Abstract
The intergenic region-internal ribosome entry site (IGR-IRES) of dicistroviruses binds to 40S ribosomal subunits in the absence of eukaryotic initiation factors (eIFs). Although the conserved loop sequences in dicistroviral IGR-IRES elements are protected from chemical modifications in the presence of the 40S subunit, molecular components in the 40S subunit, which interacts with the loop sequences in the IRES, have not been identified. Here, a chemical crosslinking study using 4-thiouridine-labeled IGR-IRES revealed interactions of the IGR-IRES with several 40S proteins but not with the 18S rRNA. The strongest crosslinking signal was identified for ribosomal protein S25 (rpS25). rpS25 is known to be a neighbor of rpS5, which has been shown to interact with a related IGR-IRES by cryo-electron microscopy. Crosslinking analysis with site-directed mutants showed that nucleotides UU6089–6090, which are located in the loop region in conserved domain 2b in the IRES, appear to interact with rpS25. rpS25 is specific to eukaryotes, which explains why there is no recognition of the IGR-IRES by prokaryotic ribosomes. Although the idea that the IGR-IRES element may be a relict of a primitive translation system has been postulated, our experimental data suggest that the IRES has adapted to eukaryotic ribosomal proteins.
INTRODUCTION
Plautia stali intestine virus (PSIV) and Cricket paralysis virus (CrPV) are members of the family Dicistroviridae (1). Known dicistroviruses contain a structurally conserved intergenic region-internal ribosome entry site (IGR-IRES) for translation of the capsid protein. Translation initiation mediated by the IRES of dicistroviruses does not require base-pair interaction between an AUG initiation codon and initiator Met-tRNA (2–4). The IRES elements of dicistroviruses share a common secondary structure domain arrangement, constructed by three pseudoknots (PK I, PK II, PK III). Because of this similarity in IRES secondary structure, it has been thought that IRES elements function via the same mechanism (5).
Usually, assembly of the eukaryotic 80S ribosome on normal mRNA is completed under the control of eIFs (6). In contrast, ribosomal assembly occurs directly on the dicistroviral IGR-IRES in the absence of eIFs (4,7,8). The nucleotides in the IRES that interact with 40S ribosomes have been attributed to the 5′ region of the IRES by chemical and enzymatic footprint analyses (7,9), whereas the 3′ region of the IRES, which consists of PK I that is responsible for determining the reading frame of the mRNA, has been shown to be exposed to the interface side at very close to the P site of the 40S ribosome (10). Although the IGR-IRES of dicistroviruses is recognized by ribosomes from various eukaryotes, such as insects, yeast, human and wheat, ribosomes from E. coli cannot recognize the IGR-IRES (Yamamoto and Uchiumi, unpublished data) and the S30 extract of E. coli cannot conduct IGR-IRES-mediated translation (11).
A cryo-electron microscopy study has reconstituted an image of the IGR-IRES of CrPV docking on the human ribosome (12). At ∼20 Å resolution, the model reveals several structural elements of the ribosome that contact the IGR-IRES, such as helices 18, 30 and 34. However, biochemical evidence showing one-to-one correspondence between nucleotides in the IRES and structural elements of the ribosome remains lacking.
Here, to identify sites on the 40S ribosomal subunit that interact with the IGR-IRES of PSIV, chemical modification and crosslinking analyses against the 18S rRNA and 40S ribosomal proteins were carried out. The crosslinking results suggest that the IGR-IRES interacts mainly with ribosomal proteins, rather than the 18S rRNA. In addition, the ribosomal protein S25 (rpS25), which was crosslinked to the conserved domain 2 region in the IRES, does not have a prokaryotic counterpart, explaining why IGR-IRES-mediated translation does not occur with bacterial ribosomes.
MATERIALS AND METHODS
40S ribosome isolation
Rabbit 40S ribosomes were prepared as described (9). Drosophila ribosomes were prepared according to Bradford and Sullivan (13) with modifications. In brief, Drosophila melanogaster strain Canton-S was maintained at 25°C and fed with Formula 4–24 medium (Carolina Biological Supply). Adult insects were homogenized in 20 mM Tris-HCl (pH 7.5), 0.25 M sucrose, 10 mM MgCl2, 25 mM KCl, 1 mM DTT and 1 mM phenylmethylsulfonylfluoride at 0°C. The slurry was adjusted to a concentration of 0.5% deoxycholate, and centrifuged at 20 000 × g for 10 min at 4°C. The supernatant was decanted and filtered through cotton gauze to exclude the lipid layer. The filtered supernatant was centrifuged at 124 000 × g for 20 min at 4°C. The precipitate was suspended in buffer A (20 mM Tris-HCl pH 7.5, 5 mM MgCl2, 100 mM NH4Cl and 1 mM DTT) containing 0.25 M sucrose. The solution was layered onto a 3-fold excess (v/v) of buffer A containing 1 M sucrose, and centrifuged at 124 000 × g for 1 h 30 min at 4°C. The crude polysomal pellet was treated with puromycin and high-salt buffer, and subjected to sucrose density gradient centrifugation to separate the 40S and 60S ribosomes (14).
IRES preparation
Plasmid pT7D1-2-3, which contains PSIV nt 6005–6192 with base substitutions to facilitate T7 transcription and HincII digestion (9), was modified by PCR to replace the HincII site immediately downstream of PK I with a HindIII site. A fragment corresponding to nt 6286–6473 of himetobi P virus (HiPV) was prepared by PCR from a plasmid containing cDNAs of the virus (15), and ligated into pT7Blue (Novagen). The HiPV IRES was also modified to facilitate T7 transcription and HindIII digestion. The PSIV and HiPV IRES elements were further truncated by PCR to delete the domain 3 regions (nts 6147–6192 and 6427–6473, respectively). Domains 1–2 of the IRES elements of black queen cell virus (BQCV) (nt 5647–5792), CrPV (nt 6029–6170) and Triatoma virus (TrV) (nt 5925–6067) were prepared by PCR using pairs of long synthetic DNAs corresponding to the first and second halves of the region; these pairs had complementary sequences at the 3′ termini, and HindIII and EcoRI sites at the 5′ termini of the former and latter fragment, respectively. The amplified fragments were digested with HindIII and EcoRI, and ligated into the corresponding sites of pT7Blue.
