Abstract
Endogenous small RNAs function in RNA interference (RNAi) pathways to control gene expression through mRNA cleavage, translational repression, or chromatin modification. Plants and animals contain many microRNAs (miRNAs) that play vital roles in development, including helping to specify cell type and tissue identity. To date, no miRNAs have been reported in unicellular organisms. Here we show that Chlamydomonas reinhardtii, a unicellular green alga, encodes many miRNAs. We also show that a Chlamydomonas miRNA can direct the cleavage of its target mRNA in vivo and in vitro. We further show that the expression of some miRNAs/Candidates increases or decreases during Chlamydomonas gametogenesis. In addition to miRNAs, Chlamydomonas harbors other types of small RNAs including phased small interfering RNAs (siRNAs) that are reminiscent of plant trans-acting siRNAs, as well as siRNAs originating from protein-coding genes and transposons. Our findings suggest that the miRNA pathway and some siRNA pathways are ancient mechanisms of gene regulation that evolved prior to the emergence of multicellularity.
Keywords: Chlamydomonas, miRNA, mRNA cleavage, small RNA, RNAi, RISC
In eukaryotes, small RNAs of ∼20–30 nucleotides (nt) function as guides for effector complexes (RNA-induced silencing complexes, RISCs) that can regulate gene expression at the transcriptional or post-transcriptional level. Small RNAs are produced by the RNase III enzyme Dicer or Dicer-like proteins (DCLs) from their precursors, which can be stem–loop RNA transcripts or long double-stranded RNAs (dsRNAs). These small RNAs directly interact with proteins in the Piwi/Argonaute (AGO) family to form the core of RISC (Hannon 2002; Meister and Tuschl 2004).
In multicellular systems, three major classes of endogenous small RNAs have been discovered: microRNAs (miRNAs), Piwi-associated small RNAs (piRNAs), and small interfering RNAs (siRNAs). miRNAs are processed from RNA transcripts that form stem–loop structures. They regulate gene expression through mRNA cleavage or translational repression based on the degree of sequence complementarity between miRNAs and their target mRNAs, many of which have demonstrated regulatory roles in development and other physiological processes (Carrington and Ambros 2003; Bartel 2004). piRNAs are a newly discovered class of small RNAs that are associated with Piwi family proteins (Aravin et al. 2006; Girard et al. 2006; Grivna et al. 2006; Lau et al. 2006). The biogenesis machinery and biological roles of piRNAs remain largely unknown. siRNAs are produced from long dsRNAs that originate from various sources including products of bidirectional transcription or RNA-dependent RNA polymerases (RDRs). siRNAs play various roles in post-transcriptional regulation of gene expression in cis or in trans (Vaucheret 2006), suppression of transposable elements, and the formation of heterochromatin (Lippman and Martienssen 2004).
Based on their origins and functions, siRNAs can be further classified into several subclasses: heterochromatic siRNAs (hc-siRNAs), repeat-associated siRNAs (ra-siRNAs), trans-acting siRNAs (ta-siRNAs), and natural antisense transcript-derived siRNAs (nat-siRNAs). In Arabidopsis, there is an abundant class of DCL3 and RDR2-dependent ∼24-nt hc-siRNAs that act through AGO4 to maintain DNA/histone methylation of transposons and repeats (Hamilton et al. 2002; Zilberman et al. 2003; Xie et al. 2004; Qi et al. 2006). Similarly, ra-siRNAs silence retrotransposons and repetitive sequences in the Drosophila germline, but their biogenesis does not seem to require Dicer (Aravin et al. 2003; Vagin et al. 2006). ta-siRNAs have been found only in plants (Peragine et al. 2004; Vazquez et al. 2004; Allen et al. 2005; Axtell et al. 2006; Talmor-Neiman et al. 2006). Their production is initiated by the miRNA-mediated cleavage of their precursor transcripts (Peragine et al. 2004; Vazquez et al. 2004; Allen et al. 2005). The cleaved transcripts serve as substrates for RDR6 to make dsRNAs that are in turn processed by DCL4 (Gasciolli et al. 2005; Xie et al. 2005; Yoshikawa et al. 2005). It was recently shown that double miRNA complementary sites are required for efficient ta-siRNA production (Axtell et al. 2006). Like miRNAs, ta-siRNAs function in trans to guide the cleavage of endogenous mRNAs, and have been shown to regulate vegetative phase changes in Arabidopsis (Peragine et al. 2004; Vazquez et al. 2004; Allen et al. 2005; Gasciolli et al. 2005; Xie et al. 2005; Yoshikawa et al. 2005) and in moss (Talmor-Neiman et al. 2006). nat-siRNAs have also been found in Arabidopsis, where they are derived from natural antisense transcripts. dsRNAs formed by the transcription of antisense overlapping gene pairs are processed by DCL2 into 24-nt nat-siRNAs, which direct the further production of 21-nt nat-siRNAs in a DCL1-dependent fashion (Borsani et al. 2005).
