Abstract
Homing endonucleases are unusual enzymes, capable of recognizing lengthy DNA sequences and cleaving site-specifically within genomes. Many homing endonucleases are encoded within group I introns, and such enzymes promote the mobility reactions of these introns. Phage T4 has three group I introns, within the td, nrdB and nrdD genes. The td and nrdD introns are mobile, whereas the nrdB intron is not. Phage RB3 is a close relative of T4 and has a lengthier nrdB intron. Here, we describe I-TevIII, the H–N–H endonuclease encoded by the RB3 nrdB intron. In contrast to previous reports, we demonstrate that this intron is mobile, and that this mobility is dependent on I-TevIII, which generates 2-nt 3′ extensions. The enzyme has a distinct catalytic domain, which contains the H–N–H motif, and DNA-binding domain, which contains two zinc fingers required for interaction with the DNA substrate. Most importantly, I-TevIII, unlike the H–N–H endonucleases described so far, makes a double-strand break on the DNA homing site by acting as a dimer. Through deletion analysis, the dimerization interface was mapped to the DNA-binding domain. The unusual propensity of I-TevIII to dimerize to achieve cleavage of both DNA strands underscores the versatility of the H–N–H enzyme family.
INTRODUCTION
Homing endonucleases are usually intron- or intein-encoded enzymes that catalyze the first step of the mobility process of their respective host elements at the DNA level (1). In the homing reaction, the endonuclease recognizes and cleaves an intronless/inteinless allele of its host gene, thereby initiating a gene conversion event through which the intron or intein is copied into the break site (2). Homing endonucleases are found in all three biological domains, the archaea, the eubacteria and the eukarya and they are remarkable in their ability to self-propagate in environments that usually select for streamlined genomes (3,4).
Phage T4 has three group I intron-containing genes: td, which encodes thymidylate synthase; nrdD (or sunY), which encodes an anerobic ribonucleotide reductase; and nrdB, which encodes the small subunit of ribonucleotide reductase (5). Not only are the ∼1-kb td and nrdD introns approximately 400 nt longer than the nrdB intron, but they are also mobile, whereas the nrdB intron is not (6). Through a PCR screen of natural phage isolates, it was discovered that phage RB3, a close relative of phage T4, has an nrdB intron larger than that of T4, with a longer open reading frame. Furthermore, the RB3 nrdB intron-encoded protein has endonuclease activity (7).
Homing endonucleases fall into distinct families based on the presence of conserved sequence elements (1,8). Comparative amino acid analysis shows that the RB3 nrdB homing endonuclease, which is called I-TevIII (intron-encoded T-even endonuclease III), is a member of the H–N–H family. The H–N–H endonucleases are part of a wider group of enzymes called ββα-Me or His-Me endonucleases (8,9). In addition, I-TevIII has a novel domain, which contains two putative zinc fingers, as discussed in detail below. The H–N–H module is found in proteins of diverse function, including bacterial colicins E7 and E9, as well as intron- and intein-encoded enzymes (10).
I-TevIII from RB3 was shown in vivo to have cleavage activity on T4 nrdB intron-minus plasmid template (7). Primer extension analysis was used to define the precise cleavage site, and the enzyme was reported to generate a 2-nt 5′ overhang, in contrast to all other characterized homing endonucleases, which generate 3′ extensions. In addition, despite the fact that the enzyme was shown to be active in vivo, mobility of the intron could not be demonstrated.
Three-dimensional crystal structures have been determined for the DNA endonuclease domains of the colicins (9,11) and of I-HmuI, an H–N–H endonuclease encoded by a group I intron found in the DNA polymerase gene of bacteriophage SPO1 (12). I-HmuI is unusual among homing endonucleases in that it can cleave both intron-plus and intron-minus alleles of its host gene, and it only nicks its substrate rather than generating a double-strand break (DSB).
H–N–H and GIY-YIG endonucleases have a common feature in that members of both families of enzymes have modular domains that separate their catalytic and DNA-binding properties (12–15). Protein modularity is found in many DNA metabolism enzymes (16), allowing for a diversification of functions by shuffling catalytic domains and DNA-binding domains, generating altered or new recognition sequences. Likewise, homing endonucleases acquire new DNA-substrate specificities (13,14).
Members of each family of homing endonuclease contain examples of enzymes that coordinate zinc for a structural rather than a catalytic purpose (17–19). Metal cofactors are sometimes essential for proper folding of proteins, and this may also lead to the proteins’ increased structural diversity and functional flexibility (20). Zinc ions, which are widely used metal cofactors, are typically found in proteins in zinc finger modules, in which cysteines or histidines are the conserved, metal-coordinating amino acids (21).
In this work, we have shown that I-TevIII can utilize specific divalent cations for cleavage activity, and that, contrary to previous results, I-TevIII generates a 2-nt 3′ overhang when it cleaves its homing site. Also, using a different strategy from that used previously, we have shown that the nrdB intron is indeed mobile and that I-TevIII catalyzes this homing process. The enzyme has distinct cleavage and DNA-binding domains, and mutagenesis revealed that the H–N–H residues have catalytic properties, whereas the zinc fingers play a role in DNA binding. Most importantly, I-TevIII, unlike H–N–H homing endonucleases so far characterized, achieves double-strand cleavage by interacting with its substrate as a dimer.
MATERIALS AND METHODS
Mobility assays
Homing of the nrdB intron was demonstrated using a plasmid donor pSURB3nrdB containing the RB3 nrdB intron and a T4 phage recipient that had each of the three introns deleted (gifted by David Shub). Crosses were carried out essentially as described previously (6). The RB3 and T4 nrdB introns were subcloned by the PCR into pSU18 as positive and negative controls, respectively. Positive (pACYtd) and negative (pACYtdΔ1-3) donor plasmid controls for td intron homing were used alongside the nrdB assays. Homing events were detected by plaque hybridization using intron-specific PCR fragments labeled with [α-32P]dCTP and the random primer labeling kit (Invitrogen). Homing frequencies were expressed as the percentage of positive plaques compared to the total number of plaques on the plate.
Cloning of I-TevIII and its domains for overexpression and purification
Overexpression plasmids for the full-length enzyme and deletion derivatives had the coding sequence for each derivative under the control of the T7 promoter. The coding sequence for each derivative was generated by the PCR using primers that did or did not incorporate the sequence for a hexa-His tag, as appropriate. The full-length enzyme was cloned into the intein-based vector pTYB2 (New England Biolabs) with a stop codon introduced upstream of the coding sequence for the intein segment in order to retain expression of the native protein. Deletion derivatives were also cloned via the PCR into either pET3a (Novagen) or pET101D (Invitrogen).
Induction and purification of I-TevIII and derivatives
Plasmids for overexpression were introduced into host strain BL21(DE3)pLysS (22). Cells were grown in TBYE (1% tryptone, 0.5% NaCl, 1% yeast extract) supplemented with ampicillin (100 µg/ml) at 37°C to A600 0.5, except for the full-length His-tagged derivative, which was grown with kanamycin (50 µg/ml). Cultures were maintained at 37°C, and expression was induced by the addition of 1 mM isopropyl β-D-1-thiogalactopyranoside. Incubation was continued at 37°C for 1.5 h before cells were harvested and stored, frozen at −80°C. All purification steps were carried out at 0–4°C. Frozen cell pellets were thawed and lysed in lysis buffer (50 mM Tris-HCl pH 8.0, 5 mM DTT) and sonicated to complete lysis and reduce viscosity. Insoluble material was removed by centrifugation (10 000 g for 20 min). Polyethyleneimine (PEI, 10%) was added to the supernatant to a final concentration of 0.2% to precipitate out nucleic acids and contaminating proteins. After removal of the precipitate by centrifugation (10 000 g for 20 min), the protein was ammonium-sulphate fractionated by the addition of solid (NH4)2SO4 to 45% saturation. The (NH4)2SO4 precipitate was harvested by centrifugation, resuspended in 50 mM Tris-HCl pH 7.5, 5 mM DTT and applied to a HiTrap S column (Amersham) equilibrated in the same buffer and connected to an ÄKTA FPLC (Pharmacia). The column was washed with six column volumes and then the proteins were eluted with a NaCl gradient (0–2 M) in the same buffer.
Derivatives with an His tag were grown and induced as for the native full-length protein and purified by immobilized metal affinity chromatography (Ni-NTA, Qiagen). The single step Ni-NTA purification scheme allowed for rapid purification to near homogeneity (∼85%), which permitted quantification of the protein and reproducibility of results (see the Discussion section).
