Abstract
Targeted disruption of the mouse TOP3α gene encoding DNA topoisomerase IIIα was carried out to study the physiological functions of the mammalian type IA DNA topoisomerase. Whereas heterozygous top3α+/− mutant mice were found to resemble phenotypically their TOP3α+/+ litermates, no viable top3α−/− homozygotes were found among over 100 progeny of top3α+/− intercrosses. Examination of embryos dissected from decidual swellings and in vitro culturing of blastocysts from top3α+/− intercrosses showed that implantation of top3α−/− embryos and the induction of decidualization could occur, but viability of these embryos was severely compromised at an early stage of development. The requirement of mouse DNA topoisomerase IIIα during early embryogenesis is discussed in terms of its plausible role in chromosome replication and its interaction with the RecQ/SGS1 family of DNA helicases, whose members include the Bloom’s syndrome and the Werner’s syndrome gene products.
Keywords: targeted gene disruption, Δtop3α mice, genome instability
Several recent studies of Bloom’s syndrome, Werner’s syndrome, and ataxia talangiectasia, three congenital diseases that exhibit elevated mitotic recombination rate and high incidence of cancer (1–3), have heightened interests on the physiological roles of mammalian DNA topoisomerase III (for reviews on DNA topoisomerases, see ref. 4 and references therein). The determinants of the first two, the BLM and WRN genes, encode proteins that are homologous to the budding yeast SGS1 gene product (1, 2), a DNA helicase that interacts with DNA topoisomerase III physically and functionally (5, 6). For human ataxia talangiectasia cells, which appear to lack a protein involved in cell-cycle regulation (7), overexpression of a truncated but not an intact DNA topoisomerase III was found to suppress their hyperrecombination phenotype (3). This finding was interpreted in terms of a recombinogenic DNA topoisomerase III in ataxia talangiectasia cells; thus overexpression of a truncated and inactive enzyme could exert a dominant negative effect (3).
Mammalian DNA topoisomerase III (8) belongs to the type IA DNA topoisomerase subfamily whose members also include bacterial DNA topoisomerases I and III, yeast DNA topoisomerase III, and the enzyme “reverse gyrase” found in hyperthermophiles (4). Information on the physiological roles of the type IA DNA topoisomerases came mostly from studies of the bacterial and yeast enzymes. The intracellular level of Escherichia coli DNA topoisomerase I is intricately regulated by transcription from multiple promoters (9, 10), and inactivation of the enzyme is lethal in the absence of a compensatory mutation (11, 12). Because several of the compensatory mutations were found to map in genes encoding the subunits of DNA gyrase, it was thought that in vivo the removal of negative supercoils by E. coli DNA topoisomerase I would counter the negative supercoiling action of gyrase to maintain the proper degree of DNA supercoiling (11–13). This interpretation was modified through the proposal of the twin-supercoiled-domain model of transcription, which postulates that both positive and negative supercoils may be generated by transcription and other processes involving the tracking of macromolecular assemblies along DNA (14). According to this model, DNA gyrase, which effectively removes positive supercoils, and DNA topoisomerase I, which effectively removes negative supercoils, can be viewed as a cooperative pair that act jointly to solve the problem of excessive supercoiling of intracellular DNA (14). In the absence of E. coli DNA topoisomerase I, excessive negative supercoiling of intracellular DNA may lead to aberrant processes such as R-loop formation between nascent RNA and the DNA template (15, 16), and the cells may die as a consequence. E. coli DNA topoisomerase III, on the other hand, is dispensable; mutants lacking the enzyme exhibit a significantly higher recombination rate between repetitive sequences, however (17).
