Abstract
The Drosophila immune system discriminates between various types of infections and activates appropriate signal transduction pathways to combat the invading microorganisms. The Toll pathway is required for the host response against fungal and most Gram-positive bacterial infections. The sensing of Gram-positive bacteria is mediated by the Pattern Recognition Receptors PGRP-SA and GNBP1 that cooperate to detect the presence of infections in the host. Here, we report that GNBP3 is a novel Pattern Recognition Receptor that is required for the detection of fungal cell wall components. Strikingly, we find that there is a second, parallel, pathway acting jointly with GNBP3. The Drosophila Persephone protease activates the Toll pathway when proteolytically matured by the secreted fungal virulence factor PR1. Thus, the detection of fungal infections in Drosophila relies both on the recognition of invariant microbial patterns and on monitoring the effects of virulence factors on the host.
Introduction
Fungi represent a threat to insects in the wild with more than 700 entomopathogenic species described (Roberts and Humber, 1981). Insects must have evolved responses to handle these infections. We have addressed here the genetically amenable fruitfly D. melanogaster to decipher the mechanisms that stimulate immune responses to fungal infections.
This host response includes both cellular and humoral arms. The analysis of the humoral immune response within the framework of a septic injury model has led to the current paradigm in which two distinct intracellular transduction pathways, Immune deficiency (IMD) and Toll, regulate the transcription of hundreds of genes by controlling the nuclear uptake of the NF-κB transcription factors Relish and Dorsal-related Immunity Factor (DIF), respectively (reviewed in Hoffmann, 2003, and references therein). The classical effector molecules of the systemic humoral response, the antimicrobial peptides, are synthesized in the fat body, a functional analogue of the mammalian liver, and are released into the hemolymph where they kill invading microorganisms. One of these peptides, Drosomycin, exhibits fungicidal activities at micromolar concentrations and is active mainly on filamentous fungi (Fehlbaum et al., 1995; Tzou et al., 2002). Others such as Cecropins, Attacins, Drosocin, and Diptericin are active mostly on Gram-negative bacteria whereas Defensin is effective against Gram-positive bacteria.
The IMD pathway is required for the host response against Gram-negative bacteria. Mutants in this pathway fail to express antibacterial peptides and are highly sensitive to such infections, yet resist fungal and Gram-positive bacterial infections as well as wild-type flies. Toll is the receptor of the second intracellular transduction pathway and is activated by the binding of a proteolytically cleaved form of the Spätzle (SPZ) cytokine. Toll pathway mutants are susceptible to infections by the entomopathogenic fungi B. bassiana and M. anisopliae or by the opportunistic pathogen Aspergillus fumigatus (Lemaitre et al., 1996; Lemaitre et al., 1997; Ligoxygakis et al., 2002; Rutschmann et al., 2000a; Tauszig-Delamasure et al., 2002). Toll is also required to resist some Gram-positive bacterial infections (Gobert et al., 2003; Michel et al., 2001; Rutschmann et al., 2002).
The Drosophila immune response is adapted to the nature of the invading microorganism (Lemaitre et al., 1997). The Toll pathway is induced by fungi and Gram-positive bacteria, whereas the IMD pathway is predominantly triggered upon Gram-negative bacterial challenges (Lemaitre et al., 1997; Rutschmann et al., 2000a). These observations imply that several receptors mediate the discrimination between various types of microbial infections. Indeed, members of the peptidoglycan recognition protein (PGRP) family have been shown to be required for these distinct events (reviewed in Ferrandon et al., 2004; Kaneko and Silverman, 2005). PGRP-LC, a receptor of the IMD pathway, can be activated by meso-diaminopimelic acid peptidoglycan (PGN), a compound characteristic of the cell wall of Gram-negative bacteria and of Gram-positive bacilli. PGRP-LE, a secreted member of the PGRP family, is also involved in sensing Gram-negative bacteria (Kaneko et al., 2006). In contrast, the circulating PGRP-SA receptor activates the Toll pathway upon detection of Lysine-type PGN which is a major component of the cell wall of many Gram-positive bacterial strains. The Gram-Negative Binding Protein 1 (GNBP1) associates with PGRP-SA and this complex is both necessary and sufficient to activate the Toll pathway upon Gram-positive challenge (Gobert et al., 2003). The circulating PGRP-SA/GNBP1 complex activates a downstream proteolytic cascade that leads to the cleavage of the Spätzle cytokine, which then activates the Toll transmembrane receptor (Jang et al., 2006; Kambris et al., 2006). Thus, PGRP-SA and GNBP1 define a Gram-positive-specific branch of Toll receptor activation. PGRP-SD also belongs to this branch and is required for the detection of other Gram-positive bacterial strains (Bischoff et al., 2004).
Here, we address the existence of a second branch devoted to the detection of fungal infections, which also activates Toll. Indeed, mutants for the persephone (psh) gene, which encodes a clip-prodomain containing protease, are characterized by an increased sensitivity to natural infections with the entomopathogenic fungus B. bassiana, whereas they are resistant to bacterial infections (Ligoxygakis et al., 2002). The psh mutations had been originally isolated as suppressors of the necrotic (nec) phenotype. nec encodes a serine protease inhibitor of the serpin family, the absence of which leads to the constitutive, psh-dependent, activation of the Toll pathway (Levashina et al., 1999; Ligoxygakis et al., 2002). Thus, psh and nec define a fungal-specific branch of Toll receptor activation. By analogy to the Gram-positive branch, it is expected that an as yet unidentified immune receptor detects fungal infections and activates in turn the psh-dependent proteolytic cascade.
GNBP1 belongs to the family of GNBP/β-Glucan Recognition Proteins (βGRP) (Kim et al., 2000). Members of this family have been reported to bind to β-(1,3)-glucan, a major component of the fungal cell wall (Lee et al., 2006; Ma and Kanost, 2000; Ochiai and Ashida, 2000). In Drosophila, three members of this family, GNBP1 to 3, have been described (Kim et al., 2000). Among these, GNBP3 shows the greatest degree of similarity to lepidopteran β-(1,3)-glucan recognition proteins and was therefore a good candidate for a fungal-specific sensor. Here, we report that GNBP3 is indeed required for Toll pathway activation in response to fungal infections. Strikingly, we also find that psh is required in a distinct yet complementary detection pathway that can be activated by fungal virulence factors.
Results
In this report, we have investigated the antifungal response of Drosophila by using two distinct models : the human opportunistic pathogenic yeast Candida albicans and the entomopathogenic fungi B. bassiana and M. anisopliae. We have monitored the immune response to these infections by two read-outs : kinetics of survival and expression levels of the Toll-dependent Drosomycin gene.
hades, a mutation of the GNBP3 locus, affects the host defense against Candida albicans
We challenged flies by pricking with a needle dipped into a concentrated C. albicans solution and monitored their survival. Whereas wild-type flies were resistant to this infection, survival of the Toll pathway mutants spz and Dif was compromised, indicating that the Toll pathway is required for defense against this opportunistic yeast (Fig. 1A) (Alarco et al., 2004). We next created a null mutation in the GNBP3 gene, which we named hades (for the generation of the mutation, see below) and observed that GNBP3hades mutants were as sensitive to C. albicans infection as Dif mutants (Fig. 1A). In contrast, GNBP1osi mutant flies, which are deficient for the activation branch of the Toll pathway by Gram-positive bacteria, survived C. albicans infection as well as wild-type flies. Flies mutant for the IMD pathway gene kenny (key) (Rutschmann et al., 2000b) also resisted this challenge (Fig. 1A), indicating that the IMD pathway is not required for the host defense against opportunistic yeast infections. The finding that both GNBP3hades and Toll pathway mutants succumb to C. albicans infection suggests that this sensitivity to yeast infection reflects a lack of activation of the Toll pathway. We next challenged wild-type, spz, and GNBP3hades mutant flies with heat-killed C. albicans (which cannot secrete virulence factors, see below). We found that Drosomycin was strongly induced in wild-type flies and that this inducibility was markedly decreased in GNBP3hades and spz mutants (Fig. 1D).
Figure 1 . GNBP3 is required in the host defense against yeast infections.
