Abstract
The synthesis of oligonucleotides containing 2′-deoxy-2′-fluoro-4′-thioarabinonucleotides is described. 2′-Deoxy-2′-fluoro-5-methyl-4′-thioarabinouridine (4′S-FMAU) was incorporated into 18-mer antisense oligonucleotides (AONs). 4′S-FMAU adopts a predominantly northern sugar conformation. Oligonucleotides containing 4′S-FMAU, unlike those containing FMAU, were unable to elicit E. coli or human RNase H activity, thus corroborating the hypothesis that RNase H prefers duplexes containing oligonucleotides that can adopt eastern conformations in the antisense strand. The duplex structure and stability of these oligonucleotides was also investigated via circular dichroism (CD)- and UV- binding studies. Replacement of the 4′-oxygen by a sulfur atom resulted in a marked decrease in melting temperature of AON:RNA as well as AON:DNA duplexes. 2′-Deoxy-2′-fluoro-4′-thioarabinouridine (4′S-FAU) was incorporated into 21-mer small interfering RNA (siRNA) and the resulting siRNA molecules were able to trigger RNA interference with good efficiency. Positional effects were explored, and synergy with 2′F-ANA, which has been previously established as a functional siRNA modification, was demonstrated.
INTRODUCTION
Gene silencing via the introduction of an antisense oligonucleotide (AON) or small interfering RNA (siRNA) into an organism is an attractive and elegant means of selectively blocking the expression of a deleterious gene (1–6). The successful development of AON and siRNA as therapeutics will undoubtedly require modification of the oligonucleotide sugar-phosphate backbone to enhance delivery, stability, efficacy and specificity. In fact, progress toward routine use in the clinic, especially for AON therapy, has been slow, partly due to limitations in the currently available chemical modifications (7).
Particularly important for antisense therapeutics are modified AONs that trigger RNase H activity when bound to an mRNA, since this leads to the specific and permanent destruction of the mRNA target (8,9). All known AONs with the ability to elicit RNase H activity (10–12) appear to share the same A–B type duplex conformation and minor groove width dimensions that are believed to be key to maintaining RNase H activity (13,14). In the case of 2′-deoxy-2′-fluoroarabinonucleic acids (2′F-ANAs), RNase H cleavage efficiency can be significantly enhanced by incorporating flexible 2′-deoxyribonucleotides, aliphatic linkers or seconucleosides within the sequence (15,16). This increased efficiency of cleavage appears to be related to the greater flexibility of the AON strand (16). 2′F-ANA–DNA chimeras are more efficient at eliciting RNase H cleavage than native DNA (17).
Much recent work has focused on the chemical modification of siRNA (18). Selective modifications within the duplex can often provide the necessary increases in nuclease stability with little loss of activity. However, there is much work to be done as currently known modifications are often very sequence- or cell-type dependent.
As part of our ongoing program to probe the substrate specificity of RNase H and the RISC complex, we chose to replace the 4′ (ring) oxygen of 2′F-ANA with a sulfur atom (19) to give 2′-deoxy-2′-fluoro-4′-thioarabinonucleic acids (4′S-FANAs). 4′-Thiolation of RNA and DNA has been shown to enhance thermal stability and exo- and endonuclease resistance (20–23). 4′-Thio-RNA is accepted by the RNAi machinery (24). The 4′-thiolation of DNA (25) or 2′F-ANA (19) induces a conformational switch to the north (RNA-like), and we would thus expect that duplexes containing 4′S-FANA would adopt an A-form helix and also be accepted by the RNAi machinery (26). Furthermore, evaluation of hybrids containing 4′S-FANA would help elucidate the structural factors that provide the optimal AON–RNA substrate for RNase H. While 2′-deoxy-2′-fluoro-4′-thioarabinonucleosides have previously been synthesized (19,27–30), they have not, to the best of our knowledge, been incorporated into oligonucleotides.
MATERIALS AND METHODS
2′-Deoxy-2′-fluoro-4′-thio-β -d-arabinouridine (1b)
To compound 4 (173 mg, 0.37 mmol) was added a 2 M solution of ammonia in cold methanol (30 ml, 60 mmol). The reaction mixture was capped with a rubber septum and allowed to stir for 48 h. It was then evaporated to dryness, adsorbed onto silica and loaded onto a short column of neutralized silica gel. Dichloromethane containing 0–5% methanol was used to elute compound 1b as a solid (92 mg, 95%). 1H NMR (400 or 500 MHz, methanol-d4): δ 8.30 (dd, 1H, JH6-F2′ = 1.6 Hz, JH6-H5 = 8.4 Hz, H6), 6.41 (dd, 1H, JH1′-H2′ = 5.6 Hz, JH1′-F2′ = 11.6 Hz, H1′), 5.71 (d, 1H, JH6-H5 = 8.4 Hz, H5), 5.00 (ddd, 1H, JH1′-H2′ = 5.6 Hz, JH2′-F2′ = 51.0 Hz, JH2′-H3′ = 5.7 Hz, H2′), 4.36 (ddd, 1H, JH3′-H4′ = 5.8 Hz, JH3′-F2′ = 11.6 Hz, JH2′-H3′ = 5.7 Hz, H3′), 3.82 (m, 2H, H5′, H5″), 3.33 (m, 1H, H4′). 13C NMR (125 MHz, methanol-d4): δ 164.75, 151.55 (C2, C4), 143.31 (d, JF2′-C6 = 2.3 Hz, C6), 101.07 (C5), 96.27 (d, JF2′-C2′ = 193.8 Hz, C2′), 73.55 (d, JF2′-C3′ = 23.6 Hz, C3′), 61.34 (d, JF2′-C5′ = 2.4 Hz, C5′), 58.93 (d, JF2-C1′ = 16.8 Hz, C1′), 51.88 (d, JF2′-C4′ = 3.8 Hz, C4′). Two pairs of NOESY crosspeaks (H6-H3′, H6-H5′) provided strong evidence for top-face uracil and therefore the β nucleoside. FAB-HRMS: calcd for C9H11N2O4SF + H+: 263.0502; found: 263.0501.
1-(2-Deoxy-2-fluoro-5-O-(4,4′-dimethoxytrityl)-4-thio-β-d-arabinofuranosyl)-thymine (2a)
2′-Deoxy-2′-fluoro-4′-thio-β-d-arabinothymidine (1a, 105 mg, 0.40 mmol) was coevaporated three times with pyridine. Dry pyridine (10 ml) was added, followed by 95% dimethoxytrityl (DMT) chloride (198 mg, 0.56 mmol). Half of the solvent was removed, heating the flask slightly on a rotary evaporator. The reaction was allowed to stir for 44 h when TLC indicated virtual completion of the reaction. It was then diluted with dichloromethane (50 ml) and washed with saturated aqueous NaHCO3 (2 × 50 ml); the aqueous layers were then washed with dichloromethane (2 × 50 ml). The organic layers were combined and concentrated. The residue was purified by preparative TLC (eluent 3.5% methanol, 0.2% triethylamine in dichloromethane) to yield 2a (260 mg, 106%). In spite of the impurities detected by the excess yield and by TLC, this product was a stable white foam and was phosphitylated directly. 1H NMR (400 MHz, acetone-d6): δ 10.20 (br s, 1H, imide H-3), 7.60 (dd, 1H, JH6-F2′ = JH6-Me5 = 1.4 Hz, H6), 7.60–6.90 (m, 14H, trityl), 6.52 (dd, 1H, JH1′-H2′ = 5.2 Hz, JH1′-F2′ = 15.2 Hz, H1′), 5.21 (br s, 1H, OH), 5.03 (ddd, 1H, JH1′-H2′ = 5.2 Hz, JH2′-F2′ = 50.8 Hz, JH2′-H3′ = 5.2 Hz, H2′), 4.49 (ddd, 1H, JH2′-H3′ = 5.2 Hz, JH3′-F2′ = 11.5 Hz, JH3′-H4′ = 4.8 Hz, H3′) 3.79 (s, 6H, 2 OCH3), 3.62–3.43 (m, 3H, H4′, H5′, H5″), 1.74 (d, 3H, JH6-Me5 = 1.4 Hz, CH3-5).
