Abstract
The scavenger receptor class B type I (SR-BI) mediates the selective uptake of cholesteryl esters from high-density lipoprotein (HDL) and cholesterol secretion into bile in the liver. In this study, we identified an SR-BI-associated protein from rat liver membrane extracts by using an affinity chromatography technique. This protein of 523 amino acids contains four PDZ domains and associates with the C terminus of SR-BI by using its N-terminal first PDZ domain. Therefore, we denoted this protein as CLAMP (C-terminal linking and modulating protein). CLAMP was located mostly in the sinusoidal membranes, whereas SR-BI was detected in both sinusoidal and canalicular membranes. After the solubilization of the liver membranes with Triton X-100, SR-BI was immunoprecipitated with anti-CLAMP monoclonal antibody, suggesting the association of these proteins in vivo. By coexpressing SR-BI with CLAMP in Chinese hamster ovary cells, we observed (i) the increase in the expression level of SR-BI, (ii) the reduction in the deacylation rate of the cholesteryl esters taken up from HDL, and (iii) the change in the intracellular distribution of fluorescent lipid 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine percholate taken up from HDL. Taken together, these data suggest that CLAMP, a four-PDZ-domain-containing protein, is associated with SR-BI in the liver sinusoidal plasma membranes and may modulate the intracellular transport and metabolism of cholesteryl esters taken up from HDL.
The level of plasma high-density lipoproteins (HDL) is inversely related to the incidence of atherosclerosis and coronary artery disease (1). The protective effect of HDL is thought to be caused by the reverse cholesterol transport from cells in the arterial wall to the liver for catabolism (2). The scavenger receptor class B type I (SR-BI) is the first HDL receptor that has been well defined at the molecular level and shown to be a mediator of selective cholesterol uptake in vitro (3). Adenovirus-mediated overexpression of SR-BI in the liver leads to reduced plasma HDL levels and increased cholesterol secretion into bile (4). Fluiter et al. (5, 6) have also reported additional strong correlative evidence for the importance of SR-BI in selective cholesterol uptake by hepatocytes in vitro and in vivo.
Recently, SR-BI has been shown to be fatty-acylated and to cluster in caveolae-like domains (7); however, the detailed mechanism of selective uptake mediated by SR-BI has not been defined. The process of selective cholesteryl ester (CE) uptake from HDL can be divided into three steps (8). The first step of selective uptake may involve receptor binding, followed by the reversible incorporation of HDL-CE into the plasma membrane pool and the subsequent transfer of the lipid to an inaccessible pool. It is uncertain whether HDL-CE uptake is achieved by SR-BI itself or whether other cellular components are involved in this process.
SR-BI is a member of the CD36 superfamily, whose members have been proposed to have similar membrane topologies (9). Topologic studies of CD36 in conjugation with sequence analysis indicate that SR-BI has two transmembrane domains that sit adjacent to relatively short cytoplasmic N-terminal (8 residues) and cytoplasmic C-terminal (45 residues) domains. SR-BI and CD36 share significant sequence homology throughout their entire extracellular loop domains; however, the cytoplasmic C-terminal domains were quite different from each other in their primary sequences. Therefore, we assumed that the cytoplasmic C-terminal domain (C45) of SR-BI is important for specific functions of the receptor or its regulations.
In this study, we used recombinant C45-glutathione S-transferase (GST) fusion protein to identify proteins that associate with the cytosolic domain of SR-BI from rat liver membrane extracts. Herein, we describe the isolation and characterization of a PDZ-domain-containing protein that interacts with the C terminus of SR-BI and name this protein CLAMP (C-terminal linking and modulating protein).
Materials and Methods
Lipoprotein Preparation and Labeling.
Human HDL (HDL3; density = 1.125–1.21 g/ml) was prepared by sequential density ultracentrifugation as described (10). HDL apolipoproteins were iodinated by the iodine monochloride method (11) to a specific radioactivity ranging from 300 to 700 cpm/ng HDL protein. HDL was also radiolabeled with [3H]CE essentially as described by Roberts et al. (12). The specific activity of HDL labeled in the CE ranged from 20,000 to 30,000 dpm/μg HDL protein. 125I (as Na125I; 17 Ci/mg of iodine) and [1,2,6,7-3H(N)]cholesteryl oleate (84 Ci/mmol) were purchased from Amersham Pharmacia and NEN, respectively; 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine percholate (DiI)-HDL was purchased from Biomedical Technologies (Stoughton, MA). Lipoprotein-deficient serum was prepared as described (13).
Preparation of Rat Liver Membrane Extracts.
Livers obtained from Wister male rats were homogenized with a homogenizing buffer (10 mM Tris⋅HCl, pH 7.4/1 mM EDTA/0.25 M sucrose), and the homogenate was centrifuged at 10,000 × g for 20 min at 4°C. The supernatant was recentrifuged at 100,000 × g for 1 h at 4°C, and the resulting precipitate was suspended in a homogenizing buffer. The proteins in this fraction were extracted with a homogenizing buffer containing 1 M KCl. The resulting pellet was solubilized with 2% (vol/vol) Triton X-100 for 1 h at 4°C and then centrifuged at 100,000 × g for 1 h at 4°C. The supernatant was used as a membrane extract. Rat liver sinusoidal and canalicular plasma membranes were prepared as described (14). According to the marker-enzyme activities, the isolated canalicular membranes were virtually devoid of sinusoidal contaminants, whereas the sinusoidal membranes were contaminated with canalicular membranes by ≈11%.
