Abstract
The complex and integrated nature of both genetic and protein level factors influencing recombinant protein production in Escherichia coli makes it difficult to predict the optimal expression strategy for a given protein. Here, two combinatorial library strategies were evaluated for their capability of tuning recombinant protein production in the cytoplasm of E. coli. Large expression vector libraries were constructed through either conservative (ExLib1) or free (ExLib2) randomization of a seven-amino-acid window strategically located between a degenerated start codon and a sequence encoding a fluorescently tagged target protein. Flow cytometric sorting and analyses of libraries, subpopulations or individual clones were followed by SDS-PAGE, western blotting, mass spectrometry and DNA sequencing analyses. For ExLib1, intracellular accumulation of soluble protein was shown to be affected by codon specific effects at some positions of the common N-terminal extension. Interestingly, for ExLib2 where the same sequence window was randomized via seven consecutive NN(G/T) tri-nucleotide repeats, high product levels (up to 24-fold higher than a reference clone) were associated with a preferential appearance of novel SD-like sequences. Possible mechanisms behind the observed effects are discussed.
INTRODUCTION
Numerous expression vector systems have been developed for efficient recombinant protein production using the bacterium Escherichia coli as host. This involves, for example, the use of different plasmid vector backbones, promoters, ribosomal-binding sites (RBS), gene fusion partners, transcriptional terminators and antibiotic resistance markers (1,2). Although a vast number of reports are describing successful use of these systems for production of recombinant proteins of different origin and characteristics, others have reported on frequently encountered problems like no or low product formation, misfolding/inclusion body formation or proteolytic degradation (3,4). If such problems are observed using a particular expression vector system, the use of different inducer concentrations, host cell mutants, growth media, cultivation temperatures and co-expression of folding factors can be tested for improved production of the target protein (1–3). Alternatively, the target protein-encoding gene can be transferred to a different expression vector system. However, with relatively few exceptions, it is difficult to predict what effects a particular system or set of conditions will have on the production of a given protein which typically leads to an empirical testing of several systems.
Many gene-specific effects on the protein production involve post-transcriptional events. For example, the mRNA sequence element denoted Shine–Dalgarno (5), located upstream of the initiation codon interacts with a complementary sequence (anti-SD) in the 3′-end of the 16S rRNA in the ribosomal 30S subunit during translation initiation complex formation (6). Strong and gene-specific mRNA secondary structures can mask the SD sequence and thereby reduce the accessibility of the RBS which influences the translation efficiency (7–12). Further, reports concerning influence on translation initiation by different mRNA determinants have emphasized the importance of the strength of the interaction between the SD sequence and the complementary anti-SD, the identity of the initiation codon and the spacing between these two sequence elements (6,13). RBS sequences can also appear within the cDNA of target proteins, compete for ribosome binding and interfere with protein translation causing low or no gene expression (14).
It has also been established that there is a bias in the codon usage in E. coli, where so-called major codons, as opposed to rare or minor codons, are more frequently represented in highly expressed genes than in genes being expressed at low levels (15). The nucleotide and codon composition in the early coding region of a gene appears to be especially important for gene expression (13,16–21), although clusters and even unique rare codons located further down in the structural gene also can have effects like ribosome stalling, frame shifting and premature translation termination (15,22,23). Codon-specific translation rates may likewise influence the in vivo protein folding, where the presence of rare (i.e. more slowly read) codons in specific regions of the structural gene can be beneficial for the folding process (24).
In this work we have investigated two related but different combinatorial strategies to obtain a post-transcriptional tuning of recombinant E. coli protein production and how this affected the soluble production of a fluorescently tagged product protein. Based on a reference eight-amino-acid translation initiation peptide (TrpL) fused to an enhanced green fluorescent protein (EGFP)-based reporter fusion protein, large expression vector libraries were constructed in which the TrpL-encoding sequence was either silently mutated into all possible genetic combinations encoding the same peptide sequence or more freely randomized allowing for the appearance of either any eight-amino-acid N-terminal extension or novel un-translated mRNA sequences which could influence the expression on the nucleotide level. Flow cytometric analyses of libraries, sorted subpopulations and individual clones were utilized to study effects on soluble protein product levels. Clonal analyses by DNA sequencing, real-time PCR, western blotting, mass spectrometry and N-terminal sequencing indicated different possible mechanisms behind observed variations.
MATERIALS AND METHODS
Bacterial strain, enzymes and oligonucleotides
The E. coli strain RRIΔM15 (25) was used both in cloning work and for recombinant protein production. Enzymes were purchased from New England BioLabs or Fermentas and used according to the suppliers’ recommendations. Oligonucleotides employed for DNA sequencing, vector and library constructions and real-time PCR were ordered from MWG Biotech or Scandinavian Gene Synthesis AB. Recombinant DNA techniques were performed according to standard methods (26).
Vector construction
pUC-TrpL-ZEGFP
A portion of plasmid pZEGFP, based on the pUC19-derivative pEGFP (Clontech, Inc.), and containing a gene encoding the IgG binding Z domain (27) in fusion with the gene encoding EGFP was PCR amplified using an upstream oligonucleotide Band-1 (5′-CTGGCACGACAGGTTTCC) encoding part of a blunt end PvuII restriction site and two downstream and partially overlapping oligonucleotides Band-3a (5′-GCTTTCATAGAGCTCGATACCCTTTGTGAAATTGTTATCCGCTC) and Band-3b (5′-CCCCAAGCTTTTCAGTACGAAAATTGCTTTCATAGAGCTCGATAC). The resulting PCR product was digested with HindIII and religated with a PvuII/HindIII restricted pZEGFP vector, to yield pUC-TrpL-ZEGFP containing the SD sequence AAGG followed by a SacI restriction site, a sequence corresponding to the eight initial amino acids of the E. coli tryptophan operon leader peptide (trpL;(28)) and a HindIII restriction site.
pBR-TrpL-ZEGFP
For construction of pBR-TrpL-ZEGFP, the low copy number plasmid pBR322 (Fermentas) was digested with EcoRI, treated with Klenow fragment to create blunt ends and subsequently restricted with Eco52I. A fragment starting upstream from the promoter region and covering the complete sequence of the ZEGFP encoding part was excised from pUC-TrpL-ZEGFP with PvuII and NotI and inserted into the prepared pBR322 vector to yield pBR-TrpL-ZEGFP (Figure 1A and B).
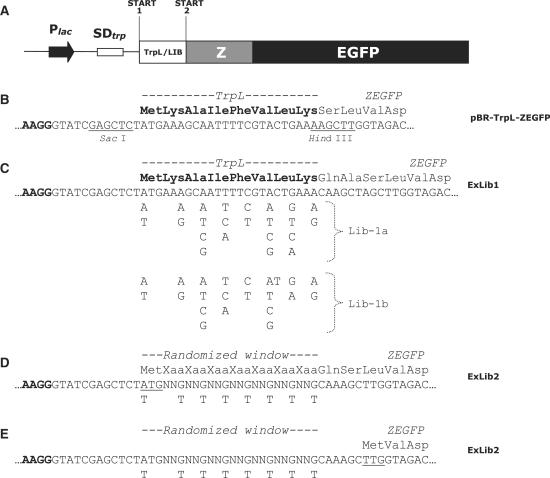
Figure 1.
Schematic representation of the expression cassette and the library designs. (A) Block diagram of the expression cassette showing (i) the E. coli lac promoter (Plac), (ii) the E. coli trp operon-derived SD sequence (SDtrp), (iii) the gene sequence corresponding to either the eight first amino acids of the wild-type TrpL peptide (reference vector), or the corresponding variegated sequence window of any library member from ExLib1 or ExLib2, (iv) the gene encoding the IgG-binding Z domain (Z) and (v) the gene encoding the EGFP. The two alternative start codons are indicated (Start 1 and Start 2); (B) DNA and deduced amino acid sequence of the translation initiation region of the reference vector pBR-TrpL-ZEGFP discussed in the text. Recognition sites for restriction enzymes discussed in the materials and methods section are indicated as underlined; (C) Description of the randomization design used to construct the ExLib1 library, showing location and nature of nucleotide variations introduced via the use of the two degenerate oligonucleotides Lib-1a and Lib-1b. Note: In order to be able to include all six codons for Leu at position +7, two different oligonucleotides (Lib-1a and Lib-1b) were used in the library construction (in a 2:1 mixture); (D, E) Description of the randomization design used to construct the ExLib2 library, showing location and nature of nucleotide variations introduced via the use of the degenerate oligonucleotide Lib-2 (N = A, G, C or T). The design of this library allowed for two in vivo scenarios, involving either a translational start at the first start codon (underlined in Figure 1D) or at the second start codon (underlined in Figure 1E). See text for details.
