Abstract
The parD operon of Escherichia coli plasmid R1 encodes a toxin–antitoxin system, which is involved in plasmid stabilization. The toxin Kid inhibits cell growth by RNA degradation and its action is neutralized by the formation of a tight complex with the antitoxin Kis. A fascinating but poorly understood aspect of the kid–kis system is its autoregulation at the transcriptional level. Using macromolecular (tandem) mass spectrometry and DNA binding assays, we here demonstrate that Kis pilots the interaction of the Kid–Kis complex in the parD regulatory region and that two discrete Kis-binding regions are present on parD. The data clearly show that only when the Kis concentration equals or exceeds the Kid concentration a strong cooperative effect exists between strong DNA binding and Kid2–Kis2–Kid2–Kis2 complex formation. We propose a model in which transcriptional repression of the parD operon is tuned by the relative molar ratio of the antitoxin and toxin proteins in solution. When the concentration of the toxin exceeds that of the antitoxin tight Kid2–Kis2–Kid2 complexes are formed, which only neutralize the lethal activity of Kid. Upon increasing the Kis concentration, (Kid2–Kis2)n complexes repress the kid–kis operon.
INTRODUCTION
Toxin–antitoxin systems in bacteria eliminate plasmid-free cells that emerge as a result of segregation or replication defects and they contribute to intra- and interspecies plasmid dissemination (1–3). Plasmid-encoded toxin–antitoxin systems and their chromosomal homologues are widespread in bacteria. Chromosomal toxin–antitoxin systems have been proposed to induce reversible cell cycle arrest or plasmid stabilization in response to nutritional and/or environmental stress. Toxin–antitoxin cassettes have a characteristic organization in which the gene encoding the toxin follows the gene encoding the antitoxin. The two loci have often a common autoregulatory mechanism exerted by both components. The toxin gene encodes a stable protein, whereas the antitoxin is either a non-translated, antisense RNA species (type I) or a labile protein (type II).
The majority of the plasmid and chromosome-encoded toxin–antitoxin loci is of the type II module (2). The toxin and antitoxin form a tight complex so that no free toxin is present in the cell. When a plasmid-free daughter cell is produced, owing to a defect in plasmid replication or maintenance, the newborn cell will still inherit the toxin–antitoxin protein complex. However, the antitoxin component is degraded easily by host proteases and is not refreshed because of the absence of the plasmid encoding for the toxin–antitoxin system. The toxin will then act on an essential host target to cause growth impairment or cell death of the plasmid-free cell. In spite of the many studies on type II toxin–antitoxin systems, only two intracellular targets have been identified. CcdB and ParE are known to act on DNA gyrase (4,5), RelE mediates cleavage of mRNA in a ribosome-dependent manner, thereby affecting the level of protein synthesis (6) and MazF, Kid and YoeB proteins have been found to show ribosome-independent RNase activity (7–10).
Toxin–antitoxin systems that have been studied so far are autoregulated at the level of transcription by binding of the antitoxin to the operator-promoter region of the operon, however, the underlying molecular mechanism of this autoregulation is poorly understood. Several toxin–antitoxin pairs repress the transcription of their toxin–antitoxin operons, such as mazEF, relBE, kid–kis, ccd, higAB and doc-phd, indicating that an autoregulation process involving one or both proteins may be a common feature for these operons (11–15). In most of the cases, the antitoxin is directly responsible for the repression, but the toxin can also assist by increasing the affinity of the regulatory complex.
In Escherichia coli, the parD operon of plasmid R1 encodes the toxin Kid (Killing determinant) and the antitoxin Kis (Killing suppressor) (16). Kid is a ribonuclease, which cleaves RNA preferentially at the 5′ side of the adenosine residue in the nucleotide sequence 5′-UA(A/C)-3′ of single-stranded regions, although cleavage in double-stranded regions and at the 3′ side of the adenosine has been observed as well (17,18). Kis prevents the inhibition of E. coli cell growth caused by Kid. Kis autoregulates parD transcription to a limited extent and this activity can be allocated to the N-terminal region of the protein (19). The coordinate action of the Kid–Kis complexes has been shown to efficiently repress parD transcription (11,18,20). In addition, synthesis of the Kid toxin is coupled to the synthesis of the Kis antitoxin and the intracellular levels of these proteins are also controlled by limited degradation of a polycistronic messenger (21). These regulatory mechanisms avoid the synthesis of the toxic component in case its antitoxin has not been translated previously and ensures a balanced production of the antitoxin relative to the toxin (22).
For mazEF and ccd addiction complexes it has been shown that the toxin and antitoxin can form various assemblies with different stoichiometries (23–25). Dao-Thi et al. (23) have proposed a model in which (CcdA2-CcdB2)n complexes interact with multiple DNA-binding sites and spiral around the 120-bp promoter region. Kis and Kid also form various complexes. The Kid2–Kis2–Kid2 heterohexamer is the most abundant species when Kid is in excess of Kis, whereas at higher concentrations of Kis, various complexes are present ranging from Kid2–Kis2 tetramer up to heterodecamers, however, the function of these complexes and especially the interactions with operator-promoter DNA has not been elucidated (M.B. Kamphuis et al., submitted for publication).
In this study, we aimed to unravel the mechanism of autoregulation at the transcriptional level of the type II toxin–antitoxin system kid–kis by analyzing the Kid–Kis complexes formed at the parD operon. We focused on the dynamic changes of the stoichiometry of Kid–Kis oligomers induced by binding of the parD operon and on their different binding affinity by using electrophoretic mobility shift assays, hydroxyl radical footprinting and macromolecular mass spectrometry. Mass spectrometry is a relatively new player in the field of structural biology of non-covalent protein–nucleic acid complexes, which allows for the analysis of multiple species in a single experiment (26,27). Moreover, in combination with gas-phase dissociation experiments, the data also provide insight in the global organization of complexes (28–30). Based on our results, we present a detailed model, which explains the transcriptional autoregulation process of the parD operon.
MATERIALS AND METHODS
Proteins and DNA
Kid toxin, 15N-labelled Kis antitoxin and His-tagged Kis were overexpressed and purified essentially as described previously (18,31, M.B. Kamphuis et al., submitted for publication). The predicted masses for these proteins on the basis of the primary sequence were 12 038, 9689 and 10 885 Da, respectively.
The dsDNA fragments used for the DNA binding and footprinting assays were obtained from PCR amplification using the Sau3A fragment from pKN1562 as DNA template, including the parD operator-promoter region, cloned into the BamHI site of pUC18. Oligonucleotides (1) 5′-TATGGAAGCAACCACGCT-3′, (2) 5′-TCAGCATAACTGAGCC-3′, (3) 5′-GTGCGTTAAAGCCTGGTGTGT-3′ and (4) 5′-CACACCAGGCTTTAACGCAC-3′ were synthesized. PCR was performed after 5′-end labelling of one of the oligonucleotides with [γ-32P] ATP and T4 polynucleotide kinase. The size of the amplification products was 175-bp (oligonucleotides 1 and 2), 81-bp (oligonucleotides 1 and 4) and 115-bp (oligonucleotides 2 and 3). The 175-bp DNA fragment contains the operator-promoter of the parD operon (including region I and II) (Figure 2C), whereas the 115-bp and 81-bp DNA fragments contain only region I or II, respectively (Figure 4A and B). Each DNA fragment was analysed on a 5% polyacrylamide native gel. The end-labelled DNA fragments were eluted from the gel at 42°C for 12 h in a buffer containing sodium chloride (200 mM), Tris/hydrochloride (20 mM) and ethylenediamine tetraacetic acid (2 mM), pH 7.4. For mass spectrometry studies, we used a 30-bp parD DNA fragment (upper strand 5′-GGATGTTATATTTAAATATAACTTTTATGG-3′), containing parD region I plus 2 bp upstream and 5 bp downstream and a 30-bp dsDNA fragment with a random sequence (upper strand 5′-AGCTGCCAGGCACCAGTGTCAGCGTCCTAT-3′).
Figure 2.
