Abstract
The spindle checkpoint delays anaphase onset until all chromosomes are correctly attached to microtubules. Ipl1 protein kinase (Aurora B) is required to correct inappropriate kinetochore–microtubule attachments and for the response to lack of tension between sister kinetochores. Here we identify residues in the checkpoint protein Mad3p that are phosphorylated by Ipl1p. When phosphorylation of Mad3p at two sites is prevented, the cell’s response to reduced kinetochore tension is dramatically curtailed. Our data provide strong evidence for a distinct checkpoint pathway responding to lack of sister kinetochore tension, in which Ipl1p-dependent phosphorylation of Mad3p is a key step.
Keywords: Mad3, Aurora kinase, tension, spindle checkpoint
Since errors in chromosome segregation lead to aneuploidy, cell death, and disease, cells have evolved mechanisms to ensure that their replicated chromosomes are accurately segregated. The spindle checkpoint, involving a conserved network of Mad and Bub proteins, acts as a surveillance system to monitor kinetochore–microtubule interactions during chromosome alignment on the mitotic spindle (Musacchio and Hardwick 2002; Lew and Burke 2003). When activated, it acts to inhibit Cdc20p, an accessory factor for the mitotic E3 ubiquitin ligase known as the Anaphase-Promoting Complex or Cyclosome (APC/C) (Zachariae and Nasmyth 1999; Yu 2002), preventing ubiquitylation of securin and mitotic cyclins and thereby delaying anaphase onset. Once biorientation is achieved (i.e., sister chromatids are attached to opposite poles of the spindle), the checkpoint is satisfied and anaphase can proceed.
Biorientation defects can be due to lack of kinetochore–microtubule attachment or result from attachment of both sister kinetochores to the same spindle pole (syntelic or monopolar attachment). Syntelic attachments are thought to result in a lack of tension between sister kinetochores, and error correction mechanisms are required to promote biorientation. It is clear from both vertebrate and yeast studies that Aurora B kinase (Ipl1p in yeast) has a crucial role to play in promoting such biorientation, and that this involves breaking incorrect (syntelic) microtubule attachments (Tanaka et al. 2002; Hauf et al. 2003; Dewar et al. 2004; Lampson et al. 2004). Aurora kinase may also link the correction of inappropriate attachments with a spindle checkpoint-dependent mitotic delay, as it is required to delay anaphase in response to the lack of cohesion at sister centromeres, reduced microtubule dynamics (taxol), and certain kinetochore defects (Biggins and Murray 2001; Ditchfield et al. 2003; Hauf et al. 2003; Pinsky et al. 2003). However, the existence of a distinct “tension checkpoint” mechanism remains controversial (for a recent review, see Pinsky and Biggins 2005). In budding yeast, although in some circumstances Ipl1p can activate the spindle checkpoint through generation of unattached kinetochores when it promotes breakage of defective microtubule attachments (Pinsky et al. 2006), it may also play a direct role in spindle checkpoint signaling in response to a lack of tension.
Studies using Aurora B kinase inhibitors in vertebrate cells have indicated a possible link between Aurora B and BubR1: BubR1 phosphorylation and localization are perturbed when Aurora B is inhibited (Ditchfield et al. 2003). Here we use budding yeast to demonstrate that the yeast BubR1-related protein Mad3p is a substrate of Ipl1p, and we have mapped multiple Ipl1p phosphorylation sites in Mad3p. Blocking phosphorylation of these sites by mutation to alanine prevents cells from delaying anaphase specifically in response to lack of sister kinetochore tension. We therefore propose that a distinct, Ipl1-dependent branch of the spindle checkpoint pathway monitors tension at sister kinetochores, and that phosphorylation of Mad3p by Ipl1p is a crucial component in this checkpoint response.
Results and Discussion
BubR1 kinetochore localization is sensitive to lack of tension (Skoufias et al. 2001; Taylor et al. 2001), and BubR1 phosphorylation is Aurora B-dependent (Ditchfield et al. 2003). In yeast, we and others have found that Mad3p shows both Polo (Cdc5p) and Aurora (Ipl1p) kinase-dependent phosphorylation in mitosis (Rancati et al. 2005; data not shown). Since Ipl1p is required for mitotic checkpoint activation in response to loss of tension (Biggins and Murray 2001), Ipl1p-dependent phosphorylation of Mad3p may be a mechanism by which Ipl1p participates in the checkpoint-dependent anaphase delay. To test this, we therefore needed to analyze yeast cells in which kinetochores remain attached to microtubules but do not come under tension, a condition proposed to occur when cells dependent on SCC1 expression from the GAL promoter undergo DNA replication following transfer to glucose to block SCC1 expression (Indjeian et al. 2005). Scc1p is a component of the cohesin complex responsible for sister chromatid cohesion, and depletion of Scc1p prevents cells from establishing cohesion as they pass through S phase.
