Abstract
To characterize the role of BRCA1 in mammary gland development and tumor suppression, a transgenic mouse model of BRCA1 overexpression was developed. Using the mouse mammary tumor virus (MMTV) promoter/enhancer, transgenic mice expressing human BRCA1 or select mutant controls were generated. Transgenic animals examined during adolescence were shown to express the human transgene in their mammary glands. The mammary glands of 13-week-old virgin homozygous MMTV-BRCA1 mice presented the morphology of moderately increased lobulo-alveolar development. The mammary ductal trees of both hemizygous and homozygous MMTV-BRCA1t340 were similar to those of control non-transgenic littermates. Interestingly, both hemi- and homozygous mice expressing a splice variant of BRCA1 lacking the N-terminal RING finger domain (MMTV-BRCA1sv) exhibited marked mammary lobulo-alveolar development, particularly terminal end bud proliferation. Morphometric analyses of mammary gland whole mount preparations were used to measure epithelial staining indices of ~35% for homozygous MMTV-BRCA1 mice and ~60% for both hemizygous and homozygous MMTV-BRCA1sv mice versus ~25% for non-transgenic mice. Homozygous MMTV-BRCA1 mice showed delayed development of tumors when challenged with 7,12 dimethylbenzanthracene (DMBA), relative to non-transgenic and homozygous BRCA1t340 expressing mice. In contrast, homozygous MMTV-BRCA1sv transgenic animals were sensitized to DMBA treatment and exhibited a very rapid onset of mammary tumor development and accelerated mortality. MMTV-BRCA1 effects on mortality were restricted to DMBA-induced tumors of the mammary gland. These results demonstrate in vivo roles for BRCA1 in both mammary gland development and in tumor suppression against mutagen-induced mammary gland neoplasia.
Keywords: BRCA1 gene, transgenic mice, mammary gland development
1. Introduction
Approximately 5 to 10% of all breast and ovarian cancers can be attributed to highly penetrant, dominantly inherited mutations in specific genes 1,2. Mutations in the BRCA1 gene are responsible for 45% of familial breast cancer and 80% of families predisposed to both breast and ovarian cancer 3,4. The ~220kD BRCA1 nuclear phosphoprotein is characterized by an N-terminal RING finger domain (Cys3-His-Cys4), two nuclear localization signals in exon 11, and an acidic C-terminal region containing BRCA1 C-terminus (BRCT) domains 5,6.
BRCA1 has been implicated in a broad range of cellular activities including DNA repair and recombination, transcriptional regulation, chromatin remodeling, cellular growth control, and genome stability 7-9. The murine BRCA1 protein consists of 1812 amino acids and exhibits approximately 60% identity and 72% similarity with the human BRCA1 protein. Northern analysis indicates that the 7.5kb Brca1 transcript is expressed in a 14.5- day whole embryo and extra-embryonic tissues, and in adult testis and thymus 10. Brca1 transcripts were detected in adult liver, uterus, kidney, lung, mammary gland, and ovary by RNase protection and in situ hybridization 10,11. In situ hybridization studies also showed that Brca1 undergoes a complex pattern of developmentally regulated expression, and is expressed in cells undergoing both proliferation and differentiation 11. In the mouse ovary, Brca1 expression is positively correlated with cellular proliferation and S-phase nuclear PCNA staining 12. Finally, the developmental pattern of Brca1 expression suggests that it is involved in proliferation and differentiation in the mouse mammary gland 11,13,14.
Studies indicate that murine Brca1 plays a crucial role in the early development of mouse embryos. Multiple laboratories have disrupted Brca1 by gene targeting 15-19. Although the phenotype of embryos homozygous for Brca1 mutations varies, all die between embryonic days 5.5-13.5 with the onset of lethality largely dependent on the region where the mutation was introduced. However, despite this variation in the age of embryonic lethality, all mutant embryos suffer similar abnormalities, including cellular proliferation defects and developmental retardation.
To better understand the function of BRCA1 in normal biological processes, we used the murine mammary gland as a model system. Transgenic mice were developed to overexpress wild type human BRCA1 and BRCA1 mutant proteins, under the control of the mouse mammary tumor virus (MMTV) LTR, during ductal and alveolar morphogenesis. The mammary gland phenotypes of these animals were characterized to provide insights into the contributions of BRCA1 to mammary development. To examine the tumor suppressor function of BRCA1 in this model, transgenic animals were treated with the carcinogen 7,12-dimethylbenzanthracene (DMBA). Orogastric instillation of DMBA results in a high incidence of mammary tumor formation among treated mice 20. Expression of wild-type BRCA1 delayed mammary tumor onset, while overexpression of selected mutant BRCA1 constructs had either no effect on mammary tumor incidence or survival, or resulted in a rapid onset of tumors and shortened survival. Our results demonstrated that overexpression of wild type human BRCA1 in the murine mammary gland provided protection against carcinogen-induced tumors. A BRCA1 construct missing the N-terminus predisposed the mammary gland to tumor development. These findings suggest that the tumor suppression and proliferation functions of BRCA1 may not be mediated by the same domains of the protein.
2. Materials and methods
Construction of MMTV-BRCA1 Plasmids
All pMMTV-BRCA1 constructs (Fig. 1) were created by subcloning the BRCA1 cDNAs into the pMMTV-LTR expression vector 21. The wild type human BRCA1 and BRCA1t340 cDNAs each contain 91bp of the 5' untranslated region (UTR) upstream of the ATG start codon. The mutant BRCA1t340 construct contains a stop codon at amino acid 340 (1010delA), resulting in protein truncation 116 amino acids into exon 11. BRCA1t340 is a known disease-causing mutation in humans. BRCA1sv was isolated from a human testis cDNA library and contains a cryptic splice site in exon 5 22,23. This alternative splicing causes translation to begin at exon 6 (amino acid 72 of wild type BRCA1), resulting in the loss of the N-terminal RING finger domain of the protein. The pMMTV vector has previously been demonstrated to be effective for transgene expression in mice 21,24-26. The entire 5.7kb BRCA1 cDNA was subcloned into the 5.9kb pMMTV LTR promoter/enhancer vector 21 at the Sal I restriction site to create the pMMTV-BRCA1 construct. The pMMTV-BRCA1sv construct was created in a similar manner. The 1.1 kb BRCA1t340 deletion mutant cDNA was created by polymerase chain reaction amplification of the N-terminal 340 amino acids from acids from the full-length BRCA1 cDNA using modified primers (5' GAA TTG TGT CGA CAT GCG GCC G 3' and 5' GGC GCC GTC GAC CTA GCT AGC TAC CTT TTT TCT GTG CTG GGA GTC C 3') containing Sal I restriction sites.
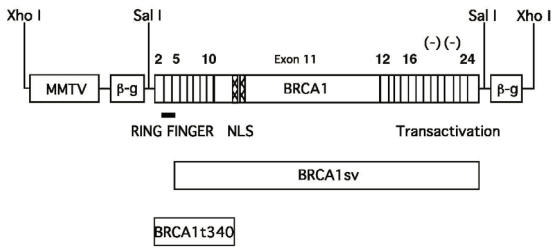
Figure 1.
