Abstract
Trichosanthin (TCS) is a type I ribosome-inactivating protein that inactivates ribosome by enzymatically depurinating the A4324 at the α-sarcin/ricin loop of 28S rRNA. We have shown in this and previous studies that TCS interacts with human acidic ribosomal proteins P0, P1 and P2, which constitute the lateral stalk of eukaryotic ribosome. Deletion mutagenesis showed that TCS interacts with the C-terminal tail of P2, the sequences of which are conserved in P0, P1 and P2. The P2-binding site on TCS was mapped to the C-terminal domain by chemical shift perturbation experiments. Scanning charge-to-alanine mutagenesis has shown that K173, R174 and K177 in the C-terminal domain of TCS are involved in interacting with the P2, presumably through forming charge–charge interactions to the conserved DDD motif at the C-terminal tail of P2. A triple-alanine variant K173A/R174A/K177A of TCS, which fails to bind P2 and ribosomal stalk in vitro, was found to be 18-fold less active in inhibiting translation in rabbit reticulocyte lysate, suggesting that interaction with P-proteins is required for full activity of TCS. In an analogy to the role of stalk proteins in binding elongation factors, we propose that interaction with acidic ribosomal stalk proteins help TCS to locate its RNA substrate.
INTRODUCTION
Trichosanthin (TCS) is a type I ribosome-inactivating protein (RIP) isolated from the root tuber of Trichosanthes kirilowii. It is used clinically to treat hydatidiform moles, trophoblastic carcinomas, ectopic pregnancies and to terminate early and mid-trimester pregnancies (1). TCS, like its homolog ricin A-chain, inactivates ribosome through its RNA N-glycosidase activity that depurinates an invariant adenine residue, A4324 (numbering according to the rat sequence), in the conserved α-sarcin/ricin loop (SRL) of 28S rRNA in eukaryotic ribosomes (2–6). Such modification prevents binding of elongation factors to the SRL, and leads to the arrest of protein synthesis (7,8).
The crystal structure of TCS (9) reveals that the protein consists of two domains: a large N-terminal domain (residues 1–172) consists of six alpha-helices, a six-stranded mixed beta-sheet and a two-stranded anti-parallel sheet, and a small C-terminal domain (residues 182–247), contains an anti-parallel beta-sheet and an alpha-helix with a bend in the middle. The two domains are connected by a loop (residues 173–181) rich in basic residues (K173, R174 and K177). The active site of TCS is located at the cleft between the N-terminal and the C-terminal domains.
Although RIP is able to cleave naked rRNA, its kcat is 105-fold slower than that for rRNA within an intact ribosome (6). This finding strongly suggests that ribosomal proteins are involved in rendering the rRNA susceptible to inactivation by RIP. However, reports on the interaction between RIP and ribosomal proteins are scarce. In an early study, ricin A-chain was cross-linked to ribosomal proteins L9 and L10e (P0) of mammalian ribosome (10). Pokeweed antiviral protein, another type I RIP, was found to interact with yeast ribosomal protein L3 (11,12). Based on a yeast two-hybrid screening and confirmed by in vitro pull-down assay, we have recently shown that TCS interacts with acidic ribosomal proteins P0 and P1 (13). In eukaryotic ribosomes, P0, P1 and P2 form a pentameric P-complex P0(P1)2(P2)2, which constitutes the ribosomal stalk (14). P-complex docks on the ribosome through the interaction with 28S rRNA, and forms the GTPase-associated centre for binding of elongation factors during protein synthesis (15,16). The P-complex determines the specificity of ribosomes for eukaryotic elongation factors. For example, swapping the prokaryotic stalk proteins (L10 · L7/124) with P-complex changes the specificity of E. coli ribosomes to use eukaryotic elongation factors for translation (17,18). Cryo-electron microscopy map shows that the ribosomal stalk extends outwards from the ribosome, and may play a role in fetching the elongation factors from the cytosol to the GTPase-associated centre of ribosome (19). All ribosomal stalk proteins, P0, P1 and P2, possess a conserved amino acid sequence rich in acidic residues in their C-termini (14). Monoclonal antibody against this consensus sequence can block the binding of elongation factors to the P-complex and the ribosome-dependent GTPase activities in protein synthesis in vitro (20). Given the sequence similarity of the C-termini of P-proteins, we hypothesize that TCS may bind P2 as well.
In this study, we first demonstrate by in vitro pull-down assay that TCS can bind P2. Deletion mutagenesis has shown that TCS interacts with the conserved C-terminal tail of P2, which explains why TCS can interact with all acidic ribosomal proteins. The P2-binding site on TCS was mapped by chemical shift perturbation to the C-terminal domain of TCS. Charge-to-alanine scanning mutagenesis has identified three basic residues, K173, R174 and K177, that are involved in binding P2, presumably forming salt bridges to conserved acidic residues of P2. We have found that triple-alanine substitutions at positions 173, 174 and 177, which abolished the interaction with P2 and with ribosomal stalk, resulted in a TCS variant with about 18-fold less active in inhibiting a rabbit reticulocyte protein synthesis system. Based on our results, the role of interaction with P-proteins in the ribosome-inactivating activity of TCS is discussed.
MATERIALS AND METHODS
Cloning and site-directed mutagenesis
TCS variants
E123A, K173A, R174A, D176A, K177A, E189A, K197A, E210A, R222A, D229A, R243A and K173A/R174A/K177A were created by polymerase chain reaction (PCR) using mutagenic primers and were subcloned to pET3d (Novagen). Expression and purification of TCS mutants were performed as described for wild-type TCS (21).
P2 and its variants
The gene encoding P2 was amplified by PCR (forward primer: 5′ CAC GCC CAT GGC GAT GCG CTA CGT CGC CTC C 3′; reverse primer: 5′ CAC AGG GAT CCT AGT CAA AGA CAC CAA ATC C 3′, with underlined NcoI and BamHI sites, respectively) from the human HTLV-transformed cDNA library. P2 C-terminal deletion mutants, P2ΔC1, P2ΔC2, P2ΔC3, P2ΔC4, P2ΔC5, P2ΔC17, P2ΔC28, P2ΔC46 and P2 DDD(106–108)AAA were created by PCR using mutagenic primers and were subcloned to pET3d. To create MBP-C7, MBP-C11, MBP-C14 and MBP-C17, synthetic double-stranded oligonucleotides encoding the last 7, 11, 14 and 17 residues of P2 were subcloned to an in-house pRSETA-MBP vector, in which the poly-histidine tag of pRSETA (Invitrogen) was replaced by maltose-binding protein. To create MBP-C29 and MBP-C36, DNA fragment encoding the last 29 and 36 residues of P2 were obtained by PCR and were subcloned to the pRSETA-MBP vector.
Preparation of protein samples
Proteins were expressed in E. coli strain C41(DE3) (22) or BL21(DE3, pLysS) (Novagen) in M9 minimal medium (6 g/l Na2HPO4, 3 g/l KH2PO4, 0.5 g/l NaCl, 2 mM MgSO4, 0.1 mM CaCl2, 1 mM thiamine-HCl) or M9ZB medium (1 g/l of NH4Cl, 3 g/l of KH2PO4, 6 g/l of Na2HPO4, 5 g/l of NaCl, 10 g/l of bacto-tryptone, 4 g/l of glucose, 1 mM MgSO4) containing the appropriate antibiotics (50 μg/ml of chloramphenicol and/or 100 μg/l of ampicillin). To prepare 15N-, 13C-, 2H-labelled NMR sample, TCS was expressed in M9 medium prepared in D2O containing 1 g/l 15NH4Cl and/or 2 g/l 13C-glucose. Bacterial cells were grown in 37°C until OD600 reached 0.4–0.8, when the expression was induced by 0.4 mM (IPTG). The cells were harvested after overnight culture at 37°C by centrifugation at 3700 g at 4°C for 10 min.
