Abstract
All group II introns known to date fold into six functional domains. However, we recently identified an intron in Bacillus cereus ATCC 10987, B.c.I4, that splices 56 nt downstream of the expected 3′ splice site in vivo (Tourasse et al. 2005, J. Bacteriol., 187, 5437–5451). In this study, we confirmed by ribonuclease protection assay that the 56-bp segment is part of the intron RNA molecule, and computational prediction suggests that it might form a stable stem-loop structure downstream of domain VI. The splicing of B.c.I4 was further investigated both in vivo and in vitro. Lariat formation proceeded primarily by branching at the ordinary bulged adenosine in domain VI without affecting the fidelity of splicing. In addition, the splicing efficiency of the wild-type intron was better than that of a mutant construct deleted of the 56-bp 3′ extension. These results indicate that the intron has apparently adapted to the extra segment, possibly through conformational adjustments. The extraordinary group II intron B.c.I4 harboring an unprecedented extra 3′ segment constitutes a dramatic example of the flexibility and adaptability of group II introns.
INTRODUCTION
Group II introns are a class of genetic retroelements that are capable of self-splicing and mobility. They are able to excise out of RNA transcripts and to ligate their flanking exons (self-splicing), and excised introns may subsequently insert (reverse splice) into identical intron-less DNA sites (process called homing) or into novel genomic locations (retrotransposition) [see (1–4) for reviews]. Group II introns are found in bacteria, archaea and the organelles of fungi, plants and lower eukaryotes, and they either interrupt genes or are inserted in intergenic regions (5–7). These retroelements typically consist of a catalytic RNA (ribozyme) containing an internal open reading frame (ORF) encoding a multifunctional reverse-transcriptase protein, although ORF-less introns do exist, especially in eukaryotic organelles (5). While some introns have been shown to self-splice in vitro in the absence of protein, demonstrating that the splicing reactions are intrinsically catalyzed by the intron RNA, the intronic protein is required for both the splicing and insertion events in vivo (1–4).
To date, known group II intron ribozymes normally fold into conserved secondary structures consisting of six domains that are linked by tertiary interactions, where domain V contains a nucleotide triad that is the presumed catalytic center (3,8,9). However, many degenerate introns in eukaryotic organelles lack RNA substructures (4,10). Differences in structure are used to divide the introns into subclasses (9). Typically, the splicing occurs via a two-step transesterification mechanism that requires magnesium (Mg2+) ions as cofactors (2–4). In the first transesterification step, the 2′ hydroxyl (2′-OH) group of a bulged adenosine (A) located in domain VI makes a nucleophilic attack on the 5′ exon–intron junction phosphodiester bond, leading to cleavage of the 5′ exon and formation of a lariat (circle with tail) with 2′–5′ linkage. Due to similarities in this splicing mechanism that involves the formation of a branched lariat RNA molecule and shared structural features group II introns are thought to be the ancestors of the nuclear spliceosomal introns of eukaryotes (11,12). In the second transesterification step, the free 3′-OH of the cleaved 5′ exon attacks the 3′ splice junction, leading to exon ligation and release of the intron lariat. However, an alternative hydrolytic pathway, in which the first splicing reaction is initiated by nucleophilic attack by a water molecule leading to release of a linear intron after transesterification in the second step of splicing has been shown to occur both in vitro and in vivo (13–15). Group II introns have also been reported to be excised as full or partial circles and several pathways have been suggested (13,16).
Splice-site selection is determined by base-pairing interactions between the intron and its flanking exons that help position the splice junctions in the active site of the intron (Figure 1). At the 5′ splice site two 5–6 nucleotide motifs, IBS1 and IBS2 (intron binding sites), located just upstream of the intron insertion site base-pair with the complementary EBS1 and EBS2 (exon binding sites) in intron domain I. Two single base pair interactions guide 3′ splice-site selection, but they differ between major intron classes. For introns of class IIB, the first nucleotide of the 3′ exon (IBS3) pairs with a nucleotide within domain I [EBS3; (2–4,17)]. The second interaction involves the last base of the intron (γ′) and a base between domain II and III (γ) (Figure 1). The location of the 3′ splice-site relative to the base of domain VI is also important. For group IIB introns, the 3′ splice site occurs 3 or 4 nt past domain VI (10,18,19). It should be noted that the correct positioning of the 3′ splice site is actually highly dependent on domain VI, which helps stabilize the intron structure and the catalytic center (3).
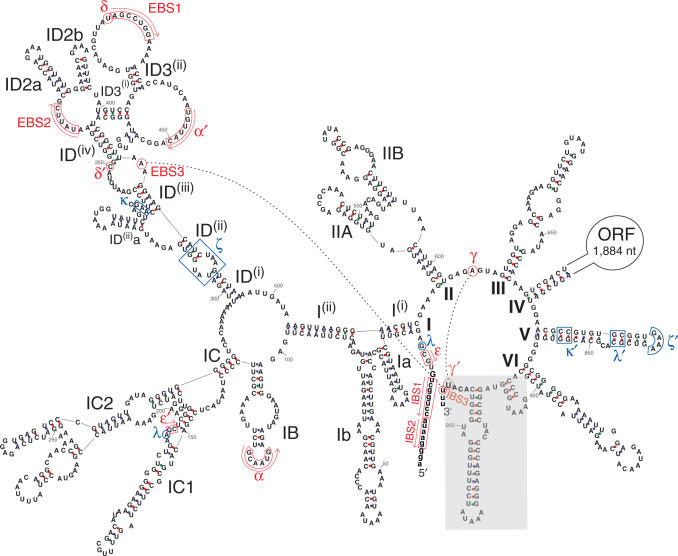
Figure 1.
Predicted secondary structure of the B.c.I4 group II intron from B. cereus ATCC 10987. Exon nucleotides are in lowercase. Roman numerals (I to VI) indicate the six typical functional RNA domains and their subdomains are designated by combinations of letters, numbers and superscripts according to the nomenclature of (10). The extra 56-nt 3′ segment harbored by B.c.I4 is boxed in grey. Sites corresponding to consensus positions involved in tertiary interactions (8,9) are indicated by pairs of Greek letters or EBS/IBS (exon binding sites and intron binding sites). Sites of tertiary base-pairing interactions are boxed or circled in red with arrows indicating the orientation of complementarity. Sites implicated in other tertiary contacts are boxed or circled in blue. The IBS3–EBS3 and γ–γ′ base-pair interactions involved in 3′ splice-site selection are indicated by black dotted lines. The δ′ nucleotide was set at the expected location (C:350) according to (17), however it is not complementary to the δ site, while nucleotide A:349 is. ORF, intron-encoded multifunctional open reading frame. Numbering of residues does not include the ORF. A–U, G–C and G–U base pairs are linked by blue, red and green dots, respectively.
We have recently reported a group IIB intron (B2-like class), B.c.I4, on the pBc10987 plasmid of Bacillus cereus ATCC 10987 that splices in vivo 56 nt downstream of domain VI and the 3′ splice site that would be expected from secondary structure predictions (19). That is, an extra 56-bp segment downstream of the intron was not present in the ligated exons, as indicated by RT-PCR and sequencing. B.c.I4 is inserted within a gene encoding a hypothetical protein with a DNA primase domain (BCEA0033 + BCEA0036), and, interestingly, the unusual splicing puts the ligated exons in-frame and would produce a protein of exactly the same size as the intron-less counterpart in other B. cereus group strains. Furthermore, the observed 3′ splice site would also give the correct γ–γ′ and IBS3–EBS3 interactions, while non-canonical pairings would form three or four bases downstream of domain VI. In the present study, we have examined the splicing of this extraordinary B.c.I4 intron in more detail using in vivo and in vitro experiments. We show that the extra 56-bp 3′ segment is an integral part of the intron RNA molecule downstream of domain VI, which represents a unique arrangement, and that 3′ splice-site selection can be more flexible than ever seen before, while branching is still maintained at the expected site.