MMTV-BRCA1 transgenic constructs. Diagram of the BRCA1 cDNAs used for the generation of transgenic animals. The expression of wild type BRCA1, BRCA1sv (70 amino acid N-terminal deletion), and BRCA1t340 (BRCA1 C-terminal truncation mutant) are controlled by the MMTV-LTR promoter. The BRCA1 cDNAs were each inserted into the third exon of the rabbit ß-globin gene (β-g). The bar indicates the RING finger motif, the hatched region corresponds to the nuclear localization signals and the negative symbols (-) indicate the negatively charged C-terminal domain with transactivation function.
Generation and Maintenance of BRCA1 Transgenic Mice
The Xho I BRCA1 fragments were microinjected into the male pronucleus of (C57BL/6x DBA/2) F1 fertilized mouse embryos and implanted into pseudopregnant ICR surrogate mice by the Vanderbilt Transgenic/ES Cell Shared Resource Facility. Founder mice were bred to C57BL/6 mice to establish transgenic lines. All animals used in these studies were handled in strict compliance with Vanderbilt University Animal Care Committee regulations.
Identification of Transgenic Founders
Transgenic animals were identified by Southern blot analysis or by PCR. For Southern analysis, 5µg of mouse tail genomic DNA (obtained from Metofane-anesthetized 3-week-old mice as described previously 27 were digested with Alu I and separated on an agarose gel. The DNA was transferred to Hybond-N Nylon membranes (Amersham) and hybridized in QuikHyb Solution (Stratagene) with a 32P-random primed-labeled 533bp Sal I / Xho I rabbit ß-globin probe, excised from the pMMTV vector.
To identify transgenic animals by PCR, 1µl of genomic tail DNA was added to a 50µl reaction volume [1x PCR buffer II, 1.5mM MgCl2, 200nM each of rabbit ß-globin forward 5' GTC TCG GAT CCT CAG AAG GTG GTG GCT GGT GTG G 3' and rabbit ß-globin reverse 5' GAA CTA GGT ACC GGC ATA TGT TGC CAA ACT CTA AAC C 3' primers, 200µM each dNTP, and 2.5 units of AmpliTaq DNA polymerase (Perkin-Elmer)]. Cycling conditions consisted of 94oC for 6 min, followed by 25 cycles of 94oC for 30 sec, 64oC for 60 sec, and 72oC for 90 sec, and a final extension at 72oC for 3 min. The PCR product was a 260bp fragment of the rabbit ß-globin exon 3 contained in the transgene construct.
The copy number of founder transgenic animals was estimated by determining the amounts of DNA equal to one copy of the integrated transgene. The approximate number of base pairs of the promoter and transgene injected (8400bp for MMTV-BRCA1 and MMTV-BRCA1sv; 3860bp for MMTV-BRCA1t340) was divided by the approximate size of the mouse haploid genome (3 x 10 9bp). Thus, 2.8pg of the promoter/transgene fragment is equal to approximately one copy of the integrated MMTV-BRCA1 or MMTV-BRCA1sv transgenes per 1µg of genomic DNA, while 1.3pg is equal to approximately one copy of the integrated MMTV-BRCA1t340 transgene per 1µg of genomic DNA. The relative intensity of probe hybridization was compared using a Bio-Rad Model GS 670 Imaging Densitometer.
Determination of Stage of Mouse Estrous Cycle
Vaginal smears were performed using 100µg 0.9% saline and a plastic transfer pipet. Sloughed cells were spread on a glass slide, stained with hematoxylin and eosin, and examined by light microscopy to determine stage of the estrous cycle (proestrus, estrus, metestrus I, metestrus II, or diestrus) 28.
Isolation of Total RNA from Murine Tissues and Northern Blot Analysis
Female transgenic (MMTV-BRCA1, MMTV-BRCA1sv, MMTV-BRCA1t340) and nontransgenic female mice were anesthetized and sacrificed by cervical dislocation to determine mammary gland phenotype. Mammary glands, salivary glands, lung, and kidneys were removed and used for RNA isolation, in TRI REAGENT 29,30 and analysis of transgene expression was performed by Northern blot. Twenty µg of total RNA was separated on a 1% agarose-formaldehyde gel. Following electrophoresis the gel was transferred to Hybond-N Nylon membranes and hybridized to 32P-random primed-labeled probes for human BRCA1 (2.3kb Kpn I / Sac I BRCA1 fragment [exon 11-15] or 1.1kb Sal I BRCA1t340 fragment [exon 2 to first 116 amino acids of exon 11], and rat cyclophilin (700bp Bam H I /Hind III 1B15 cDNA 31) with Quik Hyb Solution. Blots were washed twice in 1X SSC + 0.1% SDS at 65oC for 15 min each, followed by five washes in 0.2X SSC + 0.1% SDS at 65 oC for 30 min each. The blots were exposed to Kodak BioMax MR film and analyzed for transgene expression.
Mammary Gland Whole Mount Preparations
For morphologic analysis, dissected mouse thoracic mammary glands were placed into tissue cassettes and fixed overnight in freshly prepared 4% para-formaldehyde. The glands were defatted in acetone, hydrated in ethanol, and stained in a modified Weiger's iron hematoxylin stain. Tissues were destained and stored in methyl salicylate until photographed on an Olympus SXH10 Research Stereo Microscope using Kodak Ektachrome 64T EPY 135-36 slide film 26.
Histology
Mouse thoracic mammary glands were excised and fixed overnight in 4% para-formaldehyde. The glands were dehydrated in a series of alcohols, embedded in paraffin, sectioned, and stained with hematoxylin and eosin. Digital images of the stained mammary gland sections were captured with a Leaf Lumina brightfield digital camera attached to a Zeiss Axiophot Upright Microscope.
Quantification of Observed Morphology in Mammary Glands
Images of mammary gland whole mount preparations were imported into a Macintosh computer using a Polaroid SprintScan 35 slide scanner and the epithelial content was quantitated. The amount of ducts and terminal end buds present was scored using Photoshop-based image analysis, which allows the identification of chromogens on the basis of their RGB spectral characteristics as well as their saturation, hue, and luminosity. Quantification of the chromogen in at least three digital representative fields (10X magnification) was performed. The tolerance level of the Magic Wand tool was adjusted so that all the ducts were selected. Spatial information can be inferred for the iron hematoxylin since the number of pixels reflects a surface area on the image. The percentage of staining in the mammary gland whole mount preparations (staining index) was calculated by dividing the number of pixels of epithelial staining by the pixels of the entire sample. This method has been demonstrated to be accurately applicable for quantification and complete separation of individual chromogens.