For the I-TevIII deletion derivatives, DNA-binding and cleavage assays were carried out with crude Escherichia coli extracts of overexpressed protein. In this case, cells from 1.5 ml of induced culture were harvested by centrifugation, resuspended in 250 µl of lysis buffer and briefly sonicated. The cellular debris was removed by centrifugation, and the soluble lysate was used in experiments either directly or diluted in lysis buffer.
I-TevIII activity assays
Plasmid pSUT4nrdBΔIn, which provides the I-TevIII substrate, is a pSU18 derivative that contains a 760-bp fragment spanning the nrdB intron homing site, with the cleavage site 400 bp from the 5′ end of the fragment. For activity assays, a 100-bp PCR fragment was generated from pSUT4nrdBΔIn, using W1622 and W1623 as primers, which were end labeled with [γ-32P]ATP (Supplementary Table 1). I-TevIII was incubated with 4000 counts per minute (cpm) of the fragment in 20 μl of assay buffer (100 mM Tris-HCl pH 7.6, 150 mM NaCl and supplemented with various cations) at 37°C for 15 min. Reactions were stopped by the addition of 1 µl of proteinase K (20 mg/ml) and further incubated at 37°C for 1 h. Samples loaded on an 8% acrylamide gel were separated in 40 mM Tris-borate pH 8.0, 2 mM EDTA. Products were visualized with autoradiography.
Cleavage site mapping
Cleavage mapping of the I-TevIII homing site was carried out using a Thermosequenase Cycle-Sequencing 2.0 kit (USB) as described (23). Sequence ladders of the homing site were generated with pSUT4nrdB▵In using 2 µM of [γ-32P]ATP end-labeled primers W1171 and W1173. The primer pair W1171 and W1173 (20 µM [γ-32P]ATP end-labeled) were utilized in PCR reactions with pSUT4nrdBΔIn as template to generate a 158-bp fragment for cleavage-site mapping. Labeled substrates (10 000 cpm) were digested with purified I-TevIII in reaction mixtures (100 mM Tris-HCl pH 7.6, 10 mM MgCl2, 150 mM NaCl and 0.1 mM NiSO4) and incubated at 37°C for 15 min. To terminate the reactions, 10 µl of Sequenase (USB) stop dye was added and the products of the reactions were resolved alongside sequencing ladders on an 8% acrylamide/8M urea gel (w/v).
DNase footprinting
Footprinting was carried out essentially as described previously (24). Purified native I-TevIII or the 145C deletion derivative was incubated with DNaseI substrates that were generated by the PCR from pSUT4nrdBΔIn using [γ-32P]ATP end-labeled primers (W1374 and W1172), which yielded ∼400-bp fragment. Reactions were fractionated on an 8% acrylamide/8M urea (w/v) gel alongside a Maxam-Gilbert A + G sequencing ladder of the same fragments.
Protease digestion
Full-length native I-TevIII was digested with protease to identify proteolytically sensitive sites. Reactions (15 µl) contained 5 µg of I-TevIII in 50 mM Tris-HCl pH 8.0 and serial dilutions of chymotrypsin, thermolysin, subtilysin, or trypsin from 625:1 to 6 250 000:1 (w/w) I-TevIII:trypsin. Reaction mixtures were incubated for 15 min at room temperature and then quenched by the addition of 2 mM PMSF and 5 mM EDTA. They were stored frozen, separated on 15% SDS/polyacrylamide gels and visualized by staining with Coomassie blue.
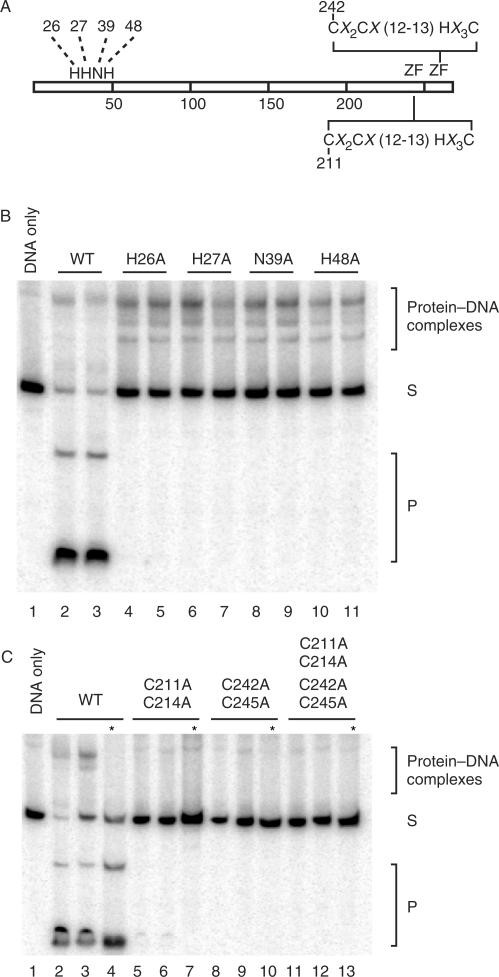
Mutational analysis of the H–N–H and CX2CX12-13HX3C motifs
To determine the contribution of individual amino acids involved in the hydrolysis of DNA we created I-TevIII with mutations in the conserved residues of the catalytic domain (H26A, H27A, N39A, H48A) using site-directed mutagenesis with the His-tagged I-TevIII as template. The oligonucleotides used in the mutagenesis are found in Supplementary Table 1. Furthermore, to determine the contribution of the putative zinc finger motif in I-TevIII to its zinc and oligomerization properties, we created mutations in the conserved cysteine residues (double mutation: C211A and C214A; double mutation: C242A and C245A; and quadruple mutation: C211A, C214A, C242A and C245A). All mutations were confirmed by nucleotide sequencing. Overexpression and purification of the mutated proteins were carried out as described above.
Determination of protein-bound zinc
A spectroscopic technique was utilized to determine the zinc content of each protein. Each protein was incubated with 4-(2-pyridylazo) resorcinol (PAR) at a concentration of 10 mM in PAR buffer (50 mM Tris-HCl pH 7.0 and 800 mM NaCl). The level of background zinc was recorded as the absorbance at 500 nm in a Beckman DU 800 spectrophotometer. To release the zinc bound to the protein, methyl methanethiolsulphonate (MMTS) was added to the reaction mixture and allowed to react for 10 min at room temperature. In this reaction, the liberated zinc forms a Zn-PAR complex, which is measured as an increase in absorbance at 500 nm. A standard curve for estimating the amount of zinc released from the I-TevIII derivatives was generated with ZnCl2.
Gel filtration chromatography
Purified I-TevIII (100 μg) in running buffer (50 mM Tris-HCl pH 7.5, 500 mM NaCl and 5 mM DTT) was applied to a Superose® 12 HR FPLC column (Pharmacia) pre-equilibrated with running buffer. A chromatograph was developed with the running buffer at a flow rate of 0.5 ml/min. Fractions of 0.5 ml were collected and analyzed by 12% SDS-PAGE (29:1 (w/w) acrylamide to bis-acrylamide). The column was calibrated by separating a set of protein standards: aldolase, 158 kDa; bovine serum albumin, 67 kDa; catalase monomer, 58 kDa; ovalbumin, 43 kDa; chymotrypsinogen A, 25 kDa; and ribonuclease A, 13.7 kDa. Mutant and deletion derivatives of I-TevIII were analyzed in the same manner as for the wild-type protein.
Gel-mobility shift analysis and Ferguson analysis
Assays were performed by incubating 1 µl of soluble lysate of I-TevIII derivatives for 15 min at room temperature with end-labeled target DNA in 20 µl reactions containing 50 mM Tris-HCl, pH 8.0, 20 µg/ml poly (dI/dC) and 10 µg/ml BSA. The substrate was a 100-bp fragment generated by the PCR with [γ-32P]ATP-end-labeled primers (W1622 and W1623) using pSUT4nrdBΔIn as substrate. Reaction mixes were loaded onto 8% polyacrylamide (29:1 (w/w) acrylamide to bis-acrylamide) non-denaturing gels in Tris-borate buffer. DNA fragments and complexes were visualized by autoradiography. For determination of the stoichiometry of I-TevIII interaction with its substrate, a Ferguson analysis was carried out as described previously (25). Purified His-tagged I-TevIII was combined with the substrate in binding reactions as above, and the substrate and complexes were resolved on a series of native polyacrylamide gels (7, 8, 9, 10 and 11% polyacrylamide in 1× TBE) alongside 10 µg of non-denatured protein molecular weight standards (Sigma). A retardation coefficient (Kr) was derived from migration of the species, as previously described (25).