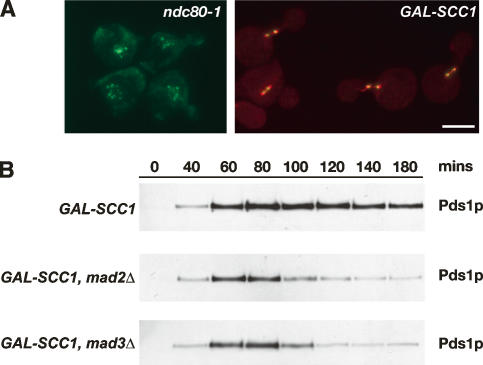
To validate such GAL-SCC1 strains as a “lack of tension assay,” we visualized cells containing fluorescently labeled spindles (Tub1-CFP) and kinetochores (Mtw1-3GFP). Such cells were presynchronized in G1 with α-factor and released in the presence of glucose to allow DNA replication in the absence of SCC1 expression, and then kinetochore–microtubule attachment was monitored by microscopy as described by Pinsky et al. (2006). We found that in all mitotic Scc1p-depleted cells analyzed (n = 650), Mtw1-3GFP localized exclusively along the spindle axis defined by Tub1-CFP fluorescence, indicative of highly efficient kinetochore attachment (Fig. 1A). We conclude that depletion of Scc1p does not generate unattached kinetochores and can therefore be used to generate sister chromatids that are attached to microtubules but that lack tension. This finding is further supported by similar analysis of cells expressing Mtw1-3GFP and Tub1-CFP but whose cohesion defect was generated using the temperature-sensitive mcd1-1 allele of SCC1 as an alternative to Scc1p depletion (data not shown).
Figure 1.
Depletion of Scc1p does not generate unattached kinetochores but promotes MAD3-dependent checkpoint activation. (A) Cultures of KH409 (GAL-SCC1) and SBY4341 (ndc80-1) were presynchronized in G1, and then depleted of Scc1p (KH409) or shifted to their restrictive temperature of 36°C (SBY4341). After release from G1, samples were taken at 15-min intervals for analysis of spindle (Tub1-CFP; red) and kinetochore (Mtw1-3GFP; green) positions in metaphase cells, which peaked 60 min following release. The images show cells from the 60-min time point and are maximum intensity projections of a stack of six Z-sections taken 0.2 μm apart. (B) Cultures of VBI545 (GAL-SCC1 MAD3), EK26 (GAL-SCC1 mad3Δ), and VBI560 (GAL-SCC1 mad2Δ) expressing Pds1-myc18 were grown in the presence of galactose, synchronized with α-factor, and then released into glucose-containing medium to allow DNA replication in the absence of SCC1 expression. Lysates were prepared from samples taken at the times indicated and immunoblotted with anti-c-myc antibody.
Mad3p is required for the ‘tension checkpoint’
Next, we examined whether Mad3p is required for the checkpoint response to lack of sister kinetochore tension following Scc1p depletion. Cells were released from G1 arrest in the absence of SCC1 expression, and the level of Pds1p (securin), degradation of which marks anaphase onset (Ciosk et al. 1998), was monitored through the following cell cycle. Wild-type MAD3 cells maintained elevated levels of Pds1p until ∼180 min after release, whereas the equivalent mad3Δ or mad2Δ cells began degradation of Pds1p after ∼100 min, consistent with the absence of any delay to anaphase in both checkpoint mutants (Fig. 1B). Thus, checkpoint-proficient cells delay anaphase onset in response to Scc1p depletion and, furthermore, MAD3 is clearly required to delay anaphase onset in response to the resulting lack of tension. These observations were confirmed using mutant strains carrying thermosensitive SCC1 alleles (mcd1-1 and scc1-73) as alternative ways to trigger loss of tension (data not shown). Lee and Spencer (2004) have argued that Mad1p and Mad2p but not Mad3p are required for the tension checkpoint. It is unclear why their findings differ from ours, although a different means was used to generate attached chromosomes not under tension. Our finding of a Mad3p requirement in the tension checkpoint is consistent with vertebrate studies, where it has been established in several experimental systems that kinetochore localization of BubR1 is associated with lack of tension at kinetochores (Skoufias et al. 2001; Taylor et al. 2001; Shannon et al. 2002; Ditchfield et al. 2003; Logarinho et al. 2004).
Ipl1p phosphorylates Mad3p in vitro
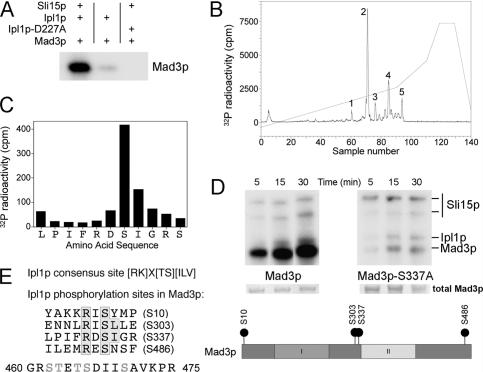
To determine whether Mad3p might be a direct substrate for Ipl1p, recombinant Mad3p was incubated with recombinant Ipl1p and its activator Sli15p (the yeast INCENP ortholog) (Kang et al. 2001), together with radiolabeled Mg2+-ATP to perform a kinase assay. Figure 2A shows that Mad3p is, indeed, phosphorylated by Ipl1p and that this is dependent on Sli15p as expected. When this 32P-labeled Mad3p was subjected to tryptic digestion and the resulting peptides separated by HPLC, five phosphopeptides were evident (Fig. 2B). Detailed phosphorylation site analysis of Mad3p identified the major Ipl1p phosphorylation site (corresponding to peak 2) on Mad3p in vitro as Ser 337 (Fig. 2C). We then replaced Ser 337 with alanine, to prevent phosphorylation at this site, and examined the ability of Mad3p-S337A to be phosphorylated by Ipl1p–Sli15p (Fig. 2D). Wild-type Mad3p was phosphorylated efficiently in comparison with the Mad3-S337A protein, confirming that Ser 337 is indeed the main site of Mad3p phosphorylation by Ipl1p in vitro. However, the low residual level of phosphorylation still evident on the mutant protein prompted us to identify the remaining phosphorylation sites using a more sensitive method employing LC-MS with precursor ion scanning. Three additional sites (Ser 10, Ser 303, and Ser 486) were definitively identified, and a fourth site was localized within a 16-amino-acid phosphopeptide (residues 460–475), although we were unable to distinguish which one of its five serines or threonines was phosphorylated.