GST Fusion Proteins.
Recombinant C45-GST fusion proteins were prepared and purified as follows. The EcoRI–SalI fragment encoding the cytoplasmic C-terminal domain of SR-BI was subcloned into a multicloning site downstream of the sequence for GST in pGEX-4T-1 (Amersham Pharmacia). This plasmid was transformed into the JM109 strain of Escherichia coli and induced with isopropyl-1-thio-β-d-galactopyranoside to produce GST fusion proteins. The bacteria were suspended in PBS , and vigorous sonication was performed before centrifugation at 10,000 × g for 20 min. The resulting supernatants were applied on the glutathione bead column and then eluted with an elution buffer (50 mM Tris⋅HCl, pH 9.6/120 mM NaCl/10 mM glutathione). Purified GST fusion proteins were dialyzed against a dialysis buffer (PBS containing 2 mM EDTA and 1 mM DTT). Recombinant C5-GST, C22-GST, and C30-GST fusion proteins were prepared and purified as described above.
C45-GST Affinity Chromatography and Peptide Sequence Analysis.
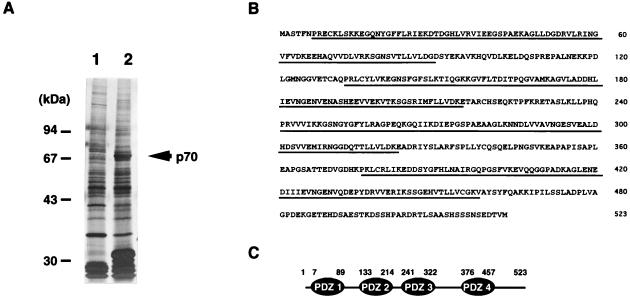
Recombinant C45-GST fusion protein, coupled to glutathione-agarose, was used to affinity-purify the C45-binding protein(s). Rat liver membrane extracts were applied on the C45-GST-glutathione bead column and then eluted with the elution buffer. The eluant was dialyzed against the dialysis buffer and loaded onto a MonoQ column equilibrated with buffer A (10 mM Tris⋅HCl, pH 7.4/0.5% Triton X-100). The adsorbed fraction was eluted with a linear gradient of 0–500 mM NaCl in buffer A. The p70 protein (see Fig. 1A) was eluted in fractions at about 200 mM NaCl, whereas most of the bands that migrate close to it were eluted in the flow-through fraction. The fractions containing the p70 protein were collected, concentrated, and subjected to SDS/PAGE. A Coomassie blue-stained band of 70 kDa was cut out and digested with lysylendopeptidase. The resulting peptides were separated onto columns of DEAE-5PW (Tosoh, Tokyo) and Mightysil RP-18 (Kanto Chemical, Tokyo) with a 0–80% gradient of acetonitrile in 0.1% trifluoroacetic acid and then sequenced.
Figure 1.
Identification and characterization of an SR-BI-interacting protein (p70). (A) Affinity purification of p70. The rat liver membrane fraction extracted with 2% (vol/vol) Triton X-100 was subjected to a GST (lane 1) or a C45-GST (lane 2) affinity column, and the bound proteins were eluted by the addition of glutathione. The eluates were subjected to SDS/PAGE, followed by silver staining. (B) Amino acid sequence of p70. p70 is a 523-amino acid protein that contains four PDZ domains and no obvious catalytic domain. The PDZ domains are indicated by underlining. (C) Schematic drawing of the domain structure of p70. Small numbers refer to amino acid residues.
cDNA Cloning of CLAMP (p70).
The cDNA fragments that contained internal amino acid sequences were amplified by PCR by using rat liver total RNA as a template and the degenerated oligonucleotide primers based on the obtained peptide sequences. The resulting PCR fragments were subcloned into a pT7Blue T-vector (Novagen), and the nucleotide sequences were determined. The DNA fragment was then used to isolate a full-length cDNA clone by plaque hybridization in a rat liver λ-phage cDNA library that was made by using the SuperScript lambda system for cDNA synthesis and λ-cloning (Life Technologies, Grand Island, NY).
Yeast Two-Hybrid Analysis.
The cDNA encoding the cytoplasmic C-terminal domain of SR-BI was fused in-frame with the lexA DNA-binding domain by using a pBTM116 expression vector to generate plexC45. A series of deletion mutants of the PDZ domain of CLAMP was constructed by using a PCR-based strategy. The cDNA encoding of various truncations was fused in-frame with the Gal4 DNA-transactivation domain by using a pGAD-C1 expression vector (15) to generate pGalPDZx. Yeast strain L40, with two reporter genes (lexA-lacZ and lexA-HIS3), was transformed with plexC45 on an SD-Trp plate and then with pGalPDZx on an SD-Trp-Leu plate. HIS3 expression or lacZ expression was examined by the method described previously (16).
Antibodies.