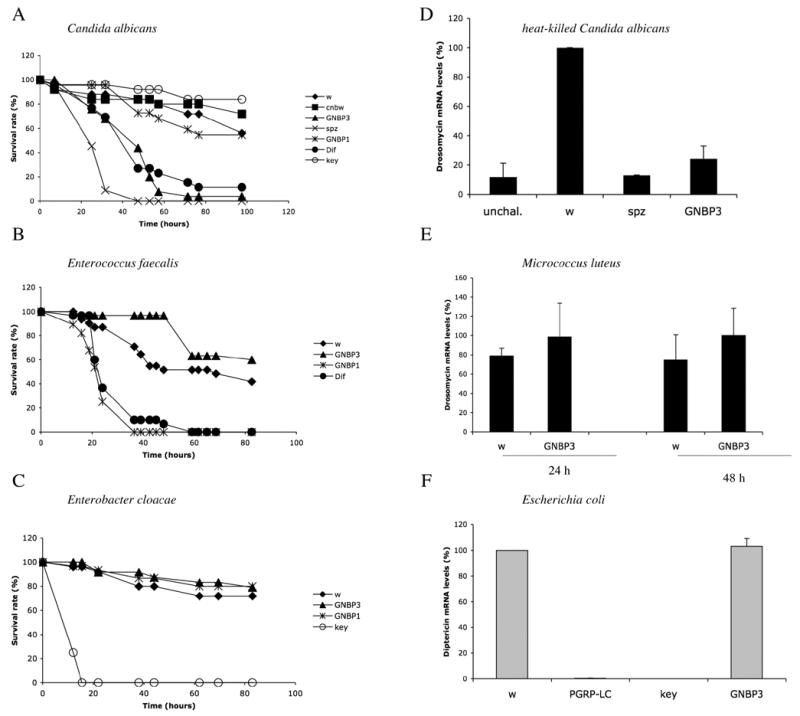
A–C : survival experiments were performed at 29 °C (C. albicans : A) or 25°C (E. faecalis : B, E. cloacae : C). The survival rate expressed in percentage is shown. w : white A5001 and cn bw flies were used as wild-type controls. D–F : expression of antimicrobial peptide genes determined by real-time PCR. Results are expressed as a percentage of the induction observed in w control flies. Drosomycin expression was measured 24 hours after a challenge with heat-killed C. albicans (D; unchal. : unchallenged control flies). Drosomycin RNA levels were monitored 24 and 48 hours after a challenge with M. luteus (E). Diptericin inducibility was checked 6 hours after a septic wound with a needle dipped into a concentrated E. coli solution. GNBP1, Dif, spz (spätzle) are mutants of the Toll pathway whereas key (kenny) and PGRP-LC are mutants of the IMD pathway.
We also tested resistance of GNBP3hades mutants to Gram-positive and Gram-negative bacterial infections and observed no susceptibility as compared to wild-type flies (Fig. 1B,C). Furthermore, the induction levels of the Drosomycin gene by Gram-positive bacteria (Fig. 1E) or those of the Diptericin gene by Gram-negative bacteria (Fig. 1F) were similar in GNBP3hades and wild-type flies. We conclude that GNBP3 is involved in the Drosophila host defense against C. albicans and not that against bacterial infections.
Rescue of the GNBP3hades mutation
We had generated the hades mutant of GNBP3 mentioned above by remobilizing a P-element transposon (d01793) located 1044 base pairs upstream of the start codon ATG of GNBP3 (Thibault et al., 2004). This resulted in an imprecise excision, a 1476 base pair deletion that removes the promoter of the gene as well as the N-terminal region of the corresponding predicted protein up to residue 144 (Fig. 2A). As expected, the GNBP3 protein was no longer detected in the mutant with a specific antibody (Matskevich et al., unpublished). We rescued the GNBP3hades mutant phenotype by expressing in this background a wild-type GNBP3 cDNA under the control of the ubiquitous heat-shock protein 70 (hsp) promoter. The low-level expression of the transgene driven by the basal activity of the hsp promoter using the UAS-GAL4 system was sufficient to restore the inducibility of Drosomycin in response to a challenge with heat-killed C. albicans (Fig. 2B, Supplementary Experimental Procedures), confirming that the effect of the mutation is due to the disruption of the GNBP3 locus.
Figure 2 . the hades mutation affects the GNBP3 locus that encodes a glucan recognition protein.
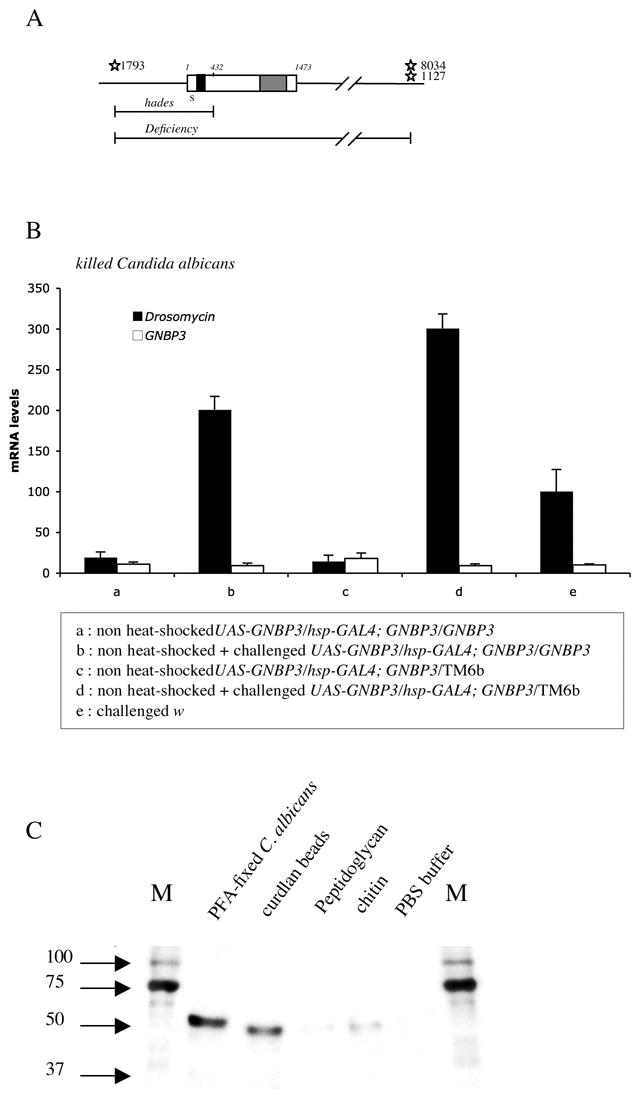
A : scheme of the GNBP3 locus. The GNBP3 gene does not contain any intron. The upper line represents the genomic organization. The open bar shows the structure of the GNBP3 gene (nucleotides 1–1473) that encodes an N-terminal β-(1,3)-glucan binding domain (black domain) and a C-terminal β-glucanase homology region (grey domain). The signal peptide (S) is indicated. The sites of insertions of modified P-elements present in the strains d01793, d08034, and d01127 are shown (stars). The extent of the hades deficiency (up to nucleotide 432 of GNBP3) as well as that of a large deficiency are depicted (lower lines). The latter deficiency was used in genetic tests to check that GNBP3hades is a null mutation.
B : rescue experiment of the GNBP3 mutant phenotype by a UAS-GNBP3 transgene after a challenge with heat-killed C. albicans. The Drosomycin RNA steady-state levels (black bars) were measured in flies of the indicated genotypes. White bars : level of expression of GNBP3 placed under the control of the hsp promoter using the UAS GAL4 system. The flies were not heat-shocked so as to avoid the constitutive activation of the Toll pathway by overexpressed GNBP3 (see Fig. 3A). TM6b is a balancer of the third chromosome; heterozygous GNBP3/TM6b sibling flies are used as controls.
C : Recombinant His-tagged GNBP3 protein was incubated with several insoluble oligosaccharides found in the cell walls of various microorganisms. After centrifugation and washing steps, the proteins associated with the precipitated glycan chains were recovered in Laemmli buffer and analyzed by Western blotting with a poly-His-specific antibody. GNBP3 binds to paraformaldehyde-fixed (PFA) C. albicans blastospores or curdlan beads (long chains of β-(1,3)-glucan) and hardly to peptidoglycan from the Gram-positive bacterium Staphylococcus aureus, or to chitin. PBS : phosphate buffer alone, M : marker (kDa).
Recombinant GNBP3 binds to β-(1,3)-glucan and to polysaccharides of the fungal cell wall
The N-terminal domain of lepidopteran βGRPs, which binds directly to β-(1,3)-glucan (Ma and Kanost, 2000; Ochiai and Ashida, 2000), displays around 65% of sequence identity to GNBP3. We expressed tagged GNBP3 in bacteria and incubated the recombinant protein with curdlan, an insoluble polymer of β-(1,3)-glucan, with killed C. albicans, and with several polymeric glycan chains. The insoluble compounds were recovered by centrifugation (Kim et al., 2000), and assessed by SDS-PAGE and Western blot analysis for the presence of GNBP3 in the pellet. We detected an association of GNBP3 with curdlan and C. albicans (Fig 2C) (Lee et al., 2006). A weaker signal was detected with chitin, a polymer of N-acetyl-D-glucosamine, whereas little GNBP3 was recovered after incubation with peptidoglycan. In addition, the injection of curdlan beads into flies elicited a variable induction of Drosomycin that was not observed with injected cellulose, which is a β-(1,4)-glucan polymer (data not shown). Because curdlan beads form aggregates that are difficult to inject quantitatively into flies, we injected instead the alkali-insoluble fraction of the A. fumigatus cell wall, which consists of polysaccharides including β-(1,3)-glucans (Fontaine et al., 2000). This fraction induced reproducibly a GNBP3-dependent Drosomycin expression (Fig. S1). We conclude that GNBP3 encodes a fungal polysaccharide-binding protein.