Figure 2.
Identification of Ipl1p-dependent phosphorylation sites in Mad3p. (A) Recombinant Mad3p was incubated with either Ipl1p or Ipl1p-D227A (kinase dead) in the presence or absence of Sli15p, together with Mg2+-[γ-32P]ATP. Phosphorylation of Mad3p was visualized following polyacrylamide gel electrophoresis and autoradiography, locating the position of the Mad3p band by Coomassie staining. (B) Following phosphorylation with Ipl1p–Sli15p complex, 32P-labeled Mad3p was digested with trypsin and separated on a C18 column, monitoring the 32P elution profile. Peaks containing 32P-labeled peptides are labeled 1–5. Peak 2 contained 57% of the applied 32P radioactivity. (C) Peak 2 was analyzed by MALDI-TOF-TOF mass spectrometry and Edman degradation: The deduced amino acid sequence from the mass spectrometric analysis is plotted against the radioactivity released in each cycle of Edman degradation. Maximal release of 32P occurred in the cycle corresponding to Ser 337. (D) Escherichia coli-expressed Mad3p and Mad3p-S337A were incubated with Ipl1p–Sli15p complex in the presence of Mg2+-[γ-32P]ATP. Phosphorylation of Mad3p and Mad3p-S337A was determined following electrophoresis on a polyacrylamide gel and autoradiography, locating the position of the Mad3p band by Coomassie staining. The relevant section of the stained gel is shown to confirm equal loadings. (E) The Ipl1p consensus phosphorylation site (Cheeseman et al. 2002) is shown together with the four Ipl1p phosphorylation sites identified by LC-MS with precursor ion scanning (one of which had been previously identified in C), highlighting the phosphorylated serine, the basic residue at −2, and the hydrophobic residue at +1 present at two of the sites. A phosphopeptide containing a fifth site that could not be unambiguously placed within the sequence is also shown (possible sites of phosphorylation are indicated in gray). The location of each defined phosphoserine within the Mad3p sequence is also indicated in relation to two conserved domains (Hardwick et al. 2000) involved in binding Cdc20p/Mad2p (I) and Bub3p (II).
We have therefore established that Mad3p is phosphorylated directly in vitro on up to five different sites by the Ipl1p–Sli15p complex. Two out of the four identified sites (Ser 303 and Ser 337) match the Ipl1p consensus phosphorylation sequence [KR]X[ST][ILV] described previously (Cheeseman et al. 2002), while the other two sites (Ser 10 and Ser 486) conform to a less strict Ipl1p/Aurora B consensus ([RK]X[TS]) found in other substrates (Fig. 2E). We also looked at conservation of these sites across the Mad3–BubR1 family in fungi and vertebrates (Supplementary Fig. 1). Ser 10 is only found in Saccharomyces cerevisiae, but the three other sites are largely conserved throughout the Saccharomyces genus, and Ser 303 is also conserved in more distant species such as Schizosaccharomyces pombe and Candida albicans. Alignment of the S. cerevisiae Mad3p amino acid sequence with Mad3p from other fungi or with BubR1 is difficult because the proteins vary enormously in primary sequence outside the two conserved regions (Hardwick et al. 2000; see Supplementary Fig. 1). However, there are at least two potential Ipl1p/Aurora sites located in the central part of BubR1 (Thr 226 and Thr 273) that could correspond to Ser 303 and Ser 337 in budding yeast Mad3p. Our data support the results of Rancati et al. (2005), who reported that Mad3p shows Ipl1p-dependent hyperphosphorylation in vivo upon entering mitosis with unreplicated chromosomes, another condition in which the spindle checkpoint is activated because kinetochores are not under tension (Stern and Murray 2001), and are consistent with data showing Aurora B-dependent phosphorylation of BubR1 in higher eukaryotes (Ditchfield et al. 2003).
Mutation of Mad3 to prevent phosphorylation of Ser 303 and Ser 337 abrogates the tension checkpoint
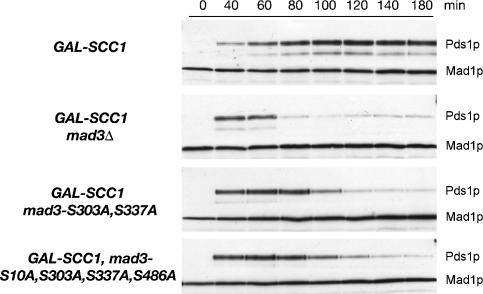
Since MAD3 is essential for the checkpoint response to lack of tension, we next investigated whether phosphorylation of Mad3p at the Ipl1p phosphorylation sites, identified in vitro, was important in vivo. We initially mutated the major in vitro site (Ser 337) to alanine and introduced the mad3-S337A allele into strains in which the tension checkpoint could be activated by SCC1 depletion. Although cells without Mad3p (mad3Δ) quickly degraded Pds1p, we found that cells carrying either MAD3 or mad3-S337A maintained elevated levels of Pds1p (data not shown), indicative of a functional checkpoint. Thus the S337A mutation alone does not impair the tension checkpoint. Although Ser 337 is the major Mad3p site phosphorylated by Ipl1p in vitro, phosphorylation of one or more of the other candidate sites may function redundantly in vivo. We therefore made a double mutant in which both Ser 303 and Ser 337 were changed to alanine (mad3-2SA), and a quadruple mutant in which serines 10, 303, 337, and 486 were similarly mutated (mad3-4SA) to prevent phosphorylation. Figure 3 shows that both the mad3-2SA and mad3-4SA mutants were largely defective in the tension checkpoint delay. Since no reproducible difference was found between the double and the quadruple mutants, we conclude that S303 and S337 are likely to be the major phosphorylation sites required for a complete tension checkpoint response and that they function redundantly in vivo. However, cells carrying mad3Δ degraded Pds1p slightly earlier than the mad3-2SA and mad3-4SA mutants. This slight residual delay may represent the contribution of Ipl1p-generated unattached kinetochores to the tension checkpoint response. Alternatively, Mad3p phosphorylation may be only part of a more complex response, and complete absence of Mad3p may additionally destabilize checkpoint protein complexes. The requirement for Ipl1p-dependent Mad3p phosphorylation for a fully functional checkpoint is consistent with a recent proposal that Aurora B activity cooperates with vertebrate spindle checkpoint proteins to inhibit the APC/C (Morrow et al. 2005).