RESULTS
The RB3 nrdB intron is mobile
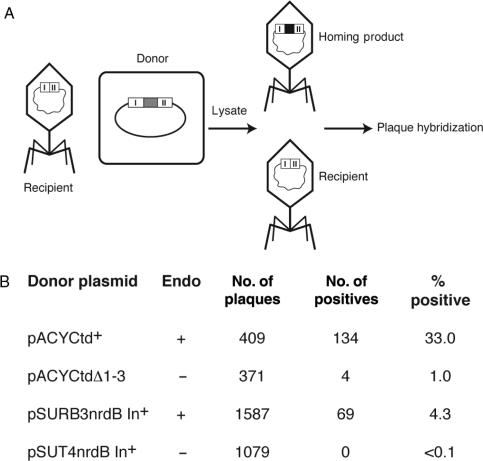
Previous work showed that the nrdB intron from phage T4 is not mobile, whereas the td and nrdD introns home into intronless targets at high efficiency (6). Also, despite the ability of the nrdB-encoded I-TevIII from RB3 to cleave the intronless gene, homing could not be demonstrated (7). To retest whether the nrdB intron from RB3 was mobile, we performed homing assays similar to those demonstrating td and nrdD intron mobility (6), whereby an intron-containing donor plasmid was crossed with a recipient phage, and the resulting lysate analyzed for homing (Figure 1A). In our assays, when the RB3 nrdB donor plasmid was used, homing levels were >40-fold higher (4.3% per recipient), than when the inactive T4 nrdB intron donor was used (<0.1% homing) (Figure 1B). As controls for the homing assay, td intron-plus (td+) and td deleted for intron-encoded protein I-TevI (tdΔ1-3) donor plasmids were used. Although homing levels were higher under these circumstances (33% and 1%, respectively), the relative boost in homing frequency was only 33-fold, as opposed to >40-fold for nrdB.
Figure 1.
Homing analysis. (A) Schematic of homing assay. After infection of cells carrying the donor plasmid, lysates were plated and scored for intron inheritance by plaque hybridization. Donor plasmids are listed in panel B. (B) Data for homing assays. Each cross was performed three times.
Purification and biochemical properties of active I-TevIII
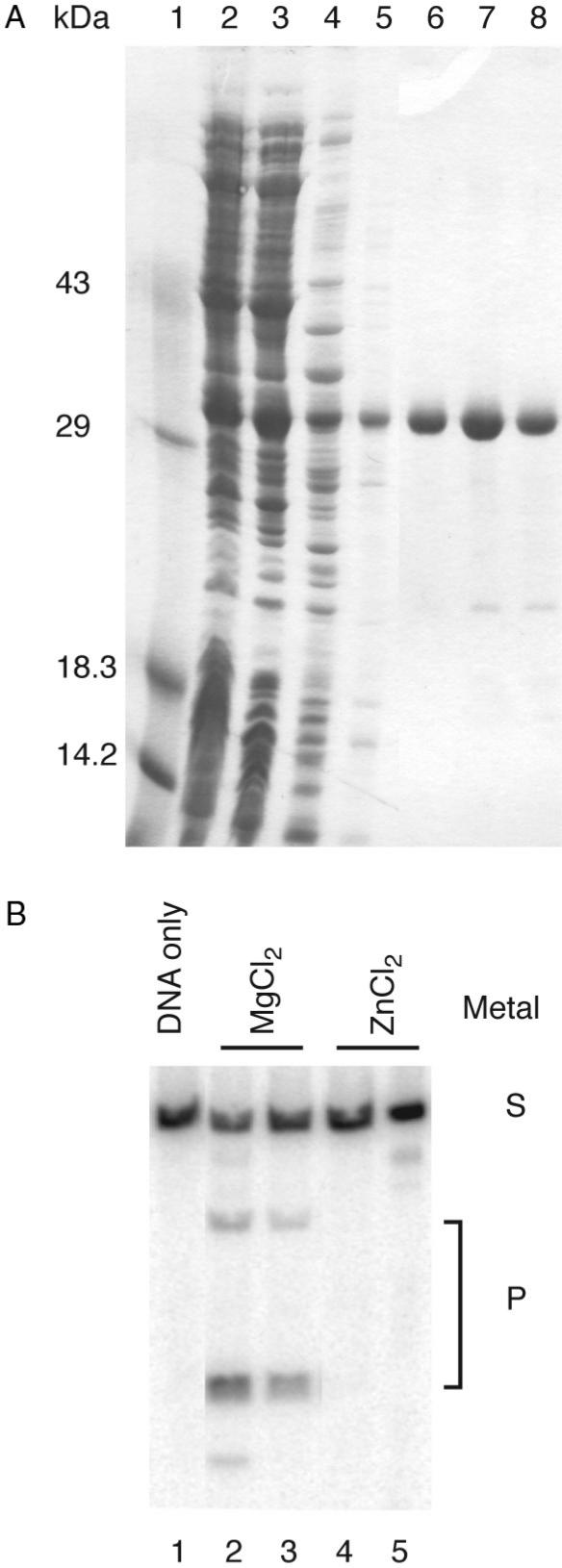
In order to carry out an in vitro characterization of I-TevIII, we overexpressed the protein in E. coli under control of the T7 expression system. The protein could be produced in a soluble form and purified using standard methods (Figure 2A). Although the purified enzyme appeared not to be very active (see below), we were able to demonstrate double-strand cleavage activity using linear DNA fragments containing the nrdB intron homing site as substrate (Figure 2B). We found that the enzyme displays optimal activity at 37°C in 100 mM Tris-HCl pH 7.6, 150 mM NaCl, with Mg2+ or Ni2+ but not Zn2+ (Figure 2B, and data not shown).
Figure 2.
Purification and activity of I-TevIII. (A) Purification. Lane 1, size markers; lane 2, crude extract; lane 3, soluble lysate; lane 4, 0.2% PEI supernatant; lane 5, 45% saturated (NH4)2SO4; lanes 6–8, NaCl gradient peak. (B) Cleavage assay. For each metal assayed, two amounts of protein were utilized, 0.2 µg or 0.4 µg. Added metals were at a concentration of 0.1 mM. Cleavage products are marked with a P and substrate is marked with an S. Cleavage in the presence of NiSO4 is not shown. The spurious band in lanes 2 and 5 are of unknown origin.
Purified enzyme has a specific activity of ∼2 U/µg, where one unit equals the amount required to cleave 250 ng of substrate to 50% completion in 1 min at 37°C. This value is three to four orders of magnitude lower than that of I-TevI, the T4 phage GIY-YIG enzyme (26), perhaps accounting for lower levels of homing in vivo (Figure 1B). Similarly, we found that I-TevIII has relatively low affinity for its DNA substrate. We estimate the Kd to be approximately 0.8 µm (data not shown), compared to that of I-TevI, which is approximately 0.9 nm (27). In addition, the activity is extremely labile, regardless of the temperature under which I-TevIII is stored. Despite many attempts to find conditions under which the enzyme would retain activity, I-TevIII activity was lost within 4 days after the cells were lysed and the protein was purified. His-tagged derivatives, which were created to facilitate purification, appeared to have equivalent cleavage activity and dimerization properties (discussed below) to native I-TevIII, and were used for many experiments. Although we found that the enzyme is active on circular or ScaI-linearized plasmid substrates, pSUT4nrdBΔIn, most of our experiments utilized PCR fragments of up to 550 bp.
