Abstract
Quantitative and mechanism-based information on differences in transfection efficiency between viral and non-viral vectors would be highly useful for improving the effectiveness of non-viral vectors. A previous quantitative comparison of intracellular trafficking between adenovirus and LipofectAMINE PLUS (LFN) revealed that the three orders of magnitude lower transfection efficiency of LFN was dominantly rate limited by the post-nuclear delivery process. In the present study, the contribution of transcription and translation processes to the overall differences in the transgene expression efficiency of nucleus-delivered DNA was independently evaluated by quantifying mRNA. As a result, transcription efficiency (Etranscript) of LFN, denoted as transgene expression divided by the amount of nuclear pDNA was about 16 times less than that for adenovirus. Furthermore, translation efficiency (Etranslate), denoted as transfection activity divided by mRNA expression was approximately 460 times less in LFN. Imaging of the decondensed form of DNA by in situ hybridization revealed that poor decondensation efficiency of LFN is involved in the inferior Etranscript. Moreover, the inferior translation efficiency (Etranslate) of LFN was mainly due to electrostatic interactions between LFN and mRNA. Collectively, an improvement in nuclear decondensation and the diminution of the interaction between vector and mRNA is essential for the development of new generations of non-viral vectors.
INTRODUCTION
Gene therapy is an ideal concept for curing intractable diseases. To realize gene therapy, an intelligent vector that satisfies the requirements for both a high transfection activity and poor cytotoxicity is essential. To date, viral vectors such as adenovirus and retrovirus have accounted for more than 70% of the clinical trials (http://www.wiley.co.uk/genmed/clinical/), mainly because transfection activities using viral vectors are typically higher than that of non-viral ones (1,2). However, many clinical trials in which viral vectors are used have been interrupted since the application of these vectors induced unexpected adverse effects such as immunogenicity (3) and oncogenicity (4). Based on these drawbacks, the development of non-viral vectors would be highly desirable.
The most severe bottleneck in the clinical use of a non-viral vector is its low transfection activity. Therefore, an improvement of transfection activity is required, if gene therapy with non-viral vectors is to be required (5). It is generally accepted that transfection activity is rate limited by intracellular processes such as cellular uptake, endosomal escape, nuclear transfer and transcription (6). Therefore, it would be useful to clarify why and to what extent the current non-vector is inferior to the viral one from the point of view of intracellular trafficking. This information would enable us to clearly recognize which of the barriers need to be overcome for improving transfection activity, and moreover, to what extent transfection activity be improved by overcoming each intracellular barrier.
A novel technique (Confocal Image-assisted Three-dimensionally Integrated Quantification (CIDIQ)) for quantifying the intracellular distribution of exogenous DNA based on confocal images (7), has recently been established. Using this technique, we compared the intracellular trafficking of adenovirus and LipofectAMINE PLUS (LFN) in cultured cells (i.e. HeLa and A549 cells), as typical of viral and non-viral vectors, respectively. Comparing the dose–response curve, LFN requires 3–4 orders of magnitude more gene copies to exhibit a transfection activity comparable to the adenovirus in dividing cells (i.e. A549 cells and HeLa cells). By measuring nuclear delivery of exogenous DNA, the contribution of intracellular trafficking and subsequent post-nuclear events to the overall difference in transgene expression could be independently evaluated. To our surprise, intracellular trafficking could not explain the difference in transfection efficiency. In contrast, the transfection efficiency of nucleus-delivered DNA (TEnuc), denoted as transfection activity divided by the amount of nuclear DNA was three orders of magnitude higher in adenovirus compared with LFN, suggesting that the post-nuclear delivery process is dominantly responsible for the difference in transfection efficiency (2). Similar results have also been reported for other types of non-viral vectors (i.e. polyplexes) (8).
Although, in a previous study, we reported that TEnuc was attributed to the nuclear transcription process (2), the transgene expression of nucleus-delivered DNA is ruled by central dogmas, consisting of transcription and translation processes. In the present study, the contributions of these two processes to the overall differences in TEnuc were quantitatively evaluated by measuring the amount of cellular mRNA, an intermediate component of the central dogma. Furthermore, mechanisms underlying the difference in each process were clarified. These studies could serve as a guideline for future studies of non-viral gene vectors, since clear answers were found in this article concerning the problem of current non-viral vectors.
MATERIALS AND METHODS
Materials
HeLa cells were obtained from the RIKEN Cell Bank (Tsukuba, Japan). To prepare the reporter gene vector encoding luciferase (GL3) and EGFP (pcDNA3.1-luciferase and pcDNA3.1-EGFP), cDNA fragment encoding these proteins were inserted into the pcDNA3.1 (Invitrogen, Carlsbad, CA, USA) respectively, as reported earlier (2,9). LFN reagents were purchased from Invitrogen (Carlsbad, CA, USA). The  ,
,  , replication-deficient serotype 5 adenovirus was used in the adenoviral vector, in which an expression cassette is inserted at the E1 position (10).
, replication-deficient serotype 5 adenovirus was used in the adenoviral vector, in which an expression cassette is inserted at the E1 position (10).
Quantification of intracellular and intranuclear mRNA
Transfection was performed following the manufacturer's instructions, as described earlier (11). Briefly, 15 µl of PLUS reagent was mixed with 482.5 µl of DMEM containing 2.5 µg of plasmid DNA (pDNA), without serum and antibiotics (DMEM(−)), and incubated for 15 min at room temperature. About 10 µl of aliquot of LipofectAMINE was suspended with 490 µl of DMEM(−). The LipofectAMINE suspension was then added to the PLUS–pDNA mixture, and further incubated for 15 min at room temperature. Samples containing 2.5 µg of pDNA suspended in 1 ml of DMEM(−) were added to 2.5 × 105 cells and incubated for 3 h at 37°C. The time when pDNA was applied to the cells is defined as time zero.
Total RNA was extracted from whole cell and isolated nuclei with an RNeasy Mini Kit with RNase-Free DNase Set (QIAGEN, Tokyo, Japan). Isolated nuclei were prepared from the suspension of the cells in lysis solution (0.5% IGEPAL CA630, 10 mM NaCl, 3 mM MgCl2 and 10 mM Tris-HCl, pH 7.4), followed by centrifugation at 9200 × g for 2 min at 4°C, as described earlier (11,12). Since nuclei can be separated during the centrifugation, the most likely contaminants would be mitochondria. A western blot analysis for the cytochrome c, a marker protein for mitochondria, confirmed that very few (less than 7%) mitochondria were present in the nuclear fraction as contaminants (data not shown). Reverse transcription was performed using an ExScript RT reagent Kit by oligo-dT primer (TAKARA, Siga, Japan), and the cDNA product encoding luciferase was then quantified by RealTime PCR with the sets of primer listed in Table 1. As a standard, a dilution series of cDNA synthesized from the known amount of luciferase mRNA (Promega, Tokyo, Japan) was used. To determine the amount of mRNA per cell, the amount of mRNA purified from whole cells was normalized by the number of cells estimated from the amount of β-actin mRNA. A linear relationship was confirmed between the numbers of cells and the amount of total cellular mRNA of β-actin. The sequences of primers used in the quantification of cDNA encoding β-actin are summarized in Table 1.