To purify TCS, cell lysate was loaded to a CM-Sepharose (Amersham) column pre-equilibrated with buffer A (20 mM phosphate buffer, pH 6.5), and eluted using a gradient of 0–0.5 M NaCl in buffer A over 240 ml. Fractions containing TCS were pooled and dialysed against buffer A, and loaded to a HiTrap™ SP column (Amersham). The protein was eluted using a gradient of 0–0.3 M NaCl in buffer A over 240 ml. To purify P2, P2ΔC1, P2ΔC2, P2ΔC3, P2ΔC4, P2ΔC5 and P2 DDD(106–108)AAA, cell lysate was loaded to a DEAE-Sepharose (Amersham) column pre-equilibrated with buffer A at pH 7.8, and was eluted using a linear gradient of 0–0.5 M NaCl in buffer A over 240 ml. Fractions were pooled and dialysed against 20 mM Tris/HCl, 1.5 M NaCl at pH 7.8 and then loaded to a 5 ml HiTrap™ Phenyl HP column (Amersham). P2 and its variants were collected in the flow-through, dialysed against buffer A, and loaded to a 5 ml HiTrap™ Q anion-exchange column pre-equilibrated with buffer A at pH 7.8. The proteins were eluted by a NaCl gradient of 0–0.3 M in 300 ml of buffer A. To purify P2ΔC17, P2ΔC28 and P2ΔC46, protein fractions from the DEAE-Sepharose column were concentrated to 5 ml before loading to a HiLoad™ 26/60 Superdex 75 column (Amersham) pre-equilibrated with 20 mM Tris/HCl, 0.2 M NaCl at pH 8.5. Fractions containing the P2 variants were pooled, dialysed against buffer A at pH 8.5, and then loaded to a 5 ml HiTrap™ ANX or Q anion exchange column. A gradient of 0–0.2 M NaCl over 200 ml was used to elute the proteins.
NMR spectroscopy
All NMR experiments were carried out using a 1 mM protein sample containing 5% D2O in a four-channel Varian Unity INOVA 750-MHz spectrometer with z-axis pulsed-field gradient capabilities for sensitivity enhancement and water suppression at 303 K. Backbone resonance assignment of TCS was achieved by the approach of triple resonance experiments. HNCACB (23), HN(CO)CACB, HNCA and HN(CO)CA (24) were acquired on a 15N, 13C, 2H triply labelled sample. The assignment was confirmed by short-range nuclear Overhauser enhancements based on NOESY-HSQC (100 ms mixing time) and TOCSY-HSQC experiments (25) acquired on a 15N-labelled sample. Data were processed using the software NMRPipe (26), and the spectra analysed by NMRView (27). Chemical shifts on the 13C, and 15N dimensions were referenced using an indirect method using the water resonances as an internal reference (28,29).
Chemical shift perturbation
Unlabelled P2 (1.5 mM sample in 20 mM Tris/HCl, pH 7.8) was titrated stepwise into 350 μl of 1.1 mM 15N-labelled TCS. The protein concentrations at five titration points were 0, 0.26, 0.44, 0.57 and 0.68 mM for P2, and 1.1, 0.92, 0.78, 0.68 and 0.60 mM for TCS, respectively. At each titration point, a 1H, 15N correlation spectrum of TCS was recorded. All spectra were processed by NMRPipe and analysed by NMRView. The changes in 1H and 15N chemical shifts of backbone amide of TCS upon interactions with P2 were followed.
In vitro pull-down assay
TCS or P2 was coupled to NHS-activated Sepharose (Amersham) according to the procedures recommended by the manufacturer. To test if P2 and its variants interact with TCS, 50 μM of protein samples were loaded to the TCS-coupled NHS column pre-equilibrated with binding buffer (20 mM Tris/HCl, 10 mM NaCl, pH 8.0). After washing extensively with the binding buffer, the protein was eluted with 0.1 M glycine, 0.5 M NaCl, pH 3.0 or 20 mM Tris/HCl, 1 M NaCl, pH 8.0. To test if TCS and its variants interact with P2, 250 μl of 0.5 mg/ml of protein sample was loaded to a P2-coupled NHS-Sepharose (200 μl resin) pre-equilibrated with the binding buffer. After washing the column extensively with the binding buffer, the protein was eluted with 20 mM Tris/HCl, 1 M NaCl, pH 8.0.
Interactions between TCS and ribosomes
Pull-down assay
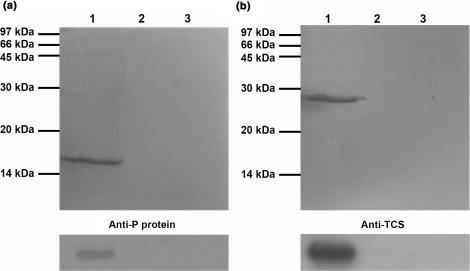
80S ribosome was prepared from rat liver as described (30). Here, 150 μl of rat ribosome (OD260 = 23.5) was loaded to a TCS-coupled NHS column pre-equilibrated with 20 mM Tris/HCl, 150 mM NaCl, 5 mM EDTA, 1% NP-40, 0.1% SDS, pH 8.0. After extensive washing, bound proteins were eluted with 20 mM Tris/HCl, 1 M NaCl, pH 8.0, and detected by western blotting using anti-P antibody.
Cross-linking
Here, 3 μg of TCS was added to 105 μl of rat ribosome (OD260 = 23.5) in 150 mM potassium phosphate buffer at pH 7.4. After incubation at room temperature for 20 min, 0.41 μg of disuccinimidyl suberate (DSS; Pierce) was added to the reaction mixture. Cross-linking reaction was allowed to occur for 30 min at room temperature before it was quenched by 50 mM Tris buffer at pH 7.5. The cross-linked product was then analysed by western blotting using anti-P and rabbit anti-TCS antibodies.
Translation inhibition assay
In vitro translation was performed as described (31) using nuclease-untreated rabbit reticulocyte lysate, which uses endogenous mRNA template for protein synthesis. In brief, 3.7 pM to 370 nM of TCS or 75 μg/ml of bovine serum albumin (BSA) was added to the rabbit reticulocyte lysate supplemented with 10 mM creatine phosphate, 35 μg/ml creatine kinase, 67 mM KCl, 0.33 mM MgCl2, 35 μg/ml haemin and 5 nCi [3H]-leucine (PerkinElmer). After incubation at 30°C for 30 min, the mixture was decolourized by NaOH/H2O2. The [3H]-labelled translation product was then precipitated by 25% (v/v) trichloroacetic acid, collected by filtration, and quantified by liquid scintillation counting. The amount of [3H]-leucine incorporation relative to that of the BSA control was plotted as a function of TCS concentration, and the values of IC50 were determined by fitting the data to a four-parameter logistic equation using non-linear regression.
