Abstract
Macroscopically realizable applications of DNA-based molecular devices require individual molecules to cooperate with each other. However, molecular crowding usually introduces disorder to the system, thus jeopardizing the molecular cooperation and slowing down their functional performance dramatically. A challenge remaining in this field is to obtain both smarter response and better cooperation simultaneously. Here, we report a swift-switching DNA nanodevice that is enhanced by an alternating electric field. The device, self-assembled from folded four-stranded DNA motifs, can robustly switch between closed and open states in smart response to pH stimulus, of which the closed state forms a nanometer-height container that is impermeable to small molecules. This character was used to directly and non-specifically catch and release small molecules emulating mechanical hand in a controllable way. The alternating electric field was used to accelerate molecular cooperative motion during the device switching, which in turn shortened the closing time remarkably to thirty seconds.
INTRODUCTION
Surface-based molecular devices have exhibited many advantages owing to their chemical designability and widespread applications (1–11). Combination of DNA materials (2–8) with surface-based ‘bottom-up’ approach has led to several important technological innovations, such as DNA microarray (9), DNA computation (10) and DNA-based nanolithography (11). Macroscopically realizable applications of DNA-based molecular devices require individual molecules to cooperate with each other. However, molecular crowding usually introduces disorder to the system, thus jeopardizing the molecular cooperation and dramatically slowing down their performance (12,13). One challenge in this field is to obtain both smarter response and better cooperation simultaneously. For instance, we recently demonstrated that single-stranded DNA (ssDNA) capped with a short piece of double-stranded DNA (dsDNA) can form a switching compartment nanostructure on surface by a compact array of DNA molecules (13). However, the switching of this nanostructure highly relies on the cooperative hybridization between complementary DNA molecules, which is usually very slow, cumbersome for operation and contaminated for cycling. In principle, the DNA-formed compartment nanostructure is still far away from an applicable DNA nanomechanical device. Here we report a swiftly switching DNA-nanocontainer device which can non-specifically catch and release small molecules emulating mechanical hand in a controllable way. In contrast to the previous studies, our new approach greatly simplifies working principle of the DNA nanodevice, in which the switching can be fulfilled by itself without assistance of complementary strands, driven by clean energy sources (pH changes), and accelerated by alternating electric fields.
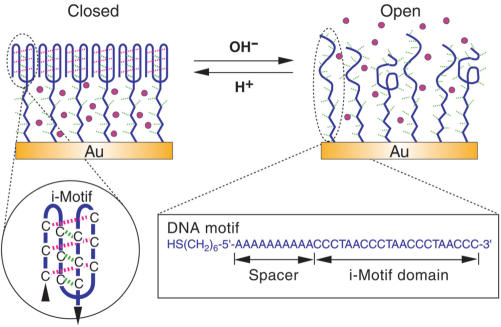
The device was implemented with the surface-supported self-assembly monolayer (SAM) of thiol-terminated DNA molecular motifs on gold surface. As shown in Figure 1, each DNA motif contains two functional domains: a 21-mer i-motif domain which can fold into a four-stranded structure via protonated cytosine–cytosine (C · C+) base-pair formation (inset of Figure 1) (14–22); a single-stranded poly-(dA)n spacer, where n is an integer in the range of 10–35. To create a monolayer with sufficient spacing between the single-stranded poly-(dA)n domain, the i-motif domain was prefolded into its four-stranded i-motif form at pH 4.5 before being applied to the gold surface, which results in a SAM that is densely packed with respect to the space-filling i-motif structures but shows low-density packing with respect to the poly-(dA)n domain. Previous studies suggest that the repulsion force between DNA molecules is too weak to prevent the dsDNA from aggregating into densely packed states (23–31). The molecular repulsion interaction between dsDNA helices was measured to be consistently exponential with 2.5–3.5 Å (24), allowing the energetically favorable separation of 25–35 Å between duplex DNA in densely packed arrays (24,25), which was further verified by theoretical calculations (26,27). It was also well established that small particles, such as ferricyanide anions, methylene blue, and so on, are impermeable through a dsDNA SAM with complete coverage (13,28–30). While the dsDNA molecules were observed approximately perpendicular to gold surface in a densely packed SAM (31), our previous studies further established that in a DNA SAM possessing the nanocompartment structure, the ssDNA spacer is sufficiently stretched and approximately perpendicular to gold surface (13), providing functioning rationality of the DNA nanocompartment. In this study, we will show that although the i-motif structure is quite different from the dsDNA helices, similar functionality can also be achieved by the use of i-motif structure instead of dsDNA. At pH 4.5, a closed nanocompartment structure is expected to form between the densely packed i-motif membrane and gold surface with an effective height (basically commensurate with the contour length of the poly-(dA)n spacer). At pH 8.0, DNA motifs are denatured into single-stranded form, causing the SAM to become much loose, and deforming the nanocompartment structure.