GNBP3 acts upstream of the Toll receptor
To provide a demonstration that GNBP3 induces Drosomycin expression through the Toll pathway, we overexpressed this gene in a wild-type and a spz mutant background. As illustrated in Fig. 3A, GNBP3 overexpression induced the challenge-independent transcription of Drosomycin, which was abolished in spz mutant flies.
Figure 3 . Fungal detection can be mediated by secreted GNBP3, which functions upstream of the Toll ligand Spätzle.
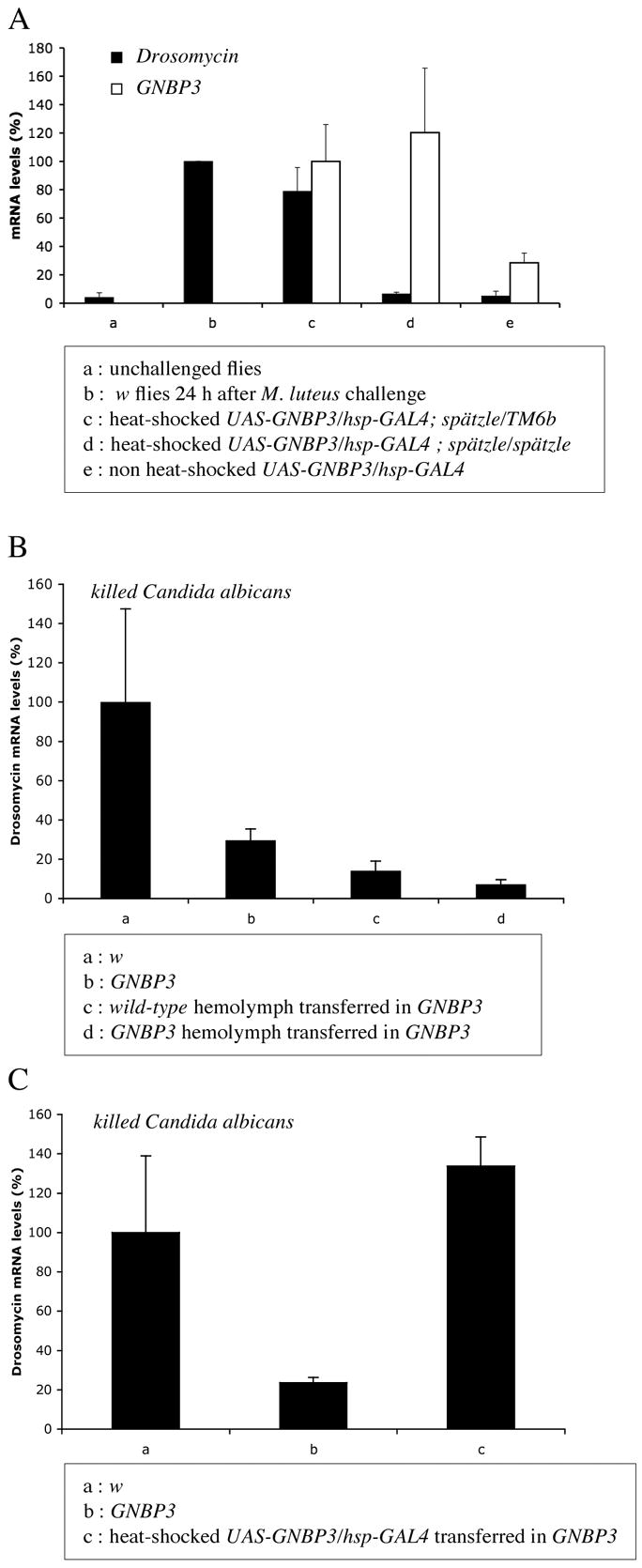
Drosomycin transcript levels as measured by quantitative RT-PCR are shown as black bars.
A : Epistatic analysis of the relationship between GNBP3 and spz. The expression of Drosomycin induced by GNBP3 overexpression under hsp promoter control is blocked in a spz mutant background. White bars represent GNBP3 mRNA levels as measured by quantitative RT-PCR.
B : Drosomycin induction by the injection of heat-killed C. albicans is not restored by the transfer of wild-type hemolymph into GNBP3hades mutant flies.
C : The transfer of hemolymph from flies overexpressing GNBP3 leads to a restored inducibility of Drosomycin expression upon the same challenge as in (B).
The Drosophila infection-sensing proteins PGRP-SA and GNBP1 were recently shown to function in circulation. To probe whether this is also valid for GNBP3, we transferred hemolymph from wild-type flies into GNBP3hades mutants. In the recipient flies, heat-killed C. albicans failed to induce Drosomycin expression (Fig. 3B). However, when flies that overexpressed GNBP3 were chosen as hemolymph donors, Drosomycin inducibility by heat-killed C. albicans was restored (Fig. 3C). Control unchallenged recipient flies did not express Drosomycin (data not shown). These data suggest that a circulating form of GNBP3 can detect infection by C. albicans.
Entomopathogenic fungi activate the Toll pathway independently from GNBP3
Entomopathogenic fungi invariably induce lethality in experimental flies, whether the challenge is achieved by injection of spores or by natural infection (deposition of spores on the cuticle). Flies mutant for genes of the Toll pathway succumb significantly earlier to such infections (Fig. 4A) and exhibit a strongly decreased inducibility of Toll pathway-dependent genes such as Drosomycin, which is no longer expressed in these mutants (Lemaitre et al., 1997) (Fig. 4C). GNBP3hades mutant flies succumbed to a natural B. bassiana infection at the same rate as Dif flies (Fig. 4A). Hemizygous and homozygous GNBP3hades flies displayed the same behavior in this assay, thus indicating that the mutation is genetically null. If GNBP3 were the sole sensor of fungal infection, one would expect Drosomycin expression to be markedly reduced in GNBP3hades mutant flies challenged with pathogenic fungi. To test this hypothesis, we carried out the following experiments : (i) we performed a B. bassiana natural infection on GNBP3hades flies and observed a strong and persistent expression of Drosomycin (Fig. 4B–C); (ii) we compared the levels of expression of Drosomycin induced by the injection of live B. bassiana spores into wild-type or GNBP3hades flies and noted a similarly strong induction of this gene in both types of flies (data not shown); (iii) however, when we injected alkali-treated spores, we detected the expression of Drosomycin in wild-type, but not in GNBP3hades mutant flies (Fig. 4D). Hardly any Drosomycin expression was detected in Dif or spz mutant flies infected with spores, whether dead or alive (Fig. 4C–D). Thus, these two types of challenge induce the transcription of Drosomycin through the intracellular part of the Toll pathway. The fact that Drosomycin induction is blocked by the GNBP3hades mutation only when dead spores are injected suggests that live spores can activate the Toll pathway by an extracellular pathway independent of GNBP3. A candidate for a gene belonging to this second pathway is psh.
Figure 4 . Distinct phenotypes of psh and GNBP3hades in response to fungal challenges.
