Abstract
Stem loop 1 (SL1) is a highly conserved hairpin in the 5′-leader of the human immunodeficiency virus type I that forms a metastable kissing dimer that is converted during viral maturation into a stable duplex with the aid of the nucleocapsid (NC) protein. SL1 contains a highly conserved internal loop that promotes the kissing–duplex transition by a mechanism that remains poorly understood. Using NMR, we characterized internal motions induced by the internal loop in an SL1 monomer that may promote the kissing–duplex transition. This includes micro-to-millisecond secondary structural transitions that cause partial melting of three base-pairs above the internal loop making them key nucleation sites for exchanging strands and nanosecond rigid-body stem motions that can help bring strands into spatial register. We show that while Mg2+ binds to the internal loop and arrests these internal motions, it preserves and/or activates local mobility at internal loop residues G272 and G273 which are implicated in NC binding. By stabilizing SL1 without compromising the accessibility of G272 and G273 for NC binding, Mg2+ may increase the dependence of the kissing–duplex transition on NC binding thus preventing spontaneous transitions from taking place and ensuring that viral RNA and protein maturation occur in concert.
INTRODUCTION
Like most retroviruses, the human immunodeficiency virus type I (HIV-1) selectively packages two copies of its RNA genome (1–4). The two copies are held together as a non-covalent dimer at the dimer linkage structure (DLS) (5–7) in the 5′-leader RNA (8,9). The RNA dimer has been shown to exert advantageous roles in reverse transcription, including in promoting recombination and generation of drug-resistant strains and its formation has been linked to packaging of the genomic RNA and viral maturation (3,4).
Stem loop1 (SL1) is a highly conserved 35-nt hairpin in the HIV-1 5′-leader RNA (Figure 1A) and a key component of the DLS structure (6,7,10). SL1 contains a self-complementary GC-rich apical loop that promotes dimerization of the HIV-1 genome by forming metastable kissing dimers that are held together by intermolecular base-pairing (Figure 1B) (1,2,6,7). During viral maturation, the metastable kissing dimer undergoes a transition into a more stable dimer in which the two genome copies are more strongly associated (11,12). This transition occurs following the proteolytic release of the N-terminus domain of nucleocapsid (NC) protein from the Gag precursor (13). The processing of Gag has also been shown to be dependent on formation of the RNA dimer (14) and on interactions with the HIV-1 RNA genome (15,16), indicating that maturation of viral proteins and RNA are tightly coupled events.
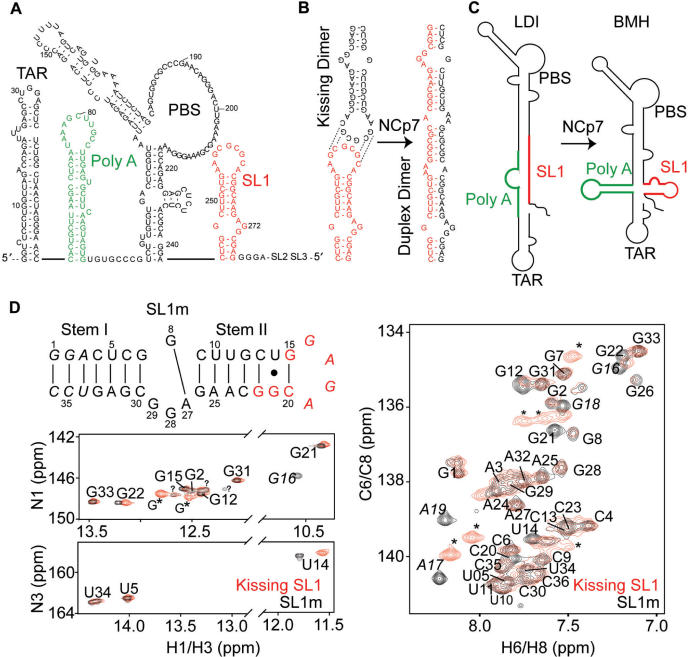
Figure 1.
Structural transitions involving SL1. (A) The 5′-leader in the HIV-1 RNA genome. Dynamical transitions between (B) metastable kissing and stable duplex SL1 dimers and (C) ‘long distance interaction’ (LDI) and ‘branched multiple hairpin’ (BMH) leader conformers. (D) Spectroscopic comparison of the SL1m mutant used in the NMR study and the wild-type kissing dimer. Base-pairs added to stem I are shown in italic. Residues exhibiting significant chemical shift differences relative to the wild-type kissing dimer are shown in red. Overlays of 2D 1H–15N and 1H–13C and HSQC spectra of SL1m (in black) on corresponding spectra of the wild-type SL1 kissing dimer (in red) in the absence of Mg2+ are shown. Peaks from the GC-rich loop in the kissing dimer are indicated using an asterisk and those belonging to the GAGA SL1m tetraloop labeled in italic. Guanine imino signals that could not be assigned are labeled with a question mark. Similar unaccounted for guanine peaks have been reported in previous NMR studies of SL1 (41). One of those peaks has a weak NOESY cross peak to G2 consistent with a G1 assignment. Through comparison with spectra of elongated SL1m, one uridine and two guanine signals could also be assigned to alternative forms of terminal residues G1, G2 and U34. In the absence of Mg2+, the U10 imino signal could not be observed while that of U11 could only be observed at low temperatures (5°C).
In vitro studies show that treatment of kissing SL1 dimers with the NCp7 protein (the maturated form of the Gag NC domain) results in the formation of a more stable duplex dimer in which two SL1 molecules are held together by extensive inter-strand base-pairing (Figure 1B) (17,18). This duplex dimer is believed to be a key component of the in vivo maturated heat-stable dimer. The conformational pathway between the kissing and duplex SL1 dimer is thought to involve the melting and reannealing of strands without disrupting the loop–loop interaction (19–25) though an alternative mechanism involving a transesterification mechanism has also been proposed (26).
There is evidence that dimerization and packaging of the HIV-1 RNA genome is further regulated by another conformational switch involving SL1 and the poly(A) hairpin in the 5′-leader (Figure 1C). The 5′-leader RNA can adopt two distinct conformations that migrate at different rates on native polyacrylamide gels (27,28). Secondary structure prediction and chemical probing suggest that the faster migrating species adopts an extended ‘long distance interaction’ (LDI) conformation that cannot be converted into dimers because the SL1 apical loop is masked by base-pairing with the poly(A) hairpin (Figure 1C) (28). The slowly migrating species is believed to be the conventional ‘branched multiple hairpin’ (BMH) conformer which is capable of dimerizing since the SL1 apical loop is exposed and available for self-pairing (Figure 1C) (28). Chemical probing experiments show that LDI can be converted into BMH by addition of NCp7 and that Mg2+ stabilizes BMH over LDI (9,27,28). This conformational switch is proposed to regulate dimerization and possibly packaging of the HIV-1 genome (27,29).
NC is known to bind single-stranded RNA particularly regions that have exposed guanine residues (30,31). In addition to the GC-rich apical loop, a candidate site in SL1 for NC binding is the highly conserved A271G272G273 × G247 internal loop (Figure 1A and B). The internal loop has been shown to be essential for the kissing–duplex transition and for proper packaging of the HIV-1 genome (22,32). Chemical probing experiments show that guanine residues G272 and G273 in the internal loop are accessible in the context of the 206 nt Ψ HIV-1 RNA (33). Footprinting data on a 401-nt fragment of the HIV-1 RNA leader shows that G272 and G273 are both strongly accessible in the free RNA and strongly protected upon binding to GST-tagged Gag or NC (34). Fluorescence experiments on isolated SL1 constructs show that NCp7 can bind to both the apical loop and internal loop (23,35). More recent studies using electrospray ionization-Fourier transform mass spectrometry show that the interaction between NC and the internal loop is key for inducing the kissing–duplex structural transition whereas the interaction with the apical loop can inhibit dimer formation (36). Five NMR structures reported for various SL1 constructs containing the internal loop show different conformations and levels of accessibility for internal loop residues G272 and G273 making it difficult to assess if they are structurally available for NC recognition (37–41). These NMR structures were all determined in the absence of divalent ions which are known to affect the SL1 structural transitions (9,23,24).
The involvement of SL1 in two distinct functionally important structural transitions raises the possibility that its structure codes for inherent plasticity that allows these conformational changes to take place in a specific and regulated manner. The kissing loop has been shown to be flexible particularly in the absence of Mg2+ (42–44) and this plasticity is believed to facilitate the exchange of strands during the kissing–duplex transition (23,45,46). There is also evidence that the internal loop introduces flexibility into SL1 that is essential for the kissing–duplex transition. SL1 kissing dimers containing the internal loop can spontaneously convert into duplex dimers in the absence of NCp7 when incubated at 55°C whereas corresponding constructs lacking the internal loop cannot even in the presence of NCp7 (22). Thus far, two NMR studies of SL1 monomers in the absence of Mg2+ provide conflicting views regarding the presence/absence of flexibility at the internal loop (37,38).
In this study, we used a combination of NMR techniques, including residual dipolar couplings (RDCs) (47,48), dynamically decoupled spin relaxation (49) and chemical shift mapping to quantitatively characterize the internal flexibility of an SL1 monomer containing the internal loop and how it varies upon Mg2+ binding. Our results suggest that the SL1 internal loop codes for internal flexibility that renders the upper stem a dynamical nucleation site for strand exchange during the kissing–duplex transition. We show that Mg2+ specifically binds to the internal loop stabilizing the overall SL1 structure while maintaining high local mobility in internal loop residues G272 and G273 making them available for NC binding. We suggest that by stabilizing SL1, Mg2+ deters spontaneous kissing–duplex transitions from taking place making the process more dependent on NC binding and thus ensuring that RNA and protein maturation occur in concert.
MATERIALS AND METHODS
Sample preparation
Uniformly 13C/15N-labeled SL1m, SL1 and elongated SL1m (E-GC-SL1m and E-AU-SL1m, see Figure 6A) NMR samples (0.6–1.0 mM) were prepared using standard in vitro transcription reactions as previously described (49). The buffer used in NMR experiments contained 15 mM sodium phosphate, 25 mM sodium chloride, 0.1 mM EDTA, and pH ∼6.4 in 8% 2H2O. NMR sample for the measurement of RDCs contained ∼15 mg/ml of Pf1 phage (Asla Biotech) (50,51). The kissing SL1 dimer was prepared following the previously described procedure (22).