Depurination assay
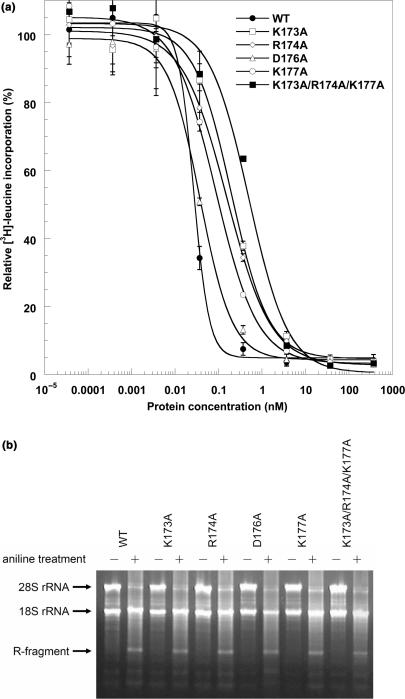
Specific depurination of A-4324 of 28S rRNA by TCS was detected by aniline treatment as described (31) with minor modification. In brief, 10 nM TCS and its variants were incubated with rabbit reticulocyte lysate at 37°C for 30 min. RNA was extracted by TRIzol® reagent (Invitrogen), treated with acidic aniline at 60°C for 5 min, and precipitated by ethanol. The RNA pellet was dissolved in water, and analysed by agarose gel electrophoresis in TAE buffer.
Molecular modelling of TCS–rRNA complex
The coordinates of the SRL were obtained from the crystal structure of the large ribosomal subunit from Haloarcula marismortui (PDB code: 1FFK) (32), and the coordinates of TCS were obtained from the crystal structure of TCS complexed with adenine (PDB code: 1MRJ). The adenine ring of A2697 (analogous to A4324 in rat 28S rRNA) was flipped to the syn conformation (as in the conformation of formycin monophosphate in the active site of ricin, PDB code: 1FMP), and was positioned to the adenine bound to the active site of TCS. The SRL was docked manually to the active site of TCS, with the guidance of a published model of pokeweed antiviral protein–SRL complex (33,34). The model was energy minimized using the program CNS (35). During the minimization, only the loop residues, G2696–A2699, were allowed to be flexible, while other parts of SRL and TCS were restrained as a rigid body.
RESULTS
TCS interacts with acidic ribosomal stalk protein P2
Based on yeast two-hybrid screening and confirmed by in vitro pull-down assay, our group has previously demonstrated that TCS interacts with human acidic ribosomal proteins P0 and P1 (13). Since P2 belongs to the same family of acidic ribosomal protein, we hypothesize that P2 can also interact with TCS. The interaction between TCS and P2 was investigated by in vitro pull-down assay. As shown in Figure 1a, P2 bound to the TCS-coupled NHS-Sepharose (lane 1), while no retention of the BSA control was observed (lane 2). The binding between TCS and P2 was further confirmed by loading TCS to P2-coupled NHS-Sepharose. It was found that TCS, but not the BSA control, bound to the P2-coupled Sepharose (Figure 1b, lanes 1 and 2). Non-specific binding of TCS and P2 to the Sepharose column was ruled out by loading TCS or P2 to an uncoupled NHS-Sepharose (Figure 1a and b, lane 3).
Figure 1.
TCS interacts with P2 in vitro. (a) P2 was loaded to TCS-coupled NHS-activated column pre-equilibrated with binding buffer. After extensive washing, P2 was eluted with 1 M NaCl (lane 1). (b) TCS was loaded to P2-coupled NHS-activated column pre-equilibrated with binding buffer. After extensive washing, TCS was eluted with 1 M NaCl (lane 1). The eluted proteins were analysed by SDS-PAGE with Coomassie blue staining (upper panel), and the identity of P2 and TCS was confirmed by western blot (lower panel) using anti-P and anti-TCS antibody, respectively. In both cases, two negative controls were carried out. First, BSA was loaded to the TCS- or P2- coupled column and no BSA was found in the elution (lane 2). Second, P2 or TCS were loaded to uncoupled NHS-Sepharose, and no P2 or TCS were found in the elution (lane 3).
Systematic deletion studies showed that TCS interacts with the conserved C-terminal tail of P2
Sequence alignment of P0, P1 and P2 reveals that the three acidic ribosomal stalk proteins share a conserved C-terminal region rich in acidic residues (Figure 2a and b). Since TCS interacts with all acidic ribosomal proteins, we hypothesize that the conserved C-terminal region of P2 is involved in binding TCS. To test this hypothesis, we created systematic deletion mutants of P2 (P2ΔC1, P2ΔC2, P2ΔC3, P2ΔC4, P2ΔC5, P2ΔC17, P2ΔC28 and P2ΔC46, where the number after the Δ sign indicates the number of residues deleted from the C-terminus). The interaction between TCS and these deletion mutants of P2 was checked by in vitro pull-down assay (Figure 2c and d). Confirming the role of C-terminus of P2 in binding TCS, our data showed that deletion of two or more residues at the C-terminus of P2 would seriously compromise or completely abolish the interaction between TCS and P2 (Figure 2c and d).
Figure 2.
TCS interacts with the conserved C-terminal region of P2. (a) The primary sequence of human P2 is aligned with the C-terminal residues of other acidic ribosomal stalk proteins P0 and P1. The last 17 residues of P0, P1 and P2 are highly conserved and are highlighted in the figure. (b) The last 11 residues of P-proteins are highly conserved. The sequences of the C-terminal residues of P-proteins from the SWISS-PROT database were aligned by the program CLUSTAL W (41), and are shown in a sequence logo representation (42) generated by a web-based program WebLogo (43). Sequences are numbered according to the human P2 sequence. (c and d) The interaction between C-terminal deletion mutants of P2 and TCS was checked by in vitro pull-down assay. In (c), deletion mutants of P2 were loaded to TCS-coupled NHS-Sepharose; while in (d), TCS was loaded to NHS-Sepharose coupled with P2 deletion mutants. The elution fractions from the pull-down assay were analysed by 15% SDS-PAGE stained with Coomassie blue. The presence of P2 mutants in (c) or TCS in (d) in the elution fractions indicates positive interaction between TCS and the P2 deletion mutants. In both (c and d), wild-type P2 was included as a positive control. (e) In vitro pull-down assay on the interaction between TCS and C-terminal tail of P2. The last 7, 11, 14, 17, 29, 36 residues of P2 were fused to MBP to create MBP-C7, MBP-C11, MBP-C14, MBP-C17, MBP-C29 and MBP-C36 fusion proteins. Bacterial lysates containing these fusion proteins were loaded to a TCS-coupled NHS-Sepharose column. The elution fractions from the pull-down assay were analysed by 15% SDS-PAGE stained with Coomassie blue. Our data indicate that MBP-C11, MBP-C14, MBP-C17, MBP-C29, MBP-C36 fusion proteins, but not the MBP control, MBP-C7 and MBP-C11 DDD(106–108)AAA, were retained by the TCS-coupled column. The results of the in vitro pull-down assay are summarized in (f).