2′-Deoxy-2′-fluoro-5′-O-(4-methoxytrityl)-4′-thio-β-d-arabinouridine (2b)
2′-Deoxy-2′-fluoro-4′-thio-β-d-arabinouridine (1b, 105 mg, 0.40 mmol) was coevaporated three times with pyridine and left in a vacuum desiccator for 48 h. Monomethoxytrityl (MMT) chloride (154 mg, 0.50 mmol, 1.25 eq) was added along with a magnetic stir bar and septum, and the flask was flushed with nitrogen. Pyridine (4 ml) was then added via syringe and the reaction was allowed to stir. TLC showed that it had progressed to about 50% completion after 5 h and did not proceed further. Another aliquot of MMT-Cl (0.6 eq.) was therefore added. After 72 h the reaction had stopped again; a few crystals of DMAP were added and the volume reduced by about half. The following day a third aliquot of MMT-Cl (0.5 eq.) was added. The reaction reached completion after 7 days. Methanol (1 ml) and a small amount of neutralized silica were then added and the reaction mixture evaporated to dryness. The product was purified by preparative TLC (eluent 5% methanol, 0.1% triethylamine in dichloromethane) to yield compound 2b as a white foam (154 mg, 74%). 1H NMR (500 MHz, acetone-d6): δ 10.3 (s, 1H, imide H-3), 7.90 (d, 1H, JH6-H5 = 7.5 Hz, H6), 7.6–6.9 (m, 14H, MMT), 6.50 (dd, 1H, JH1′-H2′ = 4.9 Hz, JH1′-F2′ = 13.7 Hz, H1′), 5.51 (d, 1H, JH6-H5 = 7.5 Hz, H5), 5.22 (br s, 1H, OH), 5.05 (ddd, 1H, JH1′-H2′ = 4.9 Hz, JH2′-F2′ = 51.0 Hz, JH2′-H3′ = 5.0 Hz, H2′), 4.54 (m, 1H, H3′) 3.79 (s, 3H, OMe), 3.57–3.52 (m, 3H, H4′, H5′, H5″) 13C NMR (125 MHz, acetone-d6): δ 162.84, 159.19, 151.16, 144.70, 144.62, 142.47, 135.26, 130.76, 128.68, 128.10, 127.30, 113.36, 101.72 (C5), 96.35 (d, JC2′-F2′ = 192.3 Hz, C2′), 87.07 (OCAr3), 74.70 (d, JC3′-F2′ = 23.7 Hz, C3′), 63.99 (d, JC5′-F2′ = 2.3 Hz, C5′), 58.86 (d, JC1′-F2′ = 16.0 Hz, C1′), 54.94 (OMe), 50.85 (d, JC4′-F2′ = 3.8 Hz, C4′). FAB-HRMS: calcd for C29H27N2O5SF + K+: 573.1262; found: 573.1261.
1-(3-O-(β-Cyanoethyl-N,N-diisopropylphosphoramidic)-2-deoxy-2-fluoro-5-O-(4,4′-dimethoxytrityl)-4-thio-β-d-arabinofuranosyl)-thymine (3a)
The crude compound 2a (260 mg) was coevaporated with dichloromethane and dried overnight over P2O5. It was then dissolved in dichloromethane (2 ml) and anhydrous diisopropylammonium tetrazolide (161 mg, 0.94 mmol) was added. Finally, 2-cyanoethyl-N,N,N′,N′-tetraisopropylphosphorodiamidite (202 μl, 184 mg, 0.61 mmol) was added via syringe under a nitrogen atmosphere. The suspension was stirred for 68 h. A column was packed using neutralized silica in hexanes, and the reaction mixture was poured directly onto it. After elution in hexanes containing 10–50% ethyl acetate and 1% triethylamine, the fractions containing product were concentrated, and the product precipitated from cold hexanes to yield 3a as a white foam (151 mg, 44% over two steps). The mixture of two diastereomers at phosphorus led to complex 1H and 13C NMR spectra. 31P NMR (81 MHz, acetone-d6): δ 151.9 (d, JF-P = 6.2 Hz), 151.3 (d, JF-P = 3.4 Hz). FAB-HRMS: Calcd for C40H48N4O7FPS + K+: 817.2602; found: 817.2606.
2′-Deoxy-2′-fluoro-3′-O-(β-cyanoethyl-N,N-diisopropylphosphoramidic)-5′-O-(4-methoxytrityl)-4′-thio-β-d-arabinouridine (3b)
Compound 2b (155 mg, 0.29 mmol) was dried over P2O5 for several days, coevaporated with dry dichloromethane halfway through this period. It was then dissolved in dichloromethane (2 ml) and anhydrous diisopropylammonium tetrazolide (102 mg, 0.60 mmol, 2.0 eq.) was added. Finally, 2-cyanoethyl-N,N,N′,N′-tetraisopropylphosphordiamidite (115 μl, 0.35 mmol) was added via syringe under a nitrogen atmosphere. The suspension was stirred for 46 h. The reaction mixture was loaded onto a column of triethylamine-neutralized silica and was purified by flash chromatography (using hexanes–ethyl acetate–triethylamine as a eluent) to yield 3b as a foam (138 mg, 65%), collected as pure amidite diastereomers. Another fraction was isolated containing a mixture of starting material and product, and was phosphitylated again to yield a further 10 mg of product, for a total yield of 70%. For the faster moving diastereomer: 31P NMR (81 MHz, acetone-d6): δ 152.2 (d, JF-P = 6.5 Hz). 1H NMR (500 MHz, acetone-d6): δ 10.19 (br s, 1H, H3 (uracil N3-H)), 7.86 (dd, 1H, JH6-H5 = 8.0 Hz, JH6-F2′ = 1.7 Hz, H6), 7.54–6.91 (m, 14H, trityl), 6.51 (dd, 1H, JH1′-H2′ = 5.0 Hz, JH1′-F2′ = 14.5 Hz, H1′), 5.49 (d, 1H, JH6-H5 = 8.0 Hz, H5), 5.14 (ddd, JH1′-H2′ = 5.0 Hz, JH2′-F2′ = 50.5 Hz, JH2′-H3′ = 4.6 Hz, H2′), 4.70 (m, 1H, H3′), 3.81 (s, 3H, OMe of MMT), 3.80–3.51 (m, 7H; H4′, H5′, H5″, OCH2 of cyanoethyl, 2 NCH(CH3)2), 2.66 (t, 2H, J = 6.2 Hz), 1.20, 1.191, 1.187, 1.17 (4s, 12H, 2 NCH(CH3)2). 13C NMR (125.7 MHz, acetone-d6): δ 162.53, 159.23, 151.04, 144.61, 144.53, 135.19 (C2, C4, 4 tertiary aromatic carbons of MMT), 142.19 (d, JC6-F2′ = 2.9 Hz, C6), 130.80, 128.72, 128.71, 128.11, 127.35 (aromatic carbons of MMT), 118.77 (CN), 113.35 (aromatic carbon of MMT), 101.86 (C5), 95.56 (dd, JC2′-F2′ = 193.8 Hz, JC2′-P = 3.6 Hz, C2′), 87.20 (OCAr3), 76.48 (dd, JC3′-P = 16.2 Hz, JC3′-F2′ = 24.3 Hz, C3′), 63.99 (d, JC5′-F2′ = 3.6 Hz, C5′), 59.18, 59.03, 58.90, 58.76 (4 signals due to iPr methyls), 54.94 (OMe), 50.53 (dd, JC4′-F2′ ∼ JC4′-P ∼ 3 Hz, C4′), 43.40 (d, JC-P = 12.6 Hz, OCH2CH2CN), 24.27, 24.21, 24.16, 24.10 (4 Me of iPr). ESI-MS: calcd for C38H44FN4O6PS + Na, 757.3; found, 757.0. Slower moving diastereomer 31P NMR (81 MHz, acetone-d6): δ 151.4 (d, JF-P = 3.7 Hz). 1H and 13C NMR very similar to those for the first diastereomer. Signals corresponding to the iPr and cyanoethyl groups were, predictably, those for which the largest differences were observed. ESI-MS: calcd for C38H44FN4O6PS + Na, 757.3; found, 757.1. NOESY spectra provided no useful information for identifying the stereochemistry of the two diastereomers.
