Abstract
Background
Tachycardia is commonly observed in hypertensive patients, predominantly mediated by regulatory mechanisms integrated within the autonomic nervous system. The genetic loci and genes associated with increased heart rate in hypertension, however, have not yet been identified.
Methods
An F2 intercross of Spontaneously Hypertensive Rats (SHR) × Brown Norway (BN) linkage analysis of quantitative trait loci mapping was utilized to identify candidate genes associated with an increased heart rate in arterial hypertension.
Results
Basal heart rate in SHR was higher compared to that of normotensive BN rats (365 ± 3 vs. 314 ± 6 bpm, p < 0.05 for SHR and BN, respectively). A total genome scan identified one quantitative trait locus in a 6.78 cM interval on rat chromosome 8 (8q22–q24) that was responsible for elevated heart rate. This interval contained 241 genes, of which 65 are known genes.
Conclusion
Our data suggest that an influential genetic region located on the rat chromosome 8 contributes to the regulation of heart rate. Candidate genes that have previously been associated with tachycardia and/or hypertension were found within this QTL, strengthening our hypothesis that these genes are, potentially, associated with the increase in heart rate in a hypertension rat model.
Background
Heart rate is primarily determined by regulatory mechanisms that are integrated within the autonomic nervous system. In arterial hypertension, an increased heart rate is commonly observed as a compensatory response to decreased stroke volume (due to either a diastolic or systolic myocardial dysfunction) [1,2]. Although an increased basal heart rate, accompanying myocardial dysfunction, might represent a secondary phenomenon in established hypertension, an early increase in heart rate may contribute directly to the pathogenesis of the disease [3-6]. In fact, tachycardia in arterial hypertension seems to be a heritable trait and independent of reflex responses [7]. Few studies, however, have focused on the identification of the genetic basis of tachycardia related to hypertension. A total genome scan to identify quantitative trait loci (QTL) provides a powerful tool to determine the chromosomal location of alleles and polygenes that contribute to complex diseases, such as autoimmune disorders [8,9], cardiovascular diseases [10,11], cancer [12,13], infectious diseases [14] and drug addiction [15,16].
Even though QTLs for complex diseases have been identified, little progress has been made towards the identification of the genes related to complex traits such as increased heart rate associated with hypertension. Recently, Jaworski et al. [17] reported a QTL in rat chromosome 2, centered around the D2Rat61/62 markers, that is related to the air-puff stimulus-induced bradycardia response. In agreement with Jaworski, Alemayehu et al. [18] described a heart rate QTL involved in bradycardia, induced by salt-load diet, on rat chromosome 2. We previously mapped five QTL's (two on chromosome 2, and one each on chromosomes 4, 8 and 16) involved in blood pressure variation in an F2 population of an intercross between Brown Norway (BN) and Spontaneously hypertensive rats (SHR) [19]. In this study, we used linkage analysis for QTL mapping and identification of candidate genes associated with basal levels of heart rate in the progenies of an F2 rat intercross between the BN and SHR strains.
Methods
Parental rat strain crosses and F2 phenotyping
To construct a detailed linkage map of all chromosomes and determine the genetic traits for tachycardia associated with hypertension we analyzed an intercross (F2) progeny (N = 188 rats) of the BN and the SHR strains, and normotensive (BN, N = 4) and hypertensive (SHR, N = 23) rats as parental strains. All animals were genotyped for 179 polymorphic markers distributed along the rat genome, according to a standard protocol [20]. Basal heart rate levels of the F2 progeny were obtained from a previously described phenotype database [19], and data from SHR and BN strains (Medical School, University of São Paulo) were evaluated in male 12–14 week old animals. Briefly, an arterial polyethylene catheter (PE-10, 0.28 mm ID, 0.61 mm OD, Biocorp Australia, Huntingdale, Victoria, Australia) was implanted under anesthesia (Pentobarbitone sodium, 30 mg/kg, i.p.) into the abdominal aorta through the left femoral artery at least 24 hours before measurements. Blood pressure was measured in non-anesthetized, freely moving rats, on a beat-to-beat basis at a 1.0 KHz of sample rate (DataQ Instruments, Inc. Ohio, USA) by connecting the arterial cannula to a pressure transducer (Statham P23Dd, Hato Rey, Puerto Rico). The pulse pressure signal from the transducer was fed to an amplifier (GPA-4 model 2, Stemtech, Inc., Wood Dale, IL, USA) and then to a 10-bit analog-to-digital converter (DataQ Instruments, Inc., Akron, OH, USA). Basal heart rate was derived from pulsatile blood pressure. All animal experimental procedures were followed in accordance to the guidelines for animal care and use of laboratory of the University of São Paulo, Brazil and the protocol approved by designated ethics committee.
Genetic marker map
The 188 animals from the F2 progeny were previously genotyped for 179 genetic markers distributed along the whole genome [19]. Although no new genotypes were incorporated for this study, we have recently re-accessed the genetic distances of each genotyped marker [21] through sequential consultation of different internet databases [22-24].
Quantitative trait loci (QTL) mapping
To assess the contribution of a putative QTL to the heart rate trait, we used the composite interval mapping method [25-28] in the F2 population (described above) using the Windows QTL Cartographer [29]. We then determined the significance of putative QTL's from the interval mapping scan based on a likelihood ratio (LR) threshold and considered 11.5 as suggestive linkage.
Criteria for candidate gene identification
Genes presented on the mapped heart rate QTL were listed by consulting the public database of the rat genome [30]. The selection of candidate genes proceeded according to the following criteria. Firstly, the known genes presented in the mapped QTL were listed; secondly, the known genes were submitted to gene ontology analysis (described below) for refinement of selection; and finally, genes were considered as candidate only if presented in a heart rate related phenotype, confirmed by citations on the NCBI PubMed browser [31].
Annotation of functional classes was carried out using information provided by the Gene Ontology database [32], accessed by NCBI's public repository [33]. The known genes found in the QTL were classified in specific function, process, or component classes through the GO terms assignation. The genes belonging to the same class were grouped and listed, and the candidate genes were elected from the most frequent gene ontology terms or according to gene ontology terms that are directly related to the heart rate trait.
Statistical analysis
Kolmogorov-Smirnov test (with Lilliefors' correction) was used to test data for normality of the estimated underlying population using SigmaStat 3.11 (Systat Software, Inc., San Jose, CA, USA). Normality was assumed for all parametric tests and regression procedures. All phenotype measurements (blood pressure and heart rate) were compared between each parental strain (SHR × BN) using the Student t test. Correlation between blood pressure and heart rate values was performed calculating the Pearson r correlation coefficient. Statistical significance was establish according to the P value (p < 0.05) for a two-tailed distribution. All data are reported as mean ± SEM.