Although the acidic ribosomal proteins share a conserved C-terminal region, their sequences differ in their N-terminal domains. It is likely that the C-terminal region of P2 alone is responsible for binding TCS. To test this hypothesis, we fused the last 7, 11, 14, 17, 29, 36 residues of P2 to the maltose binding protein (MBP) to create MBP-C7, MBP-C11, MBP-C14, MBP-C17, MBP-C29 and MBP-C36 fusion proteins, and tested the interaction between TCS and these fusion proteins by in vitro pull-down assay. Our data showed that the MBP fusion proteins with the last eleven or longer residues of P2 were retained on TCS-coupled Sepharose, while MBP-C7 failed to bind TCS (Figure 2e). These data suggested that the last eleven residues of P2 are sufficient for interacting with TCS.
The conserved DDD motif at the C-terminal tail of P2 interacts with TCS
As TCS interacted with the C11 peptide (SDDDMGFGLFD) but not the C7 peptide (MGFGLFD) of P2 (Figure 2a and f), it is likely that the conserved DDD motif of P2 interacts with TCS via formation of charge–charge interactions. To test this hypothesis, the DDD motif in P2 was substituted with alanine to create a variant of P2 DDD(106–108)AAA. In vitro pull-down assay showed that the triple-alanine substitution abolished the interaction between P2 and TCS (Figure 2c and d). When the last eleven residues (SAAAMGFGLFD) of the triple-alanine variant of P2 were fused to the MBP, the resulting MBP-C11 DDD(106–108)AAA also failed to interact with TCS (Figure 2e). These data suggest that the negative charges of the DDD motif play a crucial role in binding TCS.
Chemical shift perturbation experiments mapped the P2-binding site to the C-terminal domain of TCS
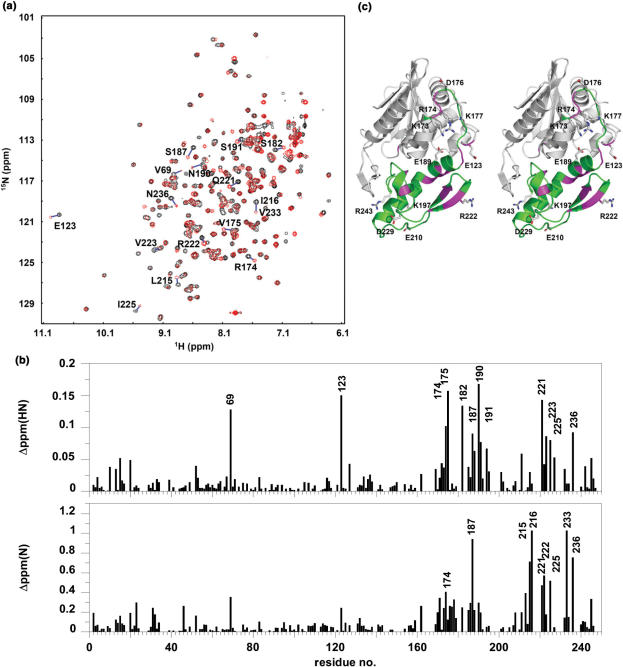
As a first step towards mapping the P2-binding site on TCS by chemical shift perturbation, we have assigned the backbone resonances of TCS based on the triple resonance experiments HNCA, HN(CO)CA, HN(CO)CACB and HNCACB, which established links between residues via their Cα and Cβ chemical shifts. Deuterated NMR samples of TCS, a 247-residue protein, were used to improve the quality of the spectra. Short-ranging nuclear Overhauser enhancements from 1H,15N NOESY-HSQC were used to resolve ambiguity and to confirm the assignment. Of 238 backbone amide resonances, 196 were assigned, which include most of the surface residues of TCS, and allow mapping the P2-binding site by chemical shift perturbation experiments.
Unlabelled P2 was titrated to 15N-labelled TCS sample, and the 1H and 15N chemical shifts of backbone amide resonances were followed. It was observed that a subset of peaks, as indicated in Figure 3a, moved gradually from the positions in the free form (black contours) to new positions (red contours) upon addition of P2 until the molar ratio of TCS:P2 reach ∼1. As shown in Figure 3b, residues with large amide chemical shift changes are predominantly clustered in the C-terminal domain (173–247) of TCS. In particular, large changes in chemical shift changes (>0.075 ppm on the 1H dimension or >0.5 ppm on the 15N dimension) were observed for residues 69, 123, 174, 175, 182, 187, 190, 191, 215, 216, 221, 222, 223, 225, 233 and 236—all of them are localized in or near the C-terminal domain of TCS (Figure 3c).
Figure 3.
P2-binding site on TCS was mapped to the C-terminal domain by chemical shift perturbation. (a) 1H–15N correlation spectra of TCS in the absence (black contours) and in the presence (red contours) of equal molar ratio of P2 were compared, and (b) changes in chemical shifts, ▵ppm(HN) and ▵ppm(N), of amide resonances of TCS were measured. Residues with ▵ppm(HN) >0.075 ppm or ▵ppm(N) >0.5 ppm are indicated in (a) and (b), and colour-coded magenta in the stereo diagram of TCS in (c). These residues are localized in or near the C-terminal domain (173–247, colour-coded green) of TCS. Scanning alanine mutagenesis was performed on all charge residues in the C-terminal domain and E123, which are indicated in (c).
Scanning charge-to-alanine mutagenesis showed that K173, R174 and K177 are involved in TCS–P2 interaction
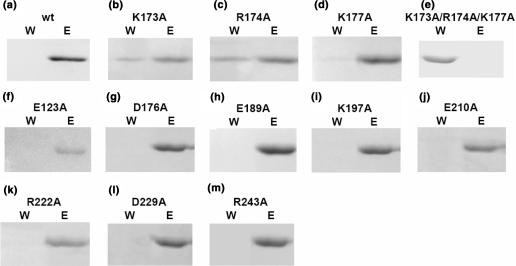
Chemical shift perturbation experiments have identified the C-terminal domain of TCS as the P2 interaction site. Since TCS interacts with all P-proteins, which contain a conserved C-terminal tail rich in charge residues (especially acidic residues), we anticipated that TCS may interact with P2 via charge–charge interactions. To identify which charge residues in the C-terminal domain interact directly with P2, we have performed scanning charge-to-alanine mutagenesis. There are 11 charge residues in this region: E123, K173, R174, D176, K177, E189, K197, E210, R222, D229 and R243 (Figure 3c). These residues were substituted by alanine to create 11 variants of TCS. To see if these charge-to-alanine substitutions would affect the TCS–P2 binding, TCS or its variants were loaded to the P2-coupled NHS-Sepharose (Figure 4). The detection of unbound K173A, R174A and K177A variants in the wash fractions (Figure 4b–d) suggests that charge-to-alanine substitution at these basic residues may weaken the binding between TCS and P2. To further investigate the role of K173, R174 and K177 in TCS–P2 interaction, we have created a triple-alanine variant (K173A/R174A/K177A) of TCS. In vitro pull-down assay showed that this triple charge-to-alanine substitution completely abolished the interaction between TCS and P2 (Figure 4e).
Figure 4.
In vitro pull-down assay on TCS variants suggests that K173, R174 and K177 are involved in binding P2. TCS (a) or its variants (b–m) were loaded to a P2-coupled NHS-Sepharose pre-equilibrated with binding buffer. Bound protein was eluted with 1 M NaCl in 20 mM Tris/HCl buffer pH 8.0. Fractions containing unbound protein collected during washing (W) and bound protein collected during elution (E) were analysed in 15% SDS-PAGE stained with Coomassie blue. As indicated by the presence of TCS in the wash fraction, substitution of alanine at K173, R174 and K177 positions decreases the binding of TCS on P2-coupled column (b–d). Triple-alanine substitutions in these residue positions resulted in a TCS variant (K173A/R174A/K177A) that was unable to bind P2 (e).