Figure 3.
Blocking phosphorylation of Mad3p abrogates the checkpoint arrest in response to lack of tension. Cultures of EK29 (GAL-SCC1 MAD3), EK26 (GAL-SCC1 mad3Δ), EK41 [GAL-SCC1 MAD3-2SA (S303A, S337A)], and EK43 [GAL-SCC1 MAD3-4SA (S10A, S303A, S337A, S486A)] expressing Pds1-myc18 were grown in the presence of galactose, synchronized with α-factor, and then released into glucose-containing medium to allow DNA replication in the absence of SCC1 expression. Lysates were prepared from samples taken at the times indicated and immunoblotted with anti-c-myc antibody to detect Pds1p.
Preventing phosphorylation of Mad3-S337 and Mad3-S303 does not abrogate the checkpoint response to unattached kinetochores
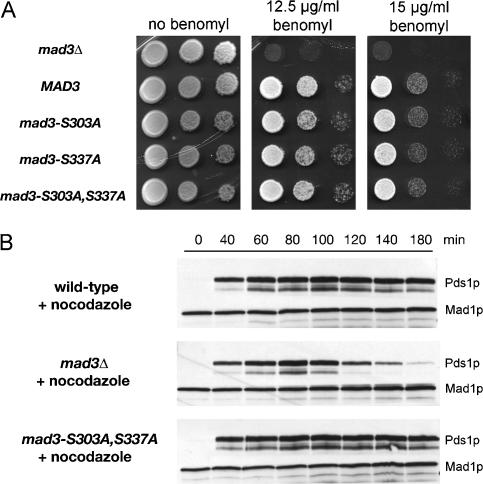
To assess the ability of strains to mount a checkpoint response to unattached kinetochores, we tested the benomyl sensitivity (Li and Murray 1991) of our alanine substitution mutants alone or in combination. Figure 4A shows that all of the mad3 mutant strains tested behaved identically to a MAD3 wild-type control and were able to grow well on rich medium containing 12.5 and 15 μg/mL benomyl, respectively. Furthermore, when synchronous mad3-2SA cells were treated with nocodazole and the stability of Pds1p was examined, they were found to display a similar mitotic arrest to wild-type cells (Fig. 4B). Thus all of these mad3 mutant alleles support a normal checkpoint response to unattached kinetochores, and the effect of the mad3-2SA allele is therefore highly specific for checkpoint activation in response to lack of tension. In support of distinct pathways responding to tension or attachment, vertebrate localization studies have shown that checkpoint components behave in a manner consistent with differential monitoring of these two properties. For example, Mad1 and Mad2 tend to dissociate from kinetochores upon stable microtubule attachment, whereas BubR1, the functional equivalent of budding yeast Mad3p in vertebrates, remains on attached chromosomes if they lack tension, such as in the presence of the microtubule-stabilizing agent taxol (Skoufias et al. 2001; Shannon et al. 2002; Logarinho et al. 2004).
Figure 4.
Blocking phosphorylation of Mad3p does not abrogate the checkpoint arrest in response to unattached kinetochores. (A) KH173 (mad3Δ) transformed with pKH535 (MAD3), pEK089 (mad3-S303A), pEK090 (mad3-S337A), and pEK091 (mad3-2SA), was suitably diluted and spotted onto YPD agar containing the indicated benomyl concentrations. Growth is shown after 3 d at 23°C. (B) Cultures of EK45 (PDS1-myc18 mad3Δ) transformed with YCplac22 (empty vector), pKH535 (MAD3), or pEK091 (mad3-2SA) were synchronized with α-factor, then released into 15 μg/mL nocodazole at 23°C to activate a checkpoint arrest in response to microtubule depolymerization. Lysates were prepared from samples taken at the times indicated and were immunoblotted with anti-c-myc antibody.