DMBA Treatments
Mice were given DMBA in 100 μl of corn oil by orogastric lavage. The dose regime was 1 mg DMBA/mouse per week for 6 weeks beginning at 8 weeks of age 20. Mice were examined weekly for palpable tumors and morbidity. Mice were sacrificed when tumors reached ~1 cm in diameter. Moribund mice were sacrificed and necropsies were carried out immediately. In nearly all cases these mice were discovered to have lymphoma at necropsy, and many had microscopic mammary tumors that were not palpable. All visible tumors and other tissues were fixed for routine histopathological examination.
Statistical Analysis
The average of the percent-stained ductal structures and buds in mammary gland whole mounts for each animal was entered into the SAS statistical package. The data was normalized by log transformation and analyzed with the GLM (general linear models) procedure using the method of least squares. In particular, the ANOVA (analysis of variance) method for unbalanced data was used to obtain p values. The time to first palpable tumor in the different groups was compared using a Kaplan-Meier test. P< 0.05 was considered statistically significant. For mammary tumor specific endpoints we considered all mice that had developed a mammary tumor whether or not it was the cause of death.
3. Results
Generation of MMTV-BRCA1 Transgenic Mice
Transgenic mice expressing wild type human BRCA1 and mutants (BRCA1t340 and BRCA1sv) under the control of the MMTV LTR promoter/enhancer were created by microinjecting the linear DNA constructs (Fig. 1) into the male pronuclei of single cell (C57BL/6 x DBA/2) F1 embryos.
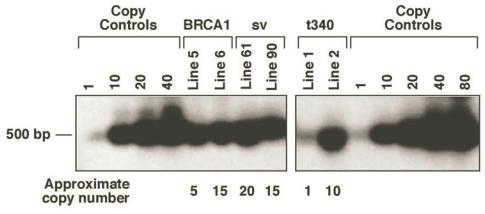
Potential founder pups were screened by either PCR or Southern blot analysis using DNA extracted from tail biopsies. Founder mice were bred to C57BL/6 mice to produce F1 progeny. Two lines per construct were established to control for insertional variation. Transgene copy number was estimated by densitometric scanning of Southern blots hybridized with a 32P-labeled rabbit ß-globin probe (Fig. 2). The MMTV-BRCA1 line 5 has approximately 5 copies of the transgene integrated, while line 6 has 15 copies. The MMTV-BRCA1t340 line 1 has approximately 1 copy of the transgene and line 2 has approximately 10 copies. The copy number of the transgene for the MMTV-BRCA1sv mice was also estimated to be 20 copies for line 61 and 15 copies for line 90.
Figure 2.
MMTV-BRCA1 transgenic mouse lines. Southern hybridization of 10µg of genomic DNA from MMTV-BRCA1 (lines 5 and 6), MMTV-BRCA1sv (lines 61 and 90), and MMTV-BRCA1t340 (lines 1 and 2) animals was performed to estimate transgene copy number. The approximate copy number of each is indicated below the lane.
Expression of the Transgene
Hemizygous and homozygous MMTV-BRCA1, MMTV-BRCA1t340, and MMTV-BRCA1sv mice appeared healthy and exhibited normal behavior. The mice were fertile and the females were able to lactate and nurse their young. To date, neither hemizygous nor homozygous transgenic mice have developed tumors.
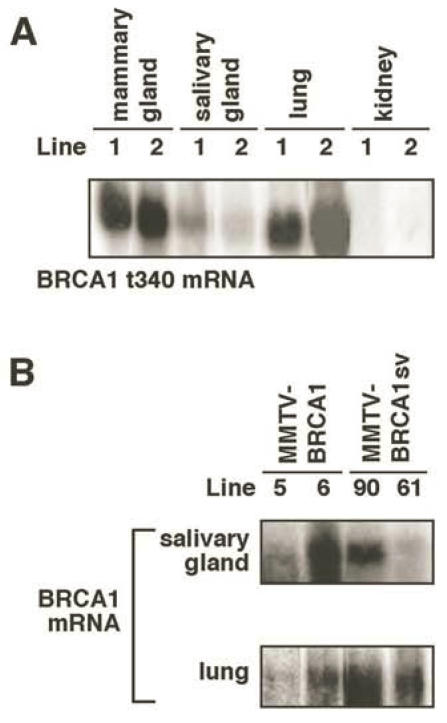
Northern hybridization analyses were performed to demonstrate expression of the transgene mRNA, which may be expected to vary with the number of copies integrated and the location of integration. To control for varying levels of expression, animals from two transgenic lines were evaluated for each construct. The ~ 5.6kb human BRCA1 transgene was expressed in the mammary glands of 13-week-old virgin MMTV-BRCA1 transgenic animals, but not in wild type littermates (Fig. 3A). No cross-reactivity with endogenous mouse Brca1 was observed with the C-terminal BRCA1 transgenic probe (~70% identity to mouse Brca1). Although both MMTV-BRCA1 (line 6) and MMTV-BRCA1sv (line 90) were estimated to each have 15 copies of transgene integrated into the mouse genome, there was significantly higher expression of transgene mRNA (Fig. 3C) in MMTV-BRCA1sv (line 90) animals.
Figure 3.
Northern analysis of MMTV-BRCA1 transgenic mice. (A) The mammary glands of 13-week-old virgin MMTV-BRCA1 (line 6) transgenic animals in diestrus were analyzed for transgene mRNA expression with a radiolabeled BRCA1 cDNA probe. The ~5.6kb BRCA1 transgene mRNA is present in transgenic animals, but not in nontransgenic controls. Homozygous transgenic mice express more human BRCA1 mRNA than hemizygous mice. Hybridization to cyclophilin was performed to control for RNA loading on the gel. (B) The mammary glands of 13-week-old hemizygous MMTV-BRCA1t340 animals (line 2) were analyzed for transgene mRNA expression by Northern hybridization with a radiolabeled BRCA1t340 cDNA probe during different stages of the estrous cycle. BRCA1t340 expression is observed in transgenic mice, but not in nontransgenic controls. (C) The mammary glands of 13-week-old virgin hemizygous MMTV-BRCA1 (line 6) and MMTV-BRCA1sv (line 90) animals in diestrus were analyzed for transgene mRNA expression with a radiolabeled BRCA1 cDNA probe. Despite similar transgene copy numbers, MMTV-BRCA1sv (line 90) mRNA is at a significantly higher level than MMTV-BRCA1 (line 6) mRNA.
Thirteen-week-old virgin MMTV-BRCA1t340 transgenic mice expressed the ~1.1kb human BRCA1t340 mRNA in their mammary glands (Fig. 3B). The human N-terminal BRCA1 cDNA probe was specific for sequences expressed from the transgene and did not recognize endogenous ~7kb mouse Brca1 mRNA. Although this N-terminal probe shows ~80% identity to mouse Brca1, the mouse transcript was not detected under the high stringency conditions used for hybridization. In an attempt to increase the expression of human BRCA1 transcripts, hemizygous transgenic mice were bred to homozygosity. Homozygous mice were identified for MMTV-BRCA1 (lines 5 and 6), MMTV-BRCA1t340 (lines 1 and 2), and MMTV-BRCA1sv (lines 61 and 90) animals using FISH performed on interphase nuclei isolated from white blood cells (data not shown) to confirm Southern blotting results.
