Abstract
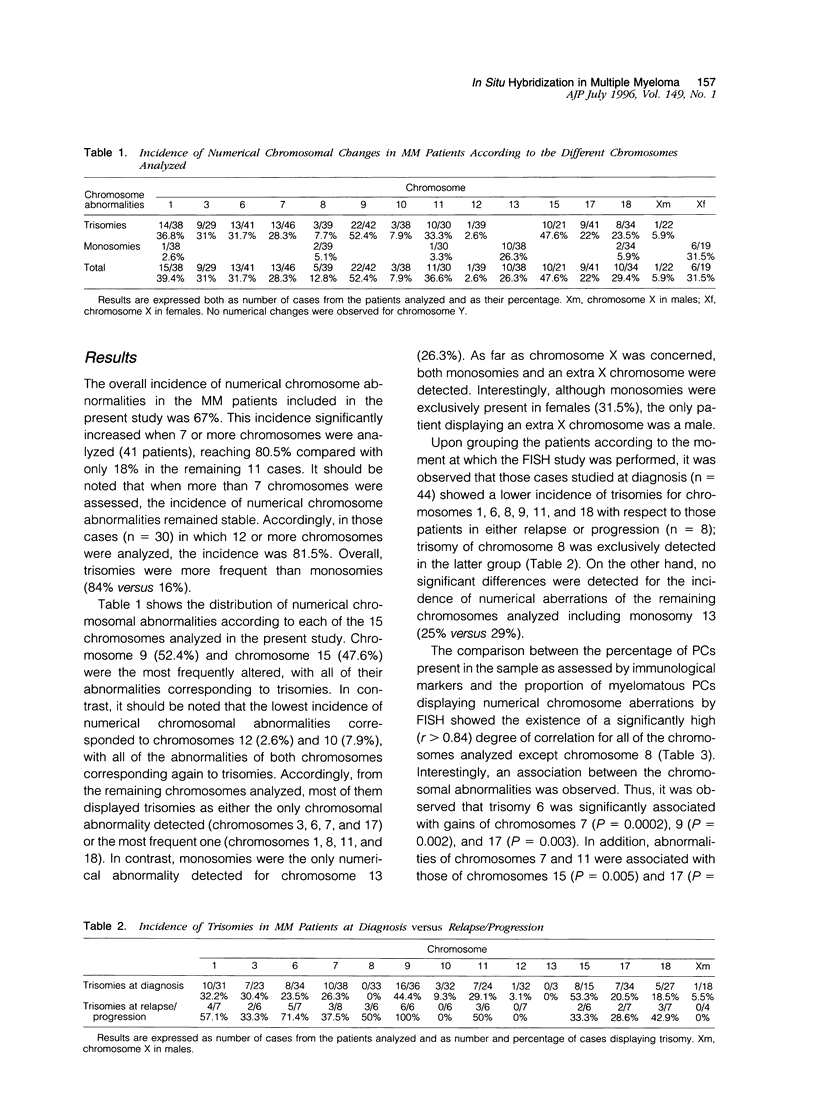
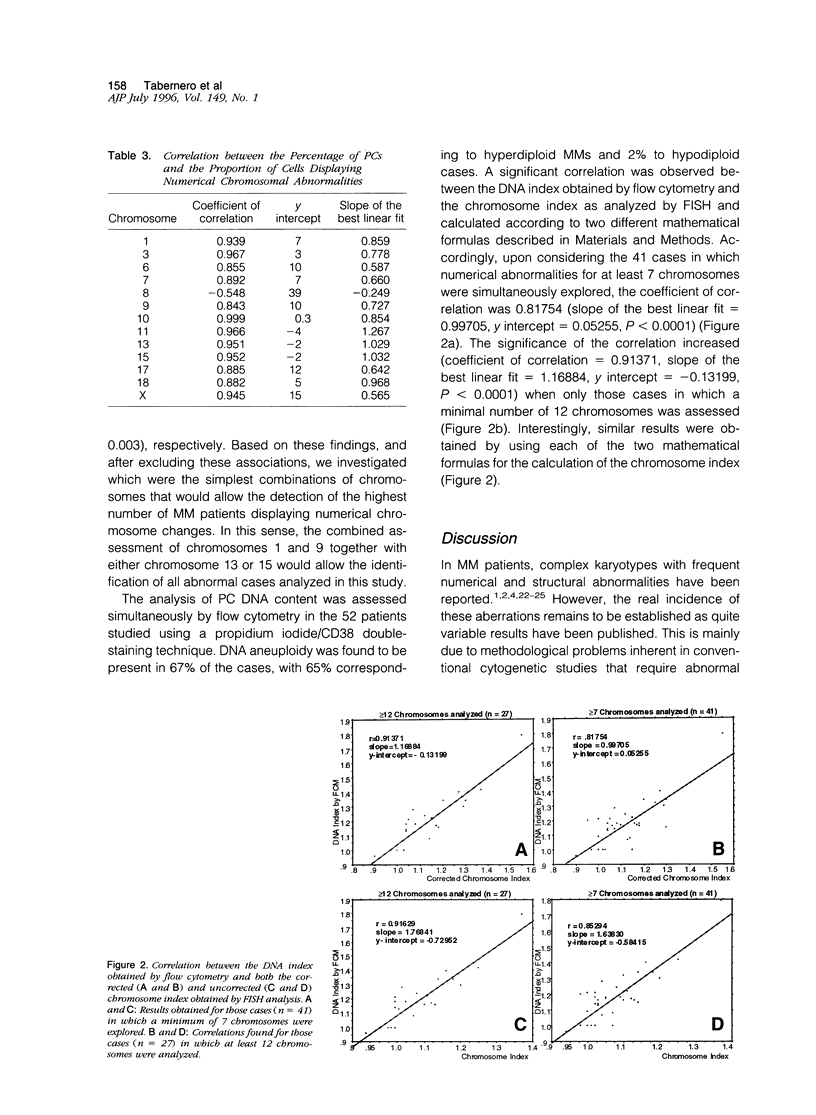
The presence of complex karotypes with frequent numerical and structural abnormalities has been reported in 20 to 50% of multiple myeloma (MM) patients. This variability is mainly due to the difficulty of conventional cytogenetics to obtain tumor metaphases representative of all possible neoplastic clones in MM. To gain insight into the real incidence of numerical chromosome changes in MM we have studied by fluorescence in situ hybridization technique 15 different human chromosomes, 1, 3, 6, 7, 8, 9, 10, 11, 12, 13, 15, 17, 18, X, and Y, in a series of 52 MM patients. In all cases, the DNA index assessed by a propidium iodide/CD38 double-staining technique with flow cytometry was simultaneously investigated for correlation, with fluorescence in situ hybridization results. Additional aims of this study were 1) to analyze whether the abnormalities detected were common to all plasma cells or were present in only a subpopulation of tumor cells, 2) to explore changes caused by disease progression, and 3) to establish possible associations among the altered chromosomes. Although the overall incidence of numerical abnormalities was 67%, this frequency increased to 80% in the 41 cases in which 7 or more chromosomes were analyzed. Trisomies were significantly more common than monosomies (84% versus 16%). Chromosomes 9 and 15 were the most frequently altered (52% and 48% of cases, respectively), with all of their abnormalities