Results
Hemodynamic correlations
As expected, inbred parental strains had different levels of basal blood pressure and heart rate (Table 1). Systolic (188 ± 2 vs. 127 ± 5 mm Hg), diastolic (138 ± 2 vs. 91 ± 4 mm Hg), and mean (162 ± 2 vs. 109 ± 5 mm Hg) blood pressure measurements were significantly increased in SHR compared to the normotensive BN strain. Moreover, the SHR strain presented an increase in heart rate levels (365 ± 3 vs. 314 ± 6 bpm, p < 0.0001) compared to BN. Although heart rate significantly correlated (p = 0.0069 for systolic blood pressure) with blood pressure levels (Table 2), the R squared value suggested that these variables are poorly correlated only (0.0335 for systolic blood pressure). Thus, we did not adjust heart rate values for systolic or diastolic blood pressure traits before QTL mapping analysis.
Table 1.
Systolic (SBP), diastolic (DBP), and mean (MBP) blood pressure, and heart rate (HR) in normotensive (Brown Norway) and hypertensive (SHR) rat strains.
|
Brown Norway (N = 04) |
SHR (N = 23) |
p | |
| SBP (mm Hg) | 127 ± 5 | 188 ± 2 | < 0.0001 |
| DBP (mm Hg) | 91 ± 4 | 138 ± 2 | < 0.0001 |
| MBP (mm Hg) | 109 ± 5 | 162 ± 2 | < 0.0001 |
| HR (bpm) | 314 ± 6 | 365 ± 3 | < 0.0001 |
Table 2.
The correlation between heart rate and mean (MBP), diastolic (DBP) and systolic (SBP) blood pressures obtained in an F2 (SHR × Brown Norway) population.
| R-squared | Pearson r | Confidence interval | P (two-tailed) | |
| vs. MBP | 0.0352 | -0.1877 | -0.3131 to -0.05587 | 0.0055 |
| vs. DBP | 0.0335 | -0.1830 | -0.3087 to -0.05098 | 0.0069 |
| vs. SBP | 0.0335 | -0.1830 | -0.3087 to -0.05105 | 0.0069 |
QTL mapping
The linkage signals for the composite interval mapping based on 179 markers along the 21 rat chromosomes (Figure 1) revealed the presence of a putative QTL responsible for an increase in heart rate in chromosome 8 (Figure 1). Figure 2 shows, in detail, the 6.78 cM heart rate QTL mapped on rat chromosome 8 (8q22–q24), flanked by APOA02 and R830 markers. In addition, the likelihood ratio statistics for the higher linkage signal of the putative QTL in chromosome 8 was 18.67. The estimated effects on heart rate are -27.3 bpm for the additive and 5.3 bpm for the dominant terms. Table 3 summarizes the allelic effects on heart rate levels for the APOA02, R1106, and R830 markers in homozygotes for either BN (BN/BN) and SHR (SHR/SHR), and heterozygotes (BN/SHR) in the F2 intercross (SHR × BN) population. Although no significant difference was observed in heart rate level for the APOA02 (p = 0.7614) and the R830 (p = 0.8963) markers, an allelic effect was observed for the R1106 marker (p = 0.0306). SHR homozygotes presented significantly lower heart rate values compared to BN homozygotes (377 ± 6 vs. 370 ± 3 vs. 354 ± 8 bpm, for BN/BN, BN/SHR, and SHR/SHR, respectively).
Figure 1.
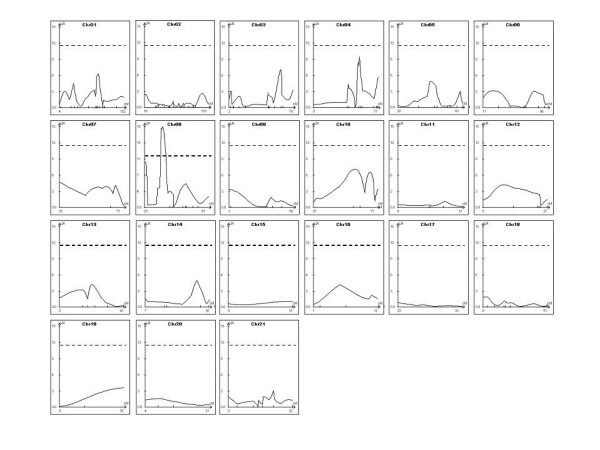
Likelihood ratio (LR) statistics for basal heart rate trait for all chromosomes in 188 rats from an F2 intercross (SHR × Brown Norway) population. Each panel represents the chromosome number with the genetic length of the chromosome (cM) on the abscissa, and the horizontal dashed line denotes an LR score of 11.5 as the threshold for suggestive linkage.
Figure 2.
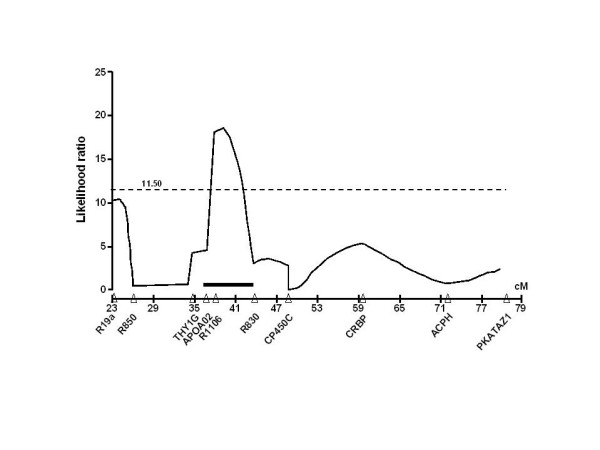
Rat chromosome 8 heart rate QTL in 188 rats from an F2 intercross (SHR × Brown Norway) population. The horizontal dashed line denotes the likelihood ratio (LR) of 11.5 as the threshold for suggestive linkage. See Table 3 for details of markers delimiting the QTL and the effects on heart rate, according to genotype.
Table 3.
Heart rate distributions, according to the genotype (BN/SHR, BN/SHR, or SHR/SHR), for the APOA02, R1106, and R830 markers that flanked the heart rate QTL mapped in a 188 F2 (SHR × Brown Norway) population.
| Markers | cM | Heart rate by genotype | One-way ANOVA (P) | Heart rate effect (bpm) | ||
| BN/BN | BN/SHR | SHR/SHR | ||||
| APOA02 | 36.87 | 370 ± 8 (N = 43) |
367 ± 3 (N = 91) |
364 ± 4 (N = 46) |
0.7614 | 5.81 |
| R1106 | 38.10 | 377 ± 6 (N = 29) |
370 ± 3 (N = 92) |
354 ± 8* (N = 37) |
0.0306 | 22.87 |
| R830 | 43.65 | 368 ± 8 (N = 41) |
368 ± 3 (N = 91) |
365 ± 4 (N = 43) |
0.8963 | 3.20 |
*p < 0.05 when compared to BN/BN.