Interaction between TCS and ribosomal stalk was reduced by triple-alanine substitutions at K173, R174 and K177 positions
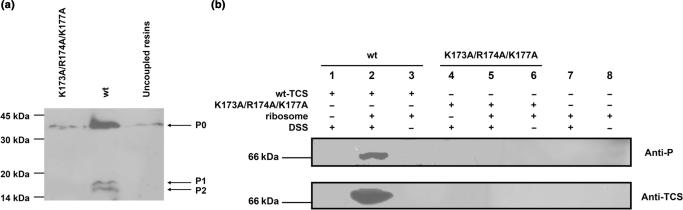
So far, we have shown that TCS interacts with the last 11 residues of P2. Since the C-terminal sequences of all stalk proteins P0, P1 and P2 are highly conserved (Figure 2a and b), it is likely that TCS interacts with the ribosome via this acidic C-terminal tails of P-proteins. If this is the case, the interactions between TCS and ribosome will be adversely affected if the three basic residues (K173/R174/K177) involved in binding P2 were substituted by alanine. To test this hypothesis, we have loaded 80S rat ribosome to NHS-Sepharose coupled with wild-type TCS or its triple-alanine variant (K173A/R174A/K177A). Our data showed that wild-type TCS was able to pull-down all acidic ribosomal proteins (P0, P1 and P2) from rat ribosome, while the interaction between the triple-alanine variant and ribosome was greatly reduced (Figure 5a). These data are consistent with the conclusion that TCS interacts with ribosome through the conserved C-terminal tails of P-proteins.
Figure 5.
Interaction between TCS and ribosome was compromised by K173A/R174A/K177A triple-alanine substitutions. (a) Pull-down assay. Rat ribosome was loaded to NHS-Sepharose coupled with TCS or its triple-alanine (K173A/R174A/K177A) variants. After extensive washing, the bound proteins were eluted with 1 M NaCl, and detected by western blot using anti-P antibody. Ribosomal proteins P0, P1 and P2 were pull-down by wild-type TCS (lane 2), while the interaction between ribosome and the triple-alanine variants (lane 1) was greatly reduced to that similar to the control (lane 3), in which the faint band of P0 was due to non-specific interactions between ribosome and the uncoupled resins. (b) Cross-linking experiments. After rat ribosome was incubated with TCS or the triple-alanine variants in room temperature for 20 min, DSS was added to induce cross-linking between TCS and ribosomal proteins, and cross-linking product was detected by western blot using anti-P or anti-TCS antibodies. A protein band at ∼66 kDa, corresponding to the size of TCS–P0 complex, was detected by both anti-P and anti-TCS antibodies when ribosome was cross-linked with wild-type TCS (lane 2), but not with the triple-alanine variants (lane 5) and in other negative controls (lanes 1 and 4: without addition of ribosome; lanes 3, 6 and 8: without addition of DSS; lanes 7 and 8: without addition of TCS or its variants).
Direct interaction between TCS and the ribosomal stalk was further confirmed by cross-linking experiments. TCS or the triple-alanine variant was cross-linked with rat ribosome using DSS, and the cross-linking between TCS and P-proteins was identified by anti-P and anti-TCS antibodies. Our data showed that wild-type TCS was able to form a ∼66 kDa cross-linking product with rat ribosome, which corresponds to the expected molecular weight of TCS–P0 complex (Figure 5b, lane 2). In contrast, no TCS–P-protein complex was observed for the triple-alanine variant (K173A/R174A/K177A) (Figure 5b, lane 5). Taken together, our data suggest that TCS can interact directly to the ribosomal stalk, and the triple-alanine substitution abolished such interaction.
It is noteworthy that no cross-linking product of TCS with P1 or P2 was observed in our experiment. We also noticed that the amount of P1 and P2 pull-down by TCS from rat ribosome was less than that for P0 (Figure 5a). Unlike P0, which forms an integral part of ribosome, P1 and P2 were also found as free proteins in the cytoplasm. It was found that the cytoplasmic pool of P1/P2 can exchange readily with those bound to the ribosomal stalk (36). It is possible that the interaction between TCS and P0 is stronger than that for P1 and P2 so that TCS was preferentially cross-linked with P0 in the stalk of rat ribosome.
Charge-to-alanine substitutions at K173, R174 and K177 weaken the inhibition of in vitro translation
Charge-to-alanine scanning mutagenesis has identified three residues (K173, R174 and K177) that are directly involved in TCS–P2 and TCS–ribosome interaction. To see if alanine substitutions at these residue positions affect the ability of TCS to inhibit in vitro translation, we have measured the IC50 values (the concentration of TCS to achieve 50% inhibition of protein synthesis in an in vitro translation system) of TCS and its variants (Figure 6a). The values of IC50 for wild-type TCS, K173A, R174A, D176A, K177A and the triple-alanine variants were 0.027 ± 0.001, 0.20 ± 0.04, 0.16 ± 0.02, 0.038 ± 0.004, 0.09 ± 0.01, 0.5 ± 0.1nM, respectively. Single alanine substitutions K173A, R174A and K177A resulted in 7-, 6- and 3-fold increases in the values of IC50, suggesting that these charge-to-alanine substitutions weaken the ability of TCS to inhibit in vitro protein synthesis. The effect of charge-to-alanine substitution was accumulative—the K173A/R174A/K177A triple-substitution resulted in 18-fold increase in the IC50 value. On the other hand, D176A substitution, which did not affect the TCS–P2 interaction, had no significant effect on the IC50. To test if the charge-to-alanine substitutions abolish the N-glycosidase activity of TCS, rabbit reticulocyte lysate was incubated with TCS or its variants at a concentration (10 nM) that is sufficient to achieve complete inhibition of in vitro translation. Depurination assay showed that all TCS variants tested were able to depurinate A4324 of 28S rRNA (Figure 6b). Taken together, our data suggest that all TCS variants inactivate ribosomes through their N-glycosidase activity, but they do differ in their potency in inhibiting in vitro protein synthesis.
Figure 6.
(a) Translation inhibition assay. The potency of TCS variants to inhibit in vitro translation was measured by adding 3.7 pM to 370 nM of protein samples to nuclease-untreated rabbit reticulocyte lysate as described in the Materials and methods section. IC50, the concentration of TCS required to achieve 50% inhibition, was determined by fitting the data to a four-parameter logistic equation. The values of IC50 for WT (filled circle), K173A (open square), R174A (open diamond), D176A (open triangle), K177A (open circle), and the triple-alanine variant (filled square) were 0.027 ± 0.001, 0.20 ± 0.04, 0.16 ± 0.02, 0.038 ± 0.004, 0.09 ± 0.01, 0.5 ± 0.1nM, respectively. (b) Depurination assay of TCS and its variants. After incubation with 10 nM of TCS or its variants, RNA from the rabbit reticulocyte lysate was extracted, treated with aniline and analysed by electrophoresis as described in the Materials and methods section. Depurination of 28S rRNA at A4324 was detected by the ∼450 bp R-fragments (indicated by an arrow). Control lanes, in which RNA samples were not treated with aniline, are labelled with the ‘−’ marks.
