Abstract
In contrast to Escherichia coli no molecular mechanism controlling the biosynthesis of ribosomal proteins has been elucidated in Gram-positive organisms. Here we show that the expression of the Bacillus subtilis infC-rpmI-rplT operon encoding translation factor IF3 and the ribosomal proteins L35 and L20 is autoregulated by a complex transcription attenuation mechanism. It implicates a 200-bp leader region upstream of infC which contains two conserved regulatory elements, one of which can act as a transcription terminator. Using in vitro and in vivo approaches we show that expression of the operon is regulated at the level of transcription elongation by a change in the structure of the leader mRNA which depends upon the presence of ribosomal protein L20. L20 binds to a phylogenetically conserved domain and provokes premature transcription termination at the leader terminator. Footprint and toeprint experiments support a regulatory model involving molecular mimicry between the L20-binding sites on 23S rRNA and the mRNA. Our data suggest that Nomura's model of ribosomal protein biosynthesis based on autogenous control and molecular mimicry is also valid in Gram-positive organisms.
INTRODUCTION
In prokaryotes, the genes for ribosomal proteins (r-proteins) are to a large extent clustered and, usually, expressed as operons. The regulation of r-protein biosynthesis has been well characterized in the Gram-negative bacterium Escherichia coli (1). In this organism, most ribosomal protein expression has been shown to be feedback regulated at the level of translation. One of the protein products of the operon acts as the key regulator. When it accumulates in amounts exceeding its available binding sites on nascent ribosomal particles, it binds to its own mRNA, at a site called the operator, causing translational repression of the first cistron. Repression is transmitted to the downstream cistrons by translational coupling (2). The r-proteins involved in this autoregulatory circuit in general are also able to bind to rRNA directly and this dual RNA-binding activity prompted Nomura and collaborators to propose the mimicry hypothesis of translational repression based on structurally similar mRNA- and rRNA-binding sites (3,4). This model has been proven correct in several cases (5–7) and explains how competition between mRNA and rRNA for binding r-proteins allows the bacterium to maintain the balance between r-protein and rRNA levels.
The primary sequences of r-proteins and to a significant extent even the genetic organization of their genes are well conserved between Gram-negative and Gram-positive organisms. For example, the largest r-protein gene cluster in Bacillus subtilis (rif-str-spc) corresponds to a combination of the E. coli rif, str and spc clusters and contains a mixture of genes for the transcriptional and translational machineries (8). However, the pattern of transcription and the regulatory mechanisms involved are different; e.g. in B. subtilis the S10 r-protein gene cluster seems to form a single 15-kb operon (9), which in E. coli corresponds to three separate transcriptional units, the S10, spc and α operons. Ribosomal protein L4 autoregulates the E. coli S10 operon at the translational level and by modulating transcription elongation via a mechanism which also requires NusA (10,11). However, L4 is not involved in the control of the corresponding B. subtilis S10 operon (9). The E. coli and Bacillus operons contain exactly the same r-protein genes with the single exception that the rpsD gene (S4) is absent from the S10-spc-alpha region in B. subtilis. Pointedly, it is S4 which controls the translation of the E. coli α operon. The Bacillus rpsD gene is monocistronic and autoregulates its own expression via a yet unknown post-transcriptional mechanism (12,13).
Thus, despite the fact that no details of the molecular mechanisms involved in the control of r-protein synthesis in Gram-positive organisms have yet been elucidated, available evidence suggests that these mechanisms are likely to be quite different from those observed in E. coli.
In this article, we focused our attention on the expression of the B. subtilis infC-rpmI-rplT gene cluster encoding translation initiation factor IF3 and the r-proteins L35 and L20, respectively. In E. coli, these genes form an operon which is regulated by two different control loops, both acting at the translational level. IF3 represses the expression of its own gene (14) and the primary r-protein L20 (rplT) acts as a repressor of rpmI expression and indirectly downregulates its own expression through translational coupling with rpmI (15). Autogenous control by L20 has been shown to involve mRNA-binding sites upstream of rpmI which are structurally similar to its binding site on 23S rRNA (6) but we could not identify a similar site upstream of the B. subtilis rpmI gene. A recent computer-based search for potential attenuation mechanisms in B. subtilis had located a putative transcription terminator structure upstream of the infC gene (16) whose function is unknown.
Here, we show that the B. subtilis rpmI and rplT genes are co-transcribed with infC forming an operon which can also include a fourth gene (ysdA) of unknown function. Control of this operon involves a dedicated leader sequence upstream of infC which does not exist in E. coli. L20 can interact with an RNA structure which can fold within the leader and which is part of a complex structural arrangement similar to metabolite controlled riboswitches (17). L20 acts as the effector molecule for the switch, so that its binding provokes premature transcription termination. We provide evidence that regulation by L20 is probably based on molecular mimicry between its mRNA and rRNA binding sites.
MATERIALS AND METHODS
Bacterial strains and culture
The B. subtilis strains used in this study are the prototrophic strain 168 1A2A (BGSC) and derivatives thereof containing an infC’-‘lacZ translational fusion (pHMI3) or a Pspac infC-‘lacZ transcriptional fusion (plasmid pHMI10) integrated at the amyE locus. E. coli JM109 was used for plasmid constructions and E. coli JM101 was used to recover concatemeric plasmid forms. For mutagenesis experiments, E. coli XL1Blue was used as host. Overexpression of intein fusion proteins from the T7 promoter was carried out in strain BL21(λDE3) (18) carrying plasmid pRARE (Novagen). E. coli was grown at 37°C in LB medium. Bacillus subtilis was grown at 37°C or at 30°C in LB medium. When required, antibiotics were added at the following concentrations: ampicillin (200 µg/ml) for E. coli, neomycin (7 µg/ml), chloramphenicol (10 µg/ml) and kanamycin (20 µg/ml) for B. subtilis. β-Galactosidase activity of lacZ fusions was measured as described previously (19).
Plasmid constructions
pHM14
A 0.8-kb EcoRI-ClaI fragment of the transcriptional lacZ fusion vector pHM2 (20) was replaced with the equivalent 0.8-kb EcoRI-ClaI fragment of the translational fusion vector pAC7 (21).
pHMI3
A 347-bp PCR fragment containing the infC promoter, the entire leader sequence and the first 53 nts of the infC structural gene was inserted as a EcoRI-BamHI fragment in frame between the respective sites of the lacZ translational fusion plasmid pHM14.
pHMI10
A 273-bp PCR fragment containing the Pspac promotor followed by the infC leader up to position 191 was inserted as a EcoRI-BamHI fragment between the respective sites of the lacZ transcriptional fusion plasmid pHM2.
pHMI11
A 443-bp PCR fragment comprising the B. subtilis rplT gene (from nt 37 upstream of the start codon to nt 44 downstream of the stop codon) was inserted as a SphI-XbaI fragment into the replicative plasmid pDG148 (22) downstream of the Pspac promoter.
pHML17
The coding sequence of the rplT gene was cloned in phase with the intein domain in the T7 expression plasmid pKYB1 (New England Biolabs) between the sites NdeI and SapI.