MATERIALS AND METHODS
Secondary structure predictions
The secondary structure of the B.c.I4 intron RNA (ORF removed) was computationally predicted by constrained folding using the MFOLD 3.1 package (20) following the consensus structure of group IIB introns from B2-like class (9,19). That is, conserved and identifiable sequence motifs corresponding to the consensus structure were forced during the folding computation.
DNA and RNA isolation
Bacillus cereus ATCC 10987 was grown on Luria Bertani (LB) agar plates at pH 7 and 30°C. An overnight culture (16 h) was inoculated for 3.5 h in 10 ml LB, and then cells were lysed with 10 mg/ml lysozyme. DNA isolation was performed using the Genomic DNA Midi kit (Qiagen) as described by the supplier. Total RNA isolation was conducted as in (19).
PCR and RT-PCR
PCR was performed with Dynazyme II using both the forward and reverse primers at a concentration of 0.4 µM, each deoxynucleoside triphosphate at a concentration of 0.8 mM, and 1 U of Dynazyme (Finnzymes). PCR was primarily run with one denaturation step at 94°C for 5 min, followed by 30–38 cycles of 1 min denaturation at 94°C, 50 s annealing at 58°C and 50 s extension at 72°C, followed by a final extension step of 7 min at 72°C. For RT-PCR, the synthesis of cDNA was initiated with the reverse primers using Superscript III (Invitrogen) and 5 µg of total RNA or 0.1 µg in vitro spliced RNA as a template according to the supplier's protocol. A negative control was conducted without addition of reverse transcriptase. A portion of the cDNA and negative control was then amplified by PCR. A complete listing of all the primers used in this study is given in Table 1.
Table 1.
List of primers used in the study
| Name | Sequence | Location |
|---|---|---|
| I4A_left | TCGGATTTTTGCCGTTAGAG | 5′ exon |
| I4A_right | ACCCCTTCTTATGTCGCAAA | intron (5′ end) |
| I4A_right_nested | CAATCTTAATTCGTTGTGGGTGT | intron (5′ end) |
| I4B_left | CCTGATGTGATCGGAGGTCT | intron (3′ end) |
| I4B_left_lariat | GACATTAAACAGTCGATGGAACG | intron (3′ end) |
| I4B_left_BamHIa | CCCGGATCCCCTGATGTGATCGGAGGTCT | intron (3′ end) |
| I4B_right | TGGTTTCGGAATGGAATCAT | 3′ exon |
| I4B_right_T7a | TAATACGACTCACTATAGGGAGAGCGGGATCCTGGTTTCGGAATGGAATCAT | 3′ exon |
| 5p_left_BamHIa | CGGGATCCCGAAATTGTGTTGGAAGATGATAATACG | 5′ exon |
| 3p_right_ClaIa | CCATCGATGGGGTCCAGATTTCACATGAACAG | 3′ exon |
| 5p_right | ATTCCATGAGCGATTGAGGT | intron |
| 3p_left | AACCTTTAGATTGAGGAAACACAAA | intron |
| d3ps_senseb | CCTTTGGGATGCGTCACAAATTATGAAATGAAGAAAGGACAAC | 3′ splice junction |
| d3ps_antisenseb | GTTGTCCTTTCTTCATTTCATAATTTGTGACGCATCCCAAAGG | 3′ splice junction |
| d56_senseb | CATCAAAGATTTACCTATCGCA[ ]ATTTTATGAAATGAAGAAAGGAC | 3′ splice junction |
| d56_antisenseb | GTCCTTTCTTCATTTCATAAAAT[ ]TGCGATAGGTAAATCTTTGATG | 3′ splice junction |
| Restore_senseb | CATCAAAGATTTACCTATCGCAATTGAAATGGGTAGGCGCTAC | intron (3′ end) |
| Restore_antisenseb | GTAGCGCCTACCCATTTCAATTGCGATAGGTAAATCTTTGATG | intron (3′ end) |
aNucleotides in boldface have been added to the primers in order to include restriction sites or promoters. The restriction sites for BamHI or ClaI are underlined. The T7 promoter sequence is in italics.
bThe sense and antisense primers are complementary to each other and were used to generate the d3ps, d56, and ‘Restore’ mutant constructs using Quikchange II (Stratagene). The mutated nucleotides are in bold. For the d56 mutant, the deletion point is indicated by brackets.
PCR performed with 2.5 U of Pfu Turbo DNA polymerase (Stratagene), with same primers and dNTP concentrations as earlier, was run with one denaturation step at 95°C for 2 min, followed by 30 cycles of 30 s denaturation at 95°C, 30 s annealing at 58°C and 1 min/kb extension at 72°C, followed by a final extension step of 10 min at 72°C.
Radioactive RT-PCR assay
The radioactive RT-PCR assay was mainly conducted as described by Robart et al. (21). Primer I4B_right was 5′-end-labeled with γ[32-P]ATP (3000 Ci/mmol, 10 mCi/ml) using T4 kinase (New England Biolabs). Labeled primer was purified with Nucleotide purification kit (Qiagen). Reverse transcription (cDNA synthesis) and PCR were conducted as described earlier, except that PCR was conducted with ∼5000 cpm of labeled primer I4B_right together with unlabeled counterpart (1 pmol) and primer I4A_left (4 pmol) (Table 1). Amplification products were phenol–CIA (25:24:1 phenol:chloroform:isoamyl alcohol) extracted, ethanol-precipitated with 0.3 M NaOAc, pH 5.2 and digested with SnaBI (New England Biolabs) to ensure homogeneous ends, and these samples were again precipitated and eluted in formamide loading buffer (Ambion). Samples were separated on 7 M urea 6% polyacrylamide gel after heating at 90°C for 2 min and cooling on ice.
Cloning and site-directed mutagenesis
RT-PCR products, either taken directly or gel-purified from 1X TAE gel (QIAquick gel extraction Kit, Qiagen), were cloned into TA cloning vector (Invitrogen) and subsequently sequenced.
Plasmid constructs for in vitro self-splicing experiments were made by cloning a PCR product covering the entire B.c.I4 intron and parts of the flanking exons, amplified with primers 5p_left_BamHI and 3p_right_ClaI (Table 1), into the BamHI site of pBluescript II KS+ (Stratagene), in orientation for T7 transcription. The intron-containing inserts were then amplified by inverse PCR with outward primers, 5p_right and 3p_left, using Pfu Turbo in order to remove the ORF encoded in domain IV of B.c.I4, and then ligated with T4 ligase (New England Biolabs), giving a wild-type construct containing 96 bp of the 5′ exon, 884 bp of intron and 151 bp of the 3′ exon.
Site-directed mutagenesis to generate point mutation and deletion constructs was performed with Quikchange II (Stratagene) according to the manufacturer's instructions using two complementary oligonucleotides (of ∼40 bases) containing the desired mutation(s) (see Table 1 for details). All constructs were verified by sequencing.