Note that heart rate effect (bpm) represents the heart rate difference between BN/BN and SHR/SHR genotypes.
Candidate gene identification
The 6.78 cM heart rate QTL segment mapped on rat chromosome 8 harbours 241 genes. When only known genes were considered (Table 4), 176 genes were initially excluded. Gene ontology analysis then revealed that most of the 65 known genes were related to "metal ion binding", "protein binding", or "synaptic transmission" functions, and some of these presented "receptor/transporter/channel activity", some of which have an ion channel activity (Chrna5, Chrna3, Chrnb4, Htr3a, Htr3b, Fxyd2, and Hcn4). After gene ontology analysis, a further 48 genes were excluded, and 17 candidate genes remained. Ten of the genes investigated (Bace1, Timm8b, Ncam1, Slc37a4, Cul5, Fxyd2, Fxyd6, Ptpn9, Scamp2 and Scamp5) did not display any association with the heart rate trait and were eliminated. Candidate genes that were associated with the heart rate trait are described in detail in Table 5: Nicotinic acetylcholine receptor subtypes α3 (Chrna3), α5 (Chrna5), and β4 (Chrnb4); hyperpolarization-activated channel (Hcn4); 5-hydroxytryptamine (serotonin) receptor 3a (Htr3a) and 3b (Htr3b); and sodium channel (Scn2).
Table 4.
Known genes present on the heart rate QTL mapped on SHR chromosome 8, and classified according to Gene Ontology (GO) terms.
| GO Terms | Gene ID | Description |
| Axon | Bace1 | beta-site APP cleaving enzyme 1 |
| Metabolism | Mpi_mapped | mannose phosphate isomerase (mapped) |
| Idh3a | isocitrate dehydrogenase 3 (NAD+) alpha | |
| Man2c1 | mannosidase, alpha, class 2C, member 1 | |
| Pkm2 | pyruvate kinase, muscle | |
| Metal Ion Binding | Pts | 6-pyruvoyl-tetrahydropterin synthase |
| Fdx1 | ferredoxin 1 | |
| Cyp19 | cytochrome P450, subfamily 19 | |
| Cyp1a2 | cytochrome P450, 1a2 | |
| Cyp11a | cytochrome P450, subfamily 11A | |
| Ireb2 | iron-regulatory protein 2 | |
| Isl2 | insulin related protein 2 | |
| Cox5a | cytochrome c oxidase, subunit Va | |
| Nucleus | Cryab | crystallin, alpha B |
| Atm_mapped | ataxia telangiectasia mutated homolog (human) (mapped) | |
| Protein Folding/Processing | Hspb2 | heat shock 27 kD protein 2 |
| Hyou1 | hypoxia up-regulated 1 | |
| Pcsk7 | proprotein convertase subtilisin/kexin, type 7 | |
| Protein Binding | Mcam | l-gicerin |
| Thy1 | thymus cell antigen 1, theta | |
| Usp2 | ubiquitin specific protease 2 | |
| Zbtb16 | zinc finger and BTB domain containing 16 | |
| Cul5 | vasopressin-activated calcium-mobilizing receptor protein | |
| Rcn2 | reticulocalbin 2 | |
| Cd3d | CD3 antigen delta polypeptide | |
| Tagln | transgelin | |
| Receptor/Transporter/Channel Activity | Blr1 | Burkitt lymphoma receptor 1 |
| Il10ra | interleukin 10 receptor, alpha | |
| Neo1 | neogenin | |
| Slc37a4 | solute carrier family 37 (glycerol-6-phosphate transporter), member 4 | |
| Fxyd2 | FXYD domain-containing ion transport regulator 2 | |
| Fxyd6 | FXYD domain-containing ion transport regulator 6 | |
| Hcn4 | hyperpolarization-activated, cyclic nucleotide-gated K+ 4 | |
| Timm8b | translocase of inner mitochondrial membrane 8 homolog b (yeast) | |
| Scamp2 | secretory carrier membrane protein 2 | |
| Scamp5 | secretory carrier membrane protein 5 | |
| Synaptic Transmission | Chrna5 | cholinergic receptor, nicotinic, alpha polypeptide 5 |
| Chrna3 | cholinergic receptor, nicotinic, alpha polypeptide 3 | |
| Chrnb4 | cholinergic receptor, nicotinic, beta polypeptide 4 | |
| Scn2b | sodium channel, voltage-gated, type II, beta polypeptide | |
| Htr3b | 5-hydroxytryptamine (serotonin) receptor 3b | |
| Htr3a | 5-hydroxytryptamine (serotonin) receptor 3a | |
| Ncam1 | neural cell adhesion molecule 1 | |
| Transferase Activity | Hmbs | hydroxymethylbilane synthase |
| Dlat | dihydrolipoamide acetyltransferase | |
| Acat1 | acetyl-coenzyme A acetyltransferase 1 | |
| Clk3 | CDC-like kinase 3 | |
| Other Terms | Rps25 | ribosomal protein s25 |
| Treh | trehalase | |
| Pafah1b2 | platelet-activating factor acetylhydrolase alpha 2 subunit (PAF-AH alpha 2) | |
| Apoa5 | apolipoprotein A-V | |
| Tmprss5 | transmembrane protease, serine 5 (spinesin) | |
| Il18 | interleukin 18 | |
| Crabp1_mapped | cellular retinoic acid binding protein I (mapped) | |
| Psma4 | proteasome (prosome, macropain) subunit, alpha type 4 | |
| Cspg4 | membrane-spanning proteoglycan NG2 | |
| Ptpn9 | protein tyrosine phosphatase, non-receptor type 9 | |
| Sdfr1 | stromal cell derived factor receptor 1 | |
| Hexa | hexosaminidase A | |
| Myo9a | myosin IXA | |
| Ube4a | ubiquitin conjugation factor E4 A | |
| Cd276 | CD276 antigen | |
| No Terms | Phldb1 | pleckstrin homology-like domain, family B, member 1 |
| Crgl2 | olfactomedin-related protein | |
| Znf291 | KIAA1454-like protein |
Table 5.
