Abstract
Mu opioid receptor (MOR) expression is under temporal and spatial controls, but expression levels of the MOR gene are relatively low in vivo. In addition to transcriptional regulations, upstream AUGs (uAUGs) and open reading frames (uORFs) profoundly affect the translation of the primary ORF and thus the protein levels in several genes. The 5′-untranslated region (UTR) of mouse MOR mRNA contains three uORFs preceding the MOR main initiation codon. In MOR-fused EGFP or MOR promoter/luciferase reporter constructs, mutating each uAUG individually or in combinations increased MOR transient heterologous expression in neuroblastoma NMB and HEK293 cells significantly. Translation of such constructs increased up to 3-fold without altering the mRNA levels if either the third uAUG or both the second and third AUGs were mutated. Additionally, these uAUG-mediated translational inhibitions were independent of their peptide as confirmed by internal mutation analyses in each uORF. Translational studies indicated that protein syntheses were initiated at these uAUG initiation sites, with the third uAUG initiating the highest translation level. These results support the hypothesis that uORFs in mouse MOR mRNA act as negative regulators through a ribosome leaky scanning mechanism. Such leaky scanning resulted in the suppression of mouse MOR under normal conditions.
INTRODUCTION
Opioid receptors (µ, δ and κ) mediate the diverse functions of endogenous opioid peptides and the opioid alkaloids such as morphine, including analgesia, reward, autonomic reflexes, and endocrine/immune regulation (1–3). The mu opioid receptor (MOR) is considered to be the main site of action for morphine (4). Many studies, including those with receptor knockout mice, have indicated that the responses to opiate agonists are dependent on the receptor level (5). MOR is first detected in rat brain at embryonic day 14 (E14) (6) followed by a slight decline during the first week of postnatal development, then increasing to peak levels 2 weeks later (7). The overall mechanisms involved in such spatial and temporal regulations of MOR have not yet been elucidated.
Generally, transcriptional control is mediated by transcription factors, RNA polymerase and a series of cis-acting elements located in the gene sequences. Such promoters, enhancers, silencers and locus-control elements are organized in a modular structure and regulate the production of pre-mRNA molecules (8). Opioid receptor expression can be regulated by multiple mechanisms, including transcriptional and post-transcriptional events (9,10). Our laboratory and others have demonstrated that MOR promoter activity is regulated by many enhancer elements and their related transcriptional factors such as SOX, SP1, AP2, NF-κB, PU.1, and NRSE (11). Post-transcriptional regulation occurs at the level of mRNA or protein. Such regulation could be due to mRNA stability, differences in translation efficiency or mRNA transport, and covalent modification of receptor molecules (10,12–14). Our recent studies indicated that the 3′-untranslated region (UTR) of MOR mRNA could affect the overall transcript's activity (15). Thus, posttranscriptional regulation of the MOR gene could have an important role in the spatial and temporal expression of the receptor proteins.
Recently, it has become increasingly clear that the 5′-UTR of eukaryotic mRNA is a key site of multiple forms of post-transcriptional regulation of gene expression, especially those containing at least one AUG codon (uAUG) upstream of the main open reading frame (ORF) (16–19). It is postulated that, in eukaryotic cells, most translation proceeds according to the ribosome scanning model (20–22), and initiates predominantly by a cap-binding/scanning mechanism (23). The scanning model predicts that an uAUG codon will interfere with translation of a downstream main ORF. However, the scanning complex may bypass such uAUG codons by ‘leaky scanning’ if the surrounding nucleotide context is suboptimal or very close to the 5′ cap region of the mRNA (24). The 43S pre-initiation complex binds to the 5′-cap region and then migrates progressively 5′ to 3′ until it recognizes an AUG start codon. The resulting complex is joined by the large subunit to form a complete ribosome, and polypeptide synthesis begins (20). Ribosomes reaching the main AUG of these mRNAs do so mainly via context-dependent leaky scanning and/or reinitiation mechanisms, although it is widely believed that these are inefficient mechanisms (24,25). The context-dependent leaky scanning mechanism accounts for the observation that some 40S subunits will fail to initiate at AUG codons with a less than optimal context and continue scanning along the 5′-UTR. The most efficient context for initiation of protein translation is known as the Kozak sequence (GCCRCCAUGG), which was identified initially as a consensus sequence in vertebrate mRNA (26). The reinitiation mechanism describes the ability of 40S subunits to continue to scan and initiate at a downstream main AUG codon after translating a small independent uORF. Reinitiation is considered to be a rare event. Although the incidence of this mechanism may be much greater, only a few mRNAs with uORFs have been examined (24). These uAUGs or uORFs are features of at least a few percent of the mRNAs in yeast, plants and mammals (26), and can be important players in negative translational control. However, in some cases, the upstream regulatory sequences stimulated translation of the major ORF (16,27).
Within the 5′-UTR region of the mouse MOR gene, between the basal proximal promoter at nucleotides −445 to −240 and the AUG of the main ORF, three uORFs with variable lengths are identified. Whether these uORFs can regulate the translation of the MOR transcripts was examined in this study. The uAUG-directed initiation and uORF-peptide-dependent regulations of the MOR transcript's translation were examined by mutational analyses of the uAUGs and internal sequences. We demonstrate that the translation of the MOR transcript is negatively regulated by these uORFs, and that such down-regulation is mediated via ribosome leaky scanning mechanism. The initiation of peptide syntheses at these uAUGs of the MOR transcript provides a novel mechanism for the regulation of expression of the mouse MOR gene product.
MATERIALS AND METHODS
Plasmid construction
All constructs were generated by PCR using Turbo Pfu polymerase (Stratagene), with forward and reverse primers containing appropriate restriction sites at their 5′ and 3′ ends or appropriate mutations at desired sequence sites. Amplified fragments were cloned into the pGL3-promoter reporter construct (Promega), using the corresponding restriction sites as described below. The wild-type uAUG(+) construct was generated by inserting the −301 to +1 PCR fragment of the 5′-UTR of mouse MOR (generated by using HindIII-ended forward primer and NcoI-ended reverse primer) in between the HindIII and NcoI restriction sites of pGL3-promoter reporter construct. Individual uORF in the mouse MOR 5′-UTR (−301 to +1) were inactivated by introducing point mutations into the start codons using an oligonucleotide-directed mutagenesis system (Stratagene) according to the manufacturer's recommendation. The following oligonucleotides were used for oligonucleotide-directed mutagenesis: uAUG #1 construct: 5′-CCTCACAGCCCACGCTCCCTCCCTT-3′ (forward) and 5′-AAGGGAGGGAGCGTGGGCTGTGAGG-3′ (reverse); uAUG #2 construct: 5′-TTTGGGGACGCTAAGGATGCGCCTC-3′ (forward) and 5′-GAGGCGCATCCTTAGCGTCCCCAAA-3′ (reverse); uAUG #3 construct: 5′-TTTGGGGATGCTAAGGACGCGCCTC-3′ (forward) and 5′-GAGGCGCGTCCTTAGCATCCCCAAA-3′ (reverse); and the 2nd/3rd uAUGs (#2, 3 construct): 5′-TTTGGGGACGCTAAGGACGCGCCTC-3′ (forward) and 5′-GAGGCGCGTCCTTAGCGTCCCCAAA-3′ (reverse). For the construct in which all three uAUGs in between the HindIII and NcoI sites were mutated [uAUG(−)], the U within each AUG sequence was substituted by C in the forward primer and with G in the reverse primer.
The uORF/luciferase (LUC) in-frame fusion constructs were generated by inserting a PCR fragment amplified from the uAUG(+) construct using the HindIII-ended forward and NcoI-ended reverse primers, 5′-CCAAGCTTGGATCCCTCACAGCCCATGCTCCC-3′ (forward) and 5′-AACCATGGAGTGGAACCAGAGAAGAGCGGCAG-3′ (reverse), where the stop codon of the 5′-UTR is replaced by the NcoI site. This cloning leads to a fusion between the stop codon of the mouse MOR uORF and the ATG codon of the LUC coding sequence. This uORF/LUC in-frame fusion construct (uORF IFR) was inactivated by introducing point mutations into the uAUG using an oligonucleotide-directed mutagenesis system (Stratagene) using primers mentioned above: #1 (uAUG#1 IFR), #2 (uAUG#2 IFR), #3 (uAUG#3 IFR), #2, 3 (uAUG#2, 3 IFR), all [uAUG(−) IFR)].