DISCUSSION
TCS, a homolog of ricin A-chain, belongs to type I RIP. TCS is an RNA N-glycosidase, the substrate of which is the α-sarcin/ricin-loop (SRL) of eukaryotic 28S rRNA. TCS inactivates ribosome by removing the adenine base specifically at A4324, which abolishes binding of elongation factors to the ribosome. Although RIP can depurinate naked rRNA, the kcat value of such reaction is 105-fold lower than that for rRNA in ribosome. This finding strongly suggests that interaction between RIP and ribosomal proteins is essential to achieve full activity of RIP. Although all RIPs can inactivate eukaryotic ribosomes, some RIPs, like pokeweed antiviral protein, Mirabilis antiviral protein, and Shiga toxin, can also inactivate prokaryotic ribosomes. The kingdom specificity of ribosome substrate for some RIPs is likely due to the interaction of RIP with ribosomal proteins.
In this study, we have demonstrated that TCS interacts with acidic ribosomal stalk protein P2. Together with our previous finding that TCS interacts with other acidic ribosomal stalk proteins P0 and P1, TCS was shown to interact with all acidic P-proteins that constitute the lateral stalk of the large 60S ribosomal subunit. The ability of TCS to bind all P-proteins can be explained by the systematic deletion study, which has mapped the TCS-binding site to the conserved C-terminal tail of P2 (Figure 2). We have fused the last eleven residues of P2 to MBP, and found that the resulting MBP–C11 fusion protein can interact with TCS (Figure 2e). Since the last eleven residues of human P-proteins are highly conserved, with the consensus sequence SDd/eDMGFGLFD (Figure 2a), our data strongly suggest that TCS can interact with all P-proteins via this consensus C-terminal tail.
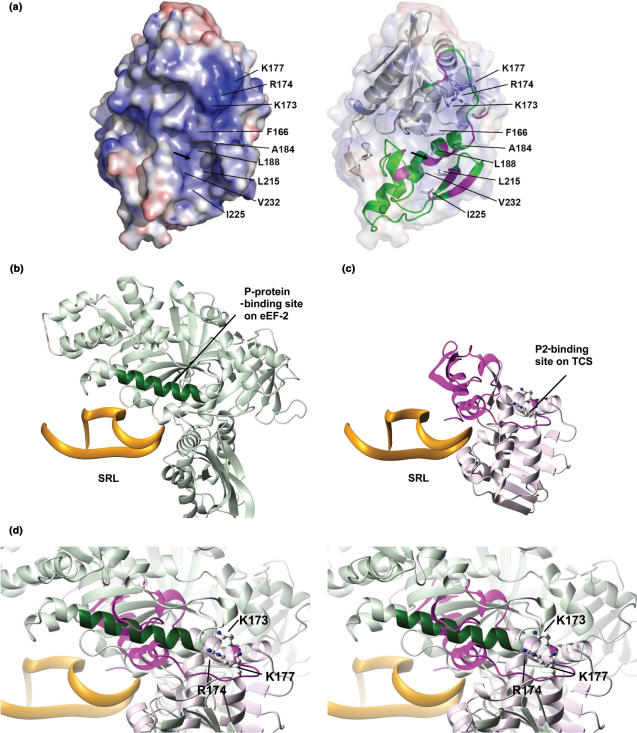
Using chemical shift perturbation experiment, we have located the P2-binding site to the C-terminal domain of TCS (Figure 3). Our finding is consistent with a previous study which showed that Saporin, another type I RIP, interacts with yeast ribosome through the C-terminal domain (37). Scanning charge-to-alanine mutagenesis has identified three basic residues, K173, R174 and K177, that are involved in P2 binding (Figure 4). Electrostatics calculation showed that the surface of TCS near these basic residues is strongly positively charged (Figure 7a), and it is likely that the acidic C-terminal tail of P2 binds to this region of TCS via charge–charge interactions. In agreement, we have shown that TCS interacts with the last eleven residues (SDDDMGFGLFD) of P2 (Figure 2a and e), and substitution of the three aspartate residues in the conserved DDD motif of the P2 C-terminal tail abolished the interaction between TCS and P2 (Figure 2c–f). This finding, together with our results that the triple-alanine variant (K173A/R174A/K177A) of TCS was unable to bind P2, strongly suggests that these basic residues form charge–charge interaction with the consensus DDD motif of P2.
Figure 7.
Comparison of P-protein-binding sites on eEF2 and TCS. (a) Putative P2-binding surface of TCS. The electrostatic calculation was performed using the program APBS (44) and visualized by PyMOL (45), where positive and negative potential surface is colour-coded blue and red, respectively, at ±10 kT. K173, R174 and K177 contribute to a strongly positive-charged surface which may interact with the DDD motif of P2. The arrow indicates the location of the hydrophobic pocket constituted by F166, A184, L188, L215, I225 and V232. A ribbon representation of TCS, with the C-terminal domain colour-coded green, is shown on the right panel. Residues with large changes in amide chemical shifts (as in Figure 3c) are colour-coded magenta. (b) The structure of eEF2–SRL complex was derived from an 11.7-Å cryo-electron microscopy map of the 80S ribosome complexed with eEF2 and sordarin (38) (PDB code 1S1H and 1S1I). Residues Q176-T191 (colour-coded dark green) of eEF2 were found to be in contact with P-proteins in the cryo-electron microscopy map (38). (c) The model of TCS–SRL complex was obtained as described in the Materials and methods section. The C-terminal domain of TCS is colour-coded magenta. (d) The model of TCS–SRL complex is superimposed onto the structure of eEF2–SRL complex. The three basic residues (K173, R174 and K177) that were found to be involved in binding P2 are shown in ball-and-stick representation in (c) and (d). Noteworthy, the P-protein-binding site (dark green) on eEF2 is in close proximity to the P-protein-binding site (magenta) of TCS.
On the other hand, electrostatic interaction is unlikely to be the only factor contributing to TCS–P2 binding. For example, deletion of the last Asp residue at P2 C-terminus (P2ΔC1) did not affect the TCS–P2 interaction. However, further deletion of non-polar residues (FGLF, P2ΔC2–P2ΔC5) seriously compromised the TCS–P2 binding. There is an exposed hydrophobic pocket at the C-terminal domain of TCS, which is constituted by F166, A184, L188, L215, I225 and V232 (Figure 7a). It is noteworthy that residues near this hydrophobic pocket were found to have larger chemical shift changes upon addition of P2 (Figures 3b and c, and 7a). It is likely that this hydrophobic pocket interacts with the non-polar residues at the C-terminal tail of P2.
Taken together, the in vitro binding data on TCS and P2 imply that TCS can bind to the stalk of ribosome through interacting with the conserved C-terminal tail of P-proteins. This hypothesis was supported by the fact that TCS can be cross-linked to P0 in rat ribosome, and such cross-linking was abolished in the triple-alanine variant (K173A/R174A/K177A) of TCS (Figure 5b). Moreover, the interaction between the triple-alanine variant and ribosome was also greatly compromised (Figure 5a). These findings suggest that the three basic residues of TCS form charge–charge interactions with the conserved acidic residues of P-proteins in ribosomal stalk. Our observation that TCS can bind to the ribosomal stalk is consistent with a previous study which showed that ricin A-chain, a homolog of TCS, was cross-linked to P0 (L10e) of ribosome from human lung carcinoma cells (10).