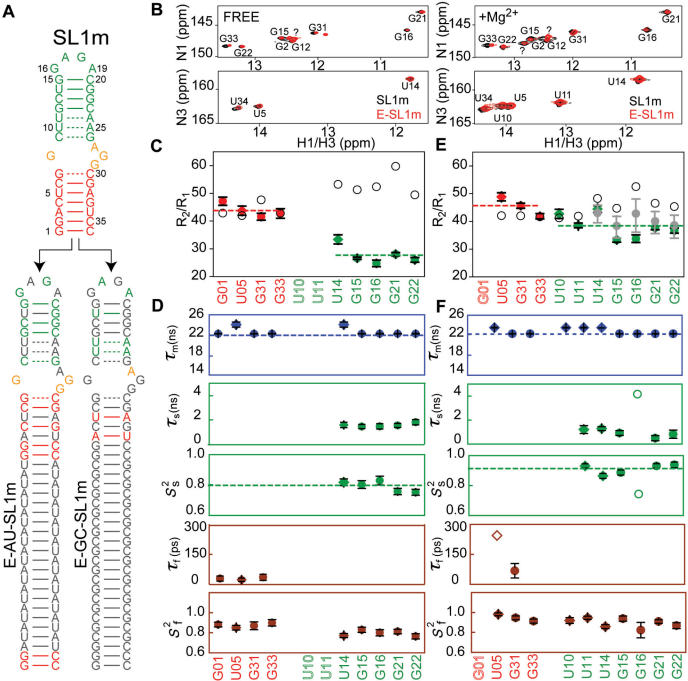
Figure 6.
Extended model-free analysis of 15N relaxation data measured in E-SL1m in the absence (49) and presence of Mg2+. (A) Secondary structure of E-GC-SL1m and E-AU-SL1m. 13C/15N-labeled and -unlabeled residues are shown in color and gray, respectively. (B) 2D 1H–15N HSQC spectra of the imino region of E-GC-SL1m + E-AU-SL1m (black) overlaid on non-elongated SL1m (red) in the absence and presence of Mg2+. Watson–Crick guanine signals that could not be directly assigned using NOESY data are labeled with ‘?’ and may correspond to either G26 or G7. (C and D) E-SL1m dynamics in the absence of Mg2+ (49). (C) R2/R1 values measured in guanine (filled circles) and uridine (filled diamonds) residues in stems I (red) and II (green) in the absence of Mg2+. Values predicted using hydrodynamic calculations are shown as black open circles. Error bars represent the measurement uncertainty. (D) Dynamical parameters for E-SL1m in the absence of Mg2+ obtained previously (49) from an extended model-free analysis (69,70). The time constants/amplitudes for fast (τf/mfcaret; Sfcaret;2) and slow (τs/mscaret; Sscaret;2) internal motions are shown. (E and F) Dynamics in E-SL1m in the presence of 10 mM Mg2+. (E) Measured R2/R1 values (filled color symbols) and those predicted hydrodynamically assuming a static conformation (black open circles) and a population-weighted average of free and Mg2+- bound states (see text) (filled gray circles). The error bar in the latter reflects different relative populations of free and bound states due to the spread of the observed Kds (0.9–7.0 mM corresponding to a free population ranging between 20 and 60%, respectively). (F) Dynamical parameters obtained in the presence of 10 mM Mg2+ using extended model-free analysis of the 15N relaxation data. Results for loop residue G16 (open symbol) varied considerably when using different input E-SL1m + Mg2+ conformations that reflect the RDC uncertainty and is therefore considered to be unreliable.
NMR spectroscopy
All NMR experiments were conducted at 298 K (unless stated otherwise) on an Avance Bruker 600 MHz NMR spectrometer equipped with a triple-resonance 5 mm cryogenic probe. Spectra were processed using NMRPipe/NMRDraw (52), analyzed using NMRView (53) and overlaid using SPARKY 3 (54). Non-exchangeable resonances were assigned using standard methods (55) and were consistent with assignments reported previously for the same construct (38). With the exception of terminal residues G1, G7 and G26, all expected exchangeable imino signals belonging to Watson–Crick base-pairs (G2, U34, G33, U5, G31, U10, U11, G12, G22, U14, G21 and G15) could be assigned using the 3D 1H–15N HSQC-NOESY experiment in the absence or presence of experiment in the absence or presence of Mg2+.
A total of 52 one bond C–H and N–H RDCs (1DC1′H1′, 1DC2H2, 1DC5H5, 1DC6H6, 1DC8H8, 1DN1H1 and 1DN3H3) were measured in duplicate in Pf1 phage (∼15 mg/ml) (50,51) in the absence and presence of Mg2+ as previously described (see Figure S4 and Table S1) (56–58). Average RDCs were used in subsequent analysis whenever possible. Imino 15N longitudinal (R1) and transverse (R2(CPMG)) relaxation rates and 1H–15N NOEs were measured in E-SL1m in the presence of 10 mM Mg2+ using 2D experiments (59,60) as previously described for free E-SL1m (49). The R1 and R2(CPMG) relaxation delays were 0.06, 0.12, 0.24, 0.48, 0.64, 0.80, 1.2 s and 0.0062, 0.0124, 0.0248, 0.0372, 0.0496, 0.0620, 0.0744 s, respectively. Resonance intensities were measured and normalized as previously described (49). Mg2+ chemical shift titrations were performed by recording 2D 1H–13C or 1H–15N HSQC spectra following incremental increases in the MgCl2 concentration (0.1, 0.2, 0.4, 1.6, 3.2 and 5.0 mM).
Data analysis
RDCs measured in non-terminal Watson–Crick base-pairs were used to compute best-fit order tensors (61–63) (Table 1) using the programs RAMAH (64) (an in-house modified version of ORDERTEN_SVD) (62) and idealized A-form helices constructed using Insight II (Molecular Simulations, Inc.) as previously described (65). The order tensor uncertainty due to RDC measurement uncertainty and A-form structural noise (Table 1) was computed using the program Aform-RDC (65). The relative orientation of stems was determined by superimposing their respective order tensor frames (62). Three out of four degenerate solutions could be excluded based on structural constraints (66). The translational disposition of stems was determined approximately by modeling in the connector internal loop from the previous NMR structure of the same GAGA SL1m mutant (38). The assembled SL1m structures were further validated through comparison of the experimental Szz direction with that predicted using the program PALES (differences ∼3°) (67). Inter-helical angles were calculated using in-house written software (68). An indirect order tensor fit was also carried out for internal loop RDCs using the previously reported NMR conformations [1N8X (38); 2GM0 (41); 2D17 (40)]. In this analysis, central base-pairs in stem II (C13–G22, G12–C23, U11–A24 and U10–A25) were used to superimpose the idealized stem II helix against each NMR structure. Next, the best-fit order tensor determined for stem II (Table 1) was used to back-predict internal loop RDCs for each conformation. This analysis was carried out on each model in a given NMR ensemble.
Table 1.
Order tensor analysis for RDCs measured in SL1m in the absence and presence of 5 mM Mg2+
| SL1m | Stem | N | CN | RMSD (Hz) | R | η | ϑ × 10−3 | ϑint |
|---|---|---|---|---|---|---|---|---|
| Free | I | 14 | 3.7 | 1.4 | 0.99 | 0.19 ± 0.04 | 1.68 ± 0.06 | 0.88 ± 0.04 |
| II | 17 | 2.9 | 0.9 | 0.99 | 0.14 ± 0.04 | 1.47 ± 0.03 | ||
| +Mg2+ | I | 14 | 6.1 | 0.7 | 0.99 | 0.15 ± 0.06 | 1.24 ± 0.06 | 0.99 ± 0.05 |
| II | 23 | 2.8 | 1.3 | 0.99 | 0.15 ± 0.04 | 1.23 ± 0.03 |
The number of measured RDCs (N), the condition number (CN) (79) describing the orientational spread of the RDC-targeted bond vectors, the root-mean-square-deviation (RMSD) and correlation coefficient (R) between measured and back-predicted RDCs, the order tensor asymmetry (η = |Syy − Sxx|/Szz), generalized degree of order (ϑ) and internal generalized degree of order (ϑint) are shown.
The 15N R1 and R2 values and their uncertainties were determined by non-linear least squares fitting of experimental data to monoexponential functions (Figure S6) as previously described (49). The relaxation data was subjected to an extended model-free analysis (69,70) using the software Modelfree (Version 4.16 for Linux) provided by Palmer and co-workers (71) closely following the procedure described previously (49). This analysis assumed (i) the decoupling approximation (69,70), (ii) an axially symmetric overall diffusion tensor, (iii) a diffusion tensor with Dratio (D///D⊥ = 5.8) and orientation computed using HYDRONMR (72) and the RDC-derived average SL1m + Mg2+ inter-stem conformation.
Insight II (Molecular Simulations, Inc.) was used to in silico elongate stem I in the RDC-derived SL1m + Mg2+ structure using a stretch of 22 bp. The resulting E-SL1m + Mg2+ structure was subjected to hydrodynamic calculations using the program HYDRONMR (72) as previously described (49). The HYDRONMR-computed diffusion tensor parameters and E-SL1m + Mg2+ structure were then used to compute 15N R2/R1 values using in-house software.
Mg2+-induced chemical shift changes were quantified using, 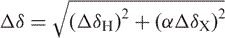 , where ΔδH and ΔδX are the chemical changes in the 1H and 13C/15N dimensions, α is the ratio of the H and C/N gyromagnetic ratio. The apparent Kds were obtained by fitting to the following equation (73):
, where ΔδH and ΔδX are the chemical changes in the 1H and 13C/15N dimensions, α is the ratio of the H and C/N gyromagnetic ratio. The apparent Kds were obtained by fitting to the following equation (73):
 |
where δobs is the observed chemical shift, ΔδT is the chemical shift difference between the free and fully bound state, [Mg2+]T is the amount of added MgCl2 and [RNA]T is the RNA concentration obtained from its absorbance at 260 nm. The data was fitted using Origin (Origin Lab Corporation) in which the unknown values ΔδT and Kd are optimized during the fit. Representative fits are shown in Figure 3C.