Figure 1.
Working principle of the switching DNA nanocontainer. At pH 4.5, the i-motif domain folds into four-stranded structure and packs into a membrane impermeable for small molecules on gold surfaces. At pH 8.0, the i-motif structures are transformed into single strands, making the packing density of the DNA SAM relatively loose to allow small molecules to diffuse freely.
MATERIALS AND METHODS
Preparation
Ultra-flat gold surface (Au(111)) was obtained by evaporating gold on newly cleaved mica surface, annealed at 350°C for 60 min and removing mica before use to minimize contamination. All DNA are purchased from Sangon Bioengineering (Shanghai, China) and PAGE purified. 5′ thiolated DNA was dissolved in 100 mM PBS buffer, pH 4.5 with 100 mM NaCl to give a final concentration of 1 μM, heated to 80°C for 5 min and let it cool to 20°C slowly; then applied to new cleaved gold surface for 24 h at 4°C to achieve a compact self-assembly monolayer of the i-motif structures. After the self-assembly procedure, the samples were washed by pH 4.5 PBS for three times, then incubated with 1 mM 2-mercaptoethanol in PBS, pH 4.5 for 15 min and washed with 10% urea in MilliQ water for three times. Once prepared, the samples were stored in pH 4.5 PBS buffer at 4°C before use.
Device operation
Switching of the device was performed in 100 mM PBS buffer with 50 mM NaCl at 20°C. pH 4.5 and pH 8.0 PBS buffers were prepared in advance respectively. For the opening process, the device was washed with pH 8.0 buffer three times and then incubated with pH 8.0 buffer for 10 min. For the closing process, the device was washed by pH 4.5 buffer three times and then incubated in pH 4.5 buffer for 10 min. The device could also be switched by alternating buffer pH in situ via adding 16 μl 1 M NaOH or 16 μl 1 M HCl alternatively into 400 μl 100 mM PBS buffer.
AFM imaging
All atomic force microscope (AFM) images were taken in phosphate buffer using Molecular Imaging PicoPlus II system (AZ, USA) with MACLever cantilevers fixed on small-range scanner controlled by Picoscan 2500. Cantilevers used (force constant: 0.1 N/m; length: 140 µm; thickness: 0.6 µm; mean width: 18 µm; resonant frequency in buffer solution: 28–30 kHz) were Type IV MACLevers purchased directly from Molecular Imaging (www.molec.com). All images are obtained under Magnetic AC mode (MAC Mode) and leveled using a plane fit to remove sample tilt. Note that cantilevers of higher force constant cannot properly image ssDNA SAM due to the insensitivity arising from the cantilever stiffness, which may disrupt the SAM surface by the cantilever tips in buffer solutions; cantilevers of lower force constant (for example, 0.01 N/m) cannot provide stable imaging due to the chaotic oscillation of cantilevers in buffer solutions. Stable imaging should have identical image contents of repeated scanning at the same places, which can proportionally scale with scaling the scanning range. In contrast, unstable imaging yields different image contents of repeated scanning at the same places.
Electrochemical analysis
Electrochemical analysis were conducted on CHI 660A electrochemical workstation (CH instruments Inc., TX, USA), with Ag/AgCl as reference electrode platinum as counter electrode, and 10 × 10 μM2 DNA-modified gold substrate as working electrode in 100 mM PBS buffer. To prepare a closed device with [Fe(CN)6]3− encaged, it was opened in pH 8.0 buffer and then closed with 10 mM [Fe(CN)6]3− in pH 4.5 buffer, washed with 1 ml pH 4.5 buffer without [Fe(CN)6]3− three times before applying to CV test. Open state of the device was obtained by incubating the device in pH 8.0 buffer and washed with 1 ml pH 8.0 buffer without [Fe(CN)6]3− three times before applying to CV test. Opening and closing procedures with different [Fe(CN)6]3− concentrations (1–100 mM), pH values, and other report small molecules are carried out accordingly.
Alternating electric fields
The alternating electric fields used in the acceleration of the device switching were generated directly by the functions (a.c. resistance, a.c. voltammetry, etc.) of CHI 660A electrochemical workstation in a three electrodes system described above. The potential amplitude is 0.1–0.5 V and absolute potential is controlled within −0.6–1.0 V. Frequency used is in the range of 1–10 kHz. In this experiment, the cell electrolyte was replaced by the PBS buffer that was used to switch the device, in which the object molecules were included. When using the electrochemical detection to trace the kinetics of the device switching (as shown in Figure 2), the electrolyte used was replaced by 100 mM PBS containing no redox species. The routine washing procedure with 1 ml PBS as described above was always carried out prior to each electrochemical measurement.