Associated phenotypes described for 13 candidate genes present on the heart rate QTL, mapped on SHR chromosome 8.
| Gene | Name | Associated phenotype | Reference |
| Chrna3 Chrna5 Chrnb4 |
Cholinergic receptor, nicotinic, alpha polypeptide 3 and 5, and beta polypeptide 4 | - Autonomic nervous system influence on heart rate; - Alzheimer's disease; - Autosomal dominant nocturnal frontal lobe epilepsy. |
Bonati et al., 2000 [46] Bonati et al., 2002 [47] Duga et al., 2001 [43] Kawamata et al., 2002 [48] Lev-Lehman et al., 2001 [44] Liang Y 2005 [49] Rassadi, S 2005 [50] Rempel et al., 1998 [45] Wang et al., 2002 [51] Wang et al., 2003 [52] |
| Hcn4 | Hyperpolarization-activated, cyclic nucleotide – gated K+ 4 | - Familial sinus bradycardia; - Generation of pacemaker potentials in the sinoatrial node; - Cardiac arrhythmia. |
Leoni et al., 2005 [53] Milanesi et al., 2006 [42] Schulze-Bahr et al., 2003 [54] Stieber et al., 2006 [55] Stieber et al., 2003 [40] Ueda et al., 2004 [41] |
| Htr3a Htr3b |
5-hydroxytryptamine (serotonin) receptor 3a and 3b | - Modulatory role in the reflex control of the heart rate by cardiopulmonary receptors; - Nausea induced by paroxetine; - Fibromyalgia; - Numerous human neuropsychiatric disorders (female major depression, bipolar affective and schizophrenic). |
Aviado et al., 2001 [56] Frank et al., 2004 [57] Yamada et al., 2006 [58] Iidaka et al., 2005 [59] Niesler et al., 2001 [60] Sevoz et al., 1997 [61] Sugai et al., 2006 [62] Thoren, 1979 [63] Whalen et al., 2000 [64] |
| Scn2b | Sodium channel, voltage-gated, type II, beta polypeptide | - Essential hypertension; - Liddle's syndrome; - Idiopathic generalized epilepsy; - Neuropathic pain; - Cell adhesion and migration; - Chronic Heart Failure. |
Baker et al., 1998 [65] Botero-Velez et al., 1994 [66] Dong et al., 2002 [67] Haug et al., 2000 [68] Kim et al., 2005 [69] Pegoraro RJ et al., 2004 [70] Pertin et al., 2005 [71] Schild et al., 1996 [72] Snyder et al., 1995 [73] Su et al., 1996 [74] Rayner et al., 2003 [75] Zicha S et al., 2004 [76] |
Discussion and conclusion
A number of reports studying hypertensive patients have demonstrated an increased basal heart rate accompanying myocardial dysfunction [1-6]. Furthermore, a positive association between heart rate and all-cause mortality or cardiovascular mortality has been observed [5,6]. As such, it is important to elucidate the genetic factors that lead to increased heart rate levels. Based on the observation of an increased basal heart rate in the SHR strain, and using a powerful genomic exploratory approach, we mapped a new QTL on the rat chromosome 8 that is associated with basal levels of heart rate in SHR and identified several genes that could potentially be responsible for heart rate modulation.
We, herein, show that the parental SHR strain displayed increased heart rate values compared to normotensive BN rats. Moreover, in the F2 intercross animals, the allelic effects on heart rate levels for the R1106 marker suggest a complex interaction of alleles. The average heart rate values were lower in the homozygote SHR allele compared with the homozygote BN allele F2 animals. Furthermore, we observed a negative allelic effect for the R1106 marker (-22.9 bpm) that is concordant with the additive effect obtained by composite interval mapping (-27.3 bpm). These data suggest that the putative QTL, mapped on rat chromosome 8, harbours gene(s) that negatively influence the heart rate levels. Moreover, it should be taken into consideration that the algorithm of the composite interval mapping method considers the influence of other chromosomal regions or genes on the effect of the mapped QTL.
Total genome scan analysis has contributed to the discovery of several QTLs associated with different phenotypes in complex diseases. However, although relative success has been achieved in the identification of monogenic diseases [34,35], limited progress has been made in the identification and confirmation of candidate genes in complex diseases. Different chromosomal regions, associated with heart rate-phenotype traits, have been mapped in both patients and experimental animals [7,17,18,36,37]. Kreutz et al. [36] mapped a QTL on rat chromosome 3 that was associated with both basal heart rate and salt-induced changes in heart rate in an F2 intercross (SHRSP × WKY) population. Jaworski et al. [17] identified independent chromosomal regions related to bradycardia in response to mild stress (airpuff startle). Alemayehu et al. [18] identified one putative heart rate locus on rat chromosome 2 and confirmed its presence in different congenic strains after the substitution of the mapped interval from the SHR for the allelic region of a normotensive strain. Interestingly, Kren et al[38] reported a blood pressure QTL on chromosome 8 which maps nearby the heart rate-related QTL reported in the present study. However, a congenic SHR-Lx strain harbouring this particular region showed only changes in basal blood pressure and cardiac mass with no heart rate effects. Therefore, it is less likely that the same gene(s) are influencing the effects on heart rate here described. In a mouse model, Sugiyama et al. [37] conducted a QTL mapping analysis in an F2 progeny (CBA/CaJ × BALB/cJ) and identified 3 loci associated with heart rate, two on the mouse chromosome 2, and one on chromosome 15. We, herein, mapped a new QTL located in chromosome 8 and associated with a negative effect on heart rate in the SHR. Kahn and collaborators [39] found the rat chromosome 2 to be involved in heart rate, but were not able to confirm their findings in a congenic strain after the substitution of the mapped interval from the SHR for the allelic region of a normotensive BN strain. With regard to the rat chromosome 8, we previously mapped a putative QTL [19], flanked by R19 and R850 markers, associated with salt-induced changes in blood pressure that is located relatively close to the QTL, demonstrated herein, that is associated with heart rate trait.
In humans, loci associated with the autonomic nervous system's control of the heart rate have also been mapped [7]. Singh et al. [7] investigated 725 patients from the Framingham Heart Study and mapped 2 QTLs (chromosomes 2 and 15) associated with the frequency domain of heart rate variability indexes. Although the authors [7] have not mapped a QTL associated specifically with basal heart rate, they proposed that the region contains genes that share function for both basal heart rate and heart rate variability traits. Interestingly, the QTL mapped on chromosome 15 in humans is a syntenic region compared with the interval we mapped for basal heart rate traits in the rat chromosome 8.
In the present investigation, interval mapping revealed 17 genes that are potentially implicated in basal heart rate in the SHR. Among these candidate genes, just 4 families of genes were identified that maintained the association with the heart rate phenotype after the use of certain selection criteria; these genes were Scn2b (Sodium channel type II, beta polypeptide); Htr3a and Htr3b (5-Hydroxytryptamine receptors type 3); Hcn4 (Hyperpolarization-activated, cyclic nucleotide-gated K+ type 4); Chrna3, Chrna5, and Chrnb4 (Neuronal nicotinic acetylcholine receptor, alpha polypeptide 3 and 5, and beta polypeptide 4). Interestingly, association studies and genetic engineered mice data support the involvement of some of the heart rate candidate genes in arterial hypertension.