Figure 3.
Chemical shift mapping of Mg2+ binding to the SL1m internal loop. (A) Overlay of 2D 1H–13C and 1H–15N HSQC spectra of SL1m recorded in the absence (in black) and presence (in red) of 5 mM Mg2+. (B) Residues undergoing the largest Mg2+-induced chemical shift perturbations (top 20% for a given type of resonance) and that yield the tightest binding (Kd < 2.0 mM) are highlighted on the SL1m secondary structure in red and using a black box, respectively. (C) Representative titration curves as a function of total Mg2+ concentration with apparent Kd values shown at the end of each curve.
RESULTS
Monomeric SL1m as a model for the stem–internal loop–stem element of the kissing SL1 dimer
We characterized the stem–internal loop–stem element of SL1 using a previously reported monomeric mutant (SL1m) (38) that is impaired from forming dimers through replacement of the wild-type apical loop with a GAGA tetraloop (Figure 1D). The lower stem I is also extended by three pairs to optimize synthesis by in vitro transcription (38). A previous NMR study used spectral comparisons to show that replacement of the wild-type apical loop with a UUCG tetraloop does not impact the structural integrity of the stem–internal loop–stem element of the SL1 kissing dimer (39). We examined if this was also the case for GAGA-loop SL1m mutant by comparing its spectra with those of the corresponding kissing SL1 dimer (Figure 1D). Aside from the expected differences at residues near the loop (Figure 1D, highlighted in red), similar chemical shifts were observed for the two constructs (Figures 1D and S1), indicating that SL1m recapitulates the stem–internal loop–stem element in the kissing SL1 dimer (38). As we discuss in subsequent sections, similar exchange broadening and Mg2+-induced chemical shift perturbations were also observed in the two constructs, indicating that SL1m also retains the dynamical and Mg2+-binding properties of the stem–internal loop–stem element in the SL1 kissing dimer.
SL1 dynamically interconverts between alternative secondary structures causing partial melting of base pairs in the upper stem in the absence of Mg2+
In a previous study of a UUCG-loop SL1 monomer, internal loop resonances exhibited severe exchange broadening characteristic of micro-to-millisecond timescale motions and this precluded detailed NMR structural characterization (37). Replacement of the wild-type AGG internal loop with a GGA internal loop yielded an SL1 construct with far less broadening and higher (∼8°C) UV melting temperatures (37). In contrast, exchange broadening was not observed in the previous study of the GAGA-loop SL1m (38). Given that similar NMR buffer conditions were used in the two studies (absence of divalent ions), the source of this discrepancy remains unresolved.
In our study, we observed exchange broadening in resonances belonging to the internal loop and neighboring residues in the absence of Mg2+ (Figure 2A and B). The exchange broadening was also observed in the SL1 kissing dimer (Figures 1D and S1) and in SL1m constructs in which stem I was elongated by 22 bp (see Figure 6A). The broadening was also observed when using the same buffer (10 mM Tris-d11, pH ∼8.0, 0.1 mM EDTA in 2H2O) used in the previous NMR study of GAGA-loop SL1m (data not shown) (38). The broadening was however strongly dependent on temperature (Figure 2A). It was very pronounced at 5°C in which the dynamic process appears to be in intermediate/slow exchange and virtually undetectable at 45°C in which the process appears to be in fast exchange (Figure 2B). In this regard, it is possible that exchange broadening was not observed in the previous study of GAGA-loop SL1m because a relatively high temperature of 37°C was used to record NMR data (38).
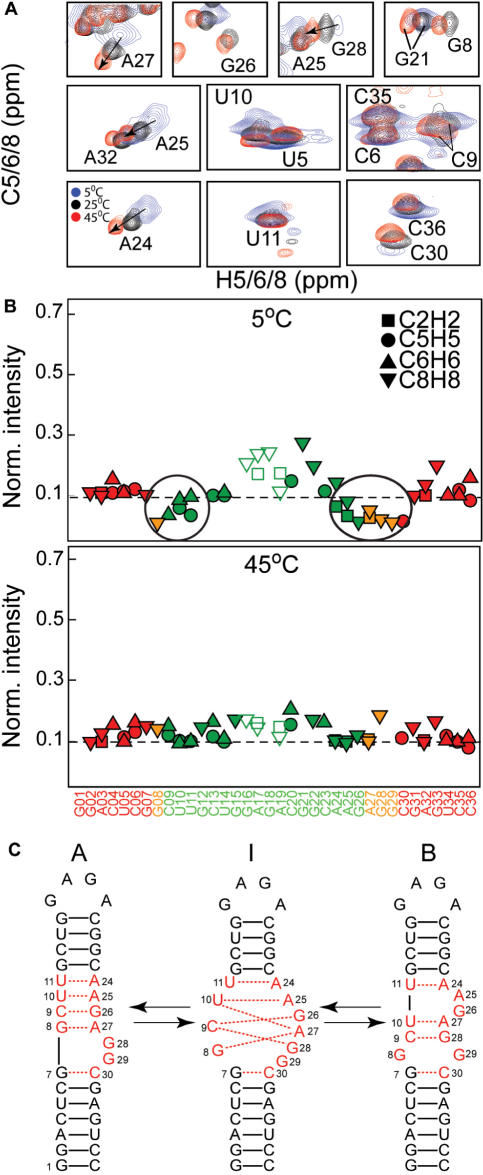
Figure 2.
Conformational exchange in SL1m. (A) Representative spectra showing the temperature dependence of the exchange broadening at 5°C (in blue), 25°C (in black) and 45°C (in red). (B) Relative resonance intensities of non-exchangeable C–H resonances measured in SL1m at 5°C and 45°C. The intensities for a given type of bond vector are normalized to a minimum value of 0.1 as previously described (49). (C) Proposed dynamical equilibrium between SL1 conformers denoted ‘A’ and ‘B’ and possible intermediates denoted ‘I’ along the pathway involving a smaller number of hydrogen bond alignments. Residues undergoing the largest degree of exchange broadening are shown in red.
The exchanged broadened residues likely sample more than one conformation. In addition to the internal loop, broadening was observed at C30 below the internal loop in stem I and the three base-pairs above the internal loop in stem II (C9-G26, U10-A25 and U11-A24) (Figure 2B). These base pairs have non-observable or weak imino signals indicating that their hydrogen alignments deviate from ideality (Figure 1D). The observed broadening and hydrogen bond melting pattern is consistent with a dynamical equilibrium involving two distinct SL1m secondary structures (referred to as ‘A’ and ‘B’, Figure 2C) that was previously proposed based on secondary structure prediction (39). Conformer A contains an asymmetric internal loop and is observed in all NMR structures reported to date (Figure 2C). Conformer B contains a symmetric internal loop and two base bulge and has not been directly observed in any structural studies. The A to B transition possibly via one or more intermediates (I) involves replacing Watson–Crick residues A25 and G26 in conformer A by internal loop residues A27 and G28 (Figure 2C). This leads to changes in the local environment of the internal loop and three upper base-pairs where exchange broadening is observed (Figure 2C, shown in red). It also leads to transient melting/reannealing of the two upper base-pairs (C9-G26 and U10-A25) explaining why their imino signals are exchange broadened beyond the limit of detection (Figure 2C). In this context, the reduced exchange broadening observed in the GGA SL1 mutant (37) can also be attributed in part to its inability to slip into the B conformer. Together, these results suggest that the SL1m internal loop transiently unzips the upper stem by stabilizing more than one conformation involving distinct hydrogen alignments.
Mg2+ binds the internal loop, arrests the dynamical exchange and stabilizes the upper stem
All NMR structures reported thus far for SL1 containing the internal loop have been determined in the absence of divalent ions. To examine if Mg2+ binds to the SL1m internal loop, we performed chemical shift titration experiments in which 2D HSQC spectra were recorded following incremental addition of Mg2+ up to 5 mM. Very large chemical shift perturbations were observed that are indicative of specific Mg2+ binding (Figure 3A). The apparent Kds range between 0.9 and 7.0 mM but for the internal loop, they cluster in a relatively narrow range (0.9–1.9 mM) indicative of tight Mg2+ binding (Figure 3C). Similar Mg2+ chemical shift perturbations were observed in the kissing SL1 dimer (Figure S1B).
Interestingly, the largest chemical shift perturbations, which reflect Mg2+ localization effects and/or Mg2+-induced changes in the SL1 conformation, were observed precisely at the residues that exhibit marked exchange broadening in the internal loop and the upper stem (Figure 3B). The exchange broadening at these sites is markedly reduced upon Mg2+ binding (Figure 3A, see also Figure 7). This is accompanied by the appearance and intensification of imino signals belonging to U10 and U11 respectively in the upper stem (Figure 3A) for which an A–U Watson–Crick hydrogen bond alignment could be established directly using the JNN-COSY experiment (Figure S3) (74,75). An additional Watson–Crick guanine imino signal appears which could not be directly assigned but which may correspond to G26 (lack of NOE connectives with U10 is not surprising given that the U10 signal remains somewhat weak). Similar spectral changes were observed for the kissing SL1 dimer (Figure S1B). These results suggest that Mg2+ binds and stabilizes the SL1m internal loop and in doing so arrests the dynamical exchange and partial melting of the upper stem.
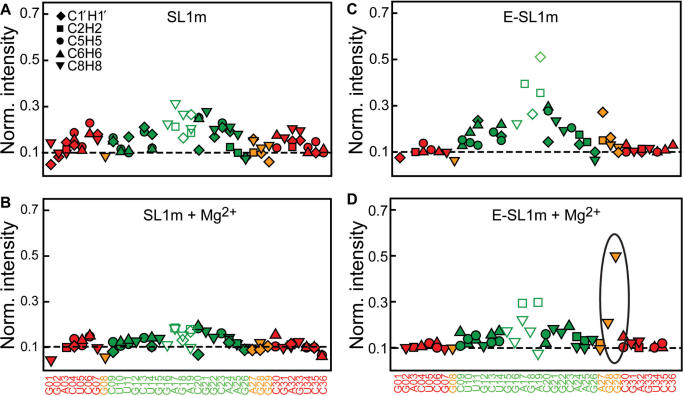
Figure 7.