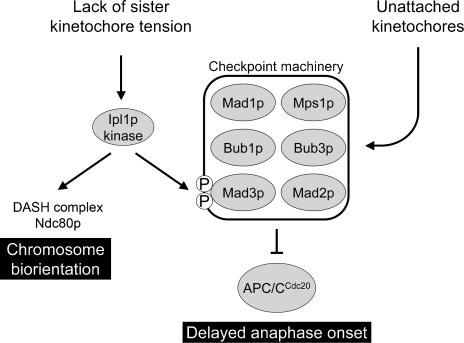
Based on our observations and previously published work, we propose a dual role for Ipl1p: resolution of maloriented sister kinetochores and phosphorylation of Mad3p so as to activate or enhance the checkpoint delay in response to lack of sister kinetochore tension (see Fig. 5). We have demonstrated that the checkpoint protein Mad3p is a substrate for Ipl1 protein kinase and shown that blocking phosphorylation at two critical sites almost completely abrogates the spindle checkpoint response to lack of sister kinetochore tension. In contrast, the same mutations have no effect on the checkpoint response to unattached kinetochores resulting from microtubule depolymerization. Our model is therefore analogous to that of Morrow et al. (2005), who have proposed a distinct, Aurora B-dependent link between kinetochores not under tension and the core checkpoint machinery. Pinsky et al. (2006) have argued that the principal role of Ipl1p kinase in the spindle checkpoint is to generate unattached kinetochores upon lack of tension, and it clearly can function in this manner when the checkpoint is activated in response to kinetochore defects produced when cells expressing thermosensitive alleles of genes encoding a range of kinetochore components are shifted to the restrictive temperature. However, using the same assay to monitor kinetochore attachment, we have failed to find any evidence for unattached kinetochores in Scc1p-depleted cells, and yet such cells show a Mad3p-dependent anaphase delay that requires the presence of two Ipl1p phosphorylation sites in Mad3p. Thus, in contrast to cells with defective kinetochore components where reattachment of microtubules may be impaired, kinetochore–microtubule interactions destabilized by Ipl1p in Scc1p-depleted cells may be rapidly reinstated and therefore remain undetected through the “attachment” pathway. Mad3p may acquire different patterns of phosphorylation when the checkpoint machinery is activated in different ways, although how this leads it to become a more potent inhibitor of the APC/C remains to be determined. Ipl1-dependent Mad3p phosphorylation may be particularly important in response to lack of tension to ensure that APC/CCdc20 is rapidly and efficiently inhibited. Such Mad3p phosphorylation may also occur more rapidly than a response to lack of tension via the “attachment” pathway, which would first require breaking the incorrect microtubule attachments, then recruitment of kinetochore-bound signaling scaffolds (Mad1p and/or Bub1p), and finally interaction with Mad2p and Mad3p to produce new anaphase inhibitors. Phosphorylated Mad3p may also be more potent as an inhibitor, acting as an amplifier of the intermittent checkpoint signal generated by rapidly detached and reattached kinetochore microtubules.
Figure 5.
Model showing the proposed dual role of Ipl1p in promoting chromosome biorientation and checkpoint activation in response to mono-oriented chromosomes. The activation of the checkpoint machinery by lack of tension is shown as a direct response mediated through Mad3p phosphorylation by Ipl1p that is separate from the pathway that activates the checkpoint in response to unattached kinetochores, as supported by the properties of the Mad3p phosphorylation site mutants described in this study.
In conclusion, our findings provide the first clear mechanistic basis for a direct role of Ipl1p kinase activity in spindle checkpoint signaling in response to a lack of sister kinetochore tension and argue strongly in favor of two distinct responses within the spindle checkpoint: an attachment response independent of Ipl1p activity and a tension response requiring Ipl1p activity and Mad3p phosphorylation (Fig. 5).
Materials and methods
Yeast strains and general methods
All yeast strains (listed in Supplementary Table 2) were derivatives of W303 (ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 ssd1-d2 Gal+). Strain and plasmid construction is described in the Supplemental Material. Basic yeast methods and growth media and routine recombinant DNA methodology were performed as previously described (Gietz et al. 1992; Kaiser et al. 1994; Sambrook and Russell 2001). Benomyl sensitivity was assessed by spotting strains, in 10-fold dilutions, onto plates containing 12.5 μg/mL or 15 μ/mL benomyl, respectively, followed by growth for 3 d at 23°C.
Biochemical and immunological techniques
Cells were synchronized in G1 using α-factor at 1 μg/mL and were released by washing three times before resuspension in the appropriate media. To prevent cells from entering the next cell cycle, α-factor was added back when small buds appeared. Where cells were released into YPD containing nocodazole, the drug was used at a concentration of 15 μg/mL at 23°C. To study tension checkpoint activation, strains expressing SCC1 from a GAL promoter, were arrested in G1 in YP medium containing raffinose and galactose for 150 min at 30°C and then transferred to YPD containing 1 μg/mL α-factor and incubated for a further 2 h at 30°C. Cells were finally released into YPD at 30°C.
Cell cycle progression was followed by monitoring Pds1p levels in strains containing PDS1-myc18. Samples taken over a 180-min time course for immunoblotting were resuspended in an equal volume of sample buffer containing 4% (w/v) SDS, 80 mM Tris-HCl (pH 6.8), 10% (v/v) glycerol, 20 mM EDTA, bromophenol blue, 0.1 M DTT, 100 μM Pefabloc, 10 μg/mL leupeptin, 10 μg/mL pepstatin, 10 μg/mL chymostatin, 2 mM sodium pyrophosphate, 50 mM NaF, and 100 μM microcystin. An equal volume of glass beads (0.5 mm diameter) was added, and cells were lysed using a Ribolyser (Hybaid) before separation of proteins by SDS-PAGE and Western blotting. Pds1p-myc18 was detected with an anti-myc antibody (c-myc A14; Santa Cruz Biotechnology) used at 1:1000 dilution followed by goat anti-rabbit-HRP secondary antibody (Pierce) at 1:1000 dilution, using ECL detection. Membranes were blotted with a rabbit anti-Mad1p antibody (Hardwick and Murray 1995) as a loading control.
