Abstract
Mammalian euchromatic gene silencing results from the combined repressive effects of histone and DNA methyltransferases. Little is known of the mechanism by which these enzymes cooperate to induce silencing. Here we show that mammalian HP1 family members mediate communication between histone and DNA methyltransferases. In vitro, methylation of histone 3 Lys 9 by G9a creates a binding platform for HP1α, β, and γ. DNMT1 interacts with HP1 resulting in increased DNA methylation on DNA and chromatin templates in vitro. The functional and physical interaction can be recapitulated in vivo. Binding of GAL4-HP1 to a reporter construct is sufficient to induce repression and DNA methylation in DNMT1 wild-type but not DNMT1-null cells. Additionally, silencing of the Survivin gene coincides with recruitment of G9a and HP1 in DNMT1 wild-type but not null cells. We conclude that direct interactions between HP1 and DNMT1 mediate silencing of euchromatic genes.
Keywords: HP1, DNMT1, G9a, histone methylation, DNA methyltransferase
Repressed chromatin is characterized by epigenetic modifications including methylation of histone 3 at Lys 9 (H3K9) and methylation of DNA at CpG dinucleotides (Wallace and Orr-Weaver 2005; Horn and Peterson 2006). These modifications serve as binding sites for factors that control chromatin accessibility and regulate gene expression (Sims et al. 2003; Maison and Almouzni 2004; Horn and Peterson 2006; Kouzarides 2007). Although the necessity of histone methyltransferases (HMTs) and DNA methyltransferases (DNMTs) in the silencing of the mammalian genome is well established, the precise mechanisms by which the two enzymes cooperate have yet to be fully elucidated.
G9a is considered to be the major euchromatic HMT responsible for dimethylation of H3K9 (H3K9me2) at transcriptionally silent regions (Tachibana et al. 2002; Peters et al. 2003; Rice et al. 2003). G9a is targeted to specific genes where it functions to repress transcription (Duan et al. 2005; Chen et al. 2006). Methylated H3K9 serves as a binding platform for heterochromatin protein 1 (HP1) (Bannister et al. 2001; Lachner et al. 2001). In turn, HP1 associates with a variety of other factors including Suv39H1, histone deacetylases (HDACs), transcriptional repressors, and chromatin remodeling enzymes (Maison and Almouzni 2004; Hiragami and Festenstein 2005).
There are three mammalian isoforms of HP1 (HP1α, β, and γ), and all associate with methylated H3K9 through their chromodomains (Jacobs et al. 2001; Cheutin et al. 2003). HP1 binds specific euchromatic loci in Drosophila and mammals during the process of gene silencing (Ayyanathan et al. 2003; Greil et al. 2003). Mammalian HP1s are recruited to specific euchromatic loci in conjunction with G9a, but it has yet to be determined if G9a dimethylation of H3K9 plays a role in this recruitment (Roopra et al. 2004; Feldman et al. 2006). Additionally, although there are many models for how HP1 could be functioning at euchromatic loci, including interactions with components of transcriptional complexes (Vassallo and Tanese 2002) or with specific repressors (Nielsen et al. 1999), the mechanism of action is not well understood.
In mammals, DNA methylation is controlled by DNMT1, DNMT3a, and DNMT3b (Goll and Bestor 2005; Turek-Plewa and Jagodzinski 2005). DNMT1 is considered to be a maintenance methyltransferase, while DNMT3a and DNMT3b are considered to be de novo methyltransferases involved in the initial wave of methylation during embryogenesis and development (Siedlecki and Zielenkiewicz 2006). However, in recent years, the distinction between these DNMTs has become less rigid. It is now thought that DNMT3b plays a role in maintenance methylation and that DNMT1 is involved in de novo methylation in specific cases (Siedlecki and Zielenkiewicz 2006).
There is evidence for interdependency between histone and DNA methylation in several organisms. Mutations of the HMTs KYPTONITE in Arabidopsis thaliana and DIM-5 in Neurospora crassa decrease genome-wide levels of DNA methylation (Jackson et al. 2002; Tamaru et al. 2003). Furthermore, G9a knockout mouse embryonic stem (ES) cells exhibit a decrease in DNA methylation at multiple sites including the Prader Willi imprinting center (Xin et al. 2003) and the Oct-3/4 gene promoter (Feldman et al. 2006). The reciprocal effect is seen in Arabidopsis, where mutation of the DNA methyltransferase gene met1 leads to a reduction in H3K9 methylation (Soppe et al. 2002). DNMT1 and 3b double-knockout mouse ES cells display altered H3K9 methylation patterns at heterochromatin and specific tumor suppressor loci (Bachman et al. 2003; Espada et al. 2004). In addition, treatment of certain breast, bladder, and colorectal cancer cell lines with the demethylating agent 5-aza-CdR results in reactivation of multiple silenced genes concomitant with a decrease in H3K9 methylation (Nguyen et al. 2002; Bachman et al. 2003; Wozniak et al. 2006).
HP1 is involved in the silencing activities of both G9a and DNMTs. Localization of HP1 proteins is disrupted in G9a-null mouse ES cells (Tachibana et al. 2005). G9a and HP1 are both recruited for silencing of Oct-3/4 (Feldman et al. 2006) and several neuronal specific genes (Roopra et al. 2004), although the precise mechanism of silencing is not understood. Additionally, HP1β interacts with DNMT1 and DNMT3a in vitro and copurifies with DNA methyltransferase activity in cell extracts (Fuks et al. 2003). However, it is currently unknown whether this interaction has functional consequences and whether HP1α or γ also directly interacts with DNMT1. However, loss of DNMT1 affects HP1 localization to pericentromeric regions (Ma et al. 2005). These studies suggest that HP1 recruitment may be functionally related to both DNA and histone methylation.
We sought to understand the relationship between HP1 family proteins and histone and DNA methyltransferases. We focused on the euchromatic HMT G9a and DNMT1 as they have recently been implicated in silencing of specific genes in euchromatin. We find that all three HP1 family members directly interact with DNMT1. This interaction results in a functional stimulation of DNMT1 methyltransferase activity. Furthermore, HP1 is sufficient to target DNMT1 activity in vivo, and HP1-dependent repression requires DNMT1. Finally, we demonstrate that HP1 is recruited to the Survivin promoter in a DNMT1-dependent manner. Our data support a model whereby HP1s mediate the cooperative silencing effects of DNA and HMTs.
Results
Association of HP1 family proteins with DNMT1 in vitro
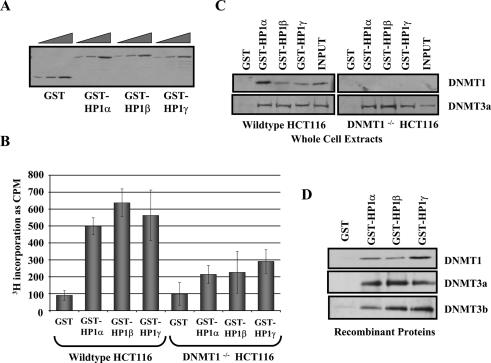
Previous studies reported that HP1β can associate with DNMT1 and DNMT3a in vitro (Fuks et al. 2003), raising the question of whether the interaction also occurs with HP1α and γ. Purified GST-HP1α, β, and γ (Fig. 1A) were used in pull-down assays (Fig. 1B) to determine if they associated with DNA methyltransferase activity from wild-type HCT116 cell extracts. We chose HCT116 cells because DNMT1 has been knocked out somatically in this cell line, and isogenic wild-type and DNMT1-null versions exist (Rhee et al. 2000). GST-HP1α, β, and γ were bound to glutathione-linked beads, incubated with whole-cell extracts (WCEs) derived from wild-type HCT116 cells and washed to remove unbound protein. The DNA methyltransferase activity was assayed by addition of DNA template and 3H-AdoMet substrate to the beads. The bar graph in Figure 1B shows that GST-HP1α, β, and γ capture a DNA methyltransferase activity from wild-type HCT116 extract. In contrast, a two- to threefold reduction in DNA methyltransferase activity was observed when the GST-HP1s were incubated with equal amounts of extract from the DNMT1-null HCT116 cell line. The GST-HP1 pull-downs were analyzed by immunoblotting with antibodies specific to DNMT1 and DNMT3a (Fig. 1C). GST-HP1 binds to DNMT1 in the wild-type extracts and binds equally well to DNMT3a in both wild-type and DNMT1-null extracts. We were unable to detect association with DNMT3b for reasons that are presently unclear. Nevertheless, these data suggest that the reduction in DNA methyltransferase activity in the null extracts is due to loss of DNMT1 association.
Figure 1.
HP1 proteins interact directly with DNMT1. (A) Coomassie blue-stained SDS-PAGE gel of full-length GST-HP1α, β, and γ. Threefold titration steps for each. (B) HP1s associate with DNA methyltransferase activity. GST-HP1α, β, and γ bound to glutathione agarose beads were incubated with WCEs from wild-type or DNMT1-null HCT116 cells. GST alone was used as a negative control. After removal of unbound proteins, DNA methylation reactions were performed by addition of 1 μg of 5S DNA template and 3H-SAM substrate. The level of DNA methyltransferase activity associated with the GST-HP1s is measured by 3H-CH3 incorporation into the DNA template. The supernatants were transferred to DE81 Whatman paper and washed with 200 mM ammonium bicarbonate, water, and 100% ethanol to remove unincorporated 3H. Samples were added to scintillation fluid and counted. Values are given in counts per minute. The error bars represent standard deviation from triplicate experiments. (C) HP1s associate with DNMT1 and 3a in cell extracts. Pull-downs were repeated and analyzed by SDS-PAGE and immunoblotting with antibodies specific for DNMT1 and DNMT3a. (D) HP1s directly bind to DNMT1. GST pull-downs were repeated with purified DNMT1, and binding was assayed by SDS-PAGE and immunoblotting with a DNMT1-specific antibody.