Dynamics in E-SL1m in the absence and presence of Mg2+ from motional narrowing of resonances. The normalized resonance intensities as a function of residue in the presence and absence of Mg2+ for (A and B) SL1m and (C and D) E-SL1m at 25°C are shown. Resonance intensities are obtained from non-constant time 1H–13C HSQC experiments. The intensity for a given type of C–H vector is normalized to a minimum nominal value of 0.1 indicated by a horizontal line. Peaks that exhibit exchange broadening as inferred from the temperature-induced perturbations were not used in the above normalization and generally have intensities <0.1.
Local conformation of the stem and internal loop in the absence and presence of Mg2+
We used RDCs (47,48,76,77) measured in Pf1 phage (50,51) to characterize the local conformation of the two stems in the absence and presence of Mg2+. In particular, we used an order tensor analysis (62,65,78,79) to examine the fit between the stem RDCs and an idealized A-form helix structure. This provided a basis for evaluating whether conformer A or B is the dominant species in solution (Figure 2C). RDC (58,63,80–84) and other studies (65) show that the idealized A-form helix is an excellent model geometry for non-terminal Watson–Crick base-pairs in different RNA contexts.
RDCs belonging to hydrogen-bonded Watson–Crick base-pairs (as inferred from the imino 2D 15N–1H HSQC spectra) were included in the order tensor fit which was carried out independently for each stem. The best-fit order tensor and idealized A-form geometry were then used to back-predict all of the measured RDCs including ones not included in the fit. For both stems, excellent agreement was obtained for RDCs measured in the hydrogen-bonded base-pairs both in the absence and presence of Mg2+ (Figure 4, filled symbols). The observed root-mean-square-deviation (RMSD) between measured and predicted RDCs (0.6–1.4 Hz) is well within the estimated RDC experimental uncertainty (∼2.5 Hz, see Figure S4). In contrast, the agreement was variable for the loop and adjacent residues that were excluded from the order tensor fit (Figure 4, open symbols).
Figure 4.
Order tensor analysis of RDCs measured in stem I in the (A) absence and (B) presence of Mg2+ and in stem II in the (C) absence and (D) presence of Mg2+. Correlation plots between measured and back-calculated RDCs when independently fitting stem order tensors to an idealized A-form geometry are shown. Data that was excluded from the order tensor fit is shown in colored open symbols. The root-mean-square-deviation (RMSD) and correlation coefficient (R) are shown on each plot. The corresponding fits obtained when including all RDCs in the order tensor fit (excluding C9–G26 in stem II) are shown in the insets.
For stem I, very poor agreement was observed for terminal residue C30 immediately below the internal loop in the absence (Figure 4A) but not in the presence of Mg2+ (Figure 4B). The same residue exhibits severe exchange broadening that is significantly reduced upon Mg2+ binding (Figure 3A). In contrast, for G7 below the internal loop which does not exhibit severe exchange broadening either in the absence or presence of Mg2+ (Figure 3A), good agreement was observed both in the absence (Figure 4A) and presence of Mg2+ (Figure 4B). Thus, Mg2+ binding appears to stabilize a Watson–Crick geometry for the G7–C30 base-pair primarily by affecting the alignment of C30. An excellent stem I order tensor fit (RMSD = 0.7 Hz) could be obtained in the presence of Mg2+ when including all RDCs from the G7–C30 base-pair (Figure 4B, inset).
The only significant deviation in stem II was observed for terminal residue C9 both in the absence (Figure 4C) and presence of Mg2+ (Figure 4D). This deviation may reflect an alternative conformation that accommodates inter-helical kinking (see below). The remaining base-pairs (U10–A25 to G15–C20) show an excellent fit both in the absence (Figure 4C, inset) and presence of Mg2+ (Figure 4D, inset). Thus, despite difficulty in observing imino signals for U10 and U11 in the absence of Mg2+, the RDCs suggest that these residues are part of an interrupted A-form helix as expected for conformer A but not conformer B both in the absence and presence of Mg2+. This however does not rule out dynamical averaging with the B conformer or other intermediates if these species are populated to a low level (<10%) such that their contributions to the population-weighted RDCs is negligible (85). The observed chemical exchange broadening and RDCs suggest that Mg2+ arrests the dynamical exchange by stabilizing the A conformer.
Mg2+ binding does not significantly affect the average internal loop conformation
The RDCs measured in the internal loop in the absence and presence of Mg2+ were very similar (Table S1). After accounting for the differences in degrees of order in the absence and presence of Mg2+ (Table 1), the RMSD between the free and Mg2+ RDCs in the internal loop is only 2.9 Hz noting that comparable differences are expected due to differences in the orientation of overall alignment (Szz direction differs by ∼5°, data not shown). This indicated that the Mg2+ binding does not significantly alter the SL1m internal loop conformation.
We further investigated the internal loop conformation by evaluating the fit between the measured RDCs and internal loop conformations reported in three previous NMR structures [1N8X (38), 2GM0 (41) and 2D17 (40)] determined in the absence of divalent ions. Although there is considerable variability in the internal loop conformation in the NMR ensembles, many of the models reproduce the RDCs. As expected, similar agreement was observed for free and Mg2+ RDCs (Figure S5). Overall, the RDCs are consistent with the G8–A27 mismatch reported in all three structures. For residues G28 and G29, which are implicated in NC binding (32,34,86), the RDCs are consistent with conformations in which G29 is extrahelical and accessible (e.g. most 2GM0 models as well as models 10, 14 and 18 in 1N8X) and G28 stacked onto A27 in a more intra-helical conformation (e.g. models 7 and 8 in 2D17, models 6 and 12 in 1N8X and models 13 and 14 in 2GM0). Interestingly, the C1′H1′ RDCs in A27, G28 and G29 were all nearly zero both in the absence and presence of Mg2+ (Figure S5). Similar values were reported in the Mg2+-free duplex structure (41). While it was possible to find static conformations that can reproduce these RDCs (Figure S5), one cannot rule out the presence of extensive dynamical averaging which goes to attenuate the measured RDCs.
Mg2+ binding arrests inter-helical motions without significantly altering the average inter-helical alignment
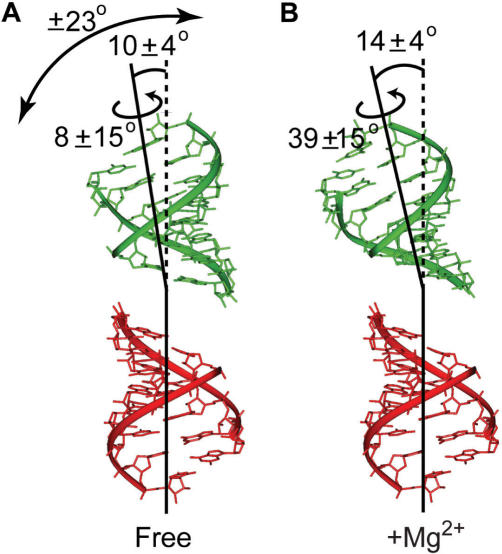
The order tensors determined for stems I and II using the idealized A-form helix were used to determine the relative orientation and dynamics of the stems in the absence and presence of Mg2+. Three order tensor elements define an order tensor frame (Sxx, Syy, Szz) that describes the average orientation of each stem relative to the applied magnetic field. The average relative orientation of stems can be determined by superimposing their respective order tensor frames (62,63,79). The inter-helical SL1m conformation determined in this manner in the absence and presence of Mg2+ is shown in Figure 5A and B, respectively. In both cases, the inter-helical angle only slightly deviates from coaxial alignment (10 and 14°, respectively). The differences between the bend/twist angles obtained in the absence (10 ± 4°/8 ± 15°) (Figure 5A) and presence (14 ± 4°/39 ± 15°) of Mg2+ is small and comparable to the experimental uncertainty which takes into account both the RDC uncertainty and A-form structural noise (Figure 5B) (65). Thus, as observed for the internal loop, Mg2+ binding does not significantly alter the average relative orientation of the two stems. The observed inter-helical bend/twist angles are in good agreement with the range of values reported in previous NMR structures of SL1 containing the same AGG internal loop (17–27°/28–65°, 14–24°/24–72°, 5–42°/48–78° for 1N8X, 2GM0 and 2D17, respectively).
Figure 5.
Relative orientation and dynamics of the two SL1m stems in (A) the absence and (B) presence of Mg2+ derived from order tensor analysis of the stem RDCs. The values of the inter-helical bend and twist angles together with the amplitude of internal motions obtained from the ratio of stem GDOs assuming an isotropic cone motional model are shown.
We previously reported evidence for nanosecond stem motions in SL1m in the absence of Mg2+ using motionally decoupled 15N spin relaxation data (49). The order tensor analysis also yields a generalized degree of order (GDO, ϑ) which can be used to assess motions between the two stems (79). Unlike spin relaxation data, which is used to probe internal motions at nanosecond and faster timescales, the GDO is sensitive to internal motions spanning a wider range of timescales (less than millisecond). The GDO describes the degree of alignment experienced by each stem (79). While rigidly held stems should experience a common degree of alignment and therefore report identical GDOs, inter-stem motions can result in different degrees of stem order and hence stem GDOs (79). The stem GDO ratio, referred to as the internal generalized degree of order (GDOint, ϑint), provides a measure of inter-stem motional amplitudes, ranging between 1 for inter-stem rigidity and 0 for maximum inter-stem motions (79).