In the budding yeast Saccharomyces cereviciae, there is only one type IA enzyme, DNA topoisomerase III. Yeast cells lacking the enzyme are viable, but their growth rate is reduced by nearly 2-fold and recombination between repetitive sequences is elevated substantially (18). In addition, yeast top3−/− diploid cells are unable to sporulate (18). Because of the low cellular level of DNA topoisomerase III in yeast and the presence of DNA topoisomerases I and II that are efficient in the removal of DNA supercoils, it is difficult to attribute the phenotypes of yeast top3 mutants to a loss of the supercoil-removal activity of DNA topoisomerase III (19–21). An alternative interpretation is that the yeast enzyme might have a significant role in the unlinking of parental strands at the final stage of chromosome replication and/or in the dissociation of structures in mitotic cells that could lead to recombination (5, 19–21). Because both the slow growth and hyperrecombination phenotype of yeast top3 mutants are suppressed by mutations in the SGS1 helicase gene, it is plausible that the DNA topoisomerase may act in conjunction with the SGS1 helicase in these processes (5). Whereas there is no direct evidence that yeast DNA topoisomerase III is a potent decatenase, it has been shown that purified E. coli DNA topoisomerase III, which resembles the yeast enzyme in several respects (21), is very efficient in unlinking intertwined parental strands in a plasmid DNA replication system (22).
Human DNA topoisomerase III was identified in 1996 and its structural gene mapped to chromosome 17p11.2–12 (8). Recent nucleotide sequencing results have suggested, however, that there is a variant of the enzyme encoded by a gene within the Ig lambda locus at chromosome 22q11–12 (23). This putative variant enzyme is tentatively designated DNA topoisomerase IIIβ and the activity reported earlier (8) as DNA topoisomerase IIIα. The likely presence of more than one type of IA enzyme in mammalian cells raises questions about their physiological functions and the possibility of their association with different DNA helicases. We therefore have initiated genetic studies of these enzymes in the mouse model. We report here our finding that inactivation of mouse DNA topoisomerase IIIα leads to early embryonic lethality.
MATERIALS AND METHODS
Construction of the Targeting Vector.
A mouse brain cDNA library in lambda-ZAPII (Stratagene) was screened by using 32P-labeled human TOP3α cDNA as the probe. Several positives were obtained, and partial nucleotide sequencing of the clones confirmed their identity as mouse TOP3α cDNA clones: they all contain ORFs encoding a protein highly homologous to human DNA topoisomerase IIIα (8) but differ significantly from the deduced amino acid sequence of DNA topoisomerase IIIβ (23). Based on the partial sequencing results, a pair of primers “YPRT” (5′-TACATCAGCTACCCCCGGACA-3′) and “RHFL” (5′-GCAGCAAGCCAGGAAATGGCG-3′), each denoted by four of the amino acids encoded by the cDNA at the location of the primer, were designed for amplification of mouse genomic DNA by PCR. This pair of primers, which are separated by about 250 bp in the cDNA, yielded a 1.2-Kb PCR product. This product was sequenced, and from the sequence a primer 5′-AACATAAACACAGATTCACTGAAG-3′ (the “FSES primer”) was synthesized. The YPRT and FSES pair of primers were found to give a clean 360-bp PCR product from mouse genomic DNA, and the pair were used for the PCR-based screening of the DuPont-Merck phage P1 library of mouse strain 129 genomic DNA (carried out by Genome Systems, St. Louis). Two clones containing overlapping DNA inserts of the TOP3α region (GS control nos. 10667 and 10668) were obtained. A 19.6-Kb region spanning the active site tyrosine region in clone 10668 was mapped for restriction sites by using a commercial kit for long template PCR (Boehringer Mannheim) and a pair of primers DYHL (5′-GACTATCATCTGTATGGCCAGAAT-3′) and GGCD (5′-CCTTCAAAGTCTCATCACATCCACC-3′). In each mapping experiment, one of the primers was 32P-labeled at its 5′-end, and partial digestion of the PCR product with a number of restriction enzymes was carried out separately to determine the positions of the sites relative to the radiolabeled end. Based on this restriction map, a 2.3-Kb EcoRI–KpnI fragment and a 6.2-Kb SspI–SspI fragment were selected for targeted deletion of a region containing the active site tyrosine of the enzyme. These fragments were inserted into a targeting vector pPGKneo/TK (kindly provided by A. McMahon, Harvard University).