Table 1.
Sequences of primers and probes used in this study
| A. Quantification of gene copies of pDNA and cDNA for luciferase | |
| Luc (+) primer | 5′-TTGACCGCCTGAAGTCTCTGA-3′ |
| Luc (−) primer | 5′-ACACCTGCGTCGAAGATGTTG-3′ |
| TaqMan probe | 5′-FAM-CCGCTGAATTGGAATCCATCTTGCTC-TAMRA-3′ |
| B. Quantification of genomic DNA of β-actin | |
| β-actin (+) primer | 5′-TGCGTGACATTAAGGAGAAGCTGTG-3′ |
| β-actin (−) primer | 5′-CAGCGGAACCGCTCATTGCCAATGG-3′ |
| C. Quantification of gene copies of pDNA for EGFP | |
| EGFP (+) primer | 5′-TCGAGCTGGACGGCGACGTA-3′ |
| EGFP (−) primer | 5′-GCTGAAGCACTGCACGCCGT-3′ |
| D. Preparation of probes used in in situ hybridization of pDNA | |
| 3076-3732 (+) primer | 5′-TCTCATGCTGGAGTTCTTCG-3′ |
| 3076-3732 (−) primer | 5′-CTCTGACTTGAGCGTCGATT-3′ |
| 3996-4525 (+) primer | 5′-CGGTAAGACACGACTTATCG-3′ |
| 3996-4525 (−) primer | 5′-CGTAGTTATCTACACGACGG-3′ |
The Luc primers were used for quantifying intranuclear and intracellular mRNA and intranuclear DNA. The TaqMan probe with FAM as a fluorescent dye on the 5′ end and TAMRA as a fluorescence quencher dye labeled to the 3′ end is designed to anneal to the target between Luc (+) and Luc (−) as described in the upper panels. The β-actin primers were used to correct the number of cells. The EGFP primers were used for the quantification of the copy number of EGFP DNA. The 3076-3732 primers and 3996-4525 primers were used in the preparation of the probe for in situ hybridization.
Similarly, the amount of luciferase mRNA in the isolated nuclei was normalized by the cell number. In order to determine cell number based on the amount of nuclear β-actin mRNA, it must be converted to the amount of total cellular β-actin mRNA that is linearly related to cell number. Therefore, the nuclear fraction of β-actin mRNA to that in the whole cell (Fnuc, β-act) was preliminarily determined as β-actin mRNA in the isolated nuclear fraction, divided by β-actin mRNA in the whole cell. As a result, Fnuc, β-act was calculated to be 0.033. In each sample, for the measurement of mRNA in an isolated nucleus, β-actin mRNA was also measured, and was then, divided by Fnuc, β-act to convert to total cellular β-actin mRNA that can be used to determine cell number.
Influence of adenovirus core proteins on transcription
To pre-load the adenovirus core proteins in the nucleus, host cells were pre-infected with adenovirus encoding EGFP for 1, 3 and 6 h at 37°C. Subsequently, pDNA encoding luciferase was transfected with LFN. After an additional incubation for 6 h at 37°C, cells were collected, and a luciferase assay was performed as reported earlier (11).
Purification of adenovirus genome and nuclear microinjection
The adenovirus genome was purified as reported earlier with minor modifications (13). Adenovirus, encoding EGFP (1 × 1012 particles) was dialyzed with TE buffer (10 mM Tris, 1 mM EDTA, pH 8.0). The adenovirus solution was then treated with an equal volume of 8 M guanidine and incubated for 5 min at 4°C on ice. After a 5–20% sucrose gradient ultracentrifugation (SW41Ti, 30 000 rpm, 4°C, 16 h), a 0.5 ml aliquot was collected from the bottom. To identify the adenovirus genome fraction, each fraction was incubated with 1 μg/ml of Hoechst33342 and the fluorescence was then measured.
The concentration of adenoviral genomic DNA and pDNA encoding EGFP was determined by RealTime PCR, as described earlier, using the set of primers shown in Table 1. Adenoviral genomic DNA and pDNA encoding EGFP was injected at a dose of 10 copies/nucleus with 0.5% of rhodamine-labeled dextran (RhoDex: M.W. 70 000) as an injection marker (9). At 24 h post-injection, EGFP expression was monitored by fluorescence microscopy, and the ratio of cells expressing EGFP to those injected with rhodamine-labeled dextran was calculated.
Fluorescence in situ hybridization (FISH) combined with tyramide signal amplification (TSA)
To address the decondensation process, intranuclear DNA was visualized by in situ hybridization. Concerning the probes for the detection of pDNA introduced by LFN, two regions (3076-3732, 3996-4525) in pcDNA 3.1 (+) were amplified by PCR with the primers shown in Table 1, and were then biotinylated using a Label IT Nucleoic Acid Modifying Kit (TAKARA, Siga, Japan). Concerning the probe for the detection of the adenovirus genome, Adenovirus Bioprobe (Enzo, NY, USA) was used. Cells were seeded in 0.002% poly-l-lysine-coated LabTek II chamber slides. After adenovirus- or LFN-mediated transfection for 10 h, the samples were washed with PBS and fixed with 4% paraformaldehyde/PBS. To activate the hybridization, the sample was treated with 5 μg/ml of Proteinase K for 15 min at room temperature. After washing with PBS, the sample was subjected to a 0.3% H2O2/methanol treatment for 20 min at room temperature to inactivate endogenous peroxidases. 0.5–1 μg/ml of each probe in hybridization buffer (50% formamid, 5× SSC, 5× Denhardts, 250 μg/ml Bakers yeast RNA, 500 μg/ml herring sperm DNA) was applied, and the sample was then denatured for 5 min at 95°C. After incubation overnight at 37°C, non-reactive probe was removed by incubation in 0.2× SSC for 20 min and 0.1× SSC for 10 min at 42°C. Hybridization signals were amplified with a TSA Biotin system (PerkinElmer, Tokyo, Japan) following the manufacturer's instructions. The signal was finally visualized with 10 μg/ml TexasRed labeled streptavidin (Molecular Probes, Eugene, OR, USA) and observed by confocal laser scanning microscopy (Carl Zeiss Co. Ltd., Jena, Germany).
Investigation of the interaction between mRNA and vectors by RealTime RT-PCR
To evaluate the interaction of adenovirus and LFN to mRNA, luciferase-encoding mRNA was subjected to reverse transcription with adenovirus and LFN encoding EGFP. If the vector strongly interacts with mRNA, reverse transcription must be inhibited. Applied adenovirus and LFN was removed by means of a GenElute Mammalian Genome DNA Miniprep kit, and the cDNA product was then quantified by RealTime PCR as described above.
In vitro translation
Adenovirus or LFN was incubated with luciferase mRNA at a concentration of 72 copies/pl for 10 min on ice. An in vitro translation was performed using a Rabbit Reticulocyte Lysate System (Promega, Tokyo, Japan) as described in the manufacturer's instructions. The amount of synthesized protein was determined by a luciferase assay as reported earlier (11).