The monoclonal antibody against CLAMP (p70) was prepared as follows. Partially purified recombinant rat CLAMP proteins were used to immunize BALB/c mice, after which the spleen cells obtained from the mice were fused with mouse myeloma cells (PAI). One of the established monoclonal antibodies was named 3B7. The SR-BI polyclonal antibody (named anti-SR-BI-110) was prepared as follows. A peptide corresponding to the extracellular domain of hamster SR-BI between amino acid residues 110 and 132 coupled to keyhole limpet hemocyanin was injected into the back of New Zealand White rabbits. The antibody was affinity-purified from the serum as previously described (17).
Cells.
Chinese hamster ovary (CHO)-K1 cells were maintained in medium A [Ham's F-12 medium supplemented with 50 units/ml penicillin, 50 μg/ml streptomycin, 2 mM glutamine, and 10% (vol/vol) FBS] at 37°C in a humidified 5% CO2, 95% air incubator. CHO-SRBI cells (18) stably expressing hamster SR-BI were maintained in medium A supplemented with 50 μg/ml G418. CHO-CLAMP cells were stably transfected with a pcDNA3.1/Hygro/CLAMP vector that was constructed by ligating the HindIII–BamHI fragment of CLAMP cDNA into pcDNA3.1/Hygro (Invitrogen). These CHO-CLAMP cells were maintained in medium A supplemented with 50 μg/ml hygromycin. Two independent clones were used in the experiments described below.
Immunoprecipitation.
CHO-CLAMP cells were transiently transfected with or without either SR-BI or SR-BIΔC15 (a deletion mutant without 15 C-terminal amino acids). Briefly, on day 0, 1.5 × 106 cells were plated on a 6-cm plate in medium A. On day 1, cells were transfected with the plasmid employing a Lipofectamine reagent (Life Technologies). On day 3, the cells were lysed with a 1.2-ml Triton X-100 solubilization buffer (10 mM Tris, pH 7.4/150 mM NaCl/1 mM EDTA/10 μg/ml leupeptin/1 mM PMSF/1% Triton X-100). The lysate was preincubated with protein G-Sepharose (Amersham Pharmacia) for 1 h at 4°C and then incubated with an anti-CLAMP antibody coupled to protein G-Sepharose. After an overnight incubation at 4°C, the beads were washed three times with a Triton X-100 solubilization buffer. The immunoprecipitates were then analyzed by immunoblot techniques with the anti-SR-BI-110 polyclonal antibody. Rat liver membrane extraction and immunoprecipitation followed an identical procedure.
Analysis of Cell Association, CE Selective Uptake, and Cellular Metabolism of [125I/3H-CE]HDL.
CHO cells and CHO-CLAMP cells were plated in a six-well plate and transiently transfected with or without either SR-BI or SR-BIΔC15. 125I/3H-double-labeled HDL particles were added at a concentration of 10 μg/ml in medium B [Ham's F-12 medium supplemented with 50 units/ml penicillin, 50 μg/ml streptomycin, 2 mM glutamine, and 6.5% (vol/vol) lipoprotein-deficient serum] in the absence or presence of a 40-fold excess of unlabeled HDL. After incubation for 4 h, the cell lysate was prepared and processed to determine trichloroacetic acid soluble, insoluble 125I-radioactivity, organic solvent-extractable 3H-radioactivity, and the cellular proteins as described (19). 3H-labeled lipids in either cells or media were extracted by the method of Bligh and Dyer (20) and subjected to TLC on a Silica-Gel G plate. After the plate was developed with hexane/ethyl ether/acetic acid (70:30:1 vol/vol), each spot [CE and unesterified cholesterol (UC)] was scraped into a liquid scintillation vial to measure the radioactivity as described (21).
Results
Identification, Purification, and Cloning of an SR-BI-Associated Protein.
To identify molecules that specifically bind to C45, the C-terminal cytoplasmic domain of SR-BI, various fractions obtained from rat liver were loaded onto a C45-GST affinity column. The proteins bound to the column were coeluted with C45-GST by the addition of glutathione. When the rat liver membrane fraction extracted with 2% (vol/vol) Triton X-100 was loaded onto a C45-GST affinity column, a protein with a molecular mass of approximately 70 kDa (p70) was detected in the glutathione eluate from a C45-GST column but not from a control GST column (Fig. 1A); therefore, it was purified further by MonoQ column chromatography and subjected to amino acid sequencing. Five peptide sequences derived from p70 were determined. On the basis of these sequences, degenerated oligonucleotides were synthesized and used in a series of PCRs with rat liver cDNA as a template. We used the amplified product to screen a rat liver λ-ZIPLOX cDNA library. Isolation of full-length p70 cDNA indicated that it was nearly 2.4 kb long with an ORF of 1,569 bp that encodes a 523-amino acid protein (Fig. 1B). The predicted molecular mass of this protein is ≈57 kDa, which is smaller than that estimated by SDS/PAGE. To determine the size of the product encoded by the isolated cDNA, translation studies were performed with a cell-free reticulocyte lysate system containing [35S]methionine. The size of the product was 70 kDa as estimated by SDS/PAGE, indicating that the isolated cDNA contained the full length of p70 (data not shown). A search of GenBank revealed that the submitted sequence shared a significant homology with human PDZK1 (22) and rat Diphor-1 (23), although the C-terminal region of the latter was shorter. It also partially shared homologies with a number of proteins containing PDZ protein interaction domains. p70 encoded four PDZ domains (Fig. 1 B and C).