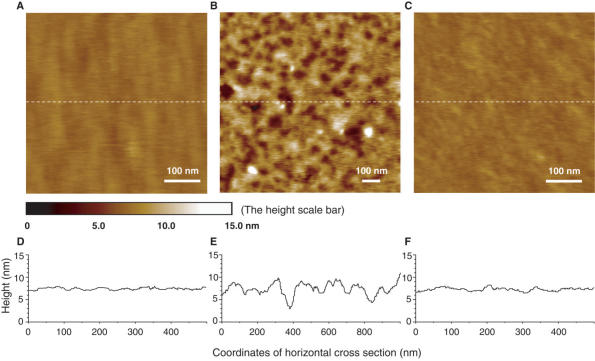
Figure 2.
AFM topographic images for the different states of the device on Au(111) surface. (A) Initial closed state at pH 4.5 after self-assembly shows 1.5 ± 0.5 nm surface roughness. (B) Open state at pH 8.0 shows 6.0 ± 1.0 nm surface roughness. (C) Repeated closed state at pH 4.5 restores the surface roughness to 1.5 ± 0.5 nm. Height scales for all images are adjusted to a uniform range of 15 nm. The color mapping to height is indicated by the height scale bar. The poly-dA spacer length of DNA motif is 10 bp. (D–F) The height analysis of the horizontal cross sections along the middle dashed white lines shown in (A–C). (D) corresponds to (A), (E) to (B), and (F) to (C), respectively.
RESULTS AND DISCUSSION
AFM Characterization
The quality of the SAM and its conformational transition of the fundamental nanostructure described above were characterized in situ by atomic force microscopy (AFM) in buffer solutions (32,33). High-resolution AFM imaging of soft matters in aqueous solution up to tens of nanometers is difficult to achieve due to disruption of sample surface by the AFM probing tips. Recent technical improvement by using magnetic field to drive oscillating cantilevers presents much reduced perturbation of AFM probing tips to the liquids near the imaging site as well as the soft sample itself, where less noise and smaller amplitude of the cantilevers can be achieved (33). This advanced technique helped us to image stably and reproducibly, the SAM topography in both folded and unfolded states without obviously perturbing or damaging the samples (see materials and methods section for technical details). As shown in Figure 2, the surface roughness of the SAM is 1–2 nm at pH 4.5 (Figure 2A and D) and 5–7 nm at pH 8.0 (Figure 2B and E), indicating that a compact SAM at the closed state transformed into low-density packing SAM at pH 8.0. The surface roughness of 1–2 nm could be recovered after switching the SAM to the closed state (Figure 2C and F), verifying the reversibility of the device switching.
The AFM image at pH 8.0 (Figure 2B) contains pore-like lower regions and mountain-like higher regions, both with diameters on the order of tens of nanometers. The cross-sectional height analysis (Figure 2E) shows that the lower regions and the higher regions are connected smoothly with height difference within 5–7 nm. We have estimated the surface coverage of the folded i-motif SAM to be about 1.2 × 1013 molecules/cm2 (see Supplementary Data), which indicates the average separation of ∼3 nm between nearest neighboring folded i-motif DNA molecules. At pH 8.0, the folded four-stranded i-motif domain was denatured into single-stranded form with cross-sectional diameter of ∼0.6 nm. Comparing with the diameter of i-motif structure, the packing of ssDNA is relatively loose and consequently leads to irregular pileup on the gold surface. These ssDNA pileups have an inhomogeneous nature, leading to a random distribution of the dense pileup regions and loose pileup regions, corresponding to the mountain-like higher regions and pore-like lower regions, respectively as observed in Figure 2B. In contrast, when the i-motif domains are in the well-folded state at pH 4.5, the same surface coverage makes the folded i-motif densely packed, as the cross-sectional diameter of i-motif structure is 1.9 nm (17), in which case the irregular pileup effect for the unfolded ssDNA is not allowed and the topography of the SAM is expected to reflect the flatness of the gold surface on which they are immobilized (Figure 2D and F).
In the large area topography of AFM observations, the SAM shows good homogeneity although there are inevitable defects, mainly pinholes on gold substrates, demonstrating that an essentially complete coverage of the DNA SAM was achievable (13,28–31). These characterizations make sure that an expected conformational transition between closed and open states was well defined in our devices, which was further confirmed by fluorescence quenching experiments (see Supplementary Data, Figure S1) and electrochemical analysis described below.