RESULTS
Systematic comparison of transcription efficiency and translation efficiency between adenovirus- and LFN-mediated transgene expression
An earlier analysis indicated that the TEnuc of LFN was 7000 times less than that of adenovirus in HeLa cells (2). These data indicate that post-nuclear events, but not intracellular trafficking, are the dominant factors responsible for the 3–4 orders of magnitude lower transgene expression efficiency in LFN. To evaluate the contribution of translation and transcription processes to overall TEnuc, the expression level of mRNA was quantified by RealTime RT-PCR at 3 h post-transfection. Under conditions where transgene expression was comparable, LFN exhibited an approximately 270 times higher mRNA expression than adenovirus (Table 2). Furthermore, it was demonstrated that the number of nuclear pDNA transfected by LFN was approximately 4000-fold more than that for adenovirus (approximately 1.7 × 104 copies/cell versus 4.1 copies/cell) (2). Transcription efficiency (Etranscript) was calculated as the number of copies of cellular mRNA divided by the nuclear amount of DNA. As a result, the Etranscript value for adenovirus was approximately 16 times higher than that for LFN (Table 2 and Figure 1).
Table 2.
Quantitative comparison of transcription and post-transcription efficiency between Ad and LFN
| Ad | LFN | |
|---|---|---|
| Nuclear DNA (copies/cell)1 | 4.1 | 1.7 × 104 |
| Cellular mRNA (copies/cell) | 1.6 × 104 | 4.3 × 106 |
| Transgene expression (RLU/mg protein)1 | 2.3 × 107 | 1.3 × 107 |
| Transcription efficiency (Etranslate) (cellular mRNA/nuclear DNA) | 4.0 × 103 | 2.5 × 102 |
| Post-transcription efficiency (Etranscript) (transgene expression/cellular mRNA) | 1.4 × 103 | 3.0 |
Nuclear DNA and transgene expression was quantified after adenovirus- and LFN-mediated transfection at 1 h, and 3 h, respectively. Cellular mRNA was quantified by RealTime RT-PCR after transfection for 3 h. Transcription efficiency was calculated as cellular mRNA divided by nuclear DNA. Post-transcription efficiency was calculated as the transgene expression divided by the cellular mRNA.
1The amount of nuclear DNA (copies/cell) and transgene expression (RLU/mg protein) are cited from reference (2).
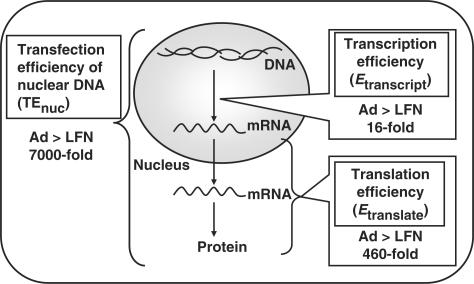
Figure 1.
Summary of the contribution of the transcription and translation processes to the overall transgene expression efficiency of nuclear DNA (TEnuc). The TEnuc for adenovirus was approximately 7000 times higher compared to that for LFN. By measuring the amount of cellular mRNA, an intermediate component of the central dogma, contributions of the differences in the transcription process and the translation process to the overall difference in TEnuc were quantitatively evaluated. In the transcription process, LFN was 16 times less efficient. In post-transcriptional processes, the translation efficiency of adenovirus was 460 times more efficient.
Similarly, we can calculate the translation efficiency (Etranslate), as transgene expression divided by the number of copies of mRNA. As a result, adenovirus represented 460 times higher efficiency compared with LFN (Table 2 and Figure 1).
Contribution of the adenovirus-derived factors to the difference of transcriptional efficiency between adenovirus and LFN
To utilize this quantitative information in developing a new-generation artificial vector, it is essential to clarify the mechanism underlying the large differences in Etranscript and Etranslate between adenovirus and LFN. Concerning the transcription process, two hypotheses were postulated, based on the assumption that activation factors exist in the adenoviral vector. One is that adenoviral core proteins delivered to the nucleus with genomic DNA may activate nuclear transcription factors. The other is that the genome structure and/or sequence derived from adenovirus were advantageous for transcription. In the present study, the sequences of a cytomegalovirus promoter/enhancer, cDNA-encoding luciferase and BGH polyadenylation used were the same between adenovirus and pDNA. However, it is possible that a unique sequence and/or linear structure in adenovirus may confer advantages to nuclear transcription.
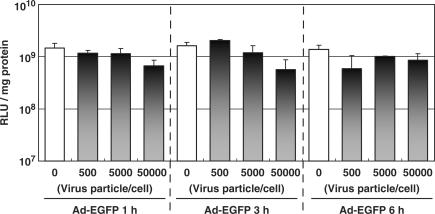
To address the contribution of the core proteins to the difference of Etranscript, they were pre-loaded to the nucleus beforehand by the infection of EGFP-encoding adenovirus. Subsequently, the pDNA-encoding luciferase was transfected with LFN. If core proteins activate nuclear transcription activity, luciferase expression of lipoplex should be improved by pre-infection with the adenovirus. However, as shown in Figure 2, the transfection activity remained essentially unchanged, compared with that in non-treatment (open bar) regardless of the pre-infection time (1, 3 and 6 h) or the dose (500–50 000 particles/cell). Unexpectedly, the transfection activity tended to decrease depending on the dose of adenovirus, presumably because of the cytotoxicity of adenovirus. This suggests that contributions of adenoviral core proteins to the difference in transcription activity are minor.
Figure 2.
Effect of the pre-loading of Ad core protein on LFN-mediated transfection activity. Cells were pre-infected by adenovirus encoding EGFP to load the adenovirus core proteins into the nucleus for 1, 3 and 6 h and at the indicated dose. pDNA encoding luciferase DNA was then transfected with LFN. At 6 h post-transfection with LFN, transgene expression was compared between non-infected control (open bar) and pre-infected cells. The vertical axis represents luciferase activity expressed as relative light units (RLUs)/mg protein. These data represent the mean values and standard deviation for triplicate experiments.
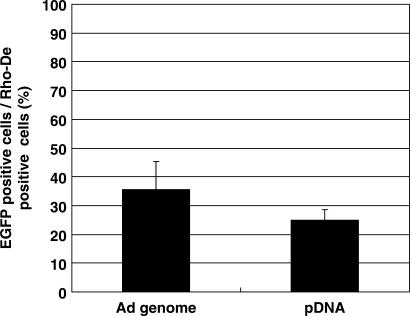
The involvement of unique genome structure and/or sequence in adenovirus in the difference of transcription efficiency was then investigated by the nuclear microinjection of adenovirus genomic DNA and pDNA encoding EGFP at a dose of 10 copies/nucleus. RhoDex was simultaneously injected as an injection marker. At 24 h post-microinjection, the expression efficiency of EGFP was evaluated as the number of EGFP-expressing cells divided by the number of RhoDex-positive cells. As a result, the expression efficiency of the adenovirus genome was not significantly different from that of pDNA (35 ± 9.8% versus 25 ± 3.7%) (Figure 3), suggesting that genomic structures or sequences derived from adenovirus are not involved in the high transcription efficiency of adenovirus. These data collectively indicate that an adenovirus-derived structure and/or sequence were not a dominant factor in the observed difference in Etranscript.
