Abstract
In humans, the poly(A)-binding proteins (PABPs) comprise a small nuclear isoform and a conserved gene family that displays at least three functional proteins : PABP1, inducible PABP (iPABP), and PABP3, plus four pseudogenes (1, 2, 3, and PABP4). In situ hybridization of PABP3 cDNA as the probe on metaphasic chromosomes have revealed five possible loci for this gene family at 2q21-q22, 13q11-q12, 12q13.3-q15, 8q22, and 3q24-q25. Amplifications of specific DNA fragments from a human-rodent somatic cell hybrid panel have allowed us to associate PABP1 and PABP3 with 8q22 and 13q11-q12, respectively. The iPABP gene has been assigned to chromosome 1. This result, compared with radiation hybrid database information, strengthens the location of this gene to 1p32-p36. The pseudogenes PABP4, 1, and 2 have been assigned to chromosomes 15, 4, and 14, respectively. Three loci detected on chromosome spreads are not associated with any amplified fragment. They might represent other related PABP genes not yet identified.
Keywords: Animals; Base Sequence; Blotting, Southern; Chromosome Mapping; Chromosomes, Human; genetics; DNA; genetics; DNA Primers; genetics; Humans; Hybrid Cells; In Situ Hybridization; Molecular Sequence Data; Multigene Family; Poly(A)-Binding Proteins; Polymerase Chain Reaction; Pseudogenes; RNA-Binding Proteins; genetics
Introduction
The poly(A)-binding proteins (PABP) bind to the poly(A) tail of eukaryotic mRNA. This interaction, among several roles (reviewed by Sachs and Wahle 1993), is notably determinant for the regulation of mRNA stability. In addition, although being anchored at the 3′ end of the mRNA, the PABPs participate in translation initiation by interacting with eIF4G, a component of the translation machinery located at the 5′ UTR of the messenger RNA (Kessler and Sachs 1998; Le et al. 1997; Tarun and Sachs 1996; Tarun et al. 1997). In humans, four PABP isoforms have been reported so far : PABP1 (70.3 kDa) is ubiquitously expressed (Blobel 1973; Grange et al. 1987), with a splicing variant PABPII (58.5 kDa) only being described in Genbank (accession no. Z48501); inducible PABP (iPABP; 72.4 kDa), which exhibits 77% identical amino-acids with PABP1, is found mainly in activated T cells and platelets (Houng et al. 1997; Yang et al. 1995); a smaller nuclear PABP isoform (PABP2; 32.7 kDa) is unrelated to PABP1 (Wahle 1991). In addition to these four functional genes, three human PABP-related pseudogenes, viz., 1, 2, and 3, display 95%, 93%, and 71% nucleotide identity with PABP1, respectively (accession nos. U64662, U60801, and U64661).
Recently, we have isolated and characterized two human testis transcripts, named PABP3 and PABP4, which exhibit 92% and 82% identical nucleotide residues with PABP1 mRNA, respectively (Feral et al., in preparation). PABP3 corresponds to a functional testis-specific protein exclusively expressed in round spermatids. In contrast, PABP4 displays a frameshift at codon 56 as compared with PABP1 and probably corresponds to a new PABP pseudogene.
Consequently, the PABP multigene family could be distributed on several loci in the human genome. Three of them have been mapped on chromosomes 3q22-q23, 12q13-q14, and 13q12-q13 (Morris and Bodger 1993). In the present study, in addition to these three locations, we have also detected specific signals on chromosomes 2 and 8 by using in situ hybridization. For the respective chromosomal localization of each PABP gene and pseudogene, we have designed sets of oligonucleotides to amplify specific fragments from a panel of somatic-cell hybrids. This has allowed us to map several PABP genes and pseudogenes and to localize precisely the functional PABP1 and PABP3 genes in 8q22 and 13q11-q12, respectively.