In the absence of Mg2+, the GDO computed for stem II (1.47 × 10−3 ± 0.03) was smaller than that computed for stem I (1.68 × 10−3 ± 0.06), yielding a GDOint value of ∼0.88 ± 0.04 (Figure 5A, Table 1). This indicates that the two stems dynamically reorient relative to one another in the absence of Mg2+. Remarkably, the amplitude of inter-stem motions obtained using RDCs are very similar to those obtained independently by 15N spin relaxation data (Ss = 0.791/2 = 0.89) (49), indicating that the inter-stem motions occur at nanosecond timescales and that little to no inter-stem motions occurs at micro-to-millisecond timescales. The larger degree of order observed for stem I is consistent with having the flexible hinge closer to stem II which is expected given the partial melting of its three lower base-pairs. Assuming isotropic motions in a cone (63,87), the observed GDOint value translates into inter-stem motional amplitudes of ∼23°. This yields an SL1m conformation with the inter-helical angle that can vary dynamically between ∼0 and ∼33°. A similar range of bend angles (5–42°) is observed in the ensemble family of three previously reported NMR structures of SL1 containing the AGG internal loop (38,40,41).
In stark contrast, the stem GDOs obtained in the presence of Mg2+ are very similar (1.24 × 10−3 ± 0.06 and 1.23 × 10−3 ± 0.03 for stems I and II, respectively), yielding a GDOint value that is close to unity (0.99 ± 5%) (Figure 5B, Table 1). This suggests that while Mg2+ binding does not significantly affect the average orientation of the two stems, it arrests their relative motions stabilizing the global SL1m conformation.
Collective and librational motions in the absence and presence of Mg2+ from dynamically decoupled 15N relaxation data
While the similar GDOs observed for the two stems in the presence of Mg2+ is consistent with a rigid inter-stem alignment, an alternative possibility is that by stabilizing the three lower base-pairs in stem II, Mg2+ equalizes their contribution to total alignment. Under such conditions, similar stem GDOs could be observed even in the presence of inter-stem motions (68). To rule out this possibility, we applied a recently described domain elongation to independently examine stem motions in SL1m using 15N relaxation data (49). Domain elongation by 22 bp is carried out using two constructs which are designed to reduce spectral overlap due to elongation residues (Figure 6A) (49). The elongation serves to slow down overall molecular tumbling providing an opportunity to probe stem motions by spin relaxation methods.
In the previous study of E-SL1m, we used spectral comparisons such as those shown in Figure 6B to show that elongation does not disrupt the structural and/or dynamical integrity of SL1m. The similar exchange-broadening pattern (Figure 6B) and Mg2+-induced chemical shift changes (Figures 6B and S2) observed in SL1m and E-SL1m further suggests that elongation does not affect the dynamical and Mg2+-binding properties of SL1m. To this end, we measured 15N relaxation data (Tables S2 and S3) in E-AU-SL1m and E-GC-SL1m (Figure 6A) in the presence of 10 mM Mg2+. Assuming two-state binding with an average Kd ∼ 2.0 mM (Figure 3C), ∼70% of E-SL1m is expected be in the Mg2+-‘bound’ state.
As shown in Figure 6E, Mg2+ binding led to a significant increase in the stem II R2/R1 values without significantly affecting the stem I values (Figure 6C and 6E). The resulting R2/R1 values measured in stems I and II are for more similar in the presence of Mg2+ (Figure 6E). This is exactly as would be expected if Mg2+ binding caused a reduction in the amplitudes of inter-stem motions. The stem II R2/R1 values remain smaller than stem I and/or values predicted assuming a static RDC-derived SL1m + Mg2+ conformation (Figure 6E, open symbols). This is not surprising given that ∼30% of E-SL1m is expected to be on average in the dynamical free state. Interestingly, very good agreement is observed between the measured stem II R2/R1 values and those back-predicted assuming a population-weighted average of free (measured experimentally) and Mg2+-bound (computed hydrodynamically) R2/R1 values (Figure 6E, in gray).
The 15N relaxation data measured in Mg2+ was analyzed using the extended model-free analysis (69,70). This analysis was repeated for a range of input E-SL1m conformations that reflect the uncertainty in the inter-stem alignment obtained by RDCs. With the exception of loop residue G16, for which the dynamical parameters obtained varied considerably depending on the input conformation, consistent dynamical parameters were obtained for all stem base-pairs (data not shown). The dynamical parameters obtained using the best-fit E-SL1m + Mg2+ RDC-derived conformation is shown in Figure 6D and Table S3. The time constant for overall rotational diffusion (τM = 22.3 ± 0.1 and 23.5 ± 0.1 ns for E-AU-SL1m and E-GC-SL1m, respectively) is very similar to the hydrodynamically predicted value (22.2 ns) (72). Relative to free E-SL1m (Figure 6D), Mg2+ binding leads to a uniform increase in the 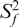 values and thus decrease in the local N–H librations suggesting local stabilization of the two stems (Figure 6F). A larger uniform increase in the
values and thus decrease in the local N–H librations suggesting local stabilization of the two stems (Figure 6F). A larger uniform increase in the 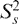 values of stem II is also observed indicating that Mg2+ binding causes a reduction in the amplitudes of stem motions (Figure 6F). Though small changes in the time constants for librations (τf) and stem motions (τs) are also observed upon Mg2+ binding, comparable variations were observed when using distinct input SL1m conformations that reflect the RDC measurement uncertainty (data not shown).
values of stem II is also observed indicating that Mg2+ binding causes a reduction in the amplitudes of stem motions (Figure 6F). Though small changes in the time constants for librations (τf) and stem motions (τs) are also observed upon Mg2+ binding, comparable variations were observed when using distinct input SL1m conformations that reflect the RDC measurement uncertainty (data not shown).
Local internal loop dynamics in G28 and G29 in the presence of Mg2+
In the previous study of free E-SL1m, the C–H resonance intensities exposed internal motions that evade detection in non-elongated SL1m because they occur at timescales approaching overall molecular tumbling (49). Ignoring chemical exchange, the intensities of non-exchangeable C–H resonances reflect the reorientation of dipolar and chemical shift anisotropy (CSA) tensors relative to the applied magnetic field. Placement of the base dipolar and 13C CSA tensors nearly perpendicular to the long axis of diffusion renders the base stem I intensities near maximum values expected from anisotropic tumbling of a rigid molecule (49). Thus, resonances that exhibit higher intensities correspond to regions with added-on internal mobility occurring at timescales faster than overall molecular tumbling.
To gain further insight into how Mg2+ binding affects the dynamics of SL1m, we measured resonance intensities in SL1m and E-SL1m in the presence of Mg2+ at 25°C and compared results with intensities observed in the free state (Figure 7). For free SL1m, small variations in intensities are observed which are reduced upon Mg2+ binding (Figure 7B). In contrast, due to the resolving power afforded by elongation, the intensities in free E-SL1m exhibit much wider variations from site to site that are indicative of variable degrees of internal motions (49). The changes in intensities following Mg2+ binding are also far more significant in E-SL1m compared to SL1m (Figure 7D). In addition to a decrease in exchange broadening in and around the internal loop, the stem II and GAGA tetraloop intensities experience a significant reduction, consistent with a long-range arrest of inter-stem motions. A significant intensity reduction is also observed for A27 possibly reflecting stabilization of the G8–A27 mismatch though new exchange-broadening contributions cannot be ruled out. Remarkably, and in stark contrast, a significant increase in the intensities is observed for the base moieties of G29 and to a smaller extent G28 (Figure 7D). This suggests that Mg2+-binding activates nanosecond local motions in the base moieties of G29 and to a lesser extent, G28. However, it is also possible that these motions were present in free E-SL1m but they were masked exchange-broadening contributions that are reduced upon Mg2+ binding. Though spectral overlap in non-elongated SL1m did not permit measurement of the corresponding resonance intensity or RDC for G29, elevated intensities are not observed for G28 in SL1m (Figure 7B) most likely because the motions occur at timescales approaching overall molecular tumbling. The high mobility observed at G29 and G28 in the presence of Mg2+ is significant given that these are precisely the internal loop residues that are implicated in NC binding (33,34).
DISCUSSION
SL1 is a highly conserved stem-loop in the HIV-1 leader RNA that is believed to be involved in functionally important structural transitions that are modulated by Mg2+ binding and that are catalyzed by NC. In our study, we examined the dynamical and Mg2+-binding properties of the SL1 structure with the goal of obtaining new insight into the molecular basis of its structural transitions.
Our study suggests that the highly conserved SL1 internal loop sequence specifically destabilizes the upper stem by allowing formation of two (or possibly more) competing secondary structures (Figure 2C). This conformational equilibrium is intricately dependent on the sequence of the AGG internal loop and neighboring residues in stem II. For example, the reduced exchange broadening observed in the GGA internal loop mutant can be explained by its inability to slip into the B conformer (37). A similar argument could be used to explain the higher stabilities of other SL1 internal loop mutants (37). Interestingly, a uridine-substituted SL1 internal loop does not disrupt dimer formation but results in a mutant virus with diminished genome packaging (86). In addition to possibly interfering with NC binding, the uridine substitution is expected to impair formation of the B conformer and thus stabilize the internal loop and upper stem.