In vitro transcription
One microgram of plasmid construct was linearized by XhoI for transcription reactions with 30 U T7 RNA polymerase (Ambion) according to the manufacturer's instructions. For radiolabeled transcripts, transcription was performed using 20 µCi[α–32P] UTP (800 Ci/mmol, Amersham Pharmacia Biosciences), 0.1 mM UTP and 0.5 mM other NTPs. After DNAse treatment, transcripts were gel-purified and resuspended in 10 mM MOPS, pH 7.5. Unlabeled transcripts were synthesized and purified as for labeled transcripts, but with concentrations of 0.5 mM for each NTP and transcripts were gel-purified after visualization with Fluor-coated TLC plates (Ambion).
Ribonuclease protection assay
RNase T1/A protection assay (RPA) was performed using the Ambion RPA III kit following the manufacturer's protocol. The RNA probe was synthesized from a PCR product made using primers I4B_right_T7, containing a T7 promoter and a 9-nt non-homologous sequence, and I4B_left_BamHI (Table 1), creating a 422-nt long product spanning over the 3′ splice junction of intron B.c.I4. The probe was synthesized by in vitro transcription as described earlier, using 0.2 µg of PCR product as template. RPA reactions were performed using 10–25 µg total RNA and ∼60 000 cpm of gel-purified probe as described by the manufacturer. After hybridization and digestion, probe was separated on a denaturing 6% polyacrylamide/7 M urea gel. For imaging, gels were exposed and analyzed using a Molecular Dynamics Storm 860 Phosphorimager.
In vitro self-splicing of ribozyme
In vitro generated transcripts were denatured and refolded using a GenAmp 2700 PCR machine (Applied Biosystems), by incubating the transcripts in 20 mM MOPS, pH 7.6 at 90°C for 1 min, 75°C for 5 min and then cooling to the splicing temperature over 5 min. Intron transcripts were spliced with 50 000 cpm RNA or ∼0.1 µg unlabeled transcripts in 40 mM MOPS, pH 7.6, 100 mM MgCl2 and 500 mM (NH4)2SO4, NH4Cl or KCl at the temperatures indicated in the text. Reactions were initiated by adding pre-warmed splicing buffer to the transcript RNA giving a total reaction volume of 40 µl. At each time point 2 µl were taken, quenched with loading buffer (Ambion) and by placing samples on dry ice. Samples were then heated to 95°C and cooled on ice before being separated on a 7 M urea 4% polyacrylamide gel. Gels were then vacuum dried, exposed and analyzed using a Molecular Dynamics Storm 860 Phosphorimager.
For subsequent RT-PCR and sequencing of these splicing products either unlabeled spliced transcripts, purified with Nucleotide purification kit (Qiagen), or labelled spliced transcript species, excised from gels, were used as templates.
For kinetic analysis the intensities of the radioactive bands were quantified using the ImageQuant 5.0 software and corrected by the number of uridines. The relative fraction of unspliced precursor RNA was computed from the intensities of the radioactive bands. Data were fitted to a biphasic exponential kinetic model [Equation (6) in (22)] by the non-linear least squares method using the GNU gretl 1.5.1 software (http://gretl.sourceforge.net/).
Reverse transcriptase primer extension
The I4B_right primer was 5′-end-labeled with 40 µCi of γ-[32P]ATP and 15 U of T4 polynucleotide kinase. Lariat, lariat with 3′ exon and precursor RNA were incubated with 4 µl of 5× first-strand buffer (Invitrogen), 0.5 µl of RNasOUT (40 U/µl), 2 pmol labeled primer at 85°C for 10 min and then transferred to 55°C for 15 min. The reaction mixture was supplemented with 1 µl of 0.1 M DTT, 40 U of SuperScript III (Invitrogen), 1 µl of 10 mM dNTPs. For primer extension reactions with precursor RNA 1 µl of 4 mM ddCTP was also added. The reaction mixtures with total volume of 20 µl were incubated at 55°C for 25 min, and stopped by heating to 70°C for 15 min. Reaction products were ethanol-precipitated in 0.3 M NaOAc, resuspended in Gel loading buffer II (Ambion), heated to 95°C for 2 min and resolved in 7.5% polyacrylamide 7 M urea gels and visualized using a Storm 860 scanner (Molecular Dynamics).
RESULTS
The extra 56-bp segment downstream of B.c.I4 is part of the intron RNA molecule that splices in vivo
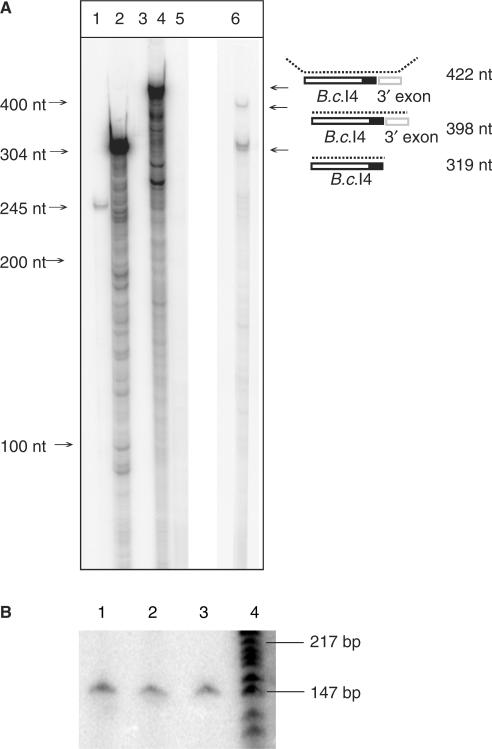
A ribonuclease protection assay was performed to investigate whether the 56-bp sequence immediately downstream of the B.c.I4 intron was part of the excised intron in vivo as it was known to be absent from the spliced exons (19). RNase digestion using a riboprobe covering the intron-3′ exon junction, including the 56-bp sequence, hybridized to total RNA from B. cereus ATCC 10987 gave two bands: (1) a band migrating at ∼400 nt, which matches the size expected for the unspliced (precursor) RNA (398 nt) and (2) a band at 319 nt that corresponds to the size of the spliced intron with the extra 3′ 56-bp segment (Figure 2A). The assay gave no indication of an excised intron without the extra 3′ segment. To detect if there could be a small amount of ligated exons that include the extra 56 nt a sensitive radioactive RT-PCR assay with total RNA was conducted where spliced exons were amplified with 5′-end-labeled primer I4B_right and unlabeled primer I4A_left (see Methods section). A single band at ∼147 bp was obtained, corresponding to the size of the splice junction sequenced previously with the same primers (19), but no band containing the extra element was detected (Figure 2B). We therefore conclude that the 56-bp sequence 3′ of B.c.I4 is part of the spliced intron RNA in vivo.
Figure 2.
RNase T1/A protection assay (A) and radioactive RT-PCR (B) showing that the extra 56-nt element 3′ of the B.c.I4 intron is part of the intron RNA and not part of the exons. In A, lanes 1, 2 and 3 show positive controls based on mouse RNA, and lanes 4, 5 and 6 show the results based on B. cereus RNA. Lane 1: digested antisense mouse β-actin RNA probe hybridized with mouse liver RNA; lane 2: same probe as in lane 1, undigested; lane 3: same probe as in lane 1, digested, without mouse liver RNA; lane 4: undigested B.c.I4-3′exon junction probe hybridized to B. cereus ATCC 10987 total RNA; lane 5: same probe as in lane 4, digested, without RNA sample; lane 6: same probe as in lane 4, digested, with RNA sample. A schematic of the experiment illustrating the location of the probe and the expected products is shown on the right. The black area represents the extra 56-nt element. In B, lanes 1, 2 and 3: RT-PCR conducted with exon-specific primers I4B_right (radiolabeled) and I4A_left (Table 1) using as template total RNA sample isolated from B. cereus ATCC 10987 at 3, 4 and 6 h of growth, respectively. Lane 4: γ[32-P]ATP 5′-end-labeled pBR322 DNA digested with MspI (New England Biolabs), as marker.