In contrast to the complex small RNA pathways found in multicellular systems, small RNA pathways in unicellular organisms appear, so far, to be relatively simple. In fission yeast, a single class of siRNAs exists in the RNA-induced transcriptional silencing complex (RITS). These siRNAs have demonstrated roles in epigenetic silencing at centromeres and the initiation of heterochromatin assembly at the mat locus (Lippman and Martienssen 2004). The ciliated protozoan Tetrahymena thermophila has ∼26- to 31-nt scan RNAs (scnRNAs) that are involved in developmentally regulated DNA elimination (Mochizuki and Gorovsky 2004a, b; Yao and Chao 2005), and ∼23- to 24-nt small RNAs with suggested roles in gene regulation at the post-transcriptional level (Lee and Collins 2006). Thus far, no miRNAs have been identified in unicellular systems, raising the question whether miRNAs exist only in multicellular systems, where regulation of cell differentiation, developmental patterning, and timing is important. Indeed, it has been suggested that miRNAs helped to drive the evolution of a multicellular state (Bartel 2004).
With the goal of probing the diversity of small RNA pathways in unicellular eukaryotes, we turned to Chlamydomonas reinhardtii. Chlamydomonas is a unicellular green alga with well-understood haploid genetics. It also occupies a unique niche in evolution. It has served as a model system to understand various aspects of cell biology including the assembly and functions of the basal body, chloroplast, and flagellum (Lefebvre and Silflow 1999). The completion of the Chlamydomonas genome sequence and substantial expression sequence tag (EST) projects has greatly increased its utility as a model system (Grossman et al. 2003; Shrager et al. 2003).
Chlamydomonas possesses a functional RNA interference (RNAi) machinery, as evident from the observation that the target gene expression can be specifically knocked down in transgenic lines expressing corresponding antisense RNAs or dsRNAs (Schroda 2006). In accord with these observations, bioinformatics analysis predicted that the Chlamydomonas genome encodes key RNAi components including three Dicer-like proteins and at least two Argonautes (Supplementary Fig. 1; Schroda 2006). The complexity of RNAi components in Chlamydomonas strongly suggested the existence of multiple small RNA pathways. However, it remains unknown whether Chlamydomonas encodes endogenous small RNAs and whether such small RNAs have regulatory roles in gene expression.
To address the aforementioned questions, we determined a large number of small RNA sequences from Chlamydomonas using a highly parallel pyrosequencing technology (Margulies et al. 2005). We found that Chlamydomonas encodes miRNAs, as well as other small RNAs derived from various genomic regions that include protein-coding genes and transposable and repetitive sequences. We also found that, like plant miRNAs, Chlamydomonas miRNAs can guide the cleavage of their target mRNAs. We further show that the accumulation of some miRNAs was either elevated or decreased in gametic cells. Our findings not only reveal the existence of multiple RNAi pathways in a unicellular eukaryotic organism, but also provide a rich resource for studying the roles of small RNAs in basic cell biology and the evolutionary transition from unicellularity to multicellularity.