Figure 3.
Comparison of transgene expression after the nuclear microinjection of adenoviral genomic DNA and pDNA. Adenovirus genomic DNA was purified by a guanidine treatment, followed by sucrose density gradient centrifugation. After quantification of the concentration of genome DNA and pDNA by RealTime PCR, genomic DNA and pDNA was microinjected into the nucleus at a dose of 10 copies/nucleus with rhodamine-labeled dextran (Rho-Dex) as an injection marker. At 24 h post-microinjection, EGFP expression efficiency was evaluated.
Comparison of the nuclear disposition of adenoviral genomic DNA and pDNA
The results obtained exclude the activation hypothesis by adenovirus-derived factors. Therefore, the nuclear disposition of adenoviral genomic DNA and pDNA was compared from two points of view. One is the difference of decondensation efficiency. To improve cellular uptake and the regulation of intracellular trafficking of DNA, it is generally condensed with a cationic polymer or liposomes. However, once inside the nucleus, it must be released from the vectors to be recognized by transcription factors. Therefore, it is possible that the low transcription efficiency of LFN is due to the inefficient dissociation of pDNA from cationic liposomes. The other concern is subnuclear localization. It is generally accepted that nuclear transcription activity is rich in the euchromatin region, and that inactivated DNA is stored in the heterochromatin region. Therefore, it is possible that differences in the localization of DNA in the nucleus could contribute to the difference in Etranscript.
To investigate the difference in decondensation efficiency, the decondensed form of pDNA was visualized. When an alkaline phosphatase-labeled oligonucleotide (ODN) probe was hybridized with naked pDNA and the lipoplex blotted onto a nylon membrane, a clear signal was detected only in naked DNA (Supplementary Figure 1A and B), presumably because interactions between pDNA and probe were prevented by LFN via steric hindrance caused by condensation. This result strongly indicates that the detection of DNA based on the hybridization technique is useful for specifically visualizing the released form of DNA. Therefore, nuclear DNA released from the vector was visualized by in situ hybridization at 10 h after transfection with adenovirus and LFN. To detect nuclear DNA with high sensitivity, a TSA was applied. As a result, a remarkable number of signals were detected in the adenoviral genomic DNA (Figure 4A), whereas only a few pDNA molecules were detected in LFN (Figure 4B). Under this condition, the nuclear delivery of pDNA was approximately 700 times higher than adenoviral genomic DNA when quantified by TaqMan PCR (Figure 4C). To compare hybridization efficiency between adenoviral genomic DNA and pDNA, known amounts of purified genomic DNA or pDNA were blotted on a nylon membrane at various dilutions, and detected by a TSA system with probes used in the in situ hybridization (Supplementary Figure 1C and D). The intensity of each signal was quantified by means of a Scion Image and plotted against the number of gene copies blotted. As a result, the slope of the regression line for the adenoviral genomic DNA was only slightly higher (less than approximately 5-fold higher in adenovirus), suggesting that a significantly higher detection of nuclear genomic DNA cannot be accounted for by a difference in hybridization efficiency. These data indicate that poor decondensation of pDNA is one of the major reasons for the decreased Etranscript of LFN.
Figure 4.
Detection of the free form of adenoviral genomic DNA and pDNA by in situ hybridization. After 10 h post-transfection with adenovirus (A) or LFN (B) at a dose of 20 copies/cell and 3.4 × 106 copies/cell, the decondensed form of DNA was detected by in situ hybridization with TSA system. (C) Quantitative comparison of nuclear DNA between adenovirus and LFN. After 10 h post-transfection with adenovirus (20 copies/cell) or LFN (3.4 × 106 copies/cell), nuclear DNA was quantified by RealTime PCR. Data are represented as copies/cell.
The subnuclear localization of adenoviral genomic DNA and LFN was further investigated. Nuclear staining with DAPI can be used to classify the intranuclear space into two regions. One was a strongly stained region, in which genomic DNA is concentrated (heterochromatin), and the other is poorly stained region, in which genomic DNA is less concentrated (euchromatin). Dual imaging of free-form DNA and DAPI staining indicated that all of the adenoviral genomic DNA was specifically located in the euchromatin region. In contrast, pDNA was detected in both regions (Figure 5). Therefore, adenovirus can deliver its genomic DNA to the euchromatin region specifically, where transcription activity is higher. These data collectively indicate that differences in nuclear disposition such as decondensation or subnuclear localization are possible mechanisms for the difference in Etranscript.
Figure 5.
Comparison of nuclear sublocalization between the adenovirus genome and pDNA. At 10 h post-transfection with Ad (20 copies/cell) or LFN (3.4 × 106 copies/cell), nuclear adenoviral genomic (A) or pDNA (B) was detected by in situ hybridization (red). Cell nuclei were stained by DAPI (blue).
Investigation of the mechanism for the different translation efficiency between adenovirus and LFN
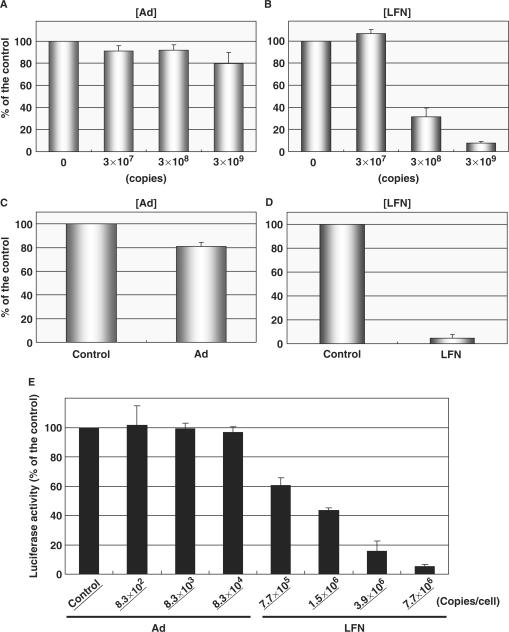
The mechanism for the differences in Etranslate was investigated. Considering that RNA, is a negatively charged molecule, like DNA, it is highly possible that LFN may interact with mRNA via electrostatic interactions. To address this issue, we first compared the interaction of LFN and adenovirus with mRNA by monitoring the inhibitory effect on the reverse transcription of mRNA. Luciferase-encoding mRNA was subjected to reverse transcription with or without adenovirus particles and LFN encoding EGFP. The cDNA product for luciferase was purified, and then quantified by RealTime PCR. cDNA production was decreased slightly in adenovirus (Figure 6A), whereas it was dramatically decreased in LFN (Figure 6B). When 3 × 109 copies of adenovirus and LFN were applied, cDNA production was decreased by 20 and 90%, respectively, compared with the non-treatment. These results suggest that LFN interacts with RNA more intensively than adenovirus.
Figure 6.