Targeted Gene Disruption in Embryonic Stem Cells.
The targeting construct was electroporated into the TC1 line (24) of mouse strain 129 embryonic stem cells (kindly provided by P. Leder and C. Deng, Harvard Medical School), and neomycin-resistant and thymidine kinase-minus clones were selected. Restriction digests of DNA samples from about 420 candidate clones were then screened by blot hybridization, by using appropriate probes described in a later section. Two of the nine correctly targeted embryonic stem cell clones were injected into C57BL/6 blastocysts to generate chimeric animals (carried out at the Center for Animal Resource and Comparative Medicine, Brigham and Women’s Hospital, Boston).
Other Methods.
DNA samples from mouse tail biopsies, embryos dissected from decidual swellings, or the inner cell mass of cultured blastocysts were prepared by exhaustive digestion of the materials with proteinase K, followed by phenol and chloroform extraction and ethanol precipitation (25). Screening for germ-line chimeras that transmitted the Δtop3α allele, breeding of the chimeras with strain C57BL/6 females, intercrosses of top3α+/− heterozygotes, and histological sectioning of formaldehyde-fixed and paraffin-embedded decidual swellings were all done according to standard procedures (25, 26). Culturing of preimplantation embryos was carried out on mouse embryonic fibroblast feeder layer cells grown in 24-well culture dishes in Hepes-buffered DMEM supplemented with 20% fetal bovine serum. Blastocysts were flushed out of uterine horns with saline at 3.5 days postcoitum (dpc), individually placed in the culture dish wells, and incubated in 5% CO2 air at 37°C, as described (25).
RESULTS
Targeted Disruption of Mouse TOP3α Gene.
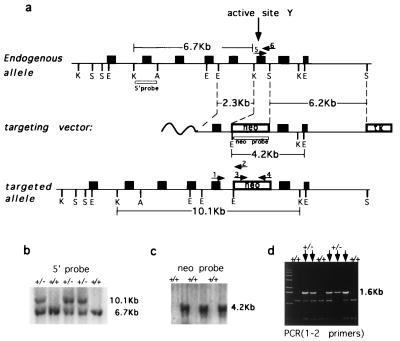
Fig. 1a depicts a region of the mouse TOP3α gene (Top), the construct used for targeted deletion of a DNA segment encoding the active site tyrosine region of the enzyme (Middle), and the expected Δtop3α allele after gene disruption (Bottom). DNA samples from tail biopsies of the F1 progeny were prepared, and these samples were used for genotyping animals with respect to the TOP3α locus by blot hybridization and PCR (Fig. 1 b–d). Heterozygous offspring were selected for intercrossing.
Figure 1.
Targeted disruption of mouse TOP3α gene. (a) Schematics of the region of mouse TOP3α containing the active site tyrosine (Top), the targeting vector (Middle), and the Δtop3α allele after gene disruption (Bottom). Filled boxes represent exons. K, S, E, and A denote KpnI, SspI, EcoRI, and ApaI restriction sites, respectively, and boxes denoted neo and TK represent the neomycin and herpes simplex virus thymidine kinase markers used. The locations of the probes used in blot hybridization (5′ probe and neo probe) and primers used in PCR (arrows numbered 1–6) also are indicated. The nucleotide sequences of primers 1–4 are, respectively, 5′-CGCGGAAAAGCTCTATACACAA-3′, 5′-TGTAGCGCCAAGTGCCAGCGGGG-3′, 5′-CGTGTTCCGGCTGTCAGCGCA-3′, 5′-ATCGCCATGGGTCACGACGAGAT-3′. Primers 5 and 6 are the same as the YPRT and the FSES primer described in Materials and Methods. (b and c) Genotyping of tail biopsies by blot hybridization. (b) KpnI digests were fractionated by agarose gel electrophoresis, and blot hybridization was carried out by using the 5′ probe. (c) EcoRI digests and the neo probe were used. The sizes of the fragments detected are in agreement with those expected from the drawings in a. (d) Genotyping of tail biopsies by PCR, using the pair of primers 1 and 2. The presence of the 1.6-kb product signified the presence of at least one copy of the mutant allele. The leftmost lane contained the “1-Kb ladder” size markers (GIBCO/BRL).