SP6 promoter-controlled uORF/LUC fused constructs were generated by cloning the luciferase gene to the SP6 promoter (SP6-LUC) construct. SP6-uORF/LUC fused constructs were generated from each uORF/LUC in-frame fusion constructs [uAUG#1 IFR, uAUG#2 IFR, uAUG#3 IFR, uAUG(−) IFR and uAUG(+) IFR] digested between HindIII and XbaI and cloned to SP6-LUC constructs [SP6-#1 IFR, SP6-#2 IFR, SP6-#3 IFR, SP6-uAUG(−) IFR and SP6-uAUG(+) IFR].
The STOP mutant constructs were generated by introducing point mutations into the start codons and/or introducing the stop codons after the start codons using the following oligonucleotides. These oligonucleotides were used for single amino acid change mutagenesis for adding the stop codons (+STOP; +S) or for mutating uAUG and adding stop codons (Mutation + STOP; M + S). uAUG#1 +S: 5′-CCTCACAGCCCATGTAGCCTCCCTT-3′ (forward) and 5′-AAGGGAGGCTACATGGGCTGTGAGG-3′ (reverse); uAUG#1 M + S: 5′-CCTCACAGCCCACGTAGCCTCCCTT-3′ (forward) and 5′-AAGGGAGGCTACGTGGGCTGTGAGG-3′ (reverse); uAUG#2 +S: 5′-TTTGGGGATGTGAAGGATGCGCCTC-3′ (forward) and 5′-GAGGCGCATCCTTCACATCCCCAAA-3′ (reverse); uAUG#2 M + S: 5′-TTTGGGGACGTGAAGGATGCGCCTC-3′ (forward) and 5′-GAGGCGCATCCTTCACGTCCCCAAA-3′ (reverse); uAUG#3 +S: 5′-TTTGGGGATGCTAAGGATGTGACTC-3′ (forward) and 5′-GAGTCACATCCTTAGCATCCCCAAA-3′ (reverse); uAUG#3 M + S: 5′-TTTGGGGATGCTAAGGACGTGACTC-3′ (forward) and 5′-GAGTCACGTCCTTAGCATCCCCAAA-3′ (reverse); uAUG#2, 3 +S: 5′-TTTGGGGATGTGAAGGATGTGACTC-3′ (forward) and 5′-GAGTCACATCCTTCACATCCCCAAA-3′ (reverse); and uAUG#2, 3 M + S: 5′-TTTGGGGACGTGAAGGACGTGACTC-3′ (forward) and 5′-GAGTCACGTCCTTCACGTCCCCAAA-3′ (reverse). The uAUG(−) +S and uAUG(−) M + S have STOP codons after each of the three uAUGs within the uAUG(+) or uAUG(−) M + S constructs. The context-improved (IMP) constructs, IMP and #2, 3_IMP, contained the sequence (ACCAUGG) and were generated by introducing point mutations around the #1 uAUG site using oligonucleotide-directed mutagenesis.
pmMUEG-fused constructs were cloned to the pEGFP-N1 vector with the mouse MOR UTR (−301 to +1). MOR coding regions (exon 1 to exon 4) and green fluorescent protein (GFP) were fused in-frame. First, mouse exon regions were produced by PCR using the following primers: 5′-CAGCAAGCATTCAGAACCATGGACAGCAGCGCCGGCCCAGGGA-3′ (forward) and 5′-TAGGCGCCAGGTACCGAGGGCAATGGAGCAGTTTCT-3′ (reverse), corresponding to −18 to +1218 bp of the mouse MOR. This PCR product was cloned to the Topo-TA vector. The mouse MOR UTR region (−301 to +1) was subsequently fused to the mouse MOR exon regions by ligation (5′-HindIII, 3′-PstI). Finally, these fused constructs were cloned into the pEGFP-N1 vector by ligation (5′-HindIII, 3′-KpnI). All constructs were confirmed by sequencing analysis.
Cell Culture, DNA transfection and reporter gene assay
Human neuroblastoma NMB cells were cultured in RPMI 1640 supplemented with 10% heat-inactivated fetal bovine serum. HEK293 cells were cultured in minimal essential medium supplemented with 10% fetal bovine serum, 100 µg/ml streptomycin, and 100 IU/ml penicillin at 37°C in a humidified atmosphere of 5% CO2. Transfection and reporter gene assays were carried out as described previously (28). Briefly, cells were plated in 6-well dishes at a concentration of 1 × 106 cells/well and cultured overnight before transfection. For luciferase reporter analysis of each promoter construct, 1 µg of the reporter plasmid was mixed with the Effectene transfection reagent (Qiagen) for 10 min before being added to the well. Forty-eight hours posttransfection, cells were washed once with 1× phosphate-buffered saline and lysed with lysis buffer (Promega). To correct for the differences in transfection efficiency, a one-fifth molar ratio of a pCH110 plasmid (Amersham) containing the β-galactosidase gene under the SV40 promoter was included in each transfection for normalization. The luciferase and galactosidase activities of each lysate were determined as according to the manufacturer's instructions (Promega and Tropix, respectively).
Quantification of LUC and LacZ transcripts by real-time PCR and reverse transcription (RT)-PCR
Total RNA was isolated according to the supplier's protocol (TRI Reagent, Molecular Research Center, Inc.). After quantification of total RNA by measuring OD at 260 nm, 1 µg of RNA was treated with 1 U of DNase I (Invitrogen). Reverse transcription using oligo-dT primer was performed with the Transcription First strand cDNA synthesis kit (Roche) according to the manufacturer's protocol. The first strand obtained was quantified using a real-time quantitative PCR system: a SYBR Green assay on the iCycler Optical System (Bio-rad). The following oligonucleotides were used for the amplification of 288 bp and 105 bp fragments of cDNAs, corresponding to LUC or LacZ, respectively: LUC primers, 5′-CTCAGAGAGTGGCGCTTTGGGGATG-3′ (forward) and 5′-CTTTATGTTTTTGGCGTCTTCC-3′ (reverse); LacZ primers, 5′-GCTGCATAAACCGACTACACAAA-3′ (forward) and 5′-GCCGCACATCTGAACTTCAG-3′ (reverse). After first strand cDNA synthesis as described above, the samples were amplified at 95°C for 30 sec, 60°C for 30 sec, 70°C for 30 sec for real-time PCR. For RT-PCR, the Qiagene one-step RT-PCR kit was used with the above primers. The relative LUC mRNA level was reported as the ratio of LUC mRNA/LacZ mRNA.
In vitro transcription/translation and autoradiography
Capped and uncapped mRNAs were synthesized in vitro with the MAXIscript In vitro Transcription Kit (Ambion) according to the manufacturer's instructions. Briefly, after linearization by XbaI digestion, DNA was gel purified (Qiagen). The resulting DNA was transcribed in vitro by SP6 RNA polymerase in the presence (capped) or absence (uncapped) of the methylated cap analog m7GpppG (1 mM; Ambion). After 1 h incubation at 37°C, DNase (2 U) was treated for 15 min at 37°C. After ethanol precipitation and a 70% ethanol wash, RNA was resuspended in DEPC-treated water. RNA integrity was confirmed by gel electrophoresis. The amounts of RNA were analyzed by spectrophotometry and ethidium bromide visualization.
Equal amounts (0.1 µg) of RNA were added to the TnT Quick Coupled Transcription/Translation System (Promega) for translation under conditions recommended by the manufacturer. In-vitro-translated proteins were labeled with l-[35S]-methionine (Amersham). Reactions were incubated at 30°C for 60 min and analyzed on 10% SDS-PAGE. The gels were dried and exposed to a Phosphorimager screen overnight at room temperature. The translated peptides were detected using a Molecular Dynamic Storm 860 Phosphorimager system.
Fluorescence-activated cell-sorting analysis (FACS)
After transfection with each reporter construct containing the GFP coding region, HEK293 cells were fixed with 3.7% formaldehyde prior to FACS analysis. Receptor fluorescence was quantitated by FACScan (Becton Dickinson, Palo Alto, CA). The fluorescence intensity of 10, 000 cells was collected for each sample. Cell Quest software (Becton Dickinson) was used to calculate the mean fluorescence intensity of the cell population. All experiments were conducted at least three times with triplicate samples.
Radioligand binding
Binding activity assays were carried out as described previously (29). Briefly, after transient transfection with the indicated constructs, confirmation of MOR expression was determined by a whole-cell binding assay using [3H]diprenorphine in 25 mM HEPES buffer, 5 mM MgCl2 (pH 7.6). Specific binding was defined as the difference between the radioactivity bound to the cells in the presence and absence of 100 µM CTOP.