corresponding to trisomies. The most frequent losses involved chromosomes 13 (26%) and X in females (32%). Other common numerical changes corresponded to chromosomes 1 (39%), 11 (37%), 6 (32%), 3 (31%), 18 (29%), 7 (28%), and 17 (22%). By contrast, chromosomes 8(13%), 10(8%), and 12(3%) were rarely altered. DNA aneuploidy by flow cytometry was detected in 67% of patients, and a high degree of correlation was observed between the DNA index obtained by flow cytometry and the chromosome index derived from fluorescence in situ hybridization studies, calculated according to two mathematical formulas (coefficient of correlation of 0.82 and 0.91 when at least 7 or 12 chromosomes were considered, respectively). The frequency of numeric chromosome aberrations was higher in those patients with progressive disease and, interestingly, trisomy of chromosome 8 was exclusively detected in this latter group of patients. Our study shows that, with the exception of chromosome 8, a possible marker of clonal evolution, the numeric chromosome changes are present in nearly all malignant plasma cells (r > 0.84). Finally, frequent associations between chromosomal aberrations were observed (ie, chromosomes 6, 7, 9, and 17; 7 and 15; and 11 and 17). By excluding them, it was found that two triple combinations of chromosome-specific probes, chromosomes 1 and 9 together with either chromosome 13 or 15, could be a useful marker for detection of residual disease, as it permits the identification of most MM patients displaying numerical changes.