Materials and methods
Southern blot analysis of human genomic DNA
Human lymphocytic genomic DNA (20 μg) was digested by using the restriction enzymes HindIII, PstI, PvuII, or EcoR1 according to the manufacturer’s specifications, fractionated on a 0.8% (w/v) agarose gel, blotted onto nitro-cellulose membrane (Hybond N, Amersham, UK), and hybridized with the [32P]-labeled PABP3 insert, by using a protocol previously described (Giuili et al. 1992). The blot was washed, twice at 65°C, with 0.1 × SSC (1 × SSC=150 mM NaCl, 15 mM sodium citrate, pH 7), 0.1% SDS. The signals were detected by using a phosphofluoroimager (STORM, Molecular Dynamics).
In situ human gene mapping
In situ hybridization was carried out on chromosome preparations obtained from phytohemagglutinin-stimulated lymphocytes cultured for 72 h, 5-bromodeoxyuridine being added for the final 7 h of culture (60 μg/ml medium), to ensure posthybridization chromosomal R-banding of good quality. The PABP3 clone, which contains a cDNA insert of 2397 bp in pGEM 7Z, was tritium-labeled by nick-translation to a specific activity of 1 × 108 dpm μg−1. The radiolabeled probe was hybridized, at a final concentration of 200 ng per ml of hybridization solution, to metaphase spreads as previously described (Mattei et al. 1985). After being coated with nuclear track emulsion (Kodak NTB2), the slides were exposed for 19 days at +4°C and then developed. To avoid any slipping of silver grains during the banding procedure, chromosome spreads were first stained with buffered Giemsa solution, and metaphases were photographed. R-banding was then performed by the fluorochrome-photolysis-Giemsa method, and metaphases were rephotographed before analysis.
Chromosomal PABP assignment on a panel of somatic cell hybrids
The sequences of the oligonucleotide primers specific for PABP1, iPABP, PABP3, and PABP4 are shown (Table 1). Polymerase chain reaction (PCR) was carried out on a Perkin-Elmer GeneAmp 9600 apparatus in a final volume of 35 μl containing: either 40 ng genomic DNA from control samples or from somatic cell hybrids (HGMP Resource Centre, MRC, UK; Kelsell et al. 1995), or 1 ng cDNA plasmid specific for the probe tested, or a mixture of 1 ng of each cDNA plasmid specific for the three other PABPs. These DNA solutions were added to 50 pmol each primer, 0.3 mM each dNTP, 1.4 mM MgSO4, and 5 U Tfl polymerase (Promega). PCRs for PABP1, 3, and 4 were performed by using the “touchdown” strategy (Don et al. 1991). Briefly, the annealing temperature of the reaction was decreased every following cycle from 55°C to a touchdown at 45°C. At this temperature, 30 cycles were carried as follows : 1 min at 94°C (denaturation), 1 min at 45°C (annealing), and 1 min at 72°C (elongation). Following the last elongation step, the incubation was continued for an additional 10 min at 72°C. Conditions for amplification of the iPABP were identical, except that the touchdown PCR was from 62°C to 52°C, and the temperature of the annealing step in each cycle was 52°C. An aliquot (10 μl) of the PCR mixture was electrophoresed on a 3% (w/v) agarose gel, blotted on HybondN+ membrane (Amersham), and hybridized with [32P]-labeled oligonucleotides (45 × 106 cpm, 90 × 106 cpm/μg) specific for each PABP (Table 1). Hybridized PCR products were visualized by using a phosphofluoroimager (STORM, Molecular Dynamics).
Table 1.
