Abstract
It has been hypothesized that a replication associated repair pathway operates on base damage and single strand breaks (SSB) at replication forks. In this study, we present the isolation from the nuclei of human cycling cells of a multiprotein complex containing most of the essential components of base excision repair (BER)/SSBR, including APE1, UNG2, XRCC1 and POLβ, DNA PK, replicative POLα, δ and ɛ, DNA ligase 1 and cell cycle regulatory protein cyclin A. Co-immunoprecipitation revealed that in this complex DNA repair proteins are physically associated to cyclin A and to DNA replication proteins including MCM7. This complex is endowed with DNA polymerase and protein kinase activity and is able to perform BER of uracil and AP sites. This finding suggests that a preassembled DNA repair machinery is constitutively active in cycling cells and is ready to be recruited at base damage and breaks occurring at replication forks.
INTRODUCTION
Both DNA replication and repair are performed by multiprotein assemblies and share common features. DNA repair must be coordinated with DNA replication in order to avoid fixation of DNA damage into heritable mutations. A fundamental level of cross-talk between DNA replication and DNA repair is ensured by the fact that the enzymes active in replicating DNA during S phase are also needed to synthesize new stretches of DNA during various types of repair including nucleotide excision repair (NER), mismatch repair (MMR), single-strand DNA break repair (SSR) and base excision repair (BER). An additional level of coordination is needed to achieve integration of the DNA repair and DNA replication protein networks within the highly sophisticated cell cycle regulatory machinery. Indeed, many components of the DNA replication machinery associate with other factors such as cyclins/Cdks in dynamic multiprotein complexes that regulate cell cycle progression. The so-called cyclin-dependent protein kinase (Cdk)-driven ‘replication switch’ model predicts that cyclin/Cdk complexes function both to activate initiation complexes assembled at the origins and to inhibit further complex assembly during S-phase, thus preventing unscheduled re-replication (reviewed in (1)).
BER counteracts the cytotoxic and mutagenic effects of most endogenously produced DNA damage. Its role must be critical when this type of damage is produced or persists at replication forks. In BER, specific DNA glycosylases are responsible for base removal followed by formation of a single strand break (SSB) by an AP endonuclease. SSB that arise directly from sugar damage usually possess non-conventional termini that need further processing to allow completion of SSB repair (SSBR). In both BER and SSBR the resulting gap is filled by DNA polymerase (POL)β (short-patch repair) or by POLβ/δ/ɛ (long-patch repair). Additional players in long-patch repair are replication factor (RF) C, proliferating cell nuclear antigen (PCNA) and flap endonuclease-1 (FEN1). The final ligation step is then operated by the XRCC1/DNA ligase IIIα (LIG3) complex or DNA ligase I (LIG1) in the short- and long-patch pathways, respectively (reviewed in (2)). Interestingly, PCNA, that is involved in the dynamic assembly and disassembly of the DNA replication machinery, has been shown to interact with several BER/SSBR proteins such as adenine DNA glycosylase (MYH) (3), uracil DNA glycosylase UNG2, 5’ AP endonuclease APE1, XRCC1, POLβ, POLδ, FEN1and LIG1 (reviewed in (4)). Based on these findings, it has been hypothesized that there is a BER/SSBR pathway that is coordinated with DNA replication and employs proteins like PCNA, FEN1, LIG1 and POLδ/ɛ that are in common with the replication machinery (5–7).
A growing body of evidence indicates that BER/SSBR proteins are regulated by post-translational modification and make physical interactions with components of other DNA transaction pathways (reviewed in (8)). One of the most compelling evidence of BER regulation via post-translation modifications is the phosphorylation in vitro and in vivo by Casein Kinase 2 of one of the central players of BER, the scaffold protein XRCC1 (9). This phosphorylated form promotes a more efficient SSBR. Another example is the homeostatic regulation of BER by a p53-induced phosphatase, PPM1D, that suppresses BER by dephosphorylation of the nuclear isoform of uracil DNA glycosylase, UNG2 (10).
It should be taken into account that most of our knowledge about BER has been derived from studies carried out in vitro by using mammalian cell extracts or purified proteins and synthetic DNA molecules containing single lesions. However, an open question is how cross-talk between DNA replication and DNA repair machineries is achieved at the cellular and molecular level. The analysis of the protein–protein interactions within BER proteins and between BER and other pathways occurring in the cell is a prerequisite to better understand the regulation of the DNA repair processes in the context of the cell cycle.
In this study, we present the isolation from the nuclei of human cycling cells of a complex containing most of the essential components of BER physically associated to cyclin A and to DNA replication proteins. This complex is endowed with a protein kinase activity and is able to perform BER of uracil residues as well as of apurinic/apyrimidinic (AP) sites via both short- and long-patch BER. The fact that this complex was isolated from human cells in the absence of any DNA-damaging treatment, suggests that a preassembled BER machinery is constitutively active in the cycling cells and is ready to be recruited to the site of damage likely to occur at the replication forks.
MATERIALS AND METHODS
Chemicals
Restriction enzymes and T4 polynucleotide kinase were from New England Biolabs. T4 DNA polymerase holoenzyme, T4 single-stranded DNA binding protein, T4 DNA ligase and dNTPs were purchased from Roche Molecular Biochemicals. Both oligodeoxyribonucleotides containing a single uracil: 5′-GATCCTCTAGAGUCGACCTGCA-3′ (for preparation of circular duplex DNA substrate) and 5’-GATCCTCTAGAGUCGACCTGCAGGCATGCA-3’ (for incision assay), were synthesized by MWG-Biotech AG. Ugi was kindly provided by S.E. Bennett (Oregon State University, Corvallis, OR, USA). [γ-32P] ATP was from GE Healthcare and [α-32P] dCTP and [α-32P]dTTP were from Perkin Elmer. Nitrocellulose membranes (HybondECL) were from GE Healthcare. GF/C filters were from Schleicher & Schuell. Olomucine and aphidicolin were from Sigma. All other reagents were purchased from BioRad, Sigma, Fluka, Gibco and BDH.