Results
Identification of Chlamydomonas small RNAs
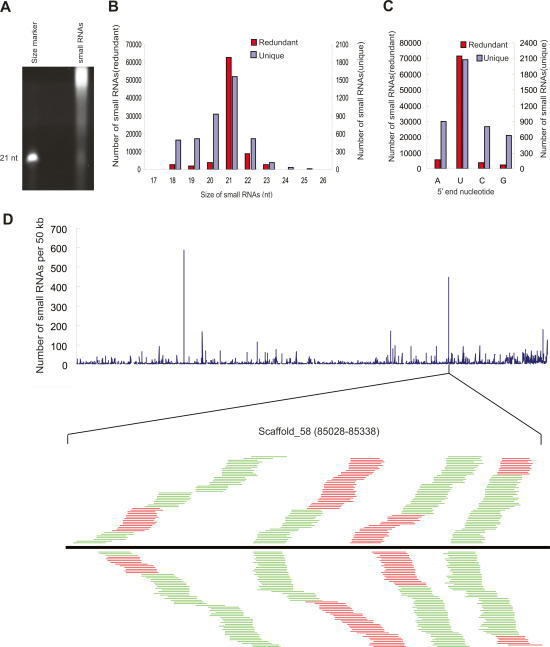
To examine whether Chlamydomonas encodes small RNAs, we prepared total RNA from Chlamydomonas cultures under vegetative growth conditions. After removal of larger RNAs by polyethylene glycol (PEG) precipitation, RNAs were separated by denaturing polyacrylamide gel electrophoresis and visualized by SYBR-gold staining. We observed an abundant class of small RNAs that are ∼21 nt in length (Fig. 1A). To identify these small RNAs, we recovered small RNAs from the gel slice at the position of ∼18–28 nt, and cloned and sequenced them by highly parallel pyrosequencing as described (Girard et al. 2006). A total of 121,274 reads were obtained. After removing unmatched nucleotides at either end of the small RNA reads, 82,350 sequences were shown to have at least one perfect match in the Chlamydomonas nuclear genome, whereas 116 and 20 sequences matched the chloroplast and mitochondrial genomes, respectively (Table 1). These sequences were analyzed further as described below. The sequences without perfect genomic matches might arise from unsequenced regions of the genomes or from sequencing errors and were not pursued further.
Figure 1.
A catalog of Chlamydomonas endogenous small RNAs. (A) SYBR-gold was used to visualize small RNAs in total RNA extracted from Chlamydomonas cultures. A 21-nt synthetic RNA oligo was used as a size reference. (B) Size distribution of Chlamydomonas small RNAs. The sets of redundant (red) and unique (blue) small RNAs were used to generate a histogram quantifying the number of sequences obtained for each size class. (C) Sequence composition of the 5′ ends of the small RNAs. (D) Genome-wide density analysis of the small RNAs on an artificial Chlamydomonas genome assembled by linked scaffolds. The number of small RNAs with perfect matches in either a direct or complementary strand within each 50-kb sliding window was plotted. A small RNA production hot spot in Scaffold_58 is shown. The black bar represents the 310-nt hot spot region. Short thin lines above the black bar represent small RNAs derived from the sense strand, and lines below the bar represent small RNAs from the antisense strand. Red lines represent small RNAs that are mapped uniquely to this hot spot, while green lines represent small RNAs that are mapped to multiple genomic sites including the hot spot.
Table 1.
Chlamydomonas small RNAs matching different categories of sequences
The 82,350 small RNAs with perfect nuclear genomic matches (the redundant data set) represent 4182 unique sequences (the unique data set) (Table 1). Of these, 2812 small RNAs were cloned only once, indicating that the small RNA population in Chlamydomonas is complex. Consistent with the SYBR-gold staining result, the sizes of the small RNAs range from 17 nt to 26 nt, with 91.5% of small RNAs in the redundant data set and 72.2% in the unique data set being 20–22 nt in length (Fig. 1B; Supplementary Table 1). In both the unique and redundant small RNA data sets, a preference for a U was observed at the 5′ end of small RNA sequences (Fig. 1C). A similar 5′ nucleotide bias has been noted in small RNAs from other organisms (Aravin et al. 2003; Bartel 2004; Girard et al. 2006). Among the 4182 unique small RNAs mapped to the nuclear genome, 2247 sequences match only one site in the genome, and 781 sequences match twice. The remaining sequences match the genome from several to a few hundred times, with one exception that has 1152 genomic matches. These small RNA-generating regions include intergenic regions, annotated protein-coding genes, repetitive sequences, and regions producing noncoding RNAs (rRNAs and tRNAs) (Table 1). The number of unique small RNAs matching the intergenic regions exceeds the number of those matching genes and repeats, similar to what has been observed for Arabidopsis small RNAs (Lu et al. 2005).