Interaction of I-TevIII with the homing site
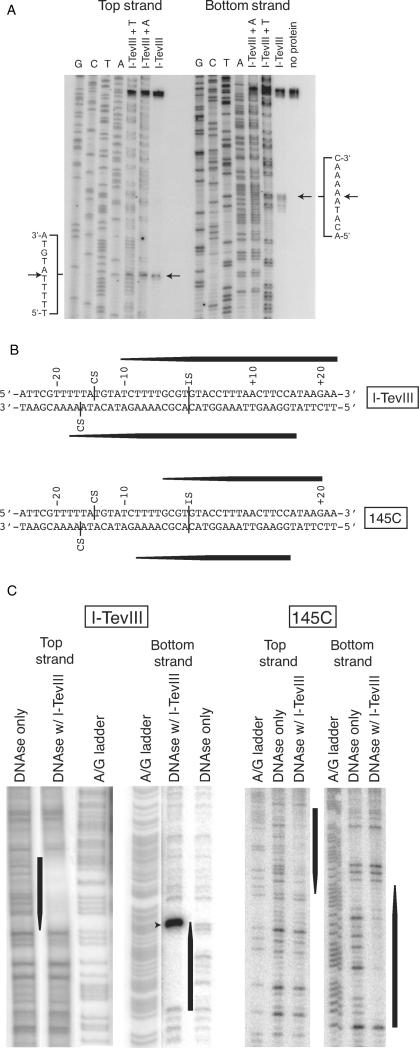
Previous experiments to characterize the cleavage activity of I-TevIII had utilized a primer extension analysis of cleavage products generated in vivo from I-TevIII expressed from a plasmid, acting on nrdBIn− T4 phage (7). These data indicated that the enzyme cuts 14 and 16 bases upstream of the insertion site, generating a 2-nt 5′ overhang. Because such ends would be unique among all known homing endonucleases, and because in vivo cleavage-site mapping is unreliable, we carried out mapping experiments in vitro. Using purified His-tagged protein and a PCR-generated fragment as substrate, repeated experiments have shown that the enzyme cleaves 14 and 16 bp upstream of the insertion site, generating 2-nt 3′ extensions (Figure 3A and B). I-TevIII is therefore consistent with other homing endonucleases in generating 3′ extensions.
Figure 3.
Characterization of the I-TevIII homing site. (A) Cleavage-site mapping for I-TevIII. Sequencing ladders were separated alongside cleavage reactions containing protein as indicated. Cleavage sites, mapped four times, are indicated by arrows and in panel B. Secondary cleavage bands on the bottom strand likely represent exonucleolytic degradation due to the large amounts of enzyme required for cleavage. (B) Schematic of homing site contains a combination of data from the cleavage-site mapping and DNaseI footprinting experiments in panels A and C, respectively. Black tapered bars correspond to footprints in panel C. (C) Interactions of I-TevIII and 145C with the homing site. DNaseI protection assays of the top and bottom strands of the homing site. A + G Maxam–Gilbert ladders were separated alongside protection reactions with homing-site DNA sequence. The black bars represent consistent protection, whereas the tapered ends represent either concentration-dependent protection, or uninterpretable protection because of the absence of DNaseI-sensitive sites. The dominant band in the left panel (marked by an arrowhead) is the cleavage product.
DNaseI footprinting experiments show that the enzyme binds an extensive segment of DNA (Figure 3B and C, left), spanning both the insertion and cleavage sites, in a manner similar to the T4 phage endonucleases I-TevI and I-TevII, which also cleave their substrate at sites distant from the insertion site (24,28). Furthermore, like these enzymes, the I-TevIII footprint shows a 3′ stagger, suggestive of minor groove interactions (29–31). The extended nature of the protein–DNA interaction is also seen in the structure of I-HmuI, the SPO1 H–N–H endonuclease that nicks its substrate (12).
Domain analysis of I-TevIII
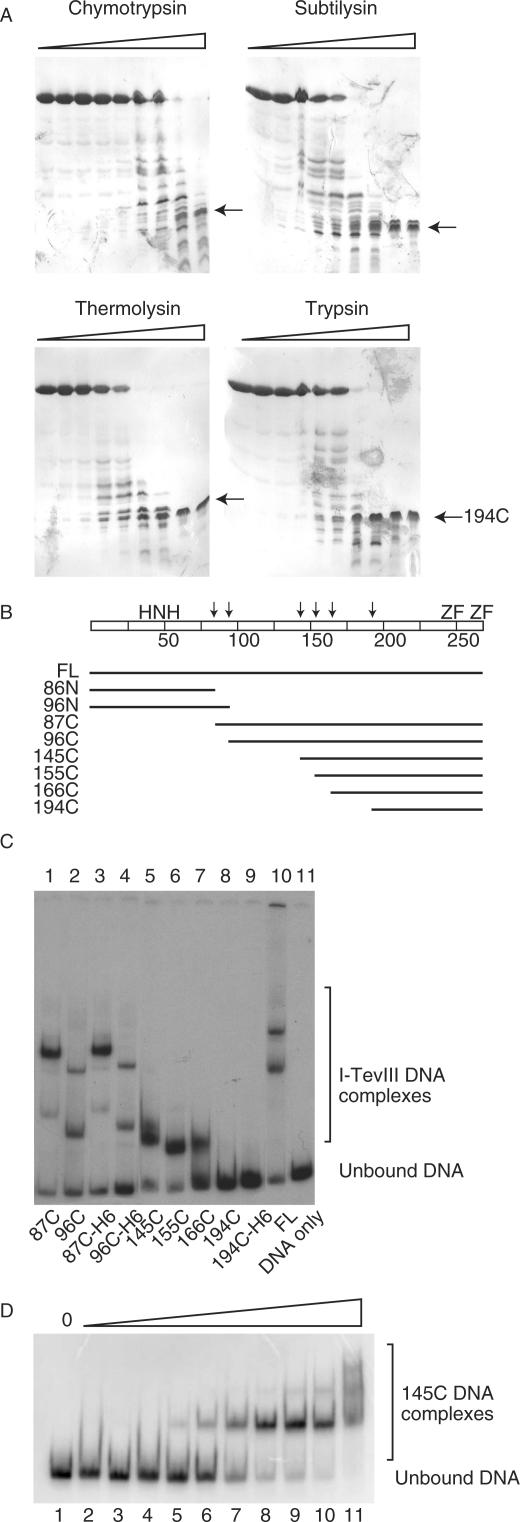
Amino acid alignments indicate that the canonical H–N–H motif for I-TevIII is at the immediate N-terminus of the protein, within the first 50 residues. However, the lack of additional sequence conservation between I-TevIII and other endonucleases did not allow us to localize other functional domains of the protein. To identify distinct structural and functional domains, we used limited proteolysis, as utilized previously for I-TevI (14).
Results of limited proteolysis with chymotrypsin, subtilysin, thermolysin and trypsin are shown in Figure 4A. In each case, the protein was digested to a series of intermediate-sized fragments down to a stable fragment of approximately 7 kDa, which remained even after all full-length protein had been degraded by the protease (arrows in Figure 4A). Trypsin digest fragments were analyzed by mass spectroscopy, and the sites of protease cleavage were identified (Figure 4B). The stable limit digest product was shown to consist of residues 194–269.
Figure 4.
I-TevIII analysis by limited proteolysis. (A) Representative SDS-PAGE gels of I-TevIII digested with proteases chymotrypsin, subtilysin, thermolysin and trypsin. The arrows indicate a proteolytically stable fragment of ∼7 kDa. (B) Schematic of protease-sensitive sites and derivatives constructed based on those sites. FL = full length. The arrows indicate proteolytically sensitive sites, and the scale of the bar is calibrated in 25-amino-acid intervals. The H–N–H motif and zinc fingers (ZF) are marked. (C) DNA-binding activity of I-TevIII and deletion derivatives. Samples were separated on native 8% polyacrylamide gels. (D) DNA-binding activity of 145C. There was a two-fold increase in the amount of protein per lane, starting with 0.001 µg and ending with 0.512 µg.
We generated a series of plasmids to overexpress deletion derivatives based on the proteolytically sensitive sites. The domains were expressed, and DNA-binding activity was assessed (Figure 4C). At least two shifted bands can be observed with the full-length protein and labeled homing site substrate (lane 10). This is also the case for 87C and 96C, which truncate the first 86 and 95 amino acids, respectively (lanes 1–4). Single-shifted species can be observed in 145C, 155C and 166C (Figure 4C, lanes 6–8), but not in 194C (lane 9), suggesting that 166C is the minimal DNA-binding domain from the constructs tested. The amino-terminal 86N and 96N constructs could not be overexpressed (see the Discussion section). We offer two possibilities to explain the multiple protein:DNA complexes. The first is that the enzyme demonstrates cooperative behavior. In titration experiments of the deletion derivative 145C, we observe multiple complexes that behave in a ladder-like manner, which is typical of cooperative binding (Figure 4D, lanes 5–11). A second possibility is in vivo degradation from the N-terminus, as was observed with both I-TevI (14) and I-BmoI (32), leaving the DNA-binding module of the protein to generate a quicker-migrating species in the gel.