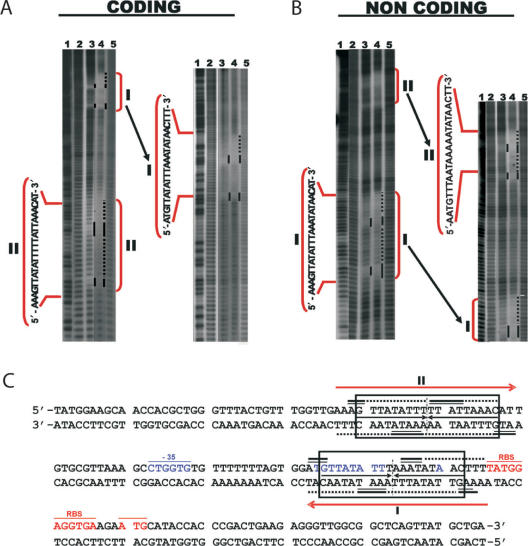
Kid–Kis and Kis interact at specific sites with parD DNA. Hydroxyl radical footprinting assays were performed on Kid–Kis mixtures (Kid–Kis ratio 1:2; Kid 2.4 and Kis 4.8 µM) and Kis (4.8 µM) alone on the 175-bp parD region I/II fragment. Protections in the coding (A) and non-coding (B) strands are indicated by black bars and dots. Lane 3 shows the protection pattern by Kis alone and lane 4 shows the protection pattern by the Kid–Kis complex. The sequences of the inverted repeats I and II that include the protected regions are indicated. Lanes 2 and 5 show the cleavage pattern of the DNA in the absence of any added protein. Lane 1 shows the Maxam–Gilbert AG ladder sequence. (C) Summary of the protected sites in parD region I/II by Kis and Kid–Kis complexes. The protected regions are indicated with numbers I and II. Region I contains an 18-bp perfect two-fold symmetry element (boxed) that includes the −10 motif. The site II includes an 18-bp pseudo-symmetric element that is also boxed. The dyad symmetry axis in each region is indicated with a broken line. Bases whose deoxyriboses are protected by Kis (thick bars) or Kid–Kis (thin bars) from cleavage by hydroxyl radical are indicated (underlined). DNA sequence of the −35 (underlined and labelled) and −10 elements of the promoter and the transcription initiation site are in blue. The ribosome-binding site (RBS) and translation initiation condon (Met) of kis are underlined and in red. The imperfect inverted repeats I and II are indicated with red arrows. The 30-bp DNA fragment used for mass spectrometry studies contains parD region I plus 2 bp upstream and 5 bp downstream.
Figure 4.
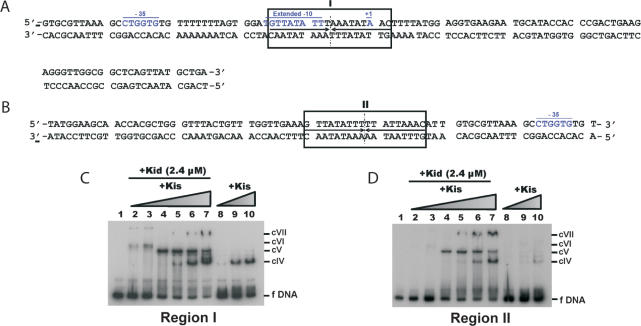
Kis and Kid–Kis complexes interact tighter to parD region I than to the parD region II. (A and B) Summary of the protected sites in region I and II of parD by Kis and Kid–Kis complexes. The 18-bp symmetric element (region I) and the 18-bp pseudo-symmetric element (region II) are boxed, and the broken lines indicate the symmetry axis. The sequence of the −35 and the extended −10 motifs and the initiation transcription (+1) are underlined and in blue. (C and D) Electrophoretic mobility shift assays were performed on the 5′-end-labelled 115-bp parD region I fragment (2 nM) or on the 5′-end-labelled 81-bp parD region II fragment (2 nM) and Kis alone (2.4, 4.8 and 9.6 μM) (lanes 8–10) or a combination of Kid (2.4 µM) and Kis (0.3, 0.6, 1.2, 2.4, 4.8 and 9.6 μM) (lanes 2–7). Lane 1 presents the negative control without proteins. The specific complexes formed are indicated with cIV, cV, cVI and cVII.
Macromolecular mass spectrometry
All mass spectrometry studies were performed in aqueous ammonium acetate (100 mM), pH 5.8. Mixtures of Kis, Kid and 30-bp parD region I DNA fragment were incubated at 20°C for 5 min before analysis. The Kis concentration was fixed at 7.5 µM when Kis alone or a Kid:Kis mixture at 2:1 molar ratio was incubated with dsDNA, whereas the Kis concentration was 15 µM when a Kid:Kis mixture at 1:1 molar ratio was incubated with parD DNA. The Kis:parD DNA molar ratios ranged from 80:1 to 20:1. Borosilicate glass capillaries (Kwik-Fil, World Precision Instruments, Inc., Sarasota, FL, USA) were used on a P-97 puller (Sutter Instrument Co., Novato, CA, USA) to prepare the nanoflow electrospray capillaries with an orifice of about 5 µm. The capillaries were subsequently coated with a thin gold layer (∼500 Å) by using an Edwards ScanCoat Six Pirani 501 sputter coater (Edwards High Vacuum International, Crawley, UK).
For native mass spectrometry experiments, samples were introduced into a nanoflow electrospray ionization orthogonal time-of-flight mass spectrometer (Micromass LC-T, Waters, Manchester, UK) modified for high mass operation and operating in positive ion mode. To generate intact ions in vacuo from protein complexes in solution, the ions were cooled by increasing the pressure in the first vacuum stages of the mass spectrometer. The pressure in the source region was adjusted to 7.0 mbar by reducing the pumping capacity of the rotatory pump by closing the speed-valve (32). In addition, nanoflow electrospray voltages were optimized for transmission of intact protein complexes and for efficient desolvation using capillary and cone voltages of 1200–1300 V and 50–60 V, respectively. All spectra were mass calibrated by using an aqueous solution of cesium iodide (5 mg/ml).
Tandem mass spectrometry is routinely used for the fragmentation of peptides, and more recently also for the gas-phase dissociation of protein complexes (29,30,33,34). In these experiments, ions of a defined m/z ratio are isolated by a (high-mass) quadrupole analyzer and subsequently dissociated by increasing the acceleration voltage in a gas-filled, usually argon or xenon, collision cell. Experimentally, it has been demonstrated that a preference is expected for the dissociation of the smallest protein and/or proteins that are at the surface of the complex. The proteins that dissociate usually take up a relatively large number of charges (35). For gas-phase dissociation experiments, samples were introduced into a Micromass Q-ToF 1 mass spectrometer (Waters, Manchester, UK) equipped with a nanoflow Z-spray source and modified for high mass-to-charge ion isolation and high mass operation (29) and operating in positive ion mode. Precursor ions of selected complex compositions were isolated in the quadrupole analyzer and accelerated into an argon-filled collision cell. Different collision energies (10–150 V) were used in combination with a gas pressure of 0.8 mbar. The voltages were optimized for transmission of intact protein complexes and for efficient desolvation using capillary and cone voltages of 1200 and 60 V, respectively. To confirm each result, at least two independent charge states were selected for each protein–parD DNA complex.
Mass spectrometry data analysis
The mass spectrometry data were semi-quantified to determine the relative amount of the protein complexes present in the different experiments. Data were accumulated over 2 min, averaged, smoothed and centred, thereby using the areas option in the software program MassLynx 4.0 (Waters). The total ion intensity for each complex was calculated by summing the intensity of all ions belonging to the Gaussian charge state envelope of the protein complex under analysis. The percentage of each protein complex was then calculated by using the total ion intensity of all identified protein complexes. The calculated relative abundances were based on two independent measurements.