We used purified DNMT1, 3a, and 3b in the GST-HP1 pull-down assay to determine if the HP1–DNMT interaction is direct. DNMT1, 3a, and 3b associated with all three HP1 isoforms as analyzed by immunoblotting (Fig. 1D). Binding to HP1 was specific as DNMT1, 3a, and 3b displayed little binding to the GST negative control. Previous studies demonstrated that HP1β interacts with the regulatory and methyltransferase domains of DNMT1 (Fuks et al. 2003). We repeated the GST pull-down using a variety of different DNMT1 domains, and our data agree with the HP1β results (data not shown).
Additionally, we analyzed the domains of HP1α, β, and γ that contact DNMT1 using GST-pull-down experiments (Supplementary Fig. 1A) with purified DNMT1. DNMT1 bound with highest affinity to the chromodomain of HP1α, β, and γ (Supplementary Fig. 1C). We conclude that interactions with DNMT1, 3a, and 3b are a general property of all mammalian HP1s and that DNMT1 interacts with a common domain of HP1α, β, and γ.
HP1s stimulate the DNA methyltransferase activity of DNMT1 on DNA
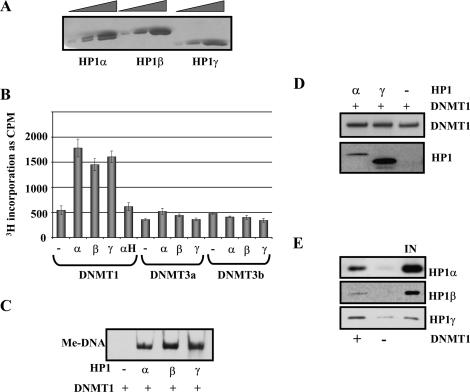
The direct interaction of HP1 and DNMT1, 3a, and 3b suggested the possibility that HP1 might influence the catalytic activity of DNA methyltransferases. This issue was explored by analyzing the activity of DNMT1, 3a, and 3b on their own and in the context of HP1 using either gel fluorography or scintillation counting assays (see Materials and Methods). Supplementary Figure 2 is a dose-response curve demonstrating that DNMT1, 3a, and 3b display de novo methylation activity of a DNA fragment in vitro. We used this strategy to determine the linearity of the DNMT response. Next, a subsaturating level of DNMT1, 3a, or 3b was incubated with DNA alone or in the presence of purified HP1α, β, or γ (Fig. 2A). HP1 binds DNA with modest avidity through the hinge domain (Meehan et al. 2003). Remarkably, all three isoforms of HP1 stimulated catalytic activity of DNMT1 by at least threefold (Fig. 2B,C) but had little effect on DNMT3a and 3b (Fig. 2B). The effect was specific, as the hinge domain from HP1α (αH), which does not interact with DNMT1 (Supplementary Fig. 1C), does not stimulate DNMT1 activity (Fig. 2B).
Figure 2.
HP1s stimulate DNMT1 DNA methyltransferase activity in vitro. (A) Coomassie blue-stained SDS-PAGE gel of purified HP1α, β, and γ. Threefold titration steps for each. (B,C) DNMT1 activity is stimulated by the addition of HP1s. In vitro DNA methyltransferase activity reaction was repeated with DNMT1, 3a, or 3b in the presence or absence of 100 ng of either HP1α, β, γ, or HP1α-hinge. Methylation levels were assayed by scintillation counting (the error bars represent standard deviation from triplicate experiments) (B) or gel fluorography (C). (D) HP1s do not alter DNMT1 recruitment in vitro. One-hundred nanograms of 5S DNA immobilized template were incubated with either HP1α or γ for 30 min at room temperature. After washing, DNMT1 was added to the template for 1 h at 37°C. DNMT1 and HP1 binding was analyzed by immunoblotting. (E) DNMT1 can increase binding of HP1s to DNA. One-hundred nanograms of immobilized template were preincubated with DNMT1. After washing, either HP1α, β, or γ was incubated with the template, and binding of HP1s was measured by immunoblotting with specific antibodies.
We considered two models to explain how HP1 stimulated DNMT1 activity. In one, HP1 helps DNMT1 bind DNA, thereby increasing DNA methylation. Alternatively, HP1 may increase the specific catalytic activity of DNMT1 independent of recruitment. We used an immobilized template approach to distinguish between these models. Various combinations of DNMT1 and HP1 were added to DNA or chromatin templates, which were biotinylated and immobilized on streptavidin-coated magnetic beads. After washing the templates with buffer, the amount of retained protein was measured by Western blotting and activity assays (Johnson et al. 2004). Although HP1α and γ were able to bind to the immobilized DNA template (Fig. 2D, bottom panel), they did not significantly increase the levels of DNMT1 associating with the template as analyzed by Western blotting (Fig. 2D, top panel). Based on the data in Figure 2, C and D, we conclude that HP1 family members are functioning to stimulate the DNA methyltransferase activity and not the recruitment of DNMT1.
Since DNMT1 binds quite robustly to DNA (Esteve et al. 2006b), we also analyzed the effect of DNMT1 on the association of HP1α, β, and γ. HP1α, β, and γ were bound to DNA in the presence or absence of prebound DNMT1. Binding of the HP1 proteins was measured by Western blotting with specific HP1 antibodies. We found that DNMT1 increased HP1α, β, and γ binding to the immobilized DNA template (Fig. 2E) by several fold. The increased binding was due to DNMT1 and not methylated DNA since addition of AdoMet did not alter HP1 recruitment (data not shown). These data illustrate a cooperative relationship in which DNMT1 increases the binding of HP1 family proteins, which in turn stimulate the activity of DNMT1.
HP1 stimulates DNMT1 activity on chromatin in a G9a-mediated histone methylation-dependent manner
In an attempt to understand whether HP1 and DNMT1 bound cooperatively in the context of chromatin, we recreated HP1 binding to methylated chromatin. HP1 proteins are known to bind H3K9me3 in the context of heterochromatin (Jacobs et al. 2001; Cheutin et al. 2003). However, the affinity of the HP1 chromodomain for di- and trimethylated H3 tail peptides differs by only twofold (Fischle et al. 2003). We therefore reasoned that dimethylation of H3K9 by G9a (Patnaik et al. 2004) would create a binding site for HP1 on the immobilized nucleosomal arrays. The arrays were assembled as described previously (Ikeda et al. 1999; Black et al. 2006) using recombinant Xenopus octamers, and binding experiments were carried out as illustrated in Figure 3A. First, we demonstrated that purified G9a methylates H3K9 in the context of nucleosomal arrays. Incubation of increasing concentrations of G9a with a nucleosomal array resulted in an increase in H3 methylation as measured by gel fluorography (Fig. 3B). The identity of the methyl mark was verified by immunoblotting for H3K9me2 (Fig. 3C). G9a-treated chromatin was also analyzed for other modifications (Supplementary Fig. 3). Although a low amount of H3K27 methylation was observed, H3K9me2 is the predominant modification. Second, we showed that the dimethylation of H3K9 increased the binding of HP1α, β, and γ (Fig. 3D). HP1 binding can be stabilized by certain factors even in the absence of H3K9Me (Li et al. 2002). In order to distinguish the contributions of G9a and H3K9me2 to HP1 recruitment, the experiment was repeated with templates incubated with G9a and in the presence and absence of AdoMet. Although HP1α binds slightly better in the presence of G9a alone, recruitment was increased greatly by methylation (Fig. 3E).
Figure 3.
HP1s stimulate DNMT1 activity on chromatin in a H3K9me2-dependent manner. (A) Schematic of HP1 binding and DNA methyltransferase activity assays. (B) G9a HMT activity in vitro. Increasing concentrations of G9a were incubated with 375 ng of chromatin array, and 3H-SAM as a substrate, for 2 h at room temperature. Samples were separated on a 10% SDS-PAGE gel, and histone methylation levels were assayed by gel fluorography. (C) G9a methylates H3K9. Two micrograms of free octamers were incubated ±G9a and AdoMet, as indicated, for 2 h at room temperature. Samples were run on an SDS-PAGE gel and immunoblotted with an antibody specific to H3K9me2. (D) Increased recruitment of HP1s to chromatin with H3K9me2. One-hundred nanograms of array were treated ±G9a and AdoMet for 2 h at room temperature, then washed to remove unbound proteins. This methylated (Me) or unmethylated (UnMe) array was incubated with either HP1α, β, or γ for 30 min, after washing amount of bound HP1 was analyzed by SDS-PAGE and immunoblotting with HP1-specific antibodies. Input represents 3% and 10% of HP1. (E) Binding of HP1α is increased by H3K9me2. One-hundred nanograms of array were preincubated with G9a and AdoMet, G9a alone, or nothing. The array was incubated with HP1α for 30 min. After washing, the amount of HP1α bound to the template was analyzed by SDS-PAGE and immunoblotting with HP1α-specific antibodies. (F) HP1s stimulate DNMT1 activity on chromatin arrays. The DNMT1 DNA methyltransferase assay was repeated as in Figure 2C with 1 μg of chromatin array, either unmethylated or methylated by G9a as in D, in the presence or absence of 100 ng of HP1α, β, or γ. DNA was isolated by phenol/chloroform extraction and ethanol precipitation. Methylation levels were assayed by scintillation counting. The error bars represent standard deviation from triplicate experiments.