Mutations in the infC leader region were introduced by using the ‘Quickchange XL1 site-directed mutagenesis’ kit (Stratagene).
Primer extension assay
Total RNA was isolated as described (19). Oligonucleotide HP667 (TACAGTGGATCCATAGTTTCTTCGAATTGATTCACAA) complementary to infC leader sequences between the terminator and the infC SD sequence was hybridized to 5 µg of total RNA and extended with Superscript III Reverse Transcriptase (Invitrogen) following the instructions of the manufacturer.
Single-round in vitro transcription
In vitro transcription was performed using PCR fragments as templates corresponding to positions −292 to −11 with respect to the infC start codon. The PCR templates were prepared using oligonucleotides HP647 (CGCTCATAGAAAACCCATGTTAC) and HP667 (TACAGTGGATCCATAGTTTCTTCGAATTGATTCACAA). The PCR fragments contained the promotor and the wild type or mutant infC leader sequence. In the first step (initiation of transcription), stable stalled transcripts were formed by incubating a mixture (40 µl) containing 0.5 nM B. subtilis RNA polymerase which was purified as described (23), template DNA (6 or 12 nM), oligoribonucleotide GpA (150 μM), GTP, ATP, UTP (10 μM) and 5 µCi [α32P] UTP for 10 min at 30°C in transcription buffer (40 mM Hepes-KOH [pH 8.0], 4 mM MgCl2, 1 mM DTT, 40 mM KCl, 50 μg/ml BSA, 12 U RNasin (Promega), 5% glycerol).
Elongation of the halted transcripts was initiated by addition of a solution (60 µl) containing 10 µg/ml rifampicin, 21 µM ATP, UTP and GTP, 25 µM CTP and 110 nM L20 protein in elongation buffer (40 mM Hepes-KOH [pH 8.0], 4 mM MgCl2, 1 mM DTT, 40 mM KCl). Elongation was carried out for 10 min at 30°C and stopped by the addition of 10 µl of 3M Na-acetate pH 5, phenolized and precipitated. The pellet was dissolved in 1 × gel loading buffer and run on a denaturing 5% polyacrylamide gel. The reaction products were visualized on a PhosphorImager and quantified.
Northern blotting
Northern analysis was carried out as described (19) using 8 µg of total RNA. A continuously α-[32P]UTP-labeled transcript generated with T7 RNA polymerase complementary to the entire leader mRNA and up to position +60 within the infC gene was used as probe. When the 5′ [32P]-labeled oligonucleotide HP1080 (GCGGGTGCTTCTGCTTGTGATTTAT) was used as a probe the hybridization temperature was lowered to 42°C.
Footprint experiments
An RNA fragment corresponding to the infC leader was synthesized from a template PCR fragment with a T7 RNA polymerase promoter (initiating at +1 of the infC transcript and ending at T residue 168. Generally, 0.5 pmol (10 nM) of transcript were incubated with 15 pmol (300 nM) of B. subtilis L20 protein for 2 min at 30° in 50 μl of the buffer supplied by the manufacturer of the RNases (Ambion). Then 6 µg of E. coli tRNA were added and the mixture incubated for 10 min at 30°C. Cleavage of the RNA was induced by the addition of RNase V1 (10−3 U/µl) or RNase T1 (0.01 U/µl). A hot phenol extraction was performed to remove the L20 protein. The same protocol was used for control reactions in the absence of L20. Cleavages were detected either by extension of a 5′-end-labeled primer HP697 (nts 168–141) or by direct detection of the cleavage products from a 5′-end-labeled substrate. Reaction products were separated on 5 or 8% denaturing polyacrylamide gels and analyzed on a PhosphorImager (Molecular Dynamics).
L20-Toeprint
The in vitro transcript used for the toeprint experiments with or without proteins (L20 or L17) was the same as described for the footprint assays. First, the infC leader mRNA (0.5 pmol) in MgCl2 (10 mM) and potassium phosphate buffer (25 mM, pH 7.7) were denatured by heating at 80°C for 2 min followed by immediate cooling on ice. After a renaturation at 30°C for 10 min, B. subtilis L20 (15 pmol) was added together with MgCl2 (10 mM final) and phosphate-potassium (25 mM final). The L20/leader infC molar ratio was 30 (final concentrations 0.75 µM and 25 nM, respectively). The samples were incubated at 30°C for 25 min and the incubation was stopped by ethanol precipitation. The L20-binding site on the mRNA was identified by extension of 5′-end-labeled primer HP697 (nt 168 to nt 141, (ACAACTCATTTTGAATGCATTTTGCAGG). Extension products were analyzed by denaturating 5% polyacrylamide gel. The reaction products were visualized on a PhosphorImager and where necessary quantified.
Purification of B. subtilis L20
Native L20 protein was overexpressed from plasmid pHML17 and isolated using the IMPACT system (New England Biolabs) according to the manufacturer's instructions. Induced self-cleavage of the intein fusion protein allowed for the isolation of untagged L20 protein.
Computer analysis
DNA sequence alignments were generated using the CLUSTAL X algorithm and manually adjusted. RNA secondary structures were predicted by using the M-Fold algorithm (24).
RESULTS
A single promoter drives expression of the four gene infC-rpmI-rplT- ysdA operon
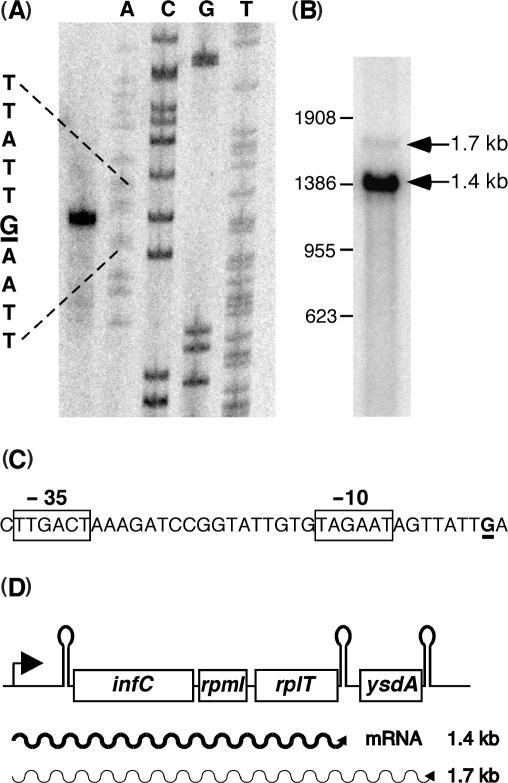
Primer extension using oligonucleotide HP667 located immediately upstream of the infC SD sequence identified a single transcription start point at the residue G –201 with respect to the potential ATT start codon of the infC gene (Figure 1A). In accordance, sigma A specific −35 and −10 promoter elements are found immediately upstream of this position (Figure 1C). Thus the infC mRNA is synthesized with a 201 nt leader. Northern analysis using a 1.3-kb PCR fragment covering the three adjacent genes infC, rpmI and rplT detected a major transcript of 1.4 kb (Figure 1B). This mRNA corresponds well to a transcript terminated at an intrinsic transcription terminator located downstream of the rplT (1.36 kb). A fourth open reading frame (ysdA), encoding a protein of unknown function, starts 60 nucleotides downstream of the rplT stop codon. A transcript including all four genes and extending to a second transcription terminator structure immediately downstream of ysdA would have a predicted length of 1.69 kb, compatible with a minor mRNA species detected by northern analysis (Figure 1B). Co-transcription of rplT and ysdA was confirmed by RT-PCR experiments (data not shown) and is corroborated by the absence of obvious promotor sequences between the two open reading frames. The four genes infC, rpmI, rplT and ysdA thus appear to form an operon transcribed from a single σA type promoter upstream of infC (Figure 1D).