Requirement of Mouse DNA Topoisomerase IIIα During Embryogenesis.
A total of 131 progeny from intercrosses of top3α+/− heterozygotes were genotyped at 4–16 weeks of age. Among these progeny, 88 were found to be top3α+/− and the remaining 43 TOP3α+/+, with a ratio close to 2:1. There was no apparent difference between the top3α+/− and the TOP3α+/+ mice in terms of their growth and overt appearance. The conspicuous absence of top3α−/− progeny, however, suggested that the homozygous mutant is inviable. This notion was confirmed by examination of embryos at various stages of gestation.
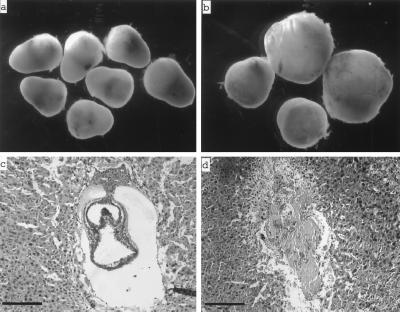
Fig. 2 (Upper) depicts individual decidual swellings dissected from uterine horns of female top3α+/− mice mated to heterozyous males at 8.5 and 10.5 dpc. The latter showed two distinct size classes (Fig. 2b). A total of seven embryos were retrieved from the 10.5-dpc decidual swellings, and all maternal tissues were removed. Genotyping of the embryonic samples by PCR showed that embryos from all five larger deciduae were either top3α+/− or TOP3α+/+, whereas the two smaller deciduae contained no organized embryos and embryonic tissues scraped off the specimens yielded no detectable DNA products. These results suggested that the smaller 10.5-dpc deciduae contained top3α−/− embryos that had undergone resorption. The 8.5-dpc decidual swellings appeared to be uniform in size (Fig. 2a). Genotyping of embryonic tissues from a total of 39 of these embryos showed, however, that 29 contained either top3α+/− or TOP3α+/+ embryos, and the remaining contained no embryonic material that could be genotyped. These and additional genotyping results of embryos at various stages of gestation are summarized in Table 1. Histological sectioning of 7.5-dpc deciduae also showed two distinct classes: those containing embryos of normal appearance that were readily identified to be either top3α+/− or TOP3α+/+ (Fig. 2c), and those containing no discernible embryonic material that could be genotyped by PCR (Fig. 2d).
Figure 2.
(a and b) Decidual swellings from intercrosses of top3α+/− mice 8.5 dpc (a) and 10.5 dpc (b). (c and d) Histological sections of 7.5-dpc deciduae from intercrosses of top3α+/− mice. Whereas these decidual swellings showed a uniform external appearance, two distinct types were revealed by sectioning: the majority contained embryos of normal appearance, such as the one shown in c, and the remaining were devoid of embryonic material, such as the one shown in d. Embryos of normal appearance were invariably found by genotyping to possess at least one copy of unaltered TOP3α. The bars correspond to a length of 100 μm.
Table 1.
Genotyping of embryos from intercrosses of top3α+/− heterozygotes
| Embryos, dpc | +/+ | +/− | −/− | Unidentified | Total |
|---|---|---|---|---|---|
| 15.5 | 4 | 6 | 0 | 3* | 13 |
| 10.5 | 3 | 2 | 0 | 2† | 7 |
| 8.5 | 8 | 21 | 0 | 10† | 39 |
| 7.5 | 2 | 7 | 0 | 6† | 15 |
*Resorption
Decidual swellings were empty or contained insufficient material for genotyping.
Culturing of Mouse top3α−/− Preimplantation Blastocysts in Vitro.