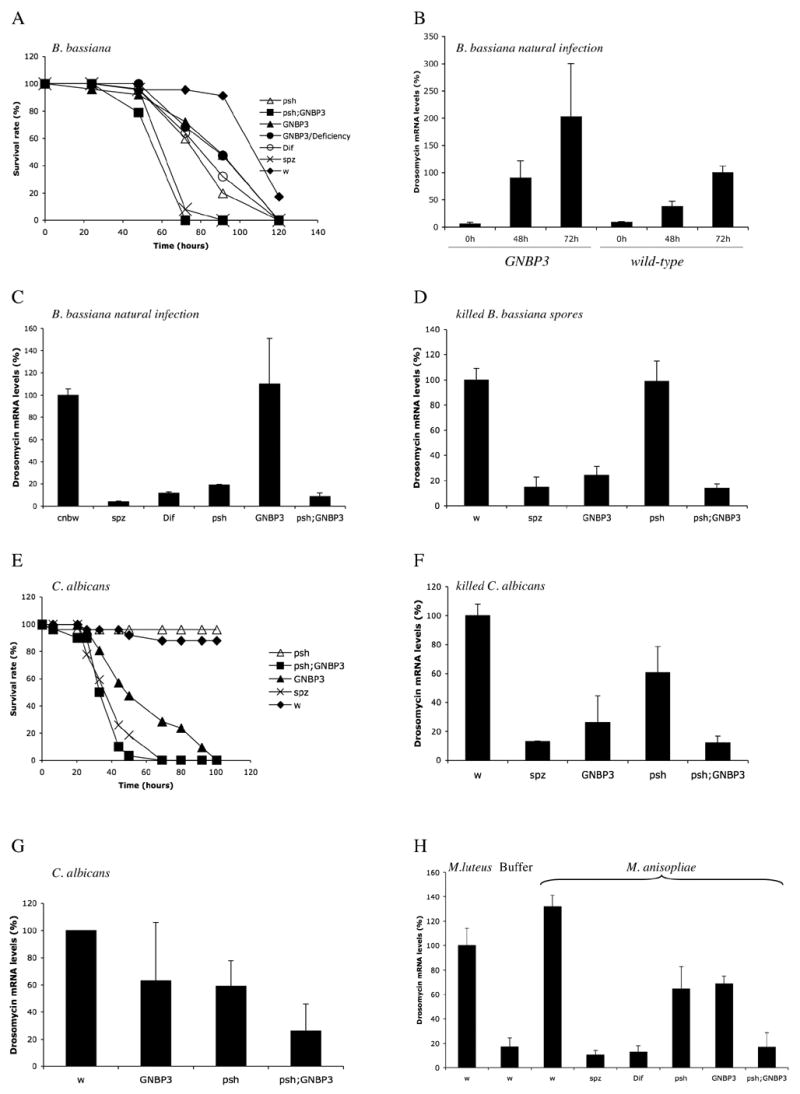
A : Survival of psh and GNBP3hades flies to natural B. bassiana infections. The genotypes of the infected flies are indicated.
B : Expression of Drosomycin as determined by quantitative RT-PCR after a natural fungal infection at 29°C with B. bassiana in wild-type and GNBP3hades flies. The expression was measured 0, 48, and 72 hours after coating the flies with fungal spores. The difference between wild-type and mutant flies at 72 hours is not statistically significant.
C : The expression of Drosomycin induced by a natural B. bassiana infection (48 hours) is blocked in psh and other mutants of the Toll pathway, but not in GNBP3hades mutants. Similar results were obtained when B. bassiana spores were injected.
D : The expression of Drosomycin induced by the injection of 9.2 nl of killed alkali-treated B. bassiana spores (48 hours) is blocked in GNBP3hades and spz mutants, but not in psh mutants.
E : Survival of psh and GNBP3hades flies to infections with C. albicans : psh is not required in the host defense against this infection.
F : psh does not have a major effect on Drosomycin inducibility by heat-killed C. albicans. Similar results were obtained with paraformaldehyde-treated C. albicans.
G : The expression of Drosomycin induced by living C. albicans yeasts is decreased significantly only in psh ; GNBP3hades double-mutant flies. The intermediate reduction of Drosomycin expression in GNBP3hades mutants in response to live C. albicans may be due to Candida proteases, which, like entomopathogenic virulence factors, may trigger partially the PSH pathway and thus bypass the GNBP3 pathway.
H : psh and GNBP3hades mutations do not block the induction of Drosomycin expression by injected M. anisopliae spores (24 hours). M. l : flies infected with M. luteus were taken as a 100% reference. Similar results were obtained in natural infections.
Persephone is required only for the host defense against live B. bassiana
The PSH protease is required for activation of the Toll pathway by the entomopathogenic fungus B. bassiana (Ligoxygakis et al., 2002) (Fig. 4A, C). In one model, psh would be required downstream of a PRR that senses fungal microbial patterns such as β-(1,3) glucan. If it were to act uniquely downstream of GNBP3, then GNBP3 and psh would be expected to share the same phenotype. In a second model, psh would act in the GNBP3-independent pathway that activates Toll after challenge with live fungi and would therefore have a phenotype distinct from that of GNBP3. In the following, we compare the psh and GNBP3hades mutant phenotypes in this light.
We first analyzed the survival rates of psh and GNBP3hades mutant flies after natural infections with B. bassiana and observed that the two mutants died with similar kinetics (Fig. 4A). Interestingly, the double-mutant flies succumbed earlier to the infection than the respective single mutants. These data suggest that GNBP3 and psh act in two complementary pathways upstream of Toll in the detection of fungal infections. We next determined whether psh is required for Drosomycin inducibility by dead fungal spores. In psh mutant flies, the injection of killed fungal spores still induced the expression of Drosomycin (Fig. 4D), in contrast to natural infections with live B. bassiana where Drosomycin inducibility was markedly reduced (Ligoxygakis et al., 2002) (Fig. 4C). Thus, live and dead fungi have distinct effects on the inducibility of the Drosomycin gene in psh and GNBP3 mutants.
Earlier studies on the psh mutant phenotype had been limited to the analysis of the response to the entomopathogenic fungus B. bassiana (Ligoxygakis et al., 2002). We therefore asked whether psh is also required for the host defense against opportunistic yeast infections. In contrast to GNBP3hades flies, we observed that psh mutants are as resistant as wild-type to C. albicans infections (Fig. 4E). These data indicate that psh and GNBP3 are indeed involved in distinct branches of Toll pathway activation. Furthermore, the level of expression of Drosomycin induced by dead C. albicans was not as strongly reduced in psh mutants as it was in GNBP3hades single mutant and in the psh ; GNBP3hades double mutant (Fig. 4F). We cannot exclude however a limited role of psh in the defense against C. albicans since psh ; GNBP3hades double mutant flies are more sensitive than the respective single mutants (Fig. 4E). In addition, only the double-mutant combination strongly blocks Drosomycin expression in response to a challenge with living C. albicans yeasts (Fig. 4G).
Taken together, our data demonstrate that GNBP3 and PSH belong to two distinct, complementary sensing pathways of fungi. Whereas GNBP3 appears to act as a classical Pattern Recognition Receptor (able to detect microbial patterns in killed fungi), PSH may be involved in the detection of fungal factors released by live entomopathogenic fungi.
The PR1 protease from the entomopathogenic fungus M. anisopliae triggers Drosomycin expression in a persephone-dependent manner
Our hypothesis is that entomopathogenic fungi secrete virulence factors that can be detected by the host through activation of the Toll pathway. Entomopathogenic hyphomycete fungi are known to secrete proteases and chitinases that perforate the cuticle barrier and allow entry of the fungus into the insect body cavity (Clarkson and Charnley, 1996). This strategy is shared by B. bassiana and M. anisopliae, which both express PR1 subtilisins when spores germinate on the cuticle of insects (58.6 % identity conservation) (Bagga et al., 2004). The PR1 protease of M. anisopliae has been shown to be a major virulence factor of this entomopathogenic fungus (St Leger et al., 1992; Wang et al., 2002). M. anisopliae behaves as B. bassiana when infecting Drosophila (Fig. S2), with the notable exception that Drosomycin expression is not blocked in a psh, but is abolished in a psh, GNBP3hades mutant background (Fig. 4H, see Discussion). To study the effects of fungal virulence factors, we generated transgenic flies that express the gene encoding the M. anisopliae PR1A protease under the control of the heat shock protein promoter using the UAS/GAL4 system. When we overexpressed PR1A in the transgenic flies, we observed a marked expression of Drosomycin in the absence of an immune challenge (Fig. 5A). Strikingly, the expression of Drosomycin induced by PR1A overexpression required the psh gene. In contrast, this expression was not dependent on the GNBP3 gene (Fig. 5A). The PR1A-dependent expression of Drosomycin requires the proteolytic activity of PR1A since transgenic constructs expressing a catalytically-inactive form of PR1A were unable to trigger the Toll pathway (Fig. 5B). The expression of PR1A in the hemolymph leads to the progressive degradation of hemolymphatic proteins (Fig. S3). PSH is a clip domain containing protease, which is thought to be activated by the cleavage of its clip prodomain. To determine whether PSH might be activated by PR1A, we have expressed either PR1A or PSH alone and the ubiquitous daughterless driver and detected a mild expression of Drosomycin 24 hours after induction (Fig. 5C). In striking contrast, the coexpression of both PR1A and psh in the absence of immune challenge led to strong Drosomycin expression, which is significantly higher than the added effect of each protease alone (Fig. 5C). This experiment indicates that PR1A either activates PSH directly or that PSH is a limiting factor in the PR1A-dependent activation branch of the Toll pathway.
Figure 5 . The proteolytic activity of Metarhizium anisopliae PR1A triggers the psh-dependent expression of Drosomycin.