PCR primers used for the chromosomal assignment of PABP genes. Name Name of the oligonucleotide primer; Position position in nucleotides of the oligonucleotide in each specific sequence, the positions referring to sequences as deposited in Genbank (PABP1: accession no. U68101, iPABP: accession no. U33818, PABP3: accession no. AF132026, PABP4: accession no. AF132027); Sequences sequence from the 5′ end to the 3′ end of each oligonucleotide
| Name | Position (nt) | Sequences | PCR product size | |
|---|---|---|---|---|
| PABP1 | 5′primer | 510–527 | 5′CCCAGCTGCTCCTAGACC3′ | 253 bp |
| 3′primer | 717–732 | 5′GAGTAGCTGCAGCGGCT3′ | ||
| probe | 581–601 | 5′CACAGCGTGTTGGTGAGTCTT3′ | ||
| iPABP | 5′primer | 2170–2191 | 5′GGACCTCAACACCAAGGATTAC3′ | 213bp |
| 3′primer | 2358–2382 | 5′GGGAAACGTAAGACTTGGGTACATC3′ | ||
| probe | 2240–2264 | 5′GCACCTAGAATTTTTCAATTATACG3′ | ||
| PABP3 | 5′primer | 20–36 | 5′GAGAATGAACCCCAGCA3′ | 178bp |
| 3′primer | 178–197 | 5′GTTCACATACGCGTAGTTGG3′ | ||
| probe | 99–120 | 5′ATGCTCTACGAGAAGTTCAGCC3′ | ||
| PABP4 | 5′primer | 1029–1046 | 5′CCTCCCCAGAAGAAGTAG3′ | 151bp |
| 3′primer | 1161–1131 | 5′GCTCGTACACTTGCCATCT3′ | ||
| prober | 1115–1131 | 5′TGCAAAGAACAGCGCCT3′ |
Results
Southern blot of human genomic DNA
Human lymphocytes genomic DNA, digested with the restriction enzymes HindIII, PstI, PvuII, or EcoR1 was hybridized with the [32P]-labeled PABP3 insert as probe. Seven to ten bands of uneven intensity, ranging from 500 bp to 20,000 bp, were observed for each digestion (Fig. 1). This pattern is compatible with a multigene family and is in accordance with the results obtained by Morris and Bodger (1993) who used the PABP1 insert as a probe on human genomic DNA.
Fig. 1.
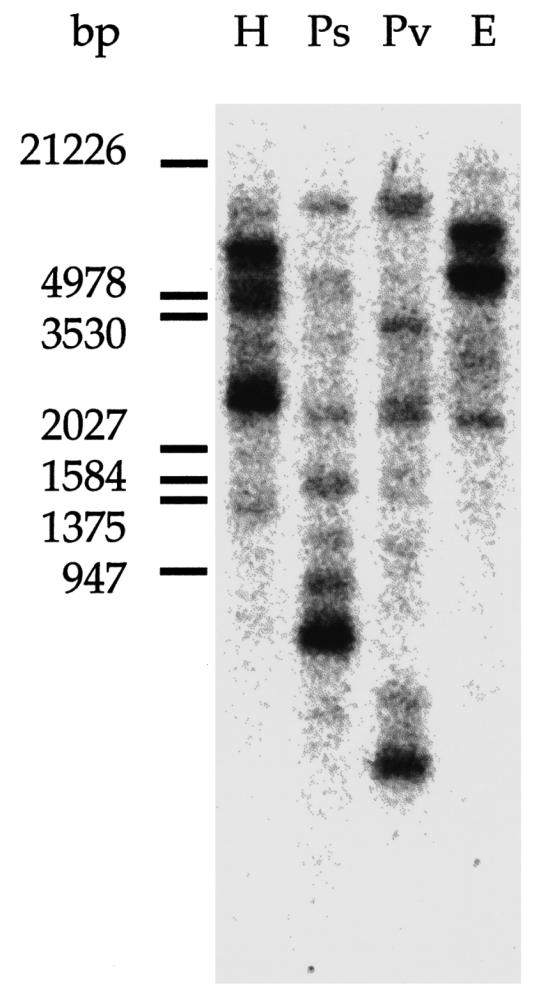
Southern blot of human genomic DNA. Human lymphocyte genomic DNA (20 μg) was digested by using either HindIII (H), PstI (Ps), PvuII (Pv), or EcoR1 (E) restriction enzymes. The blot was hybridized with the PABP3 cDNA as the probe, as described. The size of the lambda DNA/EcoRI + HindIII markers are indicated left (in bp)
Human PABP genes mapped by in situ hybridization
The loci of the PABP genes were mapped by in situ hybridization of the PABP3 cDNA to metaphase spreads. On the 150 metaphase cells examined, 356 silver grains were associated with chromosomes, with distributed hybridization signals on chromosomes 2, 13, 12, 8, and 3 of 15%, 12%, 10%, 9%, and 7%, respectively (Fig. 2A). The distribution of the grains on these chromosome was not random and ranged from 64% to 88% on the 2q21-q22, 13q11-q12, 12q13.3-q15, 8q22, and 3q24-q25 regions (Fig. 2B).