The role of histone methylation in DNA methylation was explored by analyzing the ability of HP1 to stimulate DNMT1 activity in vitro on nucleosomal arrays. The DNMT1 DNA methyltransferase activity assay was performed on nucleosomal arrays bearing either dimethylated or unmethylated H3K9 in the presence or absence of HP1. HP1α, β, and γ stimulated DNMT1 activity on the unmethylated nucleosomal arrays (Fig. 3F). Methylation by G9a alone had no stimulatory effect on DNMT1 under these assay conditions. However, in the presence of HP1α, β, and γ, DNMT1 activity was stimulated three- to fourfold (Fig. 3F). The increase in DNMT1 activity correlated well with the increase in binding of HP1 to H3K9me2 observed in Figure 3D. This result provides support for a mechanism whereby H3K9 methylation by G9a could influence the DNA methylation status of a euchromatic region via HP1 recruitment.
HP1 directs DNMT1-dependent DNA methylation in vivo
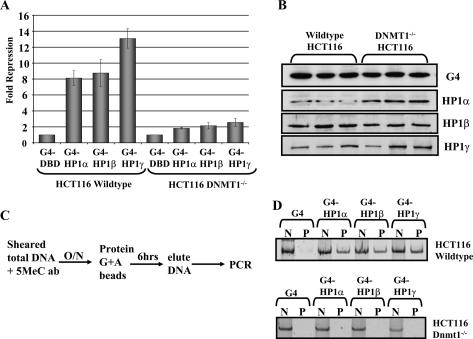
We used an HP1 tethering assay to establish a direct causal link between HP1 family proteins and DNMT1 in vivo. In this assay, HP1α, β, and γ were fused at their N termini to the DNA-binding domain (DBD) of the yeast GAL4 protein. Expression vectors bearing either GAL4-DBD alone or GAL4-DBD–HP1 were transfected into HCT116 cells along with a GAL4-responsive luciferase reporter, G5-Luc (Zhang et al. 2002). Transfection of the G5-Luc reporter into wild-type HCT116 cells with or without the GAL4-DBD elicited the same basal level of luciferase (data not shown). Addition of GAL4-DBD–HP1α, β, or γ resulted in an eightfold to 13-fold repression compared with the basal level elicited in the presence of the GAL4-DBD alone (Fig. 4A). In contrast, in the DNMT1-null cell line, GAL4-HP1α, β, or γ repressed the expression of the reporter construct by less than threefold. The levels of GAL4-HP1 expression in the two cell lines are similar as measured by Western blotting (Fig. 4B). Therefore, we conclude that DNMT1 is important for complete repression by HP1 in vivo. It is possible that association with DNMT3a and 3b could be responsible for the residual silencing by HP1α, β, or γ observed in the absence of DNMT1.
Figure 4.
HP1s and DNMT1 cooperate in gene silencing. (A) HP1 requires DNMT1 for full silencing of a luciferase reporter construct. GAL4-DBD and GAL4-DBD–HP1 expression constructs were transfected with a GAL4-responsive luciferase reporter into wild-type or DNMT1-null HCT116 cells. After 48 h, luciferase expression was assayed. The graph shows fold repression of luciferase levels, as measured by luminometer, by GAL4-DBD–HP1s as compared with the GAL4-DBD control. The error bars represent standard deviation from triplicate experiments. (B) Expression levels of GAL4-DBD and GAL4-DBD–HP1s were assayed by SDS-PAGE gel and immunoblotting with GAL4-specific antibodies. (C) Schematic of MeDIP assay. (D) GAL4-HP1s direct a DNMT1-dependent increase in DNA methylation. Total DNA was isolated from the transfected cells in A. After sonication, to shear the DNA, the samples were immunoprecipitated with a 5meC antibody overnight at 4°C. After washing, the DNA associated with the 5meC ab was harvested. The amount of methylated DNA recovered by the IP was assayed by PCR with primers specific to the GAL4-responsive luciferase reporter. (N) Input control; (P) IP.
We adapted the methylated DNA immunoprecipitation (MeDIP) assay from Zhang et al. (2006) to investigate methylation levels of the G5-Luc reporter. Total DNA was isolated from the transfection assays, sheared, and immunoprecipitated with a 5MeC antibody. PCR analysis was used to determine whether the reporter construct was enriched in methylated DNA in the presence of HP1 and DNMT1. GAL4-DBD–HP1α, β, and γ caused a dramatic increase in DNA methylation on the G5-Luc template (Fig. 4D, top panel). The effect was due to HP1 because methylation was not observed with the GAL4-DBD alone. Furthermore, methylation was not detected in the DNMT1-null cell line even in the presence of GAL4-HP1 (Fig. 4D, bottom panel). The effect of HP1 on DNA methylation levels was specific to the reporter template as there was no increase in methylation at the Actin gene in these cells (data not shown). We conclude that HP1 has the ability to stimulate DNMT1dependent DNA methylation on a promoter region in vivo.
HP1s, G9a, and DNMT1 cooperatively silence the Survivin gene in vivo
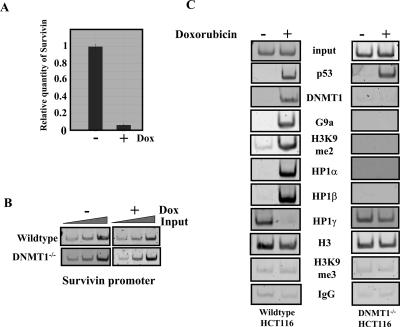
Survivin is an inhibitor of apoptosis, which is down-regulated by p53 in response to DNA damage by doxorubicin and other mutagens (Hoffman et al. 2002). Figure 5A is a qPCR experiment that quantitates Survivin expression levels in HCT116 cells after doxorubicin treatment. Previous work on the Survivin gene suggested that it might serve as a good model to study the relationship between DNMT1 and HP1 on an endogenous gene. Studies have demonstrated that G9a and DNMT1 are recruited to the Survivin gene. Additionally, DNA and H3K9 methylation correlate with silencing of expression (Esteve et al. 2005, 2006a). We used chromatin immunoprecipitation (ChIP) to explore the interdependency of HP1 and DNMT1 in this context. Formaldehyde-cross-linked chromatin from untreated and doxorubicin-treated cells was immunoprecipitated with antibodies specific for p53, DNMT1, G9a, H3, H3K9me3, and H3K9me2 along with IgG as a negative control. PCR was carried out with increasing amounts of input DNA to test the linearity of the amplification (Fig. 5B). DNMT1, G9a, and p53 do not appear to bind the promoter in the absence of doxorubicin, and only trace levels of H3K9me2 are detected (Fig. 5C, left column). However, we observed an increase in p53, DNMT1, G9a, and H3K9me2 upon doxorubicin treatment (Fig. 5C, left column). Interestingly, when we measured HP1 recruitment, HP1γ was found to be bound to the active Survivin promoter, whereas HP1α and β were not. After doxorubicin treatment HP1γ occupancy decreased, and HP1α and β occupancy increased (Fig. 5C, bottom three panels). H3 was observed in both doxorubicin-treated and untreated extracts, whereas H3K9me3 was not observed at the Survivin promoter under any conditions tested. The H3K9me3 antibody was functioning, since we detected H3K9me3 at other loci including α-satellite DNA and the IGS-rDNA (data not shown). To determine if corecruitment of DNMT1, G9a, and HP1 occurs at other genes, we performed ChIP on the Cdc2 and Cdc25 promoters, which are also repressed by p53 in response to doxorubicin treatment (Supplementary Fig. 4). We found that the HP1, G9a, and DNMT1-binding profiles, as well as the H3K9me2, were similar to Survivin. We conclude that cooperation between DNMT1, G9a, and HP1s is a general mechanism for euchromatic gene silencing.
Figure 5.
HP1s cooperate with DNMT1 to silence Survivin in response to doxorubicin treatment. (A) Survivin is down-regulated after doxorubicin treatment. HCT116 cells were treated with or without doxorubicin for 12 h. Cells were harvested, and total RNA was isolated. RNA levels were normalized by spectrometry, and RT–qPCR was performed using Survivin-specific primers to analyze expression levels. (B) PCR with primers specific to the Survivin proximal promoter region was performed on increasing levels of input DNA from parental and DNMT1-null cells to test the linearity of the amplification. (C) ChIP analysis for the presence of p53; DNMT1; G9a; H3K9me2; HP1α, β, and γ; H3; and H3K9me3 at the Survivin promoter in parental and DNMT1−/− HCT116 cells. Chromatin extracts from cells treated either with or without doxorubicin for 12 h were added to the antibodies indicated and incubated overnight at 4°C. Antibody–protein–DNA complexes were isolated by binding to Dynal protein G beads. DNA was purified and assayed by PCR with primers specific to the Survivin promoter region.
As shown in Figure 2, DNMT1 stabilizes HP1 binding in vitro. The effect of DNMT1 on HP1 binding in vivo was investigated by comparing HP1 binding by ChIP analysis in DNMT1 wild-type and null HCT116 cells. Survivin expression is unaffected by doxorubicin treatment in cells lacking DNMT1 (Esteve et al. 2005). Importantly, in the DNMT1-null cells, we could not detect binding of G9a and H3K9me2 (Fig. 5C, right column) even though p53 is recruited. Additionally, HP1α and β are no longer recruited, whereas HP1γ occupies the promoter even after doxorubicin treatment (Fig. 5C). Therefore, we conclude that silencing of a target gene in vivo requires DNMT1 to cooperatively recruit G9a and HP1.
Discussion
G9a-null mouse ES cells display DNA hypomethylation at specific loci genome-wide (Ikegami et al. 2007). It has been hypothesized that HMTs elicit such effects on DNA methylation via adapter proteins (Maison and Almouzni 2004; Feldman et al. 2006). However, little direct biochemical evidence was available to demonstrate an exact mechanism of action. The data presented here provide a mechanism by which HP1 can mediate communication between G9a and DNMT1 in the silencing of euchromatic genes. Our data support the model in Figure 6, whereby recruitment of G9a results in dimethylation of H3K9, thereby creating a binding site for mammalian HP1 proteins. HP1s stimulate DNMT1 activity and increase levels of DNA methylation in the surrounding area both in vitro and in vivo. Additionally, DNMT1 can act as an accessory factor to stabilize the binding of HP1s to chromatin templates. Previous studies have demonstrated that DNMT1 can increase G9a activity on nucleosomal substrates and G9a can increase DNMT1 recruitment (Esteve et al. 2006b). Taken together, these data describe a mechanism whereby DNMT1, HP1, and G9a form a positive feedback loop and functionally interact to coordinate the silencing of genes. Coordination of all these factors in this manner could be a mechanism to ensure the fidelity of silencing.
