Abstract
Initiation of reverse transcription of a retroviral RNA genome is strictly regulated. The tRNA primer binds to the primer binding site (PBS), and subsequent priming is triggered by the primer activation signal (PAS) that also pairs with the tRNA. We observed that in vitro reverse transcription initiation of the HIV-1 leader RNA varies in efficiency among 3′-end truncated transcripts, despite the presence of both PBS and PAS motifs. As the HIV-1 leader RNA can adopt two different foldings, we investigated if the conformational state of the transcripts did influence the efficiency of reverse transcription initiation. However, mutant transcripts that exclusively fold one or the other structure were similarly active, thereby excluding the possibility of regulation of reverse transcription initiation by the structure riboswitch. We next set out to determine the availability of the PAS element. This sequence motif enhances the efficiency of reverse transcription initiation, but its activity is regulated because the PAS motif is initially base paired within the wild-type template. We measured that the initiation efficiency on different templates correlates directly with accessibility of the PAS motif. Furthermore, changes in PAS are critical to facilitate a primer-switch to a new tRNA species, demonstrating the importance of this enhancer element.
INTRODUCTION
Reverse transcription of the human immunodeficiency virus type 1 (HIV-1) RNA genome is a critical step in the viral replication cycle. Upon infection of a host cell, the single-stranded viral RNA (vRNA) genome is reverse transcribed into a double-stranded DNA with duplicated long terminal repeats (LTR), which integrates into the host cell genome (1,2). Reverse transcription is mediated by the viral enzyme reverse transcriptase (RT) and the cellular tRNAlys3 is used as a primer (2,3). Although reverse transcription initiation can take place shortly after budding of the viral particle from the producer cell, there is evidence suggesting that reverse transcription is restricted until infection of a new host cell, possibly by limitation of dNTP building blocks (4–8).
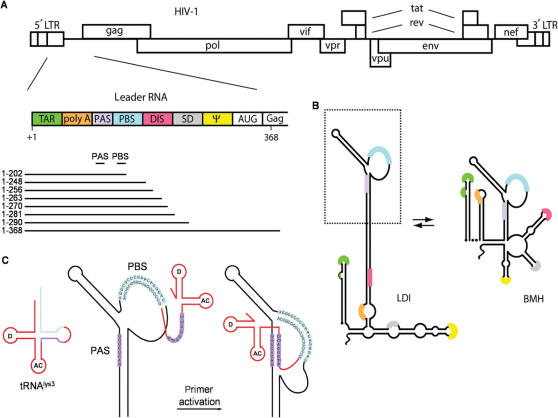
Reverse transcription initiation is mediated by multiple HIV-1 RNA–tRNA interactions. The first step in reverse transcription is the hybridization of the 3′ end of the tRNAlys3 molecule to a complementary 18-nt sequence in the 5′-untranslated leader region of the HIV-1 RNA genome termed the primer binding site (PBS) (9,10). Several additional sequence motifs in the untranslated leader have been proposed to interact with the primer tRNA, including a single-stranded A-rich motif, located in the U5 region immediately upstream of the PBS that was proposed to interact with the U-rich anticodon of tRNAlys3 (11–16). Another interaction between the anticodon stem and variable loop of tRNAlys3 and a sequence upstream of the A-rich loop in the template forms in the presence of the viral nucleocapsid protein (17). We identified another enhancer element that is located further upstream in the leader, the 8-nt PAS motif that is complementary to the TψC arm of the tRNAlys3 primer (Figure 1) (18–21). The PAS motif is not involved in tRNA placement, but enhances its usage as a primer. Hence it is termed the primer activation signal (PAS) and the complementary sequence in the TψC arm is termed anti-PAS. There is accumulating evidence for the importance of this PAS motif, e.g. its role in facilitating priming by a non-self tRNA primer (20,22,23). As the PAS sequence is masked in the HIV-1 leader RNA by base pairing within the extended PBS stem that encompasses the PBS domain (Figure 1C), this provides the virus with a mechanism to regulate the initiation of reverse transcription (18,19).
Figure 1.
Motifs in the HIV-1 RNA genome that regulate reverse transcription. (A) The HIV-1 DNA genome is shown with the nine open reading frames. The untranslated leader encoding multiple regulatory elements is schematically depicted. A 3′-nested set of transcripts used as template for reverse transcription initiation are shown with the PAS and PBS motifs. (B) Alternative structures of the HIV-1 leader RNA. The ground state conformation contains a long-distance interaction (LDI) between the polyA and DIS domains. The alternate conformation exposes the polyA and DIS hairpins and takes the form of a branched structure (BMH). (C) Model of PAS-mediated tRNA primer activation. First, the tRNAlys3 primer anneals to the PBS (indicated in blue), subsequently the anti-PAS sequence in tRNAlys3 must anneal to the PAS (purple) before efficient reverse transcription initiation can take place.
The conformational state of the HIV-1 leader RNA has also been suggested to influence reverse transcription initiation (24). The sequence of the leader RNA is highly conserved among virus isolates and contains several sequence and structure motifs that are involved in many replication steps (e.g. transcription, splicing, translation, RNA dimerization, packaging and reverse transcription) (25,26) (Figure 1A). Multiple lines of evidence indicate that the leader region can adopt two distinct structures (27–29) (Figure 1B). In the ground state conformation, the polyA and DIS regions are base paired through a long distance interaction (LDI). Disruption of this long-distance interaction rearranges the leader region into a branched structure with multiple hairpins (BMH). This BMH conformation presents the DIS hairpin that facilitates dimerization of the RNA genome through a kissing loop interaction (30,31). The RNA packaging signal ψ is also differently presented by the two leader conformations (27). This RNA switch has been proposed to regulate the appropriate timing of RNA dimerization and packaging (28,32). We now set out to test the impact of the HIV-1 leader RNA structure, either through the LDI-BMH switch or PAS accessibility, on the initiation of reverse transcription.
EXPERIMENTAL PROCEDURES
DNA synthesis
The HIV-1 LAI wild type (wt) and mutant pBluescript 5′-LTR plasmids that were used in this study as template for PCR amplification have been described earlier (18,33). The DNA templates for in vitro transcription were obtained through PCR amplification of the 5′-LTR leader region with the T7-2 sense primer, with a 5′-flanking T7 RNA polymerase promoter sequence, and the corresponding antisense primers. Sense T7-2 50-TAATACGACTCACTATAGGGTCTCCTGGTTAGACCAT-30 (T7-promotor underlined, start site transcription in bold and italic). Antisense primers: 202 5′-CAAGTCCCTGTTCGGGCGCC-3′ 248 5′-GCCGAGTCCTGCGTCGAGAG-3′ 256 5′-TTCAGCAAGCCGAGTCCTGC-3′ 263 5′-TGCGCGCTTCAGCAAGCCGA-3′ 270 5′-CTTGCCGTGCGCGCTTCAGC-3′ 281 5′-TCCCCTCGCCTCTTGCCGTG-3′ 290 5′-CCAGTCGCCTCCCCTCGCCT-3′ 368 5′-TCCCCCGCTTAATACTGACG-3′. The PCR fragments were ethanol precipitated and dissolved in water.