Northern blot analysis of total RNA from mammary glands demonstrated that homozygous MMTV-BRCA1 transgenic mice expressed higher levels of transgene RNA than hemizygous mice (Fig. 3A). Even though the mice were estrous cycle stage-matched some variability in the expression of the BRCA1 transgene within and between the various transgenic lines was observed.
Transgene expression was also detected in mouse salivary gland and lung of 13-week-old virgin transgenic mice (Fig. 4A and B). Hybridization was not observed in kidney RNA from transgenic nor wild type animals using either the 1.1kb N-terminal BRCA1t340 cDNA probe (Fig. 4A) or the 2.3kb C-terminal BRCA1 cDNA probe (data not shown). Although the MMTV LTR promoter has been shown to direct low level transgene expression in the epithelial cells of the kidney, other investigators have also reported the lack of detectable transgene mRNA in the kidney in their studies with MMTV transgenic mice 26,27.
Figure 4.
Northern analysis of transgene expression in mouse tissues. (A) The ~1.1kb BRCA1t340 mRNA is detected in mammary gland, salivary gland, and lung of 13-week-old virgin homozygous MMTV-BRCA1t340 (lines 1 and 2) transgenic mice. Transgene expression in kidney was not observed. (B) In addition to the mammary gland, the ~5.6kb BRCA1 or BRCA1sv mRNA is present in the salivary gland and lung of 13-week-old virgin homozygous MMTV-BRCA1 (lines 5 and 6) and MMTV-BRCA1sv (lines 90 and 61) transgenic animals. Transgene expression in kidney was not observed (data not shown).
Consequences of MMTV-BRCA1 Transgene Expression
Since the MMTV LTR is steroid responsive, mice were examined at the same stage of the estrous cycle. The phase of the mouse estrous cycle was determined by obtaining vaginal smears 28. The stages that vaginal cells undergo are representative of parallel changes in the entire reproductive tract. To determine the effect of expression of the different transgenes on mammary ductal structure, transgenic and nontransgenic female mice were sacrificed at 7, 13, and 24 weeks of age. The thoracic mammary glands were removed for whole mount analysis of ductal morphology. A minimum of three mice were examined in all experimental groups.
Characterization of whole mount preparations from 7- and 24-week old virgin MMTV-BRCA1 and MMTV-BRCA1t340 hemizygous transgenic animals displayed normal ductal architecture. At 24 weeks the ducts were observed to fill the entire mammary fat pad, as expected. Ductal elongation and development of the branching structure appeared to be the same in these hemizygous mice compared to nontransgenic animals. At 24 weeks, the ducts reached the periphery of the pad and the end buds regressed to form terminal ductal structures.
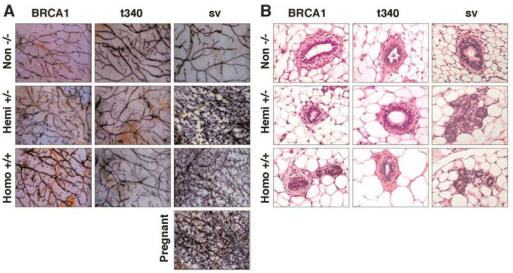
By contrast, the mammary glands of 13-week–old homozygous MMTV-BRCA1 animals appeared to have additional lateral branches and acini compared to age-matched hemizygous and nontransgenic mice. This difference in morphology was not seen in homozygous 13-week-old MMTV-BRCA1t340 mice, which had mammary glands indistinguishable from those of hemizygous and nontransgenic animals (Fig. 5A, first two columns). Histological analysis (Fig. 5B, first two columns) confirmed the results of the mammary gland whole mount preparations. Greater numbers of acini were present in homozygous MMTV-BRCA1 animals than in hemizygous MMTV-BRCA1 mice, nontransgenic controls or hemi- or homozygous MMTV-BRCA1t340 mice.
Figure 5.
Morphological appearance of mammary glands from MMTV-BRCA1 transgenic mice. (A) Iron hematoxylin-stained whole mount preparations from the thoracic mammary gland of 13-week-old virgin nontransgenic, hemizygous, and homozygous MMTV-BRCA1, MMTV-BRCA1t340, and MMTV-BRCA1sv transgenic mice in the diestrus phase of the estrous cycle are shown. Homozygous MMTV-BRCA1 mice demonstrate additional branching and end buds compared to hemizygous and wild type controls. There is an apparent morphological difference in the mammary ductal tree among homozygous and hemizygous MMTV-BRCA1t340 mice and wild type controls. The mammary glands of both hemizygous and homozygous MMTV-BRCA1sv mice show more marked proliferation, and resemble those of a 12.5-day pregnant animal. Magnification of the whole mounts is 10X. (B) Representative sections of 13-week-old virgin mammary glands from nontransgenic, hemizygous, and homozygous MMTV-BRCA1, MMTV-BRCA1t340, and MMTV-BRCA1sv mice are shown. The mammary glands of homozygous MMTV-BRCA1 mice contain more alveoli than hemizygous, nontransgenic, or MMTV-BRCA1t340 mice. The mammary glands of both hemizygous and homozygous MMTV-BRCA1sv mice contain numerous alveoli. Photographs were taken with a 20X objective.
Interestingly, the extent of lobulo-alveolar development in homozygous MMTV-BRCA1 mice was significantly less than observed in age- and estrous stage- matched MMTV-BRCA1sv mice (Fig. 5A, last column). The mammary glands of both virgin hemizygous and homozygous MMTV-BRCA1sv mice exhibited increased lateral branching and alveolar structures forming from the terminal regions of the ducts. Compared to nontransgenic littermate controls, hemizygous and homozygous MMTV-BRCA1sv mice exhibited a phenotype of secondary branching and lobulo-alveolar development similar to that observed in pregnant animals. This phenotype is observed in both 7- and 13-week-old virgin mice in the estrus and diestrus stages of the estrous cycle. These findings were also evident upon histologic staining with hematoxylin and eosin (Fig. 5B, last column). The results suggest that the epithelium of 13-week-old MMTV-BRCA1sv mammary glands resembles morphologically the state of epithelium present in a normal gland at mid-pregnancy. By 24 weeks of age, mammary glands were similar in appearance for all animals and involution of the glands with aging appeared normal. This suggests that BRCA1 overexpression did not cause mammary hyperplasia per se, but rather accelerated normal development.
Quantification of Observed Morphology in Mammary Glands
To confirm and extend qualitative observations of morphologic alterations in transgenic mice, epithelial tissue in mammary gland whole mount preparations was quantified by Photoshop-based image analysis 32,33. The areas selected for image analysis were those in which the staining intensity allowed unambiguous differentiation between mammary ducts/end buds and stroma.