Switching the device
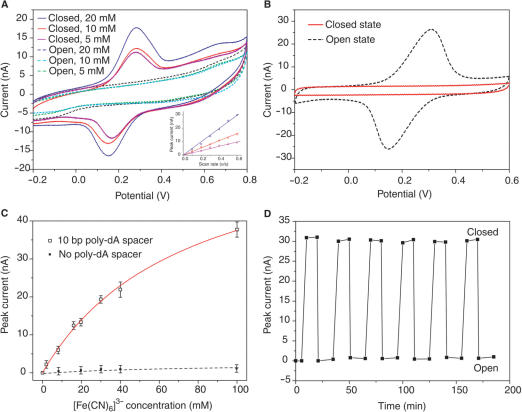
Since the diameter of i-motif structure is 1.9 nm (17), giving a 0.5–1 nm separation between the surfaces of the nearest neighboring i-motif structures at pH 4.5, the nanocompartment formed at the closed state may be impermeable for small molecules with about 0.5–3 nm sizes due to intermolecular repulsion interactions (13,28–30). This character enables the device to catch and release small molecules emulating mechanical hand on molecular level. To illustrate this process, ferricyanide anions [Fe(CN)6]3−, whose size is ∼0.8 nm, were employed as object molecules to translate the closed and open states of the device into electrochemical signals by cyclic voltammetry (CV) method (34). As shown in Figure 3A, there was obvious peak currents in CV curves when Fe[(CN)6]3− anions were confined in the closed SAM and no peak current was observed in CV curves after the device was turned open. The obvious peak currents in Figure 3A can only be attributed to the Fe[(CN)6]3− anions encaged inside the closed SAM, which refers to molecular encaging effect (13), for the following reasons. First, circular dichroism spectroscopy analysis (35) show that [Fe(CN)6]3− has no obvious intercalation with i-motif structure (see Supplementary Data, Figure S2). Second, a control experiment (Figure 3B) was performed in the presence of 10 mM [Fe(CN)6]3− in the electrolyte, without encaging any [Fe(CN)6]3− in the closed SAM in advance, showing that the closed SAM can prevent Fe[(CN)6]3− anions in surrounding buffers from diffusing to bare electrodes surface because of the electrostatic repulsive interactions between [Fe(CN)6]3− and DNA (28–30). Third, the negative charges on the sugar–phosphate backbones have higher spatial density in the i-motif structure than in the ssDNA spacer. The spatial density of the negative charges in i-motif structure versus ssDNA spacer is estimated to be roughly 8:1. Thus, the [Fe(CN)6]3− anions have much weaker electrostatic repulsion with the ssDNA spacer inside the nanocompartment than with the i-motif structure. Fourth, DNA functionalized surface without compartment nanostructure cannot confine [Fe(CN)6]3− (13,28–30). In our experiments, no obvious peak current was observed when spacer length was set to zero (Figure 3C) at both pH 4.5 and 8.0, indicating that no obvious [Fe(CN)6]3− intercalated in i-motif SAM. Finally, even though [Fe(CN)6]3− can intercalate into the i-motif membrane, the exponential decay of electron transfer across thickness ≥ 5 nm (corresponding to n ≥ 10 in poly-(dA)n domain) can result in that the electrochemical signal contributed from the intercalated reporters is not detectable (13,36–38).
Figure 3.
Using the switchable DNA nanocontainer to catch and release small molecules. (A) Cyclic voltammogram for the closed and open states of the device with different [Fe(CN)6]3− concentration. Scan rate is 0.3 Vs−1. No [Fe(CN)6]3− presented in the electrolyte of CV analysis. The linear relationship between peak currents and scan rates shown in inset confirms that the redox species were confined to the electrode surface. Note that the peak current is defined as the difference between maximum current and baseline current. (B) Control experiments show that the [Fe(CN)6]3− can not diffuse into the nanocontainer at the closed state, and can quickly diffuse to the electrode surface at the open state. (C) Isotherms for the molecular encaging effect when applied to capture [Fe(CN)6]3−. Solid line fit to Langmuir isotherm model: y = σmx/(x + K−1), where x is the concentration of the redox reporters and σm is the saturation value of the concentration and K is the association constant. K = 13.7 M−1 in the simulation. (D) Cyclic switching of the device indicated by the peak current in the cyclic voltammograms. It was found that a well-prepared device can be cycled more than twenty times without obvious performance decay.
Capacity and performance
The capacity of the device has been investigated by CV method. As shown in Figure 3C, the concentration dependence of peak current fits to a single-site occupation isotherm, suggesting that the closed SAM could be considered as a nanocontainer (13). The number of Fe[(CN)6]3− anions trapped in the closed nanocontainer has been estimated from experimental results (Figure 3) as about 100 per i-motif DNA in the case of 10-bp poly-dA spacer length (for detailed discussion please see Supplementary Data). Several other electroactive small molecules, including phenothiazinium dye (methylene blue), metal derivative ([Co(CN)6]3−) and organic reagent (p-benzoquinone), were used as object molecules to further examine the molecular encaging effect (see Supplementary Data, Figure S3). Results similar to [Fe(CN)6]3− anions have been obtained, which demonstrate that the nanocontainer is non-specific with respect to the object molecules, i.e. the molecular encaging effect is a generic property of the device.