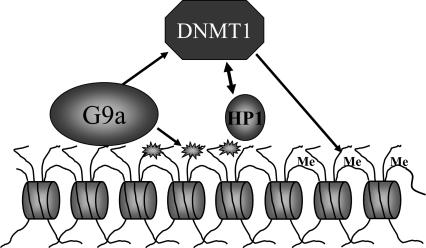
Figure 6.
Model of G9a, DNMT1, and HP1 functional interactions. G9a dimethylates the array template, creating a binding site for HP1α, β, or γ. G9a interacts with DNMT1. HP1s act to stimulate the DNA methyltransferase activity of DNMT1. HP1 binding is stabilized by DNMT1.
A previous study demonstrated that HP1β interacted with certain domains of DNMT1 and 3a in vitro and could purify a DNA methyltransferase activity from WCEs (Fuks et al. 2003). Our study has expanded upon this result and demonstrated that interaction with DNMT1, 3a, and 3b is a general property of mammalian HP1 proteins. Our data demonstrate that HP1α, β, and γ can significantly stimulate DNMT1 activity on DNA. Interestingly, this increase in DNA methyltransferase activity was not due to increased recruitment of DNMT1 by HP1 as seen for G9a stimulation of DNMT1 (Esteve et al. 2006b). Therefore, we conclude that mammalian HP1s stimulate the enzymatic activity of DNMT1. In contrast, HP1α, β, or γ had no effect on the activity of DNMT3a or 3b despite the fact that they formed a complex. DNMT1 also increases HP1 binding to our immobilized templates. It has been suggested that HP1s require auxiliary factors to stabilize binding (Eskeland et al. 2007). We propose that DNMT1 is an auxiliary factor for HP1 association with chromatin. The discovery of a functional interaction between HP1 and DNMT1 suggests a mechanism by which HP1 could mediate communication and help coordinate the action of DNMTs and HMTs.
G9a and HP1 are recruited in concert for the silencing of certain euchromatic genes (Roopra et al. 2004; Feldman et al. 2006). However, it had not been demonstrated that HP1s associate with chromatin via the H3K9me2 mark created by G9a. The data presented here show that H3K9 dimethylation increases the binding of HP1α, β, and γ to chromatin in vitro. As on DNA, HP1s also greatly stimulate DNMT1 activity on naive chromatin. Methylation by G9a increases recruitment of HP1s, resulting in a further increase in DNMT1 activity. Combined with the interaction of G9a and DNMT1 (Esteve et al. 2006b), our results describe a cooperative silencing cycle whereby G9a methylates chromatin recruiting HP1s and DNMT1. HP1 then stimulates DNMT1, which also recruits more HP1, resulting in increased DNA methylation. Interactions between G9a, HP1, and DNMT1 would lead to recruitment of each other to adjacent regions of the gene locus analogous to the spreading of histone methylation (Schotta et al. 2002; Hediger and Gasser 2006).
HP1 stimulation of DNMT1 activity suggests how DNMT1 could function as a de novo methylase. DNMT1 has de novo methyltransferase activity in vitro (Gowher et al. 2005). Additionally, DNMT1 has been implicated in de novo methylation in vivo (Esteve et al. 2005; Le Gac et al. 2006), including at CpG islands, which are hypermethylated in human cancers (Jair et al. 2006). Overexpression of DNMT1 causes de novo methylation of unmethylated genes (Biniszkiewicz et al. 2002). It is thought that the N-terminal domain is responsible for regulating DNMT1 de novo activity in vivo (Siedlecki and Zielenkiewicz 2006). HP1α, β, and γ interact with both the regulatory and methyltransferase domains of DNMT1 (Fuks et al. 2003; data not shown). Binding of hemimethylated DNA to the regulatory domain of DNMT1 has been shown to allosterically activate the enzyme (Pradhan and Esteve 2003a). Therefore, HP1s could stimulate DNMT1 activity on unmethylated DNA via a similar mechanism. DNMT3a and 3b lack the extended N-terminal region that DNMT1 possesses, which could explain the specificity of action of HP1α, β, and γ for DNMT1. Tethering of HP1s to chromatin in vivo is sufficient to cause formation of heterochromatin (Hiragami and Festenstein 2005). The formation of silenced regions was assayed by an increase in histone methyl marks and in some cases silencing of a reporter gene. However, these studies did not examine the effect of HP1 presence on DNA methylation levels. Our tethering assay demonstrated that HP1α, β, and γ function to silence a reporter gene concomitant with an increase in DNA methylation. This demonstrates that HP1s can specifically stimulate DNMT1-dependent DNA methylation in vivo.
DNMT1 and G9a are recruited to the promoter of Survivin upon silencing, causing an increase in DNA and histone methylation (Esteve et al. 2005). We have demonstrated that HP1s also play a role in the regulation of Survivin. ChIP analysis indicates that HP1γ is present when the Survivin gene is active. This is in agreement with previous studies stating that HP1γ and repressive H3K9 methyl marks are associated with the coding regions of active genes (Vakoc et al. 2005). Upon silencing, HP1γ binding decreases and HP1α and β are recruited along with G9a and DNMT1. HP1α and β appear to play a role in the inducible silencing of Survivin, whereas HP1γ does not. This disparity of action could be explained by the absence of G9a and DNMT1. Both of these factors require p53 binding to associate with the Survivin promoter, which does not occur on the active gene. Also, other chromatin modifications and activating factors could reduce the ability of HP1γ to associate with G9a and DNMT1 and hence attenuate its function in this specific situation. The loss of DNMT1 disrupts silencing of Survivin in response to doxorubicin, and this event correlates with a loss of G9a, H3K9me2, HP1α, and β binding to the promoter. Our data demonstrate that all of the factors involved are required for assembly of the repressive complex necessary for full silencing of Survivin and strengthen the idea that they interact cooperatively in a higher-order complex. In conclusion, we have presented biochemical and in vivo evidence that the interaction between HP1 and DNMT1 is important for euchromatic gene silencing.
Materials and methods
Cell culture and extracts
HCT116 wild-type cells were obtained from American Type Culture Collection and cultured in McCoy’s modified medium supplemented with 10% FBS and 5% Pen-Strep. HCT116 DNMT1-null cells were obtained from Dr. Bert Vogelstein (Johns Hopkins, Baltimore, MD) (Rhee et al. 2000) and cultured as above except supplemented with 0.1 mg/mL Hygromycin. WCEs were prepared by washing the plate once in 10 mL of 1× PBS and then harvesting them in 1 mL of PBS. Cells were resuspended in 1 mL of RIPA buffer (50 mM Tris at pH 7.4, 1% NP-40, 0.25% Na-deoxycholate, 150 mM NaCl, 1 mM EDTA) and incubated for 30 min at 4°C with rotation. Insoluble cell debris was pelleted by microcentrifugation.
Chromatin preparation
Chromatin arrays were prepared and biotinylated as described previously (Ikeda et al. 1999), using recombinant Xenopus histones assembled into octamers by stepwise salt dialysis as described previously (Luger et al. 1999). Chromatin was immobilized on M280 streptavidin beads (DYNAL) in chromatin-binding buffer (0.3 M KCl, 10% glycerol, 4 mM MgCL2, 1 mM DTT, and 200 μg/mL BSA). Chromatin assembly was assessed as described previously (Black et al. 2006).
GST constructs and pull-downs
Full-length GST-HP1 constructs were a gift from Dr. Naoko Tanese (New York University School of Medicine, New York, NY) (Vassallo and Tanese 2002). The constructs were expressed in bacteria and purified by binding to glutathione resin as described previously (Vassallo and Tanese 2002). Purified HP1 proteins were prepared by incubation of GST-HP1 glutathione beads with thrombin overnight at 4°C. The glutathione beads were collected by centrifugation, and the supernatant containing the HP1 proteins was removed and stored at −80°C. For GST pull-down experiments, 2 μg of each GST-HP1 was incubated with 500 μg of HCT116 WCE and 200 μL of binding buffer (50 mM HEPES, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 10% glycerol) overnight at 4°C. The DNA methyltransferase activity of bound protein was assayed after washing three times with binding buffer.
DNA and HMT assay
DNMT1 was purified as described previously (Pradhan and Esteve 2003b). DNA methyltransferase assays were carried out for 60 min at 37°C in duplicate with a total volume of 25 μL of reaction mix as previously described (Pradhan and Esteve 2003b). Briefly, 20 nM DNMT1, 3a, or 3b was incubated with 1 μg of 5S DNA or chromatin array template, 2 μCi of 3H-SAM in 50 mM Tris-HCl (pH 7.8), 1 mM EDTA, 5% glycerol, and 100 μg/mL BSA; 100 ng (194 nM) of HP1α, β, or γ was added with DNMT1. The reactions were spotted on to DE81 (Whatman) membrane and processed as described previously (Esteve et al. 2005); or samples were electrophoresed by native PAGE, and the gel was incubated in Amplify reagent (Amersham) for 30 min at room temperature and then dried. The dried gel was exposed to film for 2 d at −80°C (gel fluorography).