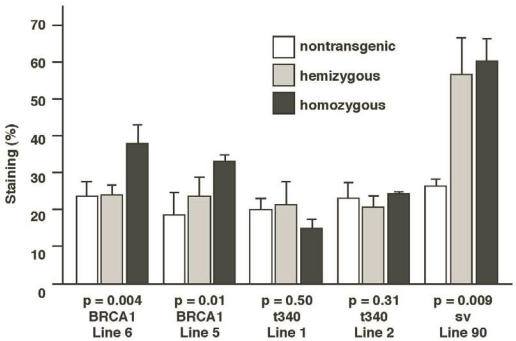
Results from this analysis revealed that the ductal structures and buds of 13-week-old homozygous MMTV-BRCA1 mice (lines 6 and 5) represented ~35% of the mammary gland, versus ~25% for nontransgenic and hemizygous mice (Fig.6; p=0.004 for line 6 and p=0.01 for line 5). MMTV-BRCA1t340 (lines 1 and 2) hemi- and homozygous transgenic mice gave quantitative results similar to those of nontransgenic littermates (Fig. 6; p=0.5 for line 1 and p=0.313 for line 2). However, the amount of lobulo-alveolar tissue in MMTV-BRCA1sv mice resembled those of normal mice during mid-pregnancy. Ductal and lobular tissue in hemi- and homozygous MMTV-BRCA1sv mice occupied ~60% (Fig. 6; p=0.009) of the mammary gland, comparable to the result of ~70% obtained for 12.5-day pregnant mice (data not shown).
Figure 6.
Quantification of epithelium in mouse mammary glands. The mammary glands of homozygous MMTV-BRCA1 mice exhibit ducts with increased branching and lobulo-alveolar structures compared to nontransgenic and hemizygous controls. Homozygous MMTV-BRCA1t340 mice have mammary glands that appear similar to those of nontransgenic and hemizygous animals. The mammary glands of homozygous MMTV-BRCA1sv mice display a phenotype resembling that observed in hemizygous mice. At least 3 animals were examined for each bar.
Since the MMTV LTR is known to be upregulated during pregnancy, we evaluated mammary gland morphology by whole mounts at various time points during pregnancy. All transgenic mice (five or three at pregnancy day 3, 7, 12, 19, and five at 1 day post-partum) and non-transgenic mice (two at day 10, five at day 13, three at day 19 and four at 2 day post-partum) were evaluated by this approach. There were no significant morphological differences (data not shown) in the mammary glands among the transgenic animals when compared to nontransgenic controls evaluated during pregnancy or lactation.
Effect of Transgene Expression on DMBA Induced Mammary Tumor Incidence
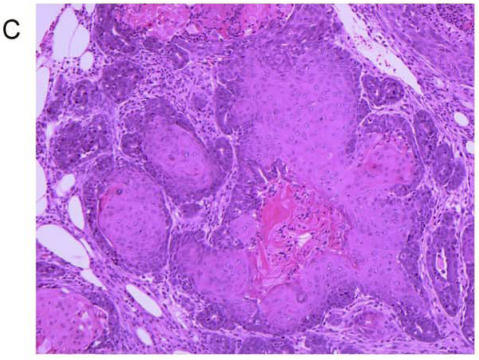
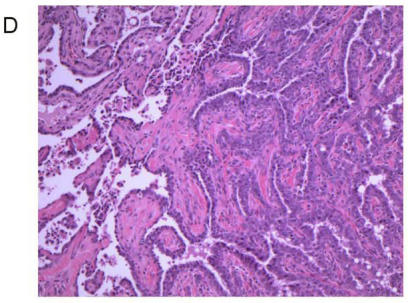
Treatment of 56-day-old mice with the chemical mutagen DMBA resulted in a high incidence of mammary tumor development (Fig. 7A). In addition to mammary tumor induction, DMBA treated mice also developed a variety of tumors in other tissue types, most notably lymphoma (Fig. 7B) as has been reported previously 20. When the mammary tumor survival data of all homozygous MMTV-BRCA1 and MMTV-BRCA1sv transgenic lines were compared against nontransgenic controls, a highly significant effect on mortality was noted (Fig. 7A, P<0.0001 for both lines versus nontransgenic controls). However, while homozygous MMTV-BRCA1 mice (lines 5 and 6) showed delayed onset against DMBA-induced mammary tumors, expression of BRCA1sv (lines 90 and 61) greatly worsened the outcome of these mice for mammary cancer. There was no significant difference in mortality due to mammary tumors between MMTV-BRCA1t340 (line 1) and nontransgenic mice (Fig. 7A). The observed effect on mortality was restricted to mammary tumors. When mortality from all cancers was pooled, there was no significant difference among any transgenic line relative to nontransgenic controls (Fig. 7B, P= 0.0715). Similar results are obtained if the MMTV-BRCA1t340 lines are omitted from the statistical analysis and only the wild-type and splice variant lines are compared against the nontransgenic controls (data not shown, mammary tumors P< 0.001; all tumor types P= 0.4199). We chose the endpoint of death or onset of palpable tumor, to insure that we captured mice that died of lymphoma and were shown to have occult mammary tumors on necropsy. The spectrum of tumors that developed was typical for DMBA carcinogen exposure. The majority of mammary tumors were adenosquamous carcinomas or adenocarcinomas as shown in Fig. 7C and 7D.
Figure 7.
Tumor specific survival and histology for MMTV-BRCA1 transgenic lines. Kaplan-Meier curves plotting tumor-free survival versus day for mice subjected to a DMBA treatment, (A) mammary tumors, (B) and all tumor types. L5 & L6 = MMTV-BRCA1 mice (n=95), NT = nontransgenic mice (n=205), L90 & L61 = MMTV-BRCA1sv mice (n=97), L1 = MMTV-BRCA1t340 mice (n=125). (C) Mammary gland adenosquamous carcinoma that developed in a DMBA-treated MMTV-BRCA1t340 female (line 2 F58). (D) Mammary gland adenocarcinoma that developed in a DMBA-treated MMTV-BRCA1 female (line 5 F185). Original magnification = 100X
4. Discussion
Wild-type Brca1 undergoes a complex pattern of regulated expression, and is expressed in cells undergoing both proliferation and differentiation such as the otic vesicle, neuroectoderm, and keratinocytes 11,13. The up-regulation of Brca1 transcripts during pregnancy is associated with ductal and glandular proliferation of the mouse mammary gland 34. In the adult male mouse, Brca1 is expressed in tissues undergoing proliferation and/or terminal differentiation such as the testis 34,35.
Previously, several independent null alleles of mouse Brca1 have been generated. Each homozygous mutant was an embryonic lethal 15-19. Homozygous Brca111 mutant embryos display abnormalities, particularly absence of mesoderm formation and gastrulation 17. Growth of homozygous mutant Brca15-6 or Brca111 blastocysts in vitro was also severely impaired 16,17. Results from the deletion of a larger portion of Brca1 exon 11 suggest that Brca1 is important for the differentiation of neuroepithelial cells during neural tube formation 15.