Cells and media
HeLa cells were grown in DMEM supplemented with 10% FCS, 50 μg/ml gentamycin and 2 mM l-Glutamine, at 37°C and 5% CO2. Cells were pelleted by centrifugation and stored in aliquots at −80°C until used.
SV40 transformed wild-type and POLβ-null mouse embryonic fibroblasts (MEFs) (a gift from Dr S.H. Wilson, NIEHS, Research Triangle Park, NC) were cultured in DMEM supplemented with Glutamax-1, 10% FCS, 100 U/ml penicillin, 100 μg/ml streptomycin and 80 μg/ml hygromycin at 34°C and 10% CO2.
Protein purification
The purification was performed essentially as described (11), with the following modifications. After the size exclusion chromatography, the pooled fractions were loaded onto a hydroxyapatite column, equilibrated in Buffer F (25 mM Bis-Tris pH 6.6, 50 mM NaCl, 0.5 mM ATP, 1 mM DTT, 5% glycerol, 1 mM PMSF and protease inhibitors). The column was eluted with a linear gradient from 0.1 to 1 M phosphate buffer pH 6.6. The active fractions were pooled, diluted in Buffer F to bring the potassium phosphate below 0.1 M and then loaded onto a Mono S column (Pharmacia) equilibrated in Buffer F. The column was eluted with a linear gradient from 0.1 to 1 M NaCl.
Immunoprecipitation experiments
The Mono S fraction 15 (5 μg) was adjusted to 10 mM Tris-HCl pH 7.4, 50 mM NaCl, 2.5 mM MgCl2 and protease inhibitors and then incubated for 1 h at 4°C with protein A-sepharose beads (BioRad) equilibrated in the same buffer and previously coupled to the appropriate antibodies, as indicated in the figure legends. After centrifugation the pellet was washed with equilibration buffer containing 80 mM NaCl and the immunoprecipitated material was analysed by western blot.
Wild-type or POLβ-null MEFs (4 × 107) were lysed in the presence of 10 mM Tris-HCl pH 7.4, 150 mM NaCl, 2.5 mM MgCl2 and protease inhibitors for 30 min on ice and then homogenized by dounce. After centrifugation at 20 000×g at 4°C, the supernatant (crude extract) was diluted two times in the same buffer without NaCl and incubated for 1 h at 4°C with protein A-sepharose beads (BioRad) equilibrated in the same buffer and previously coupled to the appropriate antibodies, as indicated in the figure legends. After centrifugation the pellet was washed with the same buffer containing 80 mM NaCl and the immunoprecipitated material was analysed by western blot.
Native gel electrophoresis
Mono S fraction 15 (5–10 μg) was resolved on a 5% native polyacrylamide gel in the absence of SDS and β-mercaptoethanol in sample loading and running buffers, at 4°C. Samples were transferred to nitrocellulose membrane at 12 V for 12–16 h at 4°C and subjected to western blot analysis.
Gel filtration
Mono S fraction 15 (20 μg) was applied to a Superdex200 10/300 GL gel filtration column equilibrated with 10 mM Tris-HCl pH 7.4, 80 mM NaCl, 2.5 mM MgCl2 and protease inhibitors. The column was eluted with the same buffer and the eluted fraction analysed by dot blot immunoassay and western blot. Molecular weight markers used for column calibration were ferritin (440 kDa), catalase (232 kDa), lactate dehydrogenase (140 kDa) and BSA (66 kDa).
Protein kinase assays
The assay was carried out in final volume of 10 μl containing 0.15 μg of Mono S fraction 14, 50 mM Tris-HCl pH 7.5, 10 mM MgCl2, 0.5 μM [γ-32P] ATP (3000 Ci/mmol). Reactions were supplemented, when indicated, with 0.4 mg/ml histone H1, 0.5 mM Olomucine or recombinant p21cip/WAF. Samples were incubated for 20 min at 37°C and loaded onto a 10% SDS-PAGE. Gels were quantified by PhosphorImager (Typhoon, GE Healthcare).
Antibodies
Anti-FEN1, anti-UDG, anti-POLα, POLδ, POLɛ, anti-cyclin B and E, anti-MCM7 and anti-APE1 were from Santa-Cruz Biotechnology. Anti-cyclin A and anti-phosphoSer/Thr were from Sigma. Anti-XRCC1 was from Trevigen. Anti-DNA PK was from Calbiochem. Anti-POLβ was kindly provided by S.H. Wilson (NIEHS, Research Triangle Park, NC, USA) and anti-POLι by T.A. Kunkel (NIEHS, Research Triangle Park, NC, USA). Anti-LIG1 was a kind gift of A. Montecucco (IGM-CNR, Pavia, Italy) and anti-POLλ was kindly provided by U. Hübscher (University of Zürich-Irchel, Switzerland).
Preparation and characterization of circular duplex DNA substrates
Closed circular DNA molecules containing a single lesion were produced as described previously (12) by priming single-stranded (+) pGem-3Zf(+) DNA (Promega) with the oligonucleotides containing the lesion of interest. In vitro DNA synthesis was performed by using T4 DNA polymerase holoenzyme, single-stranded DNA binding protein, dNTPs and T4 DNA ligase. Closed circular DNA duplex molecules were purified by cesium chloride equilibrium centrifugation. Plasmid DNA molecules containing a single uracil residue were digested with E. coli uracil DNA glycosylase (Trevigen) to produce abasic sites.