Figure 1.
Transcription profile of the B. subtilis infC operon. (A) Determination of the transcription initiation start point by extension of primer HP667; the +1 guanosine residue is underlined, the sequence ladder was generated with oligonucleotide HP667. (B) Northern analysis of infC operon transcripts with a probe spanning the four genes. (C) −35 and −10 promoter sequences upstream of the transcription start point which is underlined. (D) Schematic view of the infC operon transcripts.
The intergenic distance between B. subtilis infC and rpmI (12 bp) is considerably shorter than that between the same genes in E. coli (96 bp). As a consequence the sequences involved in the formation of the L20-dependent translational operator which controls the expression of the rpmI and rplT genes in E. coli (6) are missing in B. subtilis. We thus focused our attention on the 201-bp leader region upstream of infC potentially implicated in the control of the expression of the four-gene operon.
The leader region of the infC operon contains conserved regulatory elements
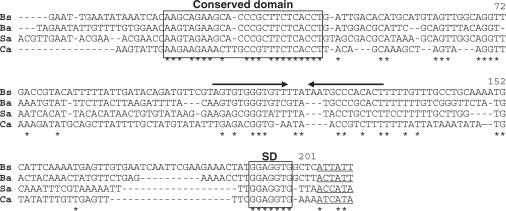
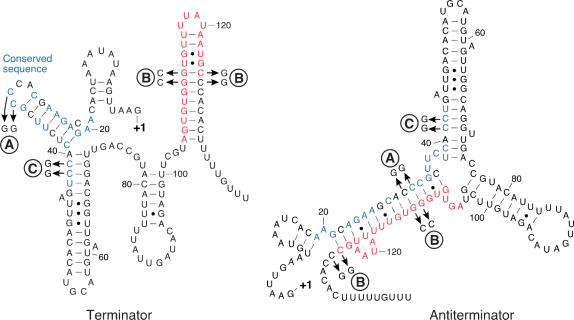
A representative alignment of the sequences upstream of the infC gene from four Gram-positive organisms highlights the presence of two conserved elements (Figure 2) that are phylogenetically conserved in most low GC Gram-positive organisms. The first comprises a 25-nucleotide stretch, labeled ‘conserved domain’ (Figure 2, positions 19–43 of the B. subtilis leader) and the second corresponds to a stem-loop structure followed by a polyT stretch typical of a factor independent transcription terminator. In the second case, the structure itself rather than the sequence is conserved. A comparison of the folding patterns of infC leader mRNAs from various Gram-positive organisms demonstrated the potential to form similar alternative structures which include the two conserved elements. Figure 3 shows the alternative folding patterns of the B. subtilis infC leader mRNA generated by the M-Fold algorithm (24) which, with minor manual adjustments, represents a terminator and antiterminator configuration. The antiterminator configuration involves a long distance interaction between bases in the conserved domain and the 5′ side of the putative terminator hairpin, producing a structure which is reminiscent of metabolite controlled riboswitches (17) (Figure 3). The significance of this observation is supported by the covariance, in different organisms, of the residues in the conserved domain and the terminator stem-loop which produce an equivalent structural switch.
Figure 2.
Alignment of the infC leader sequences from four Gram-positive organisms. A 5′ conserved domain and the Shine–Dalgarno sequences are boxed. Inverted arrows depict factor-independent transcription terminators. Potential infC translation initiation codons (which are generally not ATG/GTG) are underlined and identical residues are marked by an asterisk. Bs = Bacillus subtilis, Ba = Bacillus anthracis, Sa = Staphylococcus aureus, Ca = Clostridium acetobutylicum. The numbering corresponds to the B. subtilis sequence.
Figure 3.
Putative alternative RNA foldings within the B. subtilis infC leader. Identical nucleotides of the 5′ conserved domain (boxed in Figure 2) are in blue and residues of the terminator structure involved in the formation of the antiterminator are in red. ‘A’ and ‘B’ indicate double and quadruple point mutations, respectively, as shown in the figure. They have been introduced to analyze the folding pattern of the leader mRNA (see text). ‘C’ depicts a double mutation used for the toeprint assay (see text below).
The switch between termination and antitermination structures is functional in vitro
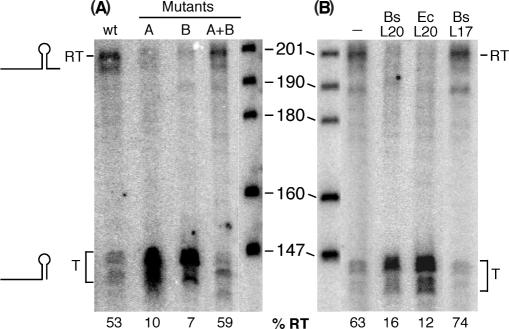
In order to demonstrate the functioning of the proposed alternative folding patterns in the infC leader, we performed single-round in vitro transcription assays (Figure 4A). With a wild-type template containing the promoter region and the entire infC leader, two different transcripts were obtained. One corresponds to the full-length transcript (200 nts), and the shorter one (∼140 nts) is due to a premature transcription arrest at the intrinsic terminator structure. Under the conditions used, 53% of the transcripts were full length indicating that the terminator structure is functional but that about half of the transcripts escapes premature termination (Figure 4, lane 1). The mutation called ‘A’ on Figure 3 (C30C31 → GG) in the loop of the conserved domain should considerably decrease the stability of the proposed antiterminator (Figure 3). In agreement, premature termination of transcription occurred in 90% of the mutant transcripts (Figure 4, lane 2). The same result was obtained with mutation ‘B’ (G111G112 → CC) in the terminator stem (Figure 4, lane 3). The concomitant transition C125C126 → GG was introduced solely to maintain base pairing of the terminator. The G111G112 nucleotides are expected to base pair with C30C31 in the antiterminator configuration and consequently have a similar effect on antiterminator formation as mutation A. Combining the A and B mutations restores base pairing within the antiterminator structure and leads to efficient read-through of the terminator to the same extent as observed with the wild-type template (Figure 4, lane 4). These data suggest that the proposed switch in RNA structure is functional in vitro and necessary for avoiding premature termination of transcription at the infC leader terminator.