A DNaseI footprint was also developed for the 145C DNA-binding domain (Figure 3B and C, right). Although the footprint did not extend as far towards the cleavage site as that for the full-length I-TevIII, the extent of the downstream binding of the two proteins was similar. Furthermore, the footprint again showed a 3′ stagger at both ends.
Mutational analysis of conserved residues in the catalytic and DNA-binding domain
Alanine substitutions were introduced into putative catalytic residues of the H–N–H motif (Figure 5A), which were identified by various alignment programs (33,34). Alanine substitutions H26A, H27A, N39A and H48A reduced cleavage activity below a detectable limit (Figure 5B, below DNA substrate). The mutants were still able to bind the homing-site DNA in gel shift assays (Figure 5B, above DNA substrate), thereby fulfilling the criteria of catalytic mutants. In contrast, interruptions to either or both of the putative zinc fingers of I-TevIII (Figure 5A), in mutants C211A and C214A; C242A and C245A; and C211A, C214A, C242A and C245A, diminished both binding and cleavage activity of the mutated protein relative to the wild type (Figure 5C).
Figure 5.
Cleavage and DNA-binding assays of alanine-substitution mutants. (A) Schematic representation of I-TevIII indicating mutated amino acids. The numbers represent amino acid positions that were changed to alanine. ZF = zinc finger with the consensus sequence. (B) H–N–H mutants. For each mutant assayed, two levels of protein were utilized, 0.2 µg or 0.4 µg. Cleavage and DNA-binding assays are shown. Samples were separated on native 8% polyacrylamide gels. Cleavage products are marked with a P, and unbound DNA with an S. (C) Zinc finger mutants. Analysis and labeling as in panel A. Asterisks represent protease-treated samples.
Binding of zinc by I-TevIII
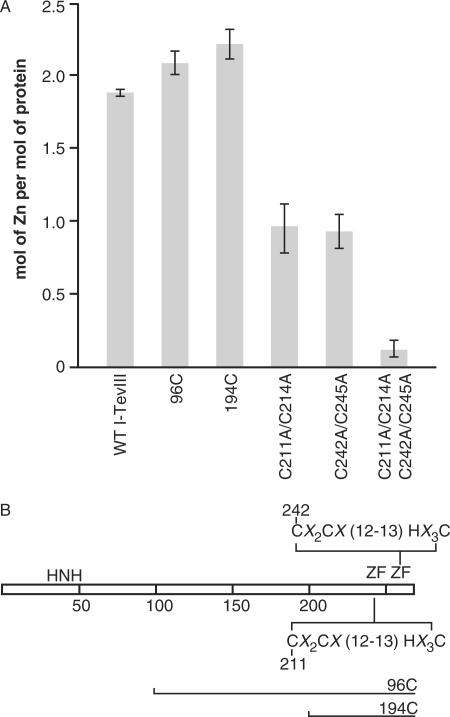
I-TevIII was investigated for the presence of zinc. Individual protein samples were incubated with PAR for 5 min to determine background levels of zinc by monitoring absorbance at 500 nm. MMTS was then added to release the bound zinc from the proteins, which resulted in an increase in absorbance at 500 nm. Absorbance values were then correlated with the standard curve to determine the amount of zinc coordinated per mole of I-TevIII, calculated in monomeric form. The amount of zinc released from the full-length I-TevIII protein indicates that ∼88% of the two zinc fingers are occupied with a zinc ion (Figure 6A). Due to the possibility that the H–N–H domain can sequester zinc and, furthermore, to demonstrate that the two putative zinc fingers contain the zinc, mutants were assessed for zinc coordination. The 96C deletion derivative (Figures 4B and 6B) has no H–N–H motif, but this deletion derivative has ∼2.0 moles of zinc per mole of protein, indicating that each zinc finger is occupied for this construct (Figure 6A). This is also the case for the 194C derivative (Figures 4B, 6A and B), indicating that the zinc finger CX2CX12-13HX3C motifs are likely coordinating the ion. To further validate the role these residues have in organizing the zinc ion, mutants C211A and C214A; C242A and C245A; and C211A, C214A, C242A, and C245A were assessed by the PAR analysis. Disruption of the first zinc finger (C211A and C214A) reduced the number of zinc moles bound to approximately one-half of the wild-type I-TevIII, and this was also the case for the second zinc finger (C242A and C245A) (Figure 6A and B). Interruption of both the zinc fingers (C211A, C214A, C242A and C245A) reduced the amount of zinc bound by the enzyme to near zero.
Figure 6.
(A) Zinc binding in wild-type and mutant derivatives of I-TevIII. The presence of zinc was determined by incubating 5 nmol of each protein with PAR, and the amount of zinc released in the presence of MMTS was measured as the increase in absorbance at 500 nm. The data represent the means ± standard errors of three independent measurements. (B) Schematic of the I-TevIII proteins used for the PAR analysis. The consensus sequence for the two tandem zinc fingers (ZF) is shown, with horizontal lines indicating the extent of proteins present in the N-terminal deletions 96C and 194C.
I-TevIII is a dimer in the presence and absence of its substrate
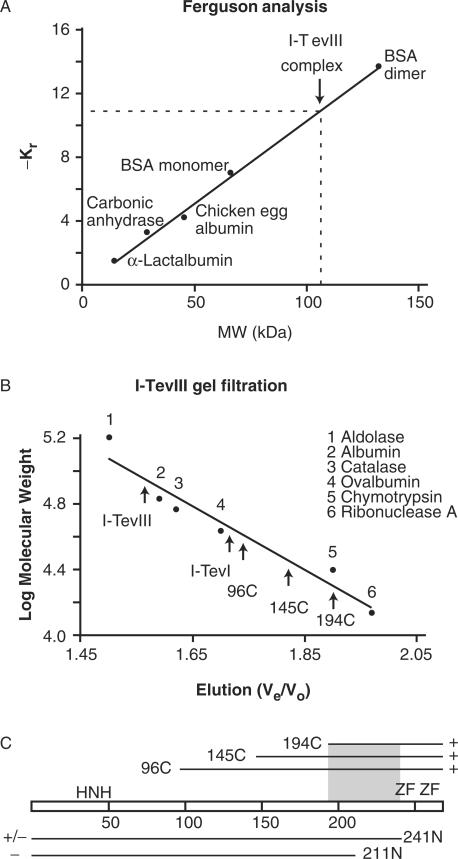
Ferguson analysis (35) was undertaken to assess the protein's ability to dimerize in the presence of the homing site. This technique, which involves separation of protein–DNA complexes alongside size standards, on polyacrylamide gels of varying concentration (see the Materials and Methods section), has been utilized in our laboratory to determine the monomeric binding of I-TevI to DNA (25). A series of native protein standards was run in parallel with I-TevIII (Figure 7A). A 100-bp duplex (molecular mass of 61.6 kDa) generated with primers W1622 and W1623 (Supplementary Table 1) and end-labeled with [γ-32P]ATP was utilized in this assay. I-TevIII in complex with the homing site has an estimated mass of 114.3 kDa. Subtracting the weight of the DNA from that of the protein–DNA complex results in an estimate of the protein component of 52.7 kDa. With a monomer mass of 30.8 kDa, this translates into ∼1.7 molecules of I-TevIII per molecule of DNA substrate. Therefore, it is most likely that I-TevIII binds its homing site as a dimer. In the case of I-TevI, which binds its substrate as a monomer (18), 0.96 molecules of protein were observed when complexed with its respective homing site substrate (25).
Figure 7.
I-TevIII stoichiometry. (A) Ferguson analysis. The retardation coefficient (Kr) values for protein standards were derived by a Ferguson analysis and plotted as a function of molecular weight (MW). Protein standards are indicated. Interpolation of -Kr for I-TevIII-DNA complex (dashed lines) indicate a molecular weight of ∼106 kDa. (B) Gel filtration analysis of I-TevIII. Elution of purified I-TevIII, deletion derivatives and I-TevI are shown relative to molecular weight standards (numbered 1–6, as indicated). Ve/Vo, elution volume divided by void volume. (C) Ability of deletion constructs to dimerize. The amino-terminal deletion constructs (96C, 145C, 194C) formed dimers (+), whereas the C-terminal construct 241N partially dimerized (+/−) and construct 211N did not dimerize (−). The shaded area represents the dimerization region.