Knockout mice for the hyperpolarization-activated, cyclic nucleotide – gated K+ 4 (HCN4) gene have cardiac cells with "mature" pacemaker potentials, demonstrating that this gene that encodes a hyperpolarization-activated channel is essential for the proper generation of pacemaker potentials in the sinoatrial node [40]. In patients, a missense D553N mutation in the HCN4 gene was associated with sinus node dysfunction, QT prolongation in the electrocardiogram, polymorphic ventricular tachycardia, and syncope [41]. In addition, the presence of the S672R mutation in the coding region of the HCN4 gene, mimics a mild vagal stimulation in the sinoatrial node, slowing the heart rate by decreasing the inward diastolic current [42]. These data suggest that loss of function of the HCN4 gene is associated with sinus nodal dysfunction.
The neuronal nicotinic acetylcholine receptor (Chrn) subtypes α3, α5, and β4 are members of a ligand-gated ion channel family that affect sodium and potassium transients. Knockout mice strains lacking the genes encoding the subtypes α5 and β4 of the nicotinic receptors showed cardiac autonomic dysfunction, affecting the heart rate response to vagal electrical stimulation. In humans, although intragenic polymorphisms on the nicotinic receptor subunits have been identified [43-45], these gene mutations have not yet been associated with cardiovascular phenotypes.
One potential study limitation is the number of genetic markers used in the genome-mapping experiment. Although a large number of F2 analyses have been reported, genes responsible for SHR hypertension and/or heart rate related phenotypes have not been yet identified. One of the possible explanations for this fact is the small number of markers genotyped in these studies. We used 179 informative markers in our mapping experiment using an intercross between the SHR and BN strains. Although genetic distances and locations of these markers were updated before conducting our analysis, it is possible that QTLs important for the phenotype studied remain unidentified.
Our data suggest that genetic markers on the rat chromosome 8 contribute to heart rate control in experimental hypertension. The recognition of genetic determinants of heart rate may help gain insights into the pathophysiology of the autonomic nervous system and offer clues as to its modulation in hypertension. Further studies will be necessary to identify the specific gene(s) in chromosome 8 that are involved in hypertension-related tachychardia and characterize the molecular mechanisms underlying the genetic influence on heart rate in hypertensive rats.
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
GJJS carried out the molecular genetic studies, participated in the sequence alignment and drafted the manuscript. ACP carried out the molecular genetic studies and drafted the manuscript. EMK conceived of the study, and participated in its design and coordination. JEK conceived of the study, and participated in its design and coordination. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Acknowledgments
Acknowledgements
This study was supported by the Fundação de Amparo a Pesquisa do Estado de São Paulo (FAPESP #2001/00009-0), and the Fundação E. J. Zerbini.
Contributor Information
Gustavo JJ Silva, Email: gjjusto@usp.br.
Alexandre C Pereira, Email: lbmpereira@incor.usp.br.
Eduardo M Krieger, Email: edkrieger@incor.usp.br.
José E Krieger, Email: krieger@incor.usp.br.
References
- Ledingham JM, Cohen RD. The Role of the Heart in the Pathogenesis of Renal Hypertension. Lancet. 1963;186:979–981. doi: 10.1016/S0140-6736(63)90677-9. [DOI] [PubMed] [Google Scholar]
- Frohlich ED, Apstein C, Chobanian AV, Devereux RB, Dustan HP, Dzau V, Fauad-Tarazi F, Horan MJ, Marcus M, Massie B, et al. The heart in hypertension. N Engl J Med. 1992;327:998–1008. doi: 10.1056/NEJM199210013271406. [DOI] [PubMed] [Google Scholar]
- Palatini P, Julius S. The physiological determinants and risk correlations of elevated heart rate. Am J Hypertens. 1999;12:3S–8S. doi: 10.1016/S0895-7061(98)00207-6. [DOI] [PubMed] [Google Scholar]
- Palatini P, Casiglia E, Julius S, Pessina AC. High heart rate: a risk factor for cardiovascular death in elderly men. Arch Intern Med. 1999;159:585–592. doi: 10.1001/archinte.159.6.585. [DOI] [PubMed] [Google Scholar]
- Palatini P, Benetos A, Julius S. Impact of increased heart rate on clinical outcomes in hypertension: implications for antihypertensive drug therapy. Drugs. 2006;66:133–144. doi: 10.2165/00003495-200666020-00001. [DOI] [PubMed] [Google Scholar]
- Palatini P, Benetos A, Grassi G, Julius S, Kjeldsen SE, Mancia G, Narkiewicz K, Parati G, Pessina AC, Ruilope LM, Zanchetti A. Identification and management of the hypertensive patient with elevated heart rate: statement of a European Society of Hypertension Consensus Meeting. J Hypertens. 2006;24:603–610. doi: 10.1097/01.hjh.0000217838.49842.1e. [DOI] [PubMed] [Google Scholar]
- Singh JP, Larson MG, O'Donnell CJ, Tsuji H, Corey D, Levy D. Genome scan linkage results for heart rate variability (the Framingham Heart Study) Am J Cardiol. 2002;90:1290–1293. doi: 10.1016/S0002-9149(02)02865-5. [DOI] [PubMed] [Google Scholar]
- Merriman TR, Cordell HJ, Eaves IA, Danoy PA, Coraddu F, Barber R, Cucca F, Broadley S, Sawcer S, Compston A, Wordsworth P, Shatford J, Laval S, Jirholt J, Holmdahl R, Theofilopoulos AN, Kono DH, Tuomilehto J, Tuomilehto-Wolf E, Buzzetti R, Marrosu MG, Undlien DE, Ronningen KS, Ionesco-Tirgoviste C, Shield JP, Pociot F, Nerup J, Jacob CO, Polychronakos C, Bain SC, Todd JA. Suggestive evidence for association of human chromosome 18q12-q21 and its orthologue on rat and mouse chromosome 18 with several autoimmune diseases. Diabetes. 2001;50:184–194. doi: 10.2337/diabetes.50.1.184. [DOI] [PubMed] [Google Scholar]