Selective disruption of Brca1 in mouse mammary epithelial cells results in increased apoptosis and aberrant ductal development during pregnancy, lactation, and involution 36. Transgenic mice expressing an MMTV-driven, truncated Brca1 display a delay in lactational mammary gland development 14. In particular, the mammary glands are smaller than normal and the ducts do not completely fill the fat pad. These findings suggest that Brca1 is required for normal morphogenesis of the mammary gland during pregnancy. It should be noted that we did not observe any delayed development in mice expressing a truncated human BRCA1 transcript.
In this study we generated transgenic mice that overexpress BRCA1 or BRCA1 mutants (BRCA1t340 and BRCA1sv) in the mammary gland under the control of the MMTV LTR promoter/enhancer. Homozygous, but not hemizygous, MMTV-BRCA1 transgenic virgin mice demonstrated moderately increased mammary lobulo-alveolar epithelium by 13 weeks of age. Both hemizygous and homozygous MMTV-BRCA1t340 mice had mammary ductal trees that were similar to those of control nontransgenic littermates. However, both hemi- and homozygous MMTV-BRCA1sv transgenic mice exhibited precocious secondary branching and lobular-alveolar development, particularly terminal end bud proliferation and lobule formation. Results of the analysis on the mammary gland whole mount preparations indicated that the lobulo-alveolar morphology observed in the hemizygous and homozygous MMTV-BRCA1sv mice was similar to that achieved by mid-pregnancy in nontransgenic mice. Homozygous MMTV-BRCA1 mice displayed an intermediate phenotype. As the mice aged, the differences became less prominent, and by 24 weeks of age, mammary glands were similar in appearance for all animals and involution of the glands with aging appeared normal.
The phenotype observed in MMTV-BRCA1 and MMTV-BRCA1sv transgenic mice may be caused by a number of factors or a combination of factors. Both MMTV-BRCA1 (line 6) and MMTV-BRCA1sv (line 90) mice are estimated to contain 15 copies of their respective transgenes. However, Northern analysis revealed that BRCA1 mRNA is expressed at significantly higher levels in MMTV-BRCA1sv (line 90) mice than in the MMTV-BRCA1 (line 6). This may be due to the activity of the integration site, as integration of a transgene into a region of heterochromatin, such as near the centromere or into satellite DNA can silence gene expression 37. Alternatively, the difference in mRNA expression may be due to differences in the structures of the transgene constructs. The MMTV-BRCA1 construct contains 91bp of the human BRCA1 5' untranslated region (UTR) upstream of the ATG start codon, while the MMTV-BRCA1sv construct contains 38bp of the UTR separated by an 89bp insert just prior to the beginning of wild type BRCA1 exon 2. The 89bp insert is part of BRCA1 alternative exons 1a and 1b 38,39. It is possible that the 89bp alternative exon contains motifs that affect the expression of BRCA1sv. Alternatively, the wild type BRCA1 5' UTR may contain motifs that allow for negative regulation of BRCA1 expression.
The observed delay in mortality among homozygous transgenic mice expressing wild-type BRCA1 following a DMBA treatment is consistent with a tumor-suppressor function for BRCA1 (Fig. 7A, 7B). In this report, we document for the first time, in vivo experimental evidence that overexpression of wild-type BRCA1 provides protection to the mammary gland against the development of mutagen-induced neoplasia. In contrast, transgenic mice expressing BRCA1sv experienced an accelerated mortality due to DMBA-induced mammary tumors. Unlike the BRCA1sv mice, the outcome of DMBA-induced mammary tumors in BRCA1 t340 homozygous transgenic lines was not affected (Fig. 7A). Although we cannot rule out dosage effects, the differential result between BRCA1sv and BRCA1t340 lines suggests that if a dominant-negative mechanism of action by BRCA1sv is the cause of the observed acceleration in mortality, then this function does not map to the BRCA1 RING domain. The modulation of DMBA-induced mortality by either the MMTV- BRCA1 or MMTV-BRCA1sv transgene was specific to the mammary gland, recognizing the relative tissue specificity of the MMTV promoter. While both the wild type and sv BRCA1 constructs promoted early development of the mammary glands, our results would suggest that an intact ring finger is essential for tumor suppression by BRCA1. Although MMTV driven-transgene expression was detected in the salivary gland and lung (Fig. 4), transgenic lines were not influenced in their response to DMBA in these or other tissues (Fig. 7B).
Results from this study demonstrate that MMTV-BRCA1 and MMTV-BRCA1sv transgenic mice exhibit accelerated mammary gland development. The finding is consistent with those previous studies, which indicate a role for Brca1 in promoting mammary epithelial cell proliferation and differentiation and suggests that the human and murine homologues of BRCA1 are, at least in some respects, functionally similar. Additionally, if proliferative effects occur through similar mechanisms in the case of both Brca1 and BRCA1, our observations on the effects of splice variant expression indicate that these do not require the presence of the N-terminal RING domain.
Our current observations on the effects of BRCA1 on both epithelial cell proliferation and mammary tumor formation stand in contrast to a previous report of effects of this splice variant in cultured tumor cells 22, in which an anti-proliferative effect of RING-deleted BRCA1 was observed. Growth inhibition in human cell lines mediated by overexpression of an additional N-terminal BRCA1 deletion mutant 40 has also been previously reported. This discrepancy may reflect differences in BRCA1 function in cultured neoplastic cells compared to normal epithelium in mice. This difference in effect for BRCA1 in normal cells is supported by conditional mammary gland Brca1 knockouts that exhibit a failure to develop in the absence of Brca1 36. In addition, our data suggest the possibility of dominant negative interactions between wild-type Brca1 and splice variant BRCA1 in transgenic animals that depend on intact C-terminal sequences, but not the intact RING domain. This hypothesis is supported by studies that have demonstrated that the tandem BRCT domains of BRCA1 can interact to form a globular unit 41-43 that shares structural similarity with that formed by the BRCT domains of 53BP1 44,45 and forms a phospho-protein recognition domain 46,47. In the case of another BRCT-containing protein, Crb2, these structures have been reported to form homo-oligomers 48. It is conceivable that the localization of these and other proteins to sites of DNA damage-associated nuclear foci may involve the formation of similar higher-order oligomer structures; however, such oligomerization could also provide the structural basis for dominant negative interactions. Abrogation of normal Brca1 function by the splice variant may not recapitulate the blunted morphogenesis observed early in Brca1 knockout mice because MMTV-directed transgene expression tends to become most pronounced in mammary epithelium only after puberty 49.
It is interesting that the naturally occurring mutant (t340) had essentially no effect on the transgenic phenotype, suggesting at least for this particular mutation and perhaps all truncating mutations, there is no dominant negative effect leading to breast cancer development in BRCA1 patients. Our results also suggest that alternatively spliced forms of BRCA1 may have modulating effects on BRCA1 function and should be a focus for future inquiry. Future studies with these transgenic animals may further define the functions of BRCA1 during mammary gland development and relate functions to models of breast cancer, its prevention, and its treatment. Additional studies will be aimed at confirming the observation that increased expression of wild type BRCA1 delays mammary tumor formation as it may have relevance to breast cancer prevention strategies.