Fig. 2.
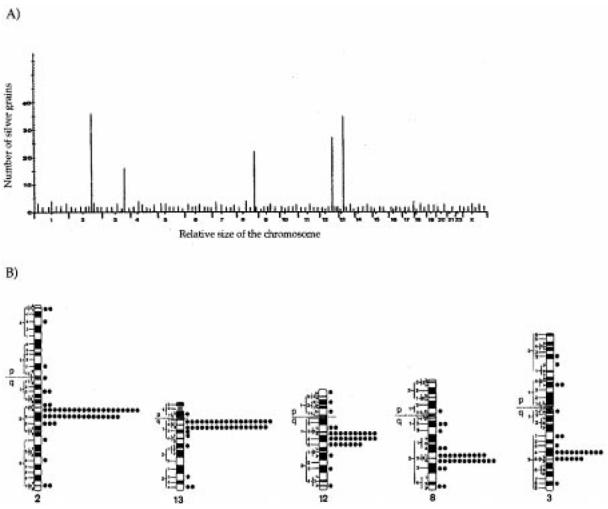
A Sublocalization of human PABP-related sequences. Histogram showing the distribution of 356 grains scored from 150 human metaphase cells following in situ hybridization of the cDNA PABP3 probe. Five significant hybridization peaks were detectable on chromosomes 2, 13, 12, 8, and 3. B Ideograms of the human R-banded chromosomes 2, 13, 12, 8, and 3, illustrating the precise distribution of labeled sites for the cDNA PABP3 probe. The distribution of the total of 356 grains was as follows: 14% chromosome 2, 69% of the grains, 36/52, in the 2q21-q22 region; 12% chromosome 13, 83% of the grains, 35/42, in the 13q11-q12 region; 10% chromosome 12, 77% of the grains, 27/35, in the 12q13.3-q15 region; 9% chromosome 8, 69% of the grains, 22/32, in the 8q22 region; 7% chromosome 3, 64% of the grains, 16/25, in the 3q24-q25 region
Chromosomal assignment for PABP genes
The assignment of each PABP gene to these loci was performed by using DNA from a panel of somatic-cell hybrid (human × mouse; human × hamster) containing single human chromosomes with, in some cases, additional fragments (see below). PABP-specific amplification products were derived from this panel by using different sets of oligonucleotide probes.
Selection of specific PCR primers
For each amplification, we designed sets of three oligonucleotides. Two oligonucleotides were used as PCR primers, and the third one, internal to the amplified sequence, was used as a probe for the identification of the PCR fragment (Table 1). In order to achieve a significant specificity, these oligonucleotides, ranging from 17 to 25 residues, were designed according to four criteria. (1) The sequence of a pair of primers specific for a PABP differed by at least five nucleotides from the corresponding sequence on the other PABPs (the PABP3 primers compared with the corresponding regions in PABP1, iPABP, and PABP4 are illustrated in Fig. 3). (2) The size of the PCR product was different for each PABP sequence. (3) Each pair of primers for PABP3, PABP4, and iPABP amplified a fragment on the corresponding cDNA plasmid and gave no product on a mixture of the plasmids related to the other PABPs (data not shown). For PABP1, we were unable to design a specific set of primers based on the mRNA sequence (accession no. Y00345) that met with these criteria. Therefore, we selected a pair of primers suitable for amplifying a fragment overlapping with the exon 10–11 sequence of the human PABP1 gene (accession no. U68101). Consequently, the PABP1-specific oligoprobe was designed from a sequence in intron 10 of the PABP1 gene (Fig. 4). This last experiment was not carried out for pseudogenes 1, 2, and 3, since no cDNA was available. (4) DNA fragments, amplified from mouse or hamster genomic DNA, could be distinguished from the human counterpart by size differences. The sequences of the sets of oligonucleotides and the amplification conditions for the seven genes or pseudogenes have been deposited in dbSTS under accession nos. G49185–G49191.