Domains Involved in CLAMP (p70) and SR-BI Association.
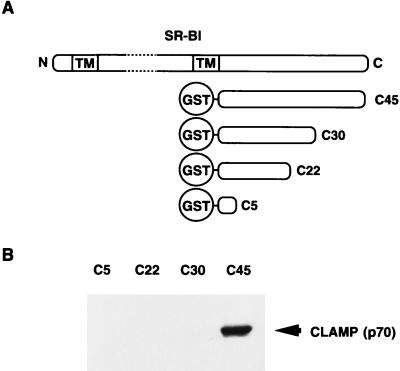
We next examined the effect of the C-terminal deletion mutants of SR-BI on its binding to p70 by in vitro binding assays (Fig. 2). Although full-length C45-GST fusion protein bound to p70, deletion constructs lacking the last 15 amino acids of SR-BI (C30-GST) abolished binding to p70. These observations indicate that the C-terminal region of SR-BI is required for interaction with p70. Therefore, we named this protein CLAMP.
Figure 2.
p70 (CLAMP) interacts with the C-terminal end of SR-BI via its first N-terminal PDZ domain (PDZ1). (A) The constructs of various versions of SR-BI are shown schematically. Various deletion mutants of SR-BI fused with GST were produced and affinity-purified with glutathione-agarose beads. N, N terminus; C, C terminus; TM, transmembrane. (B) Rat liver membrane extracts were mixed with glutathione-agarose beads with immobilized C5, C22, C30, or C45-GST. Bound proteins were eluted with the elution buffer, and the eluate was resolved by SDS/PAGE. The proteins were then transferred to nitrocellulose filters, and the filters were immunoblotted with an anti-CLAMP antibody. Proteins bound to the antibody were made visible with an enhanced chemiluminescence kit (Amersham Pharmacia).
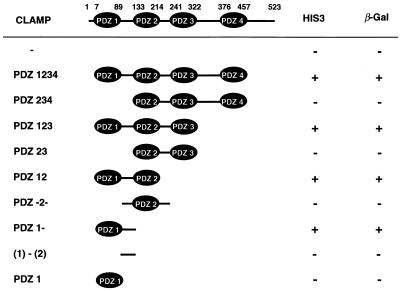
We also determined which PDZ domain or domains of CLAMP were responsible for the interaction with the C-terminal region of SR-BI with the yeast two-hybrid system. Various constructs containing different combinations of the PDZ domains were made (Table 1). The shortest construct responsible for the pairing with the SR-BI C terminus contained both PDZ1 and approximately 40 amino acids on the C-terminal side of PDZ1, indicating that the other PDZ domains are not necessary for interaction with SR-BI. In fact, PDZ234 domains did not interact with the C-terminal region of SR-BI as determined by the yeast two-hybrid system.
Table 1.
Structural determinants of CLAMP required for interaction with SR-BI
The yeast two-hybrid system was used to test the domains of CLAMP required for interaction with SR-BI as described in Materials and Methods. Positive selection on His plates and the β-galactosidase (β-Gal) activity of each construct are indicated.
Association of CLAMP with SR-BI in Vivo.
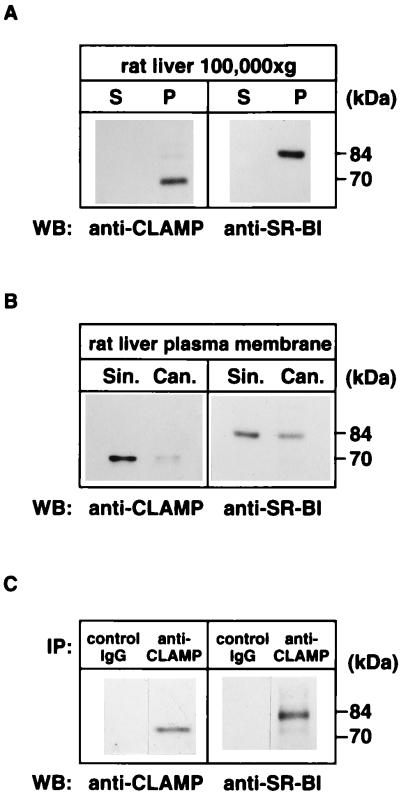
To investigate the location of CLAMP and SR-BI in the rat liver, we attempted to visualize these proteins by using the immunohistochemical technique but failed to immunostain these proteins with our antibodies, possibly because of low expression levels of these proteins in this organ. Then, we purified sinusoidal and canalicular plasma membranes from rat liver and used Western blotting to investigate on which side of the membrane these proteins are found. First, rat liver homogenates were separated into soluble (100,000 × g supernatant) and membrane (100,000 × g pellet) fractions. Immunoblotting analysis identified the presence of both SR-BI and CLAMP exclusively in the membrane fraction (Fig. 3A). When rat liver plasma membranes were fractionated into sinusoidal and canalicular membranes, SR-BI was detected in both membrane fractions, whereas CLAMP was located mainly in the sinusoidal membrane (Fig. 3B). We further examined the interaction between SR-BI and CLAMP in the liver by immunoprecipitation. The membrane fraction was solubilized with 1% Triton X-100 and immunoprecipitated with an anti-CLAMP antibody. The resulting immunoprecipitates were immunoblotted with an anti-SR-BI antibody. As shown in Fig. 3C, SR-BI could be immunoprecipitated with an anti-CLAMP antibody but not with control IgG, indicating that SR-BI associates with CLAMP in rat liver.