3′,5′-Di-O-benzoyl-2′-deoxy-2′-fluoro-4′-thio-β-d-arabinouridine (4)
To anhydrous uracil (33 mg, 0.29 mmol, 4 eq) in a 10-ml round-bottomed flask was added acetonitrile (2 ml) followed by HMDS (62 μl, 0.29 mmol, 4 eq.), with stirring. The mixture was heated to reflux, and became clear. After 4 h, the solvent was removed. A solution of 1-O-acetyl 3,5-di-O-benzoyl-2-deoxy-2-fluoro-d-arabinofuranose (30 mg, 0.072 mmol) in carbon tetrachloride (2 ml) was added followed by TMS-triflate (20 μl, 0.11 mmol, 1.5 eq.). The flask which had contained the dry sugar was then rinsed with another aliquot (1.5 ml) of carbon tetrachloride, which was added. The reaction mixture was stirred at reflux for 20 h until TLC indicated no further change. The mixture was poured onto a short column of silica gel and eluted with 0.5% triethylamine in chloroform. The separation of the anomers was achieved by a subsequent longer column of neutralized silica using chloroform as a eluent. The less-polar compound 4 was isolated as an amorphous solid (15.8 mg, 47%): 1H NMR (500 MHz, CDCl3) δ 8.78 (br s, 1H, imide-NH) 8.1–7.4 (m, 10H, 2 Bz), 6.77 (dd, 1H, JH1′-H2′ = 4.0 Hz, JH1′-F2′ = 23 Hz, H1′), 5.88 (ddd, 1H, JH2′-H3′ = 2.5 Hz, JH3′-F2′ = 9.6 Hz, JH3′-H4′ = 2.0 Hz, H3′), 5.76 (d, 1H, JH5-H6 = 8.2 Hz, H5), 5.27 (ddd, 1H, JH1′-H2′ = 4.0 Hz, JH2′-H3′ = 2.5 Hz, JH2′-F2′ = 49.6 Hz, H2′), 4.67 (m, 2H, H5′,5″), 3.99 (m, 1H, H4′). 13C NMR (125 MHz, CDCl3): δ 166.25, 164.90, 162.74, 150.94 (4 CO), 142.28 (d, JC6-F2′ = 4.7 Hz, C6), 134.37, 133.75, 130.27, 130.04, 129.53, 128.96, 128.82, 128.48 (2 OBz), 102.94 (C5), 94.66 (d, JC2′-F2′ = 189.9 Hz, C2′), 153.59 (d, JC3′-F2′ = 27.4 Hz, C3′), 64.68 (d, JC5′-F2′ = 5.3 Hz, C5′), 61.83 (d, JC1′-F2′ = 16.8 Hz, C1′), 50.96 (C4′). Two pairs of NOESY crosspeaks (H6-H3′, H6-H5′) provide strong evidence for top-face uracil and therefore the β nucleoside. FAB-HRMS: calcd for C23H19N2O6SF + H+: 471.1026; found: 471.1027.
Oligonucleotide synthesis
Standard conditions for solid-phase oligonucleotide synthesis were used for the synthesis of all oligonucleotides, at a 0.8–1.0 mmol scale. 4,5-Dicyanoimidazole (0.50 M in acetonitrile) or 5-ethylthiotetrazole (0.25 M in acetonitrile) were used as activators and 0.10 M iodine in 1:2:10 pyridine:water:THF was used as oxidant (waiting time during the oxidation step was 24 s). Phosphoramidites were prepared as 0.15 M solutions (RNA amidites) or 0.08–0.10 M solutions (DNA, 2′-fluoro and 4′-thio amidites). Coupling times were extended to 10–30 min for modified nucleotides. The oligonucleotides were treated with 3:1 ammonium hydroxide:ethanol for 16 h at 55°C to cleave them from the solid support and deprotect the phosphates and bases. Sequences containing ribonucleotides were concentrated and desilylated with Et3N · 3HF (100 μl) for 48 h at room temperature. Sequence purification was accomplished by anion exchange HPLC using 0–0.2 M LiClO4 solution as an eluent, or by preparative denaturing PAGE. Desalting was effected on Sephadex G-25 or NAP-25 columns. Sequence purity was verified using denaturing PAGE.
SVPDE/AP digestions
Phosphodiesterase I from Crotalus adamanteus venom snake venom phosphodiesterase (SVPDE) was purchased from USB Corporation (Cleveland, OH). Calf intestine alkaline phosphatase (AP) was purchased from Amersham Biosciences (Piscataway, NJ). Enzymatic digestion assays combined 1–4 units of SVPDE and 16 units AP in 50 μl of reaction buffer (100 mM Tris-HCl, pH 8.9, 100 mM NaCl, 14 mM MgCl2) along with 0.3 A260 units of the oligonucleotide to be digested. The assay was incubated at 37°C for 2–18 h, then diluted with 250 μl of water and an aliquot was injected onto a Waters Symmetry C18 5 μm 4.6 × 150 mm HPLC column. A gradient from 0–20% methanol in 20 mM NaH2PO4 buffer (pH 5.5), with a linear increase over 25 min, was used to elute the nucleosides. Fractions were desalted on Sep-Pak reverse phase columns, then analyzed by ESI-MS.
Thermal denaturation and CD studies
Equimolar amounts of complementary sequences were combined, dried and rediluted in pH 7.2 buffer containing 140 mM KCl, 1 mM MgCl2 and 5 mM NaHPO4 (1 ml). After heating to 90°C, the samples were slowly cooled to room temperature and refrigerated overnight. They were then transferred into cold cuvettes in a Cary 300 UV spectrophotometer. The change in absorbance at 260 nm was then monitored upon heating from 15 to 90°C. Melting temperatures were determined as the maxima of the first derivatives.
Circular dichroism (CD) spectra were obtained on a Jasco J-720 spectropolarimeter at 20°C using samples annealed in the same buffer and under the same conditions as for the thermal denaturation studies. Spectra were baseline-corrected with respect to a blank containing the buffer but no duplex. Smoothing and adjustment for duplex concentration were effected using the Spectra-Manager program (Jasco).
RNase H assays
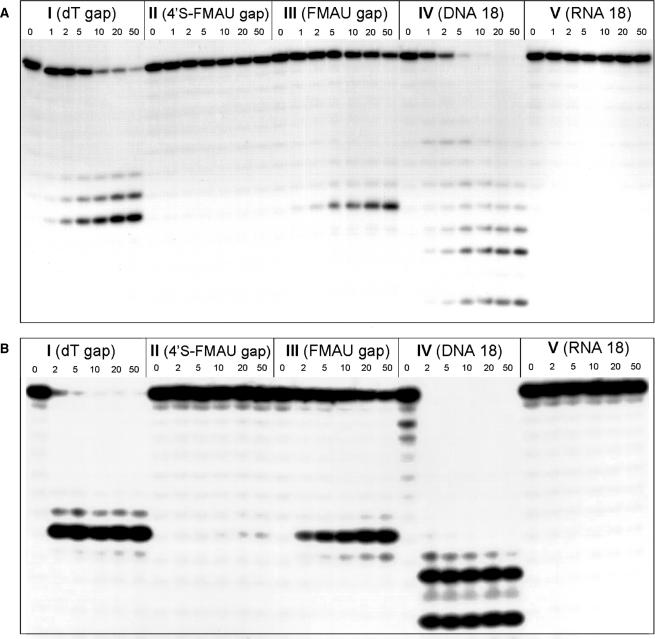
The activity of E. coli RNase HI (USB Corporation, Cleveland, OH) was tested with antisense oligonucleotides under conditions recommended by the manufacturer (50 mM Tris-HCl, pH 7.5, 50 mM KCl, 25 mM MgCl2, 0.25 mM EDTA, 0.25 mM DTT). The antisense and 5′-32P labeled sense strands were combined in a 2:1 ratio and annealed by heating to 90°C followed by slow cooling to room temperature. About 2.5 U (17 μg) of enzyme were incubated at 37°C in the described buffer for 10 min, and 100 μl final volume reactions were initiated by the addition of duplexed antisense–sense substrate to a concentration of 50 nM. Aliquots were removed at various times as indicated in Figure 3 and quenched by the addition of an equal volume of loading buffer (98% deionized formamide, 10 mM EDTA, 1 mg/ml bromophenol blue and 1 mg/ml xylene cyanol), followed by heating to 95°C for 5 min. Cleavage products were resolved on 16% denaturing PAGE and visualized by autoradiography.