Fig. 3.
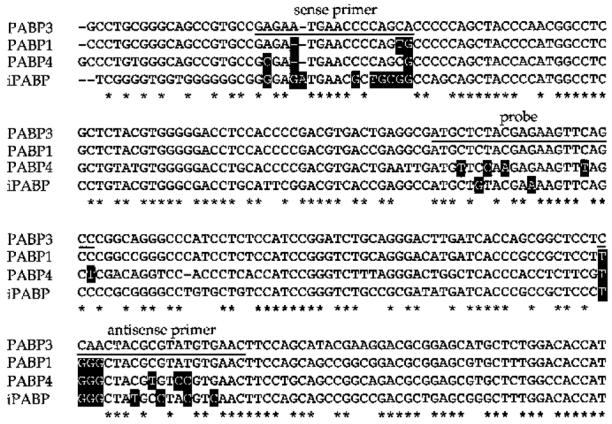
Alignment of the sequence of the PABP3-specific PCR product with the sequence of similar regions in PABP1, iPABP, and PABP4. The underlined sequences correspond to the 5′ and 3′ PCR primers and to the oligonucleotide probe. Asterisks Identical bases between the four PABP sequences. The differences in nucleotides between PABP1, iPABP, and PABP4 sequences as compared with PABP3 primers are boxed
Fig. 4.

Selection of the PABP1-specific oligonucleotide sets. The amplified fragment overlaps with the exon 10–11 sequence of the PABP1 gene (accession no. U68101). Lower case Intron sequence. The underlined sequences correspond to the 5′ and 3′ primers and control probe
DNA analysis of somatic cell hybrids panel for PABP-specific sequence
For PABP1, an intense signal of the expected size (253 bp) was observed on hybrid DNAs containing human chromosome 8. No signal of this size was observed on chromosome 22, corresponding to the contaminating fragment of the chromosomal hybrid. A weaker band at a higher size (277 bp) was also observed on hybrid DNAs containing human chromosomes 13, 16, 21, 22, and X (Fig. 5). The 141-bp band amplified from the PABP1 cDNA-specific plasmid was detected by using ethidium bromide (data not shown) but did not hybridize to the PABP1-specific oligonucleotide probe, since it was located over intron 10 (Fig. 4). Specific iPABP amplification generated a product of the correct size (213 bp) from a hybrid containing human chromosomes 1 and X. Since no specific product was detected in the hybrid DNA sample containing only the X chromosome, it can be deduced that the iPABP gene is located on chromosome 1. PABP3- and PABP4-specific amplifications each yielded a single band of the expected size (179 bp and 151 bp) from hybrids that contained human chromosomes 13 and 15, respectively (Fig. 5), and weaker bands on chromosomes 17 and 21 for PABP4. No signal was detected on the chromosomes corresponding to a contaminating fragment, so it was concluded that PABP3 and PABP4 genes are located on chromosomes 13 and 15, respectively.
Fig. 5.