Figure 3.
CLAMP interacts with SR-BI in rat liver sinusoidal membrane. (A) Presence of SR-BI and CLAMP in the membrane fraction. Rat liver homogenate was separated by centrifugation into a soluble fraction (S) and an insoluble membrane pellet fraction (P). Equal fractions (not equal protein) were separated by SDS/PAGE and processed for Western blotting (WB) with an antibody against CLAMP or SR-BI. (B) Localization of SR-BI and CLAMP in the liver sinusoidal and canalicular membranes. Rat liver membranes were separated into a sinusoidal (Sin.) and a canalicular (Can.) membrane fraction as described in Materials and Methods. Equal amounts of protein were separated by SDS/PAGE and processed for Western blotting with an antibody against CLAMP or SR-BI. (C) Coimmunoprecipitation of CLAMP with SR-BI. The rat liver membranes were extracted with Triton X-100, and CLAMP was immunoprecipitated (IP) with either an anti-CLAMP antibody or a control IgG. The immunoprecipitates were then analyzed by Western blotting with an antibody against CLAMP or SR-BI.
Coexpression of SR-BI and CLAMP in CHO Cells.
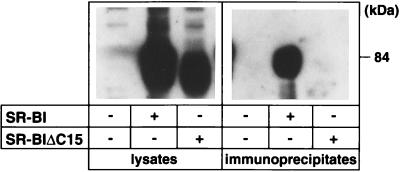
Next, we prepared CHO cells that constitutively express CLAMP, named CHO-CLAMP cells, and examined the interaction between SR-BI and CLAMP in the living cell. CHO-CLAMP cells were transiently transfected with or without either SR-BI or SR-BIΔC15 (a deletion mutant without 15 C-terminal amino acids) and then solubilized with 1% Triton X-100, and the soluble cell lysate was immunoprecipitated with an anti-CLAMP antibody. The resulting immunoprecipitates were then immunoblotted with an anti-SR-BI antibody. As shown in Fig. 4, SR-BI could be immunoprecipitated with an anti-CLAMP antibody when SR-BI was transfected into CHO-CLAMP cells, indicating that SR-BI associates with CLAMP in CHO cells. However, SR-BI was not coimmunoprecipitated with an anti-CLAMP antibody when the cells were transfected with the SR-BIΔC15 mutant, implying that the 15 C-terminal amino acids of SR-BI are necessary for the CLAMP interaction.
Figure 4.
CLAMP interacts with SR-BI in CHO cells. CHO-CLAMP cells were transiently transfected with (+) or without (−) either SR-BI or SR-BIΔC15 (a deletion mutant without 15 C-terminal amino acids). The cells were then solubilized with Triton X-100, and the CLAMP was immunoprecipitated with an anti-CLAMP antibody. The lysates and the resulting immunoprecipitates were then analyzed by Western blotting with an anti-SR-BI antibody.
Increased SR-BI Expression by CLAMP.
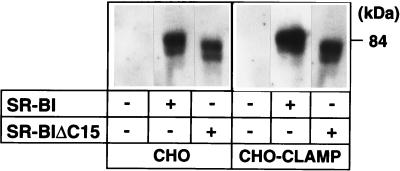
Next, we explored the biological function of CLAMP by using CHO cells. Control CHO cells and CHO-CLAMP cells were transiently transfected with or without either SR-BI or SR-BIΔC15. These cells were incubated for 4 h with 10 μg/ml double-labeled (125I-apolipoprotein/[3H]cholesteryl oleoyl ester) HDL, after which HDL cell association, selective uptake of HDL CE, and the ratio (uptake efficiency index) were determined. We found that both 125I-HDL cell association and [3H]CE selective uptake via SR-BI were increased about 2- or 3-fold by the coexpression of CLAMP; however, there was only a small decrease in the relative efficiency of CE uptake (Table 2). Deletion of the last 15 amino acids of SR-BI did not interfere either with the receptor's ability to bind 125I-HDL or with CE uptake in control CHO cells. CLAMP had no effect either on HDL cell association or on CE uptake when SR-BIΔC15 was used. We then tested the expression level of SR-BI in those cells by Western blotting and found that cells coexpressing SR-BI and CLAMP had increased expression levels of SR-BI (Fig. 5). By densitometric analysis, CLAMP was found to induce about a 2- to 3-fold increase in the level of SR-BI expression. Again, expression of SR-BIΔC15 was not affected significantly by CLAMP. The transfection efficiency didn't differ between control CHO cells and CHO-CLAMP cells (data not shown). All these data showed that CLAMP did not affect the rate of [3H]CE selective uptake but had a significant effect on the expression and/or the stability of SR-BI protein.
Table 2.