- Todd JA. From genome to aetiology in a multifactorial disease, type 1 diabetes. Bioessays. 1999;21:164–174. doi: 10.1002/(SICI)1521-1878(199902)21:2<164::AID-BIES10>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Rapp JP. Genetic analysis of inherited hypertension in the rat. Physiol Rev. 2000;80:135–172. doi: 10.1152/physrev.2000.80.1.135. [DOI] [PubMed] [Google Scholar]
- Hamet P, Pausova Z, Adarichev V, Adaricheva K, Tremblay J. Hypertension: genes and environment. J Hypertens. 1998;16:397–418. doi: 10.1097/00004872-199816040-00001. [DOI] [PubMed] [Google Scholar]
- Fijneman RJ, de Vries SS, Jansen RC, Demant P. Complex interactions of new quantitative trait loci, Sluc1, Sluc2, Sluc3, and Sluc4, that influence the susceptibility to lung cancer in the mouse. Nat Genet. 1996;14:465–467. doi: 10.1038/ng1296-465. [DOI] [PubMed] [Google Scholar]
- Nagase H, Bryson S, Fee F, Balmain A. Multigenic control of skin tumour development in mice. Ciba Found Symp. 1996;197:156–68; discussion 168-80. doi: 10.1002/9780470514887.ch9. [DOI] [PubMed] [Google Scholar]
- Lengeling A, Pfeffer K, Balling R. The battle of two genomes: genetics of bacterial host/pathogen interactions in mice. Mamm Genome. 2001;12:261–271. doi: 10.1007/s003350040001. [DOI] [PubMed] [Google Scholar]
- Bice P, Foroud T, Bo R, Castelluccio P, Lumeng L, Li TK, Carr LG. Genomic screen for QTLs underlying alcohol consumption in the P and NP rat lines. Mamm Genome. 1998;9:949–955. doi: 10.1007/s003359900905. [DOI] [PubMed] [Google Scholar]
- Grisel JE. Quantitative trait locus analysis. Alcohol Res Health. 2000;24:169–174. [PMC free article] [PubMed] [Google Scholar]
- Jaworski RL, Jirout M, Closson S, Breen L, Flodman PL, Spence MA, Kren V, Krenova D, Pravenec M, Printz MP. Heart rate and blood pressure quantitative trait loci for the airpuff startle reaction. Hypertension. 2002;39:348–352. doi: 10.1161/hy0202.103419. [DOI] [PubMed] [Google Scholar]
- Alemayehu A, Breen L, Krenova D, Printz MP. Reciprocal rat chromosome 2 congenic strains reveal contrasting blood pressure and heart rate QTL. Physiol Genomics. 2002;10:199–210. doi: 10.1152/physiolgenomics.00065.2002. [DOI] [PubMed] [Google Scholar]
- Schork NJ, Krieger JE, Trolliet MR, Franchini KG, Koike G, Krieger EM, Lander ES, Dzau VJ, Jacob HJ. A biometrical genome search in rats reveals the multigenic basis of blood pressure variation. Genome Res. 1995;5:164–172. doi: 10.1101/gr.5.2.164. [DOI] [PubMed] [Google Scholar]
- Jacob HJ, Lindpaintner K, Lincoln SE, Kusumi K, Bunker RK, Mao YP, Ganten D, Dzau VJ, Lander ES. Genetic mapping of a gene causing hypertension in the stroke-prone spontaneously hypertensive rat. Cell. 1991;67:213–224. doi: 10.1016/0092-8674(91)90584-L. [DOI] [PubMed] [Google Scholar]
- Soler JM, Pereira AC, Torres CH, Krieger JE. Gene by environment QTL mapping through multiple trait analyses in blood pressure salt-sensitivity: identification of a novel QTL in rat chromosome 5. BMC Med Genet. 2006;7:47. doi: 10.1186/1471-2350-7-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratmap http://ratmap.gen.gu.se/
- Rat Genome Database http://rgd.mcw.edu/
- Whitehead Institute - Rat Genome http://www-genome.wi.mit.edu/rat/public/
- Haley CS, Knott SA. A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity. 1992;69:315–324. doi: 10.1038/hdy.1992.131. [DOI] [PubMed] [Google Scholar]
- Zeng ZB. Precision mapping of quantitative trait loci. Genetics. 1994;136:1457–1468. doi: 10.1093/genetics/136.4.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng ZB. Theoretical basis for separation of multiple linked gene effects in mapping quantitative trait loci. Proc Natl Acad Sci U S A. 1993;90:10972–10976. doi: 10.1073/pnas.90.23.10972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- Windows QTL Cartographer http://statgen.ncsu.edu/qtlcart
- NCBI Map Viewer http://www.ncbi.nlm.nih.gov/mapview/
- NCBI PubMed http://www.pubmed.gov
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Center for Biotechnology Information http://www.ncbi.nlm.nih.gov/
- Gilliam TC, Tanzi RE, Haines JL, Bonner TI, Faryniarz AG, Hobbs WJ, MacDonald ME, Cheng SV, Folstein SE, Conneally PM, et al. Localization of the Huntington's disease gene to a small segment of chromosome 4 flanked by D4S10 and the telomere. Cell. 1987;50:565–571. doi: 10.1016/0092-8674(87)90029-8. [DOI] [PubMed] [Google Scholar]
- Kerem B, Rommens JM, Buchanan JA, Markiewicz D, Cox TK, Chakravarti A, Buchwald M, Tsui LC. Identification of the cystic fibrosis gene: genetic analysis. Science. 1989;245:1073–1080. doi: 10.1126/science.2570460. [DOI] [PubMed] [Google Scholar]
- Kreutz R, Struk B, Stock P, Hubner N, Ganten D, Lindpaintner K. Evidence for primary genetic determination of heart rate regulation: chromosomal mapping of a genetic locus in the rat. Circulation. 1997;96:1078–1081. doi: 10.1161/01.cir.96.4.1078. [DOI] [PubMed] [Google Scholar]
- Sugiyama F, Churchill GA, Li R, Libby LJ, Carver T, Yagami K, John SW, Paigen B. QTL associated with blood pressure, heart rate, and heart weight in CBA/CaJ and BALB/cJ mice. Physiol Genomics. 2002;10:5–12. doi: 10.1152/physiolgenomics.00002.2002. [DOI] [PubMed] [Google Scholar]
