of the colonies that are able to grow on an Amp/Tet plate have lost the tetracycline gene and are no longer able to grow under tetracycline selection (p < 0.01). In order to verify if all the plasmid molecules inside a single E. coli colony have undergone site-specific recombination or a few of them retain the tetracycline resistance gene, a PCR assay was performed on colonies using primer T3 and T7 (Figure 6C). A mixed plasmid population (excised and non-excised) is recovered from un-induced EL350. This is shown by the presence of two DNA fragments, one of 1.5 Kb (Tet retaining plasmids) and one of 0.2 Kb (Tet excised plasmids). After induction of EL350 with arabinose, the tetracycline resistance gene was found to be 100% excised. Plasmids that grow into DY380 do not show any sign of site-specific recombination and maintain the tetracycline resistance gene (Figure 6C, right panel).
Figure 6.
(A) pROSA26/Tet plasmid map. (B) Summary of the transformation of pROSA26/Tet plasmid into EL350. (C) PCR analysis of pROSA26/Tet transformed EL350 and DY380 under arabinose-induced and un-induced conditions using T3 and T7 primers. M1: 1 Kb Marker (New England Biolabs). M2: pBluescriptII SK+ plasmid digested with Sau3AI. The 1.5 Kb band indicates the presence of the Tet gene; the 0.2 Kb band is generated after Cre/loxP-mediated excision of the Tet gene.
Analysis of nicks in PAC111L11 DNA after incubation with Cre
The ability of Cre recombinase to introduce single or double-strand nicks at cryptic loxP sites was tested by incubation with PAC111L11 DNA. DNA fragments produced by the action of Cre were detected by Southern blot following either HindIII or EcoRVV restriction digest for the analysis of c-loxP1 nicks or c-loxP2 nicks, respectively using a c-loxP1 specific probe (P1) or c-loxP2 specific probes (P2 and P3; Figure 7).
Figure 7.
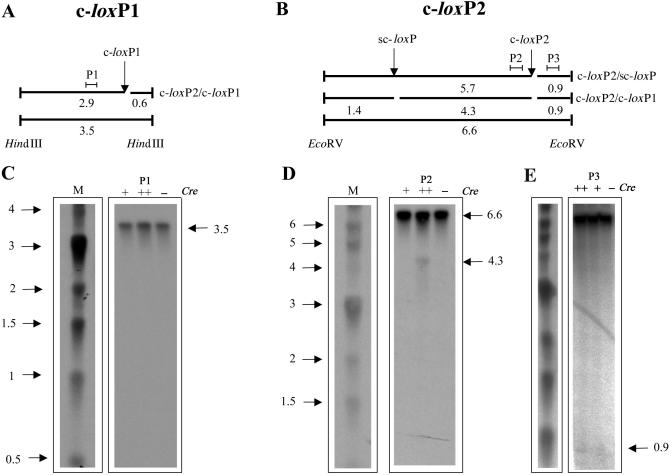
In vitro analysis of the presence of nicks in PAC111L11 insert. (A) Schematic representation of expected fragment size for DNA nicked at cryptic loxP1 site. The arrow indicates the location of the c-loxP1 site on the HindIII fragment. P1: probe 1. (B) Schematic representation of predicted fragment sizes for DNA nicked at the c-loxP2 and secondary sc-loxP sites on the EcoRVV fragment. Arrows indicate the location of the c-loxP2 and sc-loxP sites, P2 — probe 2, and P3 — probe 3. (C) Southern analysis of PAC111L11 DNA, following overnight incubation with Cre recombinase, digested with HindIII and hybridized to probe P1 to detect nicks produced at the c-loxP1 site. M:1 kb DNA ladder (NEB) Plus and minus signs on each lane refer to the presence or absence of Cre recombinase and its relative abundance. (D) and (E) Southern analysis of PAC111L11 DNA, following overnight incubation with Cre recombinase, digested with EcoRVV and hybridized with probe P2 (D) and P3 (E) to detect nicks produced by cryptic loxP site 2 and/or sc-loxP. M:1 kb DNA ladder (NEB) Plus and minus signs on each lane refer to the presence or absence of Cre recombinase and its relative concentration.
PAC111L11 DNA showed no evidence of nicking or cleavage at c-loxP1 following overnight incubation with Cre recombinase (Figure 7C). Only one band of 3.5 kb, corresponding to the intact DNA, was detected by probe P1. In contrast, hybridization of probes P2 and P3 to EcoRVV-digested PAC111L11 DNA detected fragments consistent with cleavage at both c-loxP2 and sc-loxP sites (Figure 7D and E). In the absence of Cre, the expected 6.6 kb fragment produced by EcoRVV digestion of intact PAC111L11 DNA was detected. At the higher concentration of Cre, additional fragments of 4.3 kb (with probe P2, Figure 7D) and 0.9 kb (with probe P3, Figure 7E) were detected.