ExLib2-Opt7stop
For the introduction of a stop codon between the Z target protein and the EGFP reporter in clone ExLib2-Opt7, the DNA fragment located between the sequences of the Z and EGFP genes was excised from the parental vector using BamHI and NcoI and replaced with a linker of identical length and with compatible protruding sticky ends. The linker was created by annealing oligonucleotides Band-23 (5′-GATCCCTAAAGCCCGGTCGCCAC) (in-frame stop codon underlined) and Band-24 (5′-CATGGTGGCGACCGGGCTTTAGG).
Back-transfer of library sequence fragment to an original vector preparation
For clone ExLib2-Opt7 the DNA region corresponding to the library sequence and the coding sequence for protein Z, was back-transferred to a pBR-TrpL-ZEGFP vector backbone preparation which had not been subjected to flow cytometric sorting, via digestion with SacI and NcoI. The purified DNA fragment was then re-ligated with the pBR-TrpL-ZEGFP vector fragment, previously digested with the same enzymes.
Library construction
The degenerate oligonucleotides Lib-1a (5′-CCCCAAGGGTATCGAGCTCT(a/t)TG AA(a/g)GC(a/t/c/g)AT(t/c/a)TT(c/t)GT(a/t/c/g)CT(g/t/c/a)AA(a/g)CAAGCTTGGTAGACAACCCC; the variegated nucleotides in degenerate positions are shown in small characters) and Lib-1b (5′-CCCCAAGGGTATCGAGCTCT(a/t)TGAA(a/g)GC(a/t/c/g) AT(t/c/a)TT(c/t)GT(a/t/c/g)TT(g/a)AA(a/g)CAAGCTTGGTAGACAACCCC) were used for construction of an early version of ExLib1, via mixing at a molar ratio 2:1. For the second library (ExLib2) the degenerate oligonucleotide Lib-2 (5′-CCCCAAGGGTATCGAGCTCT(a/t)TGNNKNNKNNKNNKNNKNNKNNKCAAAGCTTGGTAGACAACCC) was used. These three library oligonucleotides have surrounding constant regions containing an upstream SacI and a downstream HindIII site. To obtain double-stranded library fragments, the first library oligonucleotide strand served as a template for second strand synthesis by Klenow DNA polymerase after annealing of Band-4 (5′-GGGGTTGTCTACCAAGCTTG) (ExLib1) or Band-7 (5′-GGGTTGTCTACCAAGCTTTG) (ExLib2), respectively, to the non-variegated downstream region. The resulting DNA fragments were digested with SacI, purified with CHROMA SPIN™-30 Columns (Clontech) to remove unwanted restriction products and ligated to the linearized SacI/HindIII cleaved pBR-TrpL-ZEGFP plasmid. After HindIII digestion of the vector-anchored library fragments, the vector was recirculized using T4 DNA ligase. The resulting plasmid library pools were transformed into electrocompetent E. coli cells by electroporation with a Bio-Rad Gene Pulser™ (Bio-Rad Laboratories). Approximately 100 ng DNA ligation mixture was used for electroporation of each 100 μl vial of electrocompetent cells. Transformants were subsequently incubated for 50 min without shaking at 37°C in tryptic soy broth medium (TSB; Merck) supplemented with 5 g l−1 yeast extract (Merck), 2% glucose, 10 mM MgCl2, 10 mM MgSO4, 10 mM NaCl and 2.5 mM KCl, before portions of the libraries were titrated on ampicillin-selective agar plates. The remaining library pools were inoculated into 100 (ExLib1) or 500 (ExLib2) ml TSB supplemented with yeast extract, 2% glucose and relevant antibiotic and grown overnight at 37°C in shake flasks. Cells harboring the individual libraries were finally harvested, resuspended in phosphate-buffered saline (PBS) containing 50% glycerol and stored at −80°C until use. Plasmid preparation was carried out from cell cultures containing the early version of the ExLib1 library. The vectors were digested with HindIII, treated with Klenow DNA polymerase to fill in protruding ends and re-ligated. This rendered the degenerate sequence in the resulting library (ExLib1) in a correct reading frame with the downstream ZEGFP-encoding sequence. The total number of transformants obtained for the final version of ExLib1 and ExLib2 were 9 × 107 and 2 × 108, respectively.
DNA sequencing
DNA sequencing to verify correct cloning of constructs and for analysis of library member sequences was performed using primers Band-1 (see above), Band-5 (5′-CGCTTTGGCTTGGGTCATCT) and Band-6 (5′-CAGCATGGCCTGCAACGC) and an ABI Prism 3700 DNA analyzer (Applied Biosystems).
Real-time PCR
Oligonucleotides for real-time PCR analysis of plasmid DNA and genomic DNA were targeted at the beta-lactamase encoding bla gene and the 16S rDNA gene, respectively and were designed as described elsewhere (29). Thawed samples of induced overnight cultured cells were diluted 1:1000 in sterile water. One microliter samples of such cell suspensions were included in 25 μl PCR reactions, containing 12.5 pmol each of forward and reverse primers and 12.5 μl of 2 × iQ™ SYBR® Green Supermix (Bio-Rad Laboratories). Real-time PCR was performed with an iCycler (Bio-Rad Laboratories) instrument with the cycling protocol 2 min at 50°C, 3 min at 95°C followed by 40 cycles of 15 s at 95°C and 45 s at 60°C. At the end of the amplification reactions a melt curve analysis was performed by 20 min ramping of the temperature from 60°C to 95°C. The ΔΔCT method (30) was applied to calculate relative plasmid copy number for clones harboring library expression vectors, where the genomic DNA served as internal standard and cells containing plasmid pBR-TrpL-ZEGFP as the reference.
Culturing of libraries and individual clones
Both libraries and individual clones (for either flow cytometric analyses or for SDS-PAGE/western blotting/affinity purification/real-time PCR) were cultured according to the same basic protocol. Aliquots of thawed library stocks (covering the respective total library size) or overnight cultures of individual clones were diluted approximately 500 times by inoculation into fresh 10 or 25 ml TSB medium supplemented with 5 g l−1 yeast extract and 100 mg l−1 ampicillin and cultivated in shake flasks at 30°C or 37°C. When an OD600 nm between 0.5 and 1.0 was reached, recombinant protein production was induced by addition of isopropyl-beta-d-thiogalactopyranoside (IPTG) to a final concentration of 1 mM and the cultivation was allowed to proceed for either 4.5 h (sorting of low, medium and high clones) or 18–20 h.
Flow cytometric analysis and sorting
For flow cytometric measurements, approximately 100 μl of induced overnight cell cultures were gently harvested, washed twice with 1 ml PBS and diluted 200 times in the same buffer. Cells were analyzed and sorted using a FACSVantage SE flow cytometer (BD Biosciences). Alignment of the argon ion laser was performed with AlignFlow™ flow cytometry alignment beads for 488 nm (molecular probes). Histograms were recorded from 10 000 cells at a rate of approximately 500 cells s−1 using standard procedures. Library cells, chilled on ice, were sorted in Normal-R mode into 1 ml TSB medium supplemented with yeast extract and transferred to 30 or 37°C for approximately 1 h incubation with shaking. Sorted and incubated cells were further inoculated into 10 ml TSB medium supplemented with yeast extract and ampicillin and re-cultivated first overnight and then according to above described procedures at 30 or 37°C before next round of flow cytometric analysis. Approximately 3500 cells from the fluorescence intervals denoting low, medium and high, respectively (see results section), were sorted out at a rate of 300–500 cells s−1. For the two-round sorting of library populations conferring the highest fluorescence intensities, sorting was carried out for 1 h at a rate of 5000 cells s−1 and the resulting tubes with sorted cells were transferred to incubation conditions every 15 min. CellQuest software (BD Biosciences) was used to analyze flow cytometric data.