Repair assay
Repair reactions were carried out as described in (12). Briefly, reaction mixtures (50 μl) contained 40 mM HEPES/KOH (pH 7.9), 75 mM KCl, 5 mM MgCl2, 0.5 mM dithiothreitol, 50 μM of each dNTP, 2 μCi of [α-32] P dCTP or [α−32] P dTTP as indicated, 2 mM ATP, 40 mM phosphocreatine, 2.5 μg of creatine phosphokinase (type I, Sigma), 3.4% glycerol, 18 μg of bovine serum albumin and 5 μl of the isolated multiprotein complex (Mono S fraction 14, 1 μg of total proteins) were incubated 1 h at 30°C. Depending on the labelled dNTP used in the repair mix, the concentration of the corresponding cold dNTP was decreased to 5 μM. When the assay was performed in the presence of aphidicolin, a concentration of 300 μM was used. The repair products were digested with appropriate restriction enzymes, resolved on a denaturing 20% PAGE and quantified by electronic autoradiography (Instant Imager, Packard).
Incision assay
(γ-32P)-labelled 30-mer oligonucleotides containing a single uracil were annealed with the appropriate complementary oligonucleotide to obtain U/A-containing duplexes. Following 1 h incubation at 37°C with the complex (Mono S fraction 14, 1 μg of total proteins) in 25 mM Tris-HCl pH 7.6, 1 mM EDTA, 50 mM NaCl, the incision products were incubated for 10 min at 95°C in the loading buffer (formammide 95%, EDTA 20 mM, bromophenol blue 0.05%, xylene cyanol FF 0.05%), resolved on a denaturing 20% PAGE and quantified by electronic autoradiography (Instant Imager, Packard).
RESULTS
Isolation of a multiprotein complex from HeLa cells containing DNA replication proteins, cell cycle regulatory factors and base excision repair proteins
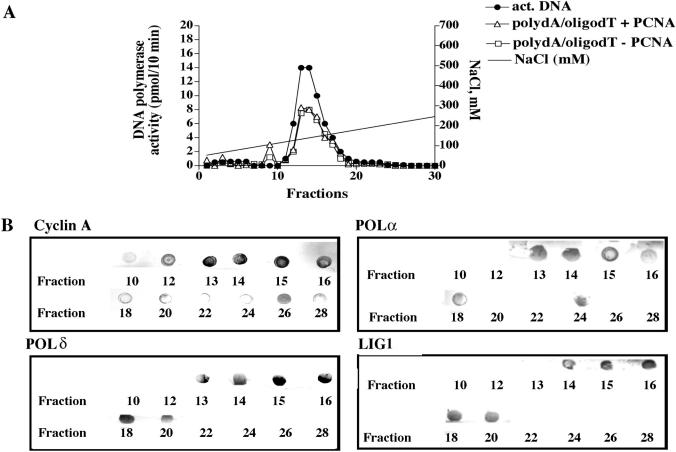
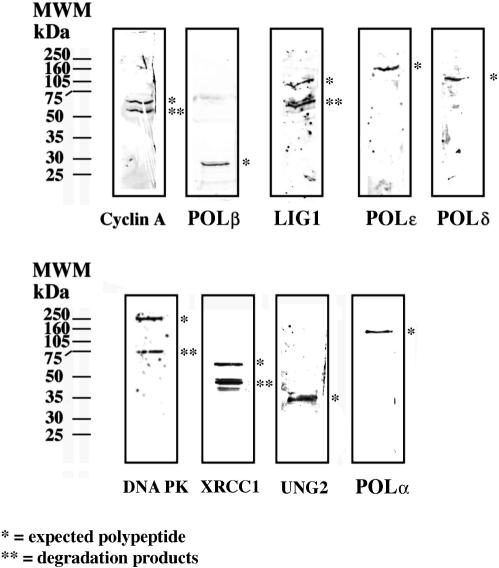
HeLa cell nuclei were isolated and fractionated according to a procedure already established in our laboratory to isolate multiprotein complexes (11). During the fractionation procedure, DNA polymerase activity was measured using three different assays, one specific for POLα (activated DNA) another one for POLδ and POLɛ (poly(dA)/oligo(dT) in the presence of PCNA) and a third one for POLα and POLɛ (poly(dA)/oligo(dT) in the absence of PCNA). Co-purification of other DNA replication and cell cycle regulatory factors was monitored by a dot blot immunoassay. Figure 1 shows the elution profile of the last purification step (Mono S column). A peak of DNA polymerase activity was observed between fraction 10 and 20 with both assays (Figure 1A). Further characterization revealed the presence of 3′–5′ exonuclease activity, DNA helicase activity and histone H1 phosphorylation activity (data not shown and Figure 2). The presence of POLα and POLδ and, in addition, LIG1 and cyclin A in the same fractions was confirmed by dot blot (Figure 1B). A slightly different elution profile was revealed at the tails of the peak (fractions 10–12 and 20–24, Figure 1) between cyclin A and the other proteins. This could be either due to different isoelectric points and/or different sensitivities of the antibodies used. The central peak fractions (14 and 15) were used for all the subsequent experiments. Next, the isolated complex (Mono S fraction 14) was analysed with a panel of poly- and monoclonal antibodies directed against a number of DNA replication and repair proteins. As shown in Figure 3, western blot analysis revealed the presence in the same fraction of four different DNA polymerases (POLα, POLβ, POLδ and POLɛ) and, in addition, LIG1, UNG2, XRCC1, DNA-dependent protein kinase (DNA PK) and cyclin A. We could not detect other factors such as cyclin B, cyclin E, PCNA, POLλ or POLι (data not shown). Analysis of the composition of the Mono S fraction 15 gave perfectly comparable results, indicating that these two peak fractions represent a homogeneous preparation (data not shown).
Figure 1.