IRES elements containing 4-thiouridine (s4U) were prepared by a Riboprobe T7 system (Promega). Because the incorporation efficiency of s4U is about 0.2 times that of U (16), a 5-fold molar excess of 4-thiouridine-5′-triphosphate (s4UTP, Trilink biotechnologies) over UTP was used. For autoradiography, [α-32P]ATP and [α-32P]CTP were also included in the reaction according to the manufacturer's recommendations. The IRES of encephalomyocarditis virus (EMCV) was prepared from EcoRV-digested pCITE4b (Novagen).
Chemical probing
Dimethyl sulfate (DMS) and 1-cyclohexyl-3-(2morpholinoethyl)carbodiimide metho-p-toluene sulfonate (CMCT) footprint analyses were done in 2 mM Mg2+ as described (9). After incubation with rabbit 40S ribosome (0.5 pmol) and renatured IRES (2.5 pmol), modified nucleotides were monitored by primer extension using 15 primers complementary to rabbit 18S rRNA nucleotides 371-351, 467-450, 576-550, 674-652, 767-750, 874-851, 971-950, 1072-1052, 1176-1150, 1262-1242, 1370-1350, 1479-1457, 1566-1550, 1669-1650 and 1863-1843.
4-Thiouridine(s4U)-mediated crosslinking
For the analysis of crosslinking between the IRES and the 18S rRNA, the rabbit 40S ribosome was incubated with 2- and 5-fold molar excess of IRES element containing s4U for 10 min at 37°C in 20 mM Tris-HCl (pH 7.5), 2 mM MgCl2, 200 mM NH4Cl and 6 mM 2-mercaptoethanol. The reaction was irradiated for 15 min at room temperature with a illuminator (λMax at 365 nm) using a light intensity of about 100 mW/cm2. The sample was adjusted to a concentration of 0.5% SDS and 5 mM EDTA, treated twice with phenol–chloroform, precipitated with ethanol, and then used for primer extension.
For the analysis of crosslinking between the IRES elements and ribosomal proteins, s4U-containing IRES elements that had been body-labeled with [α-32P]ATP and [α-32P]CTP were mixed with a 2-fold molar excess of the Drosophila 40S ribosome in 30 mM K-HEPES (pH7.0), 10 mM MgCl2 and 125 mM potassium acetate. After irradiation with UV at 365 nm, the reaction was subjected to Microcon YM-100 (Amicon) filters to concentrate the IRES–40S complexes and to remove free IRES. The solution was adjusted to 40 mM Tris-HCl (pH 7.5), 3 mM EDTA and 5000 U/ml RNase T1 before RNase A treatment. RNase A was added at a concentration of 0.2 and 2 μg/ml for weak and standard digestion, respectively, and incubated for 2.5 h at 25°C. Ribosomal proteins were prepared by the acetic acid method (17), and resolved by two-dimensional gel electrophoresis.
Two-dimensional gel electrophoresis and identification of protein spots
Radical-free and highly reducing (RFHR) two-dimensional gel electrophoresis was performed essentially as described (18) using a modified electrophoresis apparatus (Nippon eido). Zero-, first- and second-dimension polyacrylamide gels containing 8 M urea were prepared at 8, 8 and 15%, respectively, and pre-run with a radical scavenger, 2-mercaptoethylamine. Ribosomal proteins were reduced for 30 min at 40°C in 8 M urea and 0.2 M 2-mercaptoethanol, and concentrated by zero-dimension electrophoresis using anodic (200 mM glycine, 220 mM cysteine-HCl, 28 mM acetic acid, 4 M urea) and cathodic (12 mM KOH, 13 mM acetic acid) buffers. The sample band was then excised and subjected to first-dimension electrophoresis, which separated the proteins on the basis of charge using anodic (0.4 M Tris, 0.5 M boric acid, 21.5 mM EDTA, 6 M urea, 44 mM 2-mercaptoethylamine) and cathodic (0.4 M Tris, 0.5 M boric acid, 21.5 mM EDTA, 6 M urea, 87 mM acetic acid) buffers. The gel was put on the second-dimension gel, and subjected to electrophoresis using anodic (200 mM glycine, 220 mM cysteine-HCl, 28 mM acetic acid, 6 M urea) and cathodic (200 mM glycine, 28 mM HCl, 28 mM acetic acid) buffers. The 2D gel was stained with Coomassie brilliant blue or a Silver Stain Plus kit (Bio-Rad), and analyzed with ImageMaster 2D Platinum (GE Healthcare Bio-Sciences). Proteins crosslinked with 32P-labeled RNA fragments were detected using a BAS-2500 system (Fuji Photo Film).
Six protein spots in the 2D gel were excised and identified by mass spectrometry using a Mascot database search system.