G9a was obtained from New England Biolabs. The HMT assay was performed by incubating 40 nM G9a with chromatin array, 2 μCi of 3H-SAM in 50 mM Tris (pH 9), 5 mM MgCl2, 1 mM DTT, and 0.1 mM PMSF for 2 h at room temperature. Methylation was assayed by SDS-PAGE electrophoresis and gel fluorography as described above.
Immobilized template recruitment assay
Chromatin arrays were incubated in the presence or absence of G9a and the cofactor AdoMet (90 μM) for 60 min at room temperature under the reaction conditions above. After washing to remove unbound proteins, 100 ng of template were incubated with 100 ng of either HP1α, β, or γ for 30 min at room temperature. After washing, the templates were analyzed for HP1 recruitment by SDS-PAGE and immunoblotting; antibodies to HP1s were a gift of Dr. Steve Smale, University of California at Los Angeles (Los Angeles, CA).
Transfection assay
Transfection assays were carried out on wild-type and DNMT1-null HCT116 cells using Tfx-50 reagent, as per the manufacturer’s instruction (Promega). Briefly, 500 ng of G5E4T-luciferase (Zhang et al. 2002) were transfected along with 500 ng of the expression constructs pBXG0-GAL4-DBD (Emami and Carey 1992), GAL4-DBD-HP1α, β, or γ. After 48 h, the expression level of the luciferase reporter gene was assayed by luminometry according to the manufacturer’s instructions (Promega).
Methyl-dependent immunoprecipitation assay
This assay was adapted from Zhang et al. (2006). Total DNA was isolated from the transfected cells above by phenol-chloroform extraction and ethanol precipitation. DNA was incubated in FB buffer (10 mM Tris at pH 7.5, 50 mM NaCl, 1 mM EDTA) with 1 μg of 5meC antibody (Calbiochem) overnight at 4°C. Antibody–DNA complexes were isolated by addition of 20 μL each of Dynal protein G and A beads for 6 h at 4°C. Beads were washed five times in FB buffer, and DNA was eluted by vortexing in 100 μL of TE with 1.5%, 0.5%, and 0.1% SDS. Eluates were pooled, and the DNA was isolated by phenol extraction and ethanol precipitation (Zhang et al. 2006). The amount of methylated DNA enriched in immunoprecipitations (IPs) was analyzed by PCR with G5E4T-Luc vector-specific primers and compared with input DNA for each sample.
ChIP
HCT116 cells were grown on 150-mm dishes and treated with 1 μM doxorubicin. After 12 h, proteins were cross-linked with DNA by addition of 1% formaldehyde for 15 min at 37°C. Cells were washed with cold PBS, harvested, and lysed in an SDS lysis buffer (1% SDS, 50 mM Tris at pH 8, 20 mM EDTA). The lysates were sonicated to shear DNA to lengths between 150 and 700 basepairs (bp). After 10-fold dilution in ChIP dilution buffer (16.7 mM Tris, 0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 167 mM NaCl), IPs were carried out overnight at 4°C with 2 μg of DNMT1, G9a, p53, di-meH3K9, HP1α, β, γ, H3, tri-meH3K9, or 2 μg of normal IgG as a negative control. Fifty microliters of Dynal protein G beads were added to each sample for 1 h, and the beads were then washed as per the Upstate Biotechnology ChIP protocol. DNA was eluted twice with 100 μL of TE with 1% SDS for 10 min at 65°C. The cross-links were reversed overnight at 65°C. Proteinase K was added for 1 h at 65°C, then DNA was recovered by phenol extraction and ethanol precipitation. Immunoprecipitated DNA was analyzed for the presence of the Survivin (Esteve et al. 2005), the Cdc2, or the Cdc25 promoters (Le Gac et al. 2006) by PCR as described previously.
Acknowledgments
We thank Naoko Tanese for the GST-HP1 constructs, Bert Vogelstein for wild-type and DNMT1-null HCT116 cell lines, and Steve Smale (University of California at Los Angeles, Los Angeles, CA) for HP1 antibodies. This work was supported by a National Institutes of Health grant (GM074701) to M.C.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1536807
References
- Ayyanathan K., Lechner M.S., Bell P., Maul G.G., Schultz D.C., Yamada Y., Tanaka K., Torigoe K., Rauscher F.J., III, Lechner M.S., Bell P., Maul G.G., Schultz D.C., Yamada Y., Tanaka K., Torigoe K., Rauscher F.J., III, Bell P., Maul G.G., Schultz D.C., Yamada Y., Tanaka K., Torigoe K., Rauscher F.J., III, Maul G.G., Schultz D.C., Yamada Y., Tanaka K., Torigoe K., Rauscher F.J., III, Schultz D.C., Yamada Y., Tanaka K., Torigoe K., Rauscher F.J., III, Yamada Y., Tanaka K., Torigoe K., Rauscher F.J., III, Tanaka K., Torigoe K., Rauscher F.J., III, Torigoe K., Rauscher F.J., III, Rauscher F.J., III Regulated recruitment of HP1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing: A mammalian cell culture model of gene variegation. Genes & Dev. 2003;17:1855–1869. doi: 10.1101/gad.1102803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachman K.E., Park B.H., Rhee I., Rajagopalan H., Herman J.G., Baylin S.B., Kinzler K.W., Vogelstein B., Park B.H., Rhee I., Rajagopalan H., Herman J.G., Baylin S.B., Kinzler K.W., Vogelstein B., Rhee I., Rajagopalan H., Herman J.G., Baylin S.B., Kinzler K.W., Vogelstein B., Rajagopalan H., Herman J.G., Baylin S.B., Kinzler K.W., Vogelstein B., Herman J.G., Baylin S.B., Kinzler K.W., Vogelstein B., Baylin S.B., Kinzler K.W., Vogelstein B., Kinzler K.W., Vogelstein B., Vogelstein B. Histone modifications and silencing prior to DNA methylation of a tumor suppressor gene. Cancer Cell. 2003;3:89–95. doi: 10.1016/s1535-6108(02)00234-9. [DOI] [PubMed] [Google Scholar]
- Bannister A.J., Zegerman P., Partridge J.F., Miska E.A., Thomas J.O., Allshire R.C., Kouzarides T., Zegerman P., Partridge J.F., Miska E.A., Thomas J.O., Allshire R.C., Kouzarides T., Partridge J.F., Miska E.A., Thomas J.O., Allshire R.C., Kouzarides T., Miska E.A., Thomas J.O., Allshire R.C., Kouzarides T., Thomas J.O., Allshire R.C., Kouzarides T., Allshire R.C., Kouzarides T., Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Biniszkiewicz D., Gribnau J., Ramsahoye B., Gaudet F., Eggan K., Humpherys D., Mastrangelo M.A., Jun Z., Walter J., Jaenisch R., Gribnau J., Ramsahoye B., Gaudet F., Eggan K., Humpherys D., Mastrangelo M.A., Jun Z., Walter J., Jaenisch R., Ramsahoye B., Gaudet F., Eggan K., Humpherys D., Mastrangelo M.A., Jun Z., Walter J., Jaenisch R., Gaudet F., Eggan K., Humpherys D., Mastrangelo M.A., Jun Z., Walter J., Jaenisch R., Eggan K., Humpherys D., Mastrangelo M.A., Jun Z., Walter J., Jaenisch R., Humpherys D., Mastrangelo M.A., Jun Z., Walter J., Jaenisch R., Mastrangelo M.A., Jun Z., Walter J., Jaenisch R., Jun Z., Walter J., Jaenisch R., Walter J., Jaenisch R., Jaenisch R. Dnmt1 overexpression causes genomic hypermethylation, loss of imprinting, and embryonic lethality. Mol. Cell. Biol. 2002;22:2124–2135. doi: 10.1128/MCB.22.7.2124-2135.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black J.C., Choi J.E., Lombardo S.R., Carey M., Choi J.E., Lombardo S.R., Carey M., Lombardo S.R., Carey M., Carey M. A mechanism for coordinating chromatin modification and preinitiation complex assembly. Mol. Cell. 2006;23:809–818. doi: 10.1016/j.molcel.2006.07.018. [DOI] [PubMed] [Google Scholar]
- Chen H., Yan Y., Davidson T.L., Shinkai Y., Costa M., Yan Y., Davidson T.L., Shinkai Y., Costa M., Davidson T.L., Shinkai Y., Costa M., Shinkai Y., Costa M., Costa M. Hypoxic stress induces dimethylated histone H3 lysine 9 through histone methyltransferase G9a in mammalian cells. Cancer Res. 2006;66:9009–9016. doi: 10.1158/0008-5472.CAN-06-0101. [DOI] [PubMed] [Google Scholar]
- Cheutin T., McNairn A.J., Jenuwein T., Gilbert D.M., Singh P.B., Misteli T., McNairn A.J., Jenuwein T., Gilbert D.M., Singh P.B., Misteli T., Jenuwein T., Gilbert D.M., Singh P.B., Misteli T., Gilbert D.M., Singh P.B., Misteli T., Singh P.B., Misteli T., Misteli T. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science. 2003;299:721–725. doi: 10.1126/science.1078572. [DOI] [PubMed] [Google Scholar]