These results suggest that the primary (c-loxP2) and the secondary (sc-loxP) c-loxP sites that map in the PAC111L11 insert are able to bind Cre recombinase enzyme and start a recombination event, which is non-productive and results in damage to the PAC DNA molecule.
The absence of the nicked 2.9 kb at the c-loxP1 site and 5.7 kb at the c-loxP2 site suggests that these cryptic loxP site are not getting paired and defectively recombined with the proper consensus loxP site that is located in the backbone of the PAC sequence.
DISCUSSION
Although little evidence has been reported, certain BAC/PAC inserts are difficult to propagate in Cre-containing E. coli strains. It is possible this could be due to low levels of Cre promoting single strand DNA breaks or recombination through cryptic loxP sites present in the inserts. Computational analysis of the whole mouse genome has revealed an average of 1.2 primary cryptic loxP sites per Mb DNA, with few hotspots present (chromosome y has none). The new bioinformatic tool, FuzznucComparator has been made available through a Distributed Annotation System (DAS) resource to enable dynamic access to the data using http requests (i.e. a URL), with the response being returned as XML. This resource is implemented using a Dazzle [DAZZLE] server backed by an LDAS database and can be accessed at: http://wilkie226.dmed.ed.ac.uk:8080/das. Access via the DAS protocol also allows these annotations to be viewed as a track within Ensembl (Ensembl help, http://www.ensembl.org/Homo_sapiens/helpview?se?1;kw?dasconfview, documents how to achieve this).
We have tested the power of the programme to predict functional cryptic loxP sites in the genomic insert of BAC and PAC sequences, which could potentially affect their growth in Cre recombinase expressing E. coli strains. We have identified primary and secondary cryptic loxP sites in the genomic insert of three BAC/PAC clones and shown that these are substrates for Cre recombinase in vitro. Furthermore, following transformation into EL350 cells, they demonstrate a dramatically reduced growth rate in the presence of arabinose, compared to DY380 and EL250. The poor growth is ameliorated by glucose suggesting a Cre-mediated effect on bacterial growth. We suggest that, since Cre recombinase is leaky, EL350 has a growth defect when transformed with PAC111L11.
Despite the higher conservation level of c-loxP1 and c-loxP2, our data shows that c-loxP2 and sc-loxP are involved in the single-strand nicking of PAC111L11, incubated in vitro with Cre recombinase. c-loxP2 and sc-loxP are located relatively close to each other, whereas c-loxP1 lies 70 kb away from c-loxP2. The interaction of Cre recombinase with cryptic loxP sites is likely to be weaker than with genuine loxP sites. However, proximity of two cryptic loxP sites may serve to increase the local concentration of Cre recombinase, increasing the likelihood of DNA nicking at one or both sites.
All three BAC/PAC molecules we have analysed contain a loxP consensus sequence in their respective vector backbones. However, we consider it unlikely that they mediate nicking for the following reasons: (1) they occur in most if not all BAC/PAC vectors and clearly DNA nicking and poor growth is not a problem with all BAC/PACs. (2) ASBAC also encode putative cryptic loxP sites, however, their presence did not result in slower growth in EL350 cells than EL250 or DY380 cells. We cannot exclude that in other molecules DNA damage might occur if the conditions (homology in the spacer region and location of the c-loxP site close to the end of the insert, and thus proximal to the genuine loxP site) are favourable. These data provide a functional definition of active cryptic loxP sites; more relaxed mismatches allowance in comparison to the criteria previously set (12) shows that a high degree of deviation from the loxP consensus sequence is still tolerated by Cre recombinase.
The demonstration that Cre recombinase expression is leaky in this strain supports the hypothesis that the damaged PAC DNA fails to replicate, depriving the E. coli daughter cells of the selectable marker. These findings are in accordance with data in the literature that show that mammalian genomes contain active recombinase recognition sites (12) and that growth inhibition and DNA damage can be induced by the expression of Cre recombinase in mammalian cells, E. coli and yeast (14,26–28). The leakiness of Cre recombinase in EL350 cells is not surprising. Studies have demonstrated that PBAD promoters are very efficient, but the levels of repression are not always zero, because they are relative to the levels of expression at maximum induction (25).