We summed the number of unique small RNAs in a 50-kb sliding window and plotted it against the artificial genome assembled from 1266 scaffolds. A few hot spots for small RNA production were observed (Fig. 1D). One such hot spot included 427 small RNAs within a 310-nt intergenic region (Fig. 1D). The entire region was covered by small RNAs on both strands. In many cases, multiple small RNAs with identical start sites but different lengths were cloned from the same position.
We found two small RNAs with exceptionally high cloning frequency. One with a size of 21 nt (UUGUGC CGGCCGACACUGCGG) was cloned 54,353 times, the other with a size of 22 nt (UUGUGCCGGCCGACACU GCGGU) was cloned 5786 times. These two small RNAs were derived from the same genomic loci with identical start sites and differed only by one nucleotide at their 3′ ends. No identifiable homologs of these two small RNAs were found in the NCBI database. The two small RNAs matched four genomic loci with one located in an exon of a protein-coding gene of unknown function and the other three within intergenic regions. Two of the intergenic loci could also produce a 23-nt-long small RNA, whose sequence is reverse-complementary to the two small RNAs but has additional nucleotides at both ends. The cloning frequency of this small RNA ranked third among all small RNAs (cloned 1694 times) (data not shown). The exceptionally high transcription rate may suggest the unique functional significance of these small RNAs.
miRNAs in Chlamydomonas
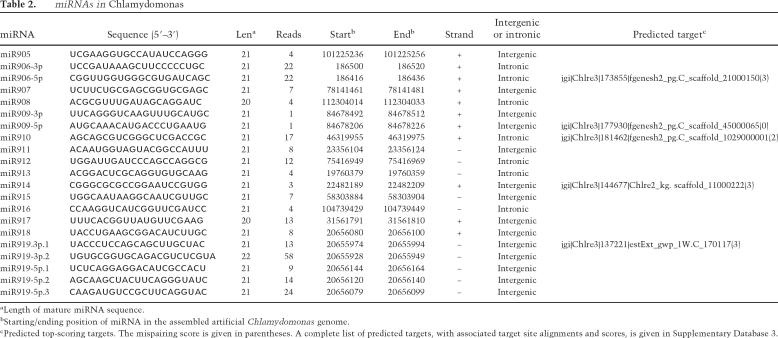
The identification of a large number of small RNAs in Chlamydomonas prompted us to examine whether some of them were miRNAs. We selected small RNAs originating from intergenic or intronic regions to examine whether their surrounding sequences could form hairpin secondary structures, a distinguishing feature of known plant and animal miRNAs.
Structural predictions identified 200 small RNA species that were derived from genomic sequences with the potential to form hairpin structures that meet the requirements as a miRNA precursor. Among these, 19 have a sequenced pairing miRNA* or close variants with lower cloning frequency, providing strong evidence that they were processed from hairpins by Dicer(s). These small RNAs were annotated as miRNAs (Table 2; Supplementary Database 1). In two cases in which the pairing small RNAs have the same or similar cloning frequency, both were annotated as miRNAs. All other small RNAs were considered as miRNA candidates (Supplementary Table 2; Supplementary Database 1).
Table 2.
miRNAs in Chlamydomonas
aLength of mature miRNA sequence.
bStarting/ending position of miRNA in the assembled artificial Chlamydomonas genome.
cPredicted top-scoring targets. The mispairing score is given in parentheses. A complete list of predicted targets, with associated target site alignments and scores, is given in Supplementary Database 3.
In order to assess the specificity of our miRNA prediction results, we randomly selected 2893 21-nt-long sequences from the Chlamydomonas intergenic and intronic regions (the same number as unique cloned small RNAs mapped to the above regions) and examined the possibility of their surrounding regions to form hairpin structures using the same criteria as applied to the cloned small RNAs. We repeated the random test 10 times, and obtained 27.5 sequences on average that meet our miRNA prediction criteria. Thus, compared with the 181 miRNAs/Candidates we predicted, we estimated that the false discovery rate of our miRNA prediction is ∼15%.