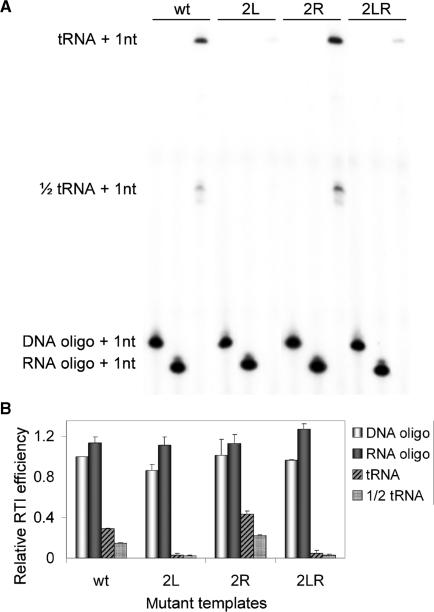
We made five sets of 3′-truncated leader RNA transcripts. The first set is based on the wt HIV-1 LAI sequence, but with 3′ truncations (1–202/248/256/263/270/281/290/368). The second set (J1–J10 1–368, Figure 8) contains mutations in the 3′-end of the leader (34). Three mutant sets have mutations in PAS (2L) or opposite to the PAS (2R) and the double mutant (2LR) (18).
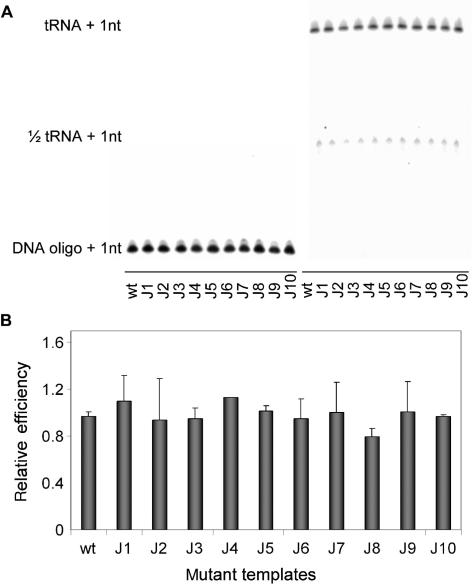
Figure 8.
DNA-, RNA- and tRNA-primed reverse transcription. (A) The 3 primers were tested on the 1–368 transcripts of wt and mutants 2L, 2R and 2LR. Primers were extended with 32P-CTP by HIV-1 RT. Extended primers were separated on a denaturing acrylamide gel. Indicated are cDNA products of DNA, RNA, tRNA and 1/2-tRNA primers. (B) Quantified data of three experiments. The efficiency of DNA-primed reverse transcription initiation of wt is set at 1.
In vitro transcription
In vitro transcription was performed with the Ambion MegaShortscript T7 transcription kit according to the manufacturer's instructions. The transcripts were separated on a 6% denaturing polyacrylamide gel containing urea, visualized by UV shadowing, subsequently excised, and eluted from the gel fragments by overnight incubation at 30°C in 1× TBE and phenol. The RNA was subsequently ethanol precipitated and dissolved in water. The dissolved RNA was quantified by UV absorbance measurement and set to an equal molarity of 0.044 pmol/μl that corresponds to 5 ng/μl RNA for the wt 1–368 transcript. Radiolabeled transcripts were synthesized by performing the transcription reaction in the presence of 1 μl α32P-dUTP and were quantified by scintillation counting.
Primer extension assay
Reverse transcription was performed by incubating the in vitro synthesized RNA templates (0.088 pmol) with either 1.5 μg calf liver tRNA (6 pmol total tRNA, of which ∼1.2 pmol tRNAlys3, Roche Molecular Biochemicals), 10 ng DNA Lys21 primer (complementary to the PBS, 5′-CAAGTCCCTGTTCGGGCGCCA-3′) or 10 ng RNA Lys18 primer (complementary to the PBS, 5′-GUCCCUGUUCGGGCGCCA-3′) in 12 μl annealing buffer (83 mM Tris-HCl (pH 7.5), 125 mM KCl). The samples were heated for 2 min at 85°C and then slowly cooled to room temperature. The primer was extended by addition of 6-μl 3× RT buffer, which contains 9 mM MgCl2, 30 mM dithiothreitol, 150 μg/ml actinomycin D, 0.33 μl of [α-32P]dCTP and 0.5 units of HIV-1 RT (Medical Research Council AIDS Reagent Project). Samples were incubated at 37°C for 30 min and the reaction was stopped by adding one volume of denaturing loading buffer containing formamide (Ambion). The samples were heated for 2 min at 95°C and subsequently separated on a 6% denaturing polyacrylamide gel (19:1 acrylamide gel containing urea, 1× TBE) at 50 W. Gels were dried and applied to a Storm PhosphorImager. Quantification of band intensities was performed with ImageQuant 5.0 (Amersham Biosciences). The data was corrected for the amount of input RNA by dividing the tRNA extension signal by the DNA-primed extension signal. The value obtained for the wt 1–368 sample was arbitrarily set at 1.
Non-denaturing polyacrylamide electrophoresis
For electrophoretic analysis, ∼200 counts/s of radiolabeled transcripts was incubated in a final volume of 20 μl TEN buffer (100 mM NaCl, 1 mM EDTA, 10 mM Tris (pH 8.0)). Samples were heated at 85°C and slowly cooled to room temperature. Samples were split (2 × 10 μl) and 2 μl of loading buffer (30% glycerol with BFB dye). Control samples were denatured at 90°C with 10-μl formamide loading buffer. Samples were analyzed on a 4% polyacrylamide gel (Sigma) in 0.25 × TBE (22.5 mM Tris, 22.5 mM boric acid, 0.625 mM EDTA), at 150 V at room temperature, dried and analyzed.
UV absorbance measurements
RNA thermal denaturation was monitored by measuring the absorbance of UV light at 260 nm with a Perkin Elmer Lambda 2 spectrophotometer. The temperature of the samples was increased from 25 to 90°C at a rate of 0.5°C/min while measuring the A260 nm at each 0.1°C. The measurements were performed in a 1-cm path length quartz cuvette containing 3.0 μg of RNA dissolved in 140 μl of 50 mM Na-cacodylate buffer (pH 7.2).