Protein purification and analysis
Induced overnight cultures were centrifuged and pelleted cells were re-suspended in a double volume of PBS and disrupted by sonication using a sonicator (Vibra cell™, Sonics and materials, Inc.) at 60% duty cycle for 3 min with 1.0 s pulses. A 5 ml IgG Sepharose™ 6 Fast Flow matrix was utilized for protein purification. Before loading of samples on the column, cell debris was removed by centrifugation and filtration (0.45 µm) and the buffer was adjusted to give a final concentration of 25 mM Tris-HCl pH 8.5, 150 mM NaCl, 1.25 mM EDTA and 0.05% Tween-20. The columns were washed with loading buffer and the absorbance at 280 nm of HAc eluted fractions was determined. Extinction coefficients were calculated using ExPASy. Fractionation of soluble and insoluble proteins for SDS-PAGE and western blot analysis was performed by centrifugation of 1 ml of previously sonicated samples at 10 000 rpm in a micro-centrifuge. PBS was used to wash the pellet twice and was then added to the soluble fraction prior to concentration by lyophilization. Samples of both soluble and insoluble materials were dissolved in a volume of beta-mercaptoethanol containing SDS-PAGE loading buffer corresponding to 1:100 of the original cultivation volume. One microliter of these samples were applied to NuPAGE™ 4–12% Bis-Tris Gels (Invitrogen™), which subsequently were stained with GelCode® Blue Stain Reagent (Pierce). For western blotting, PVDF membranes (Invitrogen™) with electrophoretically transferred proteins were blocked with 5% milk powder dissolved in PBS supplemented with 0.05% Tween-20. As primary antibody a 1:2000 dilution of anti-green fluorescent protein rabbit polyclonal antibody (Invitrogen™) was used. Polyclonal goat anti-rabbit immunoglobulin conjugated to horseradish peroxidase (DakoCytomation), diluted 1:3000 served as secondary antibody. SuperSignal West Dura Extended Duration Substrate (Pierce) was used as detection system and images of the chemiluminiscence were aquired by a Chemi Doc™ Gel Documentation system (Bio-Rad Laboratories Inc.). The five N-terminal amino acids in the IgG-affinity purified protein produced by the clone ExLib2-Opt7 were determined by Edman degradation (Protein Analysis Center, Stockholm, Sweden). Mass spectra were recorded on a MALDI-TOF MS biflexIV instrument (Bruker Daltonics).
Sequence analysis
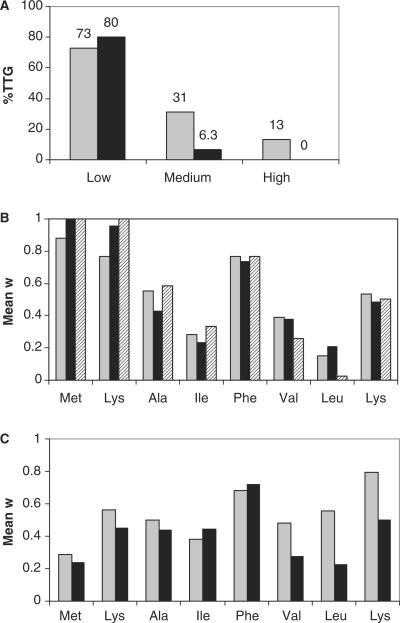
Codon adaptation index (CAI) values were calculated as described previously (31). Because no relative adaptiveness value (w) was available for the TTG codon when used as initiation codon and decoded as methionine, it was in our study given the value 0.05 to still be able to calculate CAI for the library member sequence variants. We based this on the infrequent use of TTG as initiation codon in our study (13% in ExLib1-High (30°C) and 0% in ExLib1-High (37°C), Figure 2A) and on the relative synonymous codon usage (RSCU) values assigned to so-called ‘rare’ codons in an earlier report (32).
Figure 2.
Results from statistical analyses of codons in TrpL variants corresponding to ExLib1 clones of different fluorescence intensity values. (A) Fractions (%) of investigated clones utilizing a TTG rather than an ATG codon in the first position of the randomized sequence. In conjunction with the sorting, cells were grown at either 30°C (gray bars) or 37°C (black bars). The designations low, medium and high correspond to three different fluorescence intervals (see text); (B, C) Position-specific codon biases. Position-specific mean w values were calculated for TrpL encoding sequence variants (n = 16) of categories of clones from ExLib1, sorted for the indicated fluorescence intensity intervals and grown at indicated temperatures. Individual codons with high w values reflect their overrepresentation in highly expressed E. coli proteins (31). In (B), gray, black and striped bars indicate ExLib1-High clones grown at 30°C, ExLib1-High clones grown at 37°C and ExLib1-Opt clones grown at 37°C, respectively. In (C), gray and black bars indicate ExLib1-Low clones grown at 30 and 37°C, respectively.
RESULTS
Design of expression vector libraries
Two separate E. coli expression vector libraries (ExLib1 and ExLib2) were constructed and electroporated into E. coli host cells for sorting and analysis of cell populations as well as individual clones on basis of their soluble intracellular protein product levels. All library vector constructs contained (i) an E. coli lac promoter; (ii) a trp operon-derived SD sequence followed downstream by (iii) a 21 (3 × 7) nucleotide long window of differently variegated positions (see below) following an initially placed first potential translational start codon triplet ((A/T)TG, see below) and (iv) a cassette encoding a fluorescent fusion protein product consisting of a 6 kDa IgG-binding target protein Z (27) fused to an enhanced green fluorescent protein (EGFP) reporter (33), connected via a twelve-residue linker (Figure 1). The included EGFP moiety has earlier been described as a useful reporter of intracellular levels of soluble protein (34).
For the construction of the ExLib1 library clones, the variegation of the 24 nucleotide window was designed such that all resulting library members encoded a common eight amino acid translation initiation peptide (MKAIFVLK) derived from the E. coli trp operon (TrpL leader sequence;(28)), albeit by different combinations of position-specific synonymous codons (Figure 1C). This sequence has earlier been demonstrated to promote an efficient translation initiation, leading to high product levels of recombinant protein (35). The first nucleotide of the original ATG initiation triplet in the trpL sequence was genetically doped with an equal amount of T, allowing for the less frequently used initiation codon TTG to also appear in library clones. The frequency with which this codon is used as initiation codon in E. coli is only 1% (1). Although it has been shown to be poorly efficient as initiation codon (13), it can be as favorable as ATG if followed by a certain codon context (19). Electroporation of E. coli resulted in approximately 9 × 107 transformants and the theoretical diversity of the library (4608 different possible variants) could thus be considered duly covered. DNA sequencing of 23 randomly picked colonies showed that 83% of the constructs contained correct library window sequences in reading frame with the ZEGFP fusion protein. All clones in this library would thus be expected to encode a common protein product, and any differences in cellular fluorescence intensities should be possible to attribute to the use of different codons in the N-terminal region allowing for effects related to one or several levels, including codon usage, mRNA stability, mRNA secondary structure and translation initiation efficiency.
In ExLib2, the 21 nucleotide window following the same alternative translational start (A/T)TG triplet was instead randomized using seven consecutive NN(G/T) triplets, allowing for a significantly larger genetic freedom. If recruited as codons for translation, each such triplet includes 32 possible codons covering all 20 amino acids as well as one of the stop codons (TAG), theoretically allowing for the ZEGFP protein to be extended at the N-terminal by any of 1.2 × 109 different peptide variants encoded by 6.8 × 1010 genetic variants (Figure 1D). In addition, the presence of a TTG codon downstream of the randomized window (Figure 1E) would potentially allow for the recruitment of this codon as an alternative translational start in a fraction of the ExLib2 library clones, provided the occurrence of a suitably positioned SD sequence. Thus, in comparison with the ExLib1 library, the design of the more complex ExLib2 library includes additional sequence-dependent features on both the nucleotide and protein level with potential to influence the production of the ZEGFP fusion protein. Electroporation of E. coli resulted in a library size of approximately 2 × 108 transformants, of which 82% (18/22) were found to contain correct inserts as analyzed by DNA sequencing.
Initial flow cytometric analysis of the libraries
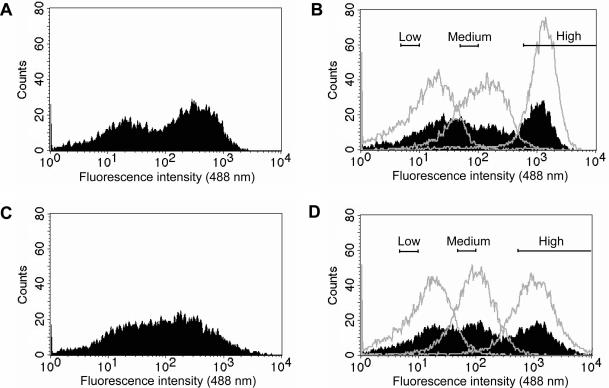
For initial library characterization cell cultures corresponding to either ExLib1 or ExLib2 were grown in shake flasks and induced for 4.5 h at 30 or 37°C, washed and analyzed for whole cell fluorescence in a flow cytometer. The results showed a wide distribution in fluorescence intensity for both ExLib1 and ExLib2 (Figure 3A and C), spanning more than three orders of magnitude. Using three different relative fluorescent intensity gate intervals, subpopulations denoting low (5–10 interval), medium (50–100 interval) and high (500–10 000 interval) from both libraries were sorted and collected in separate pools. To investigate if the observed fluorescence intensities for the subpopulations were likely to be linked to particular clonal characteristics or were merely reflecting statistical variation, the sorted pools from each library were re-cultured using the same protocol either as separate sub-pools or mixed (all three pools together), followed by flow cytometric analysis. Interestingly, the results showed that the separately re-cultured subpopulations from both libraries retained fluorescence intensities in parity with the gate values used for their isolation, indicating that these values were linked to clone-dependent characteristics (Figure 3B and D). In addition, the analysis of the co-cultured subpopulations indicated that the clonal traits had been preserved also under these conditions, in that, three subpopulations could be spotted in the flow cytometer histograms (Figure 3B and D). Sixteen individual clones from each of the twelve different populations (ExLib1/2; 30/37°C; low/medium/high) were collected and subjected to DNA sequencing (see below).