The plant and animal miRNAs contain a phosphate at the 5′ end. The small RNA cloning procedure we used has a step of ligation of an RNA adapter to the 5′ end of small RNAs by T4 RNA ligase, a process that requires a 5′ phosphate (Lau et al. 2001). Thus, the miRNAs we have cloned from Chlamydomonas should have a 5′ phosphate. We used enzymatic methods to further confirm that the Chlamydomonas miRNAs have a 5′ phosphate. The small RNAs isolated from Chlamydomonas cells were subjected to calf intestinal phosphatase (CIP) treatment followed by treatment with polynucleotide kinase in the presence of ATP. The CIP treatment caused miR912 to have a lower mobility in polyacrylamide gel (Supplementary Fig. 2), suggesting that the miRNA had one or more phosphates at the 5′ end. Subsequent treatment with polynucleotide kinase and ATP restored the original mobility (Supplementary Fig. 2). This indicates that the miRNA contains one phosphate group at the 5′ end, like plant and animal miRNAs.
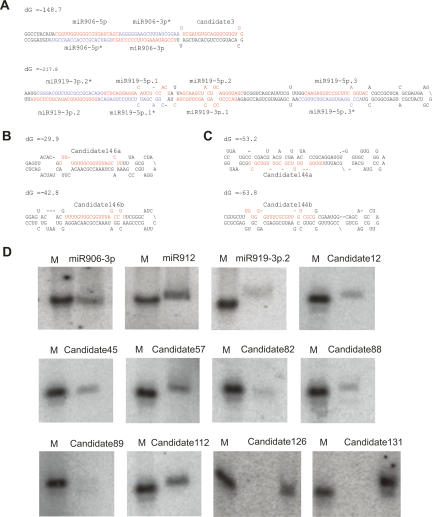
Unlike plant miRNA genes that are mostly located in intergenic regions (Jones-Rhoades et al. 2006), a considerable proportion of the Chlamydomonas miRNA/Candidate genes is intronic, while the remaining miRNA genes reside in intergenic regions (Table 2; Supplementary Table 2). Unlike instances of polycistronic microRNAs in other systems, where multiple cotranscribed stem–loops give rise to independent microRNAs, we found that multiple Chlamydomonas miRNAs/Candidates can be derived from a single stem–loop (Fig. 2A; Supplementary Database 1). For example, miR906-5p and miR906-3p as well as miR919-3p.1, miR919-3p.2, miR919-5p.1, miR919-5p.2, and miR919-5p.3 are produced from the same precursors (Fig. 2A). We also found that some miRNA genes are clustered in the genome, likely representing more conventional microRNAs polycistrons. One of the clusters (Cluster 4) only contains miRNAs in the same family (Supplementary Table 3). These two genes (Candidate160c and Candidate160d) are nearly identical, suggesting that they were recently evolved through duplication (Supplementary Database 1).
Figure 2.
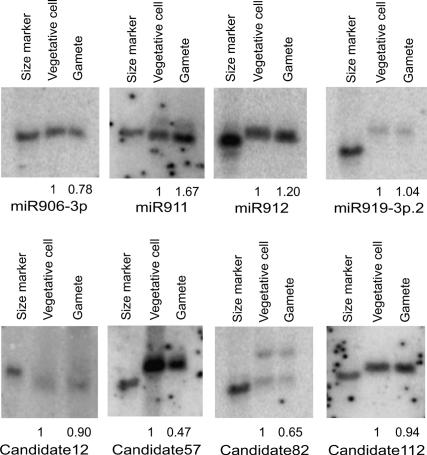
Representative miRNAs in Chlamydomonas. (A–C) Predicted fold-back structures of selected miRNA precursors. Sequences corresponding to the mature miRNAs are shown in red, and sequences corresponding to the miRNA*s are shown in blue. Protruding stem–loops in the 3′ arm of the hairpins are indicated by slashes (\). (D) Confirmation of miRNA expression by Northern blot using end-labeled oligonucleotide probes as indicated. A synthetic 32P-labeled 21-nt RNA oligo was used as a size marker.