Effect of adenovirus and LFN on the post-transcription process. (A, B) Interaction between mRNA and vectors. Adenovirus (A) or LFN (B), encoding EGFP was applied to a reverse transcription reaction using a luciferase mRNA (8.5 × 109 copies/reaction). The cDNA product was quantified by RealTime PCR. (C, D) The influence of adenovirus and LFN on an in vitro translation system. About 4.3 × 108 copies/reaction of mRNA encoding luciferase was subjected to the in vitro translation with or without adenovirus (C) or LFN (D), at a dose of 6.8 × 109 copies/reaction. Protein synthesis was quantified by a luciferase assay. (E) Influence of adenovirus and LFN on the luciferase expression stably expressed in the HeLa cells. At 10 h post-transfection with Ad or LFN at the indicated doses, transgene expression was determined by a luciferase assay. Data are represented as the percent of untreated cells. These data represent the mean values and standard deviation for triplicate experiments.
Based on these observations, two hypotheses can be postulated to explain the difference in translation efficiency. One is that LFN may entrap mRNA in the nucleus and perturb its nuclear export. The other is that the recognition of cytoplasmic mRNA by ribosomal RNA or translation-related protein is inhibited.
The nuclear distribution of mRNA was then compared for adenovirus- and LFN-mediated transfection. Three hours after the transfection, cellular and intra-nuclear mRNA was quantified by RealTime RT-PCR (Table 3). The fraction of nuclear mRNA (Fnuc, mRNA), denoted as the nuclear amount of mRNA divided by total cellular mRNA was comparable between adenovirus and LFN (approximately 5.8 and 6.3%, respectively), suggesting that electrostatic interactions between LFN and mRNA do not inhibit the nuclear export process of mRNA.
Table 3.
Quantitative comparison of the nuclear export of mRNA between Ad and LFN
| Adenovirus | LFN | |
|---|---|---|
| Nuclear mRNA (copies/cell) | 9.3 × 102 | 2.7 × 105 |
| Cellular mRNA (copies/cell) | 1.6 × 104 | 4.3 × 106 |
| Nuclear fraction (Fnuc, mRNA) (%: nuclear mRNA/cellular mRNA) | 5.8 | 6.3 |
Numbers of nuclear mRNA in Ad- and LFN-mediated transfection were quantified by nuclear isolation, followed by RealTime PT-PCR. The nuclear fraction of mRNA was calculated as nuclear mRNA divided by cellular mRNA.
To investigate the effect of vectors on the cytoplasmic translation process, mRNA encoding luciferase was subjected to in vitro translation with or without adenovirus and LFN. When adenovirus was applied to an in vitro translation system, approximately 20% of the protein synthesis was inhibited compared with non-treatment (Figure 6C). On the other hand, when LFN was added to this system, protein synthesis was drastically inhibited to less than 90% (Figure 6D). To confirm the inhibitory effect of LFN on translation process by a whole-cell system, HeLa cells stably expressing luciferase were incubated with adenovirus or LFN encoding EGFP. Luciferase activity was not affected by the adenovirus infection after 10 h, regardless of the dose (Figure 6E). In contrast, luciferase activity was decreased by incubation with LFN in a dose-dependent manner. It is noteworthy that the inhibitory effect was significant, even when a conventional dose (1.5 × 106 copies/cell) of LFN was applied to the cells (to 60%). Collectively, these data indicate that the inhibition of the translation process via electrostatic interactions of LFN and mRNA is clearly a factor in the difference in Etranslate between LFN and adenovirus.
DISCUSSION
For the development of a non-viral vector, the intracellular and intranuclear disposition of exogenous DNA must be rigorously regulated. To date, many efforts have been made to develop devices for overcoming intracellular barriers from the standpoint of strategy of viral vectors. However, in the majority of the studies, only the enhancement in transfection activity before and after the modification of the devices has been used as the definition of success, and the quantitative evaluation of intracellular trafficking has rarely been demonstrated. As a result, it is difficult to predict which of the intracellular barriers need to be improved in order to further enhance the transfection activity. Many researchers have accepted these trial-and-error approaches in the past, mainly because an appropriate methodology for evaluating the intracellular trafficking of pDNA was not available.
We recently quantified the nuclear delivery of DNA transfected by adenovirus and LFN that enabled us to recognize the contribution of intracellular trafficking (intracellular pharmacokinetics: iPK) and post-nuclear events (pharmacodynamics: PD) to the overall difference in transfection efficiency between adenovirus and LFN. As a result, the TEnuc for adenovirus, calculated as transgene expression divided by the number of gene copies in the nucleus, was approximately 7000 times higher than that of LFN, suggesting that the difference in PD is a dominant factor in the difference in transfection activity. We reported earlier that this large difference was derived from the difference of transcription process (2). However, post-nuclear process is comprised of the transcription and translation. Therefore, the mechanism underlying the difference in TEnuc was further investigated by measuring mRNA expression, an intermediate of central dogma. As a result, a three orders of magnitude difference in TEnuc was found to be attributed to the one order of magnitude difference in transcription process and to the two orders of magnitude difference in the translation process, as shown in Figure 1.
To relate the above quantitative information to the development of a non-viral vector, the mechanism underlying these differences was further investigated. Concerning the difference in the transcription process, four hypotheses were formulated. The first two are based on the assumption that specific activators are present in the adenovirus. One is a core protein that enters the nucleus with adenoviral genomic DNA and activates a nuclear transcription factor. The other is that a unique sequence or the structure of genomic DNA may be advantageous for the transcription process. The latter two hypotheses are based on the assumption that the nuclear disposition of pDNA introduced by a non-viral vector is inappropriate for the transcription. For example, the poor decondensation of pDNA in the nucleus may prevent transcription factors from recognizing the promoter region. Alternatively, the nuclear distribution of pDNA could be inappropriate for the nuclear transcription. In the present study, we clarified which hypothesis is plausible for accounting for the difference in intranuclear transcription efficiency.
Concerning the influence of adenovirus core proteins on transcription, various types of basic proteins, such as protein VII, mu and protein V have been reported as condensers of adenovirus DNA via electrostatic interactions. Non-viral vectors prepared with core proteins as a condenser of pDNA have been developed (14,15). Although it has been reported that a vector promotes the efficient delivery of exogenous gene to the nucleus, its role in transcription event has not been clarified. It has been demonstrated that terminal proteins, protein V, protein VII and mu (16) interact with nuclear subdomains (i.e. PML body, nucleoli and the nuclear matrix) (17–19). In particular, protein V releases nucleophosmin/B23 from nucleoli. Since the B23 is a chromatin-remodeling factor, which promotes transcription activity by loosening the chromatin structure of the transcriptional region, it may facilitate the transcriptional factor to gain access to the genome (20). However, as shown in Figure 2, the transfection activity of LFN was not stimulated by pre-incubation with adenovirus, suggesting that the contribution of core proteins to the difference in transcription efficiency was minor. This is consistent with the fact that transgene expression of the mu–pDNA complex does not represent a significantly higher transgene expression compared with the poly-l-lysine–pDNA complex (14). The involvement of a structure or sequence in the adenovirus genome was then evaluated. The expression cassette (i.e. cytomegalovirus promoter/enhancer, cDNA encoding luciferase and BGH polyadenylation) used in the present study was completely identical between adenovirus and pDNA. However, it is possible that a unique sequence and/or linear structure in adenovirus may confer advantages to nuclear transcription. It has been demonstrated that 5′-terminal protein is associated with transcriptional factor Oct-1, followed by recruiting the transcriptional complex on the TATA box (21,22). Furthermore, a consensus sequence is present in ITR (23) for the binding of transcription factors (e.g. SP1, ATF) which may promote the downstream CMV promoter. However, the results of microinjection experiments showed that the adenovirus genome showed only a slightly higher transcription efficiency than pDNA. Thus, a difference in the structure and sequence cannot explain the difference in transcript efficiency.