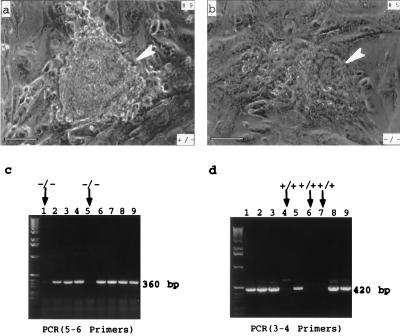
To examine further the effect of DNA topoisomerase IIIα inactivation on viability, preimplantation embryos were cultured on mouse feeder layer cells over a period of 8 days (Fig. 3). Among a total of 72 embryos examined, 63 showed normal trophoblast outgrowth and inner cell mass proliferation (Fig. 3a); 35 of these embryos were genotyped to be top3α+/− and the remaining 18 TOP3α+/+. For the other 19, 10 showed poor proliferation of inner cell mass of the embryos during the first few days after hatching, and total cessation of growth thereafter (Fig. 3b); PCR analysis of the inner cell mass of these embryos showed the pattern expected for top3α−/− cells. Of the remaining nine, seven failed to hatch or attach to the feeder layer and two showed poor trophoblast outgrowth and no identifiable inner cell mass. Taken together, these results and those presented in the section above provide strong evidence that embryos lacking DNA topoisomerase IIIα die at an early stage. Implantation of top3α−/− embryos and the induction of decidual reaction in the uterus were apparent, but viability of these embryos was severely compromised.
Figure 3.
Culturing of a top3α+/− (a) and a top3α−/− (b) blastocyst on mouse embryonic fibroblast feeder layer cells. Embryos were collected 3.5 dpc and cultured for 4 days in vitro and photographed. The heterozygote showed normal trophetoderm outgrowth and inner cell mass proliferation, and the homozygote showed poor trophoblast outgrowth and a rather small inner cell mass (indicated by an arrowhead in b). The bars correspond to a length of 100 μm. (c and d) Genotyping of nine embryos cultured in vitro, using the primers specified in Fig. 1 (a). The leftmost lane of each gel slab contained the 1-Kb ladder size markers (GIBCO/BRL). The embryos shown in a and b corresponded to samples 9 and 5, respectively.
DISCUSSION
The early lethality of mouse top3α−/− embryos shows that its cellular function can neither be substituted by its putative variant, DNA topoisomerase IIIβ, nor by the three other DNA topoisomerases I, IIα, and IIβ. The precise cellular function of mammalian DNA topoisomerase IIIα remains enigmatic. In vitro, recombinant human DNA topoisomerase III expressed in yeast cells was shown to relax only partially a highly negatively supercoiled DNA (8). Similar to the case of the budding yeast (21), the marginal relaxation activity of mammalian DNA topoisomerase IIIα, relative to the robust activities of mammalian DNA topoisomerases I, IIα, and IIβ, argues against a significant role of the enzyme in the removal of negative supercoils in vivo. The indispensability of the enzyme during embryonic development strongly suggests that the cellular process or processes catalyzed by it cannot be substituted by DNA topoisomerases I, IIα, and IIβ, and a role of the enzyme in the unlinking of parental strands during chromosome replication remains a likely function of the enzyme. If this hypothesis is true, inactivation of mammalian DNA topoisomerase IIIα is probably cell lethal. We do not know whether DNA topoisomerase IIIα and its putative variant DNA topoisomerase IIIβ are functionally distinct or differentially regulated, as the catalytic properties of the putative DNA topoisomerase IIIβ are unknown, and no information is presently available on how these enzymes are regulated during development.