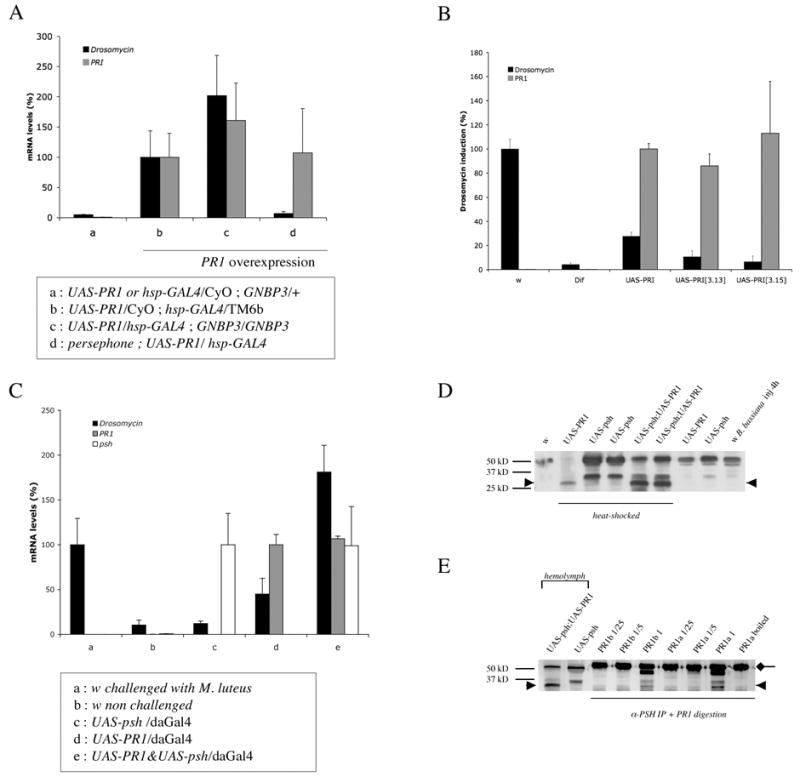
A : The overexpression of M. anisopliae PR1 in unchallenged wild-type (b) or GNBP3hades mutant flies (c) induces the expression of Drosomycin 24 hours after heat-shock as measured by quantitative RT-PCR (black bars). However, this induction is blocked in a psh background (d). Background levels of Drosomycin expression are observed in wild-type unchallenged siblings of the hsp-Gal4 to UAS-PR1 cross (a). The level of expression of the PR1 transcript is shown (grey bars). mRNA levels measured in (b) are taken as reference. The expression of the Drosomycin peptide has also been checked by MALDI-TOF mass spectrometry and coincides with the level of expression of the transcript.
B : The catalytic activity of PR1A is required for the induction of Drosomycin expression. Two transgenic lines expressing a mutated version of the PR1A (S to A mutation of the catalytic triad), [3.13] and [3.15] fail to induce Drosomycin expression. The induction observed in M. luteus-challenged w flies is taken as 100%. The difference of Drosomycin expression observed between UAS-PR1 and UAS-PR1[3,13 (or3,15)] overexpressing flies is statistically significant whereas that between M. luteus-challenged Dif flies and UAS-PR1[3,13 (or 3,15)] overexpressing flies is not.
C : Synergistic activation of the Toll pathway by the joint overexpression of PR1A and PSH. The transgenes are expressed under the control of a tub-Gal80ts; daughterless-GAL4 driver for 24 hours at 29°C, the restrictive temperature for the thermosensitive mutant GAL80, which represses GAL4 at permissive temperatures. This strategy enables the obtention of flies of the desired phenotype that would otherwise die during development as a result of PR1 overexpression.
D : Hemolymph was collected from wild-type (w) or hsp-GAL4 transgenic flies carrying also the indicated UAS transgenes and analyzed by SDS-PAGE and Western blotting with a polyclonal antibody raised against PSH. The overexpression of psh from two distinct psh trangenes leads to the appearance of a 33 kD band while the expression of fungal PR1 induces the formation of a 28 kD band (arrowhead), although a faint 33 kD band could also be observed in some experiments. Interestingly, the Bombyx mori clip protease BAEEase is processed first into a 33 kD band, and second, into a 29.5 kD band, the latter processing event being essential for activation (Jang et al., 2006). The two lanes with flies overexpressing both psh and PR1 correspond to two distinct sets of psh and PR1 transgenes.
E : Hemolymph was collected from wild-type flies and immunoprecipitated with a PSH-specific antibody. The proteins were then digested with PR1 purified preparations and analyzed by Western blotting with the PSH antibody. The 28 (arrowhead) and 33kD proteins obtained by PR1 and psh overexpression are shown on the left. The unprocessed 50 kD PSH band is masked by the Ig heavy chain.
To discriminate between these possibilities, we first monitored the PSH protein in collected hemolymph by Western blotting with a PSH-specific antibody. As shown in Fig. 5D, PSH migrates as a 50 kD band in unchallenged wild-type flies. The overexpression of PSH leads to the appearance of an additional 33 kD band, which is also detected in flies challenged by the injection of B. bassiana spores. Strikingly, the 50 kD band is mostly converted to a band of approximately 28 kD (Fig. 5D) and shorter fragments (not shown). This 28 kD band is the major band observed in flies that overexpress both PR1 and PSH. The 28 kD band may correspond to the active form of PSH since it is detected only in those transgenic flies that strongly express Drosomycin. In addition, the intensity of the band correlates with the strength of Drosomycin induction (compare Fig. 5C and D). Interestingly, the predicted size of the activated CLIP-domain protease PSH, which corresponds essentially to the trypsin catalytic domain, is 27.1 kD. Second, we incubated immunoprecipitated PSH collected from the hemolymph of unchallenged wild-type flies with purified preparations of fungal PR1. This digestion produced the 33 and 28 kD bands in a concentration-dependent manner (Fig. 5E). These bands were absent in the immunoprecipitate incubated with the heat-inactivated PR1 preparations. Taken together, our experiments demonstrate that the Toll pathway can be activated by a fungal protease, which likely activates PSH by direct proteolytic cleavage.
Discussion
The detection of infections is a crucial step in the timely initiation of an appropriate immune response. In Drosophila, the use of nonentomopathogenic bacteria such as M. luteus and E. coli has allowed the delineation of both intracellular signal transduction pathways as well as the identification of five innate receptors (PRRs), PGRP-LC and –LE for the IMD pathway, and PGRP-SA/GNBP1/PGRP-SD for the Toll pathway. To elucidate the mechanisms involved in the detection of fungi, we have first concentrated on a somewhat artificial infection system using an opportunistic human pathogenic yeast, C. albicans. We then refined our understanding of the system by using the entomopathogenic fungi B. bassiana and M. anisopliae.
GNBP3 serves as a receptor for fungal structures
In this study, we demonstrated that GNBP3 is a PRR dedicated to the detection of fungi since (i) recombinant GNBP3 is able to bind in vitro to Candida and to polymeric chains of β-(1,3)-glucan; (ii) it is required for the activation of the Toll pathway by polysaccharides of the fungal cell wall; (iii) GNBP3 is required for resistance against yeast infections, including C. albicans, C. glabrata, C. tropicalis, and against mold infections such as B. bassiana, M. anisopliae, and A. fumigatus (this work, MG, DF, unpublished data); (iv) GNBP3 triggers an adequate immune response, namely it activates the antifungal Toll pathway in a spz-dependent manner. We cannot formally exclude that another fungal receptor acts together with GNBP3 to activate the Toll antifungal host defense.
Of note is that fungi can induce the IMD pathway with short-term kinetics (Lemaitre et al., 1997). We have found that this induction is dependent on PGRP-LC and not on GNBP3 (Fig. S4). One possibility is that a PGRP-LC coreceptor senses fungal microbial patterns. Alternatively, fungal cell wall constituents might bind directly to PGRP-LC. Interestingly, Lee and coworkers have reported that a coleopteran PGRP is able, in addition to its liaison to PGN, to bind with high affinity to tetralaminariose, a tetramer of β-(1,3)-glucan (Lee et al., 2003).
As is the case for members of the PGRP family, the GNBP/βGRP proteins have evolved to recognize distinct carbohydrate chains that form the cell wall of microorganisms. Given their distribution in the arthropod lineage, it is likely that these two families form an essential part of their immunity repertoire. Whereas PGRP homologs exist in mammals, βGRP members have not been reported in vertebrates. However, the phagocytic and signalling receptor Dectin-1 detects β-(1,3)-glucans (Brown and Gordon, 2001) and may to some extent fulfill in mammals a primary function that is similar to that of GNBP3 in insects, i. e., the sensing of fungal infections.