Purification of the multiprotein DNA repair complex. (A) Elution profile of the last purification step (Mono S). POLα (black symbols), POLα/ɛ (open squares) and POLδ/ɛ (open triangles) activities coeluted in the same fractions. (B) Dot blot analysis of the Mono S fractions with specific antibodies showing coelution of cyclin A, POLα, POLδ and LIG1.
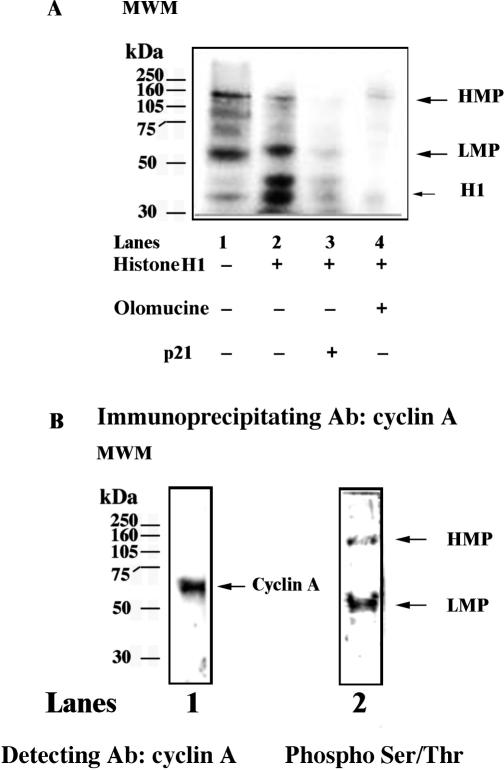
Figure 2.
Protein kinase activity of the multiprotein complex. (A) Phosphorylated polypeptides were revealed upon incubation of the Mono S fraction in the presence of [γ-32P] ATP alone (lane 1), or in combination with histone H1 (lane 2), histone plus p21 (lane 3) or histone plus Olomucine (lane 4). (B) The Mono S fraction was immunoprecipitated with anti-cyclin A antibodies and the immunoprecipitated material was probed with antibodies against cyclin A (lane 1) or anti-phosphoSer/Thr (lane 2).
Figure 3.
Western blot analysis of the Mono S fraction. The peak fraction 14 from Mono S was analysed by immunoblotting with different antibodies against DNA repair and replication proteins, as described in material and method section. A single asterisk indicates the expected polypeptide, whereas double asterisk mark indicates degradation products.
The DNA replication, DNA repair and cell cycle proteins of the isolated complex are physically associated
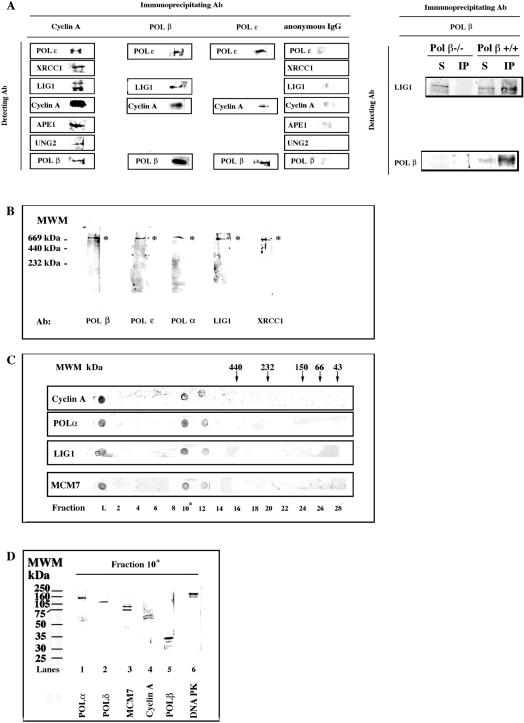
In order to test whether these proteins were physically associated, we used three different approaches: co-immunoprecipitation (co-IP), native gel electrophoresis and gel filtration. We tested all our antibodies for immunoprecipitation. The best results were obtained with anti-cyclin A monoclonal antibodies, which were then used for the analysis of the peak fraction 15. As shown in Figure 4A, anti-cyclin A antibodies were able to efficiently immunoprecipitate cyclin A from the Mono S peak fraction. In addition, several proteins were found to co-immunoprecipitate together with cyclin A: POLɛ, XRCC1, LIG1, APE1, UNG2 and POLβ. As controls, we performed the reciprocal co-IPs for some of these interactions and we found that LIG1, cyclin A and POLε were co-IPed together with POLβ by anti-POLβ antibodies, whereas POLβ and cyclin A were co-IPed together with POLɛ by anti-POLɛ antibodies. As an additional control, POLβ-null or wild-type cells (MEFs) were lysed and subjected to immunoprecipitation with anti-POLβ antibodies. Immunoprecipitated material was analysed with anti-POLβ and anti-LIG1 antibodies. As shown in Figure 4A, right panel, only in the POLβ wild-type cells LIG1 could be co-immunoprecipitated with POLβ, whereas no immunoprecipitated proteins were detected in POLβ-null cells. The Mono S fraction was analysed by native gel electrophoresis. As shown in Figure 4B, antibodies directed against POLβ, POLɛ, POLα, LIG1 and XRCC1, all reacted with a single band in the high molecular weight range (apparent Mr > 669 kDa). In order to further confirm the physical association of these proteins, the Mono S fraction 15 was subjected to gel filtration. Figure 4C shows the dot blot immunoassay of the eluted fractions, where it can be seen that POLα, LIG1 and cyclin A coeluted together in fractions 10–12, corresponding to an apparent molecular mass > 440kDa, as expected from a multiprotein complex. We tested, in addition, the presence of another DNA replication associated protein, MCM7, and we found that it was coeluting together with POLα, LIG1 and cyclin A, indicating its association to the complex. The gel filtration fraction 10 was further analysed by western blot. As shown in Figure 4D, POLα, POLβ, POLδ, LIG1, DNA PK, cyclinA and MCM7, were detected in the same fraction.