Full text
PDF

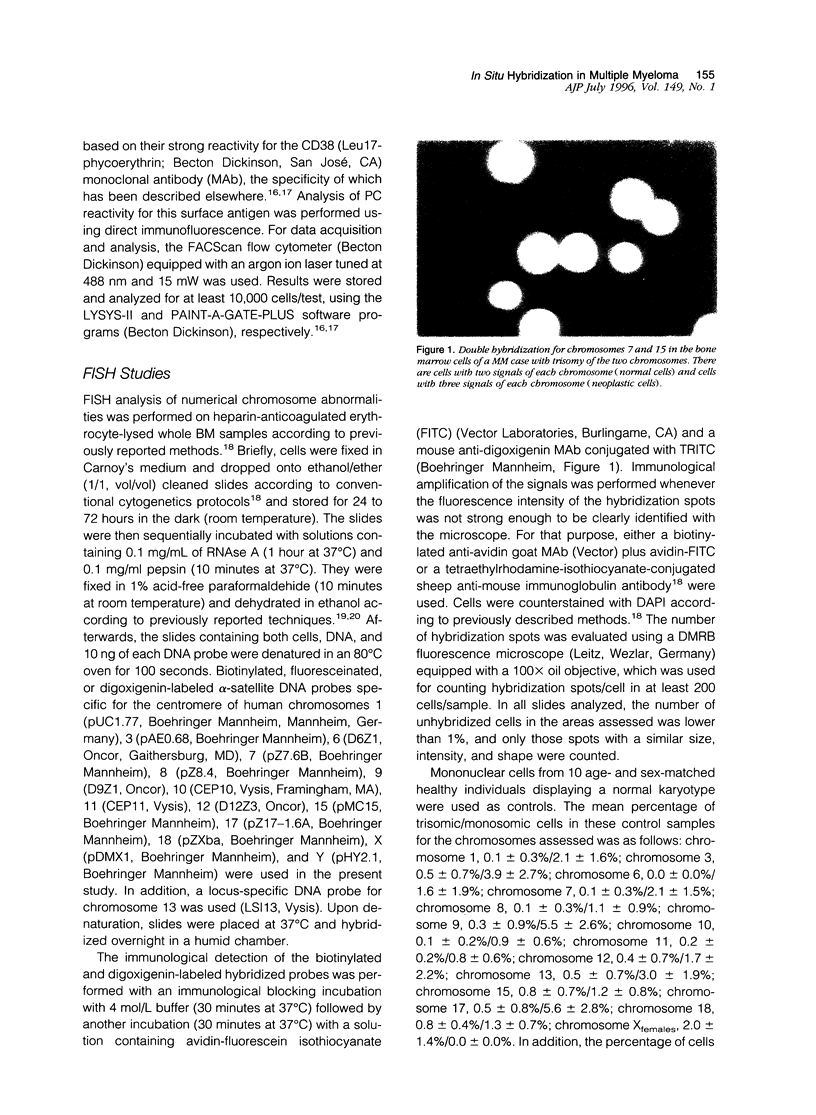




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anastasi J., Le Beau M. M., Vardiman J. W., Fernald A. A., Larson R. A., Rowley J. D. Detection of trisomy 12 in chronic lymphocytic leukemia by fluorescence in situ hybridization to interphase cells: a simple and sensitive method. Blood. 1992 Apr 1;79(7):1796–1801. [PubMed] [Google Scholar]
- Arnoldus E. P., Noordermeer I. A., Peters A. C., Voormolen J. H., Bots G. T., Raap A. K., van der Ploeg M. Interphase cytogenetics of brain tumors. Genes Chromosomes Cancer. 1991 Mar;3(2):101–107. doi: 10.1002/gcc.2870030204. [DOI] [PubMed] [Google Scholar]
- Atkin N. B. Chromosome 1 aberrations in cancer. Cancer Genet Cytogenet. 1986 Apr 15;21(4):279–285. doi: 10.1016/0165-4608(86)90206-2. [DOI] [PubMed] [Google Scholar]
- Cooke H. J., Hindley J. Cloning of human satellite III DNA: different components are on different chromosomes. Nucleic Acids Res. 1979 Jul 25;6(10):3177–3197. doi: 10.1093/nar/6.10.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewald G. W., Kyle R. A., Hicks G. A., Greipp P. R. The clinical significance of cytogenetic studies in 100 patients with multiple myeloma, plasma cell leukemia, or amyloidosis. Blood. 1985 Aug;66(2):380–390. [PubMed] [Google Scholar]
- Durie B. G., Salmon S. E. A clinical staging system for multiple myeloma. Correlation of measured myeloma cell mass with presenting clinical features, response to treatment, and survival. Cancer. 1975 Sep;36(3):842–854. doi: 10.1002/1097-0142(197509)36:3<842::aid-cncr2820360303>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Döhner H., Pohl S., Bulgay-Mörschel M., Stilgenbauer S., Bentz M., Lichter P. Trisomy 12 in chronic lymphoid leukemias--a metaphase and interphase cytogenetic analysis. Leukemia. 1993 Apr;7(4):516–520. [PubMed] [Google Scholar]
- Facon T., Lai J. L., Nataf E., Preudhomme C., Zandecki M., Hammad M., Wattel E., Jouet J. P., Bauters F. Improved cytogenetic analysis of bone marrow plasma cells after cytokine stimulation in multiple myeloma: a report on 46 patients. Br J Haematol. 1993 Aug;84(4):743–745. doi: 10.1111/j.1365-2141.1993.tb03155.x. [DOI] [PubMed] [Google Scholar]
- Facon T., Lai J. L., Nataf E., Preudhomme C., Zandecki M., Hammad M., Wattel E., Jouet J. P., Bauters F. Improved cytogenetic analysis of bone marrow plasma cells after cytokine stimulation in multiple myeloma: a report on 46 patients. Br J Haematol. 1993 Aug;84(4):743–745. doi: 10.1111/j.1365-2141.1993.tb03155.x. [DOI] [PubMed] [Google Scholar]
- Ferti A., Panani A., Arapakis G., Raptis S. Cytogenetic study in multiple myeloma. Cancer Genet Cytogenet. 1984 Jul;12(3):247–253. doi: 10.1016/0165-4608(84)90036-0. [DOI] [PubMed] [Google Scholar]