We also investigated the effect of the length and the base content of the ssDNA spacer on the performance of the encaging effect. First, the base content of the ssDNA spacer is found to be irrelevant to the performance of the device. Second, the device capability is positively correlated to the spacer length in the range of 10–30 bp, which indicates that the performance is maintained in this range; when the spacer length is longer than 35 bp, the performance decreases dramatically, as the device shows incapability of encaging object molecules (see Supplementary Data, Figure S4). We propose that the DNA nanocompartment structure breaks down when very long ssDNA spacer is applied (with the length of i-motif fixed), where the conformational entropy of very long ssDNA spacers (39,40) may overwhelm the lateral interaction energy between neighboring i-motifs (24–27), preventing the i-motifs from packing into a regular membrane.
Accelerating with alternating electric field
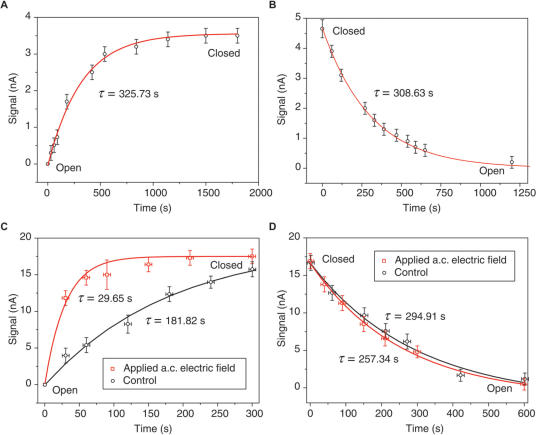
The kinetics of the device switching has been studied by tracking the stoichiometry of the encaged reporters in the compartment during the device switching. As shown in Figure 4, typical relaxation time constants of the device opening and closing were measured to be ∼300 and ∼330 s respectively, which are much slower than folding and unfolding of i-motif structure in solution phase (14,41). In this study, alternating electric field was used to modulate molecular cooperative motion during the device switching, which provides a scalable means to accelerate the device response (12,42–45). As shown in Figure 4C, appropriate alternating electric field (frequency ∼1 kHz, amplitude 0.2 V, initial potential 0.3 V, also see Supplementary Data) can remarkably shorten the closing time from 326 to 30 s. However, the same electric field did not prominently accelerate the opening kinetics (Figure 4D). In contrast, direct electric fields cannot result in any acceleration effect (data not shown).
Figure 4.
Accelerating device switching by alternating electric field. (A) Closing of the device when switching pH from 8 to 4.5 at t = 0. (B) Opening of the device when switching pH from 4.5 to 8 at t = 0. (C) Closing of the device under alternating (a.c.) electric field. (D) Opening of the device under a.c. electric field. Control in (C) and (D) is based on the same experimental setup only in the absence of a.c. electric field. Redox reporter [Fe(CN)6]3− used in the measurement is 4 mM in (A) and (B), and 40 mM in (C) and (D). Solid lines fitting the data are simulated from model y = A(1 − exp(−t/τ)) in (A) and (C), model y = Aexp(−t/τ) in (B) and (D), where τ is the relaxation time, and A is a constant parameter.
In principle, in all possible i-motif structures (intramolecular, dimeric and tetrameric), the intercalation of C · C+ base pairs between two parallel-stranded duplexes takes place in an antiparallel fashion, which means that each two neighboring strands within i-motif structure are in opposite directions (15–20). In our design, all DNA chains are immobilized on gold surface via thio-gold bonds at 5′ end; this design has ruled out the possibility of i-motif formed by four parallel ssDNA strands. Even though intermolecular i-motif structures are topologically possible by appropriate looping, the steric limitation made by the immobilization of each DNA chain may make the intermolecular i-motif structures imperfect, which contain mismatches and hanging single strands. The conformational entropy contributed by these imperfects will make them less stable than intramolecular i-motif structures (39,40). Based on these discussions, we may propose that relatively high surface coverage of DNA motifs can cause entanglement between neighboring molecules via intermolecular C · C+ base-pair formation, making the device to be trapped in the metastable states during the process of refolding, and dramatically delaying the switching process (41,46). Our recent experimental study on the detailed i-motif folding pathway and dynamics in the dense SAM suggest that the intermolecular metastable i-motif structures take place in the middle stage of the folding process and are on-pathway intermediates that disappear at the completeness of the folding procedure (unpublished data), thermodynamically consistent with the above arguments. The external alternating electric field applied may promote intramolecular diffusion (12,42–45), which could mitigate the entanglements by driving the device out of the metastable states, thus speeding up the intramolecular C · C+ base-pair matching during DNA folding. However, the opening of the device is a process of breaking the hydrogen bonds among intramolecular C · C+ base pairs, where no entanglement status is involved, so that the same electric field has nearly no effect on the opening kinetics. Detailed underlying mechanism for this phenomenon is under further theoretical investigation and will appear elsewhere.