Figure 4.
Physical association of DNA replication and repair proteins. (A) Left panel: The Mono S fraction 15 was immunoprecipitated with antibodies against cyclin A, POLβ, POLɛ or anonymous IgGs. Right panel: POLβ null (−/−) or wild type (+/+) mouse embryonic fibroblasts were lysed and the extracts used for immunoprecipitation in the presence of anti-POLβ antibodies. The immunoprecipitated material was then immunoblotted with anti-POLβ and anti-LIG1 antibodies, as indicated. S, supernatant (1:10); IP, immunoprecipitated material. (B) The Mono S fraction 15 was subjected to native gel electrophoresis, followed by immunoblotting analysis with antibodies against POLα, POLβ, POLɛ, LIG1 and XRCC1. As marked by the asterisks, all the antibodies recognized the same high molecular weight band. (C) The Mono S fraction 15 was subjected to gel filtration. Eluted proteins were analysed by dot blot with antibodies against POLα, LIG1, Cyclin A and MCM7. Arrows indicate the corresponding elution points of the molecular weight markers. (D) The gel filtration fraction 10 was analysed by western blot with antibodies against POLα, POLβ, POLδ, DNA PK, cyclin A and MCM7.
All together, these results suggested that we have isolated a multiprotein complex containing DNA replication, DNA repair and cell cycle regulatory proteins.
The multiprotein complex is active in base excision repair
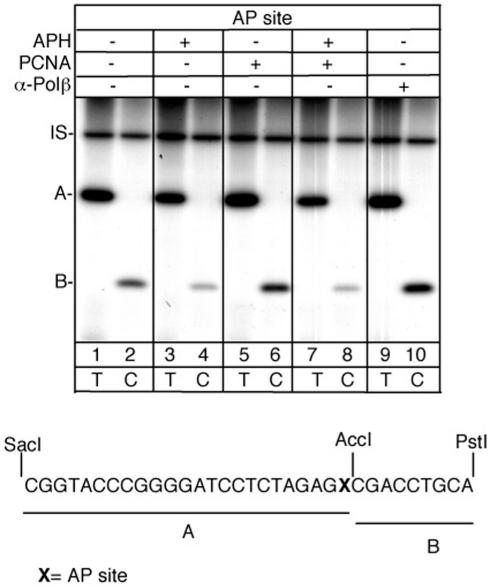
In order to verify the repair capacity of the replication complex an in vitro BER assay was performed by using, as substrate, a plasmid containing either an AP site or a single uracil residue. Following incubation of the plasmid with the multiprotein complex (Mono S fraction 14), the total DNA repair events (short- and long-patch repair events) were measured by [α-32P] dTTP incorporation in restriction fragment A, whereas incorporation of [α-32P] dCTP in fragment B corresponded exclusively to the long-patch repair events (see scheme, Figure 5 bottom). As shown in Figure 5 the AP site was repaired predominantly via short-patch pathway, with a fraction (25% of total) of long-patch repair events also occurring, which were largely aphidicolin-sensitive (lanes 2 and 4). Addition of PCNA increased incorporation in both A and B fragments (lanes 5 and 6) and this increase was abolished by aphidicolin (lanes 7 and 8), suggesting a role for the PCNA-dependent and aphidicolin sensitive enzymes POLδ/POLɛ in the repair process. Interestingly, inhibition of POLβ by specific antibodies, resulted in a switch from short- to long patch, as indicated by the increase of incorporation in fragment B (lanes 9 and 10).
Figure 5.
Mapping of the repair patches at AP site by the multiprotein complex. Top: autoradiograph of a denaturing polyacrylamide gel. Bottom: sequence of the restriction fragments A and B. Repair reactions were performed for 1 h in the presence of radiolabelled dTTP (lanes 1-3-5-7-9) or dCTP (lanes 2-4-6-8-10). Aphidicolin (APH), PCNA and α-POLβ were added as indicated. IS, internal standard.
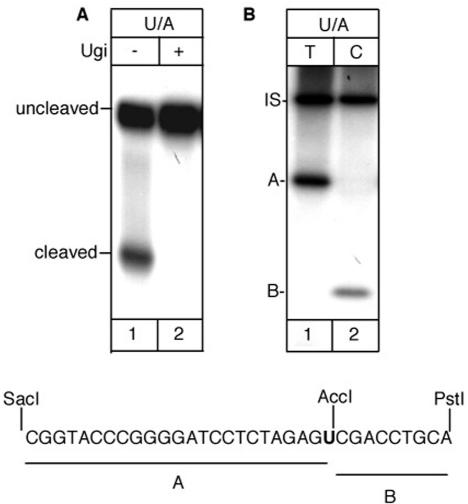
When similar experiments were performed with a uracil-containing oligonucleotide (Figure 6A), the isolated complex showed uracil excision activity (lane1). This cleavage activity was due to UNG2 as demonstrated by its specific inhibition by Ugi (lane 2) (13). The multiprotein complex performed BER of the uracil paired with adenine (Figure 6B) prevalently by short-patch pathway with 35% of the total repair events involving the replacement of more than one nucleotide (long- patch BER). The complex was unable to incise duplex oligonucleotides containing 8-oxo-7,8-dihydroguanine/A (8-OH-G/A) and 5-hydroxy-uracil (5-OHU/A) mismatches indicating the absence of MYH and NEIL1 activities (data not shown).
Figure 6.
Incision and repair activity at uracil by the multiprotein complex. (A) Incision assay in the absence (lane 1) or presence (lane 2) of the UNG2 inhibitor Ugi. (B) Repair assay in the presence of radiolabelled dTTP (lane 1) or dCTP (lane 2). Bottom: sequence of the restriction fragments A and B. IS, internal standard.