RNA secondary structure prediction by Mfold
For RNA secondary structure prediction, we used the Mfold program 3.2 version offered by Zuker. (www.bioinfo.rpi.edu/applications/mfold/old/rna/form1.cgi) (35). The folding jobs were submitted under the following standard conditions: 37°C, 1.0 M NaCl and a 5% suboptimality range. Folding restrictions were imposed for some of the folding jobs and the ΔG value of the first structure (with lowest ΔG) was used for analysis. The LDI versus BMH was defined by inspection of the region encompassing either the polyA hairpin (BMH) or the long distance interaction with the DIS domain (LDI). The ΔG value of a structure with an exposed PAS was obtained by restricting base pairing of the 8-nt PAS sequence, the masked PAS value was obtained without restriction. The PBS was restricted from base pairing in both situations.
Virus production and isolation of viral RNA
C33A cells were grown in Dulbecco's modified Eagle's medium with 10% fetal calf serum at 37°C and 5% CO2. For the production of virions, C33A cells were transiently transfected by the calcium phosphate method. Cells were grown to 60% confluence in 20 ml of culture medium in a 75-cm2 flask. Forty micrograms of the proviral construct (22) in 880 μl of water was mixed with 1 ml of 50 mM HEPES (pH 7.1), 250 mM NaCl, 1.5 mM Na2HPO4 and 120 μl of 2 M CaCl2, incubated at room temperature for 20 min, and added to the culture medium. Three days after transfection, the culture medium (20 ml) was centrifuged at 2750 × g for 15 min to remove cells. The supernatant was subsequently filtered through a 0.45-μm-pore-size filter (Schleider & Schuell), and the viruses were pelleted by centrifugation at 25 000 r.p.m. for 30 min in a Beckman SW28 rotor. Virions were resuspended in 400 μl of 10 mM Tris-HCl (pH 8.0), 100 mM NaCl, 1 mM EDTA. To isolate vRNA, the viruses were incubated for 30 min at 37°C in the presence of 100 μg of proteinase K/ml, 1% sodium dodecyl sulfate, followed by extraction with phenol–chloroform–isoamyl alcohol (25:24:1) and precipitation in 0.3 M sodium acetate (pH 5.2), ethanol at −20°C. The vRNA was pelleted by centrifugation (16 000 × g, 20 min), washed with 70% ethanol, and dried. The pellet was dissolved in 20 μl of 10 mM Tris-HCl (pH 8.0), 1 mM EDTA and stored at −70°C.
Ex vivo reverse transcription assay
The vRNA–tRNA complex corresponding to 50 ng of CA-p24 virions was incubated with or without an oligonucleotide primer (20 ng) in 12 μl of 83 mM Tris-HCl (pH 7.5), 125 mM KCl at 85°C for 2 min and at 65°C for 10 min, followed by cooling to room temperature for 1 h to allow primer annealing. The natural tRNA primer or the annealed DNA primer was extended by the addition of 6 μl of RT buffer (9 mM MgCl2; 30 mM dithiothreitol; 150 μg of actinomycin D/ml; 30 μM dGTP, dTTP; 1.5 μM dCTP), 0.5 μl of [α-32P]dCTP and 0.5 U of HIV-1 RT. The samples were incubated at 37°C for 45 min. The cDNA product was precipitated in 25 mM EDTA, 0.3 M sodium acetate (pH 5.2), 80% ethanol at −20°C. The products were analyzed on a denaturing 6% polyacrylamide-urea sequencing gel.
RESULTS
The efficiency of reverse transcription initiation varies among 3′-truncated HIV-1 leader RNA templates
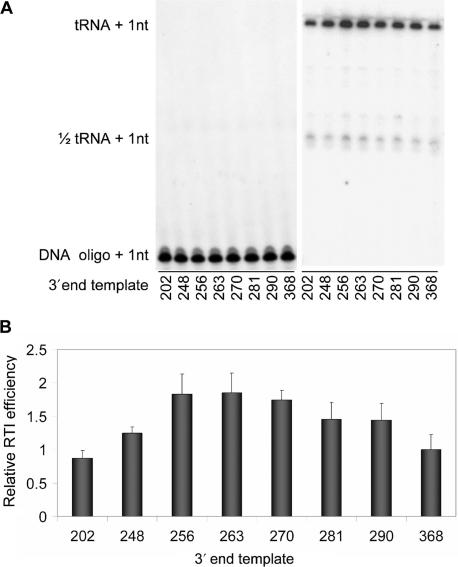
We set out to test the efficiency of reverse transcription initiation on HIV-1 leader transcripts with varying 3′ end positions. A nested set of RNA transcripts was made with the wt 5′-end (position +1) and a variable 3′ end: 202, 248, 256, 263, 270, 281, 290 and 368 (Figure 1A). The natural tRNAlys3 primer and the synthetic oligonucleotide Lys21 were used as primers for reverse transcription. To test the initiation efficiency, the annealed tRNA/DNA primer was extended by 1 nt after addition of [α-32P] dCTP and HIV-1 RT enzyme. One representative primer extension assay is shown in Figure 2A. The results of four independent assays were quantified and are summarized in Figure 2B. The DNA primer acts as a control for the amount of template because the efficiency of DNA-primed reverse transcription initiation is equal on all templates (18,36). Compared to the shortest 1–202 transcript, the efficiency of tRNA-primed reverse transcription increases for the 3′-extended transcripts, with an optimum for the 1–263 transcript, after which the activity drops gradually with transcript length. The 1–263 transcript consistently exhibits an ∼2-fold higher activity than 1–202 and 1–368. These results show that there is a difference in the efficiency of reverse transcription initiation for leader transcripts with different 3′ ends. We set out to determine the molecular reason for this effect.
Figure 2.
Reverse transcription initiation of HIV-1 leader RNA transcripts with variable 3′ ends. (A) DNA (left) or tRNAlys3 (right) primers were annealed to HIV-1 transcripts. Primers were extended by addition of 32P-CTP and HIV-1 RT enzyme. Extended primers were separated on a denaturing acrylamide gel. The 1/2 tRNA signal represents a half tRNA species that is discussed in detail in the context of Figure 8. (B) Relative reverse transcription initiation efficiency with tRNAlys3 as primer was calculated by comparison with the DNA-primed signal. The relative efficiency of reverse transcription initiation (RTI) of the 1–368 template was set at 1. Quantification and standard deviations of four independent experiments are shown.
Analysis of the leader RNA conformation
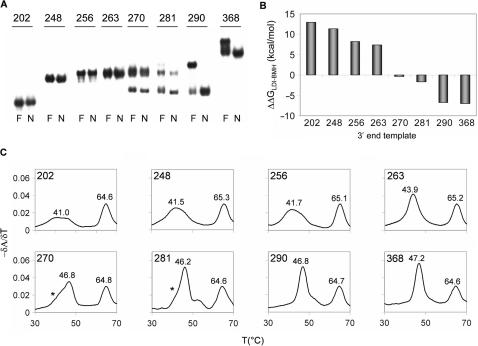
There are no known sequence motifs 3′ of the PBS that may play a positive or negative role in tRNA-primed reverse transcription. Thus, the observed effects are likely due to conformational changes of the RNA template, e.g. affecting the LDI-BMH riboswitch (Figure 1B) and/or influencing the accessibility of the PAS enhancer motif (Figure 1C). We set out to determine the LDI/BMH status of the 3′-truncated HIV-1 transcripts by three methods: non-denaturing polyacrylamide electrophoresis, secondary structure prediction by Mfold and UV absorbance melting experiments.