Since spz is required for Toll activation by GNBP3, we propose that the binding of GNBP3 to its microbial ligand leads to the activation of a proteolytic cascade that ultimately processes proSPZ into a functional Toll ligand (Fig. 6). Because psh and GNBP3hades have distinct phenotypes as regards Toll pathway activation (Fig. 4C, D, F) and because the double mutant psh; GNBP3hades displays a stronger phenotype than either mutant alone when challenged with live fungi (Fig. 4G, H), PSH cannot belong exclusively to a proteolytic cascade activated by GNBP3. However, epistatic analysis reveals that the spz-dependent expression of Drosomycin induced by GNBP3 overexpression partly requires psh function (Sup. Fig. 5). Taken together, these data indicate the existence of an alternative, psh-independent proteolytic cascade that mediates the GNBP3-dependent maturation of the Toll ligand Spätzle. This cascade is distinct from the one that activates Toll signalling during early embryogenesis (not shown).
Figure 6 . Model of Toll pathway activation.
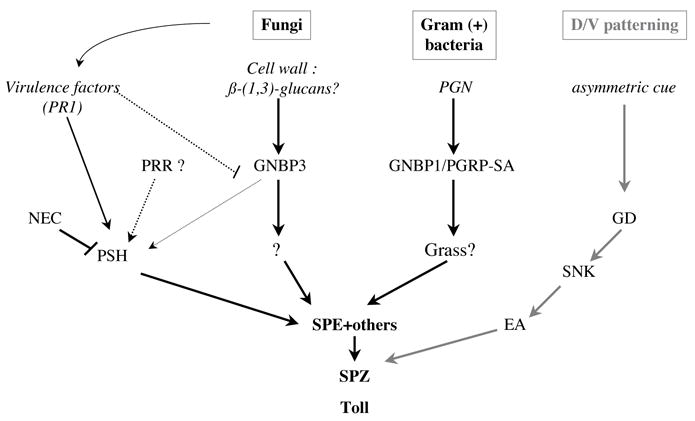
We hypothesize that at least four distinct proteolytic cascades converge to process the Toll ligand Spätzle (SPZ) (see text). Dorso-ventral (D/V) patterning occurs during early embryogenesis and involves the proteases Gastrulation Defective (GD), Snake (SNK), and Easter (EA) : this proteolytic cascade is unlikely to be involved in the activation of Drosomycin expression by fungi (MG, unpublished data). In addition to sensing virulence factors, PSH might function downstream of an unknown Pattern Recognition receptor (PRR). Indeed, epistatic analysis indicates that PSH partially functions downstream of GNBP3. PGN : peptidoglycan; SPE : Spätzle processing enzyme.
The host defense against entomopathogenic fungi does not solely rely on the Toll pathway
An unexpected finding of this study is that the Toll pathway is normally induced in GNBP3hades mutants undergoing a B. bassiana infection. Yet, these mutants are more susceptible to this pathogen than wild-type flies. These observations suggest that GNBP3 fulfills other functions required in the host defense against fungal pathogens that are independent of its role in triggering the Toll pathway. Indeed, we have some biochemical evidence that GNBP3 is involved in other aspects of host defense (AM, DF, unpublished).
PSH is involved in the sensing of fungal virulence factors
Many pathogens have adapted to their hosts and developed specific strategies to defeat their defenses. Fungi such as B. bassiana and M. anisopliae are able to infect insects following deposition of spores on the surface of the cuticle. To penetrate this physical barrier, they secrete several virulence factors such as chitinases and proteases. We found that the PR1A protease is able to activate Drosomycin expression in the absence of infection when overexpressed in flies. This effect on Toll pathway activation is specific since it can be blocked in a psh background and depends on the proteolytic activity of PR1A (Fig. 5). These data establish the proof of concept that a virulence factor can be detected by the innate immune system. Interestingly, our data indicate that PR1 can directly process PSH into its active form.
PR1A is one of ten proteases in this subtilisin family and is expressed only during cuticle penetration (Bagga et al., 2004; Wang et al., 2005). We have found that a PR1A/PR1B-deficient strain is still able to induce Drosomycin expression in a GNBP3hades mutant background, presumably through other fungal PR1 proteases (Fig. S2). Thus, further work will be required to understand the multiple pathogenic mechanisms taking place during a natural fungal infection.
Our data show that the detection of fungal infections relies on a two-pronged sensor system that constitutes a partially redundant recognition system. The psh; GNBP3 double mutant strain consistently yields a stronger phenotype than that of the respective single mutants. Since only GNBP3 is strictly required in the defense against opportunistic yeasts, it is likely that the recognition of fungal patterns represents an ancestral, basal mode of infection sensing. The psh-dependent system that monitors virulence factors may have evolved secondarily in response to the selective pressure exerted by entomopathogenic fungi. Indeed, if the psh-based and the GNBP3-based sensing systems were perfectly redundant, it would be expected that the deletion of one of these systems would not prevent the activation of the Toll pathway. This is indeed what we observe when infecting flies with live C. albicans or with M. anisopliae. In contrast, Drosomycin inducibility is abolished in psh mutants, but not in GNBP3 mutants, infected by B. bassiana. These data indicate that B. bassiana has evolved a strategy that allows it to escape or to block GNBP3 surveillance.
Future studies will reveal whether similar systems of virulence factor detection exist also to sense infection by entomopathogenic bacteria.
A general mechanism of pathogen detection ?
We surmise that some pathogens have developed strategies to inactivate the GNBP basal sensor system of Drosophila and that this led to the selection of a novel host counter-strategy : the surveillance of virulence factor activity. This theme is a central tenet of the current understanding of plant innate immunity. In plants, basal sensor systems detect the presence of microbial elicitors and trigger an immune response. Some virulence factors of the plant pathogen inhibit the elicitor-induced signaling by manipulating host proteins that regulate the host basal response (Kim et al., 2005). In some plant cultivars, a surveillance system based on R proteins “guards” the targets of virulence factors (coded by microbial avirulence (avr) genes) and triggers a strong immune response when under attack (reviewed in Dangl and Jones, 2001). One example is provided by Arabidopsis where the cleavage of the endogenous PBS1 kinase by the Pseudomonas syringae type III effector AvrPphB, a cysteine protease, leads to the activation of the hypersensitive response by the R protein RPS5 (Shao et al., 2003). A case possibly more relevant to fly immunity is provided by the tomato where the host protease Rcr3 is required for the recognition of the pathogen virulence factor Avr2 by the Cf-2 transmembrane receptor (Rooney et al., 2005 and references therein).
Fungal proteases secreted by entomopathogenic fungi have to cross the structurally invariant cuticular barrier of the insect host that thus conditions the type of proteolytic activity required to degrade the cuticular proteins. This phenomenon may have been exploited by Drosophila to detect entomopathogenic infections in a mechanism that is hence conceptually related to the guard hypothesis of plants, although in this case, PSH would monitor indirectly a passive defense mechanism, the protection provided by the bodily armor. To date, the analysis of the immune response in Drosophila has been largely limited to the study of laboratory strains in a controlled environment. By analogy to plant-pathogen interactions that involve avr genes and their cognate plant R resistance genes, a major challenge for the coming years will be to determine whether the insect-pathogen interactions in a natural environment involve several distinct virulence factors and their associated host detection systems.
The discovery of a host sensor system dedicated to the detection of virulence factor activity begs the question of the relevance of such a system to mammalian innate immunity. It has been reported that virulence factors such as the cholesterol-dependent cytolysin or pertussis toxin are able to induce immune responses through TLR4 (Kerfoot et al., 2004; Malley et al., 2003; Park et al., 2004). In these cases, the possibility remains open that TLR4 functions as a co-receptor needed for intracellular signal transduction and that the actual recognition is mediated by unknown receptors. A second class of interest is that of the Protease Activated Receptors. Indeed, PAR2 has been implicated in the induction of the HB2 defensin by bacterial proteases in epithelial cells (Chung et al., 2004). Similarly, Citrobacter rodentium induces the intestinal release of host proteases that activate the PAR2 receptor and subsequent colonic inflammation (Hansen et al., 2005). Finally, virulence factors from Salmonella and Yersinia have been shown to inhibit NF-κB and MAPK signaling (Mota and Cornelis, 2005; Viboud and Bliska, 2005). Thus, it is legitimate to ask whether receptors dedicated to the perception of virulence factor activity have been selected during the evolution of the mammalian innate immune system.
Experimental Procedures
Microbial strains
Gram-negative bacteria : Enterobacter cloacae (a kind gift of H. Monteil), Escherichia coli 1106. Gram-positive bacteria : Micrococcus luteus (CIP A270), Enterococcus faecalis. Fungi : Beauveria bassiana (80.2 strain), M. anisopliae (V275), C. albicans : a pathogenic strain isolated in Patient #3 by Pr. M. Koenig (CHU Strasbourg-Hautepierre).