Electrophoretic mobility shift assays
Binding of Kid and Kis proteins to 5′-end-labelled 175-bp parD region I/II, 81-bp parD region II or 115-bp parD region I fragment (1.4 × 106 c.p.m./mol) was determined according to a previously described method (36) with modifications. The binding reactions contained end-labelled DNA (2 nM), Tris/hydrochloride (70 mM), potassium chloride (200 mM), magnesium chloride (14 mM), sodium chloride (80 mM), ethylenediamine tetraacetic acid (40 mM), bovine serum albumin (100 µg/ml), glycerol (0.5% (v/v)) and Kid (2.4 μM) and/or Kis (0.075–9.6 μM). The assays were also performed with higher parD DNA concentrations (375 nM) and Kis (7.5 or 15 μM) and/or Kid (7.5 or 15 μM). The mixtures were incubated at 4°C for 60 min. Where indicated, poly [d(I-C)] (70 μM) as non-specific competitor DNA was added and incubation was continued for another 5 min at the same temperature. Free and bound parD DNA fragments were separated on polyacrylamide (5 or 8% (w/v)) native gels. The gels were run at 4°C in TBE buffer (Tris/hydrochloride (89 mM), boric acid (89 mM) and ethylenediamine tetraacetic acid (10 mM), pH 8.9) at 100 V for 60 min. The labelled DNA bands were visualized by autoradiography.
Hydroxyl radical footprinting
5′-End-labelled 175-bp parD region I/II or 115-bp parD region I fragment was incubated with Kid (2.4 µM) and/or Kis (4.8 µM) under the same conditions as in the electrophoretic mobility shift assays. After incubation, a solution containing iron sulphate (100 μM), ethylenediamine tetraacetic acid (200 μM), sodium ascorbate (1 mM) and hydroperoxide (0.3%) was added, and the mixture was incubated at 4°C for 5 min. The cleavage reaction was terminated by the addition of a mixture containing thiourea (8 mM) and ethylenediamine tetraacetic acid (1.7 mM). DNA–protein complexes were separated from free DNA by polyacrylamide 5% (w/v) gel electrophoresis and visualized by autoradiography. Bound DNA was eluted from gels at 42°C for 12 h, in buffer sodium chloride (200 mM), Tris/hydrochloride (20 mM) and ethylenediamine tetraacetic acid (2 mM), pH 8.0. The DNA was precipitated with ethanol, dissolved in formamide-dye solution and analysed by polyacrylamide 8% (w/v) gel electrophoresis in the presence of urea (8 M). The cleavage products were visualized by autoradiography.
RESULTS
Antitoxin Kis, but not toxin Kid, interacts with parD DNA
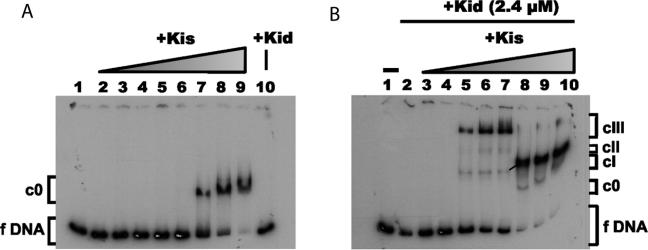
Antitoxin Kis is known to be a weak transcriptional repressor of the parD operon, however, the characteristics of its interaction with the regulatory region of this operon are unknown (19). We first analysed by electrophoretic mobility shift assays, the interaction of Kis with a 175-bp DNA fragment (parD region I/II) containing sequences that include the operator-promoter region of the parD operon and further upstream sequences (Figure 1A). At Kis:parD region I/II molar ratios lower than 1200:1 no complex was observed between the protein and the parD DNA. At molar ratios of 1200:1 and 2400:1 (lanes 7 and 8) both free DNA and a mobility-shifted complex (c0) was observed, whereas at a molar ratio of 4800:1 (lane 9) no free parD DNA was observed. These data clearly show that Kis interacts with low affinity with the parD operon and the sharp transition from unbound DNA to the c0 complex points to a strong cooperative interaction of Kis onto the operator-promoter region.
Figure 1.
Effect of Kid on the interaction of Kis to parD DNA. Electrophoretic mobility shift assays were performed on the 5′-end-labelled 175-bp parD region I/II fragment (2 nM) and Kis and/or Kid. (A) Band-shift assays in the presence of a range of concentrations of Kis (0.075, 0.150, 0.300, 0.600, 1.2, 2.4, 4.8 and 9.6 µM) (lanes 2–9). Lane 10 presents control with only Kid (2.4 μM). (B) Band-shift assays over a range of Kis concentrations identical to the ones in (A) (lanes 3–10) and in the presence of a fixed concentration of Kid (2.4 µM). Lane 2 shows a control without Kid. Lane 1 presents the negative control without proteins. The specific complexes formed are indicated with c0, cI, cII and cIII.
The specific Kis contacts on the 175-bp parD region I/II fragment were further analysed by hydroxyl radical footprinting assays. These assays showed that Kis interacts in two imperfect inverted repeats (regions I and II) of 18 bp each and separated by 33 bp (Figure 2C). Region I contains a perfect palindromic sequence whose left half overlaps with the −10 promoter element in which protection of Kis is prominent, suggesting a direct role of these interactions in parD regulation.
The binding of Kid was tested under the same conditions, however, no complexes of Kid and parD DNA were observed, confirming that the toxin does not directly interact with the parD operon (Figure 1A, lane 10).
Kis–parD DNA interactions involve dimeric Kis subunits
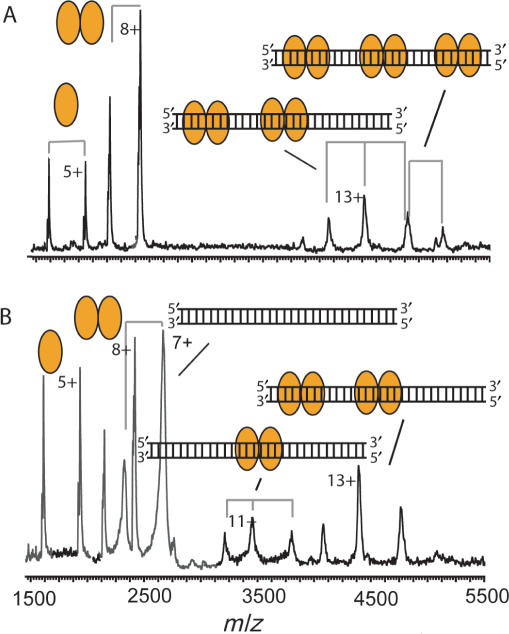
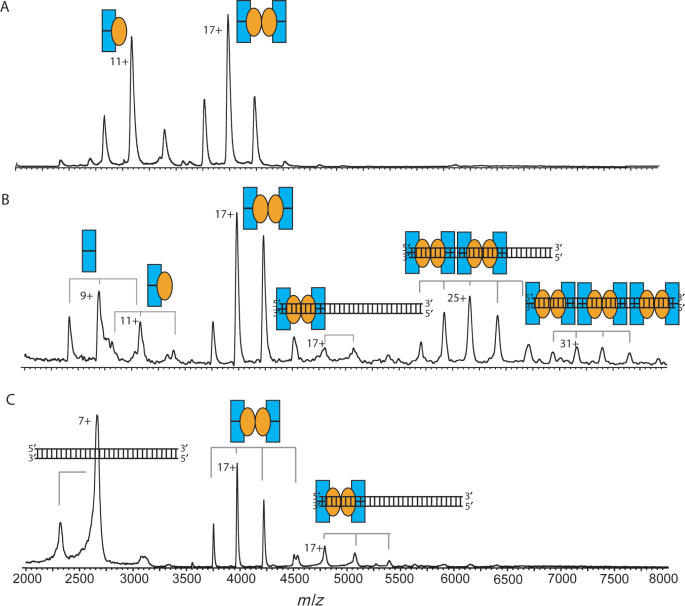
We synthesized a 30-bp palindromic DNA fragment of the 175-bp parD region I/II, comprising region I, which we could use for nanoflow electrospray ionization mass spectrometry (Figure 2C). The 175-bp fragment is not suited for mass spectrometry studies as this large fragment retains many buffer and salt molecules, which represses the ion signal and introduces many satellite ion signals. From the mass spectra of the 30-bp parD region I and Kis we determined their molecular masses: 18 408.3 Da for parD DNA and 9687 Da and 19 373 Da for monomeric and dimeric Kis, respectively (Supplementary Table 1). The determined mass of the 30-bp parD DNA matches exactly with the theoretical mass, whereas the determined mass of monomeric Kis corresponds well with the mass as calculated from the primary sequence of 15N-labelled Kis attached to a β-mercaptoethanol molecule (M.B. Kamphuis et al., submitted for publication). Upon incubation of the 30-bp parD region I with a 20-fold molar excess of Kis, the mass spectrum clearly showed four charge-state distributions (Figure 3A, Supplementary Table 1). Two ion series were detected ∼2000 m/z and could be assigned to monomeric and dimeric Kis and two other ion series were detected at m/z values ∼4500. Mass determination of these latter two ion series revealed masses of 57 074 and 76 431 Da, which corresponds with the masses of (Kis2)2–DNA1 and (Kis2)3–DNA1 complexes, respectively. Upon increasing the amount of the 30-bp parD DNA relative to Kis (5-fold molar excess of Kis) also ions originating from free DNA and Kis2–DNA1 (37 801 Da) were identified (Figure 3B). Each protein–DNA complex involves at least one dimeric Kis unit showing that the Kis dimer is required for the specific recognition process.