The indispensability of mammalian DNA topoisomerase IIIα also provides insight on the possibility of the enzyme as a target of anticancer agents. In the past two decades, the DNA topoisomerases have emerged as important targets of antimicrobial and anticancer therapeutics (see for example, ref. 27). Whereas drugs acting on type IB and type IIA DNA topoisomerases are well known, no drug acting on type IA DNA topoisomerases has been developed. The majority of anticancer drugs targeting mammalian DNA topoisomerases, including camptothecin derivatives that target topoisomerase I and doxorubicin, etoposide, and mitoxanthrone that target one or both of the type IIA enzymes, act by trapping the covalent enzyme-DNA intermediate (28). In this mode of action, the drugs act by converting a normal cellular enzyme to a DNA-damaging agent, and the cytotoxicity of the drugs increases with increasing cellular concentration of the enzyme (28). Several other drugs, including fostriecin (29), merbarone (30), and the bis(2,6-dioxopiperazine) derivatives (31), act by inhibition of DNA topoisomerase IIα and perhaps IIβ as well. Preliminary studies suggest that in mammalian cells the concentration of DNA topoisomerase IIIα is much lower than those of the type IB and IIA topoisomerases (unpublished work), and therefore the enzyme is probably not an attractive target in the search of drugs that trap the enzyme-DNA covalent intermediate. The indispensability of the enzyme and its likely involvement in chromosome segregation suggest, however, that it might be a good target for drugs that inhibit its action.
Based on the findings in yeast, it appears likely that mammalian DNA topoisomerase IIIα may interact with one or more of the mammalian homologues of the yeast SGS1 protein. These proteins are commonly referred to as the E. coli RecQ helicase family of proteins, which, in addition to the BLM and WRN proteins mentioned earlier, also include the human RecQL protein and E. coli RecQ helicase (see for example, ref. 32). In mammalian cells, the presence of multiple RecQ-type proteins and the likely presence of two type IA DNA topoisomerases offer challenging opportunities in the study of their interactions and physiological roles. In view of the essential role of DNA topoisomerase IIIα in early embryogenesis and perhaps in the propagation of genetic information, such studies are likely to provide important clues on the molecular mechanisms of genetic instability in Bloom’s and Werner’s syndrome cells, and perhaps in ataxia talangiectasia cells as well.
Acknowledgments
We are most grateful to G. C. Li, H. Ouyang, E. Robertson, A. McMahon, and L. Berg for advice, A. McMahon and B. Jacques for the gene-targeting vector pGKneoTK, P. Leder and C. Deng for the TC1 cell line, and A. Sharpe and L. Du of the Center for Animal Resource and Comparative Medicine at Brigham and Women’s Hospital for expert help in the construction of chimeric mice. This work was supported by grants from the National Institutes of Health and an award of the Lucille P. Markey Charitable Trust to the Department of Molecular and Cellular Biology of Harvard University for exploratory research.
ABBREVIATION
- dpc
days postcoitum
References
- 1.Ellis N A, Groden J, Ye T Z, Straughen J, Lennon D J, Ciocci S, Proytcheva M, German J. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 2.Yu C-E, Oshima J, Fu Y H, Wijsman E M, Hisama F, Alisch R, Mathews S, Nakura J, Miki T, Ouais S, Martin G M, Mulligan J, Schellenberg G D. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 3.Fritz E, Elsea S H, Patel P I, Meyn M S. Proc Natl Acad Sci USA. 1997;94:4538–4542. doi: 10.1073/pnas.94.9.4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang J C. Annu Rev Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 5.Gangloff S, McDonald J P, Bendixen C, Arthur L, Rothstein R. Mol Cell Biol. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu J, Mullen J R, Brill S J, Kleff S, Romeo A M, Sternglanz R. Nature (London) 1996;383:678–679. doi: 10.1038/383678a0. [DOI] [PubMed] [Google Scholar]