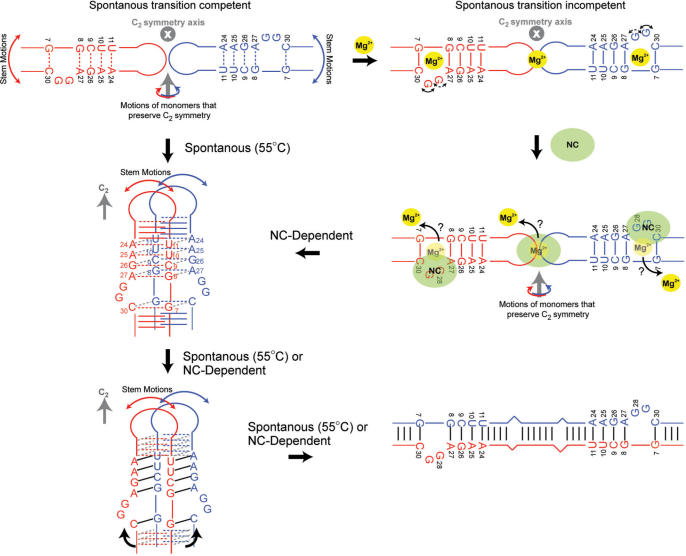
SL1 kissing dimers containing the internal loop can spontaneously convert into duplex dimers at 55°C in the absence of Mg2+ whereas constructs lacking the internal loop cannot (22). Our results show that the internal loop introduces internal flexibility into the SL1m structure that can promote the kissing–duplex transition. The transition requires the exchange of strands between monomers in the kissing dimer which in turn requires that strands from the two monomers come into close proximity. The two stems above the internal loop are likely candidates for initiating strand exchange since they are the most closely positioned in the kissing dimer (Figure 8). Studies have shown that the kissing–duplex transition can occur without disrupting the loop–loop interaction (19–25). The two monomers can be brought into close proximity without disrupting the loop–loop interaction by rotating each monomer around a direction perpendicular to the C2 axis of symmetry. Such a rotation ensures that C2 symmetry is maintained in the dimer during the transition. This leads to formation of an intermediate in which base-pairs in the upper stem are proximate and poised to form both inter- and intra-molecular hydrogen bonding. Such an intermediate has previously been proposed (23) and recently visualized by molecular dynamics simulations in the context of short kissing SL1 dimers lacking the internal loop (25). The inclusion of the internal loop is expected to destabilize the upper stem making its base-pairs a key nucleation site for initiating the melting, exchange and reannealing of strands. This would explain why constructs lacking the internal loop cannot undergo the kissing–duplex transition spontaneously (22). Due to inter-helical kinking, the lower stems are not expected to be in immediate register in the intermediate and inter-stem flexibility may play a role in bringing the lower stems into proper register for completing strand exchange (Figure 8).
Figure 8.
Proposed role for internal loop-induced dynamics and Mg2+ binding in spontaneous and NC-dependent SL1 structural transition between kissing and duplex dimers.
Our results show that Mg2+ binds to the SL1 internal loop region without significantly altering the average SL1 structure. Electrostatic calculations on the SL1m structure determined in the absence of Mg2+ (38) shows that residues that experience the largest Mg2+-induced chemical shift changes belong to regions with strong negative electrostatic potential (Figure S7). The RDCs and relaxation data do however show that Mg2+ arrests the dynamical equilibrium stabilizing the hydrogen bond alignments in the upper stem while simultaneously reducing inter-stem motions. In this regard, Mg2+ binding is expected to reduce the likelihood for spontaneous kissing–duplex transitions in SL1 constructs containing the internal loop. So far, only one study has explored the effects of Mg2+ binding on the spontaneous transition in SL1 constructs containing the internal loop. Using a ribozyme-based cleavage assay to monitor the kissing–duplex transition, this study showed that increasing the Mg2+ concentration (up to 100 mM) resulted in an increase in the probability for spontaneous kissing–duplex conversion (24). However at the low RNA concentrations used (∼0.01 mM compared to ∼1 mM used in other studies) the encounter of monomers may be rate-limiting and Mg2+ may increase the probability for encounter by stabilizing the loop–loop interaction (23).
In vivo, the transition between kissing and duplex dimers as well as between the LDI and BMH conformers is believed to be catalyzed by NC protein which is known to bind to exposed guanine residues (30,31). Previous mapping studies have shown that G28 and G29 are accessible in the context of the 206 nt Ψ HIV-1 RNA (33). Footprinting data on the 401-nt fragment of the RNA leader show that G28 and G29 are both strongly accessible in the free RNA and strongly protected upon binding to Gag and NC (34). A recent study suggests that NC binding to the SL1 internal loop is largely responsible for driving the kissing–duplex transition whereas binding at the apical loop inhibits dimer formation (36). The latter study also showed that NC binding to the SL1 internal loop is similar whether in the monomer or kissing dimer context further validating the relevance of our dynamical results on monomeric SL1m (36). Remarkably, our results show that while Mg2+ causes stabilization of the internal loop and upper stem, it increases or retains significant local flexibility in the base moieties of G28 and G29 likely making them available for NC binding. The RDCs measured in the presence and absence of Mg2+ are also consistent with an extra-helical conformation for G29 (Figure 8) (41). In this manner, Mg2+ binding may help ensure that the kissing–duplex conversion does not occur spontaneously and prematurely but rather only following NC binding (Figure 8). The conformational pathway may involve the ejection of stabilizing Mg2+ ions by the basic region of NC which has been shown to be sufficient for catalyzing the dynamical transition (88).
SUPPLEMENTARY DATA
Supplementary Data is available at NAR Online.
ACKNOWLDEGEMENT
We would like to thank Prof. Michael Summers (University of Maryland Baltimore County) for providing SL1m resonance assignments, Dr Alex Kurochkin for his expertise and for maintenance of the NMR instruments. The authors gratefully acknowledge the Michigan Economic Development Cooperation and the Michigan Technology Tri-Corridor for the support of the purchase 600 MHz spectrometer. This work was supported by funding from the NIH (RO1 AI066975-01). Funding to pay the Open Access publication charge was provided by #NIH/RO1 AI066975-01.
Conflict of interest statement. None declared.
REFERENCES
- 1.Darlix JL, Gabus C, Nugeyre MT, Clavel F, Barre-Sinoussi F. Cis elements and trans-acting factors involved in the RNA dimerization of the human immunodeficiency virus HIV-1. J. Mol. Biol. 1990;216:689–699. doi: 10.1016/0022-2836(90)90392-Y. [DOI] [PubMed] [Google Scholar]
- 2.Marquet R, Baudin F, Gabus C, Darlix JL, Mougel M, Ehresmann C, Ehresmann B. Dimerization of human immunodeficiency virus (type 1) RNA: stimulation by cations and possible mechanism. Nucleic Acids Res. 1991;19:2349–2357. doi: 10.1093/nar/19.9.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paillart JC, Shehu-Xhilaga M, Marquet R, Mak J. Dimerization of retroviral RNA genomes: an inseparable pair. Nat. Rev. Microbiol. 2004;2:461–472. doi: 10.1038/nrmicro903. [DOI] [PubMed] [Google Scholar]
- 4.D'Souza V, Summers MF. How retroviruses select their genomes. Nat. Rev. Microbiol. 2005;3:643–655. doi: 10.1038/nrmicro1210. [DOI] [PubMed] [Google Scholar]
- 5.Hoglund S, Ohagen A, Goncalves J, Panganiban AT, Gabuzda D. Ultrastructure of HIV-1 genomic RNA. Virology. 1997;233:271–279. doi: 10.1006/viro.1997.8585. [DOI] [PubMed] [Google Scholar]
- 6.Laughrea M, Jette L. A 19-nucleotide sequence upstream of the 5′ major splice donor is part of the dimerization domain of human-immunodeficiency-virus-1 genomic RNA. Biochemistry. 1994;33:13464–13474. doi: 10.1021/bi00249a035. [DOI] [PubMed] [Google Scholar]
- 7.Skripkin E, Paillart JC, Marquet R, Ehresmann B, Ehresmann C. Identification of the primary site of the human-immunodeficiency-virus type-1 RNA dimerization in-vitro. Proc. Natl. Acad. Sci. U.S.A. 1994;91:4945–4949. doi: 10.1073/pnas.91.11.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berkhout B. Structure and function of the human immunodeficiency virus leader RNA. Prog. Nucleic Acid Res. Mol. Biol. 1996;54:1–34. doi: 10.1016/s0079-6603(08)60359-1. [DOI] [PubMed] [Google Scholar]
- 9.Berkhout B, van Wamel JL. The leader of the HIV-1 RNA genome forms a compactly folded tertiary structure. RNA. 2000;6:282–295. doi: 10.1017/s1355838200991684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paillart JC, Marquet R, Skripkin E, Ehresmann C, Ehresmann B. Dimerization of retroviral genomic RNAs: structural and functional implications. Biochimie. 1996;78:639–653. doi: 10.1016/s0300-9084(96)80010-1. [DOI] [PubMed] [Google Scholar]
- 11.Fu W, Rein A. Maturation of dimeric viral RNA of Moloney murine leukemia virus. J. Virol. 1993;67:5443–5449. doi: 10.1128/jvi.67.9.5443-5449.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu W, Gorelick RJ, Rein A. Characterization of human immunodeficiency virus type 1 dimeric RNA from wild-type and protease-defective virions. J. Virol. 1994;68:5013–5018. doi: 10.1128/jvi.68.8.5013-5018.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shehu-Xhilaga M, Kraeusslich HG, Pettit S, Swanstrom R, Lee JY, Marshall JA, Crowe SM, Mak J. Proteolytic processing of the p2/nucleocapsid cleavage site is critical for human immunodeficiency virus type 1 RNA dimer maturation. J. Virol. 2001;75:9156–9164. doi: 10.1128/JVI.75.19.9156-9164.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang C, Rong L, Quan Y, Laughrea M, Kleiman L, Wainberg MA. Mutations within four distinct gag proteins are required to restore replication of human immunodeficiency virus type 1 after deletion mutagenesis within the dimerization initiation site. J. Virol. 1999;73:7014–7020. doi: 10.1128/jvi.73.8.7014-7020.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sheng N, Erickson-Viitanen S. Cleavage of p15 protein in vitro by human immunodeficiency virus type 1 protease is RNA dependent. J. Virol. 1994;68:6207–6214. doi: 10.1128/jvi.68.10.6207-6214.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sheng N, Pettit SC, Tritch RJ, Ozturk DH, Rayner MM, Swanstrom R, Erickson-Viitanen S. Determinants of the human immunodeficiency virus type 1 p15NC-RNA interaction that affect enhanced cleavage by the viral protease. J. Virol. 1997;71:5723–5732. doi: 10.1128/jvi.71.8.5723-5732.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muriaux D, De Rocquigny H, Roques BP, Paoletti J, Fosse P. NCp7 activates HIV-1Lai RNA dimerization by converting a transient loop–loop complex into a stable dimer A kissing complex together with a stable dimer is involved in the HIV-1Lai RNA dimerization process in vitro. J. Biol. Chem. 1996;271:33686–33692. doi: 10.1074/jbc.271.52.33686. [DOI] [PubMed] [Google Scholar]