The extra 3′ segment of B.c.I4 is predicted to fold into a stable stem-loop structure
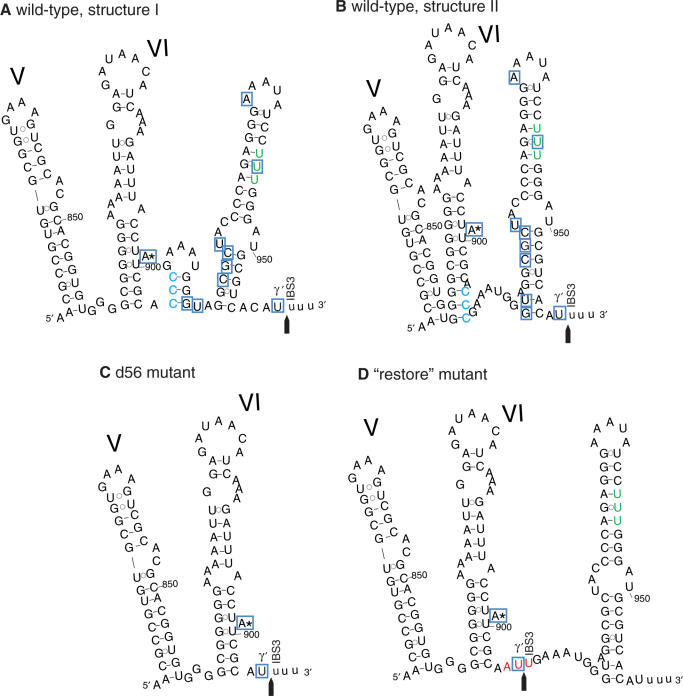
Computational secondary structure predictions suggest that the 56-bp element at the 3′ end of B.c.I4 would be able to form a long stable stem-loop structure as shown in Figure 3, although the folding of the first part of the element is unsure. If folding is conducted by constraining the structure of domain VI to be as it would be in a typical group IIB intron, then the first 12 nt of the 56-nt segment would fold into a small stem–loop element (Figure 3A), whereas they would partly extend the base of the domain VI stem if no constraint is applied on domain VI (Figure 3B). The overall secondary structure of B.c.I4 (excluding the extra segment) displays all the characteristics typical of group IIB introns belonging to the B2-like class (9), in particular two stem–loop structures inserted between subdomains I(i) and I(ii), absence of subdomain IA, a 3-nt linker between domains V and VI and a stretch of 4 bp below the bulged A (A:899; Figure 1 and ref. (19)). Furthermore, all sites involved in tertiary interactions, with the exception of γ′ and IBS3, can be predicted at the expected locations and the motifs match the consensus elements of group IIB2-like introns (Figure 1). Therefore, the structure shown in Figure 3B, which would imply important changes like the disruption of the V–VI linker and an extension of domain VI, might be less likely to form than the structure shown in Figure 3A. Interestingly, no canonical γ–γ′ and IBS3–EBS3 base-pairings could form at the expected splice site three or four bases downstream of domain VI, and both structure predictions suggest that the potential γ′ and IBS3 nucleotides (C:906 and C:907) at this site would already be base-paired and unable to interact with any other nucleotides. The stem–loop structure(s) in the extra segment might enable the 3′ end of the intron to get in closer contact with the rest of the intron core and bring the observed 3′ splice site to a location close to the site expected for typical group IIB introns.
Figure 3.
Predicted secondary structure of the 3′ end of the B.c.I4 group II intron from B. cereus ATCC 10987. Domains V and VI and the extra 56-nt 3′ segment are shown. A and B; two alternative structures for the wild-type intron. C; d56 mutant construct, in which the 56-nt element was removed. D; ‘restore’ construct, in which the 3′ splice site was restored at the expected location typical for group II introns, three bases downstream of domain VI. The nucleotides at the expected 3′ splice site in the wild-type intron, CCC, are colored in blue. These bases were mutated to AUU (colored in red) in the ‘restore’ mutant. The observed 3′ splice junction is indicated by an arrow. Exon residues are in lowercase. The three uridine residues that might create potential 3′ splice sites within the 56-nt 3′ segment, which were not observed to be used, are colored in green. Nucleotides boxed in blue were found to be linked to the intron's 5′ end in inverse RT-PCR experiments. The ordinary branchpoint bulged adenosine (A:899) in domain VI is marked by an asterisk.
In vitro splicing analysis of the B.c.I4 intron
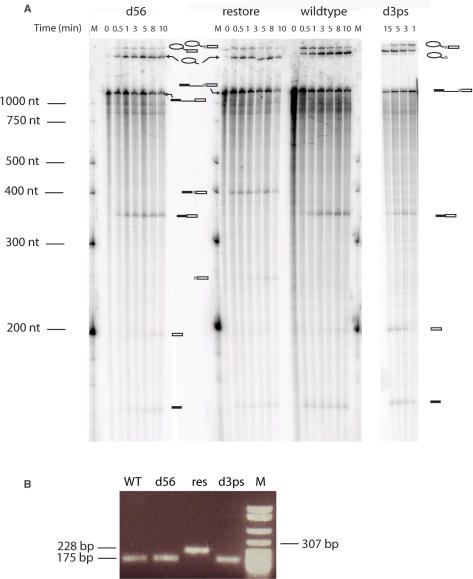
To study the role or impact of the extra 3′ segment on the splicing of the B.c.I4 intron, in vitro self-splicing experiments were conducted using wild-type and mutant constructs in the absence of intron-encoded protein. As can be seen in Figure 4A, splicing of the B.c.I4 wild-type (WT) intron done with 40 mM MOPS (pH 7.5), 500 mM (NH4)2SO4 and 100 mM MgCl2 at 45°C followed by separation of the splicing products on a polyacrylamide gel revealed that the B.c.I4 intron could splice out as a true ribozyme. The size of the ligated exon band observed on the gel (Figure 4A) and RT-PCR with primers I4B_right and 5p_left_BamHI (Table 1) and subsequent sequencing confirmed that the 56 nt 3′ of the intron were not part of the ligated exons, verifying that in vitro splicing of B.c.I4 was the same as in vivo (Figure 4B). When the extra 3′ segment was deleted from the intron (d56 construct), while maintaining the last three nucleotides before the 3′ splice site (Figure 3C), the intron could still splice. As expected the spliced exons were of the same sizes as for the wild type, while the size of the lariat was smaller than the wild-type lariat (Figure 4A). The smaller size of the d56 lariat compared to wild type also confirms that the extra element was part of the lariat form of the wild-type intron that had spliced out. RT-PCR and sequencing confirmed that the ligated exons had the same sequence as in the wild-type case and therefore the intron had spliced at the correct site (Figure 4B). These results indicate that extra 3′ element is not essential for splicing in vitro.
Figure 4.