To validate that the device is based on the conformational transition of i-motif structures, pH and temperature dependence of this device has been studied. The half transition of this device at pH 6.2 was in close agreement with the reported transition of i-motif structure in bulk phase at pH 6.3 (see Supplementary Data, Figure S5) (41,46). The melting profile of the device at pH 4.5 was also found to be well consistent to the i-motif structure in bulk solution at pH 4.5 (see Supplementary Data, Figure S5). These results indicate that enough spacing between immobilized DNA molecules was established and possible entanglements of neighboring molecules during their collective folding did not seriously demolish the performance of the device (21,22). Therefore, good cooperation between individual DNA molecules has been achieved in this system, providing an understanding to the feasibility of high closing rates of this device under alternating electric field.
In summary, we demonstrate how DNA nanocontainer devices emulating mechanical hands, which require high degree of molecular cooperation, obtain rapid switching without suffering ‘molecular crowding consequence’ by the use of alternating electric field. In contrast to the previous systems whose switching relies upon intermolecular hybridization, the DNA nanocontainers can switch without the aid of complementary strands, which substantially simplifies the system architecture. According to its functional characters, we believe that this DNA nanocontainer could be reconstituted on any substrate that is chemically available to DNA immobilization with appropriate packing density (10,13,44). It is promising to enable applications like controlled drug release via DNA molecular machinery. Moreover, it is possible to further modify the ssDNA spacer's backbone with appropriate chemical groups or design its sequences which may selectively and reversibly bind object molecules to introduce selectivity on targets. In addition, when redesigning the motif sequence to include DNA probe, the device might also be used to nucleic acids analysis and biosensors (47).
SUPPLEMENTARY DATA
Supplementary Data is available at NAR online.
ACKNOWLEDGEMENTS
The authors thank Prof. Dong Han, Mr Long Chen and Mrs Jianhua Wang for technical help in atomic force microscopy experiments, Mrs Ke Wu for help in gold substrate preparation. This work is partly funded by Science 100 Program of CAS, NSFC under grant No. 20573027, 90406024 and MOST 973 project under grant No. 2005CB724701 and 2006CB705601. Funding to pay the Open Access publication charge was provided by MOST 973 project.
Conflict of interest statement. None declared.
REFERENCES
- 1.Balzani V, Credi A, Venturi M. Molecular Devices and Machines – A Journey into Nano World. Weinheim, Germany: Wiley-VCH; 2003. [Google Scholar]
- 2.Seeman NC. DNA in a material world. Nature. 2003;421:427–431. doi: 10.1038/nature01406. [DOI] [PubMed] [Google Scholar]
- 3.Mao C, Sun W, Shen Z, Seeman NC. A nanomechanical device based on the B-Z transition of DNA. Nature. 1999;397:144–146. doi: 10.1038/16437. [DOI] [PubMed] [Google Scholar]
- 4.Yurke B, Turberfield AJ, Mills AP, Jr., Neumann JL. A DNA-fuelled molecular machine made of DNA. Nature. 2000;406:605–608. doi: 10.1038/35020524. [DOI] [PubMed] [Google Scholar]
- 5.Yan H, Zhang X, Shen Z, Seeman NC. A robust DNA mechanical device controlled by hybridization topology. Nature. 2002;415:62–65. doi: 10.1038/415062a. [DOI] [PubMed] [Google Scholar]
- 6.Kelly TR. Molecular motor: synthetic DNA-based walkers inspired by kinesin. Angew. Chem. Int. Ed. 2005;44:4124–4127. doi: 10.1002/anie.200500568. [DOI] [PubMed] [Google Scholar]
- 7.Alberti P, Mergny JL. DNA duplex-quadruplex exchange as the basis for a nanomolecular machine. Proc. Natl. Acad. Sci. U.S.A. 2003;100:1569–1573. doi: 10.1073/pnas.0335459100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y, Wang M, Mao C. A DNA nanomachine based on a duplex-triplex transition. Angew. Chem. Int. Ed. 2004;43:3554–3557. doi: 10.1002/anie.200460789. [DOI] [PubMed] [Google Scholar]