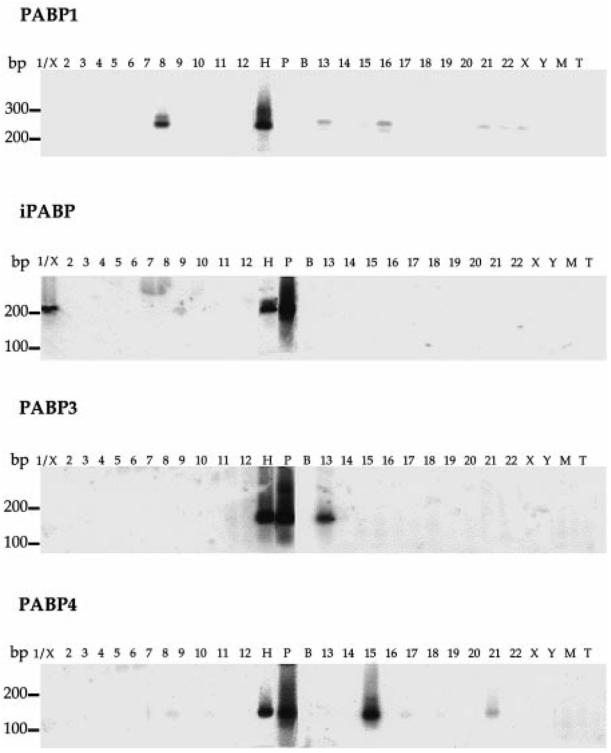
Chromosomal assignment of the PABP genes by PCR amplification of specific fragments on human-rodent hybrid cell line genomic DNA samples. Southern blot analysis of products amplified with PABP-specific oligonucleotide primers and revealed with an internal control probe, as described in Table 1. Numbers, X and Y above each lane, indicate the human chromosome contained in the hybrid cell line. Control DNAs were from human (H), mouse (M), and hamster (T) genomes or from PABP-specific plasmid (P). These specific plasmids contain cDNA corresponding to the sequences PABP1 (accession no. U68101), iPABP (accession no. U33818), PABP3 (accession no. AF132026) and PABP4 (accession no. AF132027). No DNA was present in B. The size of the DNA standards (in bp) is indicated left. In addition to the selected chromosomes, some hybrids contain small fragments of other chromosomes as follows: chromosome 1: X; chromosome 6: Xqter-Xq13; chromosome 8: fragment of 22; chromosome 10: Y; chromosome 12: 21 and X; chromosome 13: fragments of 8, 11, and 12; chromosome 14: 16p13.1-q22.1; chromosome 15: 11q and Xp; chromosome 20: X, 22q and fragments of 8 and 4; chromosome 21: X; chromosome 22: fragment of Xp
The data concerning the human assignment of PABP pseudogenes are not shown, but, using a similar approach, we obtained two pseudogene 1 amplification products from hybrids containing human chromosomes 4 and 20. Since the hybrid corresponding to chromosome 4 is apparently pure, and the one corresponding to chromosome 20 contains fragments of chromosome 4, the PABP pseudogene 1 is probably located on chromosome 4. Specific human PABP processed pseudogene 2 amplification product was observed for a hybrid containing human chromosome 14. Finally, the PCRs for human PABP processed pseudogene 3 gave four signals of the correct size from hybrids that contained human chromosomes 6, 12, 21, and X. The overall results are summarized in Table 2.
Table 2.
Summary of the chromosomal locations of the PABP gene family
| Names | Chromosomes |
|---|---|
| PABP1 | 8q22 |
| iPABP | 1p32-p36 |
| PABP3 | 13q11-q12 |
| PABP4 | 15 |
| Pseudogene 1 | 4 |
| Pseudogene 2 | 14 |
| Pseudogene 3 | 6, 12, 21, X |
Discussion
This is the first precise localization of two functional PABP genes: PABP1 and PABP3 on chromosome 8q22 and 13q11-q12, respectively. We also assign iPABP to chromosome 1, and the three pseudogenes, viz., PABP4, 1, and 2, to chromosomes 15, 4, and 14, respectively. A comparison of the results obtained by in situ hybridization and PCR amplification raises two points.
First, the specific signals detected on chromosomes 2 and 3 by in situ hybridization do not correspond to any chromosomal assignment detected by PCR on somatic cell hybrids. This observation might reflect the presence of new PABP genes or pseudogenes not yet identified. Alternatively, it might also reveal a cross hybridization of the probe to other genes, such as the gene for fibroblast activation protein alpha (FAP; accession no. U63542), located in 2q23 (Mathew et al. 1995), which contains three regions of 200–300 bp exhibiting a near 90% nucleotide identity with the PABP1 mRNA sequence, with a 300-bp sequence corresponding to the inverted non-coding strand of PABP1. Such a gene could be detected by in situ hybridization by using a PABP probe, but not by PCR amplification because of the primer selection.