Effect of CLAMP on the uptake efficiency by SR-BI
| Cell | Transfected gene | 125I-HDL cell association (a), ng HDL protein/mg cell protein | [3H]CE selective uptake (b), ng HDL protein/mg cell protein | Uptake efficiency index* |
|---|---|---|---|---|
| CHO-K1 | ||||
| Control | 38 | 152 | ||
| SR-BI | 150 | 745 | 100 | |
| SR-BIΔC15 | 139 | 661 | 95 | |
| CHO-CLAMP | ||||
| Control | 34 | 166 | ||
| SR-BI | 348 | 1,567 | 85 | |
| SR-BIΔC15 | 153 | 682 | 82 | |
Uptake efficiency index was calculated as described (12), i.e., in CHO-K1 cells = [(b − 152)/(a − 38)] × (100/5.29); for wild-type SR-BI, a = 150, b = 745, and (b − 152)/(a − 38) = 5.29; and in CHO-CLAMP cells = [(b − 166)/(a − 34)] × (100/5.29).
Figure 5.
CLAMP modulates the stability of SR-BI protein. CHO cells and CHO-CLAMP cells were transiently transfected with or without either SR-BI or SR-BIΔC15. Western blotting analysis was performed in the corresponding cellular lysates by using an anti-SR-BI antibody. Bands corresponding to SR-BI were quantified by using LAS-1000 (Fuji). Data represent samples from a typical experiment.
Effect of CLAMP on the Subsequent Metabolism of CE Taken up Through SR-BI.
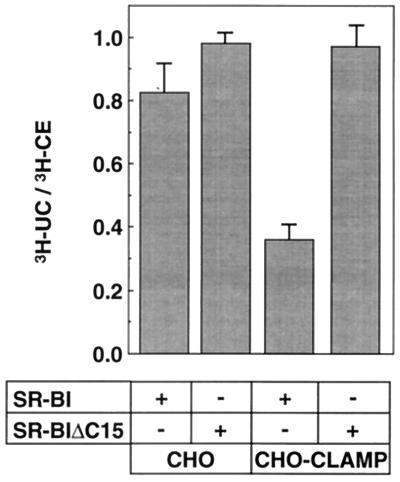
Although it is well established that SR-BI mediates the selective uptake of HDL CE, little is known about the subsequent transport and metabolism of CE in living cells. To throw light on this issue, we examined the effect of CLAMP on the metabolism of [3H]CE taken up from exogenous HDL in CHO cells. CHO cells and CHO cells constitutively expressing CLAMP were first transiently transfected with SR-BI and then incubated with [125I/3H-CE]HDL for 4 h at 37°C, and then the radioactivities of UC and CE in the cells and media were measured. Under this condition, 2% of [125I/3H-CE]HDL added to the culture medium was taken up by the cells, and the efflux of UC into medium was negligible. To compare the rate of conversion from CE to UC, the contribution from vector-transfected cells was subtracted, and the amount of UC was expressed relative to the amount of CE taken up selectively. In this way, the rate of conversion from CE to UC is normalized to the quantity of CE taken up selectively. Expression of CLAMP caused the less efficient conversion of CE to UC (about 40% to the control cells expressing only SR-BI; Fig. 6). On the other hand, CLAMP had essentially no effect on the [3H]CE metabolism in the SR-BIΔC15-expressing CHO cells. From these data, it is conceivable that CLAMP inhibits the hydrolysis of CE delivered via SR-BI. However, at present, we could not rule out the alternative possibility that the reduced conversion of CE to UC could reflect a large substrate pool of CE and near saturation of the hydrolase.
Figure 6.
Effect of CLAMP on the cellular metabolism of HDL-CE taken up via SR-BI. CHO cells and CHO-CLAMP cells were transiently transfected with either SR-BI or SR-BIΔC15 and incubated at 37°C for 4 h with [125I/3H-CE]HDL. The efficiency of conversion of HDL-CE into UC was determined by dividing the amount of cellular [3H]UC by the amount of cellular [3H]CE as described in Materials and Methods. Before calculating the ratios, the specific values for each construct were corrected by subtracting the specific background values from cells transfected with the control plasmid pcDNA3. Values represent means ± SEM (n = 3).
Effect of CLAMP on the Intracellular Movement of Fluorescent Lipid DiI Taken up Through SR-BI.
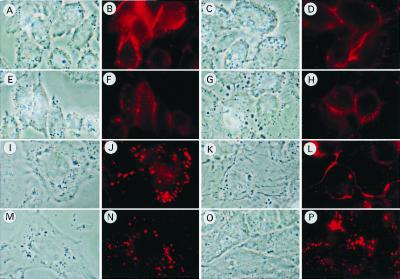
SR-BI is known to mediate the cellular uptake of not only CE but also other lipid soluble substances such as fluorescent lipid DiI (3, 8). By means of this fluorescent lipid, we examined the effect of CLAMP on the intracellular transport of lipids taken up through SR-BI. CHO cells and CHO-CLAMP cells, which transiently express SR-BI, were incubated with DiI-HDL for 5 min at 37°C, washed with a fresh medium to remove unbound DiI-HDL, and then chased for 1 h. After washing (0 h), both cells showed DiI-fluorescence on their surfaces, indicating that coexpression of CLAMP with SR-BI has no appreciable effects on the transport of DiI from HDL to the plasma membrane of CHO cells (Fig. 7 A–D). After the 1-h chase incubation, CHO cells expressing only SR-BI changed the DiI distribution in an intracellular punctate manner (Fig. 7 I and J). In contrast, the distribution pattern of DiI in CHO cells coexpressing SR-BI and CLAMP (Fig. 7 K and L) was strikingly distinct from that in CHO-SRBI cells, although the intracellular localization of DiI still remains unclear. The distribution pattern of DiI in the CHO-SRBIΔC15 cells after the 1-h chase incubation (Fig. 7 M and N) was essentially the same as that in CHO-SRBI cells and was not appreciably affected by the coexpression of CLAMP (Fig. 7 O and P).