- 9.Chee M, Yang R, Hubbel E, Berno AX, Huang C, Stern D, Winkler J, Lockhart DJ, Morris MS, et al. Accessing genetic information with high-density DNA arrays. Science. 1996;274:610–614. doi: 10.1126/science.274.5287.610. [DOI] [PubMed] [Google Scholar]
- 10.Liu Q, Wang L, Frutos AG, Condon AE, Corn RM, Smith LM. DNA computing on surfaces. Nature. 2000;403:175–179. doi: 10.1038/35003155. [DOI] [PubMed] [Google Scholar]
- 11.Deng Z, Mao C. Molecular lithography with DNA nanostructure. Angew. Chem. Int. Ed. 2004;43:4068–4070. doi: 10.1002/anie.200460257. [DOI] [PubMed] [Google Scholar]
- 12.Lahann J, Mitragotri S, Tran TN, Kaido H, Sundaram J, Choi IS, Hoffer S, Somorjai GA, Langer R. A reversibly switching surface. Science. 2003;299:371–374. doi: 10.1126/science.1078933. [DOI] [PubMed] [Google Scholar]
- 13.Mao Y, Luo C, Deng W, Jin G, Yu X, Zhang Z, Ouyang Q, Chen R, Yu D. Reversibly switchable DNA nanocompartment on surfaces. Nucleic Acids Res. 2004;32:e144. doi: 10.1093/nar/gnh145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu D, Balasubramanian S. A proton-fuelled DNA nanomachine. Angew. Chem. Int. Ed. 2003;42:5734–5736. doi: 10.1002/anie.200352402. [DOI] [PubMed] [Google Scholar]
- 15.Gehring K, Leroy J-L, Gueron M. A tetrameric DNA structure with protonated cytosine-cytosine base pairs. Nature. 1993;363:561–565. doi: 10.1038/363561a0. [DOI] [PubMed] [Google Scholar]
- 16.Gueron M, Leroy JL. The i-motif in nucleic acids. Curr. Opin. Struct. Biol. 2000;10:326–331. doi: 10.1016/s0959-440x(00)00091-9. [DOI] [PubMed] [Google Scholar]
- 17.Kang CH, Berger I, Lockshin C, Ratliff R, Moyzis R, Rich A. Crystal structure of intercalated four-stranded d(C3T) at 1.4 Å resolution. Proc. Natl. Acad. Sci. U.S.A. 1994;91:11636–11640. doi: 10.1073/pnas.91.24.11636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berger I, Egli M, Rich A. Inter-strand C-H · O hydrogen bonds stabilizing four-stranded intercalated molecules: stereoelectronic effects of O4′ in cytosine-rich DNA. Proc. Natl. Acad. Sci. U.S.A. 1996;93:12116–12121. doi: 10.1073/pnas.93.22.12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mergny JL, Lacroix L. Kinetics and thermodynamics of i-DNA formation: phosphodiester versus modified oligodeoxynucleotides. Nucleic Acids Res. 1998;26:4797–4803. doi: 10.1093/nar/26.21.4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phan AT, Leroy JL. Intramolecular i-motif structures of telomeric DNA. J. Biomol. Struct. Dyn. 2000;17:377–383. doi: 10.1080/07391102.2000.10506628. [DOI] [PubMed] [Google Scholar]
- 21.Liu D, Bruckbauer A, Abell C, Balasubramanian S, Kang DJ, Klenerman D, Zhou D. A reversible pH-driven DNA nanoswitch array. J. Am. Chem. Soc. 2006;128:2067–2071. doi: 10.1021/ja0568300. [DOI] [PubMed] [Google Scholar]
- 22.Shu W, Liu D, Watari M, Riener CK, Strunz T, Welland ME, Balasubramanian S, McKendry RA. DNA molecular motor driven micromechanical cantilever arrays. J. Am. Chem. Soc. 2005;127:17054–17060. doi: 10.1021/ja0554514. [DOI] [PubMed] [Google Scholar]
- 23.Herne TM, Tarlov MJ. Characterization of DNA probes immobilized on gold surfaces. J. Am. Chem. Soc. 1997;119:8916–8920. [Google Scholar]
- 24.Rau DC, Lee B, Parsegian VA. Measurement of the repulsive force between polyelectrolyte molecules in ionic solution: hydration forces between parallel DNA double helices. Proc. Natl. Acad. Sci. U.S.A. 1984;81:2621–2625. doi: 10.1073/pnas.81.9.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strey HH, Wang J, Podgornik R, Rupprecht A, Yu L, Parsegian VA, Sirota EB. Refusing to twist: demonstration of a line hexatic phase in DNA liquid crystals. Phys. Rev. Lett. 2000;84:3105–3108. doi: 10.1103/PhysRevLett.84.3105. [DOI] [PubMed] [Google Scholar]
- 26.Kornyshev AA, Leikin S. Electrostatic interaction between helical macromolecules in dense aggregates: an impetus for DNA poly- and meso-morphism. Proc. Natl. Acad. Sci. U.S.A. 1998;95:13579–13584. doi: 10.1073/pnas.95.23.13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kornyshev AA, Leikin S. Electrostatic zipper motif for DNA aggregation. Phys. Rev. Lett. 1999;82:4138–4141. [Google Scholar]