Western blot
To obtain antibody to Drosophila rpS25, a 13-mer peptide (KKDAKSSAKQPQK) corresponding to residues 4–16 in the amino acid sequence of Drosophila rpS25 was synthesized. An additional cysteine residue at the C-terminus of the peptide was conjugated to a carrier protein, Keyhole limpet hemocyanin, by the m-Maleimidobenzoyl-N-hydroxysuccinimide ester method, and then used to immunize mice. The polyclonal antibody obtained was purified by peptide-conjugated affinity chromatography (Hokkaido System Science) to give a total yield of 1.34 mg (1.92 mg/ml). The crosslinking reaction was performed in 20-μl reaction volumes containing 10 mM Tris-HCl, pH 7.5, 100 mM NH4Cl, 10 mM MgCl2, 3 mM 2-mercaptoethanol, 7.5 pmol Drosophila 40S ribosomal subunits and 45 pmol s4U-labeled or unlabeled RNAs. After a 2-min digestion with 40 ng of RNase A, samples were mixed with sample loading buffer and separated by 15% SDS-PAGE. The blotted PVDF membrane was treated with 1000-fold diluted polyclonal antibody directed against rpS25 and 10 000-fold diluted secondary antibody (Peroxidase-conjugated Affinipure F(ab′)2 fragment goat anti-mouse IgG, Jackson ImmunoResearch Laboratories), and then detected with ECL-Plus western blotting detection reagents (GE Healthcare Bio-Sciences) according to the manufacturer's recommendations.
RESULTS AND DISCUSSION
No interaction between the IGR-IRES and the 18S rRNA of the rabbit 40S subunit by chemical probing
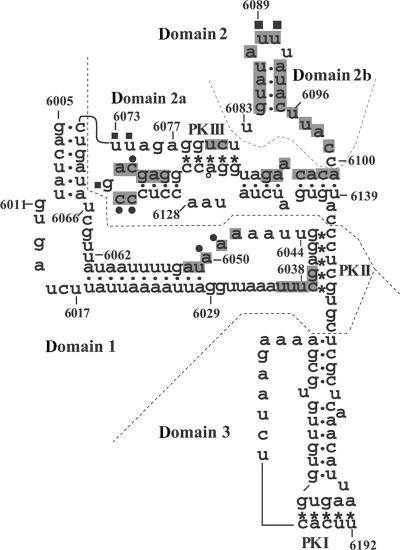
DMS modifies single-stranded adenine and cytosine, whereas CMCT modifies single-stranded uracil and guanine. Modifications by these chemicals are introduced at the site for hydrogen-bond formation for base–pair interaction, thus reverse transcription of the modified RNA is arrested at the −1 position of the modified base (19). We previously identified that nucleotides UU6089–6090 in the IGR-IRES of PSIV (Figure 1) are protected from modification by CMCT in the presence of the 40S ribosome (9). Because these two uracil residues are absolutely conserved in all known IGR-IRES elements of dicistroviruses (20), we considered that they could be candidates for sites of interaction with the 18S rRNA. Our footprint analyses using 15 primers against the rabbit 18S rRNA, however, detected no nucleotides that were obviously protected in the presence of PSIV IRES (data not shown).
Figure 1.
Secondary structure model of the PSIV IGR-IRES, showing nucleotides that are protected from chemical probes in the presence of the 40S ribosomal subunit. Dots and asterisks indicate the base–pair interaction for stems and pseudoknots, respectively. To facilitate description of the IRES structure, the IRES is divided into domain 1, domain 2 and domain 3, as indicated by the dotted lines. Domain 2 is further divided into domains 2a and 2b. Nucleotides that are protected, in the presence of the 40S ribosome, from DMS modification are marked with circles and those from CMCT are marked with squares. Nucleotides that are protected from hydroxyl-radical cleavage are shaded (Modified from Nishiyama et al. (9)).
Since we failed to detect IRES interaction sites in 18S rRNA by footprinting, we further examined the interaction by photo-crosslinking using 4-thiouridine (s4U). RNAs containing s4U forms zero-length crosslinks after irradiation with near-UV light (330–370 nm) (21). To facilitate the assignment of signals, we deleted the domain 3 region of the PSIV IRES because domains 1–2 of IGR-IRES elements, comprising nucleotides 6005–6149 (see Figure 1), can bind to the 40S ribosome independently of domain 3 (7,9,22). After extraction of 18S rRNA from the irradiated reaction mixture containing s4U-labeled domains 1–2 of the PSIV IRES, primer extension was performed to detect crosslinked sites. Because crosslinked nucleotides were barely detectable in experiments using a 1:2 molar ratio of the 40S ribosome and s4U-labeled domains 1–2, we increased the molar ratio to 1:5. In this condition, several nucleotides in the 18S rRNA were reproducibly crosslinked, however, the positions of these nucleotides appeared to be located on the solvent side of 40S ribosome, including expansion segments (23) (data not shown). In addition, at the 1:5 ratio, the nucleotides that were crosslinked with the PSIV IRES and the HiPV IRES differed. These observations indicate that the nucleotides in 18S rRNA are not major contributors to the strong binding between the IGR-IRES and the 40S ribosome. We conclude that the s4U-mediated crosslinks to 18S rRNA are secondary signals, after the authentic IRES-binding region is occupied.
Interaction between the IGR-IRES and Drosophila 40S ribosomal proteins
Next we preliminarily tested whether the conserved AUUU loop crosslink to rabbit 40S ribosomal proteins. Synthesized domain 2 RNA fragment which contains s4U at position 6090 was supposed to crosslink to a smaller protein than rpS5 (see Supplementary Data S1).
Since the amino acid sequences of the ribosomal proteins of the rabbit 40S subunit have not been determined in full, we prepared the 40S ribosome of D. melanogaster to facilitate the identification of ribosomal proteins that may crosslink to the IRES. Ribosomal proteins were separated by RFHR 2D polyacrylamide gel electrophoresis. This system is particularly suitable for small and basic proteins, such as ribosomal proteins (18). In our experimental condition, at least 30 protein spots were visible by Coomassie brilliant blue staining of gels on which a large amount of Drosophila 40S ribosome was electrophoresed (data not shown). Six protein spots were excised and identified by mass spectrometry as rpS2, rpS4, rpS6, rpS19, rpS25 and rpS26.