Electrophoresis on non-denaturing gels separates molecules according to size but also according to structure, as different structures have different migration properties. Previous research has shown that there is a relatively profound migration difference between the LDI and BMH conformations. Compared to a heat/formamide-denatured sample, the LDI-folded leader RNA migrates faster. In contrast, a BMH-folded leader RNA co-migrates with the denatured sample (28,37). The set of radiolabeled transcripts were analyzed on a non-denaturing TBE gel (Figure 3A). Transcripts 202, 248, 256 and 263 co-migrate with the denatured transcripts, indicating that they adopt the BMH conformation. Transcripts 270 and 281 show both this slow migrating BMH band and the fast migrating LDI form. Only the fast migrating LDI form is observed for 290 and 368. Thus, the shorter transcripts exclusively fold the BMH conformation, but the LDI conformation becomes possible upon inclusion of DIS sequences, which form the long-distance interaction with the polyA domain. The longest transcripts exclusively fold the LDI conformation.
Figure 3.
LDI-BMH status of the leader transcripts with variable 3′ ends. (A) Electric mobility shift assay on a 0.25 × TBE acrylamide gel. Transcripts were incubated in TEN buffer (N), or denatured in formamide containing loading buffer (F). (B) The LDI-BMH status of the transcripts as calculated by Mfold. The ΔG values of the LDI and the BMH structure were calculated as described in the Materials and methods section and subtracted to obtain the ΔΔG. (C) Thermal melting profiles of the HIV-1 transcripts with variable 3′ ends. The temperature was increased from 25 to 90°C at a rate of 0.5°C/min, the A260 nm was measured with 0.1°C intervals. The transcript length and melting temperatures (Tm) of the transitions are indicated in the plots.
We also used the RNA folding prediction program Mfold 3.2 (35). The thermodynamic stability of the LDI (ΔGLDI) and BMH (ΔGBMH) conformation was scored, and the difference (ΔΔGLDI-BMH) was calculated. A positive ΔΔG value predicts a preference for BMH folding, a negative ΔΔG value is predictive of LDI folding, and both structures are in equilibrium when ΔΔG is close to 0. The ΔΔG value gradually declines with the length of the leader transcript (Figure 3B), indicating a gradual shift from BMH to LDI structure, which is fully consistent with the gel migration results.
The secondary structure of a transcript can also be analyzed by measuring its UV absorbance at increasing temperature (T), as disruption of base pairs results in an increase of UV absorbance (hyperchromicity). This technique can efficiently discriminate between the LDI and BMH conformation (28). All transcripts form the 5′-terminal TAR hairpin that produces the characteristic peak at 65°C (Figure 3C), but this does not discriminate between LDI and BMH because both conformations have the TAR hairpin (Figure 1B). The LDI is characterized by a distinct narrow transition while the BMH is reflected by a broad transition at a lower temperature. Transcripts show either the broad peak at 41°C that is typical for BMH (202, 248, 256), or the more intense peak at 46°C that is characteristic for LDI (270, 281, 290, 368). Transcript 263 and 270 show an intermediate pattern (asterisks indicate underlying BMH peak). All conformational tests indicate that shorter transcripts are in the BMH form, with a gradual shift towards LDI upon 3′-extension. This suggests that the eventual drop in the efficiency of reverse transcription initiation from 263 to 368 may be due to LDI folding, but the LDI-BMH equilibrium does not explain the initial increase in efficiency from 202 to 263.
Reverse transcription initiation is not affected by the LDI-BMH switch
To test the impact of the leader RNA conformation on the initiation of reverse transcription initiation, we used leader mutants that specifically stabilize the LDI or BMH structure. Mutants J1–J10 were previously used to study the effect of LDI-BMH on RNA dimerization and mRNA translation (34). We selected these mutants because they contain precise mutations in the 3′-terminal region of the leader RNA and thus do not directly affect the upstream sequences that control reverse transcription initiation (PBS and PAS). The wt and mutant templates were made as 1–368 transcripts to exclusively examine the effect of LDI/BMH on the initiation of reverse transcription. The tRNA extension efficiency of the J1–J10 mutants is very similar to that of wt (Figure 4), despite the fact that some transcripts have a significantly altered LDI/BMH equilibrium. In particular, J8 and J9 that exclusively fold the BMH structure display the same efficiency as the wt transcript, which folds the LDI conformation. We conclude that the LDI/BMH conformation of the HIV-1 leader sequence has no direct effect on reverse transcription initiation.
Figure 4.
Reverse transcription initiation efficiency of the wt and J1–J10 transcripts. (A) DNA or tRNAlys3 primers were annealed to wt and mutant 1-368 transcripts and extended with 32P-CTP by the HIV-1 RT enzyme. Extended primers were separated on a denaturing acrylamide gel. The 1/2 tRNA signal represents a half tRNA species that is discussed in detail in the context of Figure 8. (B) Relative initiation of reverse transcription efficiency of the wt and mutant transcripts, the wt activity was arbitrarily set at 1. The average and standard deviations of three independent experiments were quantified.
The efficiency of reverse transcription initiation can be modulated by PAS accessibility
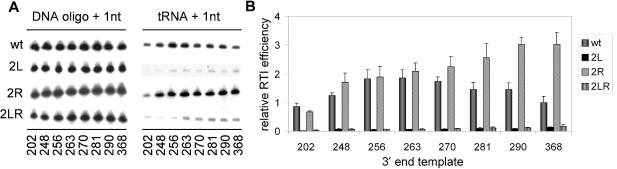
The availability of the PAS sequence could form an alternative explanation for the different efficiencies in reverse transcription initiation of the 3′-nested set of HIV-1 leader transcripts. To test this possibility, we performed the tRNA/DNA primer initiation assay with a 3′-nested set of three mutants that differ in PAS accessibility. The 2L mutant inactivates the PAS motif by a 7-nt substitution and serves as a negative control (Figure 5). The 2R mutant has a 7-nt substitution in the opposite strand, thereby opening the stem and exposing the PAS sequence. The 2LR double mutant contains both substitutions that were designed to restore base pairing, but obviously not the correct PAS sequence. Let us first discuss the behavior of the 368 transcripts. We measured very low template activity for the PAS-minus 2L and 2LR mutants, whereas the PAS-exposed 2R template showed increased activity compared to the wt transcript (Figure 6). The shortest 202 transcripts also show the defect for 2L and 2LR, but show a different pattern for the 2R mutant, which has the same activity as wt. However, this makes sense as the short wt transcript will have an exposed PAS because the sequences to which it base pairs are not present in the 202 transcript. The effect of 3′-extension of the wt transcript has been discussed before; it shows an increase up to 263, followed by a gradual decrease in efficiency. Most importantly, the PAS-exposed 2R mutant did not show the characteristic wt-pattern of reduced efficiency upon 3′-extension of the template. Both wt and 2R exhibit increased efficiency of reverse transcription initiation up to position 263, but 2R does not show the subsequent drop in activity up to the 368-nt long transcript. These results suggest that 3′ leader sequences can mask the PAS motif in the wt transcript, but not in the dominant 2R mutant with a fully exposed PAS motif.