In vitro self-splicing (A) of B.c.I4 wild-type and mutant constructs and subsequent RT-PCR (B). In A, lane M shows the marker, γ[32-P]ATP 5′-end-labeled RNA Century-Plus Marker (Ambion). Splicing was performed in 40 mM MOPS (pH 7.5), 500 mM (NH4)2SO4, and 100 mM MgCl2 at 45°C. Samples were separated on a 7 M urea 4% polyacrylamide gel. Schematic drawings are shown next to the bands corresponding to the different splicing products. The light grey box represents the extra 56-nt element. In B, RT-PCR with I4B_right and 5p_left_BamH1 primers (Table 1) using in vitro splicing products as templates, confirming the size of the ligated exons. Lane M, pBR322 DNA digested with MspI (New England Biolabs), as marker. Samples were separated on a 1% agarose gel.
We then investigated whether an intron construct carrying the extra 3′ sequence could splice at the location expected for typical group IIB introns by restoring the 3′ splice site three bases downstream of domain VI, i.e. upstream of the extra 3′ segment. In the ‘Restore’ construct, the nucleotides ACCC immediately after domain VI were changed to AATT, thus providing a potential 3′ splice site that could interact with the γ′ and EBS3 nucleotides of B.c.I4, without changing the wild-type 3′ splice-site sequence (Figure 3D). Remarkably, the intron was able to splice at the restored site, as judged by the spliced exons being ∼50 nt longer in size than the wild-type exons, but no product indicative of splicing at the wild-type site 56 bp downstream of domain VI could be observed (Figure 4A). RT-PCR and sequencing of splicing products using primers located in the exons (I4B_right and 5p_left_BamHI; Table 1) confirmed that ligated exons corresponded to splicing at the restored site (Figure 4B). Interestingly, secondary structure predictions using the restored mutant only produced one possible folding in the 3′ end of B.c.I4, whether or not constraints are applied to domain VI. In that structure (Figure 3D), domain VI would adopt the same conformation as in typical group IIB introns, which might explain why the restored site is preferred over the wild-type site. In addition, the possible long linker that may form between the restored site and the stem of the extra 56-bp element may bring the wild-type site too far away from the central core of the intron, thus preventing it from splicing there.
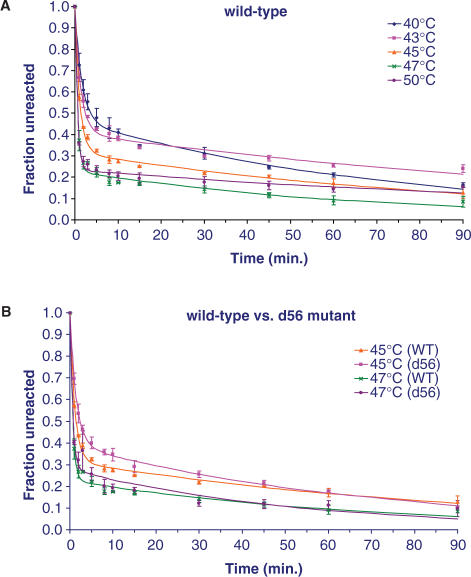
Comparing the splicing of the wild-type B.c.I4 intron with different salts such as (NH4)2SO4, NH4Cl and KCl at the same concentration (500 mM) by a 90-min quantitative time-course analysis showed that the splicing efficiency was best with (NH4)2SO4. Splicing with this latter salt at various temperatures revealed that the rate of splicing, as judged by the fraction of unreacted precursor RNA, was highest between 47 and 50°C (Figure 5A). While previous results obtained with the d56 mutant construct showed that the B.c.I4 intron could splice without the extra 56-bp segment (Figures 3C and 4), a time-course comparison under the presumed optimal conditions revealed that, overall, the d56 mutant intron was not more efficient in splicing than the wild-type B.c.I4 (Figure 5B).
Figure 5.
Time-course analysis of in vitro self-splicing of B.c.I4 wild-type (WT) and d56 mutant constructs. Splicing was performed in buffers containing 100 mM MgCl2 and 500 mM (NH4)2SO4. The relative fractions of unspliced precursor RNA were computed from the intensities of the radioactive bands using a phosphorimager. Reactions were repeated 3× for each construct, and are expressed as averages with standard deviations. Data were fitted to a biphasic exponential kinetic model (Equation (6) in (22); rate constant estimates are provided in supplementary Table 1).
3′ splice-site selection
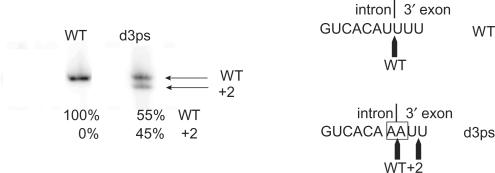
The use of an alternative 3′ splice site by a bacterial group II intron has been shown to occur both in vivo and in vitro in B. anthracis, a close relative of B. cereus (21). Interestingly, the 3′ splice junction of B.c.I4, which is delimited by two uridines (U) corresponding to the γ′ and IBS3 residues, is immediately followed by two more uridines (Figure 3A and B). These latter nucleotides could thus make two possible alternative splice sites where the γ–γ′ and EBS3–IBS3 pairings with the γ and EBS3 adenosines would be maintained. However, sequencing of eight clones of RT-PCR products from in vivo ligated exons did not reveal any use of either two possible alternative splice sites by the B.c.I4 intron, and no alternatively spliced exons could be observed in radioactive RT-PCR assays performed on splicing products from the wild-type RNA both in vivo and in vitro (Figures 2B and 6). Nevertheless, this absence of detection could be due to tiny amounts of alternative splicing. To investigate further whether use of alternative splice sites could be possible, the two first U residues at the intron-3′ exon junction of the wild-type construct were mutated to A nucleotides. Splicing of this mutant, d3ps (Figure 4A), followed by RT-PCR (Figure 4B), cloning and sequencing of five clones of ligated exon products revealed that, in one case, the B.c.I4 intron could splice in between the two uridines at the alternative 3′ site located at position +2 downstream of the wild-type splice site. Very surprising was the fact that the other four clones showed sequences corresponding to splicing at the mutated 3′ splice site, i.e. in between the two adenosines. In this case, no canonical IBS3–EBS3 pairing would form as no U is present in the EBS3 internal loop, while a uridine located in the linker between domain II and III would be the only nucleotide complementary to γ′ adenosine, the last nucleotide of the d3ps intron (Figure 1). This suggests that either splicing occurs without γ–γ′ and/or IBS3–EBS3 pairings or that non-canonical interactions form. Quantification of the 3′ splice site usage by the d3ps intron using radioactive RT-PCR indicated that the mutated and alternative sites were used in 55 and 45% of the splicing, respectively (Figure 6). It should also be noted that there is a potential 3′ splice site inside the extra element, as three uridines can be found (U:943–U:945; green bases in Figure 3), where no splicing was observed.
Figure 6.
Radioactive RT-PCR assay for detecting the use of alternative 3′ splice sites. The assay was conducted with exon-specific primers I4B_right (radiolabeled) and 5p_left_BamHI (Table 1) with wild-type (WT) and d3ps mutant constructs spliced in vitro in 40 mM MOPS (pH 7.5), 500 mM (NH4)2SO4 and 100 mM MgCl2 at 45°C. While no alternative splicing was detected for the WT B.c.I4 intron, the d3ps mutant could use a 3′ splice site at position +2 downstream of the mutated wild-type site. Quantification of the bands using a phosphorimager is shown, expressed as percentage of total radioactivity. The intron-3′ exon splice junction is shown on the right, with mutated nucleotides boxed.
Lariat and circle formation
In typical group II introns domain VI contains a bulged A, which is the branchpoint of the lariat, and has been shown to be important for guiding 3′ splice-site selection usually 7–8 nt downstream of the branchpoint (3). Since intron B.c.I4 splices 56-nt downstream of domain VI, it raises the question of whether the extra 3′ element could have an impact on the branchpoint selection.