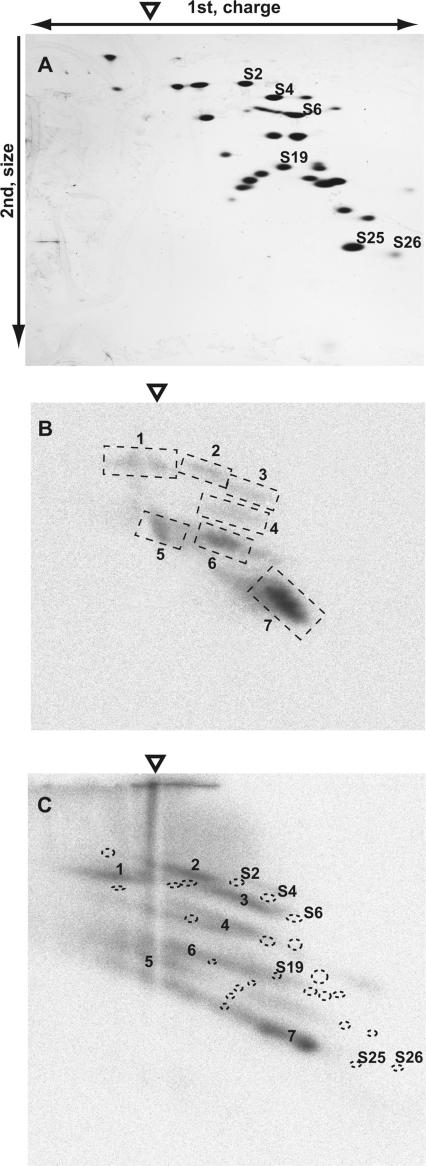
Figure 2A shows the silver-stained gel pattern of 40 pmol of Drosophila 40S ribosomal subunits. After a standard RNase A treatment (2 μg/ml) of the crosslinked reaction mixture containing the 40S ribosome and domains 1–2 of the PSIV IRES internally labeled with 32P and s4U, six weak signals (signals 1–6) and a single strong signal (signal 7) were detected in the radiograph (Figure 2B). In the 2D gel, proteins that are crosslinked should shift to positions with a more negative charge and a higher molecular mass in comparison to their original spot, because crosslinked RNA affects the electrophoretic mobility of the original proteins. Indeed, crosslinked samples digested with smaller amounts of RNase A (0.2 μg/ml) showed an extensively smeared image in a radiograph (Figure 2C). When we overlapped the images of the silver-stained protein spots and signals from the autoradiograph, extrapolation of signal 7 in Figure 2C suggested that this signal was shifted from the original spot of rpS25.
Figure 2.
Identification of a ribosomal protein crosslinked with s4U-labeled PSIV IRES domains 1–2. The origin of the first-dimension gel electrophoresis is indicated by an open triangle. (A) Silver-stained gel after separation of 40S ribosomal proteins. (B) Autoradiograph of a second-dimension gel. The seven signals that were reproducibly detected are indicated by broken squares and numbered from 1 to 7. (C) Autoradiograph of a second-dimension gel containing samples that were incompletely digested with RNase A. The locations of spots of silver-stained proteins are marked with dotted circles. Ribosomal proteins S2, S4, S6, S19, S25 and S26 were identified by mass spectrometry using protein spots excised from a similarly prepared gel.
When we carried out similar experiments using the domain 1–2 regions of the IRES elements of other dicistroviruses, including BQCV, CrPV and TrV, a similar signal corresponding to signal 7 was observed in all three (data not shown), suggesting that the protein interaction that was detected as a crosslinked component, signal 7, is conserved in IGR-IRES elements.
The ribosomal protein rpS25 interacts with IGR-IRES
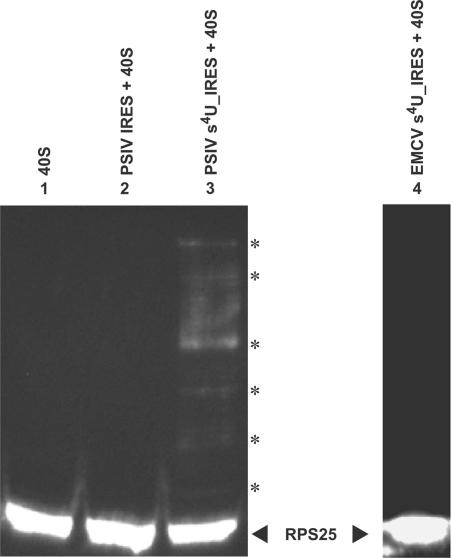
To confirm whether signal 7 was the shifted ribosomal protein rpS25, western blot analyses using antibodies raised against synthesized peptide of the N-terminal region of Drosophila rpS25 were carried out. The polyclonal antibody obtained detected a single band in Drosophila ribosome, but detected no bands in ribosomes of wheat, rabbit, brine shrimp and the brown planthopper, indicating that it does not crossreact with the other ribosomal proteins (data not shown). When samples were irradiated by near UV-light, no band larger than the RPS25 were detected in the Drosophila 40S subunit (Figure 3, lane 1) or in samples of native PSIV IRES with the 40S (Figure 3, lane 2). In contrast, samples containing the 40S subunit and the s4U-labeled PSIV-IRES showed several bands at positions above rpS25 (Figure 3, lane 3). Because irradiation to a sample containing 40S subunit and the s4U-labeled EMCV IRES did not produce any larger protein bands (Figure 3, lane 4), the larger proteins present in lane 3 must be crosslinked products specific to the s4U incorporated in the PSIV IRES. We therefore concluded that domains 1–2 of the IGR-IRES of PSIV interact with rpS25.