- García-Sanz R., Orfão A., González M., Moro M. J., Hernández J. M., Ortega F., Borrego D., Carnero M., Casanova F., Jiménez R. Prognostic implications of DNA aneuploidy in 156 untreated multiple myeloma patients. Castelano-Leonés (Spain) Cooperative Group for the Study of Monoclonal Gammopathies. Br J Haematol. 1995 May;90(1):106–112. doi: 10.1111/j.1365-2141.1995.tb03387.x. [DOI] [PubMed] [Google Scholar]
- Gould J., Alexanian R., Goodacre A., Pathak S., Hecht B., Barlogie B. Plasma cell karyotype in multiple myeloma. Blood. 1988 Feb;71(2):453–456. [PubMed] [Google Scholar]
- Grady D. L., Ratliff R. L., Robinson D. L., McCanlies E. C., Meyne J., Moyzis R. K. Highly conserved repetitive DNA sequences are present at human centromeres. Proc Natl Acad Sci U S A. 1992 Mar 1;89(5):1695–1699. doi: 10.1073/pnas.89.5.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirama T., Koeffler H. P. Role of the cyclin-dependent kinase inhibitors in the development of cancer. Blood. 1995 Aug 1;86(3):841–854. [PubMed] [Google Scholar]
- Kibbelaar R. E., van Kamp H., Dreef E. J., Wessels J. W., Beverstock G. C., Raap A. K., Fibbe W. E., den Ottolander G. J., Kluin P. M. Detection of trisomy 8 in hematological disorders by in situ hybridization. Cytogenet Cell Genet. 1991;56(3-4):132–136. doi: 10.1159/000133069. [DOI] [PubMed] [Google Scholar]
- Latreille J., Barlogie B., Johnston D., Drewinko B., Alexanian R. Ploidy and proliferative characteristics in monoclonal gammopathies. Blood. 1982 Jan;59(1):43–51. [PubMed] [Google Scholar]
- Laï J. L., Zandecki M., Mary J. Y., Bernardi F., Izydorczyk V., Flactif M., Morel P., Jouet J. P., Bauters F., Facon T. Improved cytogenetics in multiple myeloma: a study of 151 patients including 117 patients at diagnosis. Blood. 1995 May 1;85(9):2490–2497. [PubMed] [Google Scholar]
- Lee W. H., Bookstein R. E., Lee E. Y. Molecular biology of the human retinoblastoma gene. Immunol Ser. 1990;51:169–200. [PubMed] [Google Scholar]
- Nowell P. C., Besa E. C. Prognostic significance of single chromosome abnormalities in preleukemic states. Cancer Genet Cytogenet. 1989 Oct 1;42(1):1–7. doi: 10.1016/0165-4608(89)90002-2. [DOI] [PubMed] [Google Scholar]
- Oláh E., Balogh E., Kovács I., Kiss A. Abnormalities of chromosome 1 in relation to human malignant diseases. Cancer Genet Cytogenet. 1989 Dec;43(2):179–194. doi: 10.1016/0165-4608(89)90029-0. [DOI] [PubMed] [Google Scholar]
- Orfäo A., García-Sanz R., López-Berges M. C., Belén Vidriales M., González M., Caballero M. D., San Miguel J. F. A new method for the analysis of plasma cell DNA content in multiple myeloma samples using a CD38/propidium iodide double staining technique. Cytometry. 1994 Dec 1;17(4):332–339. doi: 10.1002/cyto.990170409. [DOI] [PubMed] [Google Scholar]
- Poddighe P. J., Moesker O., Smeets D., Awwad B. H., Ramaekers F. C., Hopman A. H. Interphase cytogenetics of hematological cancer: comparison of classical karyotyping and in situ hybridization using a panel of eleven chromosome specific DNA probes. Cancer Res. 1991 Apr 1;51(7):1959–1967. [PubMed] [Google Scholar]