Protein kinase assay and phosphorylation site mapping
Purified GST-Ipl1p (0.2 μg) and GST-Sli15p (0.04 μg) and Mad3p (0.6 μg) were incubated in 20 μL of reaction mixture containing 50 mM Tris-HCl (pH 7.5), 0.1% β-mercaptoethanol, 0.1 mM EGTA, 10 mM MgCl2, and 100 μM [γ-32P]ATP (826 cpm/pmol) for 15 min with 10 μM Microcystin (gift from Professor C. MacKintosh, University of Dundee, Dundee, Scotland, UK) plus recombinant substrate, as appropriate. Reactions were stopped by adding 2× sample buffer and separated by SDS-PAGE, the proteins were transferred to Immobilon-P transfer membrane (Millipore), and the membrane was exposed to film. Preparation of recombinant GST-Ipl1p, GST-Sli15p, and Mad3p is described in the Supplemental Material. To map phosphorylation sites, 32P-labeled Mad3p (6 μg) was generated in a similar manner but using high specific activity [γ-32P]ATP (5000 cpm/pmol), excised as a gel slice following SDS-PAGE, and digested with trypsin. Peptides were separated by HPLC and analyzed by MALDI-TOF-TOF mass spectrometry and Edman degradation as described previously (Lizcano et al. 2004). Alternatively, Mad3p tryptic digests prepared from in vitro kinase assays (carried out as above but with nonradiolabeled ATP) were processed and analyzed essentially as described by Williamson et al. (2006), detecting phosphopeptides by precursor ion scanning in the negative ion mode and switching to the positive ion mode to obtain the peptide sequence by an ms/ms product ion scan.
Microscopy
GAL-SCC1 cells containing Tub1-CFP and Mtw-3GFP (Pinsky et al. 2006) were synchronized by addition of α-factor and then depleted for Scc1p by growth in glucose media for 120 min followed by release from G1 into glucose medium.
At 15-min intervals over a 2-h time course, samples of cells were fixed briefly (5 min) in 3.7% (w/v) formaldehyde, washed, and then imaged using an Intelligent Imaging Innovations (3i) Marianas system, which incorporates a Zeiss Axiovert microscope, CoolSnap CCD, and Slidebook software.
Acknowledgments
We thank Sue Biggins and Andrew Murray for yeast strains; Vincent Guacci for mcd1-1 constructs; Tomo Tanaka, Paul Andrews, and Simonetta Piatti for useful discussions; and Tomo Tanaka for reading the manuscript. This work was funded by project grant 067210 from the Wellcome Trust to M.J.R.S., a BBSRC studentship to E.M.J.K., and a Wellcome Trust Senior Research Fellowship to K.G.H.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.431507
References
- Biggins S., Murray A.W., Murray A.W. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes & Dev. 2001;15:3118–3129. doi: 10.1101/gad.934801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., Anderson S., Jwa M., Green E.M., Kang J., Yates J.R., III, Chan C.S., Drubin D.G., Barnes G., Anderson S., Jwa M., Green E.M., Kang J., Yates J.R., III, Chan C.S., Drubin D.G., Barnes G., Jwa M., Green E.M., Kang J., Yates J.R., III, Chan C.S., Drubin D.G., Barnes G., Green E.M., Kang J., Yates J.R., III, Chan C.S., Drubin D.G., Barnes G., Kang J., Yates J.R., III, Chan C.S., Drubin D.G., Barnes G., Yates J.R., III, Chan C.S., Drubin D.G., Barnes G., Chan C.S., Drubin D.G., Barnes G., Drubin D.G., Barnes G., Barnes G. Phospho-regulation of kinetochore–microtubule attachments by the Aurora kinase Ipl1p. Cell. 2002;111:163–172. doi: 10.1016/s0092-8674(02)00973-x. [DOI] [PubMed] [Google Scholar]
- Ciosk R., Zachariae W., Michaelis C., Shevchenko A., Mann M., Nasmyth K., Zachariae W., Michaelis C., Shevchenko A., Mann M., Nasmyth K., Michaelis C., Shevchenko A., Mann M., Nasmyth K., Shevchenko A., Mann M., Nasmyth K., Mann M., Nasmyth K., Nasmyth K. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell. 1998;93:1067–1076. doi: 10.1016/s0092-8674(00)81211-8. [DOI] [PubMed] [Google Scholar]
- Dewar H., Tanaka K., Nasmyth K., Tanaka T.U., Tanaka K., Nasmyth K., Tanaka T.U., Nasmyth K., Tanaka T.U., Tanaka T.U. Tension between two kinetochores suffices for their bi-orientation on the mitotic spindle. Nature. 2004;428:93–97. doi: 10.1038/nature02328. [DOI] [PubMed] [Google Scholar]