Figure 3.
Evidence of contact between the s4U-labeled IGR-IRES of PSIV and the rpS25 protein of Drosophila. Drosophila 40S ribosomes were mixed with each RNA and irradiated with a near-UV lamp. After partial RNase A digestion, the reaction mixtures were separated on SDS-polyacrylamide (15%) gels and rpS25 proteins were detected by western blot. Lane 1, 40S ribosome with no irradiation; lane 2, 40S ribosome irradiated with unlabeled PSIV IRES; lane 3, 40S ribosome irradiated with s4U-labeled PSIV IRES; lane 4, 40S ribosome irradiated with s4U-labeled EMCV IRES. The position of the rpS25 protein is marked on the right. Asterisks indicate the positions of crosslinked rpS25 proteins in lane 3.
Assignment of the uridine residues in the PSIV IRES interacting with RPS25
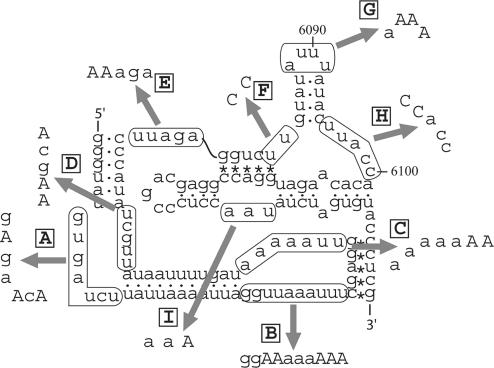
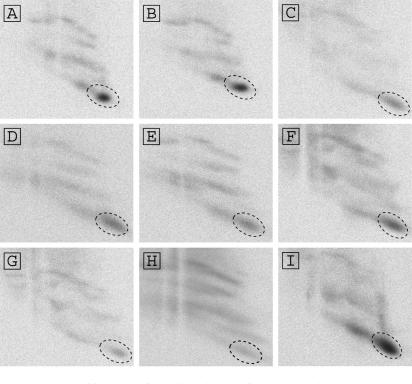
Domains 1–2 of the PSIV IRES contain 22 uridine residues in the single-stranded regions (Figure 4). To identify uridine residues that were crosslinked to rpS25, we constructed mutants of domains 1–2 in which the single-stranded uridine residues were replaced with adenine or cytosine residues (Figure 4). Previous mutational analysis of regions such as AUUU6111–6114 to UAAA in the CrPV IRES (7) slightly decreased the binding affinity of the element to the 40S subunit, suggesting that the IGR-IRES and the 40S subunit have multiple contacts site and that partial mutation of the IRES would not inhibit its binding to the 40S subunit. Figure 5 shows radiographs of 2D gels of proteins crosslinked to the mutant IRES elements. The signal of rpS25 was most intense in the radiographs of Figure 5A–F,I; however, it was weakened in those of Figure 5G and H, in which UUU6089–6091 and UU6096–6097 were mutated, respectively. Nucleotides UUU6089–6091 in PSIV are located in domain 2b (Figure 1), and all of the known dicistroviruses conserve this loop sequence (20), indicating that these uracils have an important role in IRES function. By contrast, UU6096-6097 are located in a loop region in domain 2b (Figure 1); these two uracils are conserved in this region in eight viruses, but a single uracil is present in five viruses. Because hydroxyl radical probing analysis in the presence of Mg2+ ions has indicated that the loop sequence connecting domains 2a and 2b (nt 6096–6100 in PSIV) constitutes part of the tightly packed core of the IRES (22), nucleotides UU6096–6097 are probably not exposed to the solvent. In addition, two of our previous mutational analyses in this connecting region, ACC6098–6100 to GGG or UUU, both abolished IRES-mediated translation (5,9). These experimental results implied that mutation H (Figure 4) may affect the structure of the IRES. When the secondary structure of mutant H was monitored by DMS probing, non-specific terminations were observed in the domain 2b region (see Supporting Data S1) in comparison to the wild-type IRES (9), suggesting that mutant H is structurally distorted. We thus conclude that the absolutely conserved domain 2b loop is located proximately to rpS25. Because the signal corresponding to signal 7 in Figure 5G had not completely disappeared, nucleotides UU6096–6097 must also make contacts with rpS25.
Figure 4.
Base substitutions in the single-stranded regions of domains 1–2 of the PSIV IRES to identify uridine residues that crosslink to the 40S ribosomal proteins. Mutations indicated by A–I correspond to the mutant IRES elements used in the experiments in Figure 5. To facilitate T7 transcription and HindIII digestion, the 5′ and 3′ terminal sequences were modified from the native sequence shown in Figure 1.
Figure 5.
Identification of uracil residues of the IRES that interact with rpS25. Shown are autoradiographs of second-dimension gels separating the Drosophila 40S ribosomal proteins crosslinked with domain 1–2 mutants of the PSIV IRES containing s4U. Signals corresponding to the rpS25 are circled by broken lines. (A) U6012A+U6015A+U6017A mutation; (B) UU6031AA+UUU6036-8AAA mutation; (C) UU6044-5AA mutation; (D) UU6062-3AA+U6066A mutation; (E) UU6073-4AA mutation; (F) UU6082-3CC mutation; (G) UUU6089-91AAA mutation; (H) UU6096-7CC mutation and (I) U6130A mutation.