- Prosser J., Frommer M., Paul C., Vincent P. C. Sequence relationships of three human satellite DNAs. J Mol Biol. 1986 Jan 20;187(2):145–155. doi: 10.1016/0022-2836(86)90224-x. [DOI] [PubMed] [Google Scholar]
- San Miguel J. F., García-Sanz R., González M., Moro M. J., Hernández J. M., Ortega F., Borrego D., Carnero M., Casanova F., Jiménez R. A new staging system for multiple myeloma based on the number of S-phase plasma cells. Blood. 1995 Jan 15;85(2):448–455. [PubMed] [Google Scholar]
- San Miguel J. F., González M., Gascón A., Moro M. J., Hernández J. M., Ortega F., Jimenez R., Guerras L., Romero M., Casanova F. Immunophenotypic heterogeneity of multiple myeloma: influence on the biology and clinical course of the disease. Castellano-Leones (Spain) Cooperative Group for the Study of Monoclonal Gammopathies. Br J Haematol. 1991 Feb;77(2):185–190. doi: 10.1111/j.1365-2141.1991.tb07975.x. [DOI] [PubMed] [Google Scholar]
- Shankey T. V., Rabinovitch P. S., Bagwell B., Bauer K. D., Duque R. E., Hedley D. W., Mayall B. H., Wheeless L., Cox C. Guidelines for implementation of clinical DNA cytometry. International Society for Analytical Cytology. Cytometry. 1993;14(5):472–477. doi: 10.1002/cyto.990140503. [DOI] [PubMed] [Google Scholar]
- Shimazaki C., Gotoh H., Ashihara E., Oku N., Inaba T., Murakami S., Itoh K., Ura Y., Nakagawa M., Fujita N. Immunophenotype and DNA content of myeloma cells in primary plasma cell leukemia. Am J Hematol. 1992 Mar;39(3):159–162. doi: 10.1002/ajh.2830390302. [DOI] [PubMed] [Google Scholar]
- Tabernero M. D., San Miguel J. F., Garcia J. L., Garcia-Isidoro M., Wiegant J., Ciudad J., Gonzalez M., Rios A., Raap A., Orfao A. Clinical, biological, and immunophenotypical characteristics of B-cell chronic lymphocytic leukemia with trisomy 12 by fluorescence in situ hybridization. Cytometry. 1995 Sep 15;22(3):217–222. doi: 10.1002/cyto.990220309. [DOI] [PubMed] [Google Scholar]
- Vindeløv L. L., Christensen I. J., Jensen G., Nissen N. I. Limits of detection of nuclear DNA abnormalities by flow cytometric DNA analysis. Results obtained by a set of methods for sample-storage, staining and internal standardization. Cytometry. 1983 Mar;3(5):332–339. doi: 10.1002/cyto.990030505. [DOI] [PubMed] [Google Scholar]
- Weh H. J., Gutensohn K., Selbach J., Kruse R., Wacker-Backhaus G., Seeger D., Fiedler W., Fett W., Hossfeld D. K. Karyotype in multiple myeloma and plasma cell leukaemia. Eur J Cancer. 1993;29A(9):1269–1273. doi: 10.1016/0959-8049(93)90071-m. [DOI] [PubMed] [Google Scholar]
- Wiegant J., Ried T., Nederlof P. M., van der Ploeg M., Tanke H. J., Raap A. K. In situ hybridization with fluoresceinated DNA. Nucleic Acids Res. 1991 Jun 25;19(12):3237–3241. doi: 10.1093/nar/19.12.3237. [DOI] [PMC free article] [PubMed] [Google Scholar]