Figure 3.
Dimers of Kis interact with parD region I. Macromolecular native mass spectrometry was performed on Kis-30-bp parD region I complexes in ammonium acetate (50 mM), pH 5.8. (A) Mass spectrum of the Kis:parD region I mixture at a molar ratio of 20:1 (Kis 7.5 μM). (B) Mass spectrum of Kis:parD region I mixture at a ratio of 5:1 (Kis 7.5 μM). Kis monomer and dimer are indicated with single and double orange ellipses, respectively, and the parD region I fragment with a double strand. Each complex is represented by an appropriate combination of ellipses and/or DNA double strand. Molecular masses and relative amounts of complexes are shown in Supplementary Tables 1 and 2, respectively.
Data analysis revealed that only ∼15% of Kis was bound to DNA, whereas the remaining 85% was free in solution (Supplementary Table 2). This clearly demonstrates that antitoxin Kis interacts weakly with the parD region I. It should be noted here that ionization and transmission efficiency is protein and DNA dependent, therefore, we can only semi-quantify these data. To investigate whether the binding behaviour to the parD region I was indeed specific, the antitoxin Kis was added to a solution containing a 30-bp dsDNA with a random nucleotide sequence. The mass spectra did not show any protein–DNA complexes (data not shown) confirming that the recognition of the parD region I by Kis is highly sequence specific. As expected, incubations of Kid with the 30-bp parD region I did not result in the formation of binary Kid–parD DNA complexes.
Toxin Kid enhances antitoxin Kis binding to parD DNA
We performed electrophoretic mobility shift assays on complexes between Kid and Kis mixed in different oligomeric ratios and the 175-bp parD region I/II. The retardation of the parD DNA by Kis in the presence of a fixed concentration of Kid is shown in Figure 1B. The first mobility-shifted complexes were observed at a Kid:Kis:parD region I/II molar ratio of 1200:150:1 (lane 5). At this ratio, free DNA and three different mobility-shifted complexes were observed. The high intensity band was the largest complex (cIII), whereas the two additional bands (cII and cI) with lower intensity had a lower mass. The same pattern was also observed at Kid:Kis molar ratios of 4:1 and 2:1 (lanes 6 and 7).
At an equal concentration of both proteins, however, nearly no free DNA and complex cIII were detected and the intensity of complex cI was dramatically increased (lane 8). A less intense band (c0) that may correspond to DNA bound by Kis alone was also detected. The same pattern was maintained at decreased Kid:Kis molar ratios of 1:2 and 1:4 (lanes 9 and 10). These data show that when the concentration of Kis approaches that of Kid there is a dramatic shift in the complexes formed between Kid–Kis and parD region I/II, and the complexes with the DNA fragment become more stable. The data also show that in the presence of Kid the ternary complexes formed at higher concentrations of Kis over DNA have a lower molecular mass than the complexes formed at lower concentrations of Kis, suggesting that there are multiple binding sites for the Kid–Kis complex in the operator-promoter region of the DNA and that the ratio between the two proteins plays an important role in the formation of an efficient repressor complex. We can, however, not fully exclude that migration may also be affected by DNA bending or other conformational changes.
The Kid–Kis–parD DNA interactions were further analysed by DNA footprinting assays (Figure 2). At a molar ratio Kid:Kis:parD region I/II of 1200:2400:1, in which complexes c0 and cI were present, the protection in complex cI occurred in the same two imperfect inverted repeats (regions I and II), which were also protected by Kis alone. As Kid does not bind directly to DNA these results indicate that Kis pilots the specific interaction of this complex in the operator-promoter region. Moreover, the data show that the spacer region of 33 bp is not protected by the proteins.
parD region I interacts stronger with Kis and with Kid–Kis complexes than parD region II
The regions I and II were identified as the binding sites of Kis and Kid–Kis complexes in the parD operator-promoter region. With the aim to evaluate the binding of Kis and Kid–Kis complexes to isolated regions I and II, two different DNA fragments were produced: a 115-bp fragment comprising region I and an 81-bp fragment comprising region II (Figure 4A and B). The retardation of parD region I and parD region II by Kis is shown in Figure 4C and D. When using parD region I, a mobility-shifted complex (cIV) was observed at molar ratios Kis:parD equal or higher than 2400:1 (lanes 9 and 10), whereas, when using parD region II, a mobility-shifted complex (cIV) was observed at a Kis:parD molar ratio of 4800:1 (lane 10). These results thus show that the antitoxin Kis alone interacts specifically with both parD fragments, but the interaction with region I was tighter than with region II. The electrophoretic mobility shift assays were also performed with mixtures of Kid and Kis (Figure 4C and D). As in the assays with the 175-bp parD region I/II fragment, a fixed concentration of Kid and a variable concentration of Kis was used. For the parD region I fragment the first mobility-shifted complex was observed at a Kid:Kis:parD molar ratio of 1200:150:1 (lane 2), whereas for the parD region II fragment the first mobility-shifted complex was observed at a Kid:Kis:parD molar ratio of 1200:600:1 (lane 4). Thus, the affinity of the Kid–Kis complexes for parD region I is higher than for region II, which is in full agreement with the DNA binding assays using only Kis. The type of complexes formed between parD fragments and Kid–Kis complexes was dependent on the ratios between the two proteins. When the concentration of Kis was lower than the concentration of Kid, complex cVI was most abundant, whereas when the concentration of Kis approached the concentration of Kid, complex cV became more abundant. An additional complex (cIV) observed at an excess of Kis, may represent the binding to Kis alone, as it was also present in the incubations between Kis and parD DNA. A similar pattern was also observed with the parD region I/II fragment, although the abundance of complex c0 was very low (Figure 1). These differences between the isolated parD regions I and II and parD region I/II may suggest cooperative interactions between regions I and II.
The specific contacts between parD region I fragment and Kis or Kid–Kis complexes were further studied by hydroxyl footprinting assays (Supplementary Figure 1). The protection pattern clearly showed that the interactions of either Kis (cIV) or Kid–Kis complexes (cV) occur specifically on region I and involve in both cases the same backbone DNA contacts. The protection pattern observed in region I is in accordance with the footprint observed in the same operator region I in the 175-bp parD region I/II fragment (Figure 2). It should be noted here, however, that although the affinity with region I was higher than with region II, the protected sites observed in both regions in the 175-bp parD region I/II fragment had similar intensities. This again suggests cooperative interactions between regions I and II.
A molar excess of Kid results in labile (Kid2–Kis2–Kid2)n–parD DNA1 complexes
The retardation assays showed that multiple mobility-shifted complexes can be formed between Kid, Kis and parD operon, but do not allow determination of the stoichiometry of the complexes. Therefore, we studied the Kid–Kis oligomers involved in interaction with the 30-bp parD region I by macromolecular native mass spectrometry.