IGR-IRES contact sites on the 40S ribosome
The cryo-electron microscopy study of CrPV IRES–human ribosome complex revealed that the IRES is located at the E-, and P-sites and at part of the A-site on the 40S and 80S ribosomes (12). Domain 3 mimics a tRNA-mRNA codon-anticodon pairing to determine the reading frame to be translated. Previous biochemical analyses have shown that domain 3 is functionally independent of domains 1–2, which have a major role in binding to the 40S ribosome (7,9,22). Hydroxyl radical cleavage analyses show that the domain 3 region faces the docked site of eIF1 on the 40S ribosome (10). In addition, helix 30, which is known to contain the contact site for the anticodon stem loop of the P-site tRNA (24), also contacts part of the CrPV IRES, which is extended from the head of the E-site to the P-site (12). These observations suggest that part of domain 3, which cannot be reached by hydroxyl radicals from eIF1, is likely to be close to helix 30. A few nucleotides around 6172 or 6195 in CrPV, similarly those around 6148 or 6171 in PSIV, may be candidates for interacting with helix 30.
The cryo-electron microscopy study also revealed that the CrPV-IRES interacts with rpS5 (12). To estimate whether our crosslinking signals involve rpS5, ribosomal proteins laying in positions that could be extrapolated to signals 2 and 3 were identified by mass spectrometry (Figure 2B and C); the other four signals (signals 1, 4, 5 and 6) were assumed to have molecular masses inconsistent with rpS5. A database search of the protein spots analyzed identified rpS2, rpS4 and rpS6, but not rpS5. This observation suggests that rpS5 may interact with IGR-IRES via nucleotides other than uracils. rpS5 has also been shown to interact with the IRES of hepatitis C virus (HCV) (25). Recently, studies using a reconstituted eukaryotic translation system have indicated that the HCV IRES can initiate translation in the absence of eIFs at higher concentrations of divalent ions (26). Because the IRES elements of CrPV and HCV are located at the mRNA cleft of the 40S ribosome (12,27), both IRES elements probably interact with the same ribosomal protein, rpS5.
rpS25 showed the strongest interaction with the PSIV IRES. The exact position of rpS25 on the ribosome is unknown, because there is no prokaryotic counterpart of this protein and the crystal structure of a eukaryotic ribosome has not been solved. However, rpS25 of the rat liver ribosome lies adjacent to rpS5 and is present at the decoding site of the ribosome (28). Because the domain 2 region of the PSIV IRES alone can bind to the 40S ribosome and the domain 2b stem-loop showed the strongest protection from chemical probes in the presence of the 40S ribosome (9), the rpS5 and rpS25 proteins should be two of the most important components of the 40S ribosome involved in forming the IRES–40S ribosome complex. The postulated interaction between the IRES and rpS25 is consistent with the results of the cryo-electron microscopy study and with hydroxyl radical cleavage, in which domains 1–2 of the CrPV IRES occupy the E-site of the ribosome. These observations suggest that interaction with rpS25 is likely to be important for IGR-IRES elements to dock on the ribosome.
Very recently, cryo-EM reconstruction of the CrPV IRES bound to yeast ribosome (29) and a tertiary structure of domain 1–2 of the PSIV IRES (30) are reported. The tertiary structure of the PSIV domain 1–2 shows that the two conserved loop regions, the AUUU loop and the CAGCC loop are located closely (30). A docked model with previous cryo-EM model (12) indicated that the two conserved loops contact with the rpS5 (30). While the recent cryo-EM reconstitution reports that the AUUU loop contacts with yeast rpS5, and that the CAGCC loop contacts with rpSX, a ribosomal protein with unknown identity (29). Our biochemical results indicate that the AUUU loop contacts with the rpS25, but not with rpS5. A plausible explanation for this conflict is that the rpSX in the recent cryo-EM reconstitution may be rpS25 and that rpS25 and rpS5 together form contact surfaces of 40S ribosome. This conflict would be elucidated by further biochemical experiments that identify the protein in contact with the CAGCC loop.
RNA–protein interaction is important for IRES–ribosome binding
The IGR-IRES elements of dicistroviruses form specific contacts with the ribosomal P- and E-sites in the absence of proteinous eIFs (4,7,8). In addition, after initial binding to the 40S ribosome, the IGR-IRES manipulates the structural conformation of the ribosome into an elongation mode in the presence of 60S ribosome (12). Indeed, a reconstituted translation system has shown that IGR-IRES-mediated translation can start in the absence of eIFs (8,31,32). These features raise the question of whether the IGR-IRES is a candidate relict of an ancient translation system (33,34). It has been generally accepted that the functions of RNAs have been substituted by those of proteins during the process of evolution. Indeed, structure analyses of translational apparatus suggest that some mitochondrial tRNAs lack a T-arm or D-arm and that the deletion of these structures has been compensated by part of a prolonged EF-Tu protein (35). In addition, functional replacement of shortened ribosomal RNA with additional ribosomal proteins is observed in mitochondrial 30S ribosomes (36).
We first targeted the 18S rRNA for interaction sites of the IGR-IRES because the IGR-IRES appears to exclude protein functions for translation initiation. Our chemical modifications and crosslink analyses, however, did not detect affirmative interaction sites on the 18S rRNA. In contrast, the s4U-labeled IGR-IRES interacted with several ribosomal proteins (Figure 2B). These features resemble the interactions between the 40S ribosome and the HCV IRES (37,38). Our previous analysis (9) might also suggest that binding between the IGR-IRES and 40S ribosomes probably depends on RNA–protein interactions because hydroxyl radical probing is unlikely to detect RNA–RNA interactions that are exposed to the solvent side of a packed RNA structure (22). Taken together, these facts indicate that the IGR-IRES does not require the effects of protein-based eIFs but utilizes eukaryotic ribosomal proteins for ribosome recognition.
How did the IGR-IRES emerge?