Figure 4.
Effect of infC leader mutations and the presence of L20 on the transcription profile in vitro. Single round in vitro transcription assays were performed on PCR templates comprising the promoter and 5′ noncoding region of infC using B. subtilis RNA polymerase. RT and T on the left side indicate read-through or prematurely terminated infC leader transcripts. The size of the marker fragments in bases is indicated. Numbers at the bottom indicate the percentage of read-through transcripts (%RT = (RT/(T + RT) × 100). (A) Templates were wild type (wt) or contained the A, B or A + B mutations depicted in Figure 3. (B) Where indicated proteins were added in 50-fold molar excess over the template during the elongation phase of the single-round transcription assay. The B. subtilis L20 (Bs L20) and L17 (Bs L17) r-proteins were native, E. coli L20 (Ec L20) only contained the C-terminal half of the protein.
Formation of the antiterminator structure in the infC leader is required for normal expression in vivo
In order to verify whether the alternative RNA folding observed in vitro is significant for the regulation of the infC operon in vivo we analyzed the expression of translational lacZ fusions with the infC gene. The different constructs integrated in single copy at the amyE locus contained the infC promoter and leader sequences in their wild-type configuration or harboring the same mutations already used in the in vitro transcription experiments described above. Expression of β-galactosidase activity from both A and B mutant fusions is strongly decreased (22- and 12-fold, respectively, Table 1) compared to the wild-type fusion. Combining both mutations (A + B) almost completely restores the wild-type expression level (Table 1). The in vivo expression data thus fully confirm the in vitro experiments and illustrate the importance of the switch between the alternative structures for the transcriptional control of the infC operon.
Table 1.
Expression of wild-type and mutant infC-lacZ fusions in vivo
| Fusion | β-galactosidase activitya [U/mg] | Fold change |
|---|---|---|
| WT | 1029 | |
| Mutant A | 47 | 22× |
| Mutant B | 89 | 11.6× |
| Mutant A + B | 862 | 1.2× |
aThe β-galactosidase activities shown represent mean values from at least two independent measurements.
Ribosomal protein L20 but not metabolites stimulate premature transcription termination in vitro
The alternative folding patterns within the infC leader mRNA are reminiscent of metabolite controlled riboswitches (17). We therefore decided in an initial screen to test the nucleotides ATP, GTP and (p)ppGpp for their capacity to alter premature termination at the infC leader terminator in single-round in vitro transcription assays. These metabolites are crucial co-factors in the translational process and their concentrations reflect the metabolic state of the cell. During the elongation phase of the assay, ATP and GTP (possible activators) were added in the 0.5–5 mM range and ppGpp (possible repressor) between 50 and 200 µM, while keeping all other nucleotides constant at 25 µM. In addition, the expression of an infC-lacZ fusion was compared in a wild-type and relA mutant strain following induction of the stringent response. None of these experiments revealed any role of these metabolites in the control of the infC operon (data not shown).
We then tested for a potential regulatory role of L20, the only primary ribosomal protein (i.e. r-protein binding directly to rRNA) of the operon. Single-round in vitro transcription assays were performed on a wild-type template in the presence or absence of purified L20 protein (Figure 4B). The addition of B. subtilis L20 or the C-terminal half of the E. coli L20 protein, which is sufficient for autogenous control (25), clearly inhibited transcription elongation beyond the leader terminator (Figure 4B, lanes 2 and 3, respectively). Since basic proteins like L20 might have a tendency to bind RNA nonspecifically we also tested the almost equally basic B. subtilis r-protein L17 and showed that it does not alter transcriptional termination as compared to a control reaction in the absence of exogenous proteins (Figure 4B, lane 4).
Ribosomal protein L20 causes premature transcription termination in vivo
We tested the autoregulatory potential of L20 in vivo by overexpressing it from a plasmid-borne IPTG inducible rplT gene copy. An 8-fold overexpression of L20 (estimated by western analysis) from the recombinant plasmid pHML17 caused a 3-fold repression of a wild-type transcriptional infC-lacZ fusion (data not shown). However, under these conditions cell growth was also slowed down 2- to 3-fold making it difficult to distinguish between a specific L20 related effect and a more general growth-rate-related repression. In order to reduce the high β-galactosidase activity from a continuously expressed lacZ fusion, we opted for an experimental system where the expression of the infC-lacZ fusion was inducible. For that purpose the wild-type promoter of the infC-lacZ fusion was replaced with the inducible Pspac promoter. Addition of IPTG thus simultaneously induces the expression of the infC-lacZ fusion and that of rplT from the recombinant plasmid so that we measured an effect of L20 on the ‘initial rate’ of β-galactosidase synthesis. After a 20-min expression period, during which cell growth was not significantly affected, we observed a 7-fold reduction in β-galactosidase expression when the rplT gene was overexpressed compared to a control strain harboring the parental plasmid (1.5 U/mg versus 10 U/mg of β-galactosidase activity). This indicates that ribosomal protein L20 can act as a repressor in vivo and corroborates the in vitro data described above.
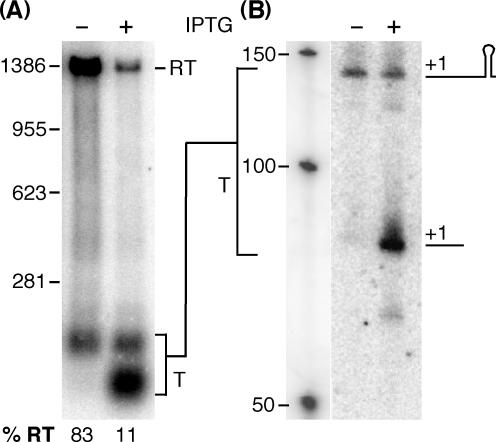
We used the same conditions of L20 overexpression as described above (induction for 20 min) to analyze its effect on the transcription profile of the wild-type infC operon. The results of the northern analysis are shown in Figure 5 and demonstrate that autogenous repression is caused by premature transcription termination. L20 overproduction (8-fold) from the recombinant plasmid caused a decrease in the amount of read-through transcription from 83% in the uninduced strain to 11% and a concomitant increase in prematurely terminated transcripts. Unexpectedly most of these terminated transcripts were actually shorter than the mRNA ending at the leader terminator (Figure 5A). We further analyzed these transcripts by northern using several oligonucleotide probes complementary to different regions of the infC leader to identify which region of the infC leader the truncated transcript corresponds to. Only probes which are specific for the 5′ half of the infC leader hybridized to the short transcript. As an example the northern blot using the oligonucleotide probe HP1080 which is complementary to positions +8 to +34 of the infC leader is shown in Figure 5B. The shorter leader transcript, which is formed in the presence of L20, is detected with the promoter proximal oligonucleotide probe showing that it starts at the original 5′ end and extends to about 80 nt which places its 3′ end a few nucleotides downstream of the second stem-loop structure which is part of the conserved domain (Figure 3). Preliminary data obtained with different endoribonuclease mutants suggest that the small transcript is generated from the normally terminated leader transcript through endonucleolytic processing by RNases J1 or J2 or both ((26), data not shown).