- Ditchfield C., Johnson V.L., Tighe A., Ellston R., Haworth C., Johnson T., Mortlock A., Keen N., Taylor S.S., Johnson V.L., Tighe A., Ellston R., Haworth C., Johnson T., Mortlock A., Keen N., Taylor S.S., Tighe A., Ellston R., Haworth C., Johnson T., Mortlock A., Keen N., Taylor S.S., Ellston R., Haworth C., Johnson T., Mortlock A., Keen N., Taylor S.S., Haworth C., Johnson T., Mortlock A., Keen N., Taylor S.S., Johnson T., Mortlock A., Keen N., Taylor S.S., Mortlock A., Keen N., Taylor S.S., Keen N., Taylor S.S., Taylor S.S. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and CENP-E to kinetochores. J. Cell Biol. 2003;161:267–280. doi: 10.1083/jcb.200208091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R.D., St Jean A., Woods R.A., Schiestl R.H., St Jean A., Woods R.A., Schiestl R.H., Woods R.A., Schiestl R.H., Schiestl R.H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick K.G., Murray A.W., Murray A.W. Mad1p, a phosphoprotein component of the spindle assembly checkpoint in budding yeast. J. Cell Biol. 1995;131:709–720. doi: 10.1083/jcb.131.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick K.G., Johnston R.C., Smith D.L., Murray A.W., Johnston R.C., Smith D.L., Murray A.W., Smith D.L., Murray A.W., Murray A.W. MAD3 encodes a novel component of the spindle checkpoint which interacts with Bub3p, Cdc20p, and Mad2p. J. Cell Biol. 2000;148:871–882. doi: 10.1083/jcb.148.5.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf S., Cole R.W., LaTerra S., Zimmer C., Schnapp G., Walter R., Heckel A., van Meel J., Rieder C.L., Peters J.M., Cole R.W., LaTerra S., Zimmer C., Schnapp G., Walter R., Heckel A., van Meel J., Rieder C.L., Peters J.M., LaTerra S., Zimmer C., Schnapp G., Walter R., Heckel A., van Meel J., Rieder C.L., Peters J.M., Zimmer C., Schnapp G., Walter R., Heckel A., van Meel J., Rieder C.L., Peters J.M., Schnapp G., Walter R., Heckel A., van Meel J., Rieder C.L., Peters J.M., Walter R., Heckel A., van Meel J., Rieder C.L., Peters J.M., Heckel A., van Meel J., Rieder C.L., Peters J.M., van Meel J., Rieder C.L., Peters J.M., Rieder C.L., Peters J.M., Peters J.M. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore–microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol. 2003;161:281–294. doi: 10.1083/jcb.200208092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indjeian V.B., Stern B.M., Murray A.W., Stern B.M., Murray A.W., Murray A.W. The centromeric protein Sgo1 is required to sense lack of tension on mitotic chromosomes. Science. 2005;307:130–133. doi: 10.1126/science.1101366. [DOI] [PubMed] [Google Scholar]
- Kaiser C., Michaelis S., Mitchell A., Michaelis S., Mitchell A., Mitchell A. Methods in yeast genetics. A Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1994. [Google Scholar]
- Kang J., Cheeseman I.M., Kallstrom G., Velmurugan S., Barnes G., Chan C.S., Cheeseman I.M., Kallstrom G., Velmurugan S., Barnes G., Chan C.S., Kallstrom G., Velmurugan S., Barnes G., Chan C.S., Velmurugan S., Barnes G., Chan C.S., Barnes G., Chan C.S., Chan C.S. Functional cooperation of Dam1, Ipl1, and the inner centromere protein (INCENP)-related protein Sli15 during chromosome segregation. J. Cell Biol. 2001;155:763–774. doi: 10.1083/jcb.200105029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson M.A., Renduchitala K., Khodjakov A., Kapoor T.M., Renduchitala K., Khodjakov A., Kapoor T.M., Khodjakov A., Kapoor T.M., Kapoor T.M. Correcting improper chromosome-spindle attachments during cell division. Nat. Cell Biol. 2004;6:232–237. doi: 10.1038/ncb1102. [DOI] [PubMed] [Google Scholar]
- Lee M.S., Spencer F.A., Spencer F.A. Bipolar orientation of chromosomes in Saccharomyces cerevisiae is monitored by Mad1 and Mad2, but not by Mad3. Proc. Natl. Acad. Sci. 2004;101:10655–10660. doi: 10.1073/pnas.0404102101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew D.J., Burke D.J., Burke D.J. The spindle assembly and spindle position checkpoints. Annu. Rev. Genet. 2003;37:251–282. doi: 10.1146/annurev.genet.37.042203.120656. [DOI] [PubMed] [Google Scholar]
- Li R., Murray A.W., Murray A.W. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- Lizcano J.M., Goransson O., Toth R., Deak M., Morrice N.A., Boudeau J., Hawley S.A., Udd L., Makela T.P., Hardie D.G., Goransson O., Toth R., Deak M., Morrice N.A., Boudeau J., Hawley S.A., Udd L., Makela T.P., Hardie D.G., Toth R., Deak M., Morrice N.A., Boudeau J., Hawley S.A., Udd L., Makela T.P., Hardie D.G., Deak M., Morrice N.A., Boudeau J., Hawley S.A., Udd L., Makela T.P., Hardie D.G., Morrice N.A., Boudeau J., Hawley S.A., Udd L., Makela T.P., Hardie D.G., Boudeau J., Hawley S.A., Udd L., Makela T.P., Hardie D.G., Hawley S.A., Udd L., Makela T.P., Hardie D.G., Udd L., Makela T.P., Hardie D.G., Makela T.P., Hardie D.G., Hardie D.G., et al. LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J. 2004;23:833–843. doi: 10.1038/sj.emboj.7600110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logarinho E., Bousbaa H., Dias J.M., Lopes C., Amorim I., Antunes-Martins A., Sunkel C.E., Bousbaa H., Dias J.M., Lopes C., Amorim I., Antunes-Martins A., Sunkel C.E., Dias J.M., Lopes C., Amorim I., Antunes-Martins A., Sunkel C.E., Lopes C., Amorim I., Antunes-Martins A., Sunkel C.E., Amorim I., Antunes-Martins A., Sunkel C.E., Antunes-Martins A., Sunkel C.E., Sunkel C.E. Different spindle checkpoint proteins monitor microtubule attachment and tension at kinetochores in Drosophila cells. J. Cell Sci. 2004;117:1757–1771. doi: 10.1242/jcs.01033. [DOI] [PubMed] [Google Scholar]