Since the difference in transcriptional efficiency cannot be accounted for based on the hypothesis that activation factors exist in adenovirus, the mechanism was investigated with reference to another hypothesis; the nuclear disposition of pDNA was disadvantageous for transcription. One possible difference in the nuclear disposition of DNA is its decondensation profile. Therefore, a decondensed form of adenoviral genomic DNA and pDNA, was detected by in situ hybridization. The selective detection of the decondensed form of DNA by a hybridization technique was confirmed by dot blotting (Supplementary Figure 1A and B). As a result, adenoviral genomic DNA was more intensively detected in the nucleus compared to pDNA, even though the total nuclear association of pDNA in the LFN-mediated transfection was 700 times higher than that of adenovirus. These data indicate that nuclear decondensation efficiency in LFN was much less prominent compared to adenovirus. Since transgene expression after nuclear microinjection was not enhanced by changing the condensing counterpart from poly-l-lysine to mu (14), other adenoviral core proteins may be responsible for the efficient decondensation in nucleus. It has been reported that the chromatin-like structure of adenoviral genomic DNA, formed with protein VII is released by the nuclear histone chaperon, TAF (24–26). In addition, it is known that protein VII possesses a domain, that is subject to acetylation (27). Considering that adenovirus-mediated transgene expression is increased by histone deacetylase inhibitors (28,29), the interaction between protein VII and DNA may be epigenetically attenuated. Collectively, adenovirus may achieve efficient decondensation by remodeling as a the result of TAF and/or epigenetic regulation (e.g. acetylation of protein VII). Moreover, the poor decondensation in LFN was consistent with previous microinjection studies showing that the transgene expression of pDNA after nuclear microinjection was greatly inhibited compared with the microinjection of lipoplex (30,31). The driving force for the decondensation process in a lipoplex has not been clarified to date. We recently found that only a part of the nuclear pDNA-positive cell exhibited transgene expression, suggesting that heterogeneity in the nuclear transcription process is involved in the overall heterogeneity in transgene expression on LFN. In addition, the transcription efficiency was enhanced at the late S phase, where basic proteins such as histones are synthesized in conjunction with DNA synthesis (32). Therefore, the nuclear accumulation of histones may induce the decondensation of pDNA by replacing the counterpart cationic liposomes with histone itself. Therefore, condensation between LFN and pDNA is too tight to allow it to decondense in the nucleus, when the synthesis of histones is insufficient. The development of a decondensation system regardless of the cell cycle may improve the transcription efficiency of non-viral vectors.
It has recently been reported that the transfection activity of adenovirus in airway epithelial cells, in which adenoviral receptors are lacking, was drastically induced with the aid of cationic liposomes (33). This successful gene expression was synergistically achieved by the function of a cationic liposome (as a regulator of cellular uptake and subsequent intracellular trafficking), and the function of adenovirus (as a regulator of intranuclear disposition). Therefore, a combination of cationic liposomes and the adenoviral vector may be one of the elegant solutions for improving transfection efficiency.
Regarding subnuclear localization, all of the decondensed adenovirus was detected in the regions that are not intensively stained by DAPI, suggesting that the adenovirus genome accumulated efficiently to the euchromatin. MAR in the ITR sequence in adenoviral genomic DNA may anchor it to the nuclear matrix region (34–36). Concerning non-viral vectors, pDNA condensed with protamine exhibited a higher transcription efficiency compared with other cationic polymers (9). Considering the fact that protamine contains MAR in its sequence (37), the regulation of subnuclear localization of pDNA by a MAR-associating signal would result in an improved transcription efficiency. Alternatively, it is possible that the difference in subnuclear localization may be derived from a difference in the nuclear entry pathway (NPC-dependent versus NPC-independent pathway). Taking the size of lipoplex (>200 nm) into consideration, pDNA may enter the nucleus via a nuclear pore complex (NPC)-independent mechanism such as membrane fusion (38) since the NPC cannot accept macromolecules with a diameter of >39 nm. Therefore, DNA entering via the NPC may be more efficiently accessible to the euchromatin region.
Concerning the translation efficiency, LFN requires two orders of magnitude more mRNA to produce a transgene expression comparable to adenovirus. Since LFN and adenovirus are positively and negatively charged, respectively (39), electrostatic interactions of each vector and mRNA should be different. In fact, LFN inhibited the reverse transcription, while adenovirus rarely inhibited (Figure 6A and B). Based on this result, it would be expected that the nuclear export process, along with cytoplasmic translation may be strongly inhibited in LFN. However, this did not hold true. As shown in Table 3, the nuclear fraction of mRNA (Fnuc, mRNA) was comparable between adenovirus and LFN (Table 3). Recent studies have indicated that the formation of an exon-junction complex (EJC), is related to the transcription process (40–42), and subsequent export from the nucleus with the function of the nuclear export factor (TAP). Since these processes were coupled to each other, LFN would have no opportunity to bind to nuclear mRNA. Alternatively, LFN cannot recognize nuclear mRNA, since the intranuclear environment is rich in negatively charged molecules such as genomic DNA.
Finally, the inhibitory effect of adenovirus and LFN on in vitro translation was compared. In the present study, LFN severely inhibited in vitro translation by 95% at a concentration of 2.7 × 102 copies/pl, while adenovirus only slightly inhibited this process (Figure 6C and D). To quantify the intracellular trafficking of pDNA, 1.4 × 106 copies/cell of pDNA was applied. In this condition, 8.0 × 104 copies/cell of pDNA were taken up by the cells. Based on the assumption that the intracellular volume is 4 pl, the intracellular concentration was calculated to be approximately 2 × 104 copies/pl. This concentration is much higher than that used in the in vitro translation study. Therefore, it is plausible that LFN associates with mRNA and inhibits the translation process in living cells. Actually, the incubation of LFN inhibited the stably expressed marker genes (Figure 6E).