- Kren V, Pravenec M, Lu S, Krenova D, Wang JM, Wang N, Merriouns T, Wong A, St Lezin E, Lau D, Szpirer C, Szpirer J, Kurtz TW. Genetic isolation of a region of chromosome 8 that exerts major effects on blood pressure and cardiac mass in the spontaneously hypertensive rat. J Clin Invest. 1997;99:577–581. doi: 10.1172/JCI119198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan IM, Singletary E, Alemayehu A, Stanislaus S, Printz MP, Yaksh TL, Taylor P. Nicotinic receptor gene cluster on rat chromosome 8 in nociceptive and blood pressure hyperresponsiveness. Physiol Genomics. 2002;11:65–72. doi: 10.1152/physiolgenomics.00079.2002. [DOI] [PubMed] [Google Scholar]
- Stieber J, Wieland K, Stockl G, Ludwig A, Hofmann F. Bradycardic and proarrhythmic properties of sinus node inhibitors. Mol Pharmacol. 2006;69:1328–1337. doi: 10.1124/mol.105.020701. [DOI] [PubMed] [Google Scholar]
- Ueda K, Nakamura K, Hayashi T, Inagaki N, Takahashi M, Arimura T, Morita H, Higashiuesato Y, Hirano Y, Yasunami M, Takishita S, Yamashina A, Ohe T, Sunamori M, Hiraoka M, Kimura A. Functional characterization of a trafficking-defective HCN4 mutation, D553N, associated with cardiac arrhythmia. J Biol Chem. 2004;279:27194–27198. doi: 10.1074/jbc.M311953200. [DOI] [PubMed] [Google Scholar]
- Milanesi R, Baruscotti M, Gnecchi-Ruscone T, DiFrancesco D. Familial sinus bradycardia associated with a mutation in the cardiac pacemaker channel. N Engl J Med. 2006;354:151–157. doi: 10.1056/NEJMoa052475. [DOI] [PubMed] [Google Scholar]
- Duga S, Solda G, Asselta R, Bonati MT, Dalpra L, Malcovati M, Tenchini ML. Characterization of the genomic structure of the human neuronal nicotinic acetylcholine receptor CHRNA5/A3/B4 gene cluster and identification of novel intragenic polymorphisms. J Hum Genet. 2001;46:640–648. doi: 10.1007/s100380170015. [DOI] [PubMed] [Google Scholar]
- Lev-Lehman E, Bercovich D, Xu W, Stockton DW, Beaudet AL. Characterization of the human beta4 nAChR gene and polymorphisms in CHRNA3 and CHRNB4. J Hum Genet. 2001;46:362–366. doi: 10.1007/PL00010921. [DOI] [PubMed] [Google Scholar]
- Rempel N, Heyers S, Engels H, Sleegers E, Steinlein OK. The structures of the human neuronal nicotinic acetylcholine receptor beta2- and alpha3-subunit genes (CHRNB2 and CHRNA3) Hum Genet. 1998;103:645–653. doi: 10.1007/s004390050885. [DOI] [PubMed] [Google Scholar]
- Bonati MT, Asselta R, Duga S, Ferini-Strambi L, Oldani A, Zucconi M, Malcovati M, Dalpra L, Tenchini ML. Refined mapping of CHRNA3/A5/B4 gene cluster and its implications in ADNFLE. Neuroreport. 2000;11:2097–2101. doi: 10.1097/00001756-200007140-00008. [DOI] [PubMed] [Google Scholar]
- Bonati MT, Combi R, Asselta R, Duga S, Malcovati M, Oldani A, Zucconi M, Ferini-Strambi L, Dalpra L, Tenchini ML. Exclusion of linkage of nine neuronal nicotinic acetylcholine receptor subunit genes expressed in brain in autosomal dominant nocturnal frontal lobe epilepsy in four unrelated families. J Neurol. 2002;249:967–974. doi: 10.1007/s00415-002-0763-8. [DOI] [PubMed] [Google Scholar]
- Kawamata J, Shimohama S. Association of novel and established polymorphisms in neuronal nicotinic acetylcholine receptors with sporadic Alzheimer's disease. J Alzheimers Dis. 2002;4:71–76. doi: 10.3233/jad-2002-4201. [DOI] [PubMed] [Google Scholar]
- Liang Y, Salas R, Marubio L, Bercovich D, De Biasi M, Beaudet AL, Dani JA. Functional polymorphisms in the human beta4 subunit of nicotinic acetylcholine receptors. Neurogenetics. 2005;6:37–44. doi: 10.1007/s10048-004-0199-7. [DOI] [PubMed] [Google Scholar]
- Rassadi S, Krishnaswamy A, Pie B, McConnell R, Jacob MH, Cooper E. A null mutation for the alpha3 nicotinic acetylcholine (ACh) receptor gene abolishes fast synaptic activity in sympathetic ganglia and reveals that ACh output from developing preganglionic terminals is regulated in an activity-dependent retrograde manner. J Neurosci. 2005;25:8555–8566. doi: 10.1523/JNEUROSCI.1983-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Orr-Urtreger A, Chapman J, Rabinowitz R, Nachman R, Korczyn AD. Autonomic function in mice lacking alpha5 neuronal nicotinic acetylcholine receptor subunit. J Physiol. 2002;542:347–354. doi: 10.1113/jphysiol.2001.013456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Orr-Urtreger A, Chapman J, Rabinowitz R, Korczyn AD. Deficiency of nicotinic acetylcholine receptor beta 4 subunit causes autonomic cardiac and intestinal dysfunction. Mol Pharmacol. 2003;63:574–580. doi: 10.1124/mol.63.3.574. [DOI] [PubMed] [Google Scholar]
- Leoni AL, Marionneau C, Demolombe S, Le Bouter S, Mangoni ME, Escande D, Charpentier F. Chronic heart rate reduction remodels ion channel transcripts in the mouse sinoatrial node but not in the ventricle. Physiol Genomics. 2005;24:4–12. doi: 10.1152/physiolgenomics.00161.2005. [DOI] [PubMed] [Google Scholar]
- Schulze-Bahr E, Neu A, Friederich P, Kaupp UB, Breithardt G, Pongs O, Isbrandt D. Pacemaker channel dysfunction in a patient with sinus node disease. J Clin Invest. 2003;111:1537–1545. doi: 10.1172/JCI200316387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stieber J, Herrmann S, Feil S, Loster J, Feil R, Biel M, Hofmann F, Ludwig A. The hyperpolarization-activated channel HCN4 is required for the generation of pacemaker action potentials in the embryonic heart. Proc Natl Acad Sci U S A. 2003;100:15235–15240. doi: 10.1073/pnas.2434235100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviado DM, Guevara Aviado D. The Bezold-Jarisch reflex. A historical perspective of cardiopulmonary reflexes. Ann N Y Acad Sci. 2001;940:48–58. [PubMed] [Google Scholar]
- Frank B, Niesler B, Bondy B, Spath M, Pongratz DE, Ackenheil M, Fischer C, Rappold G. Mutational analysis of serotonin receptor genes: HTR3A and HTR3B in fibromyalgia patients. Clin Rheumatol. 2004;23:338–344. doi: 10.1007/s10067-004-0927-2. [DOI] [PubMed] [Google Scholar]