The majority of the miRNAs/Candidates have a single precursor, whereas others have more than one precursor (Fig. 2; Supplementary Table 2). For most miRNAs with multiple precursors, the mature miRNAs are located on the same arm of the stem–loops, suggesting that these genes might share a common ancestry (Fig. 2B; Supplementary Database 1). We also identified some miRNAs that are located on either the 5′ or the 3′ arm of different precursors (Fig. 2C; Supplementary Database 1), suggesting that genes encoding a particular miRNA could have evolved from more than one ancestor.
To confirm expression of some of our miRNAs/Candidates, we performed Northern analysis on RNAs prepared from Chlamydomonas vegetative cells. Many of the miRNAs/Candidates were readily detectable (Fig. 2D), whereas some are likely expressed at lower levels and were below the detection limit of our Northern blot protocol (data not shown).
Are miRNAs conserved in green algae?
Land plants are thought to be descendants of green alga-like ancestors (Lewis and McCourt 2004; Palmer et al. 2004). It was recently shown that some miRNAs are conserved in land plants and have ancient origins (Axtell and Bartel 2005). The existence of miRNAs in the green alga Chlamydomonas raised the possibility that plants and Chlamydomonas might share some common miRNAs. We compared Chlamydomonas miRNAs with all known plant (and also animal) miRNAs, and found no homologs. As a matter of fact, no Chlamydomonas small RNAs within the entire data set are homologs of known miRNAs, or of reported Arabidopsis small RNAs (data not shown). The lack of universally conserved miRNA genes among plants, animals, and green algae suggests that miRNA genes may have evolved independently in the lineages leading to animals, plants, and green algae.
Next we asked whether Chlamydomonas miRNAs are conserved in other green algae. We searched for miRNA homologs in the three other green algae—Ostreococcus tauri, Ostreococcus lucimarinus, and Volvox carteri—whose draft genome sequences are available. V. carteri is a colonial multicellular green alga in the class Chlorophyceae, while O. tauri and O. lucimarinus belong to the class Prasinophyceae. We found that most of the Chlamydomonas miRNAs/Candidates did not have identifiable orthologs in the other three green algae. When up to three nucleotide substitutions were allowed, 11, 22, and 25 miRNAs/Candidates had homologous sequences in V. carteri, O. tauri, and O. lucimarinus, respectively. However, many of these potential orthologs are likely false positives, since they are located in the nonhomologous arms of the predicted stem–loops (Supplementary Database 2).
Prediction of miRNA targets in Chlamydomonas
As an initial step toward understanding the biological function of miRNAs in Chlamydomonas, we searched for miRNA targets among annotated protein-coding transcripts and ESTs of Chlamydomonas, adapting criteria for plant miRNA target prediction (Jones-Rhoades and Bartel 2004; Allen et al. 2005).
Applying a cutoff mispairing score of ≤3.0, we predicted a total of 697 genes as miRNA targets (Table 2; Supplementary Table 2; Supplementary Database 3), with a signal/noise ratio of 5.6:1 as estimated by summing the number of targets predicted for miRNAs/Candidates and comparing with the average predicted targets for 10 shuffled cohorts. Most of the target sites are located in the coding sequences, with 40 being in the untranslated regions (UTRs). The putative target genes appear to be involved in various biological processes. However, since the Chlamydomonas genome is not fully annotated and a great proportion of Chlamydomonas protein-coding genes have no known functions, it is difficult to draw a conclusion as to whether these miRNA targets have any functional bias. Among the predicted targets with annotated functions, many genes are involved in cellular metabolism and physiological processes, whereas transcription factors appear to be underrepresented. This is in contrast to the situation in plants where miRNAs have a remarkable propensity to target transcription factor gene families (Jones-Rhoades et al. 2006). It is noteworthy that many genes encoding Flagellar-Associated Proteins (FAPs) were predicted as miRNAs/Candidate targets. We also predicted histone 2A family proteins as the targets of Candidate136. In Arabidopsis, two histone 2A.F/Z genes are the predicted targets of miR841 (Rajagopalan et al. 2006).
Chlamydomonas miRNAs can direct target mRNA cleavage
In plants and animals, miRNAs directly interact with AGOs to form nucleoprotein complexes that direct target mRNA cleavage or translational repression. Chlamydomonas encodes two AGO proteins and one protein containing a Piwi domain but not a PAZ domain (Supplementary Fig. 1). The expression of the two AGO proteins is supported by EST sequences and RT–PCR (data not shown), whereas that of the Piwi domain-only protein has yet to be verified. We reasoned that Chlamydomonas miRNAs should reside in AGO protein complexes in order to carry out their functions.