Figure 3.
Histograms from initial flow cytometric analyses of ExLib1 and ExLib2. Induced shake flask cultures were harvested after 4.5 h of induction and analyzed. (A) Analysis of ExLib1; (B) Analysis of previously isolated and here re-cultured ExLib1 subpopulations denoted low, medium and high (gates once used for the sorting are shown), cultured either in separate flasks (non-filled histograms) or together in a common flask (co-culture) (filled histogram); (C) Analysis of ExLib2; (D) Analysis of previously isolated and here re-cultured ExLib2 subpopulations denoted low, medium and high (gates once used for the sorting are shown), cultured either in separate flasks (non-filled histograms) or together in a common flask (co-culture) (filled histogram). The pre-amplifier gain was set to 799 V and the experiments were performed at least twice with similar results (data not shown).
To exclude the possibility that the observed effects in whole cell fluorescence were due to any altered properties of the bacterial host, rather than the library plasmids, control experiments were performed. Here, plasmid preparations from cells that had been sorted out from the low, medium and high intervals were re-transformed into fresh cells and the experiment was repeated. No significant differences in the resulting fluorescence properties between these cultures and the cultures analyzed without intervening plasmid preparation could be observed (data not shown). The results show that the varying fluorescence intensity displayed by different library member clones most likely reflects effects resulting from sequence differences in their respective expression cassettes. Notably, in both libraries there was an approximate 1000-fold span in whole cell fluorescence intensity between clones showing that both randomization strategies for the libraries seem to have a dramatic influence on the soluble product levels of the fusion protein.
Isolation of high-fluorescence library clones
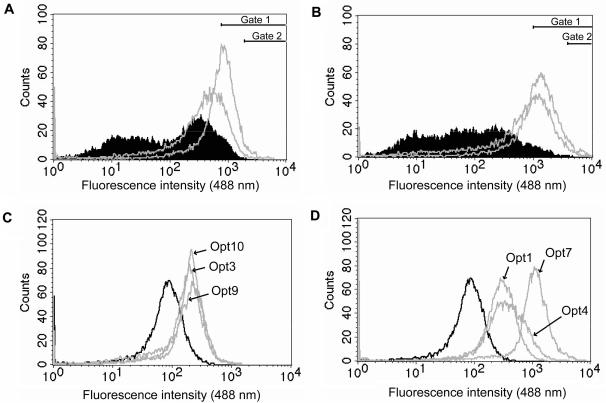
To isolate individual clones (optimized or Opt clones) from the two libraries showing markedly increased fluorescence intensities compared to the reference clone pBR-TrpL-ZEGFP, two consecutive rounds of flow cytometric sorting at relatively high gate values, with intervening re-cultivations (at 37°C) were performed (Figure 4A and B). Interestingly, after the second sorting round the resulting subpopulations of the libraries showed a distribution in whole cell fluorescence intensity similar to cultures of individual clones. Ten randomly picked individual colonies from each of these two-round sorted library pools were re-cultured according to the previously used protocol and re-analyzed in the flow cytometer (Figure 4C and D). Some of the clones from the ExLib1 library showed a 3–4-fold increase in mean whole cell fluorescence compared to the reference clone pBR-TrpL-ZEGFP (Figure 4C, Table 1). For the clones originating from the ExLib2 library, all had higher fluorescence intensities than the reference clone (typically 4–6-fold increases) and one clone (denoted ExLib2-Opt7) showed a 16-fold higher whole cell fluorescence intensity than the reference construct (Figure 4D, Table 1). Clones with high fluorescence intensity values from both libraries were subjected to DNA sequencing (see below and Table 1).
Figure 4.
Histograms from flow cytometric sortings of optimized fluorescence clones (Opt) from ExLib1 and ExLib2. (A) Overlay plots from analyses of the original ExLib1 library (filled histogram) and re-cultured subpopulations of the libraries (gray) from after the first (left) and second (right) sorting round for highly fluorescent cells using 488 nm fluorescence gate values of >800 (gate 1) and >2000 (gate 2), respectively, (B) Overlay plots from analyses of the original ExLib2 library (filled histogram) and re-cultured subpopulations of the libraries (gray) from after the first (left) and second (right) sorting round for highly fluorescent cells using 488 nm fluorescence gate values of >1000 (gate 1) and >4000 (gate 2), respectively; (C) Overlay plots from analyses of the individual clones from the ExLib1 library (gray histograms) and the reference clone pBR-TrpL-ZEGFP (black histogram); (D) Overlay plots from analyses of the individual clones from the ExLib2 library (gray histograms) and the reference clone pBR-TrpL-ZEGFP (black histogram). The pre-amplifier gain was set to 600 V (A and B) or 500 V (C and D).
Table 1.
Listing of some characteristics for a selection of clones from ExLib1 and ExLib2a
| Clone ID | Sequence (5′–3′)b | Startc | RelPCd | RelFe |
|---|---|---|---|---|
| ExLib1-Low8 | ATGAAAGCGATCT TCGTGCTGAAG | nd | ||
| ExLib1-Low9 | ATGAAGGCCATAT TCGTGCTCAAA | nd | ||
| ExLib1-Low2 | TTGAAGGCAATAT TCGTCCTCAAA | nd | ||
| ExLib1-High3 | ATGAAAGCAATAT TCGTATTAAAG | nd | ||
| ExLib1-High2 | ATGAAAGCTATTT TTGTACTCAAG | nd | ||
| ExLib1-High1 | ATGAAAGCCATCT TCGTGTTAAAG | nd | ||
| ExLib1-Opt9 | ATGAAAGCAATAT TCGTACTCAAG | 1:st | 1.1 | 4.3 |
| ExLib1-Opt10 | ATGAAAGCAATCT TTGTCTTGAAA | 1:st | 1.3 | 4.0 |
| ExLib1-Opt3 | ATGAAAGCAATAT TCGTGTTGAAG | 1:st | 1.3 | 3.0 |
| ExLib2-Low4 | ATGGTGTGGGGTA GGGAGCATCAG | nd | ||
| ExLib2-Low1 | TTGGGGGGTACGC GGGGTCAGGCT | nd | ||
| ExLib2-Low7 | ATGGCGGCTACGT CGAAGCCGGTG | nd | ||
| ExLib2-High3 | ATGAAGAATAGGT CGACGCAGCAG | 1:st | 1.3 | 2.7 |
| ExLib2-High1 | ATGTTTAAGGGGG GGGAGGGGGTT | 2:nd | ||
| ExLib2-High5 | ATGTTGGCGGCGA TTGAGGGGAAG | 2:nd | ||
| ExLib2-Opt7 | ATGGTGGATGGTC TGAAGAGGGGG | 2:nd | 2.7 | 15.8 |
| ExLib2-Opt1 | ATGAGTGATCCTA GTAGGAGGGGG | 2:nd | 1.6 | 5.5 |
| ExLib2-Opt4 | ATGAGTAGTCAGG GGTTGAGGAGT | 2:nd | 1.0 | 4.7 |
| ExLib2-Opt5 | ATGACGTAGCATC TGAATAAGGAG | nd | ||
| ExLib2-Opt6 | ATGTAGGTGAAGA TGGGGGAGGTT | nd | ||
| ExLib2-Opt10 | ATGGGTAGGGCCG TGAGGAGGAG | nd | ||
| ExLib2-Opt9 | ATGCGGGAGCGTG AGACGGGGGAG | nd | ||
| ExLib2-Opt3 | ATGAAGACGTCGC GGGGGGAGTAG | nd | ||
| ExLib2-Opt8 | TTGGCGAAGGGGA AGTTGATGATG | nd | ||
| ExLib2-Opt2 | TTGAATTGGAGGA AGGTGAGGGAG | nd |
aClones within each group are listed according to their mean fluorescence intensity values as measured by flow cytometry (first = highest fluorescence).
bSequences of the 24 nucleotide windows subjected to the variegation. Putative SD sequences are indicated in bold for clones of which purified protein products have been analyzed by mass spectrometry. The sequence giving the highest number of continuous bases complementary to the CCUCC core of the E. coli anti-SD sequence ACCUCCUUA is shown (36).
cThe nomenclature 1:st and 2:nd, refers to a translational start at the first start codon (A/T)TG or the second start codon TTG discussed in the text (Figure 1).
dRelPC is the relative plasmid copy number for a given clone (i.e. the average number of library plasmid copies/chromosome in a clone compared to the corresponding value for the pBR-TrpL-ZEGFP reference clone) (mean values from triplicate experiments) as determined in the materials and methods section.
eRelF is the relative fluorescence intensity for a given clone (i.e. the fluorescence intensity value for a clone compared to the fluorescence intensity value of the pBR-TrpL-ZEGFP reference clone) (mean value from triplicate experiments), determined by flow cytometry analyses as described in the materials and methods section.