During cleavage, Cre becomes covalently attached to the DNA through a 3′-phosphate. This type of covalent protein-DNA linkage is very similar to that observed with DNA topoisomerases (11,29–31). In the presence of cryptic loxP sites or mutant loxP sites (11), Cre protein attempts to carry out recombination, but the reaction is abortive. If the reaction progresses to the stage where nicks are introduced into the DNA, then a damaged DNA molecule is produced, which is covalently linked to a protein making it very difficult to repair by the cell repair machinery (32).
Many lines of transgenic mice have been generated that express Cre recombinase, but there are few reports of adverse effects of this protein in vivo. The potential for DNA damage is demonstrated by the infertility of transgenic mice expressing Cre in spermatids, due to illegitimate Cre-dependent chromosome rearrangements (13). It is possible that somatic mutations remain undetected, due to the high tolerance of mammals for somatic cell death and, in light of this study, a more detailed analysis of the phenotype of transgenic animals bearing the Cre recombinase gene may be informative.
Finally, these results suggest that the presence of cryptic loxP sites in BAC/PAC inserts can affect the efficiency of recombineering techniques if the host cells express Cre recombinase even in a leaky way. This problem is BAC/PAC dependent and does not diminish the usefulness of recombineering techniques. Nevertheless, these data propose and describe a mechanism, which explains why recombineering experiments sometimes don’t give the expected results and provide a bioinformatic tool, which can alert and guide in the planning process. The use of FuzznacComparator to identify cryptic loxP sites in BAC/PAC inserts may be helpful in determining which BACs may prove to be most manipulatable with the Cre/loxP system.
ACKNOWLEDGEMENTS
We are grateful to David R.F. Leach (Institute of Cell and Molecular Biology, University of Edinburgh), Janice Paterson and David Brownstein for useful discussions during the course of this work and we thank Matt Sharp for critically reading the manuscript.
We acknowledge funding from the Wellcome Trust, including the Functional Genome Initiative and the Cardiovascular Research Initiative.
J.J.M. is a Wellcome Trust PRF and S.S. is a Wellcome Trust Intermediate Research Fellow.
REFERENCES
- 1.Copeland NG, Jenkins NA, Court DL. Recombineering: a powerful new tool for mouse functional genomics. Nat. Rev. Genet. 2001;2:769–779. doi: 10.1038/35093556. [DOI] [PubMed] [Google Scholar]
- 2.Court DL, Sawitzke JA, Thomason LC. Genetic engineering using homologous recombination. Annu. Rev. Genet. 2002;36:361–388. doi: 10.1146/annurev.genet.36.061102.093104. [DOI] [PubMed] [Google Scholar]
- 3.Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murphy KC. Use of bacteriophage lambda recombination functions to promote gene replacement in Escherichia coli. J. Bacteriol. 1998;180:2063–2071. doi: 10.1128/jb.180.8.2063-2071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- 6.Sauer B, Henderson N. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc. Natl. Acad. Sci. U.S.A. 1988;85:5166–5170. doi: 10.1073/pnas.85.14.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoess RH, Ziese M, Sternberg N. P1 site-specific recombination: nucleotide sequence of the recombining sites. Proc. Natl. Acad. Sci. U.S.A. 1982;79:3398–3402. doi: 10.1073/pnas.79.11.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abremski K, Hoess R. Phage P1 Cre-loxP site-specific recombination. Effects of DNA supercoiling on catenation and knotting of recombinant products. J. Mol. Biol. 1985;184:211–220. doi: 10.1016/0022-2836(85)90374-2. [DOI] [PubMed] [Google Scholar]
- 9.Abremski K, Frommer B, Wierzbicki A, Hoess RH. Properties of a mutant Cre protein that alters the topological linkage of recombination products. J. Mol. Biol. 1988;202:59–66. doi: 10.1016/0022-2836(88)90518-9. [DOI] [PubMed] [Google Scholar]
- 10.Hoess RH, Wierzbicki A, Abremski K. The role of the loxP spacer region in P1 site-specific recombination. Nucleic. Acids. Res. 1986;14:2287–2300. doi: 10.1093/nar/14.5.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abremski K, Wierzbicki A, Frommer B, Hoess RH. Bacteriophage P1 Cre-loxP site-specific recombination. Site-specific DNA topoisomerase activity of the Cre recombination protein. J. Biol. Chem. 1986;261:391–396. [PubMed] [Google Scholar]