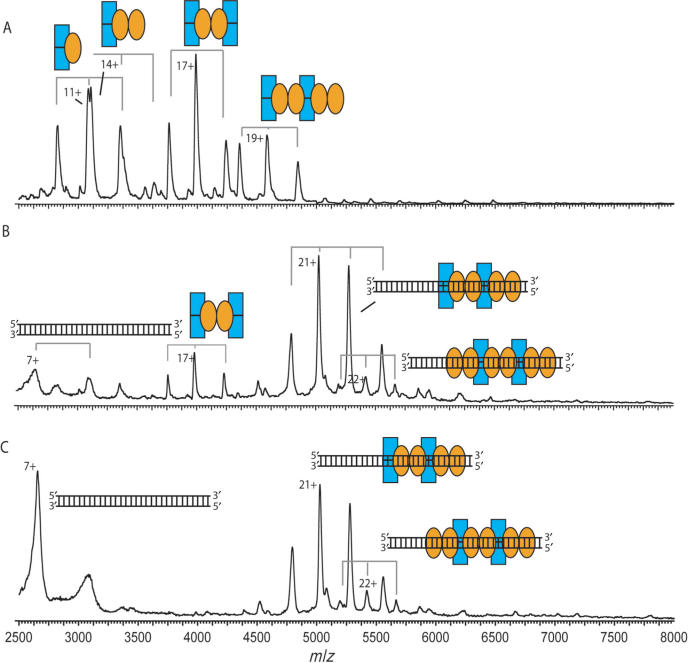
Initially, Kid and Kis were mixed at a molar ratio of 2:1, yielding the Kid2–Kis trimer (33 820 Da) and the Kid2–Kis2–Kid2 hexamer (67 716 Da) (Supplementary Table 1; Figure 5A). Upon the addition of the parD region I at a molar ratio Kid:Kis:parD of 80:40:1 several intriguing changes were observed in the mass spectrum (Figure 5B); the trimer–hexamer equilibrium was shifted towards the hexamer and the parD region I interacted with the Kid–Kis complexes. The mass spectrum showed different ion series and mass determination revealed that these ion series represented free Kid dimer, Kid2–Kis trimer, Kid2–Kis2–Kid2 hexamer, (Kid2–Kis2–Kid2)1–(DNA)1, (86 079 Da) (Kid2–Kis2–Kid2)2–(DNA)1 (153 968 Da) and (Kid2–Kis2–Kid2)3–(DNA)1 (221 957 Da) complexes. Intriguingly, in this mixture about 57% of the Kid–Kis complexes were not bound to DNA (Supplementary Table 2). These results show that at least one antitoxin Kis dimer is required for the parD DNA binding, also when the toxin Kid is present. Since this requirement holds only the hexamer and not the trimer, the shift of the trimer–hexamer equilibrium was induced by parD region I binding.
Figure 5.
Kid–Kis complexes (molar ratio of 2:1) interact with parD region I. Macromolecular native mass spectrometry was performed on Kid–Kis and on Kid–Kis–parD DNA complexes in ammonium acetate (50 mM), pH 5.8. (A) Mass spectrum of a mixture of Kid:Kis at a molar ratio of 2:1 (Kis 7.5 μM) and (B) and (C) mass spectra of mixtures of Kid:Kis:parD DNA mixtures at molar ratios of 80:40:1 and 20:10:1 (Kis 7.5 μM), respectively. Kid and Kis are indicated with blue rectangles and orange ellipses, respectively, and the parD DNA fragment with double strand. Each complex is represented by an appropriate combination of rectangles, ellipses and/or DNA double strand. Molecular masses and relative amounts of complexes are shown in Supplementary Tables 1 and 2, respectively.
Next, we mixed the parD region I with Kid:Kis in a molar ratio of Kid:Kis:parD 20:10:1 (Figure 5C). The spectrum showed two major protein ion series corresponding to free Kid2–Kis2–Kid2 hexamer and Kid2–Kis2–Kid2 bound to parD DNA at a 1:1 stoichiometry (86 077 Da). These data thus clearly show that upon increasing the DNA concentration relative to Kis there is a preference for the binding of only one hexamer to the parD DNA. Moreover, the high amount of free Kid2–Kis2–Kid2 hexamer (relative abundance of 80%) indicates a relatively low affinity between DNA and the hexameric Kid–Kis complex. This suggests that in conditions in which the Kid concentration exceeds the Kis concentration, only labile complexes are formed between parD DNA and Kis. This is consistent with the fact that Kid–Kis complexes formed on the 175-bp parD operon in excess of Kis can be easily competed by poly-(dIdC) (data not shown).
Equal concentrations of Kid and Kis induce formation of stable Kid2–Kis2–Kid2–Kis2–parD DNA1 complexes
Upon mixing Kid and Kis at a 1:1 molar ratio, we observed multiple complexes: Kid2–Kis trimer (33 841 Da), Kid2–Kis2 tetramer (43 450 Da), Kid2–Kis2–Kid2 hexamer (67 722 Da) and Kid2–Kis2–Kid2–Kis2 octamer (87 120 Da) (Supplementary Table 1; Figure 6A). Upon mixing Kid–Kis mixture with the 30-bp parD DNA at a molar ratio of 40:40:1 and subsequent mass spectrometry analysis we observed three ion series (Figure 6B). Mass determination revealed the presence of a low amount of free Kid2–Kis2–Kid2 hexamer (relative abundance of 17%) (67 675 Da) and high amounts of the DNA-bound complexes Kis2–Kid2–Kis2–Kid2–parD DNA1 (76%) (105 535 Da) and Kis2–Kid2–Kis2–Kid2–Kis2–parD DNA1 (7%) (124 563 Da) complexes (Supplementary Table 2). Thus, the addition of parD region I to a 1:1 mixture of Kid and Kis induces a strong cooperative effect between DNA binding and Kid–Kis octamerization. This cooperative effect was even stronger when the concentration of DNA in solution was increased 4-fold. Under these conditions the free hexamer completely disappeared and the most abundant complex became the Kis2–Kid2–Kis2–Kid2–parD DNA1 complex (relative abundance of 88%) (Figure 6C). We did not observe any other changes in the mass spectrum upon increasing the concentration of parD DNA. These data thus clearly demonstrate that increasing the concentration of Kis relative to Kid dramatically enhances the affinity between the DNA and the Kid–Kis complexes.
Figure 6.
Kid–Kis complexes (molar ratio of 1:1) interact tightly with parD region I. Macromolecular native mass spectrometry was performed on Kid–Kis and on Kid–Kis–parD DNA complexes in ammonium acetate (50 mM), pH 5.8. (A) Mass spectrum of a mixture of Kid:Kis at a molar ratio of 1:1 (Kis 15 μM) and (B) and (C) mass spectra of Kid:Kis:parD DNA mixtures at molar ratios of 40:40:1 and 10:10:1 (Kis 15 μM), respectively. Kid and Kis are indicated with blue rectangles and orange ellipses, respectively, and the parD DNA fragment with double strand. Each complex is represented by an appropriate combination of rectangles, ellipses and/or DNA double strand. Molecular masses and relative amounts of complexes are shown in Supplementary Tables 1 and 2, respectively.
Upon mixing Kid–Kis mixture with parD region I or II at a molar ratio of 40:40:1 or 20:20:1 and subsequent electrophoretic mobility shift analysis complexes with a similar pattern as in mass spectrometry were observed (Supplementary Figure 2). The most abundant complexes cIX and cX may represent hexameric and octameric Kid–Kis in complex with parD DNA, respectively. The assays also showed a faster migrating band with low abundance, which may represent the binding of hexameric Kis alone. Consistent with the mass spectrometry data parD region I is fully titrated when using a protein:DNA molar ratio of 20:1.
To further define the picture about the effect of the molar ratio of toxin and antitoxin on operator-promoter binding, and thus on their own expression, we added an excess of toxin or alternatively an excess of antitoxin to the Kis2–Kid2–Kis2–Kid2–parD DNA1 complex. In the first experiment, a 2-fold molar excess of Kid over Kis was added to the preformed Kis2–Kid2–Kis2–Kid2–parD DNA1 complex. The resulting mass spectrum showed three ion series corresponding to free DNA, Kid2–Kis2–Kid2 hexamer and (Kid2–Kis2–Kid2)2–DNA1 complex, thus revealing a partial reversion of the binding on the operator due to the excess of toxin over antitoxin. In contrast, the addition of a 2-fold molar excess of Kis over Kid to the Kis2–Kid2–Kis2–Kid2–parD DNA1 complex had no effect on the equilibrium binding of the Kid2–Kis2–Kid2–Kis2 octamer to the parD DNA region I.