B.c.I4 harbors a bulged A at the expected location in domain VI (A:899). To investigate lariat formation in vivo inverse nested RT-PCR experiments were performed using outward-directed primers I4A_right/I4A_right_nested and I4B_left (Table 1), located in the 5′ and 3′ end of the intron, respectively, and total RNA isolated from B. cereus ATCC 10987 (Figure 7A). Gel electrophoresis of products revealed three different bands, of which only the smallest one was clearly visible each time the assay was reproduced (Figure 7B). This band matched the size of a lariat branching at the expected bulged A:899 in domain VI and this was confirmed by cloning and sequencing. In the sequence, the bulged A residue was converted to T, which is a characteristic error due to the reverse-transcriptase which incorporates an A at the cDNA level when it passes through a branched adenosine (15,23). This result shows that B.c.I4 can branch at the typical site within domain VI despite the presence of the extra 3′ segment. The sequence of the largest inverse RT-PCR product (Figure 7B, black arrow) corresponded to molecules in which the first and last intron nucleotides were linked, potentially representing full-length intron circles, while in the middle-size product (Figure 7B, grey arrow) the first intron nucleotide was linked to an internal guanosine residue (G:921; Figure 3A and B) in the extra 3′ element, i.e. the last 38 bases of the intron were missing. In the radioactive RT-PCR experiment shown in Figure 2B, no product corresponding to ligated exons containing the last 38 bases of the intron could be observed, indicating that the G:921 residue was not used as an alternative 3′ splice site. The middle-size product could then represent an alternative lariat form or a partial circle. Therefore, from the earlier results it appears that B.c.I4 can generate a variety of heterogeneous products in vivo. The band corresponding to lariat branching in the expected bulged A:899 was by far the strongest (Figure 7B, white arrow), indicating that this pathway was the major one in vivo under the growth conditions used.
Figure 7.
Inverse RT-PCR analysis for detection of B.c.I4 lariat and other circular molecules. A; schematic drawing illustrating the inverse PCR strategy and the location of primers. B; in vivo analysis; gel electrophoresis of nested RT-PCR products obtained from total RNA isolated from B. cereus ATCC 10987 cultures. C; in vitro analysis; gel electrophoresis of RT-PCR products obtained from in vitro self-splicing of B.c.I4 wild-type (WT). Sequencing chromatograms of selected inverse RT-PCR products are shown to the right. The black, grey and white arrows indicate full-length circles, molecules linked within the extra 56-nt segment, and lariats branched in the bulged A:899, respectively. In B and C, lane M shows the size marker, pBR322 DNA digested with MspI (New England Biolabs); in B, lane NC shows a negative control done without reverse-transcriptase. Primers I4A_right/I4A_right_nested and I4B_left were used in B; I4A_right and I4B_left_lariat were used in C (Table 1). Samples were separated on a 3.5% NuSieve GTG agarose gel (Cambrex).
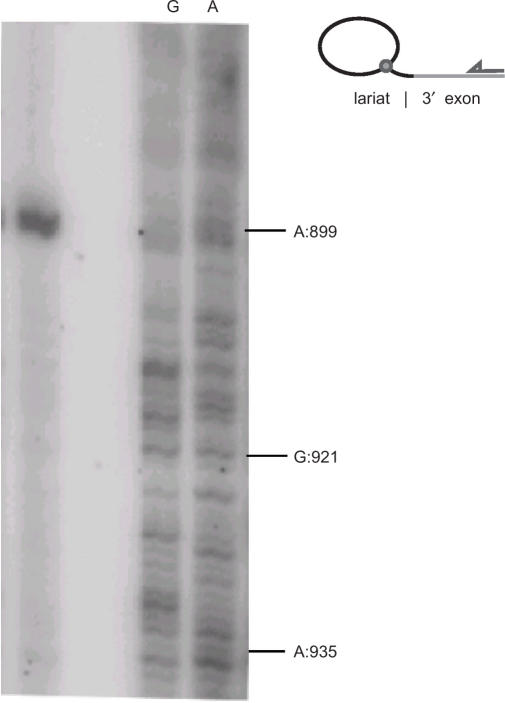
When inverse RT-PCR was conducted on the products of in vitro splicing with the WT construct using primers I4A_right and I4B_left_lariat (Table 1), similar results were obtained (Figure 7C). The three mutant introns (d3ps, d56 and restore) also gave rise to clones corresponding to full circles (data not shown). To investigate whether the molecules linked within the 56-bp 3′ segment could be lariats, inverse RT-PCR was run separately on the fractions corresponding to ‘lariat’ and ‘lariat + 3′ exon’ for the WT construct (top two bands in Figure 4A). Although most of the clones (8 of 13 in ‘lariat’ and 12 of 17 in ‘lariat + 3′ exon’) were lariats branched in A:899, molecules in which the 5′ intron nucleotide was linked to G:921 were found in both fractions, and clones linked to a variety of other sites present in either fraction (Figure 3A and B and 7). Furthermore, a primer extension analysis was conducted on the ‘lariat + 3′ exon’ fraction (top band in Figure 4A) using primer I4B_right located in the 3′ exon. The strongest stop to reverse-transcription was by far at position A:899 confirming that this position is the main branch site (Figure 8). There were no clear other bands that could be indications of stops representing minor ectopic branching events within the extra 56-bp segment of B.c.I4, although this could also be due to a very low frequency of these events, in agreement with the observed frequencies in clones of inverse RT-PCR run on the same fraction.
Figure 8.
Reverse transcriptase primer extension for detection of B.c.I4 lariat RNA. Primer extension was conducted on the ‘lariat + 3′ exon’ fraction obtained from in vitro splicing of the wild-type B.c.I4 intron (top band in Figure 4a) using primer I4B_right located in the 3′ exon. The same reaction was performed on precursor RNA using ddCTP and ddTTP to produce the G and A ladder, respectively. A schematic of the experiment is drawn to the right.
DISCUSSION
In this article, we have investigated the splicing of the atypical group IIB (B2-like class) intron B.c.I4 of B. cereus ATCC 10987 which splices, both in vivo and in vitro, 56 nt downstream of the 3′ site that would be predicted from the classical 6-domain secondary structure of group II introns (19). Here, we present data from three different types of experiments, i.e. ribonuclease protection assay, radioactive RT-PCR and in vitro self-splicing, clearly demonstrating that the extra 3′ 56-nt sequence is a part of the intron that splices out and is not part of the ligated exons (Figures 2 and 4). This is further supported by the fact that this 56-nt sequence stretch is not present in the sequence of the orthologous intron-less gene in B. anthracis (ORF pXO1-70), B. cereus ATCC 43881 and B. thuringiensis ATCC 33679 (19,24). This extraordinary arrangement raises the question of how the intron ribozyme can accommodate the extra element and be functional. In this respect, it is remarkable that the predicted secondary structure of B.c.I4 (apart from the extra segment) conforms perfectly to the consensus structure of group IIB2-like introns (Figure 1). All motifs involved in tertiary interactions are also conserved and no obvious feature deviating from the B2-like consensus is apparent. This might suggest that B.c.I4 has been able to accommodate the extra 3′ segment without major reorganization of the core structure. Use of the downstream 3′ splice site may be facilitated by the formation of a long stable stem–loop structure in the extra 56-nt element that could bring the observed 3′ site closer to the (catalytic) core of the folded intron (Figure 3). The predicted folding of this unusual sequence is supported by the fact that no splicing was observed to occur in the U:943–U:945 stretch, which could make a potential 3′ splice site with IBS3–EBS3 and γ–γ′ pairings, as would be expected if these nucleotides are part of a double-stranded stem (green bases in Figure 3A and B). Furthermore, the nucleotides at the expected 3′ splice site are probably paired with other nucleotides either from the 56-nt element or the linker between domains V and VI (blue bases in Figure 3A and B), therefore preventing splicing at the location typical of group II introns. Even though a more precise determination of the secondary structure would require experimental probing, it is likely that the structure downstream of domain VI is important for the intron to be able to splice after the 56-nt long 3′ element. It would also be interesting to determine whether this element is involved in tertiary interactions with the other intron domains and/or the intron-encoded protein.