- 7.Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, et al. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 8.Hanai R, Caron P C, Wang J C. Proc Natl Acad Sci USA. 1996;93:3653–3657. doi: 10.1073/pnas.93.8.3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qi H, Menzel R, Tse-Dinh Y-C. Mol Microbiol. 1996;21:703–711. doi: 10.1046/j.1365-2958.1996.241390.x. [DOI] [PubMed] [Google Scholar]
- 10.Qi H, Menzel R, Tse-Dinh Y-C. J Mol Biol. 1997;267:481–489. doi: 10.1006/jmbi.1997.0901. [DOI] [PubMed] [Google Scholar]
- 11.Pruss G J, Manes S H, Drlica K. Cell. 1982;31:35–42. doi: 10.1016/0092-8674(82)90402-0. [DOI] [PubMed] [Google Scholar]
- 12.DiNardo S, Voelkel S K, Sternglanz R, Reynolds A E, Wright A. Cell. 1982;31:43–55. doi: 10.1016/0092-8674(82)90403-2. [DOI] [PubMed] [Google Scholar]
- 13.Gellert M. Annu Rev Biochem. 1981;50:665–697. doi: 10.1146/annurev.bi.50.070181.004311. [DOI] [PubMed] [Google Scholar]
- 14.Liu L F, Wang J C. Proc Natl Acad Sci USA. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drolet M, Bi X, Liu L F. J Biol Chem. 1994;269:2068–2074. [PubMed] [Google Scholar]
- 16.Drolet M, Phoenix P, Menzel R, Masse E, Liu L F, Crouch R J. Proc Natl Acad Sci USA. 1996;92:3526–3530. doi: 10.1073/pnas.92.8.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schofield M A, Agbunag R, Michaels M L, Miller J H. J Bacteriol. 1992;174:5168–5170. doi: 10.1128/jb.174.15.5168-5170.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wallis J W, Chrebet G, Brodsky G, Rolfe M, Rothstein R. Cell. 1989;58:409–419. doi: 10.1016/0092-8674(89)90855-6. [DOI] [PubMed] [Google Scholar]
- 19.Wang J C, Caron P R, Kim R A. Cell. 1990;62:403–406. doi: 10.1016/0092-8674(90)90002-v. [DOI] [PubMed] [Google Scholar]
- 20.Wang J C. J Biol Chem. 1991;266:6659–6662. [PubMed] [Google Scholar]
- 21.Kim R A, Wang J C. J Biol Chem. 1992;267:17178–17185. [PubMed] [Google Scholar]
- 22.DiGate R, Marians K. J Biol Chem. 1988;263:13366–13373. [PubMed] [Google Scholar]
- 23.Kavasaki K, Minoshima S, Nakato E, Shibuya K, Shintani A, Schmeits J L, Wang J, Shimizu N. Genome Res. 1997;7:250–261. doi: 10.1101/gr.7.3.250. [DOI] [PubMed] [Google Scholar]
- 24.Deng C, Anthony W B, Zhou F, Kuo A, Leder P. Cell. 1996;84:911–921. doi: 10.1016/s0092-8674(00)81069-7. [DOI] [PubMed] [Google Scholar]
- 25.Hogan B, Beddinton R, Costatini F, Lacy E. Manipulating the Mouse Embryo: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. [Google Scholar]
- 26.Kaufman M H. The Atlas of Mouse Development. San Diego: Academic; 1992. [Google Scholar]
- 27.Liu L F. DNA Topoisomerase and Their Applications in Pharmacology. San Diego: Academic; 1994. [Google Scholar]
- 28.Liu L F. Annu Rev Biochem. 1989;58:351–375. doi: 10.1146/annurev.bi.58.070189.002031. [DOI] [PubMed] [Google Scholar]
- 29.Boritzki T J, Wolfard T S, Besserer J A, Jackson R C, Fry D W. Biochem Pharmacol. 1988;37:4063–4068. doi: 10.1016/0006-2952(88)90096-2. [DOI] [PubMed] [Google Scholar]
- 30.Drake F H, Hofmann G A, Mong S M, Bartus J O, Hertzberg R P, Johnson R K, Mattern M R, Mirabelli C K. Cancer Res. 1989;49:2578–2583. [PubMed] [Google Scholar]
- 31.Tanabe K, Ikegama Y, Ishida R, Andoh T. Cancer Res. 1991;51:4903–4908. [PubMed] [Google Scholar]
- 32.Ellis N A. Nature (London) 1995;381:110–111. doi: 10.1038/381110a0. [DOI] [PubMed] [Google Scholar]