Figure 5.
The secondary structure model of part of the HIV-1 leader RNA and the tRNAlys3 primer are shown. Mutant 2L has a 7-nt substitution in the PAS motif and mutant 2R has a substitution on the opposite side of the base-paired PAS domain. The 2L and 2R mutations were combined in the 2LR mutant. The asterisks at position 37–39 of the tRNA mark the position of a cleavage product that is referred to as 1/2 tRNAlys3 in Figure 8.
Figure 6.
Reverse transcription initiation on the 3′-nested set of wt, 2L, 2R and 2LR transcripts. (A) DNA or tRNAlys3 primers were annealed to the indicated transcripts. Primers were extended by HIV-1 RT with 32P-CTP and separated on denaturing acrylamide gels. Gel fragments of the DNA- and tRNA-primed extensions are shown. (B) Relative reverse transcription initiation efficiency of the different transcripts. Initiation of the wt368 template was set at 1. The average and standard deviation of four independent experiments was quantified.
The predicted PAS accessibility correlates with the efficiency of reverse transcription initiation
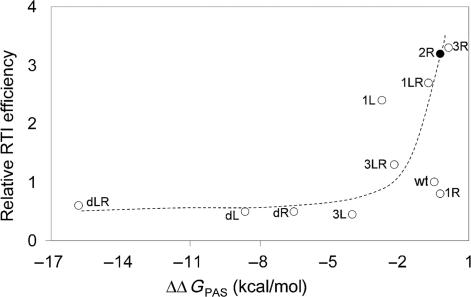
We used Mfold to calculate the availability of the PAS sequence for the different templates by calculating the ΔΔG value of PAS-masked versus PAS-exposed structures. To test and calibrate this method, we first calculated the ΔΔGPAS values of a large set of RNA structure mutants that were previously tested for tRNA-mediated reverse transcription (18). Initiation is very inefficient for transcripts with a low ΔΔGPAS value, but increases for templates with a value of ∼−3 kcal/mol or higher (Figure 7). We also plotted the 2R transcript, which displays a higher ΔΔGPAS value and increased initiation efficiency when compared to wt, consistent with the idea that the PAS is more accessible. We also analyzed the impact of 3′-end variation in this way. However, the ΔΔGPAS values were not affected by transcript length (results not shown). We conclude that effects of large mutations on reverse transcription initiation can be explained by Mfold, but the rather small effect of 3′-truncations do not show a difference in ΔΔG.
Figure 7.
Mfold-calculated PAS exposure. To calibrate this approach, we first calculated the PAS availability of a well-studied set of mutants (18). The ΔG of an open PAS structure was obtained by the restriction of base pairing. The calculated ΔΔG, which reflects the energy needed to open the PAS sequence, was plotted against the efficiency of reverse transcription initiation as described by Beerens et al. (18). The 2R mutant that is tested in this study is highlighted by a closed circle.
PAS-mediated stimulation of reverse transcription is specific for the tRNA primer
We measured differences in the efficiency of reverse transcription initiation of different templates that correlate with PAS availability. Any PAS effect should be specific for tRNA-primed reverse transcription and should not be observed with short DNA or RNA primers annealed to the PBS. We therefore directly compared these primers on the full-length wt, 2L, 2R and 2LR transcripts. DNA and RNA oligonucleotide-primed initiation is equally efficient on all templates (Figure 8). Only tRNA-primed reverse transcription varies per template according to the PAS accessibility. The 2R mutant, with a free PAS motif, shows an approximate 2-fold increase in efficiency compared to the wt template. The PAS minus 2L and 2LR mutants exhibit a very low efficiency of reverse transcription initiation. Both these positive and negative effects are specific for the tRNA primer. Interestingly, the tRNA preparation also contains a tRNA fragment that represents the 3′ half of tRNAlys3, ∼37–39 nt in length and lacking the typical tRNA cloverleaf structure (36) (Figure 5). This 1/2 tRNA primer includes the sequences complementary to the PBS and the PAS sequences. Indeed, this 1/2 tRNA primer demonstrates the same activity spectrum as the complete tRNAlys3.
Thus, we conclude that the observed tRNA-specific differences in template activity reflect their ability to present the PAS motif. We argued that differences in PAS presentation also determine the efficiency of reverse transcription initiation of the nested set of 3′ extended transcripts. Consistent with this idea, no differences were observed on these templates with the DNA primer (Figure 6A) and the RNA primer (results not shown).
Contribution of the PAS motif to a switch in tRNA usage
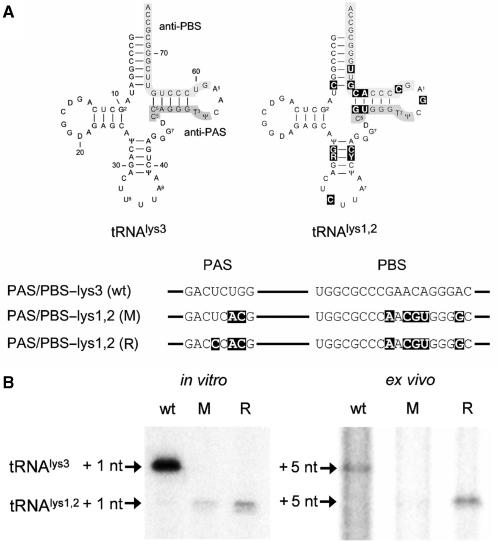
An HIV-1 variant that stably uses the alternative tRNAlys1,2 primer was recently described (22). This virus was obtained in two steps. First, we changed both the PBS and the PAS motif to complement the tRNAlys1,2 primer (Figure 9A, mutant M). Long-term culturing resulted in a revertant virus that gained replication capacity by a reversion (R) in the PAS motif. The U126C reversion in the PAS motif stabilizes the interaction with the anti-PAS. To test the effect of this mutation on reverse transcription initiation, we performed primer extension assays on in vitro transcribed wt, mutant and revertant transcripts and ex vivo on vRNA–tRNA complexes isolated from the respective virions. For the latter purpose, virions were produced in transfected C33A cells, and the vRNA–tRNA complexes were isolated. Equal amounts of wild-type, mutant and revertant complexes (based on CA-p24) were used in the reverse transcription assay. The amount of input vRNA-tRNA in the assay was verified by a control DNA primer extension reaction (19) that we routinely use in our laboratory (results not shown). The tRNA primer was extended on the in vitro transcripts with 1 radiolabeled nucleotide (Figure 9B). Because the vRNA–tRNA duplex from virions may contain tRNA primers that are already extended by 1 or 2 nt (4,7,8), we used a modified protocol to monitor the synthesis of tRNA +5 nucleotides products. The extended tRNAlys3 runs at a higher position on the denaturing gel than extended tRNAlys1,2 due to different base modifications within the tRNA backbone (7,38). tRNAlys3 is extended efficiently in vitro and ex vivo on the wt template, but not on the M and R templates (Figure 9B). A minor tRNAlys1,2 signal is apparent for the M template, but it is significantly enhanced in the R revertant. The point mutation in the PAS motif of the R template clearly enhances usage of the tRNAlys1,2 primer, both in the in vitro and ex vivo experiment. Apparently, optimization of the PAS motif enhances usage of the new tRNA primer, which underscores the importance of the PAS motif in reverse transcription initiation.