In summary, we succeeded in independently quantifying the contribution of the transcription and translation processes to the overall differences in the transgene expression efficiency of nuclear-delivered DNA. In addition, the mechanism underlying the difference in each process was clarified. As a result, the transcription efficiency of LFN was found to be 16 times less efficient than adenovirus presumably due to poor decondensation and non-selective subnuclear localization. Furthermore, the translation efficiency was approximately 460 times less in LFN, mainly due to the strong interactions between LFN and mRNA in the cytoplasm. However, these results do not exclude the importance of the regulation of intracellular pharmacokinetics. Intracellular trafficking and intranuclear transcription are connected in tandem. Therefore, if intracellular trafficking was barely regulated, transfection activity must also be poor. It should be noted that a dividing cell line (HeLa) was used in the present analysis. Therefore, in the case of non-dividing cells (i.e. primary cells), nuclear delivery must still be a severe barrier to a successful gene delivery system. Collectively, we conclude that the regulation of post-nuclear processes (improvement of intranuclear decondensation and the avoidance of electrostatic interactions between the vector and mRNA), along with intracellular trafficking is essential for developing a new generation of non-viral vector, which represents a transfection activity comparable to adenovirus.
SUPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work was supported, in part, by Grants-in-Aid for Scientific Research (B), and by Grant-in-Aid for Scientific Research on Priority Areas ‘Lifesurveyor’ from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and by Grants-in-Aid for Scientific Research on Priority Areas from the Japan Society for the Promotion of Science. We also thank Dr Milton S. Feather for helpful advice in the use of English in this manuscript. Funding to pay the Open Access publication charge was provided by Grant-in-Aid for Scientific Research on Priority Areas ‘Lifesurveyor’ from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Conflict of interest statement. None declared.
REFERENCES
- 1.Dinser R, Kreppel F, Zaucke F, Blank C, Paulsson M, Kochanek S, Maurer P. Comparison of long-term transgene expression after non-viral and adenoviral gene transfer into primary articular chondrocytes. Histochem. Cell. Biol. 2001;116:69–77. doi: 10.1007/s004180100305. [DOI] [PubMed] [Google Scholar]
- 2.Hama S, Akita H, Ito R, Mizuguchi H, Hayakawa T, Harashima H. Quantitative comparison of intracellular trafficking and nuclear transcription between adenoviral and lipoplex systems. Mol. Ther. 2006;13:786–794. doi: 10.1016/j.ymthe.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 3.Marshall E. Gene therapy death prompts review of adenovirus vector. Science. 1999;286:2244–2245. doi: 10.1126/science.286.5448.2244. [DOI] [PubMed] [Google Scholar]
- 4.Hacein-Bey-Abina S, von Kalle C, Schmidt M, Le Deist F, Wulffraat N, McIntyre E, Radford I, Villeval JL, Fraser CC, et al. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N. Engl. J. Med. 2003;348:255–256. doi: 10.1056/NEJM200301163480314. [DOI] [PubMed] [Google Scholar]
- 5.Parker AL, Newman C, Briggs S, Seymour L, Sheridan PJ. Nonviral gene delivery: techniques and implications for molecular medicine. Expert Rev. Mol. Med. 2003;2003:1–15. doi: 10.1017/S1462399403006562. [DOI] [PubMed] [Google Scholar]
- 6.Kamiya H, Akita H, Harashima H. Pharmacokinetic and pharmacodynamic considerations in gene therapy. Drug Discov. Today. 2003;8:990–996. doi: 10.1016/s1359-6446(03)02889-7. [DOI] [PubMed] [Google Scholar]
- 7.Akita H, Ito R, Khalil IA, Futaki S, Harashima H. Quantitative three-dimensional analysis of the intracellular trafficking of plasmid DNA transfected by a nonviral gene delivery system using confocal laser scanning microscopy. Mol. Ther. 2004;9:443–451. doi: 10.1016/j.ymthe.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 8.Varga CM, Tedford NC, Thomas M, Klibanov AM, Griffith LG, Lauffenburger DA. Quantitative comparison of polyethylenimine formulations and adenoviral vectors in terms of intracellular gene delivery processes. Gene. Ther. 2005;12:1023–1032. doi: 10.1038/sj.gt.3302495. [DOI] [PubMed] [Google Scholar]
- 9.Masuda T, Akita H, Harashima H. Evaluation of nuclear transfer and transcription of plasmid DNA condensed with protamine by microinjection: the use of a nuclear transfer score. FEBS Lett. 2005;579:2143–2148. doi: 10.1016/j.febslet.2005.02.071. [DOI] [PubMed] [Google Scholar]
- 10.Mizuguchi H, Koizumi N, Hosono T, Utoguchi N, Watanabe Y, Kay MA, Hayakawa T. A simplified system for constructing recombinant adenoviral vectors containing heterologous peptides in the HI loop of their fiber knob. Gene Ther. 2001;8:730–735. doi: 10.1038/sj.gt.3301453. [DOI] [PubMed] [Google Scholar]
- 11.Moriguchi R, Kogure K, Iwasa A, Akita H, Harashima H. Non-linear pharmacodynamics in a non-viral gene delivery system: positive non-linear relationship between dose and transfection efficiency. J. Control Release. 2006;110:605–609. doi: 10.1016/j.jconrel.2005.10.021. [DOI] [PubMed] [Google Scholar]
- 12.Iwasa A, Akita H, Khalil I, Kogure K, Futaki S, Harashima H. Cellular uptake and subsequent intracellular trafficking of R8-liposomes introduced at low temperature. Biochim. Biophys. Acta. 2006;1758:713–720. doi: 10.1016/j.bbamem.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 13.Sharp PA, Moore C, Haverty JL. The infectivity of adenovirus 5 DNA-protein complex. Virology. 1976;75:442–456. doi: 10.1016/0042-6822(76)90042-8. [DOI] [PubMed] [Google Scholar]
- 14.Akita H, Tanimoto M, Masuda T, Kogure K, Hama S, Ninomiya K, Futaki S, Harashima H. Evaluation of the nuclear delivery and intra-nuclear transcription of plasmid DNA condensed with micro (mu) and NLS-micro by cytoplasmic and nuclear microinjection: a comparative study with poly-L-lysine. J. Gene. Med. 2006;8:198–206. doi: 10.1002/jgm.839. [DOI] [PubMed] [Google Scholar]
- 15.Keller M, Harbottle RP, Perouzel E, Colin M, Shah I, Rahim A, Vaysse L, Bergau A, Moritz S, et al. Nuclear localisation sequence templated nonviral gene delivery vectors: investigation of intracellular trafficking events of LMD and LD vector systems. Chembiochem. 2003;4:286–298. doi: 10.1002/cbic.200390049. [DOI] [PubMed] [Google Scholar]
- 16.Rux JJ, Burnett RM. Adenovirus structure. Hum. Gene. Ther. 2004;15:1167–1176. doi: 10.1089/hum.2004.15.1167. [DOI] [PubMed] [Google Scholar]