Ribosomal stalk, which is constituted by acidic P-proteins in eukaryotic ribosomes, was shown to play an important role in binding elongation factors. For example, EF2 binding can be blocked by anti-P-protein antibodies (20). Replacement of prokaryotic stalk proteins with P-proteins changed the elongation-factor-specificity of E. coli ribosome from prokaryotic EF-G to eukaryotic EF2 (17,18). Recently, cryo-electron microscopy of EF2–sordarin–ribosome complex demonstrated that EF2 directly binds to the SRL and interacts with P-proteins (38). In an 11.7-Å cryo-electron microscopy map, residues 176–191 in domain I of EF2 was shown to be in contact with P-proteins (Figure 7b).
To compare the P-protein-binding site on TCS and on EF2, we have docked TCS to the SRL substrate as described in the Materials and methods section (Figure 7c), based on the predicted models of ricin–SRL (39) and pokeweed antiviral protein–SRL (33,34). As both TCS and EF2 bind to the SRL, it is anticipated that TCS will occupy the EF2-binding site on ribosome. Superimposition of TCS–SRL and EF2–SRL models reveals that the P-protein-binding site of TCS is close in space to that of EF2 (Figure 7d), which suggests that upon TCS binding to the ribosome, the P-proteins are able to physically reach the P-protein-binding sites on TCS.
Our results support the conclusion that interaction with P-protein is required for full activity of TCS. We showed that alanine substitutions at K173, R174 and K177 positions compromise P2 protein binding and reduce the in vitro translation inhibition of TCS. In particular, triple-alanine substitutions of these basic residues weaken the interactions between TCS and ribosomal stalk (Figure 5), and resulted in ∼18-fold increases in IC50 value (Figure 6a). As these residues are far away from the SRL-binding site of TCS, the decrease in translation inhibition is likely caused by weakening of P-protein interaction but not by direct perturbation in RNA binding.
There are several possibilities of how TCS–P-protein interaction may play a role in ribosome-inactivating activity. First, interaction with the P-proteins may guide TCS to locate the SRL. It has been proposed that the flexible stalk proteins, which are protruding out of the ribosome, increase the association rate of elongation factors by grabbing them in the solution and fetch them to the binding site on ribosome (40). Similarly, binding to the stalk proteins (P0/P1/P2) anchors TCS to a location of ribosome near the SRL substrate, and allows TCS to find its SRL substrate more efficiently. Moreover, the ability of TCS to interact with multiple copies of C-terminal tails of the stalk proteins (P0/P1/P2) may increase the association rate between TCS and ribosome. Second, interaction with P-proteins may induce conformational changes on TCS, which will increase its enzymatic activity. Alternatively, binding of TCS to the ribosome may also change the conformation of ribosome, making the SRL more susceptible to depurination by TCS. Third, binding of TCS to P-proteins may hinder the association of elongation factors to the ribosome. TCS and all RIPs, which target the SRL, occupy the elongation-factor-binding site on ribosome. Interaction with P-proteins may help TCS to compete with other elongation factors, and block the access of these factors to the SRL. For example, it has been shown that monoclonal antibody that binds specifically to the C-terminal of P-proteins inhibit protein synthesis by blocking access of elongation factors to the ribosomal stalk (20). Work is underway to clarify some of these possibilities.
ACKNOWLEDGEMENTS
This work was supported by grants (CUHK 4145/01M and CUHK 4301/03M) from the Research Grants Council of Hong Kong SAR. K.-H.S. acknowledges the financial support from the Research Grants Council of Hong Kong SAR (HKU 7350/04M) and the University of Hong Kong (UGC). Funding to pay the Open Access publication charge was provided by Department of Biochemistry, The Chinese University of Hong Kong.
Conflict of interest statement. None declared.
REFERENCES
- 1.Shaw PC, Lee KM, Wong KB. Recent advances in trichosanthin, a ribosome-inactivating protein with multiple pharmacological properties. Toxicon. 2005;45:683–689. doi: 10.1016/j.toxicon.2004.12.024. [DOI] [PubMed] [Google Scholar]
- 2.Endo Y, Tsurugi K. RNA N-glycosidase activity of ricin A-chain. Mechanism of action of the toxic lectin ricin on eukaryotic ribosomes. J. Biol. Chem. 1987;262:8128–8130. [PubMed] [Google Scholar]
- 3.Endo Y, Mitsui K, Motizuki M, Tsurugi K. The mechanism of action of ricin and related toxic lectins on eukaryotic ribosomes. The site and the characteristics of the modification in 28 S ribosomal RNA caused by the toxins. J. Biol. Chem. 1987;262:5908–5912. [PubMed] [Google Scholar]
- 4.Stirpe F, Bailey S, Miller SP, Bodley JW. Modification of ribosomal RNA by ribosome-inactivating proteins from plants. Nucleic Acids Res. 1988;16:1349–1357. doi: 10.1093/nar/16.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang JS, Liu WY. The mechanism of action of trichosanthin on eukaryotic ribosomes – RNA N-glycosidase activity of the cytotoxin. Nucleic Acids Res. 1992;20:1271–1275. doi: 10.1093/nar/20.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Endo Y, Tsurugi K. The RNA N-glycosidase activity of ricin A-chain. The characteristics of the enzymatic activity of ricin A-chain with ribosomes and with rRNA. J. Biol. Chem. 1988;263:8735–8739. [PubMed] [Google Scholar]
- 7.Sperti S, Montanaro L, Mattioli A, Stirpe F. Inhibition by ricin of protein synthesis in vitro: 60 S ribosomal subunit as the target of the toxin. Biochem. J. 1973;136:813–815. doi: 10.1042/bj1360813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nilsson L, Nygard O. The mechanism of the protein-synthesis elongation cycle in eukaryotes. Effect of ricin on the ribosomal interaction with elongation factors. Eur. J. Biochem. 1986;161:111–117. doi: 10.1111/j.1432-1033.1986.tb10130.x. [DOI] [PubMed] [Google Scholar]
- 9.Xiong JP, Xia ZX, Wang Y. Crystal structure of trichosanthin-NADPH complex at 1.7 Å resolution reveals active-site architecture. Nat. Struct. Biol. 1994;1:695–700. doi: 10.1038/nsb1094-695. [DOI] [PubMed] [Google Scholar]
- 10.Vater CA, Bartle LM, Leszyk JD, Lambert JM, Goldmacher VS. Ricin A chain can be chemically cross-linked to the mammalian ribosomal proteins L9 and L10e. J. Biol. Chem. 1995;270:12933–12940. doi: 10.1074/jbc.270.21.12933. [DOI] [PubMed] [Google Scholar]
- 11.Hudak KA, Dinman JD, Tumer NE. Pokeweed antiviral protein accesses ribosomes by binding to L3. J. Biol. Chem. 1999;274:3859–3864. doi: 10.1074/jbc.274.6.3859. [DOI] [PubMed] [Google Scholar]