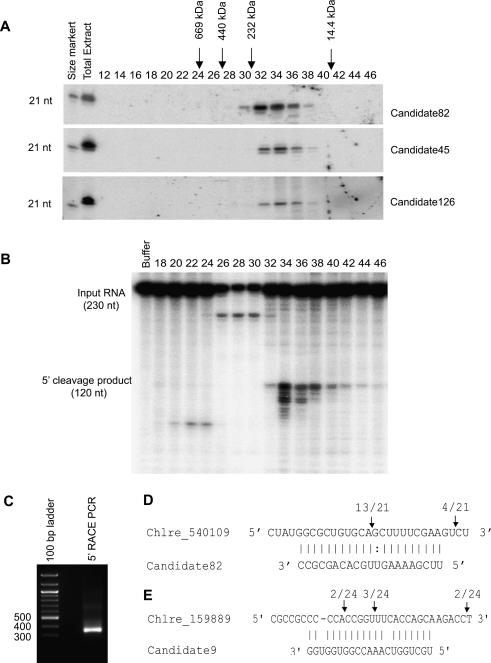
As a first step, we examined whether such a complex existed using size chromatography. We prepared total extracts from Chlamydomonas cultures, and then fractionated the extracts by gel filtration. RNAs were prepared from each fraction and used for small RNA Northern blotting. As shown in Figure 3A, several miRNAs including Candidate82 could be detected in the fractions corresponding to ∼30–440 kDa with a peak around 150 kDa as determined by standard size markers. The average size of a 21-nt RNA is ∼7 kDa; thus, the miRNAs must exist in complexes with other components, presumably including an AGO protein.
Figure 3.
Chlamydomonas miRNAs direct the cleavage of their target mRNAs. (A) Chlamydomonas miRNAs reside in complexes. Extracts from Chlamydomonas cultures were fractionated by size exclusion chromatography. RNAs were extracted from each fraction and used for Northern blotting to detect miRNAs as indicated. The fractions representing 14.4-, 232-, 440-, and 660-kDa size markers, chromatographed separately, are shown. A synthetic 21-nt RNA oligo was used as a size marker. (B) The target of Candidate82 is cleaved in vitro. Each fraction from the size exclusion chromatography was assayed for cleavage activity by incubation with 32P-labeled target transcripts (C_540109, a target of Candidate82). (C) Detection of the 3′ end Candidate82 cleavage product by 5′ RACE assay. A PCR product of the expected size is observed. (D) Identification of the Candidate82 cleavage site. The miRNA target site is aligned with Candidate82. (E) Identification of the Candidate9 cleavage site. The miRNA target site is aligned with Candidate9. Arrows indicate the ends of cleaved mRNA products as determined by 5′ RACE. Above the arrows is shown the frequency of cloned sequences corresponding to each inferred cleavage site. Only cloned sequences that matched the target gene and had a 5′ end within a 50-base-pair (bp) window centered on the miRNA complementary site were counted.
Next we sought to test through which mode (mRNA cleavage or translational repression) the Chlamydomonas miRNA complex acts on miRNA targets. We incubated an in vitro transcribed 32P-labeled RNA (jgi|Chlre3|196819|OVA_estExt_GenewiseH_1.C_540109; a putative di- or tricarboxylate translocator, the predicted target for Candidate82) with each fraction collected from size exclusion chromatography. We found that the transcripts were specifically cleaved into products of the expected size (Fig. 3B). The peak of the cleavage activity corresponded to the peak of the miRNA signal as detected by Northern blot, suggesting that Candidate82 can direct the cleavage of its target. To examine whether Candidate82 can also cleave its target in vivo, we used a 5′ RACE procedure to detect the cleavage products and to map precise cleavage sites (Llave et al. 2002). We could detect a 5′ RACE PCR product of predicted size from C_540109 on an agarose gel, suggesting that the target was cleaved in vivo (Fig. 3C). Sequencing of the PCR product revealed that the majority of the cleavage sites were located between