When the downstream 3′ segment is removed, as in the d56 construct, the intron can still splice in vitro, indicating that the extra sequence is not essential for B.c.I4 to splice (Figure 4A). On the other hand, the splicing efficiency of the d56 mutant in vitro in (NH4)2SO4 buffer is not better than that of the wild-type overall (as judged from fractions of unspliced precursor during in vitro time-course analysis in Figure 5), implying that the intron has adapted to function with the extra 3′ segment. Another sign of adaptation is given by the fact that the expected branchpoint (A:899) is still the most predominant both in vivo and in vitro, and that B.c.I4 has a high fidelity in branch-site selection (Figures 7 and 8). It has been extensively demonstrated that branch-site selection occurs with a very high fidelity in typical group II introns (25). Thus, this observed fidelity of B.c.I4 is quite remarkable compared to other group II introns in which the bulged A in domain VI is normally located 7–8 nt upstream of the 3′ splice site (3), as opposed to 60 bases.
Mutation of the 3′ splice site (UU→AA) revealed that the B.c.I4 intron could use an alternative site 2 nt downstream in vitro (Figure 6), indicating that B.c.I4 retains some flexibility allowing the selection of an alternative 3′ site. This suggests that the 3′ splice site could come in close proximity to the catatylic center of B.c.I4. On the other hand, the d3ps mutant also spliced at the mutated site, where canonical IBS3–EBS3 and/or γ–γ′ interactions are not expected to occur, implying that there may be structural constraints limiting somewhat the flexibility of the B.c.I4 intron. It could also be that the IBS3–EBS3 and/or γ–γ′ interactions are not critical for 3′ splice-site selection by B.c.I4, as these interactions support correct 3′ splice-site choice only when the 3′ splice site is located near the active center of the intron (3). Alternative splicing of a bacterial group II intron has been reported in B. anthracis, a close relative of B. cereus (21). However, no evidence for such an event could be obtained for the wild-type B.c.I4 intron both in vivo and in vitro under the conditions tested (Figures 2B and 6).
Although the B.c.I4 intron follows a two-step transesterification splicing reaction using the bulged A in domain VI, results from inverse RT-PCR could indicate that minor products may be formed, as suggested by the detection of molecules in which the 5′ end of the intron was linked to a nucleotide inside the extra 56-nt 3′ element or linked to the 3′ end of the intron. These products were obtained both in vivo and in vitro, thus indicating that their formation is not due to the action of the intron-encoded protein or host factors (e.g. RNA ligase). Molecules in which the 5′ and 3′ intron ends are joined are suggestive of fully circularized intron RNAs. Alternatively, they could be the result of reverse splicing into precursor RNA (26,27) or template switch by the reverse-transcriptase during cDNA synthesis (28,29) leading to head-to-tail intron tandems. Fully circular group II intron RNAs have been reported for a number of elements from organelles of fungi and plants (13,15,16) and recently in the bacterium Sinorhizobium meliloti (30,31), either in vitro or in vivo, and some of these introns exhibit obvious unusual features in their 3′ ends. If circle formation does occur, it seems to be a minor pathway for the wild-type B.c.I4 intron under the in vivo and in vitro conditions tested in this study. With respect to the molecules that are linked within the extra 56-nt 3′ segment, whether they represent ectopic lariats or small circles is unclear. Circular products lacking 3′ terminal nucleotides of the intron have been observed in vivo in plant mitochondria (13) and chloroplasts (15). In the case of B.c.I4, this type of molecules does not seem to be a result of splicing or hydrolysis at these ligation points, as the corresponding exonic products were not observed (Figures 2B, 4A and 6). Another alternative is that these molecules are lariats branched within the extra 3′ segment. As shown by Chu et al. (25), intron mutants exhibiting ectopic branching still give correct joined exon products. Recently, ectopic branching was suggested to explain different splicing products obtained when mutating the bulged A in the Lactococcus lactis LtrB intron (32). Similar events in B.c.I4 could be due to structural interference by the 56-nt 3′ element, e.g. when domain VI docks into domain I (33), in a minority of cases. However primer extension assay did not reveal evidence for ectopic branchpoints and we cannot exclude the possibility that these products may be the result of RT-PCR artifacts or structural effects.
The intron characterized in this study, B.c.I4, carrying a 3′ extra segment gives a dramatic new example of the flexibility and adaptability of group II introns. B.c.I4 has been able to accommodate an additional 56 nt and still remains functional for splicing with high fidelity of branch-site and splice-site selection. This is probably due to some favorable conformational adjustments. The versatility of group II introns can also be seen in the splicing ability of elements lacking the branching A or exhibiting various unusual features in domains V and/or VI, which are still able to splice via alternative pathways like hydrolytic splicing or formation of various circular forms (13,15). This adaptability may have contributed to the survival and maintenance of group II introns in genomes where they have to cope with varying physiological conditions affecting splicing.
Finally, a puzzling question relates to the origin of this extra 56-bp element. How did B.c.I4 acquire it and from where? Since the 3′ element is absent from the intron-less gene orthologous to B.c.I4's host gene in other B. cereus group strains, this element probably originated from another genomic context. A PCR screening of a set of 25 B. cereus group strains from our collection using primers specific to B.c.I4 and the 3′ exon (BCEA0036) covering the intron-3′ exon junction indicated that similar B.c.I4 copies including the 56-nt segment are present in homologous host genes in three other strains from the group (results not shown). Therefore, B.c.I4 and its 3′ extra segment are not unique to B. cereus ATCC 10987. However, the latter segment does not show similarity to any other sequence in the public databases (both at the nucleotide and amino-acid levels), thus providing no clues about its origin. It would also be interesting to determine whether B.c.I4 is capable of mobility and whether the extra 56-nt segment has any impact on the ability to reverse splice into DNA sites. Indeed, the B.c.I4-encoded protein contains the endonuclease motif involved in mobility (19).
SUPPLEMENTARY DATA
Supplementary Data is available at NAR Online.