- Yamada K, Hattori E, Iwayama Y, Ohnishi T, Ohba H, Toyota T, Takao H, Minabe Y, Nakatani N, Higuchi T, Detera-Wadleigh SD, Yoshikawa T. Distinguishable Haplotype Blocks in the HTR3A and HTR3B Region in the Japanese Reveal Evidence of Association of HTR3B with Female Major Depression. Biol Psychiatry. 2006;60:192–201. doi: 10.1016/j.biopsych.2005.11.008. [DOI] [PubMed] [Google Scholar]
- Iidaka T, Ozaki N, Matsumoto A, Nogawa J, Kinoshita Y, Suzuki T, Iwata N, Yamamoto Y, Okada T, Sadato N. A variant C178T in the regulatory region of the serotonin receptor gene HTR3A modulates neural activation in the human amygdala. J Neurosci. 2005;25:6460–6466. doi: 10.1523/JNEUROSCI.5261-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niesler B, Weiss B, Fischer C, Nothen MM, Propping P, Bondy B, Rietschel M, Maier W, Albus M, Franzek E, Rappold GA. Serotonin receptor gene HTR3A variants in schizophrenic and bipolar affective patients. Pharmacogenetics. 2001;11:21–27. doi: 10.1097/00008571-200102000-00003. [DOI] [PubMed] [Google Scholar]
- Sevoz C, Callera JC, Machado BH, Hamon M, Laguzzi R. Role of serotonin3 receptors in the nucleus tractus solitarii on the carotid chemoreflex. Am J Physiol. 1997;272:H1250–9. doi: 10.1152/ajpheart.1997.272.3.H1250. [DOI] [PubMed] [Google Scholar]
- Sugai T, Suzuki Y, Sawamura K, Fukui N, Inoue Y, Someya T. The effect of 5-hydroxytryptamine 3A and 3B receptor genes on nausea induced by paroxetine. Pharmacogenomics J. 2006 doi: 10.1038/sj.tpj.6500382. [DOI] [PubMed] [Google Scholar]
- Thoren P. Role of cardiac vagal C-fibers in cardiovascular control. Rev Physiol Biochem Pharmacol. 1979;86:1–94. doi: 10.1007/BFb0031531. [DOI] [PubMed] [Google Scholar]
- Whalen EJ, Johnson AK, Lewis SJ. Functional evidence for the rapid desensitization of 5-HT(3) receptors on vagal afferents mediating the Bezold-Jarisch reflex. Brain Res. 2000;873:302–305. doi: 10.1016/S0006-8993(00)02526-9. [DOI] [PubMed] [Google Scholar]
- Baker EH, Dong YB, Sagnella GA, Rothwell M, Onipinla AK, Markandu ND, Cappuccio FP, Cook DG, Persu A, Corvol P, Jeunemaitre X, Carter ND, MacGregor GA. Association of hypertension with T594M mutation in beta subunit of epithelial sodium channels in black people resident in London. Lancet. 1998;351:1388–1392. doi: 10.1016/S0140-6736(97)07306-6. [DOI] [PubMed] [Google Scholar]
- Botero-Velez M, Curtis JJ, Warnock DG. Brief report: Liddle's syndrome revisited--a disorder of sodium reabsorption in the distal tubule. N Engl J Med. 1994;330:178–181. doi: 10.1056/NEJM199401203300305. [DOI] [PubMed] [Google Scholar]
- Dong YB, Plange-Rhule J, Owusu I, Micah F, Eastwood JB, Carter ND, Saggar-Malik AK, Cappuccio FP, Jeffery S. T594M mutation of the beta-subunit of the epithelial sodium channel in Ghanaian populations from Kumasi and London and a possible association with hypertension. Genet Test. 2002;6:63–65. doi: 10.1089/109065702760093942. [DOI] [PubMed] [Google Scholar]
- Haug K, Sander T, Hallmann K, Rau B, Dullinger JS, Elger CE, Propping P, Heils A. The voltage-gated sodium channel beta2-subunit gene and idiopathic generalized epilepsy. Neuroreport. 2000;11:2687–2689. doi: 10.1097/00001756-200008210-00016. [DOI] [PubMed] [Google Scholar]
- Kim DY, Ingano LA, Carey BW, Pettingell WH, Kovacs DM. Presenilin/gamma-secretase-mediated cleavage of the voltage-gated sodium channel beta2-subunit regulates cell adhesion and migration. J Biol Chem. 2005;280:23251–23261. doi: 10.1074/jbc.M412938200. [DOI] [PubMed] [Google Scholar]
- Pegoraro RJ, Roberts CB, Rom L, Moodley J. T594M mutation of the epithelial sodium channel beta-subunit gene in pre-eclampsia and eclampsia in Black South African women. Bjog. 2004;111:1012–1013. doi: 10.1111/j.1471-0528.2004.00221.x. [DOI] [PubMed] [Google Scholar]
- Pertin M, Ji RR, Berta T, Powell AJ, Karchewski L, Tate SN, Isom LL, Woolf CJ, Gilliard N, Spahn DR, Decosterd I. Upregulation of the voltage-gated sodium channel beta2 subunit in neuropathic pain models: characterization of expression in injured and non-injured primary sensory neurons. J Neurosci. 2005;25:10970–10980. doi: 10.1523/JNEUROSCI.3066-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schild L, Lu Y, Gautschi I, Schneeberger E, Lifton RP, Rossier BC. Identification of a PY motif in the epithelial Na channel subunits as a target sequence for mutations causing channel activation found in Liddle syndrome. Embo J. 1996;15:2381–2387. [PMC free article] [PubMed] [Google Scholar]
- Snyder PM, Price MP, McDonald FJ, Adams CM, Volk KA, Zeiher BG, Stokes JB, Welsh MJ. Mechanism by which Liddle's syndrome mutations increase activity of a human epithelial Na+ channel. Cell. 1995;83:969–978. doi: 10.1016/0092-8674(95)90212-0. [DOI] [PubMed] [Google Scholar]
- Su YR, Rutkowski MP, Klanke CA, Wu X, Cui Y, Pun RY, Carter V, Reif M, Menon AG. A novel variant of the beta-subunit of the amiloride-sensitive sodium channel in African Americans. J Am Soc Nephrol. 1996;7:2543–2549. doi: 10.1681/ASN.V7122543. [DOI] [PubMed] [Google Scholar]
- Rayner BL, Owen EP, King JA, Soule SG, Vreede H, Opie LH, Marais D, Davidson JS. A new mutation, R563Q, of the beta subunit of the epithelial sodium channel associated with low-renin, low-aldosterone hypertension. J Hypertens. 2003;21:921–926. doi: 10.1097/00004872-200305000-00016. [DOI] [PubMed] [Google Scholar]
- Zicha S, Maltsev VA, Nattel S, Sabbah HN, Undrovinas AI. Post-transcriptional alterations in the expression of cardiac Na+ channel subunits in chronic heart failure. J Mol Cell Cardiol. 2004;37:91–100. doi: 10.1016/j.yjmcc.2004.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]