Second, PCR on somatic cell hybrids revealed several assignments of PABP sequences that were not detected by in situ hybridization. This was expected for pseudogenes 1, 2, and 3, which contain only short fragments of PABP genomic sequence, fragments that are likely to be barely detectable with PABP3 cDNA as a probe on metaphase spreads. For iPABP, we assigned the gene by PCR on a hybrid containing chromosome 1. The lack of hybridization of the PABP3 probe to this chromosome is probably a consequence of the low identical nucleotide residues (69%) between the PABP3 and iPABP sequences. A similar observation is more puzzling for PABP4. We have assigned the PABP4 gene to chromosome 15, by using PCR on a somatic cell hybrid panel, whereas in situ hybridization displays no signals on this chromosome. This discrepancy might be attributable to a lack of sensitivity of in situ hybridization methods that have previously failed to localize genes detected by Southern blot analysis (Goode et al. 1986) as described for UBA 52, one of the human ubiquitin gene family (Webb et al. 1994).
Two different locations for iPABP are registered in the radiation hybrids (RH) database: one is on chromosome 1 at p32-p36 (accession no. A001T47), and the second one is on chromosome 15 at q26-qtel (accession no. WI-19844). Our assignment of the iPABP gene to chromosome 1 resolves this controversial situation and definitively establishes the location of this gene to 1p32-p36.
Brais et al. (1998) have shown that short GCG expansions in the PABP2 gene, located in 14q11, alter PABP2 function in the disease oculopharyngeal muscular dystrophy. Because of a similar role of other PABPs, we checked whether the PABP1 and PABP3 locations (8q22 and 13q11-q12, respectively) were linked with human genetic disorders registered in the OMIM database. However, we did not find any human genetic diseases relevant to PABP dysfunction and associated with these loci.
In this study, we have definitively established the chromosomal locations of the three functional PABPs, viz., PABP1, PABP3, and iPABP, on chromosomes 8q22, 13q11-q12, and 1p32-p36, respectively. We are now investigating whether alteration of the PABP3 gene, located at 13q11-q12, might be related to dysfunction in spermatogenesis.
Acknowledgments
This work was partly funded by the Association pour la Recherche contre le Cancer and the Fondation de France. We thank Y. Laperche and J. Hanoune for critical reading of the manuscript. We are very grateful to the MRC Resource Centre (Harrow, UK) for DNA samples from the panel of somatic cell hybrids. C. Féral is a recipient of a grant from the Ministère de la Recherche et de la Technologie.
References
- Blobel G. A protein of molecular weight 78,000 bound to the polyadenylate region of eukaryotic messenger RNAs. Proc Natl Acad Sci USA. 1973;70:924–928. doi: 10.1073/pnas.70.3.924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brais B, Bouchard JP, Xie YG, Rochefort DL, Chretien N, Tome FM, Lafreniere RG, Rommens JM, Uyama E, Nohira O, Blumen S, Korczyn AD, Heutink P, Mathieu J, Duranceau A, Codere F, Fardeau M, Rouleau GA. Short GCG expansions in the PABP2 gene cause oculopharyngeal muscular dystrophy [erratum in Nat Genet 1998 19:404] Nat Genet. 1998;18:164–167. doi: 10.1038/ng0298-164. [DOI] [PubMed] [Google Scholar]
- Don RH, Cox PT, Wainwright BJ, Baker K, Mattick JS. ‘Touchdown’ PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuili G, Scholl U, Bulle F, Guellaen G. Molecular cloning of the cDNAs coding for the two subunits of soluble guanylyl cyclase from human brain. FEBS Lett. 1992;304:83–88. doi: 10.1016/0014-5793(92)80594-7. [DOI] [PubMed] [Google Scholar]