ACKNOWLEDGEMENTS
This work was supported by grants from the Norwegian Research Council through the Strategic University Programme (SUP) and the Functional Genomics (FUGE) and Consortium for Advanced Microbial Sciences and Technologies (CAMST) platform. Funding to pay the Open Access publication charge was provided by CAMST.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bonen L, Vogel J. The ins and outs of group II introns. Trends Genet. 2001;17:322–331. doi: 10.1016/s0168-9525(01)02324-1. [DOI] [PubMed] [Google Scholar]
- 2.Lambowitz AM, Zimmerly S. Mobile group II introns. Annu. Rev. Genet. 2004;38:1–35. doi: 10.1146/annurev.genet.38.072902.091600. [DOI] [PubMed] [Google Scholar]
- 3.Lehmann K, Schmidt U. Group II introns: structure and catalytic versatility of large natural ribozymes. Crit. Rev. Biochem. Mol. Biol. 2003;38:249–303. doi: 10.1080/713609236. [DOI] [PubMed] [Google Scholar]
- 4.Robart AR, Zimmerly S. Group II intron retroelements: function and diversity. Cytogenet. Genome Res. 2005;110:589–597. doi: 10.1159/000084992. [DOI] [PubMed] [Google Scholar]
- 5.Dai L, Zimmerly S. Compilation and analysis of group II intron insertions in bacterial genomes: evidence for retroelement behavior. Nucleic Acids Res. 2002;30:1091–1102. doi: 10.1093/nar/30.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dai L, Zimmerly S. ORF-less and reverse-transcriptase-encoding group II introns in archaebacteria, with a pattern of homing into related group II intron ORFs. RNA. 2003;9:14–19. doi: 10.1261/rna.2126203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Toro N. Bacteria and archaea group II introns: additional mobile genetic elements in the environment. Environ. Microbiol. 2003;5:143–151. doi: 10.1046/j.1462-2920.2003.00398.x. [DOI] [PubMed] [Google Scholar]
- 8.Qin PZ, Pyle AM. The architectural organization and mechanistic function of group II intron structural elements. Curr. Opin. Struct. Biol. 1998;8:301–308. doi: 10.1016/s0959-440x(98)80062-6. [DOI] [PubMed] [Google Scholar]
- 9.Toor N, Hausner G, Zimmerly S. Coevolution of group II intron RNA structures with their intron-encoded reverse transcriptases. RNA. 2001;7:1142–1152. doi: 10.1017/s1355838201010251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michel F, Umesono K, Ozeki H. Comparative and functional anatomy of group II catalytic introns–a review. Gene. 1989;82:5–30. doi: 10.1016/0378-1119(89)90026-7. [DOI] [PubMed] [Google Scholar]
- 11.Seetharaman M, Eldho NV, Padgett RA, Dayie KT. Structure of a self-splicing group II intron catalytic effector domain 5: parallels with spliceosomal U6 RNA. RNA. 2006;12:235–247. doi: 10.1261/rna.2237806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valadkhan S. snRNAs as the catalysts of pre-mRNA splicing. Curr. Opin. Chem. Biol. 2005;9:603–608. doi: 10.1016/j.cbpa.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 13.Li-Pook-Than J, Bonen L. Multiple physical forms of excised group II intron RNAs in wheat mitochondria. Nucleic Acids Res. 2006;34:2782–2790. doi: 10.1093/nar/gkl328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Podar M, Chu VT, Pyle AM, Perlman PS. Group II intron splicing in vivo by first-step hydrolysis. Nature. 1998a;391:915–918. doi: 10.1038/36142. [DOI] [PubMed] [Google Scholar]
- 15.Vogel J, Börner T. Lariat formation and a hydrolytic pathway in plant chloroplast group II intron splicing. EMBO J. 2002;21:3794–3803. doi: 10.1093/emboj/cdf359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murray HL, Mikheeva S, Coljee VW, Turczyk BM, Donahue WF, Bar-Shalom A, Jarrell KA. Excision of group II introns as circles. Mol. Cell. 2001;8:201–211. doi: 10.1016/s1097-2765(01)00300-8. [DOI] [PubMed] [Google Scholar]
- 17.Costa M, Michel F, Westhof E. A three-dimensional perspective on exon binding by a group II self-splicing intron. EMBO J. 2000;19:5007–5018. doi: 10.1093/emboj/19.18.5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dai L, Toor N, Olson R, Keeping A, Zimmerly S. Database for mobile group II introns. Nucleic Acids Res. 2003;31:424–426. doi: 10.1093/nar/gkg049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tourasse NJ, Stabell FB, Reiter L, Kolstø AB. Unusual group II introns in bacteria of the Bacillus cereus group. J. Bacteriol. 2005;187:5437–5451. doi: 10.1128/JB.187.15.5437-5451.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robart AR, Montgomery NK, Smith KL, Zimmerly S. Principles of 3′ splice site selection and alternative splicing for an unusual group II intron from Bacillus anthracis. RNA. 2004;10:854–862. doi: 10.1261/rna.5246804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daniels DL, Michels WJ, Jr, Pyle AM. Two competing pathways for self-splicing by group II introns: a quantitative analysis of in vitro reaction rates and products. J. Mol. Biol. 1996;256:31–49. doi: 10.1006/jmbi.1996.0066. [DOI] [PubMed] [Google Scholar]
- 23.Vogel J, Hess WR, Börner T. Precise branch point mapping and quantification of splicing intermediates. Nucleic Acids Res. 1997;25:2030–2031. doi: 10.1093/nar/25.10.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pannucci J, Okinaka RT, Sabin R, Kuske CR. Bacillus anthracis pXO1 plasmid sequence conservation among closely related bacterial species. J. Bacteriol. 2002;184:134–141. doi: 10.1128/JB.184.1.134-141.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chu VT, Adamidi C, Liu Q, Perlman PS, Pyle AM. Control of branch-site choice by a group II intron. EMBO J. 2001;20:6866–6876. doi: 10.1093/emboj/20.23.6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang J, Mohr G, Perlman PS, Lambowitz AM. Group II intron mobility in yeast mitochondria: target DNA-primed reverse transcription activity of aI1 and reverse splicing into DNA transposition sites in vitro. J. Mol. Biol. 1998;282:505–523. doi: 10.1006/jmbi.1998.2029. [DOI] [PubMed] [Google Scholar]
- 27.Zimmerly S, Guo H, Eskes R, Yang J, Perlman PS, Lambowitz AM. A group II intron RNA is a catalytic component of a DNA endonuclease involved in intron mobility. Cell. 1995;83:529–538. doi: 10.1016/0092-8674(95)90092-6. [DOI] [PubMed] [Google Scholar]
- 28.Sellem CH, Begel O, Sainsard-Chanet A. Recombinant mitochondrial DNA molecules suggest a template switching ability for group-II-intron reverse transcriptase. Curr. Genet. 2000;37:24–28. doi: 10.1007/s002940050003. [DOI] [PubMed] [Google Scholar]
- 29.Tuschl T, Sharp PA, Bartel DP. Selection in vitro of novel ribozymes from a partially randomized U2 and U6 snRNA library. EMBO J. 1998;17:2637–2650. doi: 10.1093/emboj/17.9.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Costa M, Michel F, Molina-Sanchez MD, Martinez-Abarca F, Toro N. An alternative intron-exon pairing scheme implied by unexpected in vitro activities of group II intron RmInt1 from Sinorhizobium meliloti. Biochimie. 2006;88:711–717. doi: 10.1016/j.biochi.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 31.Molina-Sanchez MD, Martinez-Abarca F, Toro N. Excision of the Sinorhizobium meliloti group II intron RmInt1 as circles in vivo. J. Biol. Chem. 2006;281:28737–28744. doi: 10.1074/jbc.M602695200. [DOI] [PubMed] [Google Scholar]
- 32.Slagter-Jäger JG, Allen GS, Smith D, Hahn IA, Frank J, Belfort M. Visualization of a group II intron in the 23 s rRNA of a stable ribosome. Proc. Natl. Acad. Sci. U.S.A. 2006;103:9838–9843. doi: 10.1073/pnas.0603956103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hamill S, Pyle AM. The receptor for branch-site docking within a group II intron active site. Mol. Cell. 2006;23:831–840. doi: 10.1016/j.molcel.2006.07.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.