- 18.Feng YX, Copeland TD, Henderson LE, Gorelick RJ, Bosche WJ, Levin JG, Rein A. HIV-1 nucleocapsid protein induces ‘maturation’ of dimeric retroviral RNA in vitro. Proc. Natl. Acad. Sci. U.S.A. 1996;93:7577–7581. doi: 10.1073/pnas.93.15.7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fosse P, Motte N, Roumier A, Gabus C, Muriaux D, Darlix JL, Paoletti J. A short autocomplementary sequence plays an essential role in avian sarcoma-leukosis virus RNA dimerization. Biochemistry. 1996;35:16601–16609. doi: 10.1021/bi9613786. [DOI] [PubMed] [Google Scholar]
- 20.Theilleux-Delalande V, Girard F, Huynh-Dinh T, Lancelot G, Paoletti J. The HIV-1(Lai) RNA dimerization. Thermodynamic parameters associated with the transition from the kissing complex to the extended dimer. Eur. J. Biochem. 2000;267:2711–2719. doi: 10.1046/j.1432-1327.2000.01292.x. [DOI] [PubMed] [Google Scholar]
- 21.Takahashi K, Baba S, Hayashi Y, Koyanagi Y, Yamamoto N, Takaku H, Kawai G. NMR analysis of intra- and inter-molecular stems in the dimerization initiation site of the HIV-1 genome. J. Biochem. (Tokyo) 2000;127:681–686. doi: 10.1093/oxfordjournals.jbchem.a022657. [DOI] [PubMed] [Google Scholar]
- 22.Takahashi KI, Baba S, Chattopadhyay P, Koyanagi Y, Yamamoto N, Takaku H, Kawai G. Structural requirement for the two-step dimerization of human immunodeficiency virus type 1 genome. RNA. 2000;6:96–102. doi: 10.1017/s1355838200991635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rist MJ, Marino JP. Mechanism of nucleocapsid protein catalyzed structural isomerization of the dimerization initiation site of HIV-1. Biochemistry. 2002;41:14762–14770. doi: 10.1021/bi0267240. [DOI] [PubMed] [Google Scholar]
- 24.Windbichler N, Werner M, Schroeder R. Kissing complex-mediated dimerisation of HIV-1 RNA: coupling extended duplex formation to ribozyme cleavage. Nucleic Acids Res. 2003;31:6419–6427. doi: 10.1093/nar/gkg873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aci S, Mazier S, Genest D. Conformational pathway for the kissing complex – >extended dimer transition of the SL1 stem-loop from genomic HIV-1 RNA as monitored by targeted molecular dynamics techniques. J. Mol. Biol. 2005;351:520–530. doi: 10.1016/j.jmb.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 26.Ennifar E, Walter P, Ehresmann B, Ehresmann C, Dumas P. Crystal structures of coaxially stacked kissing complexes of the HIV-1 RNA dimerization initiation site. Nat. Struct. Biol. 2001;8:1064–1068. doi: 10.1038/nsb727. [DOI] [PubMed] [Google Scholar]
- 27.Ooms M, Huthoff H, Russell R, Liang C, Berkhout B. A riboswitch regulates RNA dimerization and packaging in human immunodeficiency virus type 1 virions. J. Virol. 2004;78:10814–10819. doi: 10.1128/JVI.78.19.10814-10819.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huthoff H, Berkhout B. Two alternating structures of the HIV-1 leader RNA. RNA. 2001;7:143–157. doi: 10.1017/s1355838201001881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abbink TE, Ooms M, Haasnoot PC, Berkhout B. The HIV-1 leader RNA conformational switch regulates RNA dimerization but does not regulate mRNA translation. Biochemistry. 2005;44:9058–9066. doi: 10.1021/bi0502588. [DOI] [PubMed] [Google Scholar]
- 30.De Guzman RN, Wu ZR, Stalling CC, Pappalardo L, Borer PN, Summers MF. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- 31.Amarasinghe GK, De Guzman RN, Turner RB, Chancellor KJ, Wu ZR, Summers MF. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the psi-RNA packaging signal. Implications for genome recognition. J. Mol. Biol. 2000;301:491–511. doi: 10.1006/jmbi.2000.3979. [DOI] [PubMed] [Google Scholar]
- 32.Clever JL, Parslow TG. Mutant human immunodeficiency virus type 1 genomes with defects in RNA dimerization or encapsidation. J. Virol. 1997;71:3407–3414. doi: 10.1128/jvi.71.5.3407-3414.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clever J, Sassetti C, Parslow TG. RNA secondary structure and binding sites for gag gene products in the 5′ packaging signal of human immunodeficiency virus type 1. J. Virol. 1995;69:2101–2109. doi: 10.1128/jvi.69.4.2101-2109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Damgaard CK, Dyhr-Mikkelsen H, Kjems J. Mapping the RNA binding sites for human immunodeficiency virus type-1 gag and NC proteins within the complete HIV-1 and -2 untranslated leader regions. Nucleic Acids Res. 1998;26:3667–3676. doi: 10.1093/nar/26.16.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shubsda MF, Paoletti AC, Hudson BS, Borer PN. Affinities of packaging domain loops in HIV-1 RNA for the nucleocapsid protein. Biochemistry. 2002;41:5276–5282. doi: 10.1021/bi016045+. [DOI] [PubMed] [Google Scholar]
- 36.Mirambeau G, Lyonnais S, Coulaud D, Hameau L, Lafosse S, Jeusset J, Justome A, Delain E, Gorelick RJ, et al. Transmission electron microscopy reveals an optimal HIV-1 nucleocapsid aggregation with single-stranded nucleic acids and the mature HIV-1 nucleocapsid protein. J. Mol. Biol. 2006;364:496–511. doi: 10.1016/j.jmb.2006.08.065. [DOI] [PubMed] [Google Scholar]
- 37.Greatorex J, Gallego J, Varani G, Lever A. Structure and stability of wild-type and mutant RNA internal loops from the SL-1 domain of the HIV-1 packaging signal. J. Mol. Biol. 2002;322:543–557. doi: 10.1016/s0022-2836(02)00776-3. [DOI] [PubMed] [Google Scholar]
- 38.Lawrence DC, Stover CC, Noznitsky J, Wu Z, Summers MF. Structure of the intact stem and bulge of HIV-1 Psi-RNA stem-loop SL1. J. Mol. Biol. 2003;326:529–542. doi: 10.1016/s0022-2836(02)01305-0. [DOI] [PubMed] [Google Scholar]
- 39.Yuan Y, Kerwood DJ, Paoletti AC, Shubsda MF, Borer PN. Stem of SL1 RNA in HIV-1: structure and nucleocapsid protein binding for a 1 × 3 internal loop. Biochemistry. 2003;42:5259–5269. doi: 10.1021/bi034084a. [DOI] [PubMed] [Google Scholar]
- 40.Baba S, Takahashi K, Noguchi S, Takaku H, Koyanagi Y, Yamamoto N, Kawai G. Solution RNA structures of the HIV-1 dimerization initiation site in the kissing-loop and extended-duplex dimers. J. Biochem. (Tokyo) 2005;138:583–592. doi: 10.1093/jb/mvi158. [DOI] [PubMed] [Google Scholar]
- 41.Ulyanov NB, Mujeeb A, Du Z, Tonelli M, Parslow TG, James TL. NMR structure of the full-length linear dimer of stem-loop-1 RNA in the HIV-1 dimer initiation site. J. Biol. Chem. 2006;281:16168–16177. doi: 10.1074/jbc.M601711200. [DOI] [PubMed] [Google Scholar]
- 42.Reblova K, Spackova N, Sponer JE, Koca J, Sponer J. Molecular dynamics simulations of RNA kissing-loop motifs reveal structural dynamics and formation of cation-binding pockets. Nucleic Acids Res. 2003;31:6942–6952. doi: 10.1093/nar/gkg880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beaurain F, Laguerre M. MD studies of the DIS/DIS kissing complex solution and X-ray structures. Oligonucleotides. 2003;13:501–514. doi: 10.1089/154545703322860816. [DOI] [PubMed] [Google Scholar]
- 44.Ennifar E, Dumas P. Polymorphism of bulged-out residues in HIV-1 RNA DIS kissing complex and structure comparison with solution studies. J. Mol. Biol. 2006;356:771–782. doi: 10.1016/j.jmb.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 45.Aci S, Gangneux L, Paoletti J, Genest D. On the stability of different experimental dimeric structures of the SL1 sequence from the genomic RNA of HIV-1 in solution: a molecular dynamics simulation and electrophoresis study. Biopolymers. 2004;74:177–188. doi: 10.1002/bip.20032. [DOI] [PubMed] [Google Scholar]
- 46.Mihailescu MR, Marino JP. A proton-coupled dynamic conformational switch in the HIV-1 dimerization initiation site kissing complex. Proc. Natl. Acad. Sci. U.S.A. 2004;101:1189–1194. doi: 10.1073/pnas.0307966100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tolman JR, Flanagan JM, Kennedy MA, Prestegard JH. Nuclear magnetic dipole interactions in field-oriented proteins – information for structure determination in solution. Proc. Natl. Acad. Sci. U.S.A. 1995;92:9279–9283. doi: 10.1073/pnas.92.20.9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tjandra N, Bax A. Direct measurement of distances and angles in biomolecules by NMR in a dilute liquid crystalline medium. Science. 1997;278:1111–1114. doi: 10.1126/science.278.5340.1111. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Q, Sun X, Watt ED, Al-Hashimi HM. Resolving the motional modes that code for RNA adaptation. Science. 2006;311:653–656. doi: 10.1126/science.1119488. [DOI] [PubMed] [Google Scholar]
- 50.Hansen MR, Mueller L, Pardi A. Tunable alignment of macromolecules by filamentous phage yields dipolar coupling interactions. Nat. Struct. Biol. 1998;5:1065–1074. doi: 10.1038/4176. [DOI] [PubMed] [Google Scholar]