Primer extension inhibition (toeprinting) assay
The toeprinting assay was performed as described previously (30), except for the ribosome binding reaction. Briefly, the deoxyoligonucleotides mToe (5′-TCAGTTTCTTACAAGGACAAG-3′) and mToe 2 (5′-CTTATGCAGTTGCTCTCCAG-3′) were labeled at the 5′-end by T4 polynucleotide kinase and [γ-32P]ATP (3000 Ci/mmol; Amersham). In the toeprinting reaction, the 32P-labeled primers were pre-annealed to the mRNA by heating for 2 min at 60°C, followed by incubation at 37°C for 10 min in 40 mM Tris-HCl (pH 7.5) and 0.2 mM EDTA. The primer–mRNA complexes were then incubated on ice for 15 min while the reticulocyte reaction mixtures were assembled.
The ribosome binding reaction used micrococcal-nuclease-treated rabbit reticulocytes (Promega). The reaction mixtures containing 45% reticulocyte lysate were supplemented with 900 µg/ml cycloheximide. In the standard assay, the mixtures were pre-incubated at 25–30°C for 2 min. After pre-incubation, 25 µl aliquots of this mixture containing 2 µl of mRNA/primer (0.1–0.2 µg of mRNA) from the toeprinting reaction were incubated at 25–30°C for 15 min and then diluted with 20 volumes of ice-cold buffer [50 mM Tris-HCl (pH 7.5), 40 mM KCl, 6 mM MgCl2, 500 µM of each of the four dNTPs].
Primer extension was initiated by adding 2 U/µl Superscript II reverse transcriptase (Invitrogen) and incubated at 25–30°C for 10 min. Reactions were terminated by extracting with phenol. Primer extension products were mixed with formamide (98%) and EDTA (10 mM) and heated at 90°C for 5 min before layering onto 4–6% polyacrylamide sequencing gels. Sequencing ladders were generated by an fmol DNA cycle sequencing system (Promega) according to the manufacturer's protocol. Autoradiograms of the dried gels were obtained using a Molecular Dynamic Storm 860 Phosphorimager system.
RESULTS
The mouse MOR 5′-UTR contains three uORFs preceding the MOR initiation codon
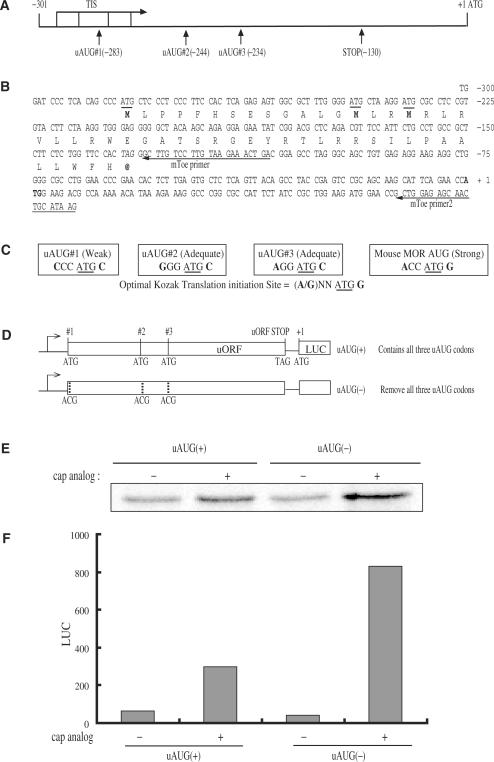
The 301-bp fragment of the mouse MOR 5′-UTR was chosen to clone into reporter constructs for the current uORF study. This 5′-UTR was within the longest major MOR transcripts observed in vivo (designated start codon to +1) (31). Computational sequence analysis of the mouse MOR 5′-region revealed the presence of three uAUGs (uAUG#1, uAUG#2 and uAUG#3) that were in the same open reading frame and shared the same stop codon (Figure 1A). Translation of the first, second and third uAUGs may give rise to uORFs containing 50, 37 and 34 amino acids, respectively, all of which terminate at −128 bp upstream of the mouse MOR initiation codon (Figure 1B). In vertebrate mRNAs, initiation sites usually conform to all or part of the sequence GCCRCCAUGG, known as the Kozak sequence rule (32). The most conserved nucleotides were the R (A or G) at −3 and the G at +4 (the A of the AUG codon was designated as +1). Strong consensus sequences contain both of these important nucleotides, whereas an adequate sequence contains only one of them, and weak sequences contain none of them. The AUG codons initiating the main ORF of a transcript have strong or adequate sequence contexts in 95–97% of the cases. This percentage was lower for the uAUGs (43–63%) (33). In the case of the mouse MOR transcript, uAUG#1 is weak (no match compared the Kozak sequence in −3 and +4), while uAUG#2 (−3 position matched) and uAUG#3 (−3 position matched) have adequate sequences. The mouse MOR main ORF has a strong consensus sequence similar to that of the luciferase main ORF (Figure 1C). To evaluate whether the translation mediated by the mouse MOR 5′-UTR was cap-dependent or -independent, we translated capped and uncapped transcripts in rabbit reticulocyte lysates (RRL) in the presence of 35S-methionine. The presence of the m7G cap increased mouse MOR translation 2- to 3-fold (Figures 1E and F), indicating that under these conditions translation initiation mainly occurred through a cap-dependent mechanism. Furthermore, a significant increase in protein synthesis was observed after elimination of the initiation codon, independent of whether capped or uncapped transcripts were used (Figure 1F). Therefore, the uORF can inhibit MOR translation in vitro.
Figure 1.
Mouse MOR 5′-UTR contains uORFs. Efficient translation of mouse MOR mRNA in vitro is inhibited by the uORF. (A) Schematic representation of the three uORFs in the mouse MOR 5′-UTR. All three uORFs are in the same open reading frame and share a termination codon. The mouse MOR translation initiation site is indicated by +1. (B) Sequence of the mouse MOR 5′-UTR. uAUG codons are underlined, the termination codon of the uORFs is in italics and the main AUG is in bold. The mouse MOR uORF encodes the putative peptide sequence represented under the sequence triplet. Bold peptide sequences represent the uAUG codons and the stop codon. Each uORF consists of 50, 37 and 34 putative amino acids, respectively. mToe and mToe2 indicate radiolabeled oligonucleotide primers for Toeprinting. (C) Each box represents an uAUG sequence compared to the Kozak sequence. The −3 and +4 positions relative to the start codon (underlined) are represented in bold. (D) Diagram of template DNAs used in vitro to generated capped and uncapped transcripts. Point mutations in uAUG(+) eliminate each of the three uAUGs [uAUG(−)]. Dotted lines indicate mutated uAUG sites. (E) Autoradiogram of capped (+) and uncapped (−) transcripts (100 ng) were translated in rabbit reticulocyte lysates (RRL) in the presence of 35S-methionine. Translation products of RRL were analyzed by 10% SDS-PAGE and autoradiography. (F) Relative luciferase expression level of capped and uncapped transcripts.