Figure 7.
Effect of CLAMP on the cellular distribution of fluorescent lipid (DiI) delivered to cells from DiI-HDL via SR-BI. CHO cells (A, B, E, F, I, J, M, and N) and CHO-CLAMP cells (C, D, G, H, K, L, O, and P) were plated in a 24-well plate containing a glass coverslip (12 × 12 mm) and transfected with either SR-BI (A–D and I–L) or SR-BIΔC15 (E–H and M–P). The cells were washed once with medium A, and medium B containing 10 μg of protein/ml of DiI-HDL was added for 5 min. The cells were then washed twice with medium A and incubated for 0 h (A–H) or 1 h (I–P) in medium A, after which, the cells were washed with PBS six times followed by fixation in 3.7% (vol/vol) formaldehyde in PBS for 20 min. The cells were examined for the localization of DiI-HDL with a fluorescence microscope (Zeiss).
Discussion
In the present study, by using the affinity chromatography technique, we isolated a protein from rat liver that interacts with the cytoplasmic C-terminal domain of SR-BI and named it CLAMP. CLAMP belongs to the PDZ protein family and contains four PDZ domains. PDZ domains are found in diverse membrane-associated proteins including signaling molecules, ion channel or receptor clustering molecules, and cytoskeletal components. CLAMP is, to our knowledge, the first example that binds to the cytoplasmic tail of a plasma lipoprotein receptor.
CLAMP is expressed in the liver but not appreciably in the adrenal gland, ovary, or testis, in which SR-BI is most abundantly expressed (unpublished data). This finding suggests that CLAMP plays a role in the SR-BI function, mainly in the liver. Moreover, CLAMP was detected in isolated rat liver parenchymal cells and the hepatoma cell line HepG2 (unpublished data), indicating that hepatocytes express CLAMP. Hepatocytes have two discrete membranes consisting of sinusoidal and canalicular membranes. We have demonstrated that CLAMP is located exclusively in the sinusoidal plasma membranes, whereas SR-BI is detected in both sinusoidal and canalicular membranes (Fig. 3). Thus, one possible role of CLAMP is to restrict the SR-BI molecule to the sinusoidal membrane, and therefore the level of CLAMP expressed in the liver may determine the expression level of SR-BI in the sinusoidal plasma membrane. Otherwise, CLAMP may serve to cluster SR-BI molecules on the cell surface or to connect the receptor to downstream machineries at the sinusoidal membrane. As mentioned above, SR-BI binds to the first N-terminal PDZ domain (PDZ1) containing ≈40 amino acids on the C-terminal side of PDZ1, and the rest of the PDZ domains (PDZ234) do not seem to interact with SR-BI. Some PDZ domains have been shown to bind directly to other PDZ domains, forming homomeric and heteromeric complexes; however, according to our preliminary data, CLAMP does not form a homodimer, suggesting that CLAMP may not act to cluster SR-BI molecules on the cell surface.
In the liver, SR-BI mediates the selective uptake of CE from HDL at the sinusoidal plasma membrane, whereas SR-BI mediates cholesterol secretion into bile at the canalicular plasma membrane. It has been shown that HDL cholesterol is a major source of substrate for biliary steroid secretion in both HDL mammals such as rats and low-density lipoprotein mammals such as humans (24). There is also considerable evidence to suggest that cholesterol in the liver is compartmentalized into different functional pools (25), with cholesterol secreted into plasma in very-low-density lipoproteins being derived from a pool that is distinct from the one used for bile acid synthesis (26). It is totally unknown at present how and where CE taken up through SR-BI is transported in the liver cells and even in the CHO cells (8). In the present study, we demonstrated that CLAMP has no effect either on HDL binding to SR-BI or on the subsequent uptake of HDL CE, but CLAMP may modulate the intracellular transport and metabolism of CE taken up from HDL (Table 2 and Fig. 7). Thus, it can be speculated that CLAMP may function to connect SR-BI with the cellular machineries for intracellular cholesterol transport and/or metabolism. Besides CLAMP, we identified several proteins that interact with the SR-BI C-terminal domain on the affinity column chromatography (Fig. 1A). Although we have not characterized those proteins yet, they may interact directly with the SR-BI C-terminal domain or indirectly with other PDZ domains of CLAMP through which they associate with the SR-BI C-terminal domain. Isolation of the protein(s) interacting with other PDZ domains of CLAMP may help to understand the mechanism of intracellular cholesterol transport in the liver more fully.
Acknowledgments
We thank Dr. Yoshimi Takai (Osaka University Medical School, Osaka) for advice on the GST fusion-protein affinity chromatography and Dr. Kazuo Kobayashi (Osaka University) for technical assistance on the preparation of rat liver sinusoidal and canalicular membranes. M.I. is a Special Postdoctoral Researcher.