- Duan Z., Zarebski A., Montoya-Durango D., Grimes H.L., Horwitz M., Zarebski A., Montoya-Durango D., Grimes H.L., Horwitz M., Montoya-Durango D., Grimes H.L., Horwitz M., Grimes H.L., Horwitz M., Horwitz M. Gfi1 coordinates epigenetic repression of p21Cip/WAF1 by recruitment of histone lysine methyltransferase G9a and histone deacetylase 1. Mol. Cell. Biol. 2005;25:10338–10351. doi: 10.1128/MCB.25.23.10338-10351.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emami K.H., Carey M., Carey M. A synergistic increase in potency of a multimerized VP16 transcriptional activation domain. EMBO J. 1992;11:5005–5012. doi: 10.1002/j.1460-2075.1992.tb05607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskeland R., Eberharter A., Imhof A., Eberharter A., Imhof A., Imhof A. HP1 binding to chromatin methylated at H3K9 is enhanced by auxiliary factors. Mol. Cell. Biol. 2007;27:453–465. doi: 10.1128/MCB.01576-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espada J., Ballestar E., Fraga M.F., Villar-Garea A., Juarranz A., Stockert J.C., Robertson K.D., Fuks F., Esteller M., Ballestar E., Fraga M.F., Villar-Garea A., Juarranz A., Stockert J.C., Robertson K.D., Fuks F., Esteller M., Fraga M.F., Villar-Garea A., Juarranz A., Stockert J.C., Robertson K.D., Fuks F., Esteller M., Villar-Garea A., Juarranz A., Stockert J.C., Robertson K.D., Fuks F., Esteller M., Juarranz A., Stockert J.C., Robertson K.D., Fuks F., Esteller M., Stockert J.C., Robertson K.D., Fuks F., Esteller M., Robertson K.D., Fuks F., Esteller M., Fuks F., Esteller M., Esteller M. Human DNA methyltransferase 1 is required for maintenance of the histone H3 modification pattern. J. Biol. Chem. 2004;279:37175–37184. doi: 10.1074/jbc.M404842200. [DOI] [PubMed] [Google Scholar]
- Esteve P.O., Chin H.G., Pradhan S., Chin H.G., Pradhan S., Pradhan S. Human maintenance DNA (cytosine-5)-methyltransferase and p53 modulate expression of p53-repressed promoters. Proc. Natl. Acad. Sci. 2005;102:1000–1005. doi: 10.1073/pnas.0407729102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteve P.O., Chin H.G., Pradhan S., Chin H.G., Pradhan S., Pradhan S. Molecular mechanisms of transactivation and doxorubicin-mediated repression of survivin gene in cancer cells. J. Biol. Chem. 2006a;282:2615–2625. doi: 10.1074/jbc.M606203200. [DOI] [PubMed] [Google Scholar]
- Esteve P.O., Chin H.G., Smallwood A., Feehery G.R., Gangisetty O., Karpf A.R., Carey M.F., Pradhan S., Chin H.G., Smallwood A., Feehery G.R., Gangisetty O., Karpf A.R., Carey M.F., Pradhan S., Smallwood A., Feehery G.R., Gangisetty O., Karpf A.R., Carey M.F., Pradhan S., Feehery G.R., Gangisetty O., Karpf A.R., Carey M.F., Pradhan S., Gangisetty O., Karpf A.R., Carey M.F., Pradhan S., Karpf A.R., Carey M.F., Pradhan S., Carey M.F., Pradhan S., Pradhan S. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes & Dev. 2006b;20:3089–3103. doi: 10.1101/gad.1463706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman N., Gerson A., Fang J., Li E., Zhang Y., Shinkai Y., Cedar H., Bergman Y., Gerson A., Fang J., Li E., Zhang Y., Shinkai Y., Cedar H., Bergman Y., Fang J., Li E., Zhang Y., Shinkai Y., Cedar H., Bergman Y., Li E., Zhang Y., Shinkai Y., Cedar H., Bergman Y., Zhang Y., Shinkai Y., Cedar H., Bergman Y., Shinkai Y., Cedar H., Bergman Y., Cedar H., Bergman Y., Bergman Y. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat. Cell Biol. 2006;8:188–194. doi: 10.1038/ncb1353. [DOI] [PubMed] [Google Scholar]
- Fischle W., Wang Y., Jacobs S.A., Kim Y., Allis C.D., Khorasanizadeh S., Wang Y., Jacobs S.A., Kim Y., Allis C.D., Khorasanizadeh S., Jacobs S.A., Kim Y., Allis C.D., Khorasanizadeh S., Kim Y., Allis C.D., Khorasanizadeh S., Allis C.D., Khorasanizadeh S., Khorasanizadeh S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes & Dev. 2003;17:1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuks F., Hurd P.J., Deplus R., Kouzarides T., Hurd P.J., Deplus R., Kouzarides T., Deplus R., Kouzarides T., Kouzarides T. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 2003;31:2305–2312. doi: 10.1093/nar/gkg332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll M.G., Bestor T.H., Bestor T.H. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- Gowher H., Stockdale C.J., Goyal R., Ferreira H., Owen-Hughes T., Jeltsch A., Stockdale C.J., Goyal R., Ferreira H., Owen-Hughes T., Jeltsch A., Goyal R., Ferreira H., Owen-Hughes T., Jeltsch A., Ferreira H., Owen-Hughes T., Jeltsch A., Owen-Hughes T., Jeltsch A., Jeltsch A. De novo methylation of nucleosomal DNA by the mammalian Dnmt1 and Dnmt3A DNA methyltransferases. Biochemistry. 2005;44:9899–9904. doi: 10.1021/bi047634t. [DOI] [PubMed] [Google Scholar]
- Greil F., van der Kraan I., Delrow J., Smothers J.F., de Wit E., Bussemaker H.J., van Driel R., Henikoff S., van Steensel B., van der Kraan I., Delrow J., Smothers J.F., de Wit E., Bussemaker H.J., van Driel R., Henikoff S., van Steensel B., Delrow J., Smothers J.F., de Wit E., Bussemaker H.J., van Driel R., Henikoff S., van Steensel B., Smothers J.F., de Wit E., Bussemaker H.J., van Driel R., Henikoff S., van Steensel B., de Wit E., Bussemaker H.J., van Driel R., Henikoff S., van Steensel B., Bussemaker H.J., van Driel R., Henikoff S., van Steensel B., van Driel R., Henikoff S., van Steensel B., Henikoff S., van Steensel B., van Steensel B. Distinct HP1 and Su(var)3-9 complexes bind to sets of developmentally coexpressed genes depending on chromosomal location. Genes & Dev. 2003;17:2825–2838. doi: 10.1101/gad.281503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hediger F., Gasser S.M., Gasser S.M. Heterochromatin protein 1: Don’t judge the book by its cover! Curr. Opin. Genet. Dev. 2006;16:143–150. doi: 10.1016/j.gde.2006.02.013. [DOI] [PubMed] [Google Scholar]
- Hiragami K., Festenstein R., Festenstein R. Heterochromatin protein 1: A pervasive controlling influence. Cell. Mol. Life Sci. 2005;62:2711–2726. doi: 10.1007/s00018-005-5287-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman W.H., Biade S., Zilfou J.T., Chen J., Murphy M., Biade S., Zilfou J.T., Chen J., Murphy M., Zilfou J.T., Chen J., Murphy M., Chen J., Murphy M., Murphy M. Transcriptional repression of the anti-apoptotic survivin gene by wild type p53. J. Biol. Chem. 2002;277:3247–3257. doi: 10.1074/jbc.M106643200. [DOI] [PubMed] [Google Scholar]
- Horn P.J., Peterson C.L., Peterson C.L. Heterochromatin assembly: A new twist on an old model. Chromosome Res. 2006;14:83–94. doi: 10.1007/s10577-005-1018-1. [DOI] [PubMed] [Google Scholar]
- Ikeda K., Steger D.J., Eberharter A., Workman J.L., Steger D.J., Eberharter A., Workman J.L., Eberharter A., Workman J.L., Workman J.L. Activation domain-specific and general transcription stimulation by native histone acetyltransferase complexes. Mol. Cell. Biol. 1999;19:855–863. doi: 10.1128/mcb.19.1.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami K., Iwatani M., Suzuki M., Tachibana M., Shinkai Y., Tanaka S., Greally J.M., Yagi S., Hattori N., Shiota K., Iwatani M., Suzuki M., Tachibana M., Shinkai Y., Tanaka S., Greally J.M., Yagi S., Hattori N., Shiota K., Suzuki M., Tachibana M., Shinkai Y., Tanaka S., Greally J.M., Yagi S., Hattori N., Shiota K., Tachibana M., Shinkai Y., Tanaka S., Greally J.M., Yagi S., Hattori N., Shiota K., Shinkai Y., Tanaka S., Greally J.M., Yagi S., Hattori N., Shiota K., Tanaka S., Greally J.M., Yagi S., Hattori N., Shiota K., Greally J.M., Yagi S., Hattori N., Shiota K., Yagi S., Hattori N., Shiota K., Hattori N., Shiota K., Shiota K. Genome-wide and locus-specific DNA hypomethylation in G9a deficient mouse embryonic stem cells. Genes Cells. 2007;12:1–11. doi: 10.1111/j.1365-2443.2006.01029.x. [DOI] [PubMed] [Google Scholar]