The host organisms of dicistroviruses are invertebrates. Most viruses infecting invertebrates, such as baculoviruses and cytoplasmic polyhedrosis viruses, have occluded proteins that protect virions from environmental inactivation. Dicistroviruses, however, do not produce occluded proteins. Although it has been indicated that CrPV can survive in soil containing suitable components (39), most dicistroviruses are likely to be forced to continue cycling between replication and transmission. This implies that dicistroviruses are forced to multiply more rapidly and abundantly. As a result, dicistroviruses have accumulated evolutional variations and finally might have armed themselves with the IGR-IRES that intensifies capsid production. In addition, the protein with the strongest interaction, rpS25, does not have prokaryotic counterparts, which explains why bacterial ribosomes cannot recognize the IGR-IRES. These facts give rise to another model in which the IGR-IRES is a translation apparatus that developed for the survival of dicistroviruses, rather than the relict of a primitive translation system.
SUPPLEMENTARY DATA
Supplementary Data is available at NAR Online.
ACKNOWLEDGEMENTS
We thank Dr Shimoda for the gift of a D. melanogaster strain and for instruction on insect rearing. This work was supported by the program for Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN), Japan. Funding to pay the Open Access publication charge was provided by National Institute of Agrobiological Sciences.
Conflict of interest statement: None declared.
REFERENCES
- 1.Christian P, Carstens E, Domier L, Johnson L, Johnson K, Nakashima N, Scotti P, van der Wilk F. Dicistroviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. Virus Taxonomy VIII. San Diego: Elsevier Academic Press; 2005. pp. 783–788. [Google Scholar]
- 2.Sasaki J, Nakashima N. Translation initiation at the CUU codon is mediated by the internal ribosome entry site of an insect picorna-like virus in vitro. J. Virol. 1999;73:1219–1226. doi: 10.1128/jvi.73.2.1219-1226.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sasaki J, Nakashima N. Methionine-independent initiation of translation in the capsid protein of an insect RNA virus. Proc. Natl. Acad. Sci. U.S.A. 2000;97:1512–1515. doi: 10.1073/pnas.010426997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilson JE, Pestova TV, Hellen CUT, Sarnow P. Initiation of protein synthesis from the A site of the ribosome. Cell. 2000;102:511–520. doi: 10.1016/s0092-8674(00)00055-6. [DOI] [PubMed] [Google Scholar]
- 5.Kanamori Y, Nakashima N. A tertiary structure model of the internal ribosome entry site (IRES) for methionine-independent initiation of translation. RNA. 2001;7:266–274. doi: 10.1017/s1355838201001741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pestova TV, Kolupaeva VG, Lomakin IB, Pilipenko EV, Shatsky IN, Agol VI, Hellen CUT. Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl. Acad. Sci. U.S.A. 2001;98:7029–7036. doi: 10.1073/pnas.111145798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jan E, Sarnow P. Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J. Mol. Biol. 2002;324:889–892. doi: 10.1016/s0022-2836(02)01099-9. [DOI] [PubMed] [Google Scholar]
- 8.Pestova TV, Hellen CUT. Translation elongation after assembly of ribosomes on the cricket paralysis virus internal ribosome entry site without initiation factors or initiator tRNA. Genes Dev. 2003;17:181–186. doi: 10.1101/gad.1040803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nishiyama T, Yamamoto H, Shibuya N, Hatakeyama Y, Hachimori A, Uchiumi T, Nakashima N. Structural elements in the internal ribosome entry site of Plautia stali intestine virus responsible for binding with ribosomes. Nucleic Acids Res. 2003;31:2434–2442. doi: 10.1093/nar/gkg336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pestova TV, Lomakin IB, Hellen CUT. Position of the CrPV IRES on the 40S subunit and factor dependence of IRES/80S ribosome assembly. EMBO Rep. 2004;5:906–913. doi: 10.1038/sj.embor.7400240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shibuya N, Nishiyama T, Kanamori Y, Saito H, Nakashima N. Conditional rather than absolute requirements of the capsid coding sequence for initiation of methionine-independent translation in Plautia stali intestine virus. J. Virol. 2003;77:12002–12010. doi: 10.1128/JVI.77.22.12002-12010.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spahn CMT, Jan E, Mulder A, Grassucci RA, Sarnow P, Frank J. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: the IRES functions as an RNA-based translation factor. Cell. 2004;118:465–475. doi: 10.1016/j.cell.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 13.Bradford CT, Sullivan DT. Isolation of polysomes from larval and adult Drosophila melanogaster. Anal. Biochem. 1981;112:176–181. doi: 10.1016/0003-2697(81)90277-3. [DOI] [PubMed] [Google Scholar]
- 14.Blobel G, Sabatini D. Dissociation of mammalian polyribosomes into subunits by puromycin. Proc. Natl. Acad. Sci. U.S.A. 1971;68:390–394. doi: 10.1073/pnas.68.2.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakashima N, Sasaki J, Toriyama S. Determining the nucleotide sequence and capsid-coding region of himetobi P virus: a member of a novel group of RNA viruses that infect insects. Arch. Virol. 1999;144:2051–2058. doi: 10.1007/s007050050726. [DOI] [PubMed] [Google Scholar]