Abbreviations
- SR-BI
scavenger receptor class B type I
- HDL
high-density lipoprotein
- UC
unesterified cholesterol
- CE
cholesteryl ester
- CHO
Chinese hamster ovary
- DiI
1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine percholate
- GST
glutathione S-transferase
Footnotes
Data deposition: The nucleotide sequence reported in this paper has been deposited in the GenBank database (accession no. AF116896).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.100114397.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.100114397
References
- 1.Gordon D J, Rifkind B M. N Engl J Med. 1989;321:1311–1316. doi: 10.1056/NEJM198911093211907. [DOI] [PubMed] [Google Scholar]
- 2.Fielding C J, Fielding P E. J Lipid Res. 1995;36:211–228. [PubMed] [Google Scholar]
- 3.Acton S, Rigotti A, Landschulz K T, Xu S, Hobbs H H, Krieger M. Science. 1996;271:518–520. doi: 10.1126/science.271.5248.518. [DOI] [PubMed] [Google Scholar]
- 4.Kozarsky K F, Donahee M H, Rigotti A, Iqbal S N, Edelman E R, Krieger M. Nature (London) 1997;387:414–417. doi: 10.1038/387414a0. [DOI] [PubMed] [Google Scholar]
- 5.Fluiter K, van Berkel T J. Biochem J. 1997;326:515–519. doi: 10.1042/bj3260515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fluiter K, van der Westhuijzen D R, van Berkel T J. J Biol Chem. 1998;273:8434–8438. doi: 10.1074/jbc.273.14.8434. [DOI] [PubMed] [Google Scholar]
- 7.Webb N R, Connel P M, Graf G A, Smart E J, de Villiers W J, de Beer F C, van der Westhuyzen D R. J Biol Chem. 1998;273:15241–15248. doi: 10.1074/jbc.273.24.15241. [DOI] [PubMed] [Google Scholar]
- 8.Gu X, Trigatti B, Xu S, Acton S, Babitt J, Krieger M. J Biol Chem. 1998;273:26338–26348. doi: 10.1074/jbc.273.41.26338. [DOI] [PubMed] [Google Scholar]
- 9.Greenwalt D E, Lipsky R H, Ockenhouse C F, Ikeda H, Tandon N N, Jamieson G A. Blood. 1992;80:1105–1115. [PubMed] [Google Scholar]
- 10.Basu S K, Goldstein J L, Anderson G W, Brown M S. Proc Natl Acad Sci USA. 1976;73:3178–3182. doi: 10.1073/pnas.73.9.3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McFarlane A S. Nature (London) 1948;182:53–54. [Google Scholar]
- 12.Roberts D C K, Miller N E, Price S G L, Crook D, Cortese C, LaVille A, Masana L, Lewis I. Biochem J. 1985;226:319–322. doi: 10.1042/bj2260319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldstein J L, Basu S K, Brown M S. Methods Enzymol. 1983;98:241–260. doi: 10.1016/0076-6879(83)98152-1. [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi K, Sogame Y, Hara H, Hayashi K. J Biol Chem. 1990;265:7737–7741. [PubMed] [Google Scholar]
- 15.James P, Halladay J, Craig E A. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finkelstein D B, Strausberg S. Mol Cell Biol. 1983;3:1625–1633. doi: 10.1128/mcb.3.9.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shiratsuchi A, Kawasaki Y, Ikemoto M, Arai H, Nakanishi Y. J Biol Chem. 1999;274:5901–5908. doi: 10.1074/jbc.274.9.5901. [DOI] [PubMed] [Google Scholar]
- 18.Fukasawa M, Adachi H, Hirota K, Tsujimoto M, Arai H, Inoue K. Exp Cell Res. 1996;222:246–250. doi: 10.1006/excr.1996.0030. [DOI] [PubMed] [Google Scholar]
- 19.Connely M A, Klein S M, Azhar S, Abumrad N A, Williams D L. J Biol Chem. 1999;274:41–47. doi: 10.1074/jbc.274.1.41. [DOI] [PubMed] [Google Scholar]
- 20.Bligh E G, Dyer W J. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 21.Nishikawa K, Arai H, Inoue K. J Biol Chem. 1990;265:5226–5231. [PubMed] [Google Scholar]
- 22.Kocher O, Comella N, Tognazzi K, Brown L F. Lab Invest. 1998;78:117–125. [PubMed] [Google Scholar]
- 23.Custer M, Spindler B, Verrey F, Murer H, Biber J. Am J Physiol. 1997;273:F801–F806. doi: 10.1152/ajprenal.1997.273.5.F801. [DOI] [PubMed] [Google Scholar]
- 24.Schwartz C C, Zech L A, van der Broek J M, Cooper P S. J Clin Invest. 1993;91:923–938. doi: 10.1172/JCI116314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stange E F. Biochem Soc Trans. 1987;15:189–192. doi: 10.1042/bst0150189. [DOI] [PubMed] [Google Scholar]
- 26.Nervi F, Marinovic I, Rigotti A, Ulloa N. J Clin Invest. 1988;82:1818–1825. doi: 10.1172/JCI113797. [DOI] [PMC free article] [PubMed] [Google Scholar]