Figure 9.
Reverse transcription assays with in vitro transcribed transcripts and vRNA–tRNA complexes isolated from virions. (A) The differences in the anti-PBS and anti-PAS sequences between tRNAlys3 and tRNAlys1,2 are boxed in the latter molecule. The anti-PAS and anti-PBS motifs are boxed in gray. The HIV-1 PAS and PBS motifs were mutated to complement the tRNAlys1,2 primer in the mutant PAS/PBS-lys1,2 (M). Long-term culturing of the mutant virus resulted in a revertant virus that gained replication capacity by a point mutation in the PAS motif (R). The U to C mutation stabilizes the interaction of the anti-PAS with the PAS motif. (B) Reverse transcription initiation of HIV-1 leader RNA transcripts. tRNA primers were annealed to the in vitro transcribed wt, mutant (M) and revertant (R) transcripts or the vRNA–tRNA complexes were isolated from the corresponding virions. Primers were extended by addition of HIV-1 RT enzyme and 32P-dCTP for the in vitro extension assay or dTTP, dGTP and 32P-dCTP for ex vivo extension. This results in the extension of the tRNA primer with 1 and 5 nt, respectively. The extended primers were separated on a denaturing polyacrylamide gel. The tRNAlys3 product runs at a higher position in the gel than the tRNAlys1,2 product due to different base modification.
DISCUSSION
Initiation of HIV-1 reverse transcription is a highly regulated process in which the tRNAlys3 primer has to interact with different sequence motifs in the template; the PBS for binding and the upstream PAS motif for initiation of reverse transcription. In this study, we observed that sequences downstream of the PBS also affect the initiation of reverse transcription. The 3′ leader domain could encode additional regulatory sequence motifs, but it is more likely that the observed effects are due to conformational changes in the leader RNA. First, 3′ sequences may affect the LDI-BMH riboswitch and consequently PBS or PAS availability. We ruled out this possibility by testing specific LDI-BMH mutants. Second, 3′ sequences could affect the accessibility of the PAS motif, and thus affect reverse transcription initiation. Several lines of evidence support this explanation. The 3′ sequences inhibit the initiation of reverse transcription for the wt template, but not for a designed PAS-exposed mutant. Furthermore, the variation in efficiency was observed exclusively for the tRNA and 1/2 tRNA primers, and not for DNA/RNA oligonucleotide primers. These combined results demonstrate that differential PAS accessibility is underlying differences in the efficiency of reverse transcription initiation of 3′-extended templates.
We were unable to define a precise structural rearrangement of the PAS motif that causes the inhibitory effect of 3′ sequences. Mfold was used to calculate the exposure of the PAS motif, but this did not provide an explanation in terms of base pairing. This could be due to the fact that Mfold is a secondary structure prediction program that will miss effects at the tertiary structure level. It also remains unclear why the shortest 202 transcript displays the lowest efficiency in reverse transcription initiation. This has in fact also been observed in previous studies, and it seems that sequences 3′ of the PBS are required for efficient reverse transcription initiation (17,39,40). We argue that the stimulatory effect of the exposed PAS motif is nullified because the U5-leader stem cannot fold properly (Figure 5).
We also analyzed the role of the PAS motif in the context of a tRNA-switch in an evolved virus variant (22). An acquired mutation in the PAS motif stabilizes base pairing with the anti-PAS motif of the new tRNAlys1,2primer. This PAS adaptation increased tRNAlys1,2-primed reverse transcription initiation in vitro and ex vivo, confirming the importance of the PAS motif in tRNA usage.
Several HIV-1 RNA–tRNAlys3 interactions have been proposed (11–20). Chemical and enzymatic RNA structure probing suggested that the tRNAlys3 anticodon loop pairs with the A-rich loop located upstream and adjacent to the PBS (41–43). However, a recent publication by Goldschmidt et al. argues that this interaction is exclusive for the unusual MAL isolate (44). The same authors performed chemical probing of the PBS structure with and without an annealed tRNA primer, and obtained no evidence for the PAS–anti-PAS interaction (45). However, we argued that the PAS–tRNA interaction is transient in nature (21). Furthermore, it is quite possible that this relative late initiation step is facilitated by the RT enzyme, which was lacking in these experiments.
The results obtained in the current study seem to contradict the results described in two other studies. In our assays, the efficiencies of reverse transcription initiation on different templates are equal for an RNA oligonucleotide primer (and a DNA primer). This contrasts with the results of Goldschmidt et al. (45) and Iwatani et al. (17). Goldschmidt et al. showed that tRNA-primed (−)strand synthesis on PAS-mutated templates is equal to RNA oligonucleotide-primed (−)strand synthesis, which seems to contradict the proposed role of the PAS motif. However, close examination of their results indicates that the elongation phase of reverse transcription is affected rather than initiation. Similarly, Iwatani et al. observed equal (−)strand synthesis on a 3′-truncated set of transcripts with a tRNA or RNA primer. They also measured full-length (−)strand synthesis, which is the sum of initiation and elongation efficiencies. We think that primer extension measured by single nucleotide addition, as used in our assays, is a better measure for the efficiency of reverse transcription initiation.
Previous research has presented evidence for the regulation of reverse transcription initiation by occlusion and exposure of the PAS sequence (18–21,46). The importance of the PAS motif is underscored by its conservation among all retroviridae (20,47), and the PAS motif turned out to be critical in attempts to modify tRNA-usage of HIV-1 by alteration of the PBS motif (20,22). Here we present evidence that HIV-1 transcript length affects the initiation of reverse transcription via variation in the accessibility of the PAS element. These results emphasize the important role of the PAS motif for interaction with the tRNAlys3 primer in HIV-1 reverse transcription.