To complement the Ferguson studies, I-TevIII was subjected to gel filtration analysis, to assess quaternary organization in the absence of substrate. To estimate the size of I-TevIII in solution, markers of known size were used to calibrate a Superose® 12 HR FPLC column (Figure 7B). Fractions were examined on SDS-PAGE from the beginning to the end of the peak to verify the protein elution position. Single-peak elutions were observed at 280 nm for multiple replicates of I-TevIII samples, indicating that I-TevIII elutes with a size of 69.8 ± 2.0 kDa (Figure 7B). The predicted molecular mass from the amino acid sequence is 30.8 kDa, suggesting that I-TevIII forms a dimer in solution under the conditions tested. For comparative purposes, the 28.2-kDa I-TevI, which is known to bind as a monomer (18,25), was also subjected to gel filtration analysis, and this protein eluted with an estimated mass of 38.8 kDa (Figure 7B). These results are consistent with I-TevI existing as a monomer in solution under the conditions tested. Deviations from theoretical numbers for both I-TevI and I-TevIII are thought to result from the extended nature of these types of proteins (12,18) relative to the globular standards.
His-tagged and non-tagged I-TevIII eluted from the Superose® column at a similar volume (data not shown). Therefore, His-tagged I-TevIII was utilized to examine some I-TevIII mutants, because of the facile purification procedure. Each of the zinc-finger mutant proteins was subjected to gel filtration analysis, and their elution volumes corresponded to the following molecular masses: C211A and C214A, 72.1 kDa; C242A and C245A, 73.5 kDa; C211A, C214A, C242A and C245A, 70.4 kDa. Thus, mutating either or both zinc fingers did not alter the oligomerization properties of the protein.
The dimerization properties of a series of deletion derivatives, 96C, 145C, 194C, 211N and 241N (Figure 4B and C) were examined with gel filtration in the same manner as for I-TevIII. Interestingly, the truncated forms of the protein eluted with estimated masses of 43.5 kDa for 96C; 33.3 kDa for 145C; and 18.7 kDa for 194C. Their predicted molecular masses of 19.7, 14.1 and 8.7 kDa, respectively, suggested that these deletion derivatives are also dimers in solution (Figure 7B). Thus, removal of the first 194 amino acids, containing the H–N–H module, did not abolish the ability of the protein to oligomerize, suggesting that the dimerization interface is within the C-terminal ∼75 amino acids of the protein. Consistent with this hypothesis, deletions from the carboxy terminus disrupted dimerization. Construct 241N, in which the C-terminal zinc finger is deleted, eluted in part with an estimated mass of ∼60 kDa, and in part as a broad smaller peak, suggesting that the 27.7 kDa protein forms unstable dimers (data not shown). In contrast, the elution profile of construct 211N, with both zinc fingers removed, was irregular, with multiple peaks, corresponding to neither monomers nor dimers, suggesting an inability to dimerize (Figure 7C).
DISCUSSION
The nrdB intron is mobile
Previous work in our laboratory examined the mobility of the nrdB intron of phage T4 and found that, unlike the other T4 introns, the nrdB intron was not mobile (6). An examination of the relative sizes of the ORFs of the td, nrdD and nrdB introns revealed that the nrdB ORF was approximately one-third the size of the other two intron ORFs. Eddy and Gold's discovery of a longer nrdB intron from phage RB3 has allowed us to demonstrate mobility for the enigmatic nrdB intron, a finding that is contrary to their report (7). Comparing the amino acid sequences for I-TevIII between the phages T4 and RB3 reveals that the N-terminus has been deleted in T4, removing the catalytic H–N–H domain and leaving the putative C-terminal zinc fingers. It is interesting to note that the genomes of phage T2 and T6, close relatives of T4, do not contain an intron interrupting the nrdB genes (36), suggesting the possibility that the eroded nrdB intron of phage T4 is in the process of being lost from the genome.
Biochemical properties of the I-TevIII dimer
The reason for the low catalytic efficiency of I-TevIII as compared to other H–N–H endonucleases is unknown, but is reminiscent of the differences between two closely related GIY-YIG enzymes. Specifically, I-TevI is three orders of magnitude more active than I-BmoI (37). The low specific activity of I-TevIII compared to I-TevI may explain why this homing endonuclease can be overexpressed in vivo in an E. coli system, in contrast to I-TevI (26). However, the free catalytic domain of I-TevIII cannot be expressed in E. coli, as is the case for I-TevI (14). Presumably, free nucleases unconstrained by a site-specific DNA-binding domain are toxic to cells because they cause indiscriminate DNA damage.
Mapping of recombinant I-TevIII cleavage on the homing site revealed that the enzyme cuts 14 and 16 bp upstream of the insertion site, generating 2-nt 3′ overhangs (see Figure 3B for schematic). I-TevIII is therefore consistent with other homing endonucleases in generating 3′ extensions, again in contrast to a previous report (7). Interestingly, the H–N–H endonuclease from Chlamydomonas moewusii, I-CmoeI, which also makes a DSB, creates 4-nt 3′ extensions (38). These variant cleavage patterns suggest that there is a fundamental difference in the way these two enzymes approach their cleavage sites. This difference may well relate to our observation that I-TevIII binds as a dimer, whereas I-CmoeI acts as a monomer (38). In any event, it is noteworthy that a classification of restriction endonuclease structures on the basis of cleavage pattern (i.e. the nature and length of single-strand extensions) has been proposed (39,40). Clearly, homing endonucleases evolve independently of restriction enzymes, and classification of the homing enzymes on the basis of the length of single-strand DNA extensions is not possible. These findings speak to the diversity of members of the H–N–H endonuclease family, a topic elaborated below.
Metal ion preferences of the H–N–H family are variable. I-CmoeI was shown to have optimal activity with the cation Mg2+, whereas substitution with Zn2+ ions abolished DNA cleavage (38). In contrast, crystal structures of the H–N–H domain of colicin E9 contained Ni2+, while the H–N–H colicin E7 enzyme coordinated Zn2+ (41,42). We show here that, like I-CmoeI, I-TevIII cleaves in the presence of Mg2+ or Ni2+, but not Zn2+.
Modularity of I-TevIII and evolution of the H–N–H endonucleases
The modularity of I-TevIII was revealed through the domain analysis studies (Figures 4 and 8). Alanine substitutions to the predicted H–N–H residues and loss of cleavage (but not binding activity) confirmed that these residues are key to catalysis. The biochemical and structural studies of bacillus-phage SPO1 I-HmuI, a member of the H–N–H family of endonucleases, reveals that these residues are involved in the activation of water and divalent metal binding and are essential for hydrolysis of the phosphate backbone (12).
Figure 8.
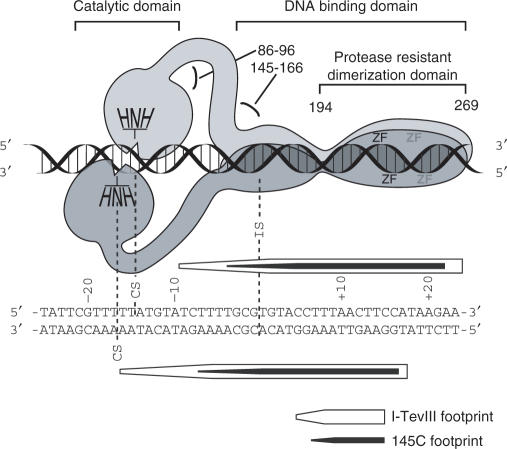
Dimer model for I-TevIII interacting with its cognate homing site. The model is based on domain analysis, footprinting and stoichiometry data. The sequence below the model represents the homing site, aligned with the model, and showing a summary of the footprint data from Fig. 3C. The numbers near the protein represent amino acid residues. The figure is not strictly to scale, nor do we know the relative orientations of the DNA-binding domains.
Interruptions of either zinc finger within I-TevIII reduced the protein's ability to bind or cleave the homing site, implicating a role for the zinc fingers in DNA binding. In contrast, the zinc finger of I-TevI is not involved in DNA binding, but rather it orders the protein's linker region. Mutations to the I-TevI zinc finger still allow DNA binding, but result in faulty distance determination by I-TevI (27). The recruitment of zinc fingers to perform different roles in two homing endonucleases of the same phage family provides an interesting example of how modularity enables functional diversification.