Figure 3.
Ribonuclease H degradation of various hybrid duplexes. An 18-nt 5′-32P-labeled target RNA (5′-ACG UGA AAA AAA AUG UCA-3′) was preincubated with complementary 18-nt I–V, and then added to reaction assays containing either (A) E. coli RNase HI or (B) human RNase HII (110 nM assay shown here). Aliquots were removed as listed in the figure (in minutes). Base sequences of antisense oligomers are given in Table 1. See the Results and Discussion section for detailed assay conditions. Gels from human RNase H assay at lower enzyme concentration and shorter exposure times are available as Supplementary Data.
Human RNase HII was expressed and purified using a slight modification of the published procedure (31). The assays were performed analogously to that described above, using a 3:1 antisense:sense strand ratio, a buffer containing 60 mM Tris-HCl, pH 7.8, 60 mM KCl, 2.5 mM MgCl2 and 2 mM DTT and enzyme concentrations of 37 and 110 nM. A gel from the assay at lower enzyme concentration is given in the Supplementary Data.
siRNA assays
HelaX1/5 cells that stably express firefly luciferase were grown as described earlier (32). The day prior to transfection, 0.5 × 105 cells were plated in each well of a 24-well plate. The next day, the cells were incubated with increasing amounts of siRNAs premixed with lipofectamine-plus reagent (Invitrogen) using 1 μl of lipofectamine and 4 μl of the plus reagent per 20 pmol of siRNA (for the highest concentration tested). For the siRNA titrations, each siRNA was diluted into dilution buffer (30 mM HEPES-KOH, pH 7.4, 100 mM KOAc, 2 mM MgOAc2) and the amount of lipofectamine-plus reagent used relative to the siRNAs remained constant. Twenty-four hours after transfection, the cells were lysed in hypotonic lysis buffer (15 mM K3PO4, 1 mM EDTA, 1% Triton, 2 mM NaF, 1 mg/ml BSA, 1 mM DTT, 100 mM NaCl, 4 μg/ml aprotinin, 2 μg/ml leupeptin and 2 μg/ml pepstatin) and the firefly light units were determined using a Fluostar Optima 96-well plate bioluminescence reader (BMG Labtech) using firefly substrate as described earlier (33). The luciferase counts were normalized to the protein concentration of the cell lysate as determined by the DC protein assay (BioRad). Error bars represent the standard deviation of at least four transfections. Cotransfecting the siRNAs and the plasmid pCI-hRL-con expressing the Renilla luciferase mRNA (34) in the same cell line showed no difference in expression of this reporter, demonstrating the specificity of the RNAi effects (data not shown).
RESULTS AND DISCUSSION
Preparation of the nucleoside phosphoramidite
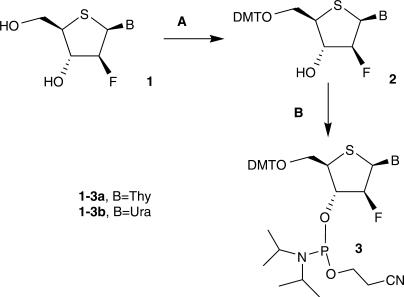
2′-Fluoro-4′-thioarabino nucleoside 1a was prepared as described earlier (19). Its uracil analog 1b was prepared analogously (experimental details and characterization are given above). The 5′-hydroxyl group was protected using either 4-MMT or 4,4′-DMT chloride, but the latter required significantly shorter reaction times and was preferred (Figure 1). Phosphitylation of the tritylated compounds 2 using bis(diisopropylamino)-β-cyanoethylphosphoramidite in the presence of diisopropylammonium tetrazolide, followed by precipitation from cold hexanes, gave the phosphoramidites 3 of suitable purity for solid-phase oligonucleotide synthesis. The tritylation and phosphitylation reactions were in general much slower for these modified nucleosides than for standard deoxyribo- or ribonucleosides.
Figure 1.
Synthesis of 4′S-FANA phosphoramidites. Reagents and conditions: (A) DMTrCl, Pyridine, rt, 44 h; (B) (N(iPr2))2P(OCH2CH2CN), Diisopropylammonium tetrazolide, CH2Cl2, rt, 68 h.
Oligonucleotide design
It has been shown that an 18-mer chimera containing six central 2′-deoxyribonucleotides (35–37) or 2′F-ANA (38) nucleotides surrounded by native RNA wings is a substrate for RNase H. The RNA wings serve to ensure tight binding, and the central section is adequate to elicit RNase H activity. In this way, new modifications can be tested for a true effect on RNase H activity without compromising the binding properties of the oligonucleotide. Oligonucleotides I–V (Table 1) were therefore synthesized by adopting this approach in the oligomer design, using standard solid-phase methodology. The three gapmers I, II and III (containing six central dT, (4′S-FMAU) 2′-deoxy-2′-fluoro-5-methyl-4′-thioarabinouridine or FMAU residues, respectively) and two 18-mers IV and V (DNA and RNA) were chosen to provide a wide range of duplex structure and thus varying degrees of RNase H hydrolysis. The RNA wings of the sequences chosen were of mixed base composition to minimize the formation of partial duplexes that can occur when polyuridylates are hybridized to RNA. siRNA sequences were chosen for their continuity with previous studies on 2′F-ANA-modified siRNA (32).
Table 1.
Antisense oligonucleotide sequences and thermal denaturation studiesa
| Sequence | Tm (RNA target) | Tm (DNA target) | ||||||
|---|---|---|---|---|---|---|---|---|
| I | 5′-UGA | CAU | ttt | ttt | UCA | CGU-3′ | 60.0 | 51.0 |
| II | 5′-UGA | CAU | TTT | TTT | UCA | CGU-3′ | 51.0 | 36.0 |
| III | 5′-UGA | CAU | TTT | TTT | UCA | CGU-3′ | 62.0 | 50.1 |
| IV | 5′-tga | cat | ttt | ttt | tca | cgt-3′ | 42.1 | 55.5 |
| V | 5′-UGA | CAU | UUU | UUU | UCA | CGU-3′ | 59.1 | 40.2 |
aLegend: RNA, DNA, 4′S-FANA, 2′F-ANA. Complementary strands were as follows: RNA, 5′-ACG UGA AAA AAA AUG UCA-3′; DNA, 5′-acg tga aaa aaa atg tca-3′.
Oligonucleotide characterization
The modified nucleoside phosphoramidites 3 behaved well during the solid-phase synthesis, as described in detail in the experimental section. All 4′S-FANA-modified oligonucleotides were examined by MALDI and ESI mass spectrometry and were of the correct mass (Table S1, Supplementary Data).
As a further characterization, we carried out a snake venom phosphodiesterase/alkaline phosphatase (SVPDE/AP) digestion on strand II, purified the resulting nucleosides by HPLC, and confirmed by ESI-MS that the isolated 4′S-FMAU was the authentic nucleoside. This provided further evidence that the 4′S-FMAU is stable to oxidation under solid-phase coupling conditions.
Binding and CD studies
The ‘antisense’ sequences I–V were hybridized to complementary ssRNA and ssDNA, and their thermal stability was examined by monitoring the change in hyperchromicity at 260 nm. All sequences and melting temperatures of the duplexes are shown in Table 1.