Fly strains
Stocks were raised on standard cornmeal-agar medium at 25°C. w A5001 flies were used as wild-type throughout the experiments since the GNBP3hades mutant was generated in this background. In experiments involving the Dif1 and key1 mutants, the original cn bw stock was used as a further wild-type control (Jung et al., 2001; Rutschmann et al., 2000a; Rutschmann et al., 2000b). GNBP1osi, PGRP-LCE12, psh4, UAS-psh, and hsp-Gal4 stocks have been described previously (Gobert et al., 2003; Gottar et al., 2002; Ligoxygakis et al., 2002). The hades deficiency was generated by crossing the modified P-element-containing strain d01793 to a strain carrying the Delta2–3 transposase. Revertants that had lost the dominant mini-white marker were tested by PCR for imprecise excision events. The exact extent of the deletion was determined by sequencing of PCR products. Stocks used for epistatic analysis and overexpression analysis were generated using standard crosses. We checked the overexpression of GNBP3 by Q-RT-PCR using the relevant primer set (see Supp. Mat). Heat-shocks were performed as follows : 20 minutes at 37°C, 20 minutes at 18°C, 20 minutes at 37°C, 1 hour at 29°C and then 24 hours incubation at 25°C. Two large deficiencies that remove about 40 kb of the genomic region were generated by crossing d01793 flies to either d08034 or d01127 flies. These strains carry modified P-elements that contain FRT recombination sites and selected progeny of the cross also carried a transgene expressing the yeast flippase. The offspring was screened by PCR to isolate the desired recombinants.
Binding assay
Curdlan (insoluble polymeric β-(1,3)-glucan), a kind gift of JP. Latgé, peptidoglycan from S. aureus, a kind gift of Y. G. Boneca (Paris), chitin (β-(1,4)-N-acetyl-D-glucosamine) from crab shells, Aldrich Chem. Comp, Inc, and paraformaldehyde-treated Candida Albicans yeast were used for in vitro binding assay of GNBP3. All preparations were resuspended in pyrogen-free water (ACILA GMN). 100 μg of each insoluble polysaccharide or 10 μl of PFA-fixed yeast was added to 5 μg of purified GNBP3 and incubated in 200 μl of binding buffer (10mM Tris-HCl, pH 7.5, 500mM NaCl) at room temperature with mild agitation for 1 hour. The mixture was centrifuged (14,000g for 5 min) and the pellet was washed three times with 0.5 ml of washing buffer (10mM Tris, pH 7.5, 500 mM NaCl, 0.02% Tween 20). The proteins bound on insoluble polysaccharide or yeast cells were detached by adding SDS-PAGE sample buffer and analyzed by Western blot using affinity-purified mouse monoclonal anti-His antibody coupled to horseradish peroxidase following the manufacturer’s instructions (Penta-His HRP Conjugate Kit, Qiagen).
The PGN and PBS (phosphate buffer saline, 10 μl, as a control) binding assay was carried out by essentially the same method as described above except the binding mixture was centrifuged at 14,000g for 30 min at each step to precipitate small particles of PGN. Further experimental information is found in the Supplementary online Material section.
Supplementary Material
Acknowledgments
We thank M.-C. Lafarge, S. Ozkhan, and A. Meunier for expert technical help, V. Schott (Entomed) for yeast strains, Dr. L. Joshi for the Metarhizium anisopliae PR1 cDNA, Drs. Y. G. Boneca, JP. Latgé for microbial cell wall compounds, Dr. Alain Roussel for the gift of bacterial recombinant PSH, Dr. J.P. Latgé for discussions and advice, David Rabel for mass spectrometry experiments. This work was supported by CNRS and the Ministère de l’Education Nationale de la Recherche et de la Technologie (ACI Biologie du Développement et Physiologie Integrative). Financial support from the NIH is acknowledged (PO1 AI44220). MG and VG were supported by the Association pour la Recherche sur le Cancer. DF is the leader of a “Equipe Fondation Recherche Médicale”.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alarco AM, Marcil A, Chen J, Suter B, Thomas D, Whiteway M. Immune-deficient Drosophila melanogaster: a model for the innate immune response to human fungal pathogens. J Immunol. 2004;172:5622–5628. doi: 10.4049/jimmunol.172.9.5622. [DOI] [PubMed] [Google Scholar]
- Bagga S, Hu G, Screen SE, St Leger RJ. Reconstructing the diversification of subtilisins in the pathogenic fungus Metarhizium anisopliae. Gene. 2004;324:159–169. doi: 10.1016/j.gene.2003.09.031. [DOI] [PubMed] [Google Scholar]
- Bischoff V, Vignal C, Boneca IG, Michel T, Hoffmann JA, Royet J. Function of the drosophila pattern-recognition receptor PGRP-SD in the detection of Gram-positive bacteria. Nat Immunol. 2004;5:1175–1180. doi: 10.1038/ni1123. [DOI] [PubMed] [Google Scholar]
- Brown GD, Gordon S. Immune recognition. A new receptor for beta-glucans. Nature. 2001;413:36–37. doi: 10.1038/35092620. [DOI] [PubMed] [Google Scholar]
- Chung WO, Hansen SR, Rao D, Dale BA. Protease-activated receptor signaling increases epithelial antimicrobial peptide expression. J Immunol. 2004;173:5165–5170. doi: 10.4049/jimmunol.173.8.5165. [DOI] [PubMed] [Google Scholar]
- Clarkson JM, Charnley AK. New insights into the mechanisms of fungal pathogenesis in insects. Trends Microbiol. 1996;4:197–203. doi: 10.1016/0966-842x(96)10022-6. [DOI] [PubMed] [Google Scholar]
- Dangl JL, Jones JD. Plant pathogens and integrated defence responses to infection. Nature. 2001;411:826–833. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- Fehlbaum P, Bulet P, Michaut L, Lagueux M, Brockaert WF, Hétru C, Hoffmann JA. Septic injury of Drosophila induces the synthesis of a potent antifungal peptide with sequence homology to plant antifungal peptides. J Biol Chem. 1995;269:33159–33163. [PubMed] [Google Scholar]
- Ferrandon D, Imler JL, Hoffmann JA. Sensing infection in Drosophila: Toll and beyond. Semin Immunol. 2004;16:43–53. doi: 10.1016/j.smim.2003.10.008. [DOI] [PubMed] [Google Scholar]
- Fontaine T, Simenel C, Dubreucq G, Adam O, Delepierre M, Lemoine J, Vorgias CE, Diaquin M, Latge JP. Molecular organization of the alkali-insoluble fraction of Aspergillus fumigatus cell wall. J Biol Chem. 2000;275:27594–27607. doi: 10.1074/jbc.M909975199. [DOI] [PubMed] [Google Scholar]
- Gobert V, Gottar M, Matskevich A, Rutschmann S, Royet J, Belvin M, Hoffmann JA, Ferrandon D. Dual Activation of the Drosophila Toll Pathway by Two Pattern Recognition Receptors. Science. 2003;302:2126–2130. doi: 10.1126/science.1085432. [DOI] [PubMed] [Google Scholar]
- Gottar M, Gobert V, Michel T, Belvin M, Duyk G, Hoffmann JA, Ferrandon D, Royet J. The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature. 2002;416:640–644. doi: 10.1038/nature734. [DOI] [PubMed] [Google Scholar]
- Hansen KK, Sherman PM, Cellars L, Andrade-Gordon P, Pan Z, Baruch A, Wallace JL, Hollenberg MD, Vergnolle N. A major role for proteolytic activity and proteinase-activated receptor-2 in the pathogenesis of infectious colitis. Proc Natl Acad Sci U S A. 2005;102:8363–8368. doi: 10.1073/pnas.0409535102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann JA. The immune response of Drosophila. Nature. 2003;426:33–38. doi: 10.1038/nature02021. [DOI] [PubMed] [Google Scholar]
- Jang IH, Chosa N, Kim SH, Nam HJ, Lemaitre B, Ochiai M, Kambris Z, Brun S, Hashimoto C, Ashida M, et al. A Spatzle-processing enzyme required for toll signaling activation in Drosophila innate immunity. Dev Cell. 2006;10:45–55. doi: 10.1016/j.devcel.2005.11.013. [DOI] [PubMed] [Google Scholar]
- Jung A, Criqui MC, Rutschmann S, Hoffmann JA, Ferrandon D. A microfluorometer assay to measure the expression of β-galactosidase and GFP reporter genes in single Drosophila flies. Biotechniques. 2001;30:594–601. doi: 10.2144/01303rr04. [DOI] [PubMed] [Google Scholar]