Repression of mouse MOR translation by the uORF in the MOR 5′-UTR
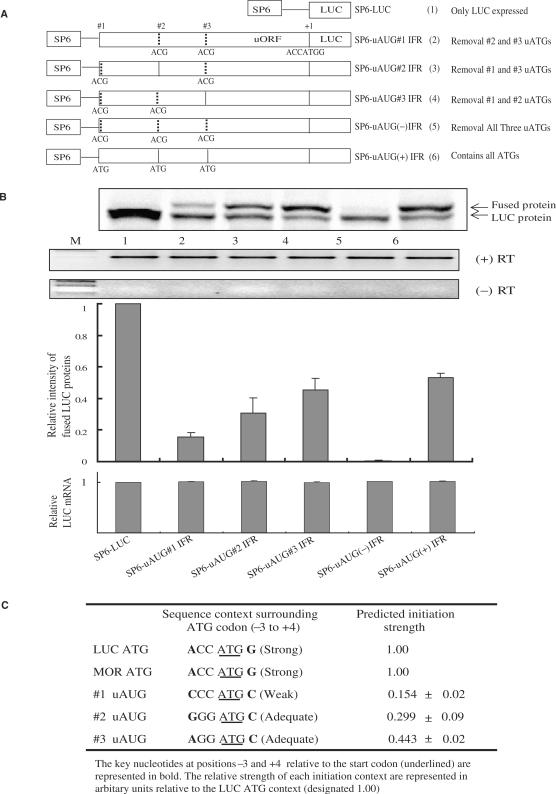
As technical difficulties prevent visualization of the translational products of these uORFs in vivo, we instead examined the initiation of peptide synthesis at individual uAUGs by in vitro translation studies. Several constructs under the control of the SP6 promoter were generated (Figure 2A). In this experiment, LUC synthesis should occur if ribosomes initiate translation at the uAUG codons of the uORF and/or the LUC main initiation site with the TnT Quick Coupled Transcription/Translation System (Promega). The autoradiography results (Figure 2B) show that both the uORF-LUC fusion and LUC proteins were expressed at different levels. As expected, the uORF-LUC fusion protein had a molecular weight slightly higher than the control LUC protein. These data demonstrate that the uORF is efficiently translated. The most effective initiation site was uAUG#3, and the least effective initiation site was uAUG#1. The above results revealed that each of the uORFs were successfully initiated and translated, but these effects were not reflected in any differences in the transcription levels (Figure 2B). These in vitro translation results were similar to those observed with LUC activities in NMB cells transfected with similar constructs. To understand how the translation of the first initiation codon can affect subsequent initiation events, we characterized the relative efficiency of all the initiation codons. Figure 2C shows the relative strength of each initiation context represented in arbitrary units relative to the LUC ATG context (i.e., a Strong Kozak sequence, designated 1.00), which has one of the closest matches to the strong context. The results show that translation is most effectively initiated at uAUG#3 and most weakly at uAUG#1.
Figure 2.
The expression levels from each respective initiation codon within the mouse MOR uORF region. (A) A schematic summary of various in-frame constructs. Mouse MOR uORF regions were fused in-frame to the luciferase gene. Initiation codons with vertical dotted lines indicate point mutations of ATG to ACG. (B) Autoradiogram of the proteins translated by a coupled transcription/translation system in the presence of [35S]-methionine from representative in vitro translations. Proteins were separated by 10% SDS-PAGE (LUC, Fused Protein). Quantification of mRNA levels from the cell-free transcription/translation system was performed by RT-PCR (+/− RT). Expression levels from each initiation codon were normalized against those of SP6-LUC. Normalized mRNA levels (lower panel). M: 100 bp molecular weight markers. (C) Relative strength of each initiation context within the mouse MOR UTR as determined by in vitro translation. The LUC ATG context is designated 1.00.
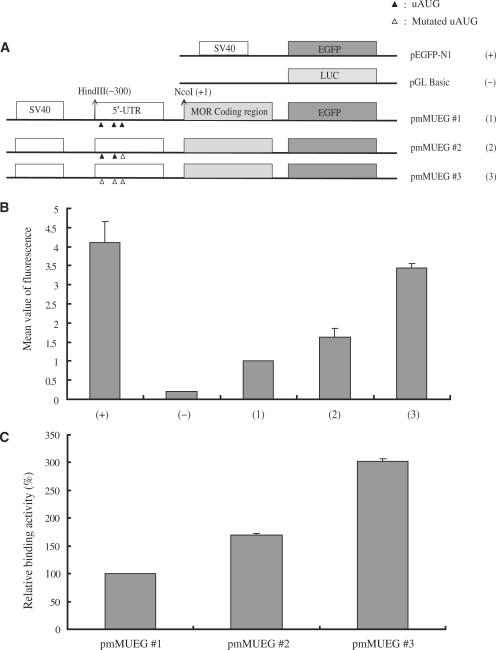
FACS is a type of flow cytometry, a method for sorting a suspension of biological cells based on specific light scattering and fluorescent characteristics of each cell. We prepared GFP-fused mouse MOR constructs (Figure 3A) and confirmed that uORFs mediated mouse MOR expression. As shown in Figure 3B, removal of all three uAUGs in cells transfected with pmMUEG #3 resulted in increased fluorescence levels up to 3.5-fold compared to pmMUEG #1 (wild-type). In cells transfected with pmMUEG #2 (only mutated at uAUG#3), the fluorescence level increased up to 1.7-fold compared to pmMUEG #1. These data suggest that uORFs of the mouse MOR can be repressed downstream of MOR expression with the most effective repression activity containing uAUG#3. Additionally, these three uAUGs could synergistically repress the downstream of MOR gene expression.
Figure 3.
The uORFs can affect the expression of mouse mu opioid receptor. (A) Structures of fused constructs. Each construct contains mouse MOR promoter. The coding regions were fused in-frame to the GFP gene except in the case of the negative control vector (pGL Basic). (B) Flow cytometry analysis of mouse MOR fusion constructs in transiently transfected mammalian cells. EGFP mean value (fluorescence intensity) in HEK293 cells was measured by flow cytometry. Mutation of uAUGs greatly increased the level of GFP expression. The data represent an average of three independent experiments. (C) Radioligand binding assay. Opioid-receptor-expressing cells were transiently transfected with the above-reported DNA constructs. Binding activity of [3H]diprenorphine to NMB cells was carried out as described in Materials and Methods. All values correspond to mean ± SEM calculated from at least three independent experiments.
Because mouse uORFs could repress downstream MOR gene expression, we performed opioid radioligand binding assays (Figure 3C). The binding activity of MOR was monitored by measuring [3H]diprenorphine-specific binding with or without CTOP, a selective antagonist for MOR. After transfection of the constructs (Figure 3A) into NMB cells, pmMUEG #3-transfected cells showed binding activity up to three times higher than that observed in cells transfected with the other uAUG-mutated constructs. The repression effect exhibited in the cells transfected with the uAUG#3-mutated construct confirmed the results found in the flow cytometry studies. These results suggest that uORFs can negatively regulate MOR expression.
Regulation of MOR translation by uORF initiation
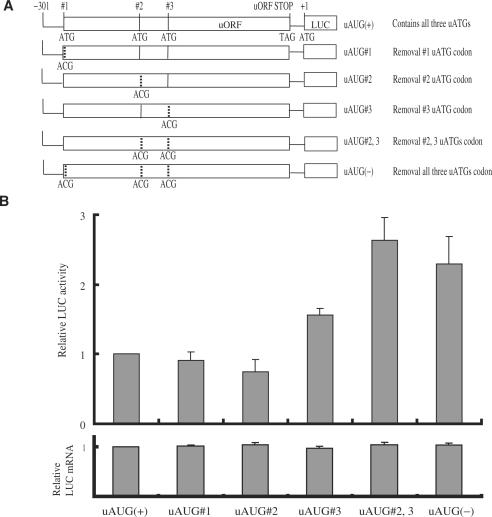
To determine whether these uORFs affect the expression of the downstream main ORF, we constructed a series of plasmids [derived from uAUG(+); see Figure 4A] containing corresponding point mutations for each uAUG (i.e. ATG changed to ACG). These wild-type and mutant constructs were transiently transfected in NMB cells that expressed MOR endogenously. After transfection, cellular extracts were prepared and assayed for translational (luciferase assay, LUC/ß-gal relative activity) and transcriptional (real-time RT-PCR, LUC/LacZ mRNA) levels (Figure 4B). The levels of transcripts were very similar among all constructs, indicating that the point mutations did not alter transcription levels. In contrast, LUC activity was differentially affected by the mutations of these uAUGs. Particularly, mutations within the third uAUG or second/third uAUG (uAUG#3 or uAUG#2, 3) caused a 1.6- and 2.9-fold increase in LUC activity, respectively. Mutations of the uAUG#1 or uAUG#2 had no significant effect. When all the uAUGs were mutated [uAUG(−)], the luciferase activity was not significantly different from that observed with the uAUG#2, 3 mutated constructs. These results suggest that the repression of the LUC activity is due to the presence of the MOR 5′-UTR occurring at a translational level, and that the second/third uAUGs are involved in this repressive effect synergistically.
Figure 4.
Translation of mouse MOR gene is controlled by three uORFs. (A) Schematic representation of reporter constructs with wild-type and mutant mouse 5′-UTRs. Vertical dotted lines represent ATGs converted to ACGs by point mutations. (B) Transient transfection of each mutant construct in NMB cells. After transfection, cells were trypsinized and half were used for luciferase and ß-galactosidase activity assays, while the other half were examined by RNA extraction and transcript quantification. Relative LUC activity and mRNA levels were determined as the ratio LUC/ß-gal and LUC/LacZ as described in Materials and Methods. The error bars indicate the standard errors of triplicate LUC assays.