- 16.Dubreuil YL, Expert-Bezançon A, Favre A. Conformation and structural fluctuations of a 218 nucleotides long rRNA fragment: 4-thiouridine as an intrinsic photolabelling probe. Nucleic Acids Res. 1991;19:3653–3660. doi: 10.1093/nar/19.13.3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hardy SJ, Kurland CG, Voynow P, Mora G. The ribosomal proteins of Escherichia coli. I. Purification of the 30S ribosomal proteins. Biochemistry. 1969;8:2897–2905. doi: 10.1021/bi00835a031. [DOI] [PubMed] [Google Scholar]
- 18.Wada A. Analysis of Escherichia coli ribosomal proteins by an improved two dimensional gel electrophoresis. I Detection of four new proteins. J. Biochem. (Tokyo) 1986;100:1583–1594. doi: 10.1093/oxfordjournals.jbchem.a121866. [DOI] [PubMed] [Google Scholar]
- 19.Moazed D, Noller HF. Transfer RNA shields specific nucleotides in 16S ribosomal RNA from attack by chemical probes. Cell. 1986;47:985–994. doi: 10.1016/0092-8674(86)90813-5. [DOI] [PubMed] [Google Scholar]
- 20.Hatakeyama Y, Shibuya N, Nishiyama T, Nakashima N. Structural variant of the intergenic internal ribosome entry site elements in dicistroviruses and computational search for their counterparts. RNA. 2004;10:779–786. doi: 10.1261/rna.5208104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Favre A, Saintome C, Fourrey JL, Clivio P, Laugaa P. Thionucleobases as intrinsic photoaffinity probes of nucleic acid structure and nucleic acid-protein interactions. J. Photochem. Photobiol. B. 1998;42:109–124. doi: 10.1016/s1011-1344(97)00116-4. [DOI] [PubMed] [Google Scholar]
- 22.Costantino D, Kieft JS. A preformed compact ribosome-binding domain in the cricket paralysis-like virus IRES RNAs. RNA. 2005;11:332–343. doi: 10.1261/rna.7184705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van de Peer Y, de Rijk P, Wuyts J, Winkelmans T, de Wachter R. The European small subunit ribosomal RNA database. Nucleic Acids Res. 2000;28:175–176. doi: 10.1093/nar/28.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:868–869. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 25.Fukushi S, Okada M, Stahl J, Kageyama T, Hoshino HB, Katayama K. Ribosomal protein S5 interacts with the internal ribosomal entry site of hepatitis C virus. J. Biol. Chem. 2001;276:20824–20826. doi: 10.1074/jbc.C100206200. [DOI] [PubMed] [Google Scholar]
- 26.Lancaster AM, Jan E, Sarnow P. Initiation factor-independent translation mediated by the hepatitis C virus internal ribosome entry site. RNA. 2006;12:894–902. doi: 10.1261/rna.2342306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spahn CMT, Kieft JS, Grassucci RA, Penczek PA, Zhou K, Doudna JA, Frank J. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40S ribosomal subunit. Science. 2001;291:1959–1962. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- 28.Uchiumi T, Terao K, Ogata K. Identification of neighboring protein pairs cross-linked with dimethyl 3,3′-dithiobispropionimidate in rat liver 40S ribosomal subunits. J. Biochem. (Tokyo) 1981;90:185–193. doi: 10.1093/oxfordjournals.jbchem.a133449. [DOI] [PubMed] [Google Scholar]
- 29.Schüler M, Connell SR, Lescoute A, Giesebrecht J, Dabrowski M, Schroeer B, Mielke T, Penczek PA, Westhof E, et al. Structure of the ribosome-bound cricket paralysis virus IRES RNA. Nat. Struct. Mol. Biol. 2006;13:1092–1096. doi: 10.1038/nsmb1177. [DOI] [PubMed] [Google Scholar]
- 30.Pfingsten JS, Costantino D, Kieft JS. Structural basis for ribosome recruitment and manipulation by a viral IRES RNA. Science. 2006;314:1450–1454. doi: 10.1126/science.1133281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jan E, Kinzy TG, Sarnow P. Divergent tRNA-like element supports initiation, elongation and termination of protein biogenesis. Proc. Natl. Acad. Sci. U.S.A. 2003;100:15410–15415. doi: 10.1073/pnas.2535183100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pestova TV, Hellen CUT. Reconstitution of eukaryotic translation elongation in vitro following initiation by internal ribosomal entry. Methods. 2005;36:261–269. doi: 10.1016/j.ymeth.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 33.Lehto K, Karentikov A. Relicts and models of the RNA world. Int. J. Astrobiol. 2005;4:33–41. [Google Scholar]
- 34.Atkins JF, Gesteland RF, Jackson RJ, Wills NM. The shapely mRNA: Knotting ventured, knotting gained. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. Third. NY, USA: Cold Spring Harbor Laboratory Press, Cold Spring Harbor; 2006. pp. 437–467. [Google Scholar]
- 35.Otsuki T, Sato A, Watanabe Y, Watanabe K. A unique serine-specific elongation factor Tu found in nematode mitochondria. Nat. Struct. Biol. 2002;9:669–673. doi: 10.1038/nsb826. [DOI] [PubMed] [Google Scholar]
- 36.Zhao F, Ohtsuki T, Yamada K, Yoshinari S, Kita K, Watanabe Y, Watanabe K. Isolation and physiochemical properties of protein-rich nematode mitochondrial ribosomes. Biochemistry. 2005;44:9232–9237. doi: 10.1021/bi047833c. [DOI] [PubMed] [Google Scholar]
- 37.Otto GA, Lukavsky PJ, Lancaster AM, Sarnow P, Puglisi JD. Ribosomal proteins mediate the hepatitis C virus IRES-HeLa 40S interaction. RNA. 2002;8:913–923. doi: 10.1017/s1355838202022057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laletina E, Graifer D, Malygin A, Ivanov A, Shatsky I, Karpova G. Proteins surrounding hairpin IIIe of the hepatitis C virus internal ribosome entry site on the human 40S ribosomal subunit. Nucleic Acids Res. 2006;34:2027–2036. doi: 10.1093/nar/gkl155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Christian PD, Richards AR, Williams T. Differential adsorption of occluded and nonoccluded insect-pathogenic viruses to soil-forming minerals. Appl. Environ. Microbiol. 2006;72:4648–4652. doi: 10.1128/AEM.00254-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.