- Jackson J.P., Lindroth A.M., Cao X., Jacobsen S.E., Lindroth A.M., Cao X., Jacobsen S.E., Cao X., Jacobsen S.E., Jacobsen S.E. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416:556–560. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- Jacobs S.A., Taverna S.D., Zhang Y., Briggs S.D., Li J., Eissenberg J.C., Allis C.D., Khorasanizadeh S., Taverna S.D., Zhang Y., Briggs S.D., Li J., Eissenberg J.C., Allis C.D., Khorasanizadeh S., Zhang Y., Briggs S.D., Li J., Eissenberg J.C., Allis C.D., Khorasanizadeh S., Briggs S.D., Li J., Eissenberg J.C., Allis C.D., Khorasanizadeh S., Li J., Eissenberg J.C., Allis C.D., Khorasanizadeh S., Eissenberg J.C., Allis C.D., Khorasanizadeh S., Allis C.D., Khorasanizadeh S., Khorasanizadeh S. Specificity of the HP1 chromo domain for the methylated N-terminus of histone H3. EMBO J. 2001;20:5232–5241. doi: 10.1093/emboj/20.18.5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jair K.W., Bachman K.E., Suzuki H., Ting A.H., Rhee I., Yen R.W., Baylin S.B., Schuebel K.E., Bachman K.E., Suzuki H., Ting A.H., Rhee I., Yen R.W., Baylin S.B., Schuebel K.E., Suzuki H., Ting A.H., Rhee I., Yen R.W., Baylin S.B., Schuebel K.E., Ting A.H., Rhee I., Yen R.W., Baylin S.B., Schuebel K.E., Rhee I., Yen R.W., Baylin S.B., Schuebel K.E., Yen R.W., Baylin S.B., Schuebel K.E., Baylin S.B., Schuebel K.E., Schuebel K.E. De novo CpG island methylation in human cancer cells. Cancer Res. 2006;66:682–692. doi: 10.1158/0008-5472.CAN-05-1980. [DOI] [PubMed] [Google Scholar]
- Johnson K.M., Wang J., Smallwood A., Carey M., Wang J., Smallwood A., Carey M., Smallwood A., Carey M., Carey M. The immobilized template assay for measuring cooperativity in eukaryotic transcription complex assembly. Methods Enzymol. 2004;380:207–219. doi: 10.1016/S0076-6879(04)80010-7. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Lachner M., O’Carroll D., Rea S., Mechtler K., Jenuwein T., O’Carroll D., Rea S., Mechtler K., Jenuwein T., Rea S., Mechtler K., Jenuwein T., Mechtler K., Jenuwein T., Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Le Gac G., Esteve P.O., Ferec C., Pradhan S., Esteve P.O., Ferec C., Pradhan S., Ferec C., Pradhan S., Pradhan S. DNA damage-induced down-regulation of human Cdc25C and Cdc2 is mediated by cooperation between p53 and maintenance DNA (cytosine-5) methyltransferase 1. J. Biol. Chem. 2006;281:24161–24170. doi: 10.1074/jbc.M603724200. [DOI] [PubMed] [Google Scholar]
- Li Y., Kirschmann D.A., Wallrath L.L., Kirschmann D.A., Wallrath L.L., Wallrath L.L. Does heterochromatin protein 1 always follow code? Proc. Natl. Acad. Sci. 2002;99:16462–16469. doi: 10.1073/pnas.162371699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K., Rechsteiner T.J., Richmond T.J., Rechsteiner T.J., Richmond T.J., Richmond T.J. Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol. Biol. 1999;119:1–16. doi: 10.1385/1-59259-681-9:1. [DOI] [PubMed] [Google Scholar]
- Ma Y., Jacobs S.B., Jackson-Grusby L., Mastrangelo M.A., Torres-Betancourt J.A., Jaenisch R., Rasmussen T.P., Jacobs S.B., Jackson-Grusby L., Mastrangelo M.A., Torres-Betancourt J.A., Jaenisch R., Rasmussen T.P., Jackson-Grusby L., Mastrangelo M.A., Torres-Betancourt J.A., Jaenisch R., Rasmussen T.P., Mastrangelo M.A., Torres-Betancourt J.A., Jaenisch R., Rasmussen T.P., Torres-Betancourt J.A., Jaenisch R., Rasmussen T.P., Jaenisch R., Rasmussen T.P., Rasmussen T.P. DNA CpG hypomethylation induces heterochromatin reorganization involving the histone variant macroH2A. J. Cell Sci. 2005;118:1607–1616. doi: 10.1242/jcs.02291. [DOI] [PubMed] [Google Scholar]
- Maison C., Almouzni G., Almouzni G. HP1 and the dynamics of heterochromatin maintenance. Nat. Rev. Mol. Cell Biol. 2004;5:296–304. doi: 10.1038/nrm1355. [DOI] [PubMed] [Google Scholar]
- Meehan R.R., Kao C.F., Pennings S., Kao C.F., Pennings S., Pennings S. HP1 binding to native chromatin in vitro is determined by the hinge region and not by the chromodomain. EMBO J. 2003;22:3164–3174. doi: 10.1093/emboj/cdg306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen C.T., Weisenberger D.J., Velicescu M., Gonzales F.A., Lin J.C., Liang G., Jones P.A., Weisenberger D.J., Velicescu M., Gonzales F.A., Lin J.C., Liang G., Jones P.A., Velicescu M., Gonzales F.A., Lin J.C., Liang G., Jones P.A., Gonzales F.A., Lin J.C., Liang G., Jones P.A., Lin J.C., Liang G., Jones P.A., Liang G., Jones P.A., Jones P.A. Histone H3-lysine 9 methylation is associated with aberrant gene silencing in cancer cells and is rapidly reversed by 5-aza-2′-deoxycytidine. Cancer Res. 2002;62:6456–6461. [PubMed] [Google Scholar]
- Nielsen A.L., Ortiz J.A., You J., Oulad-Abdelghani M., Khechumian R., Gansmuller A., Chambon P., Losson R., Ortiz J.A., You J., Oulad-Abdelghani M., Khechumian R., Gansmuller A., Chambon P., Losson R., You J., Oulad-Abdelghani M., Khechumian R., Gansmuller A., Chambon P., Losson R., Oulad-Abdelghani M., Khechumian R., Gansmuller A., Chambon P., Losson R., Khechumian R., Gansmuller A., Chambon P., Losson R., Gansmuller A., Chambon P., Losson R., Chambon P., Losson R., Losson R. Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. EMBO J. 1999;18:6385–6395. doi: 10.1093/emboj/18.22.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patnaik D., Chin H.G., Esteve P.O., Benner J., Jacobsen S.E., Pradhan S., Chin H.G., Esteve P.O., Benner J., Jacobsen S.E., Pradhan S., Esteve P.O., Benner J., Jacobsen S.E., Pradhan S., Benner J., Jacobsen S.E., Pradhan S., Jacobsen S.E., Pradhan S., Pradhan S. Substrate specificity and kinetic mechanism of mammalian G9a histone H3 methyltransferase. J. Biol. Chem. 2004;279:53248–53258. doi: 10.1074/jbc.M409604200. [DOI] [PubMed] [Google Scholar]
- Peters A.H., Kubicek S., Mechtler K., O’Sullivan R.J., Derijck A.A., Perez-Burgos L., Kohlmaier A., Opravil S., Tachibana M., Shinkai Y., Kubicek S., Mechtler K., O’Sullivan R.J., Derijck A.A., Perez-Burgos L., Kohlmaier A., Opravil S., Tachibana M., Shinkai Y., Mechtler K., O’Sullivan R.J., Derijck A.A., Perez-Burgos L., Kohlmaier A., Opravil S., Tachibana M., Shinkai Y., O’Sullivan R.J., Derijck A.A., Perez-Burgos L., Kohlmaier A., Opravil S., Tachibana M., Shinkai Y., Derijck A.A., Perez-Burgos L., Kohlmaier A., Opravil S., Tachibana M., Shinkai Y., Perez-Burgos L., Kohlmaier A., Opravil S., Tachibana M., Shinkai Y., Kohlmaier A., Opravil S., Tachibana M., Shinkai Y., Opravil S., Tachibana M., Shinkai Y., Tachibana M., Shinkai Y., Shinkai Y., et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell. 2003;12:1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- Pradhan S., Esteve P.O., Esteve P.O. Allosteric activator domain of maintenance human DNA (cytosine-5) methyltransferase and its role in methylation spreading. Biochemistry. 2003a;42:5321–5332. doi: 10.1021/bi034160+. [DOI] [PubMed] [Google Scholar]
- Pradhan S., Esteve P.O., Esteve P.O. Mammalian DNA (cytosine-5) methyltransferases and their expression. Clin. Immunol. 2003b;109:6–16. doi: 10.1016/s1521-6616(03)00204-3. [DOI] [PubMed] [Google Scholar]
- Rhee I., Jair K.W., Yen R.W., Lengauer C., Herman J.G., Kinzler K.W., Vogelstein B., Baylin S.B., Schuebel K.E., Jair K.W., Yen R.W., Lengauer C., Herman J.G., Kinzler K.W., Vogelstein B., Baylin S.B., Schuebel K.E., Yen R.W., Lengauer C., Herman J.G., Kinzler K.W., Vogelstein B., Baylin S.B., Schuebel K.E., Lengauer C., Herman J.G., Kinzler K.W., Vogelstein B., Baylin S.B., Schuebel K.E., Herman J.G., Kinzler K.W., Vogelstein B., Baylin S.B., Schuebel K.E., Kinzler K.W., Vogelstein B., Baylin S.B., Schuebel K.E., Vogelstein B., Baylin S.B., Schuebel K.E., Baylin S.B., Schuebel K.E., Schuebel K.E. CpG methylation is maintained in human cancer cells lacking DNMT1. Nature. 2000;404:1003–1007. doi: 10.1038/35010000. [DOI] [PubMed] [Google Scholar]
- Rice J.C., Briggs S.D., Ueberheide B., Barber C.M., Shabanowitz J., Hunt D.F., Shinkai Y., Allis C.D., Briggs S.D., Ueberheide B., Barber C.M., Shabanowitz J., Hunt D.F., Shinkai Y., Allis C.D., Ueberheide B., Barber C.M., Shabanowitz J., Hunt D.F., Shinkai Y., Allis C.D., Barber C.M., Shabanowitz J., Hunt D.F., Shinkai Y., Allis C.D., Shabanowitz J., Hunt D.F., Shinkai Y., Allis C.D., Hunt D.F., Shinkai Y., Allis C.D., Shinkai Y., Allis C.D., Allis C.D. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol. Cell. 2003;12:1591–1598. doi: 10.1016/s1097-2765(03)00479-9. [DOI] [PubMed] [Google Scholar]