Acknowledgments
We would like to thank Dr. Brigid L. M. Hogan for her critical review of this work. Dr. L. Renee Bailey from the Program in Human Genetics at Vanderbilt University for performing the statistical analysis. We would like to acknowledge Kimberly Newsom-Johnson from the Human Tissue Acquisition and Pathology Shared Resource Lab for the preparation of the histological sections. The imaging equipment used was from the Vanderbilt Cell Imaging Shared Resource Lab.
References
- 1.Ford D, Easton DF, Peto J. Estimates of the gene frequency of BRCA1 and its contributions to breast and ovarian cancer incidence. American Journal of Human Genetics. 1995;57:1457–1462. [PMC free article] [PubMed] [Google Scholar]
- 2.Claus EB, Schildkraut JM, Thompson WD, Risch NJ. The genetic attributable risk of breast and ovarian cancer. Cancer. 1996;77:2318–2324. doi: 10.1002/(SICI)1097-0142(19960601)77:11<2318::AID-CNCR21>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 3.Easton DF, Bishop DT, Ford D, Crockford GP. Genetic linkage analysis in familial breast and ovarian cancer: results from 214 families: The breast cancer linkage consortium. American Journal of Human Genetics. 1993;52:678–701. [PMC free article] [PubMed] [Google Scholar]
- 4.Castilla LH, Couch FJ, Erdos MR et al. Mutations in the BRCA1 gene in families with early-onset breast and ovarian cancer. Nature Genetics. 1994;8:387–391. doi: 10.1038/ng1294-387. [DOI] [PubMed] [Google Scholar]
- 5.Miki Y, Swensen J, Shattuck C et al. A strong candidate for the breast and ovarian cancer susceptibilty gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 6.Thakur S, Zhang HB, Peng Y et al. Localization of BRCA1 and a splice variant identifies the nuclear localization signal. Molecular and Cellular Biology. 1997;17:444–452. doi: 10.1128/mcb.17.1.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng CX, Wang RH. Roles of BRCA1 in DNA damage repair: a link between development and cancer. Human Molecular Genetics. 2003;12:R113–R123. doi: 10.1093/hmg/ddg082. [DOI] [PubMed] [Google Scholar]
- 8.Scully R, Livingston DM. In search of the tumour-suppressor functions of BRCA1 and BRCA2. Nature. 2000;408:429–432. doi: 10.1038/35044000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Venkitaraman AR. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell. 2002;108:171–182. doi: 10.1016/s0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- 10.Abel KJ, Xu J, Yin GY, Lyons RH, Meisler MH, Weber BL. Mouse Brca1: localization, sequence analysis, and identification of evolutionarily conserved domains. Human Molecular Genetics. 1995;4:2265–2273. doi: 10.1093/hmg/4.12.2265. [DOI] [PubMed] [Google Scholar]
- 11.Marquis ST, Rajan JV, Wynshaw-Boris A et al. The developmental pattern of Brca1 expression implies a role in differentiation of the breast and other tissues. Nature Genetics. 1995;11:17–26. doi: 10.1038/ng0995-17. [DOI] [PubMed] [Google Scholar]
- 12.Phillips KW, Goldsworthy SM, Bennett LM, Brownlee HA, Wiseman RW, Davis BJ. Brca1 is expressed independently of hormonal stimulation in the mouse ovary. Laboratory Investigation. 1997;76:419–425. [PubMed] [Google Scholar]
- 13.Lane TF, Deng CX, Elson A, Lyu MS, Kozak CA, Leder P. Expression of Brca1 is associated with terminal differentiation of ectodermally and mesodermally derived tissues in mice. Genes and Development. 1995;9:2712–2722. doi: 10.1101/gad.9.21.2712. [DOI] [PubMed] [Google Scholar]
- 14.Brown MA, Nicolai H, Howe K et al. Expression of a truncated Brca1 protein delays lactational mammary development in transgenic mice. Transgenic Research. 2002;11:467–478. doi: 10.1023/a:1020348025139. [DOI] [PubMed] [Google Scholar]
- 15.Gowen LC, Johnson BL, Latour AM, Sulik KK, Koller BH. Brca1 deficiency results in early embryonic lethality characterized by neuroepithelial abnormailities. Nature Genetics. 1996;12:191–194. doi: 10.1038/ng0296-191. [DOI] [PubMed] [Google Scholar]
- 16.Hakem R, de la Pompa JL, Sirard C et al. The tumor suppressor gene Brca1 is required for embryonic cellular proliferation in the mouse. Cell. 1996;85:1009–1023. doi: 10.1016/s0092-8674(00)81302-1. [DOI] [PubMed] [Google Scholar]
- 17.Liu CY, Flesken-Nikitin A, Li S, Zeng YX, Lee WH. Inactivation of the mouse Brca1 gene leads to failure in the morphogenesis of the egg cylinder in early implantation development. Genes and Development. 1996;10:1835–1843. doi: 10.1101/gad.10.14.1835. [DOI] [PubMed] [Google Scholar]
- 18.Ludwig T, Chapman DL, Papaioannou VE, Efstratiadis A. Targeted mutations of breast cancer susceptibility gene homologs in mice: lethal phenotypes of Brca1, Brca2, Brca1/Brca2, Brca1/p53, and Brca2/p53 nullizygous embryos. Genes and Development. 1997;11:1226–1241. doi: 10.1101/gad.11.10.1226. [DOI] [PubMed] [Google Scholar]
- 19.Shen SX, Weaver Z, Xu X et al. A targeted disruption of the murine Brca1 gene causes gamma-irradiation hypersensitivity and genetic instability. Oncogene. 1998;17:3115–3124. doi: 10.1038/sj.onc.1202243. [DOI] [PubMed] [Google Scholar]
- 20.Coffey RJ, Meise KS, Matsui Y, Hogan BLM, Dempsey PJ, Halter SA. Acceleration of mammary neoplasia in transforming growth factor alpha transgenic mice by 7,12 dimethylbenzanthracene. Cancer Research. 1994;54:1678–1683. [PubMed] [Google Scholar]
- 21.Matsui Y, Halter SA, Holt JT, Hogan BLM, Coffey RJ. Developmental mammary hyperplasia and neoplasia in MMTV-TGF alpha transgenic mice. Cell. 1990;61:1147–1155. doi: 10.1016/0092-8674(90)90077-r. [DOI] [PubMed] [Google Scholar]
- 22.Holt JT, Thompson ME, Szabo C et al. Growth retardation and tumor inhibition by BRCA1. Nature Genetics. 1996;12:298–302. doi: 10.1038/ng0396-298. [DOI] [PubMed] [Google Scholar]
- 23.Tait DL, Obermiller PS, Redlin-Frazier S et al. A phase I trial of retroviral BRCA1sv gene therapy in ovarian cancer. Clinical Cancer research. 1997;11:1959–1968. [PubMed] [Google Scholar]