ACKNOWLEDGEMENTS
We thank Dr D. Stammers for the gift of purified HIV-1 RT enzyme (obtained through the MRC AIDS Reagent Project). RNA studies in the Berkhout lab are sponsored by ZonMw (VICI-grant) and NWO-Chemical Sciences (TOP-grant). The Open Access publication charge was waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Telesnitsky A, Goff SP. Reverse transcriptase and the generation of retroviral DNA. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1997. pp. 121–160. [PubMed] [Google Scholar]
- 2.Varmus H, Swanstrom R. Replication of retroviruses. In: Weiss R, Teich N, Varmus H, Coffin J, editors. RNA tumor viruses. Molecular biology of tumor viruses. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory; 1991. pp. 369–512. [Google Scholar]
- 3.Leis J, Aiyar A, Cobrinik D. Regulation of initiation of reverse transcription of retroviruses. In: Skalka AM, Goff SP, editors. Reverse Transcriptase. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1993. pp. 33–48. [Google Scholar]
- 4.Trono D. Partial reverse transcripts in virions from human immunodeficiency and murine leukemia viruses. J. Virol. 1992;66:4893–4900. doi: 10.1128/jvi.66.8.4893-4900.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lori F, Veronese FDM, De Vico AL, Lusso P, Reitz MS, Jr, Gallo RC. Viral DNA carried by human immunodeficiency virus type 1 virions. J. Virol. 1992;66:5067–5074. doi: 10.1128/jvi.66.8.5067-5074.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arts EJ, Mak J, Kleiman L, Wainberg MA. DNA found in human immunodeficiency virus type 1 particles may not be required for infectivity. J. Gen. Virol. 1994;75:1605–1613. doi: 10.1099/0022-1317-75-7-1605. [DOI] [PubMed] [Google Scholar]
- 7.Oude Essink BB, Das AT, Berkhout B. HIV-1 reverse transcriptase discriminates against non-self tRNA primers. J. Mol. Biol. 1996;264:243–254. doi: 10.1006/jmbi.1996.0638. [DOI] [PubMed] [Google Scholar]
- 8.Huang Y, Wang J, Shalom A, Li Z, Khorchid A, Wainberg MA, Kleiman L. Primer tRNALys3 on the viral genome exists in unextended and two-base extended forms within mature human immunodeficiency virus type 1. J. Virol. 1997;71:726–728. doi: 10.1128/jvi.71.1.726-728.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oude Essink BB, Das AT, Berkhout B. Structural requirements for the binding of tRNA Lys3 to reverse transcriptase of the human immunodeficiency virus type 1. J. Biol. Chem. 1995;270:23867–23874. doi: 10.1074/jbc.270.40.23867. [DOI] [PubMed] [Google Scholar]
- 10.Tisne C, Roques BP, Dardel F. The annealing mechanism of HIV-1 reverse transcription primer onto the viral genome. J. Biol. Chem. 2004;279:3588–3595. doi: 10.1074/jbc.M310368200. [DOI] [PubMed] [Google Scholar]
- 11.Isel C, Westhof E, Massire C, Le Grice SFJ, Ehresmann B, Ehresmann C, Marquet R. Structural basis for the specificity of the initiation of HIV-1 reverse transcription. EMBO J. 1999;18:1038–1048. doi: 10.1093/emboj/18.4.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Z, Kang S-M, Li Y, Morrow CD. Genetic analysis of the U5-PBS of a novel HIV-1 reveals multiple interactions between the tRNA and RNA genome required for initiation of reverse transcription. RNA. 1998;4:394–406. [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, Zhang Z, Wakefield JK, Kang S-M, Morrow CD. Nucleotide substitutions within U5 are critical for efficient reverse transcription of human immunodeficiency virus type 1 with a primer binding site complementary to tRNAHis. J. Virol. 1997;71:6315–6322. doi: 10.1128/jvi.71.9.6315-6322.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lanchy J-M, Isel C, Keith G, Le Grice SFJ, Ehresmann C, Marquet R. Dynamics of the HIV-1 reverse transcription complex during initiation of DNA synthesis. J. Biol. Chem. 2000;275:12306–12312. doi: 10.1074/jbc.275.16.12306. [DOI] [PubMed] [Google Scholar]
- 15.Arts EJ, Stetor SR, Li Y, Rausch JW, Howard KJ, Ehresmann B, North TW, Wohrl BM, Goody RS, et al. Initiation of (-) strand DNA synthesis from tRNALys3 on lentiviral RNAs: implications of specific HIV-1 RNA-tRNALys3 interactions inhibiting primer utilization by retroviral reverse trancriptases. Proc. Natl. Acad. Sci. U.S.A. 1996;93:10063–10068. doi: 10.1073/pnas.93.19.10063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang Y, Shalom A, Li Z, Wang J, Mak J, Wainberg MA, Kleiman L. Effects of modifying the tRNALys3 anticodon on the initiation of human immunodeficiency virus type 1 reverse transcription. J. Virol. 1996;70:4700–4706. doi: 10.1128/jvi.70.7.4700-4706.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iwatani Y, Rosen AE, Guo J, Musier-Forsyth K, Levin JG. Efficient initiation of HIV-1 reverse transcription in vitro. Requirement for RNA sequences downstream of the primer binding site abrogated by nucleocapsid protein-dependent primer-template interactions. J. Biol. Chem. 2003;278:14185–14195. doi: 10.1074/jbc.M211618200. [DOI] [PubMed] [Google Scholar]
- 18.Beerens N, Groot F, Berkhout B. Initiation of HIV-1 reverse transcription is regulated by a primer activation signal. J. Biol. Chem. 2001;276:31247–31256. doi: 10.1074/jbc.M102441200. [DOI] [PubMed] [Google Scholar]
- 19.Beerens N, Berkhout B. The tRNA primer activation signal in the HIV-1 genome is important for initiation and processive elongation of reverse transcription. J. Virol. 2002;76:2329–2339. doi: 10.1128/jvi.76.5.2329-2339.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beerens N, Berkhout B. Switching the in vitro tRNA usage of HIV-1 by simultaneous adaptation of the PBS and PAS. RNA. 2002;8:357–369. doi: 10.1017/s1355838202028194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huthoff H, Bugala K, Barciszewski J, Berkhout B. On the importance of the primer activation signal for initiation of tRNA(lys3)-primed reverse transcription of the HIV-1 RNA genome. Nucleic Acids Res. 2003;31:5186–5194. doi: 10.1093/nar/gkg714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abbink TEM, Beerens N, Berkhout B. Forced selection of a human immunodeficiency virus type 1 variant that uses a non-self tRNA primer for reverse transcription: involvement of viral RNA sequences and the reverse transcriptase enzyme. J. Virol. 2004;78:10706–10714. doi: 10.1128/JVI.78.19.10706-10714.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freund F, Boulme F, Litvak S, Tarrago-Litvak L. Initiation of HIV-2 reverse transcription: a secondary structure model of the RNA-tRNA(Lys3) duplex. Nucleic Acids Res. 2001;29:2757–2765. doi: 10.1093/nar/29.13.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berkhout B, Ooms M, Beerens N, Huthoff H, Southern E, Verhoef K. In vitro evidence that the untranslated leader of the HIV-1 genome is an RNA checkpoint that regulates multiple functions through conformational changes. J. Biol. Chem. 2002;277:19967–19975. doi: 10.1074/jbc.M200950200. [DOI] [PubMed] [Google Scholar]