Topology of the octameric Kid–Kis–par D DNA complex
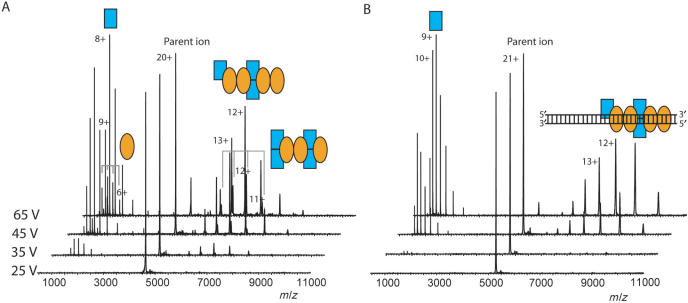
To further investigate the effect of parD DNA binding on the Kid–Kis oligomers, octameric Kid–Kis complexes free and bound to parD DNA were analyzed by macromolecular tandem mass spectrometry. The 20+ ion of the free octamer (m/z 4127) was isolated by the high-mass quadrupole and subsequently accelerated in the argon-filled collision cell. Already at a low acceleration voltage (35 V) we observed dissociation of the octameric ion into highly charged monomers and moderately charged heptameric complexes (Figure 7A). At low m/z values, two charge-state envelopes were observed corresponding to Kid and Kis monomer, whereas at high m/z values, the corresponding Kis–Kid2–Kis2–Kid2 and Kis2–Kid2–Kis2–Kid heptamer were detected. The relative abundances of the ion peaks showed that dissociation of Kid from the octamer was somewhat favoured.
Figure 7.
Macromolecular tandem mass spectrometry reveals topology of Kid–Kis–parD DNA region I complexes. Tandem mass spectrometry was performed on Kid–Kis and Kid–Kis–parD DNA in ammonium acetate (50 mM), pH 5.8. (A) Tandem mass spectra of Kis2–Kid2–Kid2–Kis2 after selection of the 20+ ion and (B) tandem mass spectra of Kis2–Kid2–Kid2–Kis2–parD DNA complex after selection of the 21+ ion. Acceleration voltages varied between 25 and 65 V. Kid and Kis are indicated with blue rectangles and orange ellipses, respectively and the parD DNA fragment with double strand. Each complex is represented by an appropriate combination of rectangles, ellipses and/or DNA double strand.
From the octameric Kid–Kis–parD DNA complex we selected the 21+ (m/z 5290). At an acceleration voltage of 45 V, the Kid monomer, but not Kis monomer or parD region I, started to dissociate from the octamer–parD DNA complex (Figure 7B). Also further increasing the acceleration voltage did not result in dissociation of Kis. Thus, in comparison with free octamer, the dissociation was initiated at an increased acceleration voltage and only Kid dissociated from the DNA bound complex. This not only shows that the DNA bound complex is more stable, but also confirms the direct interaction between Kis and parD region I, thereby protecting the protein from dissociation. Such a phenomenon has also been observed by van Duijn et al. (30) for GroEL-substrate complexes, for which the substrate molecules are buried within the hydrophobic ring structure of GroEL and do not dissociate upon increasing the acceleration voltage. We did not observe any acceleration voltage-induced covalent fragmentation of parD DNA up to voltages of 150 V when bound to Kid–Kis octamer. In contrast, free 30-bp parD DNA already fragmented at an acceleration voltage of 25 V (data not shown). This data, therefore, strongly indicates that the DNA tightly interacts with the protein complex.
DISCUSSION
The parD operon of plasmid R1 encodes the toxin Kid and the antitoxin Kis. It has been demonstrated that in vivo efficient autoregulation of the parD operon requires the consorted action of both proteins (20). Recent in vitro studies have shown that Kid and Kis can form multiple complexes with different stoichiometries and oligomeric states, depending on the molar ratio between Kid and Kis (18). The Kid2–Kis2–Kid2 hexamer is the most abundant species when Kid exceeds the concentration of Kis, whereas various Kid–Kis complexes are present when the concentration of Kis equals or exceeds the concentration of Kid.
Our DNA binding and mass spectrometry data presented here show that the antitoxin Kis interacts with the parD operon with low affinity. These data are in line with previous studies, which have shown that Kis alone is a poor repressor in vivo (11). The addition of toxin Kid to Kis enhances the binding affinity with parD DNA, however, the tightness of this interaction with parD DNA is determined by the molar ratio between Kid and Kis. We demonstrated that when Kid and Kis were mixed in a molar ratio of 2:1, the interaction between the resulting Kid2–Kis2–Kid2 hexamer and the parD DNA is weak. Thus, in these conditions transcriptional repression is expected to be limited. On the contrary, when the complex mixture of Kid–Kis oligomers, obtained at an equimolar ratio of Kid and Kis, was added to the parD DNA a strong cooperative effect of DNA binding and Kid2–Kis2–Kid2–Kis2 octamerization was observed and the parD DNA interacted tightly to this octamer. We also observed that the addition of extra toxin (up to a 2-fold molar excess of Kid) to the Kid–Kis octamer–parD DNA complex weakened the interaction with the DNA. From these data we conclude that different molar ratios of Kid and Kis can either enhance or diminish the parD DNA-binding activity of Kis. Therefore, the transcriptional repression of the parD operon and thus the expression of Kid and Kis is critically dependent on the molar ratio of Kid and Kis. It should be noted here that the Kid–Kis octamer can interact with the two half-sites of the specific operator region (region I and II) using the two dimers of the antitoxin, whereas the Kid–Kis hexameric can interact with the two half-sites using only one dimer. This is likely to explain the more efficient binding of the octamer. Alleviation of the repression modulated by toxin and antitoxin complexes in excess of the toxin has also been reported for the ccd system (25,37) as well as for the phD-doc system (15).
Our data also show that Kis and Kid–Kis complexes interact in two imperfect inverted repeats (region I and II). Region I contains an 18-bp symmetric element and region II a pseudo-symmetric element. Moreover, by using separated fragments containing parD regions I or II, we found that Kis and Kid–Kis complexes interact with higher affinity to region I. The lower affinity to region II is probably due to the four non-conserved bases in this element (5′-GTTATATTTTTATTAAAC-3′, in italic non-conserved residues). However, cooperative interactions between regions I and II potentially play an important role in the transcriptional regulation of the parD operon.
Are the physical parameters of the Kid–Kis complexes sufficient to form multiple interactions over the full length of the 30-bp parD DNA region I? Calculation of the length of the 30-bp parD region I and the Kid–Kis octamer, assuming a similar topology as the MazF–MazE hexamer (18,24), revealed lengths of ∼100 Å for the DNA and ∼150 Å for the Kid–Kis octameric complex. Although no 3D structural model is available for a Kid–Kis–DNA complex or a related toxin–antitoxin–DNA complex, it can be speculated that the octamer can fully cover the 30-bp DNA, which is in line with the DNA footprinting and tandem mass spectrometry data. Very recent nuclear magnetic resonance chemical-shift mapping data have revealed that the antitoxin CcdA alone interacts with duplex DNA comprising a 6-bp palindromic sequence (37). In here, it is also shown that a 33-bp DNA fragment, containing three potential CcdA binding sites, can bind in a cooperate manner with three CcdA dimers. The antitoxins of the ccd and mazEF systems have amino terminal regions that dimerize to form the DNA-binding region, and contain an unstructured C-terminal part, which interacts with the toxins. The toxins of these systems have also substantial structural homology (24,31), however the N-terminal domains of the CcdA and MazE antitoxin adopt different protein folds. As pointed by these authors, this surprising result confirms the proposal that gene shuffling or partner switching has been important in the evolution of the toxin–antitoxin systems (1).