- Morrow C.J., Tighe A., Johnson V.L., Scott M.I., Ditchfield C., Taylor S.S., Tighe A., Johnson V.L., Scott M.I., Ditchfield C., Taylor S.S., Johnson V.L., Scott M.I., Ditchfield C., Taylor S.S., Scott M.I., Ditchfield C., Taylor S.S., Ditchfield C., Taylor S.S., Taylor S.S. Bub1 and aurora B cooperate to maintain BubR1-mediated inhibition of APC/CCdc20. J. Cell Sci. 2005;118:3639–3652. doi: 10.1242/jcs.02487. [DOI] [PubMed] [Google Scholar]
- Musacchio A., Hardwick K.G., Hardwick K.G. The spindle checkpoint: Structural insights into dynamic signalling. Nat. Rev. Mol. Cell Biol. 2002;3:731–741. doi: 10.1038/nrm929. [DOI] [PubMed] [Google Scholar]
- Pinsky B.A., Biggins S., Biggins S. The spindle checkpoint: Tension versus attachment. Trends Cell Biol. 2005;15:486–493. doi: 10.1016/j.tcb.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Pinsky B.A., Tatsutani S.Y., Collins K.A., Biggins S., Tatsutani S.Y., Collins K.A., Biggins S., Collins K.A., Biggins S., Biggins S. An Mtw1 complex promotes kinetochore biorientation that is monitored by the Ipl1/Aurora protein kinase. Dev. Cell. 2003;5:735–745. doi: 10.1016/s1534-5807(03)00322-8. [DOI] [PubMed] [Google Scholar]
- Pinsky B.A., Kung C., Shokat K.M., Biggins S., Kung C., Shokat K.M., Biggins S., Shokat K.M., Biggins S., Biggins S. The Ipl1–Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nat. Cell Biol. 2006;8:78–83. doi: 10.1038/ncb1341. [DOI] [PubMed] [Google Scholar]
- Rancati G., Crispo V., Lucchini G., Piatti S., Crispo V., Lucchini G., Piatti S., Lucchini G., Piatti S., Piatti S. Mad3/BubR1 phosphorylation during spindle checkpoint activation depends on both Polo and Aurora kinases in budding yeast. Cell Cycle. 2005;4:972–980. doi: 10.4161/cc.4.7.1829. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russell D., Russell D. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- Shannon K.B., Canman J.C., Salmon E.D., Canman J.C., Salmon E.D., Salmon E.D. Mad2 and BubR1 function in a single checkpoint pathway that responds to a loss of tension. Mol. Biol. Cell. 2002;13:3706–3719. doi: 10.1091/mbc.E02-03-0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoufias D.A., Andreassen P.R., Lacroix F.B., Wilson L., Margolis R.L., Andreassen P.R., Lacroix F.B., Wilson L., Margolis R.L., Lacroix F.B., Wilson L., Margolis R.L., Wilson L., Margolis R.L., Margolis R.L. Mammalian Mad2 and Bub1/BubR1 recognize distinct spindle-attachment and kinetochore-tension checkpoints. Proc. Natl. Acad. Sci. 2001;98:4492–4497. doi: 10.1073/pnas.081076898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern B.M., Murray A.W., Murray A.W. Lack of tension at kinetochores activates the spindle checkpoint in budding yeast. Curr. Biol. 2001;11:1462–1467. doi: 10.1016/s0960-9822(01)00451-1. [DOI] [PubMed] [Google Scholar]
- Tanaka T.U., Rachidi N., Janke C., Pereira G., Galova M., Schiebel E., Stark M.J., Nasmyth K., Rachidi N., Janke C., Pereira G., Galova M., Schiebel E., Stark M.J., Nasmyth K., Janke C., Pereira G., Galova M., Schiebel E., Stark M.J., Nasmyth K., Pereira G., Galova M., Schiebel E., Stark M.J., Nasmyth K., Galova M., Schiebel E., Stark M.J., Nasmyth K., Schiebel E., Stark M.J., Nasmyth K., Stark M.J., Nasmyth K., Nasmyth K. Evidence that the Ipl1–Sli15 (Aurora kinase–INCENP) complex promotes chromosome bi-orientation by altering kinetochore–spindle pole connections. Cell. 2002;108:317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- Taylor S.S., Hussein D., Wang Y., Elderkin S., Morrow C.J., Hussein D., Wang Y., Elderkin S., Morrow C.J., Wang Y., Elderkin S., Morrow C.J., Elderkin S., Morrow C.J., Morrow C.J. Kinetochore localisation and phosphorylation of the mitotic checkpoint components Bub1 and BubR1 are differentially regulated by spindle events in human cells. J. Cell Sci. 2001;114:4385–4395. doi: 10.1242/jcs.114.24.4385. [DOI] [PubMed] [Google Scholar]
- Williamson B.L., Marchese J., Morrice N.A., Marchese J., Morrice N.A., Morrice N.A. Automated identification and quantification of protein phosphorylation sites by LC/MS on a hybrid triple quadrupole linear ion trap mass spectrometer. Mol. Cell. Proteomics. 2006;5:337–346. doi: 10.1074/mcp.M500210-MCP200. [DOI] [PubMed] [Google Scholar]
- Yu H. Regulation of APC–Cdc20 by the spindle checkpoint. Curr. Opin. Cell Biol. 2002;14:706–714. doi: 10.1016/s0955-0674(02)00382-4. [DOI] [PubMed] [Google Scholar]
- Zachariae W., Nasmyth K., Nasmyth K. Whose end is destruction: Cell division and the anaphase-promoting complex. Genes & Dev. 1999;13:2039–2058. doi: 10.1101/gad.13.16.2039. [DOI] [PubMed] [Google Scholar]