Sequences I, III and V (DNA and 2′F-ANA gapmers, and an all RNA strand) had very similar affinity towards their common ssRNA target. In fact, this observation is precedented (15,39) and related to the purine-rich complementary RNA strand, 5′-ACG UGA AAA AAA AUG UCA-3′, which predominantly dictates, in each case, adoption of the high-melting A-form helical conformation. In contrast, a drop in Tm of −1.4°C per insert with respect to V:RNA is observed for sequence II (4′S-FANA gapmer) and implies either that this modified AON cannot easily adopt the classical A-form geometry upon duplexation, or that there are unfavorable interactions within an A-form structure that weaken the association. Given that the nucleoside favors a northern conformation, and that the II:RNA hybrid does display an A-like conformation (CD studies described below), the second option seems more likely. It is interesting to observe that 2′-fluoroarabino (40), 4′-thio-RNA (23) and 4′thio-DNA (25) modifications, taken alone, usually provide an increase in stability toward complementary strands. Thus it may be that the ‘combination’ of the longer S4′–C1′ bond (and the larger van der Waals radius of S versus O) with the top-face fluorine is responsible for the destabilization of the hybrid structure. This likely influences the N-glycosidic torsion angle in such a way as to cause destabilization. Indeed, this is manifest in the CD spectrum (discussed below), in which the long-wavelength band (generally associated with nucleobase stacking) is quite weak for this duplex, and even more so when DNA is the target (Figure 2). Since both sulfur and fluorine are more hydrophobic than oxygen, another possible explanation for the lower thermal stability of the 4′S-FANA(II):RNA duplex relative to RNA(V):RNA is the more complete and favorable hydration of 4′-O and 2′-OH groups in the latter. Either modification (4′S or 2′F) taken alone may have enough hydrophilic character to avoid this destabilization.
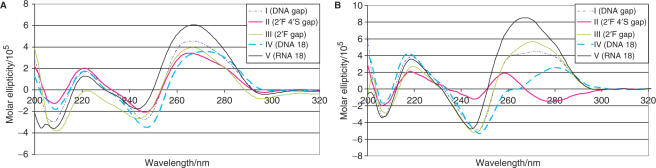
Figure 2.
Circular dichroism spectra (A) I–V, ssRNA target; (B) I–V, ssDNA target. Spectra were run at 20°C after annealing the duplexes under the same conditions described for the binding studies.
For the ssDNA target, it is again worth comparing the binding behavior of the various gapmers. AONs containing DNA or 2′F-ANA gaps (I and III) had the highest affinity towards the ssDNA target. The 4′S-FANA gapmer II provided the least stable AON:DNA duplex; however, its Tm was comparable to that of the RNA 18-mer V. This is consistent with the well-known order of stability of oligonucleotide duplexes, where dPy:dPu > rPy:dPu (40,41) and the proposal that 2′F-ANA mimics the DNA structure (southeast pucker), whereas 4′S-FANA more closely resembles RNA (north pucker). The 4′S-FANA gapmer (II) and RNA (V) exhibit similar affinity towards the ssDNA target (ΔTm = 4°C; Table 1). The same trend does not hold when the target is ssRNA; in this case, the RNA (V):RNA duplex is significantly more stable than the 4′S-FANA (II):RNA hybrid (ΔTm = 8.1°C), suggesting that the 4′S-FANA modification prefers an alternative conformation that is available only when duplexed to the more flexible DNA target. In support of this notion, the CD spectrum of II:DNA is neither A-form nor B-form, whereas that of II:RNA is characteristic of A-form duplexes (discussed below).
Hybrids comprising any one of the sequences I–V bound to either ssRNA or ssDNA targets were further evaluated for possible variations in duplex structure via CD spectroscopy, in the region from 320 to 200 nm (Figure 2). The spectra of all AON:RNA hybrids exhibit the characteristic A-form pattern, with the largest changes evident in the magnitude and positions of the positive Cotton effect at ∼265 nm. The highest Cotton effect (molar ellipticity) observed corresponds to that of the pure RNA:RNA duplex (V:RNA). The Cotton effects of the 4′S-FANA gapmer (II):RNA duplex are blue shifted, but the overall CD trace similarly indicates an A-form global geometry. The spectra of the AON:DNA hybrids, however, are much more varied in comparison. Most striking is the CD signature of the II:DNA duplex, which bears no similarity to either A- or B-form reference spectra. Of note, for example, are the negative peak at 280 nm, the cross-over at 270 nm and the positive peak at 257 nm, all of which are unique to the II:DNA spectrum. The helical structure of this hybrid is apparently quite different from either A-form or B-form helices, thus supporting the notion that the increased S–C bond length, the smaller C–S–C bond angle or the more puckered ring causes a divergence from the classical helix structure, or might perturb the N-glycosidic orientation around the nucleotide sugars, thereby destacking the helix. The fact that greater structural distortions are observed with ssDNA instead of ssRNA targets (as measured by CD) suggests that the 4′S-FANA-modified strand only reveals its preferred conformation in a duplex when not overwhelmed by the A-form preference of RNA.
RNase H assays
The RNase H family of enzymes recognize and cleave the RNA strand of AON:RNA hybrids having a conformation that is intermediate between the pure A- or B-form conformations adopted by dsRNA and dsDNA, respectively. Sugar geometries that fall within the eastern (O4′-endo) range within the AON have been postulated to actively induce RNase H-assisted RNA strand cleavage (42,43). Chemical changes of the sugar constituents or alterations in the sugar conformation (e.g. orientation of the sugar to the base) or flexibility (e.g. DNA versus the more rigid 2′F-ANA analog) can all dramatically affect RNase H activation (16).
Oligomers I–V were assessed for their ability to elicit E. coli RNase HI and human RNase HII activity. As shown in Figure 3, the control DNA oligomer IV and DNA gap I both promoted essentially complete degradation of the 5′-32P-labeled RNA. As expected, the RNA duplex was not a substrate of RNase H. With the 2′F-ANA gap (III) the enzyme activity was somewhat lower compared to DNA, although significant cleavage (>50%) occurred after 50 min under these conditions, as observed earlier (15). Negligible or no cleavage was observed for the 4′S-FANA modified (II):RNA hybrid. The ability of the various gaps to elicit E. coli RNase HI activity followed the order: DNA > 2′F-ANA ≫ 4′S-FANA ∼ RNA (Figure 3A). The same trend was observed with the human enzyme (Figure 3B). The lack of RNase H activity supported by 4′S-FANA is fully consistent with the anticipated northern conformation (C3′-endo) of this modification.
RNA interference
2′-Fluoro-4′-thioarabinouridine was introduced at various positions into both strands of an siRNA sequence (32) targeting the firefly luciferase gene (Table 2). siRNAs containing FMAU at the same positions were used as controls, along with native RNA. The resulting modified duplexes were transfected into HeLa cells stably expressing firefly luciferase (details in Materials and methods section). Results are summarized in Tables 2 and 3, and Figures 4–6.
Table 2.
siRNA sequences and thermal denaturation studiesa
| Duplex | Tm (°C) | IC50 (nM) | |
|---|---|---|---|
| Ctl | 5′-GCUUGAAGUCUUUAAUUAAtt-3′ | 62.3 | 0.16 |
| 3′-ggCGAACUUCAGAAAUUAAUU-5′ | |||
| Ctl-p | 5′-GCUUGAAGUCUUUAAUUAAtt-3′ | n.d. | 0.04 |
| 3′-ggCGAACUUCAGAAAUUAAUUp-5′ | |||
| T1 | 5′-GCUUGAAGUCUUUAAUUAAtt-3′ | 60.2 | 0.10 |
| 3′-ggCGAACUUCAGAAAUUAAUU-5′ | |||
| F1 | 5′-GCUUGAAGUCUUUAATTAAtt-3′ | 63.0 | 0.20 |
| 3′-ggCGAACUUCAGAAAUUAAUU-5′ | |||
| T2 | 5′-GCUUGAAGUCUUUAAUUAAtt-3′ | 57.2 | 0.17 |
| 3′-ggCGAACUUCAGAAAUUAAUU-5′ | |||
| F2 | 5′-GCUUGAAGUCUUUAAUUAAtt-3′ | 60.0 | 0.31 |
| 3′-ggCGAACUTCAGAAAUUAAUU-5′ | |||
| T3 | 5′-GCUUGAAGUCUUUAAUUAAtt-3′ | 62.0 | 3.6 |
| 3′-ggCGAACUUCAGAAAUUAAUU-5′ | |||
| T3p | 5′-GCUUGAAGUCUUUAAUUAAtt-3′ | n.d. | 0.16 |
| 3′-ggCGAACUUCAGAAAUUAAUUp-5′ | |||
| F3 | 5′-GCUUGAAGUCUUUAAUUAAtt-3′ | 62.1 | 1.0 |
| 3′-ggCGAACUUCAGAAAUUAATT-5′ | |||
| F3p | 5′-GCUUGAAGUCUUUAAUUAAtt-3′ | n.d. | 0.04 |
| 3′-ggCGAACUUCAGAAAUUAATTp-5′ | |||
aLegend: RNA, DNA, 4′S-FANA, 2′F-ANA. Sense strands are listed on top and antisense strands below. Duplexes with names ending in ‘p’ were 5′-phosphorylated on the antisense strand (see text for details).