Protein production analysis
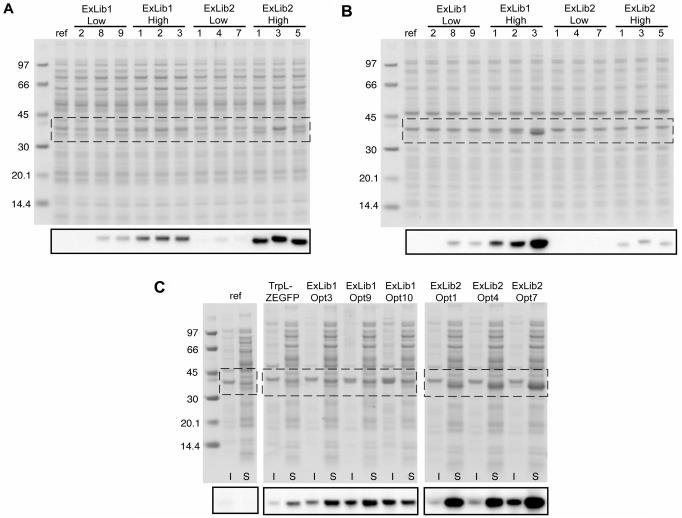
To verify the results from the different sorting experiments, and also to estimate the relative proportions of soluble and insoluble fractions of the ZEGFP fusion protein in the cells, SDS-PAGE and western blot (anti-EGFP) analyses were performed. Homogenates of cell cultures from three separate clones from the fluorescence intervals low and high, respectively, as well as from clones identified during the two-round sorting of the libraries (Opt clones) were divided into soluble and insoluble fractions. The results shown in Figure 5 indicate that cultures of clones originating from the low and high fluorescence intervals of both libraries contained correlating relative amounts of soluble ZEGFP fusion protein (Figure 5A). Noteworthy, the levels of soluble ZEGFP fusion protein appeared to be higher for ExLib2 clones than for clones from ExLib1 (Figure 5A). Interestingly, whereas only very low levels of co-existing insoluble ZEGFP fusion protein was observed for ExLib2-Low and -High clones, clones from ExLib1 showed to generate equal or even higher (e.g. ExLib1-High3) amounts of insoluble compared to soluble target protein (Figure 5B).
Figure 5.
Analysis of sorted ExLib1 and ExLib2 clones by SDS-PAGE and western blotting. Clones of indicated identities were cultivated and treated as described in the material and methods section for fractionation of soluble and insoluble materials, which were subsequently analyzed by SDS-PAGE under reducing conditions with protein staining or western blotting (smaller insets) using a polyclonal anti-GFP rabbit IgG reagent as primary antibody. The regions of the SDS-PAGE gels corresponding to the western blotting analyses are indicated by the boxes. (A) Soluble fractions from ExLib1-low/high and ExLib2-low/high clones; (B) Insoluble fractions from ExLib1-low/high and ExLib2-low/high clones; (C) Insoluble (I) and soluble (S) fractions from ExLib1-Opt and ExLib2-Opt clones. The lanes designated ‘ref’ refers to samples prepared from plasmid-less host cells (grown without added antibiotic). The numbers indicate molecular weights of reference proteins in kDa (Amersham Biosciences). In (C), samples from the pBR-TrpL-ZEGFP clone was included as additional reference.
The corresponding results obtained for cultures of the Opt clones, isolated for their high fluorescence intensities in two consecutive sorting rounds, also showed higher intracellular amounts of soluble ZEGFP reporter protein for ExLib2 clones than for ExLib1 clones (Figure 5C). Further, ExLib2 clones also displayed higher apparent soluble to insoluble product ratios than ExLib1 clones.
Both the SDS-PAGE and western blotting data suggested that the amounts of soluble ZEGFP protein product in the ExLib2 clones had been significantly increased compared to the pBR-TrpL-ZEGFP reference, and that observed whole cell fluorescence values had indeed been indicative of amounts of soluble ZEGFP fusion proteins for both libraries. To further investigate this notion, soluble ZEGFP fusion proteins were purified and quantified by IgG-affinity chromatography from lysed shake-flask cultures corresponding to the pBR-TrpL-ZEGFP and ExLib2-Opt7 clones. The results showed that the average yield of soluble ZEGFP fusion protein from the ExLib2-Opt7 library clone was approximately 24-fold higher (1.2 × 10−5 moles/l culture or 410 mg/l culture; MW = 34.9 kDa) than for the pBR-TrpL-ZEGFP reference (4.7 × 10−7 moles/l culture or 17 mg/l; MW = 36.0 kDa). This dramatic increase in soluble protein expression was in fact higher than could be expected from the flow cytometric analysis where a fluorescence intensity ratio of 15.8 was observed between cells corresponding to these two clones (Figure 4D, Table 1).
Further, to investigate if this positive effect on soluble Z protein product level was retained after functional removal of the C-terminally fused reporter moiety, a stop codon was genetically introduced between the gene fragments encoding the Z target protein and the EGFP reporter protein in the ExLib2-Opt7 clone. Interestingly, a quantification of the Z protein product via IgG-affinity chromatography showed that a similar amount of soluble Z product protein (1.3 × 10−5 mole/l culture or 94 mg/l culture; MW = 7.3 kDa) was obtained without the EGFP fusion partner present which indicated a neutral net effect for the EGFP reporter on the product expression, stability and solubility under the conditions used.
Additional analysis of ExLib1 clones
To further investigate possible reasons for the observed differences in ZEGFP protein expression between clones belonging to ExLib1, 16 clones from each of the fluorescence intensity intervals low, medium and high from library cultures grown at either 30 or 37°C were subjected to DNA sequencing. In addition, IgG-affinity purified soluble protein from cultures of three clones was subjected to mass spectrometric analyses. The fact that all investigated clones showed to produce an IgG-binding protein product of the molecular mass of 36.19 kDa (±0.015 kDa), showed that the assumption that different members of the library encoded identical TrpL-ZEGFP gene products (theoretical mass of 36.19 kDa), albeit via different sets of synonymous codons for the N-terminal region was correct (data not shown).
In the construction of the ExLib1 library, the two alternative initiation codons (ATG or TTG) were included in equal proportions. The data from an analysis of clones belonging to different sorted categories presented in Figure 2A, shows a clear correlation between lower fluorescence intensity values and the use of TTG as the initiation codon at both investigated temperatures. Interestingly, none of the ExLib1-High (37°C) or the three isolated ExLib1-Opt clones (Figure 2A and B) used a TTG triplet as the initiation codon.
To further address the possible influence from alternative codon choices, codon adaptation index (CAI) values were calculated for the N-terminal sequence of eight amino acids of each of the library members included in the analysis. Using an index for each of the 20 amino acids (denoted w values) based on their appearance in a set of highly expressed proteins in E. coli (31), a given sequence using solely the theoretically most preferred codons would result in a CAI value of 1.0. The gene encoding the first eight amino acids of the wild-type TrpL-sequence included as reference in this work uses relatively highly biased/optimized codons and has a CAI value of 0.69. Interestingly, the mean CAI values, calculated for the eight amino acid ExLib1 library member sequences in each of the categories low, medium, high and Opt, all ranged between approximately 0.2 and 0.3 (data not shown). Thus, no obvious correlation between fluorescence intensity values and codon bias could be seen.