- Kambris Z, Brun S, Jang IH, Nam HJ, Romeo Y, Takahashi K, Lee WJ, Ueda R, Lemaitre B. Drosophila immunity: a large-scale in vivo RNAi screen identifies five serine proteases required for Toll activation. Curr Biol. 2006;16:808–813. doi: 10.1016/j.cub.2006.03.020. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Silverman N. Bacterial recognition and signalling by the Drosophila IMD pathway. Cell Microbiol. 2005;7:461–469. doi: 10.1111/j.1462-5822.2005.00504.x. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Yano T, Aggarwal K, Lim JH, Ueda K, Oshima Y, Peach C, Erturk-Hasdemir D, Goldman WE, Oh BH, et al. PGRP-LC and PGRP-LE have essential yet distinct functions in the drosophila immune response to monomeric DAP-type peptidoglycan. Nat Immunol. 2006;7:715–723. doi: 10.1038/ni1356. [DOI] [PubMed] [Google Scholar]
- Kerfoot SM, Long EM, Hickey MJ, Andonegui G, Lapointe BM, Zanardo RC, Bonder C, James WG, Robbins SM, Kubes P. TLR4 contributes to disease-inducing mechanisms resulting in central nervous system autoimmune disease. J Immunol. 2004;173:7070–7077. doi: 10.4049/jimmunol.173.11.7070. [DOI] [PubMed] [Google Scholar]
- Kim MG, da Cunha L, McFall AJ, Belkhadir Y, DebRoy S, Dangl JL, Mackey D. Two Pseudomonas syringae type III effectors inhibit RIN4-regulated basal defense in Arabidopsis. Cell. 2005;121:749–759. doi: 10.1016/j.cell.2005.03.025. [DOI] [PubMed] [Google Scholar]
- Kim YS, Ryu JH, Han SJ, Choi KH, Nam KB, Jang IH, Lemaitre B, Brey PT, Lee WJ. Gram-negative bacteria-binding protein, a pattern recognition receptor for lipopolysaccharide and beta-1,3-glucan that mediates the signaling for the induction of innate immune genes in Drosophila melanogaster cells. J Biol Chem. 2000;275:32721–32727. doi: 10.1074/jbc.M003934200. [DOI] [PubMed] [Google Scholar]
- Lee MH, Osaki T, Lee JY, Baek MJ, Zhang R, Park JW, Kawabata SI, Soderhall K, Lee BL. J Biol Chem. 2003. Peptidoglycan recognition proteins involved in 1,3-beta-D-glucan-dependent prophenoloxidase activation system of insect. [DOI] [PubMed] [Google Scholar]
- Lee SH, Carpenter JF, Chang BS, Randolph TW, Kim YS. Effects of solutes on solubilization and refolding of proteins from inclusion bodies with high hydrostatic pressure. Protein Sci. 2006;15:304–313. doi: 10.1110/ps.051813506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- Lemaitre B, Reichhart JM, Hoffmann JA. Drosophila host defense: differential display of antimicrobial peptide genes after infection by various classes of microorganisms. Proc Natl Acad Sci USA. 1997;94:14614–14619. doi: 10.1073/pnas.94.26.14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levashina EA, Langley E, Green C, Gubb D, Ashburner M, Hoffmann JA, Reichhart JM. Constitutive activation of Toll-mediated antifungal defense in serpin-deficient Drosophila. Science. 1999;285:1917–1919. doi: 10.1126/science.285.5435.1917. [DOI] [PubMed] [Google Scholar]
- Ligoxygakis P, Pelte N, Hoffmann JA, Reichhart JM. Activation of Drosophila Toll during fungal infection by a blood serine protease. Science. 2002;297:114–116. doi: 10.1126/science.1072391. [DOI] [PubMed] [Google Scholar]
- Ma C, Kanost MR. A beta1,3-glucan recognition protein from an insect, Manduca sexta, agglutinates microorganisms and activates the phenoloxidase cascade. J Biol Chem. 2000;275:7505–7514. doi: 10.1074/jbc.275.11.7505. [DOI] [PubMed] [Google Scholar]
- Malley R, Henneke P, Morse SC, Cieslewicz MJ, Lipsitch M, Thompson CM, Kurt-Jones E, Paton JC, Wessels MR, Golenbock DT. Recognition of pneumolysin by Toll-like receptor 4 confers resistance to pneumococcal infection. Proc Natl Acad Sci U S A. 2003;100:1966–1971. doi: 10.1073/pnas.0435928100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel T, Reichhart J, Hoffmann JA, Royet J. Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature. 2001;414:756–759. doi: 10.1038/414756a. [DOI] [PubMed] [Google Scholar]
- Mota LJ, Cornelis GR. The bacterial injection kit: type III secretion systems. Ann Med. 2005;37:234–249. doi: 10.1080/07853890510037329. [DOI] [PubMed] [Google Scholar]
- Ochiai M, Ashida M. A pattern-recognition protein for beta-1,3-glucan. The binding domain and the cDNA cloning of beta-1,3-glucan recognition protein from the silkworm, Bombyx mori. J Biol Chem. 2000;275:4995–5002. doi: 10.1074/jbc.275.7.4995. [DOI] [PubMed] [Google Scholar]
- Park JM, Ng VH, Maeda S, Rest RF, Karin M. Anthrolysin O and other gram-positive cytolysins are toll-like receptor 4 agonists. J Exp Med. 2004;200:1647–1655. doi: 10.1084/jem.20041215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DW, Humber RA. Biology of Conidial Fungi. Academic Press Inc; 1981. Entomogenous fungi; pp. 201–236. [Google Scholar]
- Rooney HC, Van’t Klooster JW, van der Hoorn RA, Joosten MH, Jones JD, de Wit PJ. Cladosporium Avr2 inhibits tomato Rcr3 protease required for Cf-2-dependent disease resistance. Science. 2005;308:1783–1786. doi: 10.1126/science.1111404. [DOI] [PubMed] [Google Scholar]
- Rutschmann S, Jung AC, Hetru C, Reichhart JM, Hoffmann JA, Ferrandon D. The Rel protein DIF mediates the antifungal, but not the antibacterial, response in Drosophila. Immunity. 2000a;12:569–580. doi: 10.1016/s1074-7613(00)80208-3. [DOI] [PubMed] [Google Scholar]
- Rutschmann S, Jung AC, Rui Z, Silverman N, Hoffmann JA, Ferrandon D. Role of Drosophila IKKγ in a Toll-independent antibacterial immune response. Nat Immunology. 2000b;1:342–347. doi: 10.1038/79801. [DOI] [PubMed] [Google Scholar]
- Rutschmann S, Kilinc A, Ferrandon D. The Toll pathway is required for resistance to Gram-positive bacterial infections in Drosophila. J Immunol. 2002;168:1542–1546. doi: 10.4049/jimmunol.168.4.1542. [DOI] [PubMed] [Google Scholar]
- Shao F, Golstein C, Ade J, Stoutemyer M, Dixon JE, Innes RW. Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science. 2003;301:1230–1233. doi: 10.1126/science.1085671. [DOI] [PubMed] [Google Scholar]
- St Leger RJ, Frank DC, Roberts DW, Staples RC. Molecular cloning and regulatory analysis of the cuticle-degrading-protease structural gene from the entomopathogenic fungus Metarhizium anisopliae. Eur J Biochem. 1992;204:991–1001. doi: 10.1111/j.1432-1033.1992.tb16721.x. [DOI] [PubMed] [Google Scholar]
- Tauszig-Delamasure S, Bilak H, Capovilla M, Hoffmann JA, Imler JL. Drosophila MyD88 is required for the response to fungal and Gram-positive bacterial infections. Nat Immunology. 2002;3:91–97. doi: 10.1038/ni747. [DOI] [PubMed] [Google Scholar]
- Thibault ST, Singer MA, Miyazaki WY, Milash B, Dompe NA, Singh CM, Buchholz R, Demsky M, Fawcett R, Francis-Lang HL, et al. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat Genet. 2004;36:283–287. doi: 10.1038/ng1314. [DOI] [PubMed] [Google Scholar]
- Tzou P, Reichhart JM, Lemaitre B. Constitutive expression of a single antimicrobial peptide can restore wild-type resistance to infection in immunodeficient Drosophila mutants. Proc Natl Acad Sci U S A. 2002;99:2152–2157. doi: 10.1073/pnas.042411999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viboud GI, Bliska JB. Yersinia outer proteins: role in modulation of host cell signaling responses and pathogenesis. Annu Rev Microbiol. 2005;59:69–89. doi: 10.1146/annurev.micro.59.030804.121320. [DOI] [PubMed] [Google Scholar]
- Wang C, Hu G, St Leger R. Differential gene expression by Metarhizium anisopliae growing in root exudate and host (Manduca sexta) cuticle or hemolymph reveals mechanisms of physiological adaptation. Fungal Genet Biol. 2005;42:704–718. doi: 10.1016/j.fgb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Wang C, Typas MA, Butt TM. Detection and characterisation of pr1 virulent gene deficiencies in the insect pathogenic fungus Metarhizium anisopliae. FEMS Microbiol Lett. 2002;213:251–255. doi: 10.1111/j.1574-6968.2002.tb11314.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