Figure 5.
Effect of L20 overproduction on the transcription profile of the infC operon. Northern analysis of total RNA of a B. subtilis wild-type strain harboring plasmid pHML17 carrying an IPTG inducible copy of the rplT (L20) gene was performed using two different probes. (A) RNA was separated on a 0.8% agarose gel and probed with an infC specific probe including the entire leader sequence (see Materials and methodssection). (B) RNA separated on a 8% polyacrylamide gel was probed with oligonucleotide HP1080 complementary to positions +8 to +34 of the infC leader. Where indicated IPTG (1 mM) was added to mid-log cultures for 20 min prior to isolation of the RNA. RT = read-through transcript, T = terminated transcript. The positions of the molecular size markers are indicated. Percentages of read-through transcripts are indicated below the gel.
Control of terminator read-through by L20 likely involves molecular mimicry between mRNA and rRNA
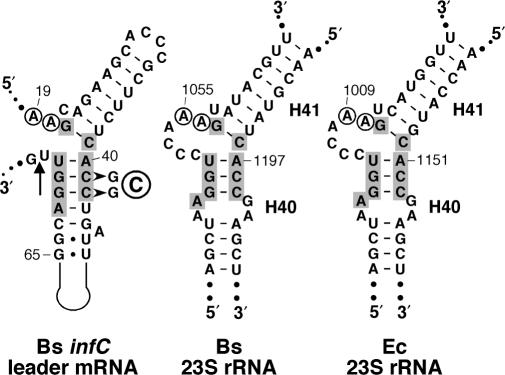
A close analysis of the leader mRNA in the terminator conformation revealed another potentially important secondary structure. The first two (major) helices in the 5′ half of the infC leader (positions 19–71) including the conserved domain (see Figure 3) resemble the L20-binding site on eubacterial 23S rRNA. This is most apparent when this region is redrawn as shown in Figure 6. High-resolution ribosome structures from a Gram-positive and Gram-negative bacterium (27,28) localized the L20-binding site to the interhelical junction of helices 40 and 41 in 23S rRNA. In addition, a phylogenetic analysis (6) identified two conserved adenosine residues (positions 19 and 20 in the infC leader) adjacent to the upper helix (Figure 6).
Figure 6.
Secondary structure diagrams of the L20-binding sites on B. subtilis and E. coli 23S rRNA and a region of the B. subtilis infC leader. A segment of the infC leader (nts 19–73) comprising the conserved 5′ domain (Figure 2) is drawn to highlight the structural resemblance between the mRNA fold and the L20-binding sites at the junction of helices 40 and 41 (H40/41) on 23S rRNA from B. subtilis and E. coli. Numbering corresponds to the infC mRNA and the respective mature 23S rRNA molecules. The arrow indicates the position of the reverse transcription arrest observed in the presence of L20 (see text and Figure 7). The two A residues conserved in eubacterial 23S rRNA are encircled, nucleotides at the interhelical junction conserved between the structures are shown in grey boxes.
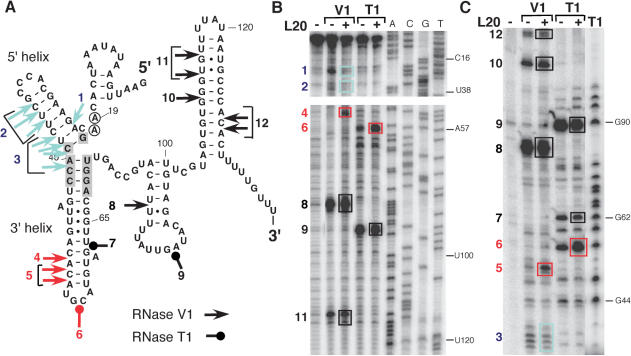
Binding of B. subtilis L20 to the infC leader structure depicted in Figure 6 is an attractive hypothesis to explain its role as a repressor of transcription elongation. In fact, L20 binding should stabilize the terminator conformation by inhibiting the formation of the competing antiterminator structure (Figure 3). In order to determine which region of the infC leader is recognized by L20 we performed footprint experiments using RNase V1 which cleaves double-stranded RNA and RNase T1 which cleaves single-stranded RNA 3′ to G residues. The infC leader transcript (positions 1–168) was digested with either RNase V1 or RNase T1 in the presence or absence of B. subtilis L20, and the cleavages were located by primer extension on the cleaved product (Figure 7B) or analyzed directly on a gel using a 5′ end labeled transcript (Figure 7C). We observed multiple protections from cleavage by RNase V1 within the conserved 5′ domain (5′ helix) and the interhelical junction between the 5′ and the adjacent 3′ helix when the infC leader mRNA was incubated in the presence of purified B. subtilis L20 protein prior to addition of the RNase (Figure 7, blue arrows positions 1–3). Binding of L20 also leads to a stabilization of the 3′ adjacent helix as judged by the appearance of strong RNase V1 cleavages under these conditions (Figure 7, red arrows, positions 4,5). As expected, no significant alterations of the RNase cleavage pattern was observed for the 3′ half of the infC leader which contains the terminator structure (positions 8–12). This suggests that L20 actually binds to the region resembling the 23S rRNA-binding site illustrated in Figure 6 that in the context of the regulatory mechanism described here can also be considered as an anti-antiterminator structure.
Figure 7.
RNase probing of the infC leader mRNA structure in the absence or presence of L20 protein. An in vitro infC leader transcript (nts 1–168) was subjected to cleavage by RNases V1 or T1. Where indicated purified B. subtilis L20 protein was added prior to RNase cleavage. (A) Cleavages by RNase V1 and RNase T1 are shown on the infC leader mRNA structure. Colors indicate the change in cleavage efficiency observed in the presence of L20: black (no change), red (increase), blue (decrease). The numbers next to the symbols locate the corresponding cleavages on the gels (boxes). Shaded and encircled nucleotides correspond to positions conserved at the L20-binding site on 23S rRNA as shown in Figure 6. (B) Cleavages on an unlabeled transcript were detected by primer extension with labeled oligonucleotide HP697 (positions 168–141 on infC leader). (C) Cleavages on a 5′ labeled transcript were analyzed directly.