However, if the eight codons of the sequences were treated separately and average values were calculated for each group of clones (i.e. low, medium and high), a different pattern was observed. The results from a series of analyses involving two different cultivation temperatures, are shown in Figure 2B and C. ExLib1 clones belonging to the high category (grown at 30 or 37°C) show similar and highly biased codon usage in positions +1, +2 and +5, which is similar to the pattern observed for Opt clones from ExLib1 (Figure 2B). It should be noted though, that there only exists two codons for phenylalanine and that the average w values of the high and Opt clones in position +5 reflects an approximate equal usage of these two codons (w(UUC) = 1.0 and w(UUU) = 0.296). The lysine codon AAA was exclusively used in position +2 in the three ExLib1-Opt clones and in 15 out of 16 clones belonging to the ExLib1-High category (37°C). In contrast, ExLib1 clones sorted for low fluorescence intensity values showed more varying codon usage in positions +1 and +2 (Figure 2C).
To investigate possible contributions to the increased product levels from changes in the gene dosage, the relative plasmid copy numbers (i.e. the number of library plasmid copies per chromosome observed for a clone compared to the corresponding value for the pBR-TrpL-ZEGFP reference clone) were determined for the ExLib1-Opt3, -Opt9 and -Opt10 clones via a real-time PCR-based method. Here, mean values from three independent cultures of each clone were calculated using plasmid and chromosome-specific primer-pairs in separate experiments, the genomic DNA serving as internal cellular reference. The results showed that all the three investigated ExLib1-Opt clones had relative plasmid copy numbers in the range 1.1–1.3 (Table 1), thus showing only marginally higher gene dosages than the reference which suggested that other factors (as discussed above) were mainly responsible for the high relative fluorescence intensities observed for these clones (in the range 3.0–4.3; Table 1).
Additional analysis of ExLib2 clones
For the ExLib2 library, 16 clones originating from each of the fluorescence intensity intervals low, medium and high (grown at either 30 or 37°C) and a number of Opt clones were subjected to DNA sequencing. As mentioned above, the design of this library opened up for the possible isolation of clones for which the translation had started at a second alternative start codon (TTG), located immediately in front of the ZEGFP fusion protein-encoding sequence (Figure 1E). Indeed, an analysis of ExLib2-High and -Opt clones (Table 1) revealed several SD-like sequence regions within the randomized window. The sequence context GGAG or GAGG was present in the majority of ExLib2-High and -Opt clones (Table 1) but was less frequent in the ExLib2-Medium and ExLib2-Low clones (data not shown). Thus, with the exception of the sequence for ExLib2-High3, all ExLib2-High and -Opt clone sequences listed in Table 1, contain at least one sub-sequence showing an apparent complementarity to the E. coli 16S rRNA anti-SD sequence.
An analysis by mass spectrometry of IgG-affinity purified proteins from three Opt clones showing the highest fluorescence intensity values (ExLib2-Opt1, ExLib2-Opt4, ExLib2-Opt7) and two of the ExLib2-High clones (ExLib2-High1 and ExLib2-High5) showed molecular masses consistent with translational starts at the second alternative initiation codon. For the clone ExLib2-Opt7, an N-terminal sequencing of the purified protein yielding the sequence Val-Asp-Asn-Lys-Phe further confirmed a translational start at the second initiation site. Thus, this suggests that the putative SD-like sequences shown in Table 1 for these five clones were productively positioned to promote a translational start at the TTG start codon placed downstream. The aligned spacing between the SD-like sequences and the TTG start codon varies between 5 and 10 nucleotides, which is in accordance with literature values for productive arrangements (36). For the ExLib2-High3, containing no apparent SD-like sequence within the randomized window, the mass spectrometric analysis showed an N-terminal extension consistent with a translational start at the first initiation codon. This is consistent with the small upward shift of the bands visualized in the western blotting for this clone relative to the neighboring bands corresponding to two clones for which the translation was confirmed to start at the second start codon (Figure 5A and B). The ExLib2-High3 also showed to result in a higher yield of ZEGFP protein product than the TrpL-ZEGFP, as evaluated from IgG affinity purification (60.6 mg/l culture and 17.2 mg/l culture, respectively). Thus, the use of the ExLib2 library showed that increased levels of soluble ZEGFP protein, relative to the reference, could be obtained both with and without an N-terminal extension.
As described above for some of the ExLib1 clones, the relative plasmid copy numbers were also determined for four ExLib2 clones (-High3, -Opt1, -Opt4 and -Opt7) to be able to correlate obtained values to the relative fluorescence intensities and soluble product yields (Table 1). For the three clones ExLib2-High3, ExLib2-Opt1 and ExLib2-Opt4, the relative plasmid copy numbers were found to be in the range 1.0–1.6 (Table 1), suggesting that the contribution from a gene dosage effect to the elevated soluble product levels and higher relative fluorescence intensities (in the range 2.7–5.5) was limited.
Interestingly, the ExLib2-Opt7 clone, showing the highest soluble product levels and the highest relative fluorescence intensity (15.8; Table 1) had a relative plasmid copy number value of 2.7, suggesting that a gene dosage effect, in addition to sequence-related effects, had contributed to the overall clonal characteristics.
As an additional investigation of the ExLib2-Opt7 clone, to be able to rule out that the library work, including sorting had resulted in spurious genetic changes in non-addressed parts of the expression plasmid and influenced the results, a fragment containing the randomized sequence window and the Z protein encoding gene was transferred to a fresh lot of the reference vector backbone (containing the EGFP gene cassette). In flow cytometry analyses, this re-constructed clone showed as high fluorescence intensities as the originally sorted ExLib2-Opt7 clone. This indicated that the basis for the elevated product levels seen for this clone could be specifically located to the studied sequence region.
DISCUSSION
The translation initiation efficiency is often described as the rate-limiting event in the translational process and is considered to influence the overall expression level of a gene (8,37). Sequence-related features affecting the initiation efficiency in E. coli include the initiation codon, the region downstream of the initiation codon, the SD sequence and its spacing to the initiation codon as well as variations in mRNA secondary structures (6). The integrated nature of these features makes it difficult, if not impossible, to perform truly isolated studies of any one of these single factors.
The aim of this study was to investigate the possibilities to influence the soluble product levels of a recombinant protein from varying the sequence of a 5′-end-located mRNA element by either conservative or free combinatorial randomization. The recruitment of an easily detected fluorescent gene product as reporter opened up for efficient flow cytometric monitoring of thousands to tens of millions different variants in single experiments. Both constructed libraries showed to result in wide distributions of the cellular fluorescence intensity, spanning up to three orders of magnitude. The stable expression characteristics for isolated subpopulations and individual clones indicated that the traits were due to inherent properties related to particular expression cassette sequence contexts.
For the ExLib1 library, corresponding to a conservative combinatorial variation (silent mutations) of the eight amino acid TrpL translation initiation peptide, a clear correlation between the start codon triplet and the cellular fluorescence intensities was seen, in accordance with the earlier findings showing that TTG generally is a less efficient initiation codon than ATG (13,19). Some of the sorted TrpL-ZEGFP encoding constructs of ExLib1 conferred up to 4-fold increases in fluorescence intensity relative to the wild-type reference. The library sequences in these constructs generally have low CAI values and accordingly contain high frequencies of rare codons. This is intriguing since translation of rare codons has been suggested to be slower than that of frequent codons (38). Furthermore, rare codons appearing in clusters or in the N-terminal part of the protein have been reported to be particularly important (22). However, an analysis of individual positions revealed that in position +2 there was a highly biased prevalence of an AAA codon in comparison to the alternative lysine codon AAG. Interestingly, AAA has been shown to be the most frequent +2 codon in E. coli genes, a position where it is also almost 4-fold as frequent as AAG (39). Further, an AUC to AAA codon substitution at the +2 position of E. coli dhfr gene resulted in a more than 2-fold increased yield using an in vitro expression system (39). The biased codon usage in position +2 was also evident when studying a subset of 3540 E. coli genes with an AUG as start codon (21). Several reports have noted significantly higher expression levels of reporter protein when AAA is used in position +2 as compared to AAG (18,21,39) and consequently our results are in agreement with these findings.
The increase in fluorescence intensity relative to the reference, reflecting the increase in amount of soluble protein products, was up to 4-fold in ExLib1-Opt clones. Although this corresponds to a significantly increased yield, it is not as pronounced as the increase observed for ExLib2-Opt clones. However, the amounts of TrpL-ZEGFP protein existing as insoluble inclusion bodies seen for these clones (Figure 5B) should also be taken into account when considering the effect on the production and further indicate that the overall increase in gene expression was considerable from the relatively minor changes at the nucleotide level.