Table 3.
Effect of significantly modified sense strands with FAU point modifications in the antisense stranda
| Duplex | Tm (°C) | IC50 (nM) | |
|---|---|---|---|
| Ctl | 5′-GCUUGAAGUCUUUAAUUAAtt-3′ | 62.1 | 0.16 |
| 3′-ggCGAACUUCAGAAAUUAAUU-5′ | |||
| Ctl-gg | 5′-GCUUGAAGUCUUUAAUUAAgg-3′ | 61.7 | 0.87 |
| 3′-ggCGAACUUCAGAAAUUAAUU-5′ | |||
| Ctl-f | 5′-GCTTGAAGTCTTTAATTAAGG-3′ | 66.0 | 7.8 |
| 3′-ggCGAACUUCAGAAAUUAAUU-5′ | |||
| Ctl-fr | 5′-GCTTGAAGTCTTTAATTAATT-3′ | 63.0 | 2.4 |
| 3′-ggCGAACUUCAGAAAUUAAUU-5′ | |||
| Ctl-fm | 5′-GCTTGAAGTCTTTATTAAA3′ | 62.1 | >20 |
| 3′-ggCGAACUUCAGAAAUUAAUU-5′ | |||
| T2-f | 5′-GCTTGAAGTCTTTAATTAAGG-3′ | 61.0 | 17 |
| 3′-ggCGAACUUCAGAAAUUAAUU-5′ | |||
| T2-fr | 5′-GCTTGAAGTCTTTAATTAATT-3′ | 56.6 | 0.60 |
| 3′-ggCGAACUUCAGAAAUUAAUU-5′ | |||
| T2-fm | 5′-GCTTGAAGTCTTTATTAAA-3′ | 54.1 | >20 |
| 3′-ggCGAACUUCAGAAAUUAAUU-5′ | |||
| F2-f | 5′-GCTTGAAGTCTTTAATTAAGG-3′ | 65.6 | 10.8 |
| 3′-ggCGAACUTCAGAAAUUAAUU-5′ | |||
| F2-fr | 5′-GCTTGAAGTCTTTAATTAATT-3′ | 61.6 | 0.87 |
| 3′-ggCGAACUTCAGAAAUUAAUU-5′ | |||
| F2-fm | 5′-GCTTGAAGTCTTTATTAAA-3′ | 61.1 | >20 |
| 3′-ggCGAACUTCAGAAAUUAAUU-5′ | |||
aLegend: RNA, DNA, 4′S-FANA, 2′F-ANA. Mismatches are indicated with gray text. Duplexes with names ending in ‘f’ contained fully 2′F-ANA sense strands, ‘fr’ indicated a 2′F-ANA sense strand containing five RNA inserts near the sense 3′-end, and ‘fm’ indicated a 2′F-ANA sense strand with two internal mismatches near the sense 3′-end (see text for details).
Figure 4.
Activity of 4′S-FANA-modified siRNA (sequences given in Table 2).
Figure 5.
Effect of phosphorylation on siRNAs modified at the 5′-terminal of the antisense strand (sequences given in Table 2).
Figure 6.
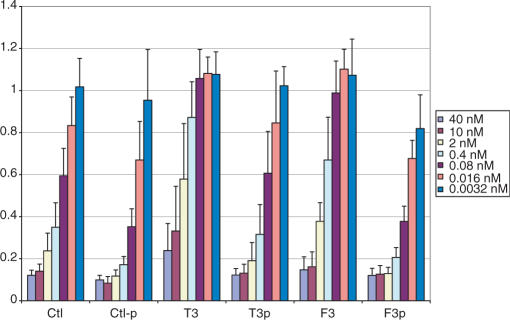
Activity of 4′S-FANA in combination with various heavily modified sense strands (sequences given in Table 3).
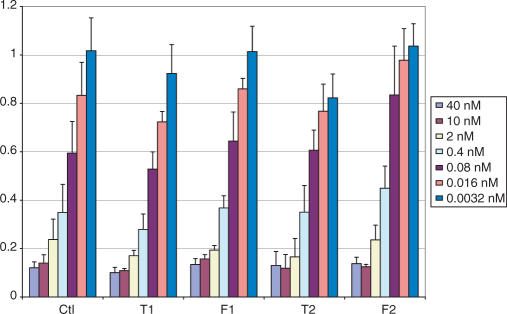
The 4′S-FANA modification is generally well tolerated by the RNAi machinery, with some dependence on position. The potencies of the 4′-S-FANA- and 2′F-ANA- modified strands are comparable.
When the terminal pair of nucleotides of the antisense strand is modified by either one of the nucleotides under investigation in this study, the activity is significantly reduced. We hypothesized that this was related to the inability of the strand to be phosphorylated in vivo, an essential step for incorporation into RISC. Indeed, chemical or enzymatic phosphorylation prior to transfection dramatically increased the activity of terminally modified strands (Figure 5). Even the control strand showed an improvement in potency upon 5′-phosphorylation. This is understandable, given the fact that although the RISC complex can phosphorylate exogenous siRNAs, the natural process involves fragments cleaved by Dicer and possessing a 5′-phosphate (44).
It is significant that this 5′-terminal pair of nucleotides can be modified, if phosphorylated, with no significant loss in activity for the 2′S-FANA or 4′S-FANA. These positions have recently been shown to play a key role in reducing miRNA-type off-target effects when modified with 2′-O-methylribonucleotides (45). Given that the very small structural perturbation of a 2′-O-methyl group is effective in reducing off-target effects, we are optimistic that the larger conformational changes introduced by 4′S-FANA moieties may cause an even greater decrease in undesired miRNA-type silencing.
The 4′S-FANA antisense modifications were tested in combination with various heavily modified sense strands. We reported earlier that an all-2′F-ANA sense strand was tolerated by the RNAi machinery. We therefore included such a duplex in our assays (duplexes Ctl-f, T2-f and F2-f). To improve upon the activity of this strand, however, we made two other modifications. Functional siRNAs exhibit strand bias; at the 5′-end of the duplex (defined by the antisense strand), the duplex-binding affinity should be reduced (46,47). Hohjoh (48) showed that it is possible to introduce this strand bias and thus improve siRNA activity by introducing 1-4 internal mismatches near the 3′-end of the sense strand. Thus we attempted a similar design and made a fully modified 2′F-ANA sense strand containing two appropriately placed mismatches (duplexes Ctl-fm, T2-fm and F2-fm). Finally, in an attempt to both lower the affinity and maintain a more authentic A-form helical structure, a sense strand was made containing five RNA inserts at its 3′-end (duplexes Ctl-fr, T2-fr and F2-fr). The 2-nucleotide 3′-overhang was left as 2′F-ANA to help provide 3′-exonuclease resistance. Results are given in Figure 6.
In all cases, the ‘fr’ type sense strand was the best heavily modified sense strand, reaching levels of potency close to that of the control. It is interesting to note the synergy between 4′S-FANA and 2′F-ANA in the T2-fr duplex, which gave particularly good results. Neither antisense strand modification was helpful in the case of the ‘f’ type sense strand. The ‘fm’ type sense strand showed very poor activity. Apparently the deviation in structure from an RNA duplex is great enough in this case that it is not well recognized by RISC. This significant difference in potency is in spite of the fact that the binding affinities of the ‘fm’ and ‘fr’ strands are similar (Table 3); thus in this case the duplex distortion is more important than the required strand bias discussed above.