- 28.Kelley SO, Barton JK, Jackson NM, Hill MG. Electrochemistry of methylene blue bound to a DNA-modified electrode. Bioconjugate Chem. 1997;8:31–37. doi: 10.1021/bc960070o. [DOI] [PubMed] [Google Scholar]
- 29.Boon EM, Ceres DM, Drummond TG, Hill MG, Barton JK. Mutation detection by electrocatalysis at DNA-modified electrodes. Nature Biotechnol. 2000;18:1096–1100. doi: 10.1038/80301. [DOI] [PubMed] [Google Scholar]
- 30.Aoki H, Buhlmann P, Umezawa Y. Electrochemical detection of a one-base mismatch in an oligonucleotide using ion-channel sensors with self-assembled PNA monolayers. Electroanalysis. 2000;12:1272–1276. [Google Scholar]
- 31.Kelley SO, Barton JK, Jackson NM, McPherson LD, Potter AB, Spain EM, Allen MJ, Hill MG. Orienting DNA helices on gold using applied electric fields. Langmuir. 1998;14:6781–6784. [Google Scholar]
- 32.Binnig G, Quate CF, Gerber C. Atomic force microscope. Phys. Rev. Lett. 1986;56:930–933. doi: 10.1103/PhysRevLett.56.930. [DOI] [PubMed] [Google Scholar]
- 33.Han W, Lindsay SM, Jing T. A magnetically driven oscillating probe microscope for operation in liquids. Appl. Phys. Lett. 1996;69:4111–4113. [Google Scholar]
- 34.Bard AJ, Fulkner LR. Electrochemical Methods. 2nd. New York: John Wiley & Sons; 1980. [Google Scholar]
- 35.Berova N, Nakanishi K, Woody RW. Circular Dichroism: Principles and Application. 2nd. New York: John Wiley & Sons; 2000. [Google Scholar]
- 36.Mao Y, Luo C, Ouyang Q. Studies of temperature-dependent electronic transduction on DNA hairpin loop sensor. Nucleic Acids Res. 2003;31:e108. doi: 10.1093/nar/gng108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fan C, Plaxco KW, Heeger AJ. Electrochemical interrogation of conformational changes as a reagentless method for the sequence-specific detection of DNA. Proc. Natl. Acad. Sci. U.S.A. 2003;100:9134–9137. doi: 10.1073/pnas.1633515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Drummond TG, Hill MG, Barton JK. Electron transfer rates in DNA films as a function of tether length. J. Am. Chem. Soc. 2004;126:15010–15011. doi: 10.1021/ja044910i. [DOI] [PubMed] [Google Scholar]
- 39.de Gennes PG. Scaling Concepts in Polymer Physics. Ithaca and London: Cornell University Press; 1979. [Google Scholar]
- 40.Pande VS, Grosberg AY, Tanaka T. Heteropolymer freezing and design: towards physical models of protein folding. Rev. Mod. Phys. 2000;72:259–314. [Google Scholar]
- 41.Zhao Y, Zeng ZX, Kan ZY, Hao YH, Tan Z. The folding and unfolding kinetics of the i-motif structure formed by the C-rich strand of human telomere DNA. Chembiochem. 2005;6:1957–1960. doi: 10.1002/cbic.200500175. [DOI] [PubMed] [Google Scholar]
- 42.Bermudez V, Capron N, Gase T, Gatti FG, Kajzar F, Leigh DA, Zerbetto F, Zhang S. Influencing intramolecular motion with an alternating electric field. Nature. 2000;406:608–611. doi: 10.1038/35020531. [DOI] [PubMed] [Google Scholar]
- 43.Rant U, Arinaga K, Fujita S, Yokoyama N, Abstreiter G, Tornow M. Dynamic electric switching of DNA layers on a metal surface. Nano Lett. 2004;4:2441–2445. [Google Scholar]
- 44.Wei F, Chen C, Zhai L, Zhang N, Zhao XS. Recognition of single nucleotide polymorphisms using scanning potential hairpin denaturation. J. Am. Chem. Soc. 2004;127:5306–5307. doi: 10.1021/ja043661v. [DOI] [PubMed] [Google Scholar]
- 45.White SS, Ying L, Balasubramanian S, Klenerman D. Individual molecules of dye-labeled DNA act as a reversible two-color switch upon application of an electric field. Angew. Chem. Int. Ed. 2004;43:5926–5930. doi: 10.1002/anie.200460323. [DOI] [PubMed] [Google Scholar]
- 46.Leroy JL, Gehring K, Kettani A, Gueron M. Acid multimers of oligodeoxycytidine strands: stoichiometry, base-pair characterization, and proton exchange properties. Biochemistry. 1993;32:6019–6031. doi: 10.1021/bi00074a013. [DOI] [PubMed] [Google Scholar]
- 47.Bourdoncle A, Torres AE, Gosse C, Lacroix L, Vekhoff P, Le Saux T, Jullien L, Mergny J-L. Quadruplex-based molecular beacons as tunable DNA probes. J. Am. Chem. Soc. 2006;128:11094–11105. doi: 10.1021/ja0608040. [DOI] [PubMed] [Google Scholar]