- 24.Pierce DF, Gorska AE, Chytil A et al. Mammary tumor suppression by transforming growth factor beta transgene expression. Proceedings of the National Academy of Sciences USA. 1995;92:4254–4258. doi: 10.1073/pnas.92.10.4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rudolph-Owens LA, Cannon P, Matrisian LM. Overexpression of the matrix metalloproteinase matrilysin results in premature mammary gland differentiation and male infertility. Molecular Biology of the Cell. 1998;9:421–435. doi: 10.1091/mbc.9.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Witty JP, Wright JH, Matrisian LM. Matrix metalloproteinases are expressed during ductal and alveolar mammary morphogenesis and misregulation of stromelysin-1 in transgenic induces unscheduled alveolar development. Molecular Biology of the Cell. 1995;6:1287–1303. doi: 10.1091/mbc.6.10.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pierce DF, Johnson MD, Matsui Y et al. Inhibition of mammary duct development but not alveolar outgrowth during pregnancy in transgenic mice expressing active TGF-beta 1. Genes and Development. 1993;7:2308–2317. doi: 10.1101/gad.7.12a.2308. [DOI] [PubMed] [Google Scholar]
- 28.Rugh R. The mouse: Its reproduction and development. Minneapolis, MN: Burgess Pub Co; 1968. [Google Scholar]
- 29.Chomczynski P, Sacchi N. Single-step method of RNA isolation by a guanidinium thiocyanate-phenol-chloroform extraction. Analytical Chemistry. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 30.Chomczynski P. A reagent for the single-step simultaneous isolation of RNA, DNA, and proteins from cell and tissue samples. Biotechniques. 1993;15:532–537. [PubMed] [Google Scholar]
- 31.Danielson PE, Forss-Petter S, Brow MA et al. p1B15: a cDNA clone of the rat mRNA encoding cyclophilin. DNA. 1988;7:261–267. doi: 10.1089/dna.1988.7.261. [DOI] [PubMed] [Google Scholar]
- 32.Lehr HA, Mankoff DA, Corwin D, Santeusanio G, Gown AM. Application of Photoshop-based image analysis to quantification of hormone receptor expression in breast cancer. Journal of Histochemistry and Cytochemistry. 1997;45:1559–1565. doi: 10.1177/002215549704501112. [DOI] [PubMed] [Google Scholar]
- 33.Lehr HA, van der Loos CM, Teeling P, Gown AM. Complete chromogen separation and analysis in double immunohistochemical stains using Photoshop-based image analysis. Journal of Histochemistry and Cytochemistry. 1999;47:119–126. doi: 10.1177/002215549904700113. [DOI] [PubMed] [Google Scholar]
- 34.Blackshear PE, Goldsworthy SM, Foley JF et al. Brca1 and Brca2 expression patterns in mitotic and meiotic cells of mice. Oncogene. 1998;16:61–68. doi: 10.1038/sj.onc.1201506. [DOI] [PubMed] [Google Scholar]
- 35.Zabludoff SD, Wright WW, Harshman K, Wold BJ. BRCA1 mRNA is expressed highly during meiosis and spermiogenesis but not during mitosis of male germ cells. Oncogene. 1996;13:649–653. [PubMed] [Google Scholar]
- 36.Xu X, Wagner KU, Larson D et al. Conditional mutation of Brca1 in mammary epithelial cells results in blunted ductal morphogenesis and tumour formation. Nature Genetics. 1999;22:37–43. doi: 10.1038/8743. [DOI] [PubMed] [Google Scholar]
- 37.Garrick D, Fiering S, Martin DI, Whitelaw E. Repeat-induced gene silencing in mammals. Nature Genetics. 1998;18:56–59. doi: 10.1038/ng0198-56. [DOI] [PubMed] [Google Scholar]
- 38.Xu CF, Brown MA, Chambers JA, Griffiths BL, Nicolai H, Solomon E. Distinct transcriptional start sites generate two forms of BRCA1 mRNA. Human Molecular Genetics. 1995;4:2259–2264. doi: 10.1093/hmg/4.12.2259. [DOI] [PubMed] [Google Scholar]
- 39.Brown MA, Xu CF, Nicolai H et al. The 5' end of the BRCA1 gene lies within a duplicated region of human chromosome 17. Oncogene. 1996;12:2507–2513. [PubMed] [Google Scholar]
- 40.You F, Chiba N, Ishioka C, Parvin JD. Expression of an amino-terminal BRCA1 deletion mutant causes a dominant growth inhibition in MCF10A cells. Oncogene. 2004;23:5792–5798. doi: 10.1038/sj.onc.1207739. [DOI] [PubMed] [Google Scholar]
- 41.Derbyshire DJ, Basu BP, Serpell LC et al. Crystal Structure of human 53BP1 BRCT domains bound to p53 tumour suppressor. EMBO Journal. 2002;21:3863–3872. doi: 10.1093/emboj/cdf383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Joo WS, Jeffrey PD, Cantor SB, Finnin MS, Livingston DM, Pavletich NP. Structure of the 53BP1 BRCT region bound to p53 and its comparison to the Brca1 BRCT structure. Genes and Development. 2002;16:583–593. doi: 10.1101/gad.959202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Williams RS, Green R, Glover JN. Crystal structure of the BRCT repeat region from the breast cancer-associated protein BRCA1. Nature Structural Biology. 2001;8:838–842. doi: 10.1038/nsb1001-838. [DOI] [PubMed] [Google Scholar]
- 44.Wilson J, Wilson S, Warr N, Watts FZ. Isolation and characterization of the Schizosaccharomyces pombe rhp9 gene: a gene required for the DNA damage checkpoint but not the replication checkpoint. Nucleic Acids Research. 1997;25:2138–2146. doi: 10.1093/nar/25.11.2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saka Y, Esashi F, Matsusaka T, Mochida S, Yanagida M. Damage and replication checkpoint control in fission yeast is ensured by interactions of Crb2, a protein with BRCT motif, with Cut5 and Chk1. Genes and Development. 1997;11:3387–4000. doi: 10.1101/gad.11.24.3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu X, Chini CC, He M, Mer G, Chen J. The BRCT domain is a phospho-peptide binding domain. Science. 2003;302:639–642. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- 47.Manke IA, Lowery DM, Nguyen A, Yaffe MB. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science. 2003;302:636–639. doi: 10.1126/science.1088877. [DOI] [PubMed] [Google Scholar]
- 48.Du LL, Moser BA, Russell P. Homo-oligomerization is the essential function of the tandem BRCT domains in the checkpoint protein Crb2. Journal of Biological Chemistry. 2004;279:38409–38414. doi: 10.1074/jbc.M403326200. [DOI] [PubMed] [Google Scholar]
- 49.Munarini N, Jager R, Abderhalden S et al. Altered mammary epithelial development, pattern formation and involution in transgenic mice expressing the EphB4 receptor tyrosine kinase. Journal of Cell Science. 2002;115:25–37. doi: 10.1242/jcs.115.1.25. [DOI] [PubMed] [Google Scholar]