- 17.Lee TW, Blair GE, Matthews DA. Adenovirus core protein VII contains distinct sequences that mediate targeting to the nucleus and nucleolus, and colocalization with human chromosomes. J. Gen. Virol. 2003;84:3423–3428. doi: 10.1099/vir.0.19546-0. [DOI] [PubMed] [Google Scholar]
- 18.Lee TW, Lawrence FJ, Dauksaite V, Akusjarvi G, Blair GE, Matthews DA. Precursor of human adenovirus core polypeptide Mu targets the nucleolus and modulates the expression of E2 proteins. J. Gen. Virol. 2004;85:185–196. doi: 10.1099/vir.0.19352-0. [DOI] [PubMed] [Google Scholar]
- 19.Matthews DA. Adenovirus protein V induces redistribution of nucleolin and B23 from nucleolus to cytoplasm. J. Virol. 2001;75:1031–1038. doi: 10.1128/JVI.75.2.1031-1038.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okuwaki M, Matsumoto K, Tsujimoto M, Nagata K. Function of nucleophosmin/B23, a nucleolar acidic protein, as a histone chaperone. FEBS Lett. 2001;506:272–276. doi: 10.1016/s0014-5793(01)02939-8. [DOI] [PubMed] [Google Scholar]
- 21.de Jong RN, Mysiak ME, Meijer LA, van der Linden M, van der Vliet PC. Recruitment of the priming protein pTP and DNA binding occur by overlapping Oct-1 POU homeodomain surfaces. EMBO J. 2002;21:725–735. doi: 10.1093/emboj/21.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsumoto K, Nagata K, Yamanaka K, Hanaoka F, Ui M. Nuclear factor I represses the reverse-oriented transcription from the adenovirus type 5 DNA terminus. Biochem. Biophys. Res. Commun. 1989;164:1212–1219. doi: 10.1016/0006-291x(89)91798-1. [DOI] [PubMed] [Google Scholar]
- 23.Hatfield L, Hearing P. The NFIII/OCT-1 binding site stimulates adenovirus DNA replication in vivo and is functionally redundant with adjacent sequences. J. Virol. 1993;67:3931–3939. doi: 10.1128/jvi.67.7.3931-3939.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haruki H, Gyurcsik B, Okuwaki M, Nagata K. Ternary complex formation between DNA-adenovirus core protein VII and TAF-Ibeta/SET, an acidic molecular chaperone. FEBS Lett. 2003;555:521–527. doi: 10.1016/s0014-5793(03)01336-x. [DOI] [PubMed] [Google Scholar]
- 25.Haruki H, Okuwaki M, Miyagishi M, Taira K, Nagata K. Involvement of template-activating factor I/SET in transcription of adenovirus early genes as a positive-acting factor. J. Virol. 2006;80:794–801. doi: 10.1128/JVI.80.2.794-801.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gyurcsik B, Haruki H, Takahashi T, Mihara H, Nagata K. Binding modes of the precursor of adenovirus major core protein VII to DNA and template activating factor I: implication for the mechanism of remodeling of the adenovirus chromatin. Biochemistry. 2006;45:303–313. doi: 10.1021/bi051248+. [DOI] [PubMed] [Google Scholar]
- 27.Fedor MJ, Daniell E. Acetylation of histone-like proteins of adenovirus type 5. J. Virol. 1980;35:637–643. doi: 10.1128/jvi.35.3.637-643.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taura K, Yamamoto Y, Nakajima A, Hata K, Uchinami H, Yonezawa K, Hatano E, Nishino N, Yamaoka Y. Impact of novel histone deacetylase inhibitors, CHAP31 and FR901228 (FK228), on adenovirus-mediated transgene expression. J. Gene. Med. 2004;6:526–536. doi: 10.1002/jgm.546. [DOI] [PubMed] [Google Scholar]
- 29.Gaetano C, Catalano A, Palumbo R, Illi B, Orlando G, Ventoruzzo G, Serino F, Capogrossi MC. Transcriptionally active drugs improve adenovirus vector performance in vitro and in vivo. Gene Ther. 2000;7:1624–1630. doi: 10.1038/sj.gt.3301296. [DOI] [PubMed] [Google Scholar]
- 30.Pollard H, Remy JS, Loussouarn G, Demolombe S, Behr JP, Escande D. Polyethylenimine but not cationic lipids promotes transgene delivery to the nucleus in mammalian cells. J. Biol. Chem. 1998;273:7507–7511. doi: 10.1074/jbc.273.13.7507. [DOI] [PubMed] [Google Scholar]
- 31.Zabner J, Fasbender AJ, Moninger T, Poellinger KA, Welsh MJ. Cellular and molecular barriers to gene transfer by a cationic lipid. J. Biol. Chem. 1995;270:18997–19007. doi: 10.1074/jbc.270.32.18997. [DOI] [PubMed] [Google Scholar]
- 32.Akita H, Ito R, Kamiya H, Kogure K, Harashima H. J. Gene. Med. Cell-cycle dependent transcription, a determinant factor of heterogeneity in cationic lipid-mediated transgene expression. in press. [DOI] [PubMed] [Google Scholar]
- 33.Price A, Limberis M, Gruneich JA, Wilson JM, Diamond SL. Targeting viral-mediated transduction to the lung airway epithelium with the anti-inflammatory cationic lipid dexamethasone-spermine. Mol. Ther. 2005;12:502–509. doi: 10.1016/j.ymthe.2005.03.033. [DOI] [PubMed] [Google Scholar]
- 34.Hatfield L, Hearing P. Redundant elements in the adenovirus type 5 inverted terminal repeat promote bidirectional transcription in vitro and are important for virus growth in vivo. Virology. 1991;184:265–276. doi: 10.1016/0042-6822(91)90843-z. [DOI] [PubMed] [Google Scholar]
- 35.Fredman JN, Engler JA. Adenovirus precursor to terminal protein interacts with the nuclear matrix in vivo and in vitro. J. Virol. 1993;67:3384–3395. doi: 10.1128/jvi.67.6.3384-3395.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schaack J, Ho WY, Freimuth P, Shenk T. Adenovirus terminal protein mediates both nuclear matrix association and efficient transcription of adenovirus DNA. Genes Dev. 1990;4:1197–1208. doi: 10.1101/gad.4.7.1197. [DOI] [PubMed] [Google Scholar]
- 37.Martins RP, Ostermeier GC, Krawetz SA. Nuclear matrix interactions at the human protamine domain: a working model of potentiation. J. Biol. Chem. 2004;279:51862–51868. doi: 10.1074/jbc.M409415200. [DOI] [PubMed] [Google Scholar]
- 38.Kamiya H, Fujimura Y, Matsuoka I, Harashima H. Visualization of intracellular trafficking of exogenous DNA delivered by cationic liposomes. Biochem. Biophys. Res. Commun. 2002;298:591–597. doi: 10.1016/s0006-291x(02)02485-3. [DOI] [PubMed] [Google Scholar]
- 39.Li QG, Lindman K, Wadell G. Hydropathic characteristics of adenovirus hexons. Arch. Virol. 1997;142:1307–1322. doi: 10.1007/s007050050162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo ML, Zhou Z, Magni K, Christoforides C, Rappsilber J, Mann M, Reed R. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature. 2001;413:644–647. doi: 10.1038/35098106. [DOI] [PubMed] [Google Scholar]
- 41.Strasser K, Hurt E. Splicing factor Sub2p is required for nuclear mRNA export through its interaction with Yra1p. Nature. 2001;413:648–652. doi: 10.1038/35098113. [DOI] [PubMed] [Google Scholar]
- 42.Strasser K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondon AG, Aguilera A, Struhl K, et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002;417:304–308. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]