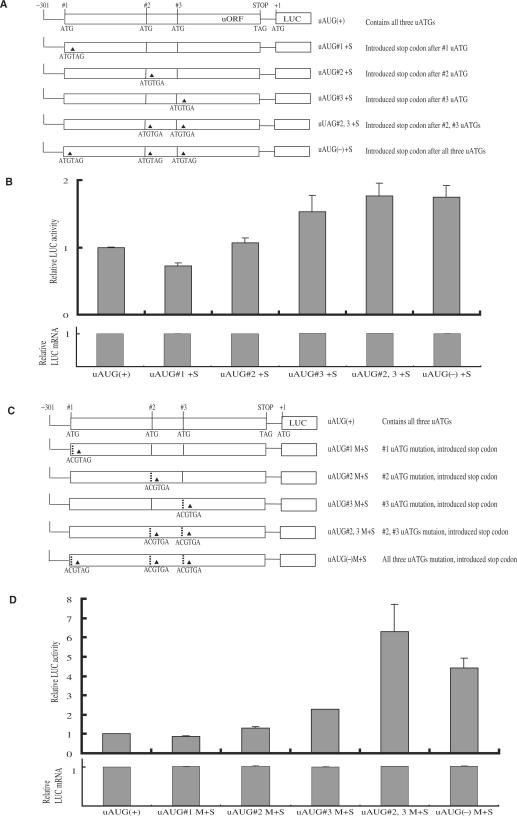
Repression by uORFs in some eukaryotic genes is dependent on the peptide-coding sequence of their uORF (34,35). To assess the importance of these uORF peptides in the translation of the downstream ORF, the uORF sequences were mutated by introducing a stop codon within the sequence (Figure 5A). Introducing the stop codon after uAUG#3 either alone (uAUG#3 +S) or in combination with a stop codon inserted after uAUG#2 (uAUG#2, 3 +S) increased LUC activity by about 1.5- and 1.7-fold, respectively (Figure 5B). Again, these increases in LUC activity were not caused by alterations in transcriptional activity. In order to examine the full repression activity of individual uORFs, constructs with uAUG mutations combined with a stop codon mutation within each uORF were generated (M + S) (Figure 5C). The third uAUG plus stop construct mutation (uAUG#3 M + S) increased LUC activity more than the uAUG#3 +S construct but did not affect the mRNA content. The uAUG#2, 3 M + S and uAUG(−) M + S constructs also increased LUC activity to a greater degree than that observed with the uAUG#3 M + S construct (Figure 5D). It is therefore possible that the MOR uORFs negatively regulate expression independent of their peptide-coding sequences.
Figure 5.
Inhibition of translation by the uORF is independent of its peptide sequence (A and C). Schematic representations of the DNA constructs used in this study. Shaded triangles indicate point mutations to the Stop (S) codon; vertical dotted lines indicate point mutations of ATG to ACG (M) (B and D). Forty-eight hours after transfection, cells were trypsinized and examined for luciferase and ß-galactosidase activities, as well as by RNA extraction and transcript quantification. Relative LUC activity and mRNA levels were expressed as the ratio LUC/ß-gal and LUC/LacZ, as described in Materials and Methods.
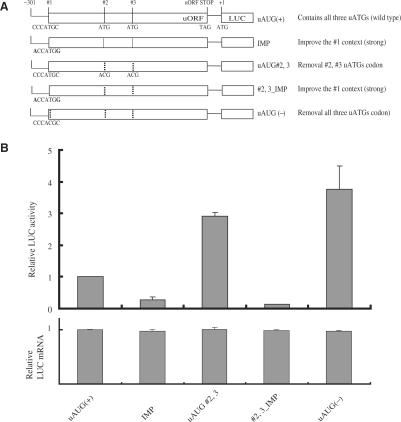
To confirm our hypothesis of context-dependent leaky scanning, we designed 5′-UTR constructs fused with luciferase as a reporter (Figure 6A). These constructs contain the original context, or an improved uAUG context based on the Kozak sequence rule. The strengths of the AUG context sequences ranged from ‘strong’ to ‘weak’ and in descending order were A−3 + G+4 > G−3 + G+4 > A−3 + A+4 > G−3 + A+4 > U−3+ G+4 > U−3 + A+4 (19). We modified the context of the first uAUG (weak) to an improved (IMP) sequence (ACCAUGG; strong). After transfection of these constructs into NMB cells, expression was analyzed by luciferase assays. The IMP construct decreased reporter activity about five-fold relative to that of the wild-type construct [uAUG (+)] (Figure 6B). Furthermore, introduction of the stronger uAUG context around the first uAUG, combined with mutation within the second/third uAUG (#2, 3_IMP) decreased reporter activity 24-fold, relative to the activity of uAUG#2, 3 constructs. The levels of transcripts were very similar among all constructs, indicating that these mutations did not alter transcription levels (Figure 6B, lower panel). These data suggest that a stronger AUG context at the first uAUG position could recruit ribosome complexes more efficiently to initiate translation, and represses the main AUG initiation site via a context-dependent leaky scanning mechanism. Taken together, these data suggest that the original weak context of the first uAUG (#1 uAUG) has a lower potential for initiation of translation and inhibition of the main AUG initiation site compared to those of the second and third uAUG (#2 and #3 uAUG).
Figure 6.
uAUG sequence contexts of different strengths can regulate the ribosome initiation activity of the main AUG. (A) Schematic of constructs containing three mouse MOR uAUGs with contexts of various strengths at the #1 uAUG site. One set of constructs (IMP) contains an improved uAUG context (‘strong’) at the first uAUG position. For another set of constructs, after removal of the #2 and #3 uAUG (uAUG#2, 3), the improved contexts were also added to the first uAUG position (#2, 3_IMP). The context improvements at the first uAUG (ACCATGG) in each construct differ from each other at positions −3 and +4, as indicated in bold. (B) Graphic representation of relative luciferase activity determined by luciferase assay (Promega) of the constructs shown in (A). Relative LUC activity and mRNA levels were determined as the ratio LUC/ß-gal and LUC/LacZ, as described in Materials and Methods. The error bars indicate the standard errors of triplicate LUC assays.
Ribosome leaky scanning regulates mouse MOR gene expression
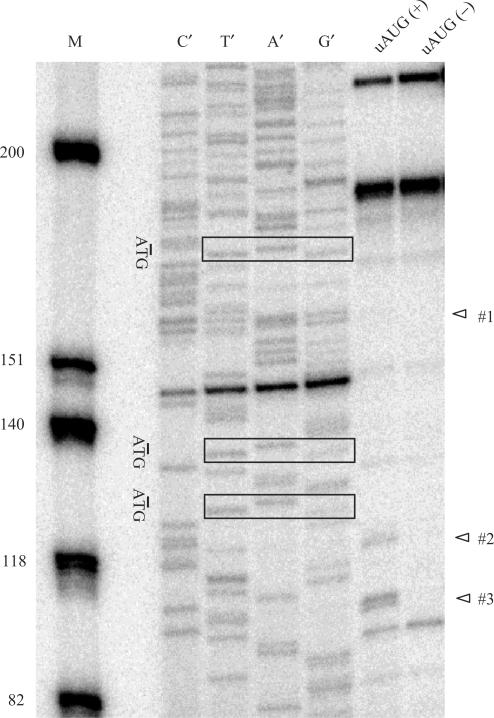
Toeprinting assays have been known to reveal the effects of initiation context and regulated-ribosome stalling on the association of ribosomes with mRNA (36). We therefore examined two mRNAs: the uAUG(+) construct contains the uORFs in their wild-type initiation context, while the uAUG(−) construct was mutated in all three uAUG sites. Toeprints corresponding to ribosomes at each of the MOR uORF start codons were observed, except for uAUG#1 (Figure 7). Toeprints corresponding to ribosomes at the start codon of uAUG#2 and uAUG#3 were also observed in rabbit reticulocyte lysates by radiolabeled mToe primer. The uAUG#2 and uAUG#3 initiation-codon toeprint maps showed 16 nucleotides downstream of the AUG codons, indicating ribosomes with the initiation codon in their P-site (37,38). As shown in the preceding experiments, both the uAUG#2 and uAUG#3 sequences were sufficient by the Kozak sequence rule, but stronger inhibition was observed in uAUG#3. Figure 7 shows similar results: the uAUG#3 toeprint was stronger than the uAUG#2, while the uAUG#1 toeprint was not detected in uAUG(+) transcripts. However, in transcripts where all three uAUGs were mutated [uAUG(−)], no toeprints were detected. These data indicate that ribosomes scanned linearly from the 5′-end of the mRNA to initiate translation at each of these start codons rather than being loaded internally. Minor bands were also seen; these minor products might represent either authentic truncated mRNAs or false priming (36).