- Goode ME, vanTuinen P, Ledbetter DH, Daiger SP. The anonymous polymorphic DNA clone D1S1, previously mapped to human chromosome 1p36 by in situ hybridization, is from chromosome 3 and is duplicated on chromosome 1. Am J Hum Genet. 1986;38:437–446. [PMC free article] [PubMed] [Google Scholar]
- Grange T, de Sa CM, Oddos J, Pictet R. Human mRNA polyadenylate binding protein: evolutionary conservation of a nucleic acid binding motif. Nucleic Acids Res. 1987;15:4771–4787. doi: 10.1093/nar/15.12.4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houng AK, Maggini L, Clement CY, Reed GL. Identification and structure of activated-platelet protein-1, a protein with RNA-binding domain motifs that is expressed by activated platelets. Eur J Biochem. 1997;243:209–218. doi: 10.1111/j.1432-1033.1997.0209a.x. [DOI] [PubMed] [Google Scholar]
- Kelsell DP, Rooke L, Warne D, Bouzyk M, Cullin L, Cox S, West L, Povey S, Spurr NK. Development of a panel of monochromosomal somatic cell hybrids for rapid gene mapping. Ann Hum Genet. 1995;59:233–241. doi: 10.1111/j.1469-1809.1995.tb00743.x. [DOI] [PubMed] [Google Scholar]
- Kessler SH, Sachs AB. RNA recognition motif 2 of yeast Pab1p is required for its functional interaction with eukaryotic translation initiation factor 4G. Mol Cell Biol. 1998;18:51–57. doi: 10.1128/mcb.18.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le H, Tanguay RL, Balasta ML, Wei CC, Browning KS, Metz AM, Goss DJ, Gallie DR. Translation initiation factors eIF-iso4G and eIF-4B interact with the poly(A)-binding protein and increase its RNA binding activity. J Biol Chem. 1997;272:16247–16255. doi: 10.1074/jbc.272.26.16247. [DOI] [PubMed] [Google Scholar]
- Mathew S, Scanlan MJ, Mohan Raj BK, Murty VV, Garin-Chesa P, Old LJ, Rettig WJ, Chaganti RS. The gene for fibroblast activation protein alpha (FAP), a putative cell surface-bound serine protease expressed in cancer stroma and wound healing, maps to chromosome band 2q23. Genomics. 1995;25:335–337. doi: 10.1016/0888-7543(95)80157-h. [DOI] [PubMed] [Google Scholar]
- Mattei MG, Philip N, Passage E, Moisan JP, Mandel JL, Mattei JF. DNA probe localization at 18p113 band by in situ hybridization and identification of a small supernumerary chromosome. Hum Genet. 1985;69:268–271. doi: 10.1007/BF00293038. [DOI] [PubMed] [Google Scholar]
- Morris CM, Bodger MP. Localization of the human poly(A)-binding protein gene (PAB1) to chromosomal regions 3q22-q25, 12q13-q14, and 13q12-q13 by in situ hybridization. Genomics. 1993;15:209–211. doi: 10.1006/geno.1993.1037. [DOI] [PubMed] [Google Scholar]
- Sachs A, Wahle E. Poly(A) tail metabolism and function in eucaryotes. J Biol Chem. 1993;268:22955–22958. [PubMed] [Google Scholar]
- Tarun SZ, Jr, Sachs AB. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- Tarun SZ, Jr, Wells SE, Deardorff JA, Sachs AB. Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc Natl Acad Sci USA. 1997;94:9046–9051. doi: 10.1073/pnas.94.17.9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahle E. A novel poly(A)-binding protein acts as a specificity factor in the second phase of messenger RNA polyadenylation [erratum in Cell 1991 67:following 639] Cell. 1991;66:759–768. doi: 10.1016/0092-8674(91)90119-j. [DOI] [PubMed] [Google Scholar]
- Webb GC, Baker RT, Coggan M, Board PG. Localization of the human UBA52 ubiquitin fusion gene to chromosome band 19p13.1-p12. Genomics. 1994;19:567–569. doi: 10.1006/geno.1994.1108. [DOI] [PubMed] [Google Scholar]
- Yang H, Duckett CS, Lindsten T. iPABP, an inducible poly(A)-binding protein detected in activated human T cells. Mol Cell Biol. 1995;15:6770–6776. doi: 10.1128/mcb.15.12.6770. [DOI] [PMC free article] [PubMed] [Google Scholar]