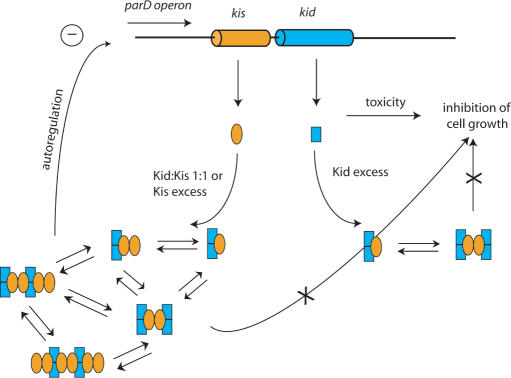
Our results lead us to propose a model in which the repression of the parD operon is tightly regulated by the molar ratio of the toxin and the antitoxin present in the cell (Figure 8). When the level of the toxin exceeds the one of the antitoxin, stable (Kid2–Kis)n non-covalent complexes are formed, which are able to completely neutralize Kid lethal activity (M.B. Kamphuis et al., submitted for publication), but do not tightly interact with the parD operator-promoter region. On the contrary, when the concentration of Kis is enhanced relative to Kid, such that both proteins have similar concentrations, (Kid2–Kis2)n or (Kid2–Kis2–Kid2–Kis2)n oligomers are formed capable of strongly interacting with the parD operator-promoter region. These stoichiometric complexes have a very strong capacity to interact with the specific parD sequences and to repress the transcription pathway. When the expression is repressed, no replacement can occur of the labile Kis antitoxin, which is prone to degradation by Lon protease. The level of the toxin will, therefore, exceed the one of the antitoxin. At this stage, the equilibrium between (Kid2–Kis2)n oligomers will shift towards (Kid2–Kis)n oligomers, thereby reducing the affinity for the parD operon. Subsequently, the inhibition of transcription will be alleviated. The tight interaction with parD region I suggests that this region plays a prominent role in the regulation of parD repression, however, interactions in region II could be required for fine adjustment in this regulation. As mentioned above, cooperative interactions between the two regions could introduce additional complexity in this regulation. The neutralization of the negative charges of the antitoxin by the toxin may stabilize interaction of the repressor complexes with the DNA. How this particular configuration contributes to the efficient binding of the repressor complex and thus to the fine-tuning of the promoter activity remains to be established.
Figure 8.
Schematic model of the transcription autoregulation of the parD operon. The kid gene and the Kid protein are shown in blue and the kis gene and the Kis protein in orange. Each protein complex is represented by an appropriate combination of blue rectangles (Kid) and orange ellipses (Kis). Free Kid inhibits cell growth. In conditions in which the concentration of Kid is higher than that of Kis Kid2–Kis1 and Kid2–Kis2–Kid2 complexes are formed. These complexes repress ribonuclease activity of Kid, but allow efficient transcription. When the concentration of Kid is equal or lower than that of Kis, mostly 1:1 complexes are formed. These complexes do not only repress the ribonuclease activity of Kid, but also the transcription process.
Similar mechanisms of transcription autoregulation have been proposed for the ccd and the mazEF addiction systems (23–25,37). Electrophoretic mobility shift assays have shown that multimers of CcdA2CcdB2 have multiple DNA-binding sites and spirals around the promoter region (25). It has also been proposed that when CcdB is present in a molar excess over CcdA, binding of a CcdB2 dimer to a (CcdB2-CcdA2) – DNA complex causes steric hindrance and, therefore, loosens the interaction of the protein complex with DNA. This will alleviate the inhibition of transcription (37). On the other hand, for the parD system, the binding of Kis or Kid–Kis complexes to the parD operator-promoter region occurs only in two discrete regions (I and II) spaced by 33 bp; the distribution of the specific contacts that Kis and Kid–Kis complexes make on each of the DNA regions are spaced 11–13 bp, indicating that the proteins bind on the same face of the DNA. It should be noted here that CcdB and Kid have different activities: CcdB acts as a toxin and inhibitor for DNA gyrase, an essential enzyme that catalyses negative supercoiling of DNA (4), whereas Kid functions as a ribosome-independent RNase (10,18).
Although no stoichiometric complexes of MazF–MazE on the DNA have been identified, it has been postulated that MazE mediates assembly of heterocomplexes on DNA. The resulting higher order complexes would then form stoichiometric MazF–MazE complexes. In the mazEF system, the promoter region contains three antitoxin-binding regions (11–12-bp long) that can form an ‘alternating palindrome’. It has been proposed that MazE antitoxin binds to these sites and that two MazF dimers can bridge the MazE dimers in a highly cooperative interaction (38). The situation in the kid–kis system is different in that the promoter region contains two binding regions, I and II, of 18-bp.
SUPPLEMENTARY DATA
Supplementary Data is available at NAR Online.
ACKNOWLEDGEMENTS
M.C.M. was supported by a Short Term FEBS Fellowship and R.H.H.vdH. by a VENI fellowship (700.54.402) from the Netherlands Organization for Scientific Research (NWO). R.D.O. was supported by Project BFU2005-03911 from the Spanish Ministry of Education and Science (MEC) and by Project PIF 200420F0332 (CSIC) and M.B.K. by the Centre for Biomedical Genetics. We thank the Netherlands Proteomics Centre for financial support. Funding to pay the Open Access publication charge was provided by Netherlands Proteomics Centre.
Conflict of interest statement. None declared.
REFERENCES
- 1.Anantharaman V, Aravind L. New connections in the prokaryotic toxin-antitoxin network: relationship with the eukaryotic nonsense-mediated RNA decay system. Genome Biol. 2003;4:81. doi: 10.1186/gb-2003-4-12-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hayes F. Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science. 2003;301:1496–1499. doi: 10.1126/science.1088157. [DOI] [PubMed] [Google Scholar]
- 3.Gerdes K, Christensen SK, Lobner-Olesen A. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 2005;3:371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- 4.Bernard P, Couturier M. Cell killing by the F-plasmid Ccdb protein involves poisoning of DNA-topoisomerase-Ii complexes. J. Mol. Biol. 1992;226:735–745. doi: 10.1016/0022-2836(92)90629-x. [DOI] [PubMed] [Google Scholar]
- 5.Jiang Y, Pogliano J, Helinski DR, Konieczny I. ParE toxin encoded by the broad-host-range plasmid RK2 is an inhibitor of Escherichia coli gyrase. Mol. Microbiol. 2002;44:971–979. doi: 10.1046/j.1365-2958.2002.02921.x. [DOI] [PubMed] [Google Scholar]
- 6.Pedersen K, Zavialov AV, Pavlov MY, Elf J, Gerdes K, Ehrenberg M. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell. 2003;112:131–140. doi: 10.1016/s0092-8674(02)01248-5. [DOI] [PubMed] [Google Scholar]
- 7.Kamada K, Hanaoka F. Conformational change in the catalytic site of the ribonuclease YoeB toxin by YefM antitoxin. Mol. Cell. 2005;19:497–509. doi: 10.1016/j.molcel.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 8.Zhang YL, Zhang JJ, Hoeflich KP, Ikura M, Qing GL, Inouye M. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol. Cell. 2003;12:913–923. doi: 10.1016/s1097-2765(03)00402-7. [DOI] [PubMed] [Google Scholar]