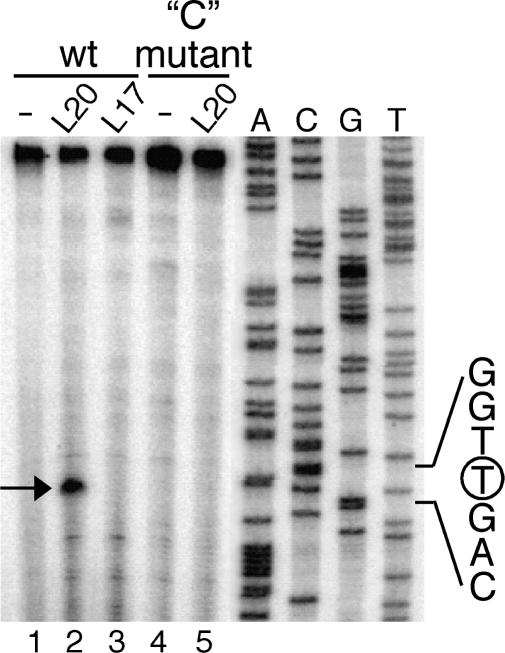
We then carried out a toeprint experiment and, in the presence of L20, observed a single strong stop signal at position U72 (Figures 6 and 8). This signal is completely absent in control reactions without added proteins or in the presence of B. subtilis ribosomal protein L17 (Figure 8, lanes 1–3). This result corroborates the RNase probing data and indicates that L20 binds in the region covering the conserved domain and the lower half of the 3′ adjacent helix. The integrity of the two helices is probably crucial for the binding of L20 as can be inferred from studies on the E. coli translational operator upstream of the rpmI (L35) gene which also binds L20 (29). We introduced a double mutation called ‘C’ (C41C42 → GG, Figures 3 and 6) in order to destabilize the 3′ helix. As shown in Figure 8 (lanes 4 and 5) L20 is no longer able to induce the reverse transcription arrest observed with the wild-type substrate.
Figure 8.
Toeprint caused by L20 binding to the B. subtilis infC leader mRNA. Oligonucleotide HP697 was extended on a wild type and ‘C’ mutant (CC41,42 → GG, see Figure 5) in vitro transcript of the infC leader region. Where indicated the transcript was incubated with the B. subtilis r-proteins L20 or L17 prior to the extension reaction. The reverse transcriptase stop caused by L20 bound to the transcript is indicated by an arrow; it corresponds to a primer extension arrest at uridine residue 72 (see Figure 6). The sequence ladder was generated with the same primer HP697, the encircled nucleotide indicates the arrest position on the cDNA.
In conclusion, our data suggest that binding of L20 to the infC leader within the phylogenetically conserved region (Figure 6) probably involves molecular mimicry between rRNA and mRNA structures recognized by the protein.
DISCUSSION
The data presented here describe the first molecular mechanism controlling the expression of ribosomal proteins in a Gram-positive bacterium. As already suggested by the observed autoregulation of the monocistronic rpsD (S4) gene (13), the general principle of autogenous control of ribosomal protein biosynthesis, as observed in E. coli, appears to be maintained in B. subtilis. However, at the molecular level the mechanisms involved in this regulation turn out to be quite different.
L20-mediated autogenous control probably involves molecular mimicry
The presence of purified B. subtilis L20 protein in single-round transcription assays greatly stimulates premature transcription termination at the leader terminator (Figure 4B). Addition of the C-terminal half of E. coli L20 containing the 23S rRNA-binding domain has the same effect. This observation suggests that both E. coli and B. subtilis L20 proteins are recognizing a sequence similar to their primary binding site on 23S rRNA. As illustrated in Figure 6, a region encompassing two helices including the conserved domain near the 5′ end of the infC leader (referred to as the operator) strongly resembles the L20-binding site on the E. coli 23S rRNA. L20 essentially recognizes the interhelical region between helices 40 and 41 on 23S rRNA that is almost identical in E. coli and B. subtilis (Figure 6). Footprint experiments located the L20-binding site between the two helices that constitute the proposed operator region. The 5′ helix containing the phylogenetically conserved sequence is well protected by the binding of L20 as can be concluded by the reduction of RNase V1 cleavages (Figure 7). In addition, the 3′ helix is stabilized as indicated by the appearance of strong RNase V1 cleavages in the upper part of the helix. This suggests that L20 actually binds to both helices. We observed no significant changes in the cleavage patterns in other regions of the infC leader mRNA which suggests that the proposed operator is the principal site that is recognized by L20. The toeprint signal observed immediately downstream of the 3′ helix in the presence of L20 (Figure 8) confirms the notion that the binding of L20 stabilizes the 3′ helix. The toeprint was abolished when the 3′ helix was destabilized by mutation (Figure 8). However, we cannot distinguish whether L20 actually needs a fully base-paired 3′ helix for binding or if it can still recognize the operator in the mutant construct but that the interaction between L20 and the operator is not strong enough to block reverse transcriptase.
A recent phylogenetic comparison of the E. coli translational operator upstream of rpmI and eubacterial 23S rRNA suggested some structural requirements for L20 binding (6). The basic requirements consist of two adjacent helices, connected by an internal loop with no strict topology but containing two unpaired A residues immediately 5′ to the upper helix. The primary sequence and length of the internal loop show no conservation except for the two conserved A nucleotides (6). This suggests that the open configuration of the Bacillus operator without a bulge loop connecting the two helices (Figure 6), also allows L20 binding. The high-resolution 3D structure of the E. coli ribosome (28) revealed that L20 interacts primarily with the phosphodiester backbone around the junction of helices 40 and 41. This explains the requirement for both stems to form and is consistent with our observation that L20 no longer constitutes a ‘road block’ for reverse transcriptase when the base of the lower helix is destabilized by mutation (Figures 3 and 6).
It should be mentioned that the correct folding of the operator domain is extremely sensitive to alterations of its base composition. We have tried to introduce single base substitutions at the putative L20-binding site to further validate the proposed model. However, these experiments would only be conclusive if complementary changes that restore base pairing also enable the correct folding of the operator. This is not the case, even complementary base changes in this region lead to a complete rearrangement of the local secondary structure when analyzed by the M-fold algorithm (data not shown). For this reason we have not included more data on operator mutations.
The ‘conserved domain’ primary sequence spanning 26 bases near the 5′ end of the infC leader (Figure 2) is an integral part of the operator, forming essentially the upper helix (Figures 3 and 6). Given the extent of sequence conservation in diverse organisms within the firmicutes it cannot be excluded that the primary sequence of the upstream domain might also be important for the binding of L20 to the infC operator.