Ten out of ten investigated ExLib2-Opt clones and the majority of ExLib2-High clones were found to contain one or more SD-like sequences, and all three mass-determined protein products originating from ExLib2-Opt clones proved to be translated from the second start codon (TTG). Thus, the appearance of alternative, or additional, SD sequences seemed to account for a more efficient mechanism to increase the amount of soluble ZEGFP protein product, than via N-terminal extension by a particular peptide sequence. Genetic engineering approaches specifically directed to an already present SD region have earlier been demonstrated useful for the development of vector variants yielding increased product levels (40,41). Even if the randomized windows of ExLib2-Opt and ExLib2-High clones contain novel SD-like sequences, open reading frames starting from the original translation initiation site and continuing through the same windows can in many cases still be deduced. This could potentially have led to the production of two protein products of different lengths from a single mRNA species. Nevertheless, in the determination of translational start positions of purified proteins by mass spectrometry analysis, only masses corresponding to initiation from one of the two start codons were detected. This was also confirmed by an N-terminal sequence analysis of the IgG-affinity purified product from clone ExLib2-Opt7. The notion that the occurrence of novel SD-like sequences was the predominant mechanism for the increased product levels is also strengthened from sequence features observed in Opt clones for which a protein-level analysis was not performed. Amber (TAG) stop codons are seen in frame with the first start codon for clones ExLib2-Opt3, -Opt5, -Opt6 (Table 1). Although amber stop codons to some extent can be suppressed in the supE 44 strain used in the study, their presence suggest that the high fluorescence intensities observed for these clones originate from translation initiations at codons different from the first start codon. A one-base base deletion within the variegated sequence is seen for clone ExLib2-Opt10, which if translation was initiated at the first start codon would lead to an out-of-frame translation of the ZEGFP protein.
In a previous study concerning translational activation of the Qβ coliphage maturation cistron, a model was suggested where a strong downstream coat protein gene RBS nearly always out-competed a nearby weaker upstream maturation protein initiation site (42). It has further been reported that ribosome-binding site-like sequences present in the cDNA of the target protein are able to interfere with vector encoded ribosome-binding site sequences, affecting the expression of the target gene (14). Thus, sufficiently strong SD sequences appearing in ExLib2 clones could potentially out-compete the upstream SD sequence and promote translational start from the second start position. Nuclease protection studies have shown that bacterial ribosomes cover approximately 15 nucleotides on each side of the initiation codon during translation initiation (37). The region of lac mRNA which was protected in RNA footprinting experiments was 14 bases preceding the initiation codon and 20 following it (43). Taking these observations into account and considering the close proximity between the two alternative translation initiation sites in our constructs, the binding of a ribosome to one site, presumably the stronger, would probably block binding to the other site. In future work, it would be interesting to eliminate the upstream ribosome binding site in one of the library clones, e.g. ExLib2-Opt7, to investigate if the efficient protein production seen for this construct is caused by cooperative effects from two neighboring ribosome-binding sites or is solely the result of the library-derived initiation region. In an earlier version of the ExLib1 library, the randomized TrpL encoding window was placed out-of-frame relative the ZEGFP gene located downstream. Analysis of induced cells from this library showed no fluorescence corresponding to ZEGFP protein expression (data not shown). This indicates that efficient SD-like sequences are not readily formed based on the limited genetic diversity used for the construction of this conserved library. Accordingly, three ExLib1-Opt derived protein products had masses corresponding to a translational start at the first position (Table 1).
It should be noted that all experiments in the study were performed under identical induction conditions (1 mM IPTG), and that observed effects therefore most likely were not related to differences on the transcriptional level. However, a single clone in the study (ExLib2-Opt7) showed to have significantly higher plasmid copy number than the reference, suggesting an influence on the product mRNA level. However, in view of the results for the other investigated clones, the 3-fold higher gene dosage seen for the ExLib2-Opt7 should only partially explain the higher levels of soluble product protein.
Mass spectrometry analysis of purified protein from the ExLib2-High3 clone, isolated for its high fluorescence intensity value, showed a translational start at the first initiation codon. Interestingly, the deduced amino acid sequence of the randomized window for this clone contained codon signatures previously associated with low gene expression. The CAI for the N-terminal extension peptide of this high expression construct is low (0.12), due to the presence of several consecutive rare codons. Consequently, there must be other compensating effects for the ExLib2-High3 clone, either on the nucleotide or amino acid level, contributing to the observed high amounts of soluble protein.
A strength of the presented combinatorial library approach coupled to a powerful screening strategy for expression optimization, is that it allows for a massive testing of very large collections of variants in a relatively short time. In the present study, many parameters of the cellular protein production machinery capable of influencing the desired trait were addressed simultaneously, and in a given clone some parameters may have been affected positively and others negatively. However, the functional screening using EGFP as reporter for soluble product protein allowed for the isolation of rare clones for which the net effects for soluble production of the target protein were the most positive. Future work should reveal if the clonal traits seen for the optimization of the production of the rapidly folded and highly soluble target protein Z (44,45) will be similar to when target proteins of other intrinsic characteristics are investigated using the described combinatorial vector library methodology.
ACKNOWLEDGEMENTS
We thank the VINNOVA and Swedish biotech-industry-supported Center for BioProcess Technology (CBIOPT) for support. We are also grateful to Olof Nord and John Löfblom for guidance and support related to the flow cytometric analyses and sortings. Funding to pay the Open Access publication charge was provided by KTH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Jana S, Deb JK. Strategies for efficient production of heterologous proteins in Escherichia coli. Appl. Microbiol. Biotechnol. 2005;67:289–298. doi: 10.1007/s00253-004-1814-0. [DOI] [PubMed] [Google Scholar]
- 2.Jonasson P, Liljeqvist S, Nygren PA, Stahl S. Genetic design for facilitated production and recovery of recombinant proteins in Escherichia coli. Biotechnol. Appl. Biochem. 2002;35:91–105. doi: 10.1042/ba20010099. [DOI] [PubMed] [Google Scholar]
- 3.Baneyx F, Mujacic M. Recombinant protein folding and misfolding in Escherichia coli. Nat. Biotechnol. 2004;22:1399–1408. doi: 10.1038/nbt1029. [DOI] [PubMed] [Google Scholar]
- 4.Sorensen HP, Mortensen KK. Advanced genetic strategies for recombinant protein expression in Escherichia coli. J. Biotechnol. 2005;115:113–128. doi: 10.1016/j.jbiotec.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 5.Shine J, Dalgarno L. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hartz D, McPheeters DS, Gold L. Influence of mRNA determinants on translation initiation in Escherichia coli. J. Mol. Biol. 1991;218:83–97. doi: 10.1016/0022-2836(91)90875-7. [DOI] [PubMed] [Google Scholar]
- 7.Chang JT, Green CB, Wolf RE., Jr Inhibition of translation initiation on Escherichia coli gnd mRNA by formation of a long-range secondary structure involving the ribosome binding site and the internal complementary sequence. J. Bacteriol. 1995;177:6560–6567. doi: 10.1128/jb.177.22.6560-6567.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Smit MH, van Duin J. Translational initiation on structured messengers. Another role for the Shine-Dalgarno interaction. J. Mol. Biol. 1994;235:173–184. doi: 10.1016/s0022-2836(05)80024-5. [DOI] [PubMed] [Google Scholar]