Figure 7.
Toeprint analyses of initiation of uAUGs. Synthetic RNA transcripts (100 ng) were used to program translation mixtures derived from RRL (Promega). Mouse MOR-LUC transcripts containing all three uORFs [i.e. wild-type; uAUG(+)] or mutated at all three uAUGs [uAUG(−)] were incubated at 30°C for 15 min in micrococcal-nuclease-treated RRL. Radiolabeled mToe primer was used for primer extension analyses and for sequencing of the uAUG(+) templates. The nucleotide complementary to the dideoxynucleotide added to each sequencing reaction is indicated above the corresponding lane (C′, T′, A′ and G′), so that the sequence of the template can be directly deduced by 5′ to 3′ sequence reads from top to bottom. M, dephosphorylated Φ174 HinfI markers (Promega); Rectangles, sequence position of uAUGs; Open triangles, indicated toeprint of each uAUG.
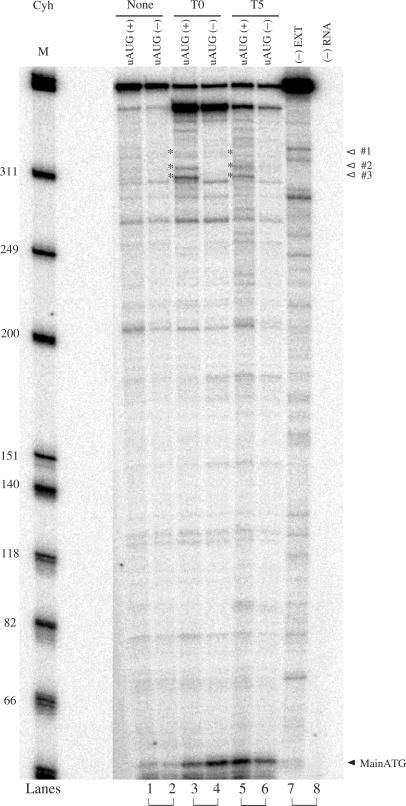
By adding the translational elongation inhibitor cycloheximide to rabbit reticulocyte lysates under steady-state conditions, toeprints corresponding to ribosomes at each of the uORF start codons were observed (Figure 8). As controls, toeprinting was performed on reaction mixtures lacking either RNA or extract. Without added RNA, negligible cDNA synthesis from the radiolabeled primer was observed (Figure 8, lane 8). When RNA is added without extract, the major cDNA product is the full-length cDNA (Figure 8, lane 7). Additional common premature termination products occurred in all of the lanes. Many of these were also observed in primer extension analyses of RNA in the absence of extract but not in primer extension analyses of extract in the absence of exogenously added mRNA. We compared toeprints when cycloheximide was added to extract prior to adding RNA template (T0) to those obtained when cycloheximide was added after translation of the RNA was underway (T5). Cycloheximide at T0 should show where ribosomes first initiate translation, since the cycloheximide interferes with elongation, not initiation. When cycloheximide was added to rabbit reticulocyte lysates programmed with mouse MOR-LUC mRNA, ribosomes were observed at both the uORF and LUC initiation codons (Figure 8, lanes 3 and 4). Thus, when cycloheximide was added early (T0), significant leaky scanning past the MOR uORF initiation codon was observed. Samples containing the uAUG#2 and uAUG#3 codons (Figure 8, lane 3) exhibit toeprints for those sequences as well as for the LUC start codon. In contrast, samples deleted of all the uAUGs (Figure 8, lane 4) exhibited only the LUC start codon, and at higher levels than in the wild-type construct. When cycloheximide was added later (T5), toeprints were observed for both the uAUGs and LUC start codons (Figure 8, lanes 5 and 6). If the uORF mechanism functions by reinitiation, scanning ribosomes would load only at the uORF start codon at T0 state while later (T5) ribosomes would load at both uORF and LUC start codon. However, if the uORF mechanism functions by leaky scanning, ribosomes at both the uORF and LUC start codons would load at both T0 and T5 (36). The results obtained here support the notion that ribosomes reach the downstream start codon in the MOR mRNA by the leaky scanning mechanism, rather than by reinitiation.
Figure 8.
Toeprinting analyses of mouse MOR containing uORFs that regulate ribosome leaky scanning. Synthetic RNA (100 ng) transcripts were used to program translation mixtures derived from RRL. Mouse MOR-LUC RNA containing the wild-type uORFs [uAUG(+)] or deleted of all three uORFs [uAUG(−)] were incubated at 30°C for 15 min in reaction mixtures without cycloheximide (None) or supplemented with cycloheximide (Cyh) either prior to incubation (T0) or after 5 min of incubation at 30°C (T5). Control samples of RNA in reaction mixtures without extract (-EXT) or reaction mixtures containing extract without mRNA (-RNA) are indicated. The positions of the cDNA extension products corresponding to the mRNA 5′ end, uORF initiation (open triangles, asterisks) and LUC initiation codon (filled triangle) by radiolabeled mToe2 primer are indicated. During leaky scanning, ribosomes scan and load at either the upstream or downstream start codons. Adding cycloheximide at T0 traps ribosomes at the positions where they first load on the mRNA. Adding it during steady-state translation (T5) traps ribosomes where they are loaded, following the primary initiation event and subsequent reinitiation events.
DISCUSSION
The distribution of opioid receptors and the expression patterns of their mRNAs have been extensively examined (39,40). Opioid receptor proteins are also regulated by variations in the mRNA sequences (10). These mRNA variants differ mainly in the 5′- or 3′-UTR regions. As such, regulation of opioid receptor expression involves not only transcriptional control at the DNA level, but also posttranscriptional control at the RNA level (11). We have characterized a novel regulatory mechanism of mouse MOR expression mediated by uAUG/uORF in this study. The 301-bp extension of the mouse MOR 5′-UTR region includes the major MOR mRNA transcription sequence (derived from the proximal MOR promoter), and contains three uORFs. These uORFs use the same termination codon (Figure 1). In most well-characterized examples of regulatory uORFs, mutation of the uAUG codon alters protein expression without affecting mRNA abundance. However, uORFs do not always inhibit translation; cases have been described in which uORFs stimulated expression of downstream ORFs (41,42). If removal of uAUGs alters mRNA levels, then the AUG nucleotides may alter transcription or RNA stability, independent of translation of the uORF (16). We found that the presence of the uORF region suppressed translation without changing MOR transcription levels. This was due to three uAUGs present in the region from nucleotides −301 to +1 bp in the MOR 5′-UTR. As uORF mechanisms usually function in a cap-dependent manner (23), we investigated this well-characterized regulatory mechanism by using capped and uncapped transcripts for in vitro translation (Figure 1E and F). We determined that mouse MOR containing uORFs do indeed function in a cap-dependent manner.
The initiation efficiencies of each uAUG are important features of the codons in MOR 5′-UTR leaky scanning models (43,44). The relative strengths of the initiation codons were tested by in vitro translation (Figure 2). The uAUG#1 had a very weak initiation context, while uAUG#2 had a slightly stronger initiation context. The uAUG#3 had the strongest initiation context. This likely facilitates higher levels of leaky scanning, enabling peptide synthesis from the uAUG#3 initiation codon on the MOR 5′-UTR. When the sequence was mutated at all three uAUGs [uAUG(−)], only peptide products initiated from the main uAUG were produced. These results indicate that all three uAUGs can effectively be initiated and produce peptides, and that uORFs can negatively regulate downstream MOR ORF initiation.
We also performed FACS analyses and receptor binding assays to examine cell surface expression of the receptor (45,46). The results confirmed that these uORFs mediated down-regulation of MOR expression. Indeed, the presence of the uORFs decreased expression by up to 3.5-fold relative to samples lacking the uORFs (Figure 3).
We further confirmed the context-dependent initiation activity of each uAUG by mutation. Translation initiation strength was most efficient in the third uAUG (Figure 4B). If the uAUGs reside in a non-optimal context, the scanning ribosomal complex may bypass possible starting AUGs by leaky scanning. Transient transfection results suggest that the inhibition of the main AUG imposed by one of the uORF (uAUG#3) was greater than that produced by the other uORFs (uAUG#1 or uAUG#2).