Our data suggest a regulatory model based on molecular mimicry between the L20-binding sites on ribosomal and messenger RNA (Figure 9). When rRNA is synthesized at high rate (i.e. rapid growth) L20 is titrated by rRNA leaving little protein available for binding to the operator. On the contrary, when the rRNA concentration decreases excess L20 will bind to the operator (Figure 9A). This renders the nucleotides necessary for the long-range antiterminator interaction inaccessible thereby provoking premature transcription termination. Thus, the operator can also be viewed as an anti-antiterminator, a secondary structure trapped by the binding of L20, hence favoring termination.
Figure 9.

Regulatory model for the control of the B. subtilis infC operon. (A) L20 is primarily recruited by 23S rRNA to integrate the ribosome (in grey). When in excess, free L20 will bind at the operator in the infC leader which is similar to its binding site on 23S rRNA and inhibit formation of the antiterminator structure causing premature transcription termination. (B) In the absence of free L20 (i.e. rapid growth) a part of the operator (blue line) will hybridize to bases included in the terminator (red line) to form an antiterminator structure and provoke transcriptional read-through.
The regulatory model implies that the default state of the transcription attenuation system is read-through. In fact, expression of infC-lacZ fusions is greatly reduced in mutant constructs affecting the formation of the antiterminator (Table 1). Moreover, efficient read-through of the terminator is also observed in in vitro transcription assays in the absence of exogenous L20. Under these conditions the operator is free to engage in a long-range base-pair interaction with the 5′ half of the terminator stem (Figure 9B). The existence of this antiterminator structure is consistent with the in vivo and in vitro effects of complementary mutations which maintain the proposed interaction (Figure 3). The antiterminator is thermodynamically slightly more stable than the terminator (ΔG values of −16.9 versus −15 kcal/mol for the respective helices) but more importantly its formation is kinetically favored (it can form before transcription of the terminator structure is complete).
Based on sequence comparisons and analysis of alternative folding patterns we found that the control system identified in B. subtilis is probably also operative in most low GC Gram-positive bacteria. The phylogenetically most conserved element is the operator itself which corroborates the regulatory model presented here.
Molecular mimicry involving ribosomal protein L20 seems to constitute an important aspect in the regulation of the rpmI-rplT operon in both E. coli (29) and B. subtilis even though the control mechanisms are totally different in the two organisms. Nomura's model (3,4) of the regulation of r-protein biosynthesis based on autogenous control and molecular mimicry is thus valid not only in E. coli but also in B. subtilis and probably other Gram-positive organisms as well. However, while in the original model autocontrol was proposed to take place at the translational level, the mechanism decribed here shows that ribosomal protein expression can also be regulated effectively at the level of transcription. Our data seem to confirm that the concept of molecular mimicry albeit based only on a few examples is very powerful and capable of being integrated into a wide variety of control mechanisms.
A regulatory mechanism that controls the entire operon
Since all four genes of the Bacillus infC operon are co-transcribed from a single promoter this implies that L20-mediated control influences the expression of the entire operon, infC, rpmI, rplT and ysdA. However with very few exceptions (e.g. Oceanobacillus) the ysdA gene is not conserved as part of the infC operon even in closely related species and in B. subtilis the short intergenic region between the rplT and ysdA genes contains a sequence capable of folding into an intrinsic transcription terminator. The YsdA protein has no assigned or suspected function. Using the B. subtilis YsdA protein sequence for a BLAST search we found orthologs essentially in Bacilli, Clostridia and proteobacteria but also in some Euryarcheota and Dyctostelium. In many cases, most notably in the beta-, gamma- and delta-subfamilies of the proteobacteria the ysdA ortholog is part of a larger protein predicted to have nucleic-acid-binding characteristics.
In E. coli translational feedback regulation by L20 only represses the expression of the gene for L35 (rpmI) and that of its own gene by translational coupling (6,15). In principle, it makes sense to co-regulate the expression of translation factor IF3 (infC) and ribosomal proteins; efficient translation of proteins requires a good coordination of the synthesis of ribosomes and translation factors. However, the latter are present in the cell at 0.2–0.3 molecules per ribosome that is enough to saturate free 30S particles (30). The B. subtilis infC gene like that of E. coli has an atypical AUU start codon which, at least in E. coli, is sufficient to autoregulate its translation (31). It is possible but remains to be verified whether the B. subtilis infC gene is also subject to translational autoregulation. This would allow to fine-tune the biosynthesis of IF3 independently of the downstream rpmI (L35) and rplT(L20) genes.
ACKNOWLEDGEMENTS
We thank J. Plumbridge for useful discussions and critical reading of the manuscript. We thank H. Isambert for helpful discussions. We are grateful to N. Mathy for help in the initial phase of the project and to F. Allemand and C. Chiaruttini for the gift of E. coli L20 protein. S.E. benefited from financial support from INRA (ASC). This work was supported by funds from the CNRS (UPR 9073), MRE (Contract 92C0315), Université Paris VII (Contract DRED) and PRFMMIP from the Ministère de l’Education Nationale. N.C. was recipient of a studentship from the French Ministry of Research. Funding to pay the Open Access publication charge was provided by CNRS (UPR9073).
Conflict of interest statement. None declared.