The multiprotein complex is endowed with a protein kinase activity and contains two major phosphorylated species
Since the multiprotein complex isolated here contained cyclin A, which is the regulatory subunit of cyclin-dependent protein kinases (cdks), we tested whether this complex displayed cdk activity. As a reference substrate we used histone H1. As shown in Figure 2A, lane 1, incubation of the purified complex (Mono S fraction 14) in the presence of [γ-32P] ATP resulted in the appearence of two major phosphorylated polypeptides, one with an apparent MW of 110–120 kDa (high molecular weight phosphopeptide, HMP) and one with an apparent MW of 50 kDa (low molecular weight phosphopeptide, LMP). Addition of histone H1 to the reaction resulted in the appearance of an additional product at the expected position for phosphorylated H1 (lane 2). When the reaction was complemented with either the kinase inhibitor Olomucine (lane 4) or the specific cdk inhibitor protein p21 (lane 3), all phosphorylated products were greatly reduced, suggesting that they were phosphorylated by a cyclin A/cdk complex. In order to prove the physical association of the HMP and LMP proteins with the complex, a co-immunoprecipitation experiment was performed with anti-cyclin A antibodies. As shown in Figure 2B, lane 1, cyclin A was successfully immunoprecipitated. When the same sample was probed with a cocktail of antiphosphoserine–threonine antibodies, two bands appeared, with apparent MW identical to the LMP and HMP proteins detected by the kinase assay (Figure 2B, lane 2). According to its apparent electrophoretic mobility, the LMP protein appears to be distinct from cyclin A (Figure 2B, compare lane 1 with lane 2). These results seem to indicate that two major phosphopeptides are physically associated to the isolated complex and are likely phosphorylated by cyclin A/cdk. The precise identity of these two proteins is under investigation. In summary, the multiprotein complex presented here contains an active cyclinA/cdk complex which is able to phosphorylate some components of the complex itself.
DISCUSSION
The mammalian genome is constantly subjected to chemical alterations (∼106 modifications per day) that have the potential to cause genome instability. Particular vulnerability exists during replication when attempted replication across a damaged template can lead to dangerous lesions such as double strand breaks. To counteract these threats, cells are provided with sophisticated systems that sense DNA damage and coordinate its repair. Of particular importance for the removal of damaged bases from the DNA is the BER system (2). Although BER can be reconstituted in vitro with a few essential components, numerous studies have revealed that physical and/or functional protein–protein interactions occur at virtually every step of the BER process. These interactions involve not only the classical BER proteins, but also proteins associated with other DNA transaction pathways. These interactions could play different roles including: (i) stabilizing the associated repair protein; (ii) recruiting specific partners to the damaged site for lesion repair; (iii) altering enzymatic function or activity; and (iv) coordinating BER with other pathways of DNA metabolism. Thus, physical contact between proteins does not always result in a change in the enzymatic activity of the proteins involved, and for this reason it might even go undetected by in vitro enzymatic studies.
Here, we demonstrated the existence in human cells of a preassembled multiprotein complex containing cyclin A, DNA replication proteins and BER/SSBR components. Also one component of DSB repair, DNA PK, is present in this complex. Interestingly, the DNA PK/Ku70/Ku80 heterotrimeric complex that contributes to the resolution of DSB has been recently shown to interact with XRCC1 and stimulates its phosphorylation (14). Since we isolated this complex from nuclei of proliferating human cells without prior treatment with DNA- damaging agents, it seems that the presence of this complex is independent of damaged DNA. However, we cannot exclude that the basal physiological level of DNA damage might be enough to trigger complex formation. The isolation from undamaged cells of multiprotein BER/SSBR complexes by different methods has been previously reported (15–18). In 1996, Prasad et al. (15) reported the isolation by a POLβ affinity method of a complex of a molecular mass of ∼180 kDa from bovine testis that contained POLβ, LIG1 and UNG2. Whitehouse et al. and Luo et al. demonstrated that XRCC1 protein is in complex with LIG3, PNK and POLβ (16,17). An alternative complex comprises XRCC1, LIG3 and aprataxin (16). Akbari et al. (18) isolated a repair complex (UNG2–ARC) that contains UNG2, APE1, POLβ, XRCC1, PCNA and LIG1 by using antibodies specific for the N-terminal non- catalytic domain of UNG2. More recently an interphase-specific XRCC1 complex has been described: it comprises PARP1, FEN1, POLδ and condensin, a factor well known to be essential in mitotic chromosome organization (19). Including this study, BER complexes have been isolated by six different methods indicating that these complexes are stable and DNA damage recognition by the DNA glycosylase is not required to trigger complex formation. In vivo large foci containing XRCC1, a key player of BER/SSBR, have been described both in undamaged (20) and irradiated (21–22) cells. As expected, there is significant overlapping between the BER components contained in the UNG2-ARC complex (18) and those identified in our complex. Our complex contains UNG2 as the only uracil DNA glycosylase, as shown by the complete inhibition of uracil cleavage in the presence of Ugi, as well as APE1, LIG1 and XRCC1. However, in addition to UNG2, APE1, LIG1, XRCC1, POLβ and POLδ, we identified two additional DNA polymerases, namely POLα and POLɛ POLδ and POLɛ have been already suggested to take part in the long-patch BER (reviewed in (2)). Accordingly, our isolated complex was able to perform both short- and long-patch BER at AP sites with the short patch BER as the predominant pathway. Addition of PCNA and/or inhibition of Polβ switched the equilibrium in favour of long-patch repair products, whereas addition of aphidicolin resulted in the opposite effect. This reflects an intrinsic ability of the complex to respond to exogeneous stimuli (such as the presence of PCNA and/or limiting POLβ activity). We did not find PCNA associated to our complex. It must be stressed that we used an extraction protocol which preserves the chromatin intact. We hypothesize that PCNA is not stably part of the complex, but rather could act as a recruiting factor for chromatin binding through physical interaction with one or more of the components of the complex.