Moreover, some peptides produced by translation of uORFs have been reported to interact with the ribosome, further diminishing the efficiency of protein translation (47,48). In contrast, regulation of yeast GCN4 translation by nutrient levels was independent of the peptide sequences encoded by its uORFs (49,50). Our current study showed that introduction of stop codon constructs (+S), thereby changing the sequence of peptide encoded by the uORF, exhibited changes similar to those seen in the translation of the downstream cistron (Figure 5B). Furthermore, mutating the uAUGs while also introducing the stop codon (M + S) caused a marked increase in the translation of the downstream cistron (Figure 5D). These data show that regulation of the MOR uORF is independent of the peptide sequence of the uORF. Thus, the inhibitory effect of the uORF during MOR translation is a protein-independent mechanism. Although the mechanisms involved in explaining the inhibitory effects of the uORF peptide on translation are not understood, several models could be proposed. For example, the peptide of the uORF could be synthesized and have the ability to inhibit translation only at high concentrations in the local microenvironment (51).
As shown in Figure 6, differences in LUC levels were detected between constructs with different uAUG contexts. This indicates that ribosomes initiating at the main AUG must have scanned the uAUGs, especially when these uAUGs contain strong or adequate contexts. Leaky scanning is an extension of the scanning model and suggests that an AUG might be bypassed by scanning ribosomes if it resides in a suboptimal context. This mechanism could only occur by context-dependent leaky scanning.
It is known that uAUGs can inhibit translation in several ways. When they are recognized by translational machinery, a futile cycle can occur, such that only the ribosomes skipping the uAUG (leaky scanning) can reach the main ORF. Accordingly, the level of inhibition is directly related to the context for translation initiation: the better an uAUG is recognized, the greater the resulting inhibition (23). Even in conditions of optimal uAUG recognition, a small percentage of ribosomes, after translation of the uORF, can continue scanning for start sites (reinitiation), eventually reaching the AUG of the main ORF (44,52).
Despite efficient translation of the uORF, the main cistron remains translated. Several mechanisms can explain the translation of the downstream cistron by internal ribosome entry, ribosome reinitiation and leaky scanning. The stronger suppression of uAUGs with respect to uORFs can be partially explained by the fact that initiation at the main ORF may occur only by leaky scanning in uAUG-containing transcripts, whereas both leaky scanning and ribosome reinitiation may allow initiation at the AUG of main uORF-containing transcripts (53). Figure 7 shows that the uAUG toeprint binds exactly to each AUG site, confirming the prior experiments. These toeprints show the same Kozak sequence rule. In the case of reinitiation, they must first initiate translation at an uAUG codon. Therefore, when cycloheximide is added to arrest translation elongation before ribosomes can initiate translation, ribosomes should collect at the uAUG codon but not at the downstream AUG. However, in the case of leaky scanning, they will load at the downstream start codon without prior translation of an uORF (50). Therefore, whether Cyh is added to the translation reaction prior to initiation or under steady-state conditions, ribosomes will stall at both the uAUG and main AUG codons.
In conclusion, we provide evidence that mouse MOR expression is inhibited at the translational level by the presence of uORFs. Mainly, the uAUG#3 in the 5′-UTR of the MOR mRNA functions efficiently as a translation initiation site. However, all three uAUGs synergistically regulate translation of the main AUG. The MOR uORF uses a mechanism independent of peptide sequence, and translational repression of MOR uORF is not dependent on the intercistronic region. Furthermore, leaky scanning is involved in inhibition of physiological AUG-initiated MOR translation, resulting in weak expression of MOR under normal conditions. These uORFs have the potential to exert a major impact on MOR gene expression, and some, but not all, serve as important regulatory elements under normal conditions.
ACKNOWLEDGEMENTS
This work was supported by National Institutes of Health Research Grants DA000564, DA001583, DA011806, K05-DA070554, DA011190, DA013926, and by the A&F Stark Fund of the Minnesota Medical Foundation. The costs of publication of this article were defrayed in part by the payment of page charges. The article must therefore be marked as ‘advertisement’ in accordance with 18 U.S.C. Section 1734, solely to indicate this fact. Funding to pay the Open Access Publication charge was provided by the grants listed above.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kieffer BL. Recent advances in molecular recognition and signal transduction of active peptides. Cell. Mol. Neurobiol. 1995;15:615–635. doi: 10.1007/BF02071128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loh HH, Smith AP. Molecular characterization of opioid receptors. Annu. Rev. Pharmacol. 1990;30:123–147. doi: 10.1146/annurev.pa.30.040190.001011. [DOI] [PubMed] [Google Scholar]
- 3.Olson GA, Olson RD, Kastin AJ. Endogenous opiates. Peptides. 1997;18:1651–1688. doi: 10.1016/s0196-9781(97)00264-7. [DOI] [PubMed] [Google Scholar]
- 4.Wood PL, Iyengar S. The Opiate Receptors. Clifton, NJ: Humana Press Inc.; 1988. pp. 307–356. [Google Scholar]
- 5.Loh HH, Liu CH, Cavalli A, Yang W, Chen YF, Wei LN. mu Opioid receptor knockout in mice: effects on ligand-induced analgesia and morphine lethality. Mol. Brain Res. 1998;54:321–326. doi: 10.1016/s0169-328x(97)00353-7. [DOI] [PubMed] [Google Scholar]
- 6.Harrison LM, Kastin AJ, Zadina JE. Opiate tolerance and dependence: receptors, G-proteins, and antiopiates. Peptides. 1998;19:1603–1630. doi: 10.1016/s0196-9781(98)00126-0. [DOI] [PubMed] [Google Scholar]
- 7.Law PY, Wong YH, Loh HH. Molecular mechanisms and regulation of opioid receptor signaling. Annu. Rev. Pharmacol. Toxicol. 2000;40:389–430. doi: 10.1146/annurev.pharmtox.40.1.389. [DOI] [PubMed] [Google Scholar]
- 8.Mignone F, Gissi C, Liuni S, Pesole G. Untranslated regions of mRNAs. Genome Biol. 2002;2:1–10. doi: 10.1186/gb-2002-3-3-reviews0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei LN, Loh HH. Regulation of opioid receptor expression. Curr. Opin. Pharmacol. 2002;2:69–75. doi: 10.1016/s1471-4892(01)00123-0. [DOI] [PubMed] [Google Scholar]
- 10.Wei LN, Law PY, Loh HH. Post-transcriptional regulation of opioid receptors in the nervous system. Frontiers in Bioscience. 2004;9:1665–1679. doi: 10.2741/1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Law PY, Loh HH, Wei LN. Insights into the receptor transcription and signaling implications in opioid tolerance and dependence. NeuroPharmacology. 2004;47:300–311. doi: 10.1016/j.neuropharm.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 12.Van der Veiden AW, Thomas AA. The role of the 5′ untranslated region of an mRNA in translation regulation during development. Int. J. Biochem. Cell. Biol. 1999;31:87–106. doi: 10.1016/s1357-2725(98)00134-4. [DOI] [PubMed] [Google Scholar]
- 13.Jansen RP. mRNA localization: message on the move. Nat. Rev. Mol. Cell. Biol. 2001;2:247–256. doi: 10.1038/35067016. [DOI] [PubMed] [Google Scholar]
- 14.Bashirullah A, Cooperstock RL, Lipshitz HD. Spatial and temporal control of RNA stability. Proc. Natl. Acad. Sci. U. S. A. 2001;98:7025–7028. doi: 10.1073/pnas.111145698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu Q, Hwang CK, Yao S, Law PY, Loh HH, Wei LN. A major species of mouse mu-opioid receptor mRNA and its promoter-dependent functional polyadenylation signal. Mol. Pharmacol. 2005;68:279–285. doi: 10.1124/mol.105.012567. [DOI] [PubMed] [Google Scholar]