The H–N–H endonucleases comprise a strikingly diverse family of proteins. They exist not only as freestanding enzymes such as bacterial colicins, but are also embedded within introns and inteins, and can function both in free form and in association with intron RNA (10,43,44). Their role in introns is to cleave the DNA target; but, interestingly, many of the enzymes characterized to date nick the DNA, rather than making a DSB. In both the phage SPO1 group I intron and the bacterial group II introns, the single-strand cleavage facilitates a repair event that leads to intron inheritance (44,45). However, I-TevIII, like its two T-even phage counterparts, I-TevI and I-TevII, and like I-CmoeI, makes a DSB to mobilize its intron. Both I-CmoeI, an H–N–H enzyme, and I-TevI, a GIY-YIG enzyme, bind as a monomer, the latter in a similar manner to the H–N–H nicking enzyme I-HmuI (12,18,38). How these monomeric enzymes make a DSB remains an open question. This question has now been answered for I-TevIII, where it is likely that each subunit of the dimer nicks one of the two strands, 14 and 16 bp upstream of the intron insertion site (Figure 8).
Endonucleases that form dimers usually bind to palindromic or at least pseudosymmetric sites. In view of the asymmetry of the homing site, we can only speculate how dimerization achieves a DSB for I-TevIII. Given that the two identical H–N–H active sites likely have the same polarity of phosphodiester bond cleavage of two anti-parallel DNA strands, we have drawn the H–N–H domains in an opposing head-to-head arrangement (Figure 8). The remainder of the model is based on the domain analysis, footprinting and dimerization studies. The protease cleavage sites occurr in the 86- to 96-amino acid region, in the 145- to 166-residue region and at amino acid 194 (Figure 4B). The first sites of proteolysis occur after the catalytic domain, residue 166 defines the minimum DNA-binding domain (Figure 4C) and residue 194 demarcates the start of the protease-resistant regions (Figure 4A). The point on the homing site at which the DNA-binding domain extends into a flexible region, to allow the appropriate orientation of the H–N–H module, corresponds to the end of the footprint of the N-terminal deletion 145C. The observation that the footprint does not extend to the cleavage site on the top strand implies a transience of interactions at the catalytic site, as is the case for I-TevI (24,29). Furthermore, as for I-TevI and I-TevII, the 3′ staggers in the footprints are indicative of minor groove interactions, likely reflecting the presence of modifications to DNA in the major groove of T-even phage DNA (46).
Our analysis of the N- and C-terminal deletions suggests that the dimerization interface is between residues 194 and 241 (Figure 7C), at the start of the zinc fingers, which are not directly involved in dimerization. Regardless, it appears that the C-terminal end of this modular protein has a determinant for dimerization that is reflected in the cleavage pattern mediated by the opposite N-terminal end of I-TevIII.
It is apparent that the modularity built into the GIY-YIG and H–N–H endonucleases allows them to evolve specificity by shuffling conserved catalytic domains with variant DNA-binding domains, as well as facilitating functional diversity, as exemplified by alternative exploitation of zinc fingers. It would now appear that the stoichiometry of an H–N–H enzyme can also evolve to meet the needs for DNA cleavage, to promote gene conversion in particular circumstances. Given that phage T4 uses DSBs as a means to initiate recombination and replication (46), it is tempting to speculate that the DSB created by I-TevIII, as that of I-TevI and I-TevII, is integrated into the T-even phage life cycle to maximize intron inheritance. One might then hypothesize that the nicks introduced by I-HmuI are similarly consistent with the mode of replication of bacillus phage, reported to proceed by discontinuous DNA synthesis of both strands (47). Thus, the combination of modularity and quaternary structure would allow these different enzymes to evolve features that meet the requirements for the spread of the mobile intron in the host that it invades.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by NIH grant numbers GM44844 and GM39422 to M.B. Plasmid pSURB3nrdB and intronless T4 phage were kindly provided by D.A. Shub, University at Albany (Albany, NY). Phage RB3 was graciously provided by S. Eddy and L. Gold, Washington University (St. Louis, MO) and University of Colorado (Boulder, CO), respectively. The authors wish to thank two anonymous reviewers for their insightful contributions to the model in Figure 8, and Maryellen Carl for expert manuscript preparation. We thank D.A. Shub and members of the Belfort laboratory for useful scientific discussions. Funding to pay the Open Access publication charge was provided by grants cited in acknowledgement and by Institutional Membership.
Conflict of interest statement. None declared.
REFERENCES
- 1.Belfort M, Roberts RJ. Homing endonucleases: keeping the house in order. Nucleic Acids Res. 1997;25:3379–3388. doi: 10.1093/nar/25.17.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chevalier BS, Stoddard BL. Homing endonucleases: structural and functional insight into the catalysis of intron/intein mobility. Nucleic Acid Res. 2001;29:3757–3774. doi: 10.1093/nar/29.18.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Darnell JE, Doolittle WF. Speculations on the early course of evolution. Proc. Natl. Acad. Sci. USA. 1986;83:1271–1275. doi: 10.1073/pnas.83.5.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ochman H, Davalos LM. The nature and dynamics of bacterial genomes. Science. 2006;311:1730–1733. doi: 10.1126/science.1119966. [DOI] [PubMed] [Google Scholar]
- 5.Belfort M. Phage T4 introns: self-splicing and mobility. Annu. Rev. Genet. 1990;24:363–385. doi: 10.1146/annurev.ge.24.120190.002051. [DOI] [PubMed] [Google Scholar]
- 6.Quirk SM, Bell-Pedersen D, Belfort M. Intron mobility in the T-Even phages: high frequency inheritance of group I introns promoted by intron open reading frames. Cell. 1989;56:455–465. doi: 10.1016/0092-8674(89)90248-1. [DOI] [PubMed] [Google Scholar]
- 7.Eddy SR, Gold L. The phage T4 nrdB intron: a deletion mutant of a version found in the wild. Genes Dev. 1991;5:1032–1041. doi: 10.1101/gad.5.6.1032. [DOI] [PubMed] [Google Scholar]
- 8.Stoddard BL. Homing endonuclease structure and function. Q. Rev. Biophys. 2005;38:49–95. doi: 10.1017/S0033583505004063. [DOI] [PubMed] [Google Scholar]
- 9.Kleanthous C, Kuhlmann UC, Pommer AJ, Ferguson N, Radford SE, Moore GR, James R, Hemmings AM. Structural and mechanistic basis of immunity toward endonuclease colicins. Nature Struct. Biol. 1999;6:243–252. doi: 10.1038/6683. [DOI] [PubMed] [Google Scholar]
- 10.Keeble AH, Mate MJ, Kleanthous C. In: Homing Endonucleases and Inteins. Belfort M, Derbyshire V, Stoddard BL, Wood DW, editors. Springer-Verlag; 2005. pp. 49–65. [Google Scholar]
- 11.Doudeva LG, Huang H, Hsia KC, Shi Z, Li CL, Shen Y, Cheng YS, Yuan HS. Crystal structural analysis and metal-dependent stability and activity studies of the ColE7 endonuclease domain in complex with DNA/Zn2+ or inhibitor/Ni2+ Protein Sci. 2006;15:269–280. doi: 10.1110/ps.051903406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen BW, Landthaler M, Shub DA, Stoddard BL. DNA binding and cleavage by the HNH homing endonuclease I-HmuI. J. Mol. Biol. 2004;342:43–56. doi: 10.1016/j.jmb.2004.07.032. [DOI] [PubMed] [Google Scholar]
- 13.Landthaler M, Shub DA. The nicking homing endonuclease I-BasI is encoded by a group I intron in the DNA polymerase gene of the Bacillus thuringiensis phage Bastille. Nucleic Acids Res. 2003;31:3071–3077. doi: 10.1093/nar/gkg433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Derbyshire V, Kowalski JC, Dansereau JT, Hauer CR, Belfort M. Two-domain structure of the td intron-encoded endonuclease I-TevI correlates with the two-domain configuration of the homing site. J. Mol. Biol. 1997;265:494–506. doi: 10.1006/jmbi.1996.0754. [DOI] [PubMed] [Google Scholar]