- 9.Munoz-Gomez AJ, Santos-Sierra S, Berzal-Herranz A, Lemonnier M, Diaz-Orejas R. Insights into the specificity of RNA cleavage by the Escherichia coli MazF toxin. FEBS Lett. 2004;567:316–320. doi: 10.1016/j.febslet.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 10.Munoz-Gomez AJ, Lemonnier M, Santos-Sierra S, Berzal-Herranz A, Diaz-Orejas R. RNase/anti-RNase activities of the bacterial parD toxin-antitoxin system. J. Bacteriol. 2005;187:3151–3157. doi: 10.1128/JB.187.9.3151-3157.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruiz-Echevarria MJ, Berzal-Herranz A, Gerdes K, Diaz-Orejas R. The kis and kid genes of the parD maintenance system of plasmid R1 form an operon that is autoregulated at the level of transcription by the co-ordinated action of the Kis and Kid proteins. Mol. Microbiol. 1991;5:2685–2693. doi: 10.1111/j.1365-2958.1991.tb01977.x. [DOI] [PubMed] [Google Scholar]
- 12.de Feyter R, Wallace C, Lane D. Autoregulation of the ccd operon in the F plasmid. Mol. Gen. Genet. 1989;218:481–486. doi: 10.1007/BF00332413. [DOI] [PubMed] [Google Scholar]
- 13.Gronlund H, Gerdes K. Toxin-antitoxin systems homologous with relBE of Escherichia coli plasmid P307 are ubiquitous in prokaryotes. J. Mol. Biol. 1999;285:1401–1415. doi: 10.1006/jmbi.1998.2416. [DOI] [PubMed] [Google Scholar]
- 14.Tam JE, Kline BC. Control of the Ccd operon in plasmid-F. J. Bacteriol. 1989;171:2353–2360. doi: 10.1128/jb.171.5.2353-2360.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Magnuson R, Yarmolinsky MB. Corepression of the P1 addiction operon by Phd and Doc. J. Bacteriol. 1998;180:6342–6351. doi: 10.1128/jb.180.23.6342-6351.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bravo A, Ortega S, de Torrontegui G, Diaz R. Killing of Escherichia coli cells modulated by components of the stability system ParD of plasmid R1. Mol. Gen. Genet. 1988;215:146–151. doi: 10.1007/BF00331316. [DOI] [PubMed] [Google Scholar]
- 17.Zhang J, Zhang Y, Zhu L, Suzuki M, Inouye M. Interference of mRNA function by sequence-specific endoribonuclease PemK. J. Biol. Chem. 2004;279:20678–20684. doi: 10.1074/jbc.M314284200. [DOI] [PubMed] [Google Scholar]
- 18.Kamphuis MB, Bonvin AM, Monti MC, Lemonnier M, Munoz-Gomez A, van den Heuvel RH, Diaz-Orejas R, Boelens R. Model for RNA binding and the catalytic site of the RNase kid of the bacterial parD toxin-antitoxin system. J. Mol. Biol. 2005;357:115–126. doi: 10.1016/j.jmb.2005.12.033. [DOI] [PubMed] [Google Scholar]
- 19.Santos-Sierra S, Pardo-Abarrio C, Giraldo R, Diaz-Orejas R. Genetic identification of two functional regions in the antitoxin of the parD killer system of plasmid R1. Fems Microbiol. Lett. 2002;206:115–119. doi: 10.1111/j.1574-6968.2002.tb10995.x. [DOI] [PubMed] [Google Scholar]
- 20.Lemonnier M, Santos-Sierra S, Pardo-Abarrio C, Diaz-Orejas R. Identification of residues of the kid toxin involved in autoregulation of the parD system. J. Bacteriol. 2004;186:240–243. doi: 10.1128/JB.186.1.240-243.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruiz-Echevarria MJ, de la Cueva G, Diaz-Orejas R. Translational coupling and limited degradation of a polycistronic messenger modulate differential gene expression in the parD stability system of plasmid R1. Mol. Gen. Genet. 1995;248:599–609. doi: 10.1007/BF02423456. [DOI] [PubMed] [Google Scholar]
- 22.de la Cueva-Mendez G, Mills AD, Clay-Farrace L, Diaz-Orejas R, Laskey RA. Regulatable killing of eukaryotic cells by the prokaryotic proteins Kid and Kis. EMBO J. 2003;22:246–251. doi: 10.1093/emboj/cdg026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dao-Thi MH, Charlier D, Loris R, Maes D, Messens J, Wyns L, Backmann J. Intricate interactions within the ccd plasmid addiction system. J. Biol. Chem. 2002;277:3733–3742. doi: 10.1074/jbc.M105505200. [DOI] [PubMed] [Google Scholar]
- 24.Kamada K, Hanaoka F, Burley SK. Crystal structure of the MazE/MazF complex: Molecular bases of antidote-toxin recognition. Mol. Cell. 2003;11:875–884. doi: 10.1016/s1097-2765(03)00097-2. [DOI] [PubMed] [Google Scholar]
- 25.Afif H, Allali N, Couturier M, Van Melderen L. The ratio between CcdA and CcdB modulates the transcriptional repression of the ccd poison-antidote system. Mol. Microbiol. 2001;41:73–82. doi: 10.1046/j.1365-2958.2001.02492.x. [DOI] [PubMed] [Google Scholar]
- 26.Hanson CL, Robinson CV. Protein-nucleic acid interactions and the expanding role of mass spectrometry. J. Biol. Chem. 2004;279:24907–24910. doi: 10.1074/jbc.R300037200. [DOI] [PubMed] [Google Scholar]
- 27.van den Heuvel RHH, Gato S, Versluis C, Gerbaux P, Kleanthous C, Heck AJR. Real-time monitoring of enzymatic DNA hydrolysis by electrospray ionization mass spectrometry. Nucleic Acids Res. 2005;33:96. doi: 10.1093/nar/gni099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharon M, Taverner T, Ambroggio XI, Deshaies RJ, Robinson CV. Structural organization of the 19S proteasome Lid: insights from MS of intact complexes. PLoS Biol. 2006;4:e267. doi: 10.1371/journal.pbio.0040267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van den Heuvel RHH, van Duijn E, Mazon H, Synowsky SA, Lorenzen K, Versluis C, Brouns SJJ, Langridge D, van der Oost J, et al. Improving the performance of a quadrupole time-of-flight mass spectrometer for macromolecular mass spectrometry. Anal. Chem. 2006;78:7473–7483. doi: 10.1021/ac061039a. [DOI] [PubMed] [Google Scholar]
- 30.van Duijn E, Simmons DA, van den Heuvel RHH, Bakkes PJ, van Heerikhuizen H, Heeren RMA, Robinson CV, van der Vies SM, Heck AJR. Tandem mass spectrometry of intact GroEL-substrate complexes reveals substrate-specific conformational changes in the trans ring. J. Am. Chem. Soc. 2006;128:4694–4702. doi: 10.1021/ja056756l. [DOI] [PubMed] [Google Scholar]
- 31.Hargreaves D, Santos-Sierra S, Giraldo R, Sabariegos-Jareno R, de la Cueva-Mendez G, Boelens R, Diaz-Orejas R, Rafferty JB. Structural and functional analysis of the Kid toxin protein from E-coli plasmid R1. Structure. 2002;10:1425–1433. doi: 10.1016/s0969-2126(02)00856-0. [DOI] [PubMed] [Google Scholar]
- 32.Tahallah N, Pinkse M, Maier CS, Heck AJ. The effect of the source pressure on the abundance of ions of noncovalent protein assemblies in an electrospray ionization orthogonal time-of-flight instrument. Rapid Commun. Mass Spectrom. 2001;15:596–601. doi: 10.1002/rcm.275. [DOI] [PubMed] [Google Scholar]
- 33.Sobott F, Hernandez H, McCammon MG, Tito MA, Robinson CV. A tandem mass spectrometer for improved transmission and analysis of large macromolecular assemblies. Anal. Chem. 2002;74:1402–1407. doi: 10.1021/ac0110552. [DOI] [PubMed] [Google Scholar]
- 34.Videler H, Ilag LL, McKay ARC, Hanson CL, Robinson CV. Mass spectrometry of intact ribosomes. FEBS Lett. 2005;579:943–947. doi: 10.1016/j.febslet.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 35.Benesch JLP, Aquilina JA, Ruotolo BT, Sobott F, Robinson CV. Tandem mass spectrometry reveals the quaternary organization of macromolecular assemblies. Chem. Biol. 2006;13:597–605. doi: 10.1016/j.chembiol.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 36.Garner MM. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions - application to components of the Escherichia coli operon regulatory system. Nucleic Acids Res. 1981;9:3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Madl T, van Melderen L, Mine N, Responsdek M, Oberer M, Keller W, Khatai L, Zangger K. Structural basis for nucleic acid and toxin recognition of the bacterial antitoxin CcdA. J. Mol. Biol. 2006;364:170–185. doi: 10.1016/j.jmb.2006.08.082. [DOI] [PubMed] [Google Scholar]
- 38.Buts L, Lah J, Dao-Thi MH, Wyns L, Loris R. Toxin-antitoxin modules as bacterial metabolic stress managers. Trends Biochem. Sci. 2005;30:672–679. doi: 10.1016/j.tibs.2005.10.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.