- 51.Clore GM, Starich MR, Gronenborn AM. Measurement of residual dipolar couplings of macromolecules aligned in the nematic phase of a colloidal suspension of rod- shaped viruses. J. Am. Chem. Soc. 1998;120:10571–10572. [Google Scholar]
- 52.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. Nmrpipe – a multidimensional spectral processing system based on unix pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 53.Johnson BA, Blevins RA. NMR view – a computer-program for the visualization and analysis of NMR data. J. Biomol. NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 54.Goddard TD, Kneller DG. SPARKY 3. San Franscisco: University of California; [Google Scholar]
- 55.Furtig B, Richter C, Wohnert J, Schwalbe H. NMR spectroscopy of RNA. Chembiochem. 2003;4:936–962. doi: 10.1002/cbic.200300700. [DOI] [PubMed] [Google Scholar]
- 56.Meissner A, Duus JO, Sorensen OW. Spin-state-selective excitation. Application for E.COSY-type measurement of J(HH) coupling constants. J. Magn. Reson. 1997;128:92–97. [Google Scholar]
- 57.Meissner A, Sorensen OW. The role of coherence transfer efficiency in design of TROSY-type multidimensional NMR experiments. J. Magn. Reson. 1999;139:439–442. doi: 10.1006/jmre.1999.1788. [DOI] [PubMed] [Google Scholar]
- 58.Pitt SW, Majumdar A, Serganov A, Patel DJ, Al-Hashimi HM. Argininamide binding arrests global motions in HIV-1 TAR RNA: comparison with Mg2+-induced conformational stabilization. J. Mol. Biol. 2004;338:7–16. doi: 10.1016/j.jmb.2004.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yip GN, Zuiderweg ER. A phase cycle scheme that significantly suppresses offset-dependent artifacts in the R2-CPMG 15N relaxation experiment. J. Magn. Reson. 2004;171:25–36. doi: 10.1016/j.jmr.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 60.Yip GN, Zuiderweg ER. Improvement of duty-cycle heating compensation in NMR spin relaxation experiments. J. Magn. Reson. 2005;176:171–178. doi: 10.1016/j.jmr.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 61.Saupe A, Englert G. High-resolution nuclear magnetic resonance spectra of oriented molecules. Phys. Rev. Lett. 1963;11:462–464. [Google Scholar]
- 62.Losonczi JA, Andrec M, Fischer MWF, Prestegard JH. Order matrix analysis of residual dipolar couplings using singular value decomposition. J. Magn. Reson. 1999;138:334–342. doi: 10.1006/jmre.1999.1754. [DOI] [PubMed] [Google Scholar]
- 63.Al-Hashimi HM, Gosser Y, Gorin A, Hu W, Majumdar A, Patel DJ. Concerted motions in HIV-1 TAR RNA may allow access to bound state conformations: RNA dynamics from NMR residual dipolar couplings. J. Mol. Biol. 2002;315:95–102. doi: 10.1006/jmbi.2001.5235. [DOI] [PubMed] [Google Scholar]
- 64.Hansen AL, Al-Hashimi HM. Insight into the CSA tensors of nucleobase carbons in RNA polynucleotides from solution measurements of residual CSA: towards new long-range orientational constraints. J. Magn. Reson. 2006;179:299–307. doi: 10.1016/j.jmr.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 65.Musselman C, Pitt SW, Gulati K, Foster LL, Andricioaei I, Al-Hashimi HM. Impact of static and dynamic a-form heterogeneity on the determination of RNA global structural dynamics using NMR residual dipolar couplings. J. Biomol. NMR. 2006;36:235–249. doi: 10.1007/s10858-006-9087-9. [DOI] [PubMed] [Google Scholar]
- 66.Al-Hashimi HM, Valafar H, Terrell M, Zartler ER, Eidsness MK, Prestegard JH. Variation of molecular alignment as a means of resolving orientational ambiguities in protein structures from dipolar couplings. J. Magn. Reson. 2000;143:402–406. doi: 10.1006/jmre.2000.2049. [DOI] [PubMed] [Google Scholar]
- 67.Zweckstetter M, Bax A. Predicition of sterically induced alignment in a dilute liquid crystalline phase; aid to protein structure determination by NMR. J. Am. Chem. Soc. 2000;122:3791–3792. [Google Scholar]
- 68.Zhang Q, Throolin R, Pitt SW, Serganov A, Al-Hashimi HM. Probing motions between equivalent RNA domains using magnetic field induced residual dipolar couplings: accounting for correlations between motions and alignment. J. Am. Chem. Soc. 2003;125:10530–10531. doi: 10.1021/ja0363056. [DOI] [PubMed] [Google Scholar]
- 69.Lipari G, Szabo A. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 1. Theory and range of validity. J. Am. Chem. Soc. 1982;104:4546–4559. [Google Scholar]
- 70.Clore GM, Szabo A, Bax A, Kay LE, Driscoll PC, Gronenborn AM. Deviations from the simple two-parameter model-free approach to the interpretation of nitrogen-15 nuclear magnetic relaxation of proteins. J. Am. Chem. Soc. 1990;112:4989–4991. [Google Scholar]
- 71.Mandel AM, Akke M, Palmer AG. Backbone dynamics of Escherichia-coli ribonuclease Hi - correlations with structure and function in an active enzyme. J. Mol. Biol. 1995;246:144–163. doi: 10.1006/jmbi.1994.0073. [DOI] [PubMed] [Google Scholar]
- 72.Garcia de la Torre J, Huertas ML, Carrasco B. HYDRONMR: prediction of NMR relaxation of globular proteins from atomic-level structures and hydrodynamic calculations. J. Magn. Reson. 2000;147:138–146. doi: 10.1006/jmre.2000.2170. [DOI] [PubMed] [Google Scholar]
- 73.Gonzalez RLJ, Tinoco IJ. Solution structure and thermodynamics of a divalent metal ion binding site in an RNA pseudoknot. J. Mol. Biol. 1999;289:1267–1282. doi: 10.1006/jmbi.1999.2841. [DOI] [PubMed] [Google Scholar]
- 74.Dingley AJ, Grzesiek S. Direct observation of hydrogen bonds in nucleic acid base pairs by internucleotide (2)J(NN) couplings. J. Am. Chem. Soc. 1998;120:8293–8297. [Google Scholar]
- 75.Pervushin K, Ono A, Fernandez C, Szyperski T, Kainosho M, Wuthrich K. NMR scaler couplings across Watson–Crick base pair hydrogen bonds in DNA observed by transverse relaxation optimized spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 1998;95:14147–14151. doi: 10.1073/pnas.95.24.14147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Latham MP, Brown DJ, McCallum SA, Pardi A. NMR methods for studying the structure and dynamics of RNA. Chembiochem. 2005;6:1492–1505. doi: 10.1002/cbic.200500123. [DOI] [PubMed] [Google Scholar]
- 77.Al-Hashimi HM. Dynamics-based amplification of RNA function and its characterization by using NMR spectroscopy. Chembiochem. 2005;6:1506–1519. doi: 10.1002/cbic.200500002. [DOI] [PubMed] [Google Scholar]
- 78.Saupe A. Recent results in the field of liquid crystals. Angew. Chem. Int. Ed. Engl. 1968;7:97–112. [Google Scholar]
- 79.Tolman JR, Al-Hashimi HM, Kay LE, Prestegard JH. Structural and dynamic analysis of residual dipolar coupling data for proteins. J. Am. Chem. Soc. 2001;123:1416–1424. doi: 10.1021/ja002500y. [DOI] [PubMed] [Google Scholar]
- 80.Mollova ET, Hansen MR, Pardi A. Global structure of RNA determined with residual dipolar couplings. J. Am. Chem. Soc. 2000;122:11561–11562. [Google Scholar]
- 81.Sibille N, Pardi A, Simorre JP, Blackledge M. Refinement of local and long-range structural order in theophylline-binding RNA using C-13-H-1 residual dipolar couplings and restrained molecular dynamics. J. Am. Chem. Soc. 2001;123:12135–12146. doi: 10.1021/ja011646+. [DOI] [PubMed] [Google Scholar]
- 82.Getz M, Andrews AJ, Fierke CA, Al-Hashimi HM. Structural plasticity and Mg2+ binding properties of RNase P P4 from combined analysis of NMR residual dipolar couplings and motionally decoupled spin relaxation. RNA. 2006;13:251–266. doi: 10.1261/rna.264207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pitt SW, Zhang Q, Patel DJ, Al-Hashimi HM. Evidence that electrostatic interactions dictate the ligand-induced arrest of RNA global flexibility. Angew. Chem. Int. Ed. Engl. 2005;44:3412–3415. doi: 10.1002/anie.200500075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Al-Hashimi HM, Pitt SW, Majumdar A, Xu W, Patel DJ. Mg2+-induced variations in the conformation and dynamics of HIV-1 TAR RNA probed using NMR residual dipolar couplings. J. Mol. Biol. 2003;329:867–873. doi: 10.1016/s0022-2836(03)00517-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tolman JR, Al-Hashimi HM. In: Ann. Rep. NMR. Spec. Webb GA, editor. Vol. 51. Academic Press; 2003. pp. 105–166. [Google Scholar]
- 86.Clever JL, Wong ML, Parslow TG. Requirements for kissing-loop-mediated dimerization of human immunodeficiency virus RNA. J. Virol. 1996;70:5902–5908. doi: 10.1128/jvi.70.9.5902-5908.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tolman JR, Flanagan JM, Kennedy MA, Prestegard JH. NMR evidence for slow collective motions in cyanometmyoglobin. Nat. Struct. Biol. 1997;4:292–297. doi: 10.1038/nsb0497-292. [DOI] [PubMed] [Google Scholar]
- 88.Takahashi K, Baba S, Koyanagi Y, Yamamoto N, Takaku H, Kawai G. Two basic regions of NCp7 are sufficient for conformational conversion of HIV-1 dimerization initiation site from kissing-loop dimer to extended-duplex dimer. J. Biol. Chem. 2001;276:31274–31278. doi: 10.1074/jbc.M104577200. [DOI] [PubMed] [Google Scholar]
- 89.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, et al. A 2nd generation force-field for the simulation of proteins, nucleic-acids, and organic-molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 90.Chin K, Sharp KA, Honig B, Pyle AM. Calculating the electrostatic properties of RNA provides new insights into molecular interactions and function. Nat. Struct. Biol. 1999;6:1055–1061. doi: 10.1038/14940. [DOI] [PubMed] [Google Scholar]
- 91.Nicholls A, Sharp K, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.