- 15.Van Roey P, Derbyshire V. In: Homing Endonucleases and Inteins. Belfort M, Derbyshire V, Stoddard BL, Wood DW, editors. Springer-Verlag; 2005. pp. 67–83. [Google Scholar]
- 16.Stauffer ME, Chazin WJ. Structural mechanisms of DNA replication, repair, and recombination. J. Biol. Chem. 2004;279:30915–30918. doi: 10.1074/jbc.R400015200. [DOI] [PubMed] [Google Scholar]
- 17.Bakhrat A, Jurica MS, Stoddard BL, Raveh D. Homology modeling and mutational analysis of Ho endonuclease of yeast. Genetics. 2004;166:721–728. doi: 10.1534/genetics.166.2.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Roey P, Waddling CA, Fox KM, Belfort M, Derbyshire V. Intertwined structure of the DNA-binding domain of intron endonuclease I-TevI with its substrate. EMBO J. 2001;20:3631–3637. doi: 10.1093/emboj/20.14.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flick KE, Jurica MS, Monnat RJ, Jr, Stoddard BL. DNA binding and cleavage by the nuclear intron-encoded homing endonuclease I-PpoI. Nature. 1998;394:96–101. doi: 10.1038/27952. [DOI] [PubMed] [Google Scholar]
- 20.Grishin NV. Treble clef finger—a functionally diverse zinc-binding structural motif. Nucleic Acids Res. 2001;29:1703–1714. doi: 10.1093/nar/29.8.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krishna SS, Majumdar I, Grishin NV. Structural classification of zinc fingers: survey and summary. Nucleic Acids Res. 2003;31:532–550. doi: 10.1093/nar/gkg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. In: Methods in Enzymology. Goeddel DV, editor. Vol. 185. New York: Academic Press; 1990. pp. 60–89. [DOI] [PubMed] [Google Scholar]
- 23.Edgell DR, Stanger MJ, Belfort M. Coincidence of cleavage sites of intron endonuclease I-TevI and critical sequences of the host thymidylate synthase gene. J. Mol. Biol. 2004;343:1231–1241. doi: 10.1016/j.jmb.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 24.Bell-Pedersen D, Quirk SM, Bryk M, Belfort M. I-TevI, the endonuclease encoded by the mobile td intron, recognizes binding and cleavage domains on its DNA target. Proc. Natl. Acad. Sci. USA. 1991;88:7719–7723. doi: 10.1073/pnas.88.17.7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mueller JE, Smith D, Bryk M, Belfort M. Intron-encoded endonuclease I-TevI binds as a monomer to effect sequential cleavage via conformational changes in the td homing site. EMBO J. 1995;14:5724–5735. doi: 10.1002/j.1460-2075.1995.tb00259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu W, Wood DW, Belfort G, Derbyshire V, Belfort M. Intein-mediated purification of cytotoxic endonuclease I-TevI by insertional inactivation and pH-controllable splicing. Nucleic Acids Res. 2002;30:4864–4871. doi: 10.1093/nar/gkf621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dean AB, Stanger MJ, Dansereau JT, Van Roey P, Derbyshire V, Belfort M. Zinc finger as distance determinant in the flexible linker of intron endonuclease I-TevI. Proc. Natl. Acad. Sci. USA. 2002;99:8554–8561. doi: 10.1073/pnas.082253699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bell-Pedersen D, Quirk S, Clyman J, Belfort M. Intron mobility in phage T4 is dependent upon a distinctive class of endonucleases and independent of DNA sequences encoding the intron core: mechanistic and evolutionary implications. Nucleic Acids Res. 1990;18:3763–3770. doi: 10.1093/nar/18.13.3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bryk M, Quirk SM, Mueller JE, Loizos N, Lawrence C, Belfort M. The td intron endonuclease I-TevI makes extensive sequence-tolerant contacts across the minor groove of its DNA target. EMBO J. 1993;12:2141–2149. doi: 10.1002/j.1460-2075.1993.tb05862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loizos N, Silva GH, Belfort M. The intron-encoded endonuclease I-TevII binds across the minor groove and induces two distinct conformational changes in its DNA substrate. J. Mol. Biol. 1996;255:412–424. doi: 10.1006/jmbi.1996.0034. [DOI] [PubMed] [Google Scholar]
- 31.Oakley MG, Dervan PB. Structural motif of the GCN4 DNA binding domain characterized by affinity cleaving. Science. 1990;248:847–850. doi: 10.1126/science.2111578. [DOI] [PubMed] [Google Scholar]
- 32.Edgell DR, Shub DA. Related homing endonucleases I-BmoI and I-TevI use different strategies to cleave homologous recognition sites. Proc. Natl. Acad. Sci. USA. 2001;98:7898–7903. doi: 10.1073/pnas.141222498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shub DA, Goodrich-Blair H, Eddy SR. Amino acid sequence motif of group I intron endonucleases is conserved in open reading frames of group II introns. Trends Biochem. Sci. 1994;19:402–404. doi: 10.1016/0968-0004(94)90086-8. [DOI] [PubMed] [Google Scholar]
- 34.Gorbalenya AE. Self-splicing group I and group II introns encode homologous (putative) DNA endonucleases of a new family. Protein Sci. 1994;3:1117–1120. doi: 10.1002/pro.5560030716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferguson KA. Starch electrophoresis - application to the classification of pituitary proteins and polypeptides. Metabolism. 1964;13:985–1002. doi: 10.1016/s0026-0495(64)80018-4. [DOI] [PubMed] [Google Scholar]
- 36.Pedersen-Lane J, Belfort M. Variable occurrences of the nrdB intron in T-even phages suggests intron mobility. Science. 1987;237:182–184. doi: 10.1126/science.3037701. [DOI] [PubMed] [Google Scholar]
- 37.Edgell DR, Stanger MJ, Belfort M. Importance of a single base pair for discrimination between intron-containing and intronless alleles by endonuclease I-BmoI. Curr. Biol. 2003;13:973–978. doi: 10.1016/s0960-9822(03)00340-3. [DOI] [PubMed] [Google Scholar]
- 38.Drouin M, Lucas P, Otis C, Lemieux C, Turmel M. Biochemical characterization of I-CmoeI reveals that this H–N–H homing endonuclease shares functional similarities with H–N–H colicins. Nucleic Acids Res. 2000;28:4566–4572. doi: 10.1093/nar/28.22.4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Athanasiadis A, Vlassi M, Kotsifaki D, Tucker PA, Wilson KS, Kokkinidis M. Crystal structure of PvuII endonuclease reveals extensive structural homologies to EcoRV: partial folding and unfolding on DNA binding. Struct.Biol. 1994;1:469–475. doi: 10.1038/nsb0794-469. [DOI] [PubMed] [Google Scholar]
- 40.Newman M, Strzelecka T, Dorner LF, Schildkraut I, Aggarwal AK. Structure of restriction endonuclease BamHI and its relationship to EcoRI. Nature. 1994;368:660–664. doi: 10.1038/368660a0. [DOI] [PubMed] [Google Scholar]
- 41.Ku W-Y, Liu Y-W, Hsu Y-C, Liao C-C, Liang P-H, Yuan HS, Chak K-F. The zinc ion in the N–H–N motif of the endonuclease domain of colicin E7 is not required for DNA binding but is essential for DNA hydrolysis. Nucleic Acids Res. 2002;30:1670–1678. doi: 10.1093/nar/30.7.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pommer AJ, Kuhlmann UC, Cooper A, Hemmings AM, Moore GR, James R, Kleanthous C. Homing in on the role of transition metals in the HNH motif of colicin endonucleases. J. Biol. Chem. 1999;274:27153–27160. doi: 10.1074/jbc.274.38.27153. [DOI] [PubMed] [Google Scholar]
- 43.Pietrokovski S. Modular organization of inteins and C-terminal autocatalytic domains. Protein Sci. 1998;7:64–71. doi: 10.1002/pro.5560070106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsuura M, Saldanha R, Ma H, Wank H, Yang J, Mohr G, Cavanagh S, Dunny GM, Belfort M, et al. A bacterial group II intron encoding reverse transcriptase, maturase, and DNA endonuclease activities: biochemical demonstration of maturase activity and insertion of new genetic information within the intron. Genes Dev. 1997;11:2910–2924. doi: 10.1101/gad.11.21.2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goodrich-Blair H, Shub DA. Beyond homing: competition between intron endonucleases confers a selective advantage on flanking genetic markers. Cell. 1996;84:211–221. doi: 10.1016/s0092-8674(00)80976-9. [DOI] [PubMed] [Google Scholar]
- 46.Mosig G. Recombination and recombination-dependent DNA replication in bacteriophage T4. Annu. Rev.Genet. 1998;32:379–413. doi: 10.1146/annurev.genet.32.1.379. [DOI] [PubMed] [Google Scholar]
- 47.Stewart C. In: The Bacteriophages. Calendar R, editor. Vol. 1. New York and London: Plenum Press; 1988. pp. 477–515. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.