Several BER proteins have been shown to undergo post-translational modifications. For example, FEN1, XRCC1, APE1 and LIG1 are phosphorylated (reviewed in (8)). Interestingly, in our complex we detected a protein kinase activity, which was inhibited by p21WAF/CIP and was able to phosphorylate endogeneous polypeptides, suggesting that phosphorylation can occur within the complex itself.
The fact that the major S-phase specific cyclin A, was physically associated to the complex together with the major replicative enzyme POLα and with the DNA replication protein MCM7 immediately suggests a possible link of the BER/SSBR pathway with the DNA replication process. Recent data indicated that the DSBR pathway is controlled by the cyclin A–cdk complex (23) suggesting a possible model of co-regulation of repair pathways during the cell cycle (24). A preassembled complex containing DNA replication, cell cycle and BER components specifically involved in endogenous DNA damage processing (i.e. UNG2 and APE1) might indicate the need of a repairosome for these lesions frequently encountered by the replication machinery. Indeed, UNG2 has been found associated to DNA replication proteins (25–27). The present data, however, does not allow to conclude that the isolated repairosome is specifically formed in S-phase or exists also in other phases of the cell cycle, such as G1 or G2. Analysis of the dynamics of the repairosome as a function of the cell cycle is currently underway in our laboratory. Interestingly, the DSB repair enzyme DNAPK is also present in our BER complex, along with XRCC1. XRCC1 has been shown to coordinate the assembly of SSBR and BER components at damaged sites and is essential for cells survival after SSB induction (reviewed in (28)). In addition, recent results suggest that XRCC1 phosphorylation by DNA PK in response to ionizing radiation, might trigger a signal cascade leading to NHEJ-dependent DSBR (14). Thus, XRCC1 appears to act as an early responder of DNA breaks at stalled replication forks. In light of these results, our findings raise the intriguing hypothesis that XRCC1 and DNA- PK might be stably associated to a repairosome, which is linked to the DNA replication fork through association with POLα and MCM7. Depending on the cellular context, cell cycle phase and type of damage, this repairosome will have the ability to quickly respond to a stalled replication fork, activating different DNA repair pathways.
ACKNOWLEDGEMENTS
Grant support: Associazione Italiana per la Ricerca sul Cancro (AIRC), Compagnia di S. Paolo (programma ‘Oncologia’, coordinator Guido Frosina), MIUR/FIRB (RBNE01RNN7), CARIPLO Foundation project ‘Oncogenetica e Proteomica della Replicazione’ (2003.1663/10.8441). Funding to pay the Open Access publication charge was provided by CARIPLO Foundation Project (2003.1663/10.8441).
Conflict of interest statement. None declared.
REFERENCES
- 1.Frouin I, Montecucco A, Spadari S, Maga G. DNA replication: a complex matter. EMBO Rep. 2003;4:666–670. doi: 10.1038/sj.embor.embor886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dogliotti E, Fortini P, Pascucci B, Parlanti E. Multiple pathways for DNA base excision repair. The mechanism of switching among multiple BER pathways. Prog. Nucleic Acid Res. Mol. Biol. 2001;68:1–28. doi: 10.1016/s0079-6603(01)68086-3. [DOI] [PubMed] [Google Scholar]
- 3.Parker A, Gu Y, Mahoney W, Lee SH, Singh KK, Lu AL. Human homolog of the MutY repair protein (hMYH) physically interacts with proteins involved in long patch DNA base excision repair. J. Biol. Chem. 2001;276:5547–5555. doi: 10.1074/jbc.M008463200. [DOI] [PubMed] [Google Scholar]
- 4.Maga G, Hubscher U. Proliferating cell nuclear antigen (PCNA): a dancer with many partners. J. Cell. Sci. 2003;116:3051–3060. doi: 10.1242/jcs.00653. [DOI] [PubMed] [Google Scholar]
- 5.Caldecott KW. Mammalian DNA single-strand break repair: an X-ra(y)ted affair. Bioessays. 2001;23:447–455. doi: 10.1002/bies.1063. [DOI] [PubMed] [Google Scholar]
- 6.Mitra S, Izumi T, Boldogh I, Bhakat KK, Hill JW, Hazra TK. Choreography of oxidative damage repair in mammalian genomes. Free Radic. Biol. Med. 2002;33:15–28. doi: 10.1016/s0891-5849(02)00819-5. [DOI] [PubMed] [Google Scholar]
- 7.Pascucci B, Russo MT, Crescenzi M, Bignami M, Dogliotti E. The accumulation of MMS-induced single strand breaks in G1 phase is recombinogenic in DNA polymerase beta defective mammalian cells. Nucleic Acids Res. 2005;33:280–288. doi: 10.1093/nar/gki168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fan J, Wilson DM., 3rd Protein-protein interactions and posttranslational modifications in mammalian base excision repair. Free Radic. Biol. Med. 2005;38:1121–1138. doi: 10.1016/j.freeradbiomed.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 9.Loizou JI, El-Khamisy SF, Zlatanou A, Moore DJ, Chan DW, Qin J, Sarno S, Meggio F, Pinna LA, et al. The protein kinase CK2 facilitates repair of chromosomal DNA single-strand breaks. Cell. 2004;117:17–28. doi: 10.1016/s0092-8674(04)00206-5. [DOI] [PubMed] [Google Scholar]