- 16.Morris DR, Geballe AP. Upstream open reading frames as regulators of mRNA translation. Mol. Cell. Biol. 2000;20:8635–8642. doi: 10.1128/mcb.20.23.8635-8642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peri S, Pandey A. A reassessment of the translation initiation codon in vertebrates. Trends Genet. 2001;17:685–687. doi: 10.1016/s0168-9525(01)02493-3. [DOI] [PubMed] [Google Scholar]
- 18.Pesole G, Gissi C, Grillo G, Licciulli F, Liuni S, Saccone C. Analysis of oligonucleotide AUG start codon context in eukaryotic mRNAs. Gene. 2000;261:85–91. doi: 10.1016/s0378-1119(00)00471-6. [DOI] [PubMed] [Google Scholar]
- 19.Wang XQ, Rothnagel JA. 5′-untranslated regions with multiple upstream AUG codons can support low-level translation via leaky scanning and reinitiation. Nucleic Acids Res. 2004;32:1382–1391. doi: 10.1093/nar/gkh305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pestova TV, Klupaeva VG. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Gene Dev. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kozak M, Shatkin AJ. Migration of 40 S ribosomal subunits on messenger RNA in the presence of edeine. J. Biol. Chem. 1978;253:6568–6577. [PubMed] [Google Scholar]
- 22.Kozak M. The scanning model for translation: an update. J. Cell. Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- 24.Meijer HA, Thomas AA. Ribosomes stalling on uORF1 in the Xenopus Cx41 5′ UTR inhibit downstream translation initiation. Nucleic Acids Res. 2003;31:3174–3184. doi: 10.1093/nar/gkg429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozak M. Pushing the limits of the scanning mechanism for initiation of translation. Gene. 2002;299:1–34. doi: 10.1016/S0378-1119(02)01056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kozak M. An analysis of vertebrate mRNA sequences: intimations of translational control. J. Cell Biol. 1991;115:887–903. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vilela C, McCarthy JE. Regulation of fungal gene expression via short open reading frames in the mRNA 5′-untranslated region. Mol. Microbiol. 2003;49:859–867. doi: 10.1046/j.1365-2958.2003.03622.x. [DOI] [PubMed] [Google Scholar]
- 28.Kim CS, Hwang CK, Choi HS, Song KY, Law PY, Wei LN, Loh HH. Neuron-restrictive silencer factor (NRSF) functions as a repressor in neuronal cells to regulate the mu opioid receptor gene. J. Biol. Chem. 2004;279:46464–46473. doi: 10.1074/jbc.M403633200. [DOI] [PubMed] [Google Scholar]
- 29.Choi HS, Kim CS, Hwang CK, Song KY, Wang W, Qiu Y, Law PY, Wei LN, Loh HH. The opioid ligand binding of human l-opioid receptor is modulated by novel splice variants of the receptor. Biochem. Biophys. Res. Commun. 2006;343:1132–1140. doi: 10.1016/j.bbrc.2006.03.084. [DOI] [PubMed] [Google Scholar]
- 30.Kozak M. Primer extension analysis of eukaryotic ribosome-mRNA complexes. Nucleic Acids Res. 1998;26:4853–4859. doi: 10.1093/nar/26.21.4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Min BH, Augustin LB, Felsheim RF, Fuchs JA, Loh HH. Genomic structure analysis of promoter sequence of a mouse mu opioid receptor gene. Proc. Natl. Acad. Sci. U. S. A. 1994;91:9081–9085. doi: 10.1073/pnas.91.19.9081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kozak M. An analysis of 5′-noncoding sequence from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meijer HA, Thomas AA. Control of eukaryotic protein synthesis by upstream open reading frames in the 5′-untranslated region of an mRNA. J. Biochem. 2002;367:1–11. doi: 10.1042/BJ20011706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao J, Geballe AP. Coding sequence-dependent ribosomal arrest at termination of translation. Mol. Cell. Biol. 1998;16:603–608. doi: 10.1128/mcb.16.2.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reynold K, Zimmer AM, Zimmer A. Regulation of RAR beta 2 mRNA expression: evidence for an inhibitory peptide encoded in the 5′-untranslated region. J. Biol. Chem. 1996;272:255–261. doi: 10.1083/jcb.134.4.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sachs MS, Wang Z, Gaba A, Fang P, Belk J, Ganesan R, Amrani N, Jacobson A. Toeprint analysis of the positioning of translation apparatus components at initiation and termination codons of fungal mRNAs. Methods. 2006;26:105–114. doi: 10.1016/S1046-2023(02)00013-0. [DOI] [PubMed] [Google Scholar]
- 37.Wang Z, Sachs MS. Arginine-specific regulation mediated by the Neurospora crassa arg-2 upstream open reading frame in a homologous, cell-free in vitro translation system. J. Biol. Chem. 1997;272:255–261. doi: 10.1074/jbc.272.1.255. [DOI] [PubMed] [Google Scholar]
- 38.Pestova TV, Hellen CU, Shatsky IN. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosome entry. Mol. Cell. Biol. 1996;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elde R, Arvidsson U, Riedl M, Vulchanova L, Lee JH, Dado R, Nakano A, Chakrabarti S, Zhang X, Loh HH, Law PY, Hokfelt T, Wessendorf M. Distribution of neuropeptide receptors. New views of peptidergic neurotransmission made possible by antibodies to opioid receptor. Annals NY Acad. Sci. 1995;757:390–404. doi: 10.1111/j.1749-6632.1995.tb17497.x. [DOI] [PubMed] [Google Scholar]
- 40.Mansour A, Fox CA, Akil H, Watson SJ. Opioid receptor mRNA expression in the rat CNS: anatomical and functional implications. Trends in Neurosci. 1995;18:22–29. doi: 10.1016/0166-2236(95)93946-u. [DOI] [PubMed] [Google Scholar]
- 41.Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 42.Pooggin MM, Hohn T, Futterer J. Role of a short open reading frame in ribosome shunt on the cauliflower mosaic virus RNA leader. J. Biol. Chem. 2000;275:17288–17296. doi: 10.1074/jbc.M001143200. [DOI] [PubMed] [Google Scholar]
- 43.Fouillot N, Tlouzeau S, Rossignol JM, Jean-Jean O. Translation of the hepatitis B virus P gene by ribosomal scanning as an alternative to internal initiation. J. Virol. 1993;67:4886–4895. doi: 10.1128/jvi.67.8.4886-4895.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin CG, Lo SJ. Evidence for involvement of a ribosomal leaky scanning mechanism in the translation of the hepatitis B virus pol gene from the viral pregenome RNA. Virology. 1992;188:342–352. doi: 10.1016/0042-6822(92)90763-f. [DOI] [PubMed] [Google Scholar]
- 45.Law PY, Erickson-Herbrandson LJ, Zha QQ, Solberg J, Chu J, Sarre A, Loh HH. Heterodimerization of mu- and delta-opioid receptors occurs at the cell surface only and requires receptor-G protein interactions. J. Biol. Chem. 2005;280:11152–11164. doi: 10.1074/jbc.M500171200. [DOI] [PubMed] [Google Scholar]
- 46.Wang W, Loh HH, Law PY. The intracellular trafficking of opioid receptors directed by carboxyl tail and a di-leucine motif in Neuro2A cells. J. Biol. Chem. 2003;278:36848–36858. doi: 10.1074/jbc.M301540200. [DOI] [PubMed] [Google Scholar]
- 47.Jousse C, Bruhat A, Carraro V, Urano F, Ferrara M, Ron D, Fafournoux P. Inhibition of CHOP translation by a peptide encoded by an open reading frame localized in the chop 5′UTR. Nucleic Acids Res. 2001;29:1864–1867. doi: 10.1093/nar/29.21.4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gong F, Yanofsky C. Instruction of translating ribosome by nascent peptide. Science. 2002;297:1864–1867. doi: 10.1126/science.1073997. [DOI] [PubMed] [Google Scholar]
- 49.Hinnebusch AG. In: Translational Control. Sonenberg N, Hershey JWB, Mathews MB, editors. NY: Cold Spring Harbor Press, Cold Spring Harbor, NY; 1996. [Google Scholar]
- 50.Gaba A, Wang Z, Krishnamoorthy T, Hinnebusch AG, Sachs MS. Physical evidence for distinct mechanisms of translational control by upstream open reading frames. EMBO J. 2001;20:6453–6463. doi: 10.1093/emboj/20.22.6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parola AL, Kobilka BK. The peptide product of a 5′ leader cistron in the beta 2 adrenergic receptor mRNA inhibits receptor synthesis. J. Biol. Chem. 1994;269:4497–4505. [PubMed] [Google Scholar]
- 52.Griffin E, Re A, Hamel N, Fu C, Bush H, McCaffrey T, Asch AS. A link between diabetes and atherosclerosis: Glucose regulates expression of CD36 at the level of translation. Nat. Med. 2001;7:840–846. doi: 10.1038/89969. [DOI] [PubMed] [Google Scholar]
- 53.Iacono M, Mignone F, Pesole G. uAUG and uORFs in human and rodent 5′untranslated mRNAs. Gene. 2005;349:97–105. doi: 10.1016/j.gene.2004.11.041. [DOI] [PubMed] [Google Scholar]