- 9.Griswold KE, Mahmood NA, Iverson BL, Georgiou G. Effects of codon usage versus putative 5′-mRNA structure on the expression of Fusarium solani cutinase in the Escherichia coli cytoplasm. Protein Expr. Purif. 2003;27:134–142. doi: 10.1016/s1046-5928(02)00578-8. [DOI] [PubMed] [Google Scholar]
- 10.Pfleger BF, Fawzi NJ, Keasling JD. Optimization of DsRed production in Escherichia coli: effect of ribosome binding site sequestration on translation efficiency. Biotechnol. Bioeng. 2005;92:553–558. doi: 10.1002/bit.20630. [DOI] [PubMed] [Google Scholar]
- 11.Schauder B, McCarthy JE. The role of bases upstream of the Shine-Dalgarno region and in the coding sequence in the control of gene expression in Escherichia coli: translation and stability of mRNAs in vivo. Gene. 1989;78:59–72. doi: 10.1016/0378-1119(89)90314-4. [DOI] [PubMed] [Google Scholar]
- 12.Wang G, Liu N, Yang K. High-level expression of prochymosin in Escherichia coli: effect of the secondary structure of the ribosome binding site. Protein Expr. Purif. 1995;6:284–290. doi: 10.1006/prep.1995.1037. [DOI] [PubMed] [Google Scholar]
- 13.Ringquist S, Shinedling S, Barrick D, Green L, Binkley J, Stormo GD, Gold L. Translation initiation in Escherichia coli: sequences within the ribosome-binding site. Mol. Microbiol. 1992;6:1219–1229. doi: 10.1111/j.1365-2958.1992.tb01561.x. [DOI] [PubMed] [Google Scholar]
- 14.Hrzenjak A, Artl A, Knipping G, Kostner G, Sattler W, Malle E. Silent mutations in secondary Shine-Dalgarno sequences in the cDNA of human serum amyloid A4 promotes expression of recombinant protein in Escherichia coli. Protein Eng. 2001;14:949–952. doi: 10.1093/protein/14.12.949. [DOI] [PubMed] [Google Scholar]
- 15.Kane JF. Effects of rare codon clusters on high-level expression of heterologous proteins in Escherichia coli. Curr. Opin. Biotechnol. 1995;6:494–500. doi: 10.1016/0958-1669(95)80082-4. [DOI] [PubMed] [Google Scholar]
- 16.Deana A, Ehrlich R, Reiss C. Silent mutations in the Escherichia coli ompA leader peptide region strongly affect transcription and translation in vivo. Nucleic Acids Res. 1998;26:4778–4782. doi: 10.1093/nar/26.20.4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzalez de Valdivia EI, Isaksson LA. A codon window in mRNA downstream of the initiation codon where NGG codons give strongly reduced gene expression in Escherichia coli. Nucleic Acids Res. 2004;32:5198–5205. doi: 10.1093/nar/gkh857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Looman AC, Bodlaender J, Comstock LJ, Eaton D, Jhurani P, de Boer HA, van Knippenberg PH. Influence of the codon following the AUG initiation codon on the expression of a modified lacZ gene in Escherichia coli. EMBO J. 1987;6:2489–2492. doi: 10.1002/j.1460-2075.1987.tb02530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stenstrom CM, Holmgren E, Isaksson LA. Cooperative effects by the initiation codon and its flanking regions on translation initiation. Gene. 2001;273:259–265. doi: 10.1016/s0378-1119(01)00584-4. [DOI] [PubMed] [Google Scholar]
- 20.Stenstrom CM, Isaksson LA. Influences on translation initiation and early elongation by the messenger RNA region flanking the initiation codon at the 3′ side. Gene. 2002;288:1–8. doi: 10.1016/s0378-1119(02)00501-2. [DOI] [PubMed] [Google Scholar]
- 21.Stenstrom CM, Jin H, Major LL, Tate WP, Isaksson LA. Codon bias at the 3′-side of the initiation codon is correlated with translation initiation efficiency in Escherichia coli. Gene. 2001;263:273–284. doi: 10.1016/s0378-1119(00)00550-3. [DOI] [PubMed] [Google Scholar]
- 22.Gustafsson C, Govindarajan S, Minshull J. Codon bias and heterologous protein expression. Trends Biotechnol. 2004;22:346–353. doi: 10.1016/j.tibtech.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 23.Rosenberg AH, Goldman E, Dunn JJ, Studier FW, Zubay G. Effects of consecutive AGG codons on translation in Escherichia coli, demonstrated with a versatile codon test system. J. Bacteriol. 1993;175:716–722. doi: 10.1128/jb.175.3.716-722.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cortazzo P, Cervenansky C, Marin M, Reiss C, Ehrlich R, Deana A. Silent mutations affect in vivo protein folding in Escherichia coli. Biochem. Biophys. Res. Commun. 2002;293:537–541. doi: 10.1016/S0006-291X(02)00226-7. [DOI] [PubMed] [Google Scholar]
- 25.Ruther U. pUR 250 allows rapid chemical sequencing of both DNA strands of its inserts. Nucleic Acids Res. 1982;10:5765–5772. doi: 10.1093/nar/10.19.5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 27.Nilsson B, Moks T, Jansson B, Abrahmsen L, Elmblad A, Holmgren E, Henrichson C, Jones TA, Uhlen M. A synthetic IgG-binding domain based on staphylococcal protein A. Protein Eng. 1987;1:107–113. doi: 10.1093/protein/1.2.107. [DOI] [PubMed] [Google Scholar]
- 28.Yanofsky C, Platt T, Crawford IP, Nichols BP, Christie GE, Horowitz H, VanCleemput M, Wu AM. The complete nucleotide sequence of the tryptophan operon of Escherichia coli. Nucleic Acids Res. 1981;9:6647–6668. doi: 10.1093/nar/9.24.6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee CL, Ow DS, Oh SK. Quantitative real-time polymerase chain reaction for determination of plasmid copy number in bacteria. J. Microbiol. Methods. 2006;65:258–267. doi: 10.1016/j.mimet.2005.07.019. [DOI] [PubMed] [Google Scholar]
- 30.Winer J, Jung CK, Shackel I, Williams PM. Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal. Biochem. 1999;270:41–49. doi: 10.1006/abio.1999.4085. [DOI] [PubMed] [Google Scholar]
- 31.Sharp PM, Li WH. The codon adaptation index-a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharp PM, Li WH. Codon usage in regulatory genes in Escherichia coli does not reflect selection for ‘rare’ codons. Nucleic Acids Res. 1986;14:7737–7749. doi: 10.1093/nar/14.19.7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cormack BP, Valdivia RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 34.Hedhammar M, Stenvall M, Lonneborg R, Nord O, Sjolin O, Brismar H, Uhlen M, Ottosson J, Hober S. A novel flow cytometry-based method for analysis of expression levels in Escherichia coli, giving information about precipitated and soluble protein. J. Biotechnol. 2005;119:133–146. doi: 10.1016/j.jbiotec.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 35.Jonasson P, Nygren PA, Johansson BL, Wahren J, Uhlen M, Stahl S. Gene fragment polymerization gives increased yields of recombinant human proinsulin C-peptide. Gene. 1998;210:203–210. doi: 10.1016/s0378-1119(98)00026-2. [DOI] [PubMed] [Google Scholar]
- 36.Chen H, Bjerknes M, Kumar R, Jay E. Determination of the optimal aligned spacing between the Shine-Dalgarno sequence and the translation initiation codon of Escherichia coli mRNAs. Nucleic Acids Res. 1994;22:4953–4957. doi: 10.1093/nar/22.23.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- 38.Sorensen MA, Kurland CG, Pedersen S. Codon usage determines translation rate in Escherichia coli. J. Mol. Biol. 1989;207:365–377. doi: 10.1016/0022-2836(89)90260-x. [DOI] [PubMed] [Google Scholar]
- 39.Sato T, Terabe M, Watanabe H, Gojobori T, Hori-Takemoto C, Miura K. Codon and base biases after the initiation codon of the open reading frames in the Escherichia coli genome and their influence on the translation efficiency. J. Biochem. (Tokyo) 2001;129:851–860. doi: 10.1093/oxfordjournals.jbchem.a002929. [DOI] [PubMed] [Google Scholar]
- 40.Wilson BS, Kautzer CR, Antelman DE. Increased protein expression through improved ribosome-binding sites obtained by library mutagenesis. Biotechniques. 1994;17:944–953. [PubMed] [Google Scholar]
- 41.Zhelyabovskaya OB, Berlin YA, Birikh KR. Artificial genetic selection for an efficient translation initiation site for expression of human RACK1 gene in Escherichia coli. Nucleic Acids Res. 2004;32:e52. doi: 10.1093/nar/gnh050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Priano C, Arora R, Jayant L, Mills DR. Translational activation in coliphage Qbeta: on a polycistronic messenger RNA, repression of one gene can activate translation of another. J. Mol. Biol. 1997;271:299–310. doi: 10.1006/jmbi.1997.1194. [DOI] [PubMed] [Google Scholar]
- 43.Murakawa GJ, Nierlich DP. Mapping the lacZ ribosome binding site by RNA footprinting. Biochemistry. 1989;28:8067–8072. doi: 10.1021/bi00446a015. [DOI] [PubMed] [Google Scholar]
- 44.Arora P, Oas TG, Myers JK. Fast and faster: a designed variant of the B-domain of protein A folds in 3 microsec. Protein Sci. 2004;13:847–853. doi: 10.1110/ps.03541304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Samuelsson E, Moks T, Nilsson B, Uhlen M. Enhanced in vitro refolding of insulin-like growth factor I using a solubilizing fusion partner. Biochemistry. 1994;33:4207–4211. doi: 10.1021/bi00180a013. [DOI] [PubMed] [Google Scholar]