The need for strand bias may explain why the 4′S-FANA-modified duplex T1 is more potent than the control, since the duplex has lower affinity at the antisense 5′-end (Table 2). However, it remains to be seen if this strand bias will be generally applicable or will be overwhelmed by the helical distortions associated with 4′S-FANA inserts. For example, the 4′S-FANA-modified duplex T3p shows slightly reduced activity, even though the modification is appropriately placed at the antisense 5′-end of the duplex. Thus we believe that the slightly reduced activity of T3p with respect to Ctl-p can be attributed to the helical distortion observed and discussed above: although the nucleoside puckers in the north, the helix seems to be prevented from adopting a perfectly A-form duplex by other factors. When given a flexible target strand, as discussed above for the II:DNA duplex, 4′S-FANA seems to induce a duplex that is neither A-form nor B-form. Deviations of helical structure from the A-form (whether through unsuitable chemical modifications or antisense-strand mismatches) are known to divert RNAs from the siRNA pathway (26). 2′F-ANA, on the other hand, prefers to adopt a southeast conformation (19), but its CD spectra discussed above correspond much more closely to an A-form helix; thus, it is more able to allow the RNA to dictate the helical conformation.
CONCLUSIONS
Modified duplexes containing 4′S-FANA units were thermodynamically less stable than unmodified duplexes and, as expected, they were unable to elicit E. coli or human RNase H activity. Since 4′S-FANA nucleosides are RNA-like (north) in conformation, these results lend further credence to the hypothesis that RNase H enzymes recognize duplexes in which the nucleoside building blocks of the antisense strand can adopt an eastern conformation (42). They also highlight important structural differences that give rise to proficient enzymatic activity and further clarify the role of substrate conformation on the discriminatory properties of RNase H.
While 4′S-FMAU adopts a north conformation, CD spectra and thermal denaturation studies of oligomers containing this modification indicate that it perturbs the RNA structure. We believe that the main sources of this perturbation are steric hindrance and lower hydration compared to the native RNA. The combination of the 2′ ‘up’ fluorine and the 4′ sulfur substitution lead to destabilizing interactions not observed for either modification alone.
siRNAs containing 4′S-FANA units were able to enter the RNAi pathway. One or two inserts internally in either strand gave duplexes of potency comparable to that of the control. The 5′-end of the antisense strand can be modified as long as it is 5′-phosphorylated prior to the siRNA assay. The 4′S-FANA modification was also able to work with good efficiency in a duplex with a 75%-modified 2′F-ANA—RNA sense strand, demonstrating that 2′F-ANA (with its preference for southern and eastern conformations) can achieve synergy with its northern cousin, 4′S-FANA, in RNAi gene silencing.
SUPPLEMENTARY DATA
Supplementary Data is available at NAR Online.
ACKNOWLEDGEMENTS
We are grateful to the Natural Sciences and Engineering Research Council of Canada (NSERC) and the Canadian Institutes for Health Research (CIHR) for financial support in the form of grants (to B.M.P. and M.J.D., respectively). J.K.W. is grateful to NSERC and Dr Richard H. Tomlinson for postgraduate fellowships. M.J.D. is the recipient of a James McGill professorship and the 2007 Bernard Belleau Award (CSC). Funding to pay the Open Access publication charge was provided by CIHR.
Conflict of interest statement. None declared.
REFERENCES
- 1.Stephenson ML, Zamecnik PC. Inhibition of Rous sarcoma viral RNA translation by a specific oligodeoxyribonucleotide. Proc. Natl. Acad. Sci. U.S.A. 1978;75:285–288. doi: 10.1073/pnas.75.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Uhlmann E, Peyman A. Antisense oligonucleotides: a new therapeutic principle. Chem. Rev. 1990;90:543–584. [Google Scholar]
- 3.Braasch DA, Corey DR. Novel antisense and peptide nucleic acid strategies for controlling gene expression. Biochemistry. 2002;41:4503–4510. doi: 10.1021/bi0122112. [DOI] [PubMed] [Google Scholar]
- 4.Opalinska JB, Gewirtz AM. Nucleic-acid therapeutics: basic principles and recent applications. Nature Rev. Drug Discov. 2002;1:503–514. doi: 10.1038/nrd837. [DOI] [PubMed] [Google Scholar]
- 5.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 6.Dorsett Y, Tuschl T. siRNAs: applications in functional genomics and potential as therapeutics. Nature Rev. Drug Discov. 2004;3:318–329. doi: 10.1038/nrd1345. [DOI] [PubMed] [Google Scholar]
- 7.Stein CA, Cheng YC. Antisense oligonucleotides as therapeutic agents - Is the bullet really magical? Science. 1993;261:1004–1012. doi: 10.1126/science.8351515. [DOI] [PubMed] [Google Scholar]
- 8.Minshull J, Hunt T. The use of single-stranded DNA and RNase H to promote quantitative ‘hybrid arrest of translation’ of mRNA/DNA hybrids in reticulocyte lysate cell-free translations. Nucleic Acids Res. 1986;14:6433–6451. doi: 10.1093/nar/14.16.6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haeuptle MT, Frank R, Dobberstein B. Translation arrest by oligodeoxynucleotides complementary to mRNA coding sequences yields polypeptides of predetermined length. Nucleic Acids Res. 1986;14:1427–1448. doi: 10.1093/nar/14.3.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Damha MJ, Wilds CJ, Noronha A, Brukner I, Borkow G, Arion D, Parniak MA. Hybrids of RNA and arabinonucleic acids (ANA and 2′F-ANA) are substrates of ribonuclease H. J. Am. Chem. Soc. 1998;120:12976–12977. [Google Scholar]
- 11.Verbeure B, Lescrinier E, Wang J, Herdewijn P. RNase H mediated cleavage of RNA by cyclohexene nucleic acid (CeNA) Nucleic Acids Res. 2001;29:4941–4947. doi: 10.1093/nar/29.24.4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wahlestedt C, Salmi P, Good L, Kela J, Johnsson T, Hokfelt T, Broberger C, Porreca F, Lai J, et al. Potent and nontoxic antisense oligonucleotides containing locked nucleic acids. Proc. Natl. Acad. Sci. U.S.A. 2000;97:5633–5638. doi: 10.1073/pnas.97.10.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fedoroff OY, Salazar M, Reid BR. Structure of a DNA: RNA hybrid duplex: why RNase H does not cleave pure RNA. J. Mol. Biol. 1993;233:509–523. doi: 10.1006/jmbi.1993.1528. [DOI] [PubMed] [Google Scholar]
- 14.Salazar M, Fedoroff OY, Miller JM, Ribeiro NS, Reid BR. The DNA strand in DNA: RNA hybrid duplexes is neither B-form nor A-form in solution. Biochemistry. 1993;32:4207–4215. doi: 10.1021/bi00067a007. [DOI] [PubMed] [Google Scholar]
- 15.Lok C-N, Viazovkina E, Min K-L, Nagy E, Wilds CJ, Damha MJ, Parniak MA. Potent gene-specific inhibitory properties of mixed-backbone antisense oligonucleotides comprised of 2′-deoxy-2′-fluoro-d-arabinose and 2′-deoxyribose nucleotides. Biochemistry. 2002;41:3457–3467. doi: 10.1021/bi0115075. [DOI] [PubMed] [Google Scholar]
- 16.Mangos MM, Min K-L, Viazovkina E, Galarneau A, Elzagheid MI, Parniak MA, Damha MJ. Efficient RNase H-directed cleavage of RNA promoted by antisense DNA or 2′F-ANA constructs containing acyclic nucleotide inserts. J. Am. Chem. Soc. 2003;125:654–661. doi: 10.1021/ja025557o. [DOI] [PubMed] [Google Scholar]