- 10.Lu X, Bocangel D, Nannenga B, Yamaguchi H, Appella E, Donehower LA. The p53-induced oncogenic phosphatase PPM1D interacts with uracil DNA glycosylase and suppresses base excision repair. Mol. Cell. 2004;15:621–634. doi: 10.1016/j.molcel.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 11.Frouin I, Montecucco A, Biamonti G, Hubscher U, Spadari S, Maga G. Cell cycle-dependent dynamic association of cyclin/Cdk complexes with human DNA replication proteins. EMBO J. 2002;21:2485–2495. doi: 10.1093/emboj/21.10.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frosina G, Fortini P, Rossi O, Carrozzino F, Raspaglio G, Cox LS, Lane DP, Abbondandolo A, Dogliotti E. Two pathways for base excision repair in mammalian cells. J. Biol. Chem. 1996;271:9573–9578. doi: 10.1074/jbc.271.16.9573. [DOI] [PubMed] [Google Scholar]
- 13.Nilsen H, Haushalter KA, Robins P, Barnes DE, Verdine GL, Lindahl T. Excision of deaminated cytosine from the vertebrate genome: role of the SMUG1 uracil-DNA glycosylase. EMBO J. 2001;20:4278–4286. doi: 10.1093/emboj/20.15.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levy N, Martz A, Bresson A, Spenlehauer C, de Murcia G, Menissier-de Murcia J. XRCC1 is phosphorylated by DNA-dependent protein kinase in response to DNA damage. Nucleic Acids Res. 2006;34:32–41. doi: 10.1093/nar/gkj409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prasad R, Singhal RK, Srivastava DK, Molina JT, Tomkinson AE, Wilson SH. Specific interaction of DNA polymerase beta and DNA ligase I in a multiprotein base excision repair complex from bovine testis. J. Biol. Chem. 1996;271:16000–16007. doi: 10.1074/jbc.271.27.16000. [DOI] [PubMed] [Google Scholar]
- 16.Whitehouse CJ, Taylor RM, Thistletwaite A, Zhang H, Karimi-Busheri F, Lasko DD, Weinfeld M, Caldecott KW. XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair. Cell. 2001;104:107–117. doi: 10.1016/s0092-8674(01)00195-7. [DOI] [PubMed] [Google Scholar]
- 17.Luo H, Chan DW, Yang T, Rodriguez M, Chen BP, Leng M, Mu JJ, Chen D, Songyang Z, Wang Y, Qin J. A new XRCC1-containing complex and its role in cellular survival of methyl methanesulfonate treatment. Mol. Cell Biol. 2004;24:8356–8365. doi: 10.1128/MCB.24.19.8356-8365.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akbari M, Otterlei M, Pena-Diaz J, Aas PA, Kavli B, Liabakk NB, Hagen L, Imai K, Durandy A, Slupphaug G, et al. Repair of U/G and U/A in DNA by UNG2-associated repair complexes takes place predominantly by short-patch repair both in proliferating and growth-arrested cells. Nucleic Acids Res. 2004;32:5486–5498. doi: 10.1093/nar/gkh872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heale JT, Ball AR, Jr, Schmiesing JA, Kim JS, Kong X, Zhou S, Hudson DF, Earnshaw WC, Yokomori K. Condensin I interacts with the PARP-1-XRCC1 complex and functions in DNA single-strand break repair. Mol. Cell. 2006;21:837–848. doi: 10.1016/j.molcel.2006.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan J, Otterlei M, Wong HK, Tomkinson AE, Wilson DM., 3rd XRCC1 co-localizes and physically interacts with PCNA. Nucleic Acids Res. 2004;32:2193–2201. doi: 10.1093/nar/gkh556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okano S, Lan L, Caldecott KW, Mori T, Yasui A. Spatial and temporal cellular responses to single-strand breaks in human cells. Mol. Cell Biol. 2003;23:3974–3981. doi: 10.1128/MCB.23.11.3974-3981.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lan L, Nakajima S, Oohata Y, Takao M, Okano S, Masutani M, Wilson SH, Yasui A. In situ analysis of repair processes for oxidative DNA damage in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2004;101:13738–13743. doi: 10.1073/pnas.0406048101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muller-Tidow C, Ji P, Diederichs S, Potratz J, Baumer N, Kohler G, Cauvet T, Choudary C, van der Meer T, Chan WY, Nieduszynski C, Colledge WH, Carrington M, Koeffler HP, Restle A, Wiesmuller L, Sobczak-Thepot J, Berdel WE, Serve H. The cyclin A1-CDK2 complex regulates DNA double-strand break repair. Mol. Cell Biol. 2004;24:8917–8928. doi: 10.1128/MCB.24.20.8917-8928.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scully R, Xie A. In my end is my beginning: control of end resection and DSBR pathway 'choice' by cyclin-dependent kinases. Oncogene. 2005;24:2871–2876. doi: 10.1038/sj.onc.1208609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Otterlei M, Warbrick E, Nagelhus TA, Haug T, Slupphaug G, Akbari M, Aas PA, Steinsbekk K, Bakke O, et al. Post-replicative base excision repair in replication foci. EMBO J. 1999;18:3834–3844. doi: 10.1093/emboj/18.13.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mer G, Bochkarev A, Gupta R, Bochkareva E, Frappier L, Ingles CJ, Edwards AM, Chazin WJ. Structural basis for the recognition of DNA repair proteins UNG2, XPA, and RAD52 by replication factor RPA. Cell. 2000;103:449–456. doi: 10.1016/s0092-8674(00)00136-7. [DOI] [PubMed] [Google Scholar]
- 27.Ko R, Bennett SE. Physical and functional interaction of human nuclear uracil-DNA glycosylase with proliferating cell nuclear antigen. DNA Repair (Amst) 2005;4:1421–1431. doi: 10.1016/j.dnarep.2005.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caldecott KW. XRCC1 and DNA strand break repair. DNA Repair (Amst) 2003;2:955–969. doi: 10.1016/s1568-7864(03)00118-6. [DOI] [PubMed] [Google Scholar]