- Roopra A., Qazi R., Schoenike B., Daley T.J., Morrison J.F., Qazi R., Schoenike B., Daley T.J., Morrison J.F., Schoenike B., Daley T.J., Morrison J.F., Daley T.J., Morrison J.F., Morrison J.F. Localized domains of G9a-mediated histone methylation are required for silencing of neuronal genes. Mol. Cell. 2004;14:727–738. doi: 10.1016/j.molcel.2004.05.026. [DOI] [PubMed] [Google Scholar]
- Schotta G., Ebert A., Krauss V., Fischer A., Hoffmann J., Rea S., Jenuwein T., Dorn R., Reuter G., Ebert A., Krauss V., Fischer A., Hoffmann J., Rea S., Jenuwein T., Dorn R., Reuter G., Krauss V., Fischer A., Hoffmann J., Rea S., Jenuwein T., Dorn R., Reuter G., Fischer A., Hoffmann J., Rea S., Jenuwein T., Dorn R., Reuter G., Hoffmann J., Rea S., Jenuwein T., Dorn R., Reuter G., Rea S., Jenuwein T., Dorn R., Reuter G., Jenuwein T., Dorn R., Reuter G., Dorn R., Reuter G., Reuter G. Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J. 2002;21:1121–1131. doi: 10.1093/emboj/21.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siedlecki P., Zielenkiewicz P., Zielenkiewicz P. Mammalian DNA methyltransferases. Acta Biochim. Pol. 2006;53:245–256. [PubMed] [Google Scholar]
- Sims R.J., III, Nishioka K., Reinberg D., Nishioka K., Reinberg D., Reinberg D. Histone lysine methylation: A signature for chromatin function. Trends Genet. 2003;19:629–639. doi: 10.1016/j.tig.2003.09.007. [DOI] [PubMed] [Google Scholar]
- Soppe W.J., Jasencakova Z., Houben A., Kakutani T., Meister A., Huang M.S., Jacobsen S.E., Schubert I., Fransz P.F., Jasencakova Z., Houben A., Kakutani T., Meister A., Huang M.S., Jacobsen S.E., Schubert I., Fransz P.F., Houben A., Kakutani T., Meister A., Huang M.S., Jacobsen S.E., Schubert I., Fransz P.F., Kakutani T., Meister A., Huang M.S., Jacobsen S.E., Schubert I., Fransz P.F., Meister A., Huang M.S., Jacobsen S.E., Schubert I., Fransz P.F., Huang M.S., Jacobsen S.E., Schubert I., Fransz P.F., Jacobsen S.E., Schubert I., Fransz P.F., Schubert I., Fransz P.F., Fransz P.F. DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J. 2002;21:6549–6559. doi: 10.1093/emboj/cdf657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M., Sugimoto K., Nozaki M., Ueda J., Ohta T., Ohki M., Fukuda M., Takeda N., Niida H., Kato H., Sugimoto K., Nozaki M., Ueda J., Ohta T., Ohki M., Fukuda M., Takeda N., Niida H., Kato H., Nozaki M., Ueda J., Ohta T., Ohki M., Fukuda M., Takeda N., Niida H., Kato H., Ueda J., Ohta T., Ohki M., Fukuda M., Takeda N., Niida H., Kato H., Ohta T., Ohki M., Fukuda M., Takeda N., Niida H., Kato H., Ohki M., Fukuda M., Takeda N., Niida H., Kato H., Fukuda M., Takeda N., Niida H., Kato H., Takeda N., Niida H., Kato H., Niida H., Kato H., Kato H., et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes & Dev. 2002;16:1779–1791. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M., Ueda J., Fukuda M., Takeda N., Ohta T., Iwanari H., Sakihama T., Kodama T., Hamakubo T., Shinkai Y., Ueda J., Fukuda M., Takeda N., Ohta T., Iwanari H., Sakihama T., Kodama T., Hamakubo T., Shinkai Y., Fukuda M., Takeda N., Ohta T., Iwanari H., Sakihama T., Kodama T., Hamakubo T., Shinkai Y., Takeda N., Ohta T., Iwanari H., Sakihama T., Kodama T., Hamakubo T., Shinkai Y., Ohta T., Iwanari H., Sakihama T., Kodama T., Hamakubo T., Shinkai Y., Iwanari H., Sakihama T., Kodama T., Hamakubo T., Shinkai Y., Sakihama T., Kodama T., Hamakubo T., Shinkai Y., Kodama T., Hamakubo T., Shinkai Y., Hamakubo T., Shinkai Y., Shinkai Y. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes & Dev. 2005;19:815–826. doi: 10.1101/gad.1284005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru H., Zhang X., McMillen D., Singh P.B., Nakayama J., Grewal S.I., Allis C.D., Cheng X., Selker E.U., Zhang X., McMillen D., Singh P.B., Nakayama J., Grewal S.I., Allis C.D., Cheng X., Selker E.U., McMillen D., Singh P.B., Nakayama J., Grewal S.I., Allis C.D., Cheng X., Selker E.U., Singh P.B., Nakayama J., Grewal S.I., Allis C.D., Cheng X., Selker E.U., Nakayama J., Grewal S.I., Allis C.D., Cheng X., Selker E.U., Grewal S.I., Allis C.D., Cheng X., Selker E.U., Allis C.D., Cheng X., Selker E.U., Cheng X., Selker E.U., Selker E.U. Trimethylated lysine 9 of histone H3 is a mark for DNA methylation in Neurospora crassa. Nat. Genet. 2003;34:75–79. doi: 10.1038/ng1143. [DOI] [PubMed] [Google Scholar]
- Turek-Plewa J., Jagodzinski P.P., Jagodzinski P.P. The role of mammalian DNA methyltransferases in the regulation of gene expression. Cell. Mol. Biol. Lett. 2005;10:631–647. [PubMed] [Google Scholar]
- Vakoc C.R., Mandat S.A., Olenchock B.A., Blobel G.A., Mandat S.A., Olenchock B.A., Blobel G.A., Olenchock B.A., Blobel G.A., Blobel G.A. Histone H3 lysine 9 methylation and HP1γ are associated with transcription elongation through mammalian chromatin. Mol. Cell. 2005;19:381–391. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Vassallo M.F., Tanese N., Tanese N. Isoform-specific interaction of HP1 with human TAFII130. Proc. Natl. Acad. Sci. 2002;99:5919–5924. doi: 10.1073/pnas.092025499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace J.A., Orr-Weaver T.L., Orr-Weaver T.L. Replication of heterochromatin: Insights into mechanisms of epigenetic inheritance. Chromosoma. 2005;114:389–402. doi: 10.1007/s00412-005-0024-6. [DOI] [PubMed] [Google Scholar]
- Wozniak R.J., Klimecki W.T., Lau S.S., Feinstein Y., Futscher B.W., Klimecki W.T., Lau S.S., Feinstein Y., Futscher B.W., Lau S.S., Feinstein Y., Futscher B.W., Feinstein Y., Futscher B.W., Futscher B.W. 5-Aza-2′-deoxycytidine-mediated reductions in G9A histone methyltransferase and histone H3 K9 di-methylation levels are linked to tumor suppressor gene reactivation. Oncogene. 2006;26:77–90. doi: 10.1038/sj.onc.1209763. [DOI] [PubMed] [Google Scholar]
- Xin Z., Tachibana M., Guggiari M., Heard E., Shinkai Y., Wagstaff J., Tachibana M., Guggiari M., Heard E., Shinkai Y., Wagstaff J., Guggiari M., Heard E., Shinkai Y., Wagstaff J., Heard E., Shinkai Y., Wagstaff J., Shinkai Y., Wagstaff J., Wagstaff J. Role of histone methyltransferase G9a in CpG methylation of the Prader-Willi syndrome imprinting center. J. Biol. Chem. 2003;278:14996–15000. doi: 10.1074/jbc.M211753200. [DOI] [PubMed] [Google Scholar]
- Zhang L., Adams J.Y., Billick E., Ilagan R., Iyer M., Le K., Smallwood A., Gambhir S.S., Carey M., Wu L., Adams J.Y., Billick E., Ilagan R., Iyer M., Le K., Smallwood A., Gambhir S.S., Carey M., Wu L., Billick E., Ilagan R., Iyer M., Le K., Smallwood A., Gambhir S.S., Carey M., Wu L., Ilagan R., Iyer M., Le K., Smallwood A., Gambhir S.S., Carey M., Wu L., Iyer M., Le K., Smallwood A., Gambhir S.S., Carey M., Wu L., Le K., Smallwood A., Gambhir S.S., Carey M., Wu L., Smallwood A., Gambhir S.S., Carey M., Wu L., Gambhir S.S., Carey M., Wu L., Carey M., Wu L., Wu L. Molecular engineering of a two-step transcription amplification (TSTA) system for transgene delivery in prostate cancer. Mol. Ther. 2002;5:223–232. doi: 10.1006/mthe.2002.0551. [DOI] [PubMed] [Google Scholar]
- Zhang X., Yazaki J., Sundaresan A., Cokus S., Chan S.W., Chen H., Henderson I.R., Shinn P., Pellegrini M., Jacobsen S.E., Yazaki J., Sundaresan A., Cokus S., Chan S.W., Chen H., Henderson I.R., Shinn P., Pellegrini M., Jacobsen S.E., Sundaresan A., Cokus S., Chan S.W., Chen H., Henderson I.R., Shinn P., Pellegrini M., Jacobsen S.E., Cokus S., Chan S.W., Chen H., Henderson I.R., Shinn P., Pellegrini M., Jacobsen S.E., Chan S.W., Chen H., Henderson I.R., Shinn P., Pellegrini M., Jacobsen S.E., Chen H., Henderson I.R., Shinn P., Pellegrini M., Jacobsen S.E., Henderson I.R., Shinn P., Pellegrini M., Jacobsen S.E., Shinn P., Pellegrini M., Jacobsen S.E., Pellegrini M., Jacobsen S.E., Jacobsen S.E., et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]