REFERENCES
- 1.Zengel JM, Lindahl L. Diverse mechanisms for regulating ribosomal protein synthesis in Escherichia coli. Prog. Nucleic Acid Res. Mol. Biol. 1994;47:331–370. doi: 10.1016/s0079-6603(08)60256-1. [DOI] [PubMed] [Google Scholar]
- 2.Mattheakis LC, Nomura M. Feedback regulation of the spc operon in Escherichia coli: translational coupling and mRNA processing. J. Bacteriol. 1988;170:4484–4492. doi: 10.1128/jb.170.10.4484-4492.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fallon AM, Jinks CS, Strycharz GD, Nomura M. Regulation of ribosomal protein synthesis in Escherichia coli by selective mRNA inactivation. Proc. Natl. Acad. Sci. U.S.A. 1979;76:3411–3415. doi: 10.1073/pnas.76.7.3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nomura M, Yates JL, Dean D, Post LE. Feedback regulation of ribosomal protein gene expression in Escherichia coli: structural homology of ribosomal RNA and ribosomal protein MRNA. Proc. Natl. Acad. Sci. U.S.A. 1980;77:7084–7088. doi: 10.1073/pnas.77.12.7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Merianos HJ, Wang J, Moore PB. The structure of a ribosomal protein S8/spc operon mRNA complex. RNA. 2004;10:954–964. doi: 10.1261/rna.7030704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guillier M, Allemand F, Dardel F, Royer CA, Springer M, Chiaruttini C. Double molecular mimicry in Escherichia coli: binding of ribosomal protein L20 to its two sites in mRNA is similar to its binding to 23S rRNA. Mol. Microbiol. 2005;56:1441–1456. doi: 10.1111/j.1365-2958.2005.04644.x. [DOI] [PubMed] [Google Scholar]
- 7.Stelzl U, Zengel JM, Tovbina M, Walker M, Nierhaus KH, Lindahl L, Patel DJ. RNA-structural mimicry in Escherichia coli ribosomal protein L4-dependent regulation of the S10 operon. J. Biol. Chem. 2003;278:28237–28245. doi: 10.1074/jbc.M302651200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henkin TM. Ribosomes, protein synthesis factors, and tRNA synthetases. In: Sonenshein AL, editor. Bacillus subtilis and its Closest Relatives: from Genes to Cells. Washington D.C.: ASM Press; 2002. pp. 313–322. [Google Scholar]
- 9.Li X, Lindahl L, Sha Y, Zengel JM. Analysis of the Bacillus subtilis S10 ribosomal protein gene cluster identifies two promoters that may be responsible for transcription of the entire 15-kilobase S10-spc-alpha cluster. J. Bacteriol. 1997;179:7046–7054. doi: 10.1128/jb.179.22.7046-7054.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zengel JM, Lindahl L. Escherichia coli ribosomal protein L4 stimulates transcription termination at a specific site in the leader of the S10 operon independent of L4-mediated inhibition of translation. J. Mol. Biol. 1990;213:67–78. doi: 10.1016/S0022-2836(05)80122-6. [DOI] [PubMed] [Google Scholar]
- 11.Zengel JM, Sha Y, Lindahl L. Surprising flexibility of leader RNA determinants for r-protein L4-mediated transcription termination in the Escherichia coil S10 operon. RNA. 2002;8:572–578. doi: 10.1017/s1355838202026237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grundy FJ, Henkin TM. The rpsD gene, encoding ribosomal protein S4, is autogenously regulated in Bacillus subtilis. J. Bacteriol. 1991;173:4595–4602. doi: 10.1128/jb.173.15.4595-4602.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grundy FJ, Henkin TM. Characterization of the Bacillus subtilis rpsD regulatory target site. J. Bacteriol. 1992;174:6763–6770. doi: 10.1128/jb.174.21.6763-6770.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Butler JS, Springer M, Dondon J, Graffe M, Grunberg-Manago M. Escherichia coli protein synthesis initiation factor IF3 controls its own gene expression at the translational level in vivo. J. Mol. Biol. 1986;192:767–780. doi: 10.1016/0022-2836(86)90027-6. [DOI] [PubMed] [Google Scholar]
- 15.Lesage P, Chiaruttini C, Graffe M, Dondon J, Milet M, Springer M. Messenger RNA secondary structure and translational coupling in the Escherichia coli operon encoding translation initiation factor IF3 and the ribosomal proteins, L35 and L20. J. Mol. Biol. 1992;228:366–386. doi: 10.1016/0022-2836(92)90827-7. [DOI] [PubMed] [Google Scholar]
- 16.Merino E, Yanofsky C. Transcription attenuation: a highly conserved regulatory strategy used by bacteria. Trends Genet. 2005;21:260–264. doi: 10.1016/j.tig.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 17.Winkler WC, Breaker RR. Regulation of bacterial gene expression by riboswitches. Annu. Rev. Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- 18.Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 19.Putzer H, Gendron N, Grunberg-Manago M. Co-ordinate expression of the two threonyl-tRNA synthetase genes in Bacillus subtilis: control by transcriptional antitermination involving a conserved regulatory sequence. EMBO J. 1992;11:3117–3127. doi: 10.1002/j.1460-2075.1992.tb05384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gendron N, Putzer H, Grunberg-Manago M. Expression of both Bacillus subtilis threonyl-tRNA synthetase genes is autogenously regulated. J. Bacteriol. 1994;176:486–494. doi: 10.1128/jb.176.2.486-494.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weinrauch Y, Msadek T, Kunst F, Dubnau D. Sequence and properties of comQ, a new competence regulatory gene of Bacillus subtilis. J. Bacteriol. 1991;173:5685–5693. doi: 10.1128/jb.173.18.5685-5693.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stragier P, Bonamy C, Karmazyn-Campelli C. Processing of a sporulation sigma factor in Bacillus subtilis: how morphological structure could control gene expression. Cell. 1988;52:697–704. doi: 10.1016/0092-8674(88)90407-2. [DOI] [PubMed] [Google Scholar]
- 23.Moran CP. Measuring gene expression in Bacillus. In: Harwood CR, Cutting SM, editors. Molecular Biological Methods for Bacillus. Chichester: John Wiley & Sons Ltd; 1990. pp. 267–293. [Google Scholar]
- 24.Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]
- 25.Guillier M, Allemand F, Graffe M, Raibaud S, Dardel F, Springer M, Chiaruttini C. The N-terminal extension of Escherichia coli ribosomal protein L20 is important for ribosome assembly, but dispensable for translational feedback control. RNA. 2005;11:728–738. doi: 10.1261/rna.7134305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Even S, Pellegrini O, Zig L, Labas V, Vinh J, Brechemmier-Baey D, Putzer H. Ribonucleases J1 and J2: two novel endoribonucleases in B. subtilis with functional homology to E. coli RNase E. Nucleic Acids Res. 2005;33:2141–2152. doi: 10.1093/nar/gki505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harms J, Schluenzen F, Zarivach R, Bashan A, Gat S, Agmon I, Bartels H, Franceschi F, Yonath A. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell. 2001;107:679–688. doi: 10.1016/s0092-8674(01)00546-3. [DOI] [PubMed] [Google Scholar]
- 28.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 29.Guillier M, Allemand F, Raibaud S, Dardel F, Springer M, Chiaruttini C. Translational feedback regulation of the gene for L35 in Escherichia coli requires binding of ribosomal protein L20 to two sites in its leader mRNA: a possible case of ribosomal RNA-messenger RNA molecular mimicry. RNA. 2002;8:878–889. doi: 10.1017/s1355838202029084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bremer H, Dennis PP. Modulation of chemical composition and other parameters of the cell by growth rate. In: Neidhardt FC, Curtiss R, Ingraham JL, Lin EC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. Vol. 2. Washington, D.C.: American Society for Microbiology; 1996. pp. 1553–1569. [Google Scholar]
- 31.Sacerdot C, Chiaruttini C, Engst K, Graffe M, Milet M, Mathy N, Dondon J, Springer M. The role of the AUU initiation codon in the negative feedback regulation of the gene for translation initiation factor IF3 in Escherichia coli. Mol. Microbiol. 1996;21:331–346. doi: 10.1046/j.1365-2958.1996.6361359.x. [DOI] [PubMed] [Google Scholar]