- 12.Rajamohan F, Ozer Z, Mao C, Uckun FM. Active center cleft residues of pokeweed antiviral protein mediate its high-affinity binding to the ribosomal protein L3. Biochemistry. 2001;40:9104–9114. doi: 10.1021/bi002851p. [DOI] [PubMed] [Google Scholar]
- 13.Chan SH, Hung FS, Chan DS, Shaw PC. Trichosanthin interacts with acidic ribosomal proteins P0 and P1 and mitotic checkpoint protein MAD2B. Eur. J. Biochem. 2001;268:2107–2112. doi: 10.1046/j.1432-1327.2001.02091.x. [DOI] [PubMed] [Google Scholar]
- 14.Tchorzewski M. The acidic ribosomal P proteins. Int. J. Biochem. Cell Biol. 2002;34:911–915. doi: 10.1016/s1357-2725(02)00012-2. [DOI] [PubMed] [Google Scholar]
- 15.Uchiumi T, Kominami R. Binding of mammalian ribosomal protein complex P0.P1.P2 and protein L12 to the GTPase-associated domain of 28 S ribosomal RNA and effect on the accessibility to anti-28 S RNA autoantibody. J. Biol. Chem. 1997;272:3302–3308. doi: 10.1074/jbc.272.6.3302. [DOI] [PubMed] [Google Scholar]
- 16.Uchiumi T, Kominami R. Direct evidence for interaction of the conserved GTPase domain within 28 S RNA with mammalian ribosomal acidic phosphoproteins and L12. J. Biol. Chem. 1992;267:19179–19185. [PubMed] [Google Scholar]
- 17.Uchiumi T, Honma S, Nomura T, Dabbs ER, Hachimori A. Translation elongation by a hybrid ribosome in which proteins at the GTPase center of the Escherichia coli ribosome are replaced with rat counterparts. J. Biol. Chem. 2002;277:3857–3862. doi: 10.1074/jbc.M107730200. [DOI] [PubMed] [Google Scholar]
- 18.Uchiumi T, Hori K, Nomura T, Hachimori A. Replacement of L7/L12.L10 protein complex in Escherichia coli ribosomes with the eukaryotic counterpart changes the specificity of elongation factor binding. J. Biol. Chem. 1999;274:27578–27582. doi: 10.1074/jbc.274.39.27578. [DOI] [PubMed] [Google Scholar]
- 19.Gomez-Lorenzo MG, Spahn CM, Agrawal RK, Grassucci RA, Penczek P, Chakraburtty K, Ballesta JP, Lavandera JL, Garcia-Bustos JF, et al. Three-dimensional cryo-electron microscopy localization of EF2 in the Saccharomyces cerevisiae 80S ribosome at 17.5 Å resolution. EMBO J. 2000;19:2710–2718. doi: 10.1093/emboj/19.11.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uchiumi T, Traut RR, Kominami R. Monoclonal antibodies against acidic phosphoproteins P0, P1, and P2 of eukaryotic ribosomes as functional probes. J. Biol. Chem. 1990;265:89–95. [PubMed] [Google Scholar]
- 21.Zhu RH, Ng TB, Yeung HW, Shaw PC. High level synthesis of biologically active recombinant trichosanthin in Escherichia coli. Int. J. Pept. Protein Res. 1992;39:77–81. doi: 10.1111/j.1399-3011.1992.tb01558.x. [DOI] [PubMed] [Google Scholar]
- 22.Miroux B, Walker JE. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 23.Wittekind M, Mueller L. HNCACB, a high-sensitivity 3D NMR experiment to correlate amide-protein and nitrogen resonances with the alpha- and beta-carbon resonances in proteins. J. Magn. Reson. B. 1993;101:201–205. [Google Scholar]
- 24.Grzesiek S, Bax A. Correlating backbone amide and side-chain resonances in larger proteins by multiple relayed triple resonance NMR. J. Am. Chem. Soc. 1992;114:6291–6293. [Google Scholar]
- 25.Marion D, Driscoll PC, Kay LE, Wingfield PT, Bax A, Gronenborn AM, Clore GM. Overcoming the overlap problem in the assignment of 1H NMR spectra of larger proteins by use of three-dimensional heteronuclear 1H-15N Hartmann-Hahn-multiple quantum coherence and nuclear Overhauser-multiple quantum coherence spectroscopy: application to interleukin 1 beta. Biochemistry. 1989;28:6150–6156. doi: 10.1021/bi00441a004. [DOI] [PubMed] [Google Scholar]
- 26.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 27.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 28.Bax A, Subramanian S. Sensitivity-enhanced two-dimensional heteronuclear shift correlation NMR-spectroscopy. J. Magn. Reson. 1986;67:565–569. [Google Scholar]
- 29.Live DH, Davis DG, Agosta WC, Cowburn D. Long-range hydrogen-bond mediated effects in peptides – N-15 NMR-study of gramicidin-S in water and organic-solvents. J. Am. Chem. Soc. 1984;106:1939–1941. [Google Scholar]
- 30.Spedding G. Isolation and analysis of ribosomes from prokaryotes, eukaryotes, and organelles. In: Spedding G, editor. Ribosomes and Protein Synthesis. Oxford: Oxford University Press; 1990. pp. 9–12. [Google Scholar]
- 31.Wong KB, Ke YB, Dong YC, Li XB, Guo YW, Yeung HW, Shaw PC. Structure/function relationship study of Gln156, Glu160 and Glu189 in the active site of trichosanthin. Eur. J. Biochem. 1994;221:787–791. doi: 10.1111/j.1432-1033.1994.tb18792.x. [DOI] [PubMed] [Google Scholar]
- 32.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 33.Rajamohan F, Pugmire MJ, Kurinov IV, Uckun FM. Modeling and alanine scanning mutagenesis studies of recombinant pokeweed antiviral protein. J. Biol. Chem. 2000;275:3382–3390. doi: 10.1074/jbc.275.5.3382. [DOI] [PubMed] [Google Scholar]
- 34.Rajamohan F, Mao C, Uckun FM. Binding interactions between the active center cleft of recombinant pokeweed antiviral protein and the alpha-sarcin/ricin stem loop of ribosomal RNA. J. Biol. Chem. 2001;276:24075–24081. doi: 10.1074/jbc.M011406200. [DOI] [PubMed] [Google Scholar]
- 35.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 36.Tsurugi K, Ogata K. Evidence for the exchangeability of acidic ribosomal proteins on cytoplasmic ribosomes in regenerating rat liver. J. Biochem. (Tokyo) 1985;98:1427–1431. doi: 10.1093/oxfordjournals.jbchem.a135410. [DOI] [PubMed] [Google Scholar]
- 37.Savino C, Federici L, Ippoliti R, Lendaro E, Tsernoglou D. The crystal structure of saporin SO6 from Saponaria officinalis and its interaction with the ribosome. FEBS Lett. 2000;470:239–243. doi: 10.1016/s0014-5793(00)01325-9. [DOI] [PubMed] [Google Scholar]
- 38.Spahn CM, Gomez-Lorenzo MG, Grassucci RA, Jorgensen R, Andersen GR, Beckmann R, Penczek PA, Ballesta JP, Frank J. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J. 2004;23:1008–1019. doi: 10.1038/sj.emboj.7600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Monzingo AF, Robertus JD. X-ray analysis of substrate analogs in the ricin A-chain active site. J. Mol. Biol. 1992;227:1136–1145. doi: 10.1016/0022-2836(92)90526-p. [DOI] [PubMed] [Google Scholar]
- 40.Diaconu M, Kothe U, Schlunzen F, Fischer N, Harms JM, Tonevitsky AG, Stark H, Rodnina MV, Wahl MC. Structural basis for the function of the ribosomal L7/12 stalk in factor binding and GTPase activation. Cell. 2005;121:991–1004. doi: 10.1016/j.cell.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 41.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad.Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]