- 25.Berkhout B. Structure and function of the human immunodeficiency virus leader RNA. Prog. Nucleic Acid Res. Mol. Biol. 1996;54:1–34. doi: 10.1016/s0079-6603(08)60359-1. [DOI] [PubMed] [Google Scholar]
- 26.Baudin F, Marquet R, Isel C, Darlix JL, Ehresmann B, Ehresmann C. Functional sites in the 5′ region of human immunodeficiency virus type 1 RNA form defined structural domains. J. Mol. Biol. 1993;229:382–397. doi: 10.1006/jmbi.1993.1041. [DOI] [PubMed] [Google Scholar]
- 27.Abbink TEM, Berkhout B. A novel long distance base-pairing interaction in human immunodeficiency virus type 1 RNA occludes the gag start codon. J. Biol. Chem. 2003;278:11601–11611. doi: 10.1074/jbc.M210291200. [DOI] [PubMed] [Google Scholar]
- 28.Huthoff H, Berkhout B. Two alternating structures for the HIV-1 leader RNA. RNA. 2001;7:143–157. doi: 10.1017/s1355838201001881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huthoff H, Berkhout B. Mutations in the TAR hairpin affect the equilibrium between alternative conformations of the HIV-1 leader RNA. Nucleic Acids Res. 2001;29:2594–2600. doi: 10.1093/nar/29.12.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laughrea M, Jette L. Kissing-loop model of HIV-1 genome dimerization: HIV-1 RNAs can assume alternative dimeric forms, and all sequences upstream or downstream of hairpin 248-271 are dispensable for dimer formation. Biochemistry. 1996;35:1589–1598. doi: 10.1021/bi951838f. [DOI] [PubMed] [Google Scholar]
- 31.Paillart J-C, Skripkin E, Ehresmann B, Ehresmann C, Marquet R. A loop-loop “kissing” complex is the essential part of the dimer linkage of genomic HIV-1 RNA. Proc. Natl. Acad. Sci. U.S.A. 1996;93:5572–5577. doi: 10.1073/pnas.93.11.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ooms M, Huthoff H, Russell R, Liang C, Berkhout B. A riboswitch regulates RNA dimerization and packaging in human immunodeficiency virus type 1 virions. J. Virol. 2004;78:10814–10819. doi: 10.1128/JVI.78.19.10814-10819.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Das AT, Klaver B, Klasens BIF, van Wamel JLB, Berkhout B. A conserved hairpin motif in the R-U5 region of the human immunodeficiency virus type 1 RNA genome is essential for replication. J. Virol. 1997;71:2346–2356. doi: 10.1128/jvi.71.3.2346-2356.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abbink TEM, Ooms M, Haasnoot PCJ, Berkhout B. The HIV-1 leader RNA conformational switch regulates RNA dimerization but does not regulate mRNA translation. Biochemistry. 2005;44:9058–9066. doi: 10.1021/bi0502588. [DOI] [PubMed] [Google Scholar]
- 35.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oude Essink BB, Berkhout B. The fidelity of reverse transcription differs in reactions primed with RNA versus DNA primers. J. Biomed. Sci. 1999;6:121–132. doi: 10.1007/BF02256443. [DOI] [PubMed] [Google Scholar]
- 37.Berkhout B, van Wamel JL. The leader of the HIV-1 RNA genome forms a compactly folded tertiary structure. RNA. 2000;6:282–295. doi: 10.1017/s1355838200991684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Das AT, Berkhout B. Efficient extension of a misaligned tRNA-primer during replication of the HIV-1 retrovirus. Nucleic Acids Res. 1995;23:1319–1326. doi: 10.1093/nar/23.8.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li X, Liang C, Quan Y, Chandok R, Laughrea M, Parniak MA, Kleiman L, Wainberg MA. Identification of sequences downstream of the primer binding site that are important for efficient replication of human immunodeficiency virus type 1. J. Virol. 1997;71:6003–6010. doi: 10.1128/jvi.71.8.6003-6010.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang C, Rong L, Gotte M, Li X, Quan Y, Kleiman L, Wainberg MA. Mechanistic studies of early pausing events during initiation of HIV-1 reverse transcription. J. Biol. Chem. 1998;273:21309–21315. doi: 10.1074/jbc.273.33.21309. [DOI] [PubMed] [Google Scholar]
- 41.Isel C, Marquet R, Keith G, Ehresmann C, Ehresmann B. Modified nucleotides of tRNA(3Lys) modulate primer/template loop-loop interaction in the initiation complex of HIV-1 reverse transcription. J. Biol. Chem. 1993;268:25269–25272. [PubMed] [Google Scholar]
- 42.Elgavish T, VanLoock MS, Harvey SC. Exploring three-dimensional structures of the HIV-1 RNA/tRNALys3 initiation complex. J. Mol. Biol. 1999;285:449–453. doi: 10.1006/jmbi.1998.2310. [DOI] [PubMed] [Google Scholar]
- 43.Harrich D, Hooker B. Mechanistic aspects of HIV-1 reverse transcription initiation. Rev. Med. Virol. 2002;12:31–45. doi: 10.1002/rmv.339. [DOI] [PubMed] [Google Scholar]
- 44.Goldschmidt V, Paillart JC, Rigourd M, Ehresmann B, Aubertin AM, Ehresmann C, Marquet R. Structural variability of the initiation complex of HIV-1 reverse transcription. J. Biol. Chem. 2004;279:35923–35931. doi: 10.1074/jbc.M404473200. [DOI] [PubMed] [Google Scholar]
- 45.Goldschmidt V, Ehresmann C, Ehresmann B, Marquet R. Does the HIV-1 primer activation signal interact with tRNA(3)(Lys) during the initiation of reverse transcription? Nucleic Acids Res. 2003;31:850–859. doi: 10.1093/nar/gkg187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beerens N, Berkhout B. Strict regulation of HIV-1 reverse transcription. Curr. Top. Virol. 2002;2:115–127. [Google Scholar]
- 47.Aiyar A, Cobrinik D, Ge Z, Kung HJ, Leis J. Interaction between retroviral U5 RNA and the T psi C loop of the tRNA(Trp) primer is required for efficient initiation of reverse transcription. J. Virol. 1992;66:2464–2472. doi: 10.1128/jvi.66.4.2464-2472.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]