- 17.Min K-L, Viazovkina E, Galarneau A, Parniak MA, Damha MJ. Oligonucleotides comprised of alternating 2′-deoxy-2′-fluoro-β-d-arabinonucleosides and 2′-deoxyribonucleosides (2′F-ANA/DNA ‘Altimers’) induce efficient RNA cleavage mediated by RNase H. Bioorg. Med. Chem. Lett. 2002;12:2651–2654. doi: 10.1016/s0960-894x(02)00439-0. [DOI] [PubMed] [Google Scholar]
- 18.Manoharan M. RNA interference and chemically modified small interfering RNAs. Curr. Opin. Chem. Biol. 2004;8:570–579. doi: 10.1016/j.cbpa.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 19.Watts JK, Sadalapure K, Choubdar N, Pinto BM, Damha MJ. Synthesis and conformational analysis of 2′-fluoro-5-methyl-4′-thioarabinouridine (4′S-FMAU) J. Org. Chem. 2006;71:921–925. doi: 10.1021/jo051844+. [DOI] [PubMed] [Google Scholar]
- 20.Bellon L, Barascut JL, Maury G, Divita G, Goody R, Imbach JL. 4′-Thio-oligo-β-d-ribonucleotides: synthesis of β-4′-thio-oligouridylates, nuclease resistance, base pairing properties, and interaction with HIV-1 reverse transcriptase. Nucleic Acids Res. 1993;21:1587–1593. doi: 10.1093/nar/21.7.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leydier C, Bellon L, Barascut JL, Morvan F, Rayner B, Imbach JL. 4′-Thio-RNA: synthesis of mixed base 4′-Thio-oligoribonucleotides, nuclease resistance, and base pairing properties with complementary single and double strand. Antisense Res. Dev. 1995;5:167–174. doi: 10.1089/ard.1995.5.167. [DOI] [PubMed] [Google Scholar]
- 22.Jones GD, Lesnik EA, Owens SR, Risen LM, Walker RT. Investigation of some properties of oligodeoxy-nucleotides containing 4′-thio-2′-deoxynucleotides: duplex hybridization and nuclease sensitivity. Nucleic Acids Res. 1996;24:4117–4122. doi: 10.1093/nar/24.21.4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoshika S, Minakawa N, Matsuda A. Synthesis and physical and physiological properties of 4′-thio-RNA: application to post-modification of RNA aptamer toward NF-κB. Nucleic Acids Res. 2004;32:3815–3825. doi: 10.1093/nar/gkh705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoshika S, Minakawa N, Kamiya H, Harashima H, Matsuda A. RNA interference induced by siRNAs modified with 4′-thioribonucleosides in cultured mammalian cells. FEBS Lett. 2005;579:3115–3118. doi: 10.1016/j.febslet.2005.04.073. [DOI] [PubMed] [Google Scholar]
- 25.Inoue N, Minakawa N, Matsuda A. Synthesis and properties of 4′-ThioDNA: unexpected RNA-like behavior of 4′-ThioDNA. Nucleic Acids Res. 2006;34:3476–3483. doi: 10.1093/nar/gkl491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiu YL, Rana TM. siRNA function in RNAi: a chemical modification analysis. RNA. 2003;9:1034–1048. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoshimura Y, Kitano K, Yamada K, Satoh H, Watanabe M, Miura S, Sakata S, Sasaki T, Matsuda A. A novel synthesis of 2′-modified 2′-deoxy-4′-thiocytidines from d-glucose. J. Org. Chem. 1997;62:3140–3152. doi: 10.1021/jo9700540. [DOI] [PubMed] [Google Scholar]
- 28.Yoshimura Y, Endo M, Miura S, Sakata S. An alternative synthesis of the antineoplastic nucleoside 4′-ThioFAC and its application to the synthesis of 4′-ThioFAG and 4′-thiocytarazid. J. Org. Chem. 1999;64:7912–7920. [Google Scholar]
- 29.Yoshimura Y, Endo M, Sakata S. An alternative synthesis of antineoplastic 4′-thiocytidine analogue 4′-thioFAC. Tetrahedron Lett. 1999;40:1937–1940. [Google Scholar]
- 30.Yoshimura Y, Kitano K, Yamada K, Sakata S, Miura S, Ashida N, Machida H. Synthesis and biological activities of 2′-deoxy-2′-fluoro-4′-thioarabinofuranosylpyrimidine and -purine nucleosides. Bioorg. Med. Chem. 2000;8:1545–1558. doi: 10.1016/s0968-0896(00)00065-1. [DOI] [PubMed] [Google Scholar]
- 31.Wu H, Lima WF, Crooke ST. Properties of cloned and expressed human RNase H1. J. Biol. Chem. 1999;274:28270–28278. doi: 10.1074/jbc.274.40.28270. [DOI] [PubMed] [Google Scholar]
- 32.Dowler T, Bergeron D, Tedeschi A-L, Paquet L, Ferrari N, Damha MJ. Improvements in siRNA properties mediated by 2′-deoxy-2′-fluoro-β-d-arabinonucleic acid (FANA) Nucleic Acids Res. 2006;34:1669–1675. doi: 10.1093/nar/gkl033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Novac O, Guenier A-S, Pelletier J. Inhibitors of protein synthesis identified by a high throughput multiplexed translation screen. Nucleic Acids Res. 2004;32:902–915. doi: 10.1093/nar/gkh235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 35.Baker BF, Lot SS, Condon TP, Cheng-Flournoy S, Lesnik EA, Sasmor HM, Bennett CF. 2′-O-(2-methoxy)ethyl-modified anti-intercellular adhesion molecule 1 (ICAM-1) oligonucleotides selectively increase the ICAM-1 mRNA level and inhibit formation of the ICAM-1 translation initiation complex in human umbilical vein endothelial cells. J. Biol. Chem. 1997;272:11994–12000. doi: 10.1074/jbc.272.18.11994. [DOI] [PubMed] [Google Scholar]
- 36.Freier SM, Altmann K-H. The ups and downs of nucleic acid duplex stability: structure-stability studies on chemically-modified DNA:RNA duplexes. Nucleic Acids Res. 1997;25:4429–4443. doi: 10.1093/nar/25.22.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishizaki T, Iwai S, Ohtsuka E, Nakamura H. Solution structure of an RNA · 2′-O-methylated RNA hybrid duplex containing an RNA · DNA hybrid segment at the center. Biochemistry. 1997;36:2577–2585. doi: 10.1021/bi962297c. [DOI] [PubMed] [Google Scholar]
- 38.Viazovkina E, Min K-L, Galarneau A, Damha MJ. Synthesis and properties of oligonucleotide chimeras containing 5′-amino-2′-deoxy-2′-fluoroarabinonucleosides. Nucleosides Nucleotides Nucleic Acids. 2003;22:1335–1338. doi: 10.1081/ncn-120022959. [DOI] [PubMed] [Google Scholar]
- 39.Tedeschi A-L. M.Sc. Thesis. Montreal: McGill University; 2004. [Google Scholar]
- 40.Wilds CJ, Damha MJ. 2′-Deoxy-2′-fluoro-β-d-arabinonucleosides and oligonucleotides (2′F-ANA): synthesis and physicochemical studies. Nucleic Acids Res. 2000;28:3625–3635. doi: 10.1093/nar/28.18.3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ratmeyer L, Vinayak R, Zhong YY, Zon G, Wilson WD. Sequence specific thermodynamic and structural properties for DNA · RNA duplexes. Biochemistry. 1994;33:5298–5304. doi: 10.1021/bi00183a037. [DOI] [PubMed] [Google Scholar]
- 42.Minasov G, Teplova M, Nielsen P, Wengel J, Egli M. Structural basis of cleavage by RNase H of hybrids of arabinonucleic acids and RNA. Biochemistry. 2000;39:3525–3532. doi: 10.1021/bi992792j. [DOI] [PubMed] [Google Scholar]
- 43.Trempe J-F, Wilds CJ, Denisov AY, Pon RT, Damha MJ, Gehring K. NMR solution structure of an oligonucleotide hairpin with a 2′F-ANA/RNA stem: implications for RNase H specificity toward DNA/RNA hybrid duplexes. J. Am. Chem. Soc. 2001;123:4896–4903. doi: 10.1021/ja003859p. [DOI] [PubMed] [Google Scholar]
- 44.Rose SD, Kim D-H, Amarzguioui M, Heidel JD, Collingwood MA, Davis ME, Rossi JJ, Behlke MA. Functional polarity is introduced by Dicer processing of short substrate RNAs. Nucleic Acids Res. 2005;33:4140–4156. doi: 10.1093/nar/gki732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jackson AL, Burchard J, Leake D, Reynolds A, Schelter J, Guo J, Johnson JM, Lim L, Karpilow J, et al. Position-specific chemical modification of siRNAs reduces ‘off-target’ transcript silencing. RNA. 2006;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 47.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 48.Hohjoh H. Enhancement of RNAi activity by improved siRNA duplexes. FEBS Lett. 2004;557:193–198. doi: 10.1016/s0014-5793(03)01492-3. [DOI] [PubMed] [Google Scholar]