- 12.Thyagarajan B, Guimaraes MJ, Groth AC, Calos MP. Mammalian genomes contain active recombinase recognition sites. Gene. 2000;244:47–54. doi: 10.1016/s0378-1119(00)00008-1. [DOI] [PubMed] [Google Scholar]
- 13.Schmidt EE, Taylor DS, Prigge JR, Barnett S, Capecchi MR. Illegitimate Cre-dependent chromosome rearrangements in transgenic mouse spermatids. Proc. Natl. Acad. Sci. U.S.A. 2000;97:13702–13707. doi: 10.1073/pnas.240471297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loonstra A, Vooijs M, Beverloo HB, Allak BA, van Drunen E, Kanaar R, Berns A, Jonkers J. Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2001;98:9209–9214. doi: 10.1073/pnas.161269798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Missirlis PI, Smailus DE, Holt RA. A high-throughput screen identifying sequence and promiscuity characteristics of the loxP spacer region in Cre-mediated recombination. BMC Genomics. 2006;7:73. doi: 10.1186/1471-2164-7-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Court DL, Swaminathan S, Yu D, Wilson H, Baker T, Bubunenko M, Sawitzke J, Sharan SK. Mini-lambda: a tractable system for chromosome and BAC engineering. Gene. 2003;315:63–69. doi: 10.1016/s0378-1119(03)00728-5. [DOI] [PubMed] [Google Scholar]
- 17.Oinn T, Addis M, Ferris J, Marvin D, Senger M, Greenwood M, Carver T, Glover K, Pocock MR, Wipat A, et al. Taverna: a tool for the composition and enactment of bioinformatics workflows. Bioinformatics. 2004;20:3045–3054. doi: 10.1093/bioinformatics/bth361. [DOI] [PubMed] [Google Scholar]
- 18.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 19.Senger M, Rice P, Oinn T. In: Cox SJ, editor. Proceedings, UK e-Science, All Hands Meeting; 2003; 2003. pp. 509–513. [Google Scholar]
- 20.Sinn PL, Davis DR, Sigmund CD. Highly regulated cell type-restricted expression of human renin in mice containing 140- or 160-kilobase pair P1 phage artificial chromosome transgenes. J. Biol. Chem. 1999;274:35785–35793. doi: 10.1074/jbc.274.50.35785. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook J, Maniatis T. Molecular Cloning: A Laboratory Manual. New York: Cold Spring Harbor; 1989. [Google Scholar]
- 22.Lee N. In: The Operon. Miller JH, Reznikoff WS, editors. New York: Cold Spring Harbor; 1980. pp. 389–410. [Google Scholar]
- 23.Lee N, Francklyn C, Hamilton EP. Arabinose-induced binding of AraC protein to araI2 activates the araBAD operon promoter. Proc. Natl. Acad. Sci. U.S.A. 1987;84:8814–8818. doi: 10.1073/pnas.84.24.8814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyada CG, Stoltzfus L, Wilcox G. Regulation of the araC gene of Escherichia coli: catabolite repression, autoregulation, and effect on araBAD expression. Proc. Natl. Acad. Sci. U.S.A. 1984;81:4120–4124. doi: 10.1073/pnas.81.13.4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sternberg N, Hamilton D, Hoess R. Bacteriophage P1 site-specific recombination. II. Recombination between loxP and the bacterial chromosome. J. Mol. Biol. 1981;150:487–507. doi: 10.1016/0022-2836(81)90376-4. [DOI] [PubMed] [Google Scholar]
- 27.Sauer B. Identification of cryptic lox sites in the yeast genome by selection for Cre-mediated chromosome translocations that confer multiple drug resistance. J. Mol. Biol. 1992;223:911–928. doi: 10.1016/0022-2836(92)90252-f. [DOI] [PubMed] [Google Scholar]
- 28.Sauer B. Multiplex Cre/lox recombination permits selective site-specific DNA targeting to both a natural and an engineered site in the yeast genome. Nucleic Acids Res. 1996;24:4608–4613. doi: 10.1093/nar/24.23.4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tse Y, Wang JC. E. coli and M. luteus DNA topoisomerase I can catalyze catenation of decatenation of double-stranded DNA rings. Cell. 1980;22:269–276. doi: 10.1016/0092-8674(80)90174-9. [DOI] [PubMed] [Google Scholar]
- 30.Been MD, Champoux JJ. Breakage of single-stranded DNA by rat liver nicking-closing enzyme with the formation of a DNA-enzyme complex. Nucleic Acids Res. 1980;8:6129–6142. doi: 10.1093/nar/8.24.6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Champoux JJ. DNA is linked to the rat liver DNA nicking-closing enzyme by a phosphodiester bond to tyrosine. J. Biol. Chem. 1981;256:4805–4809. [PubMed] [Google Scholar]
- 32.Connelly JC, Leach DR. Repair of DNA covalently linked to protein. Mol. Cell. 2004;13:307–316. doi: 10.1016/s1097-2765(04)00056-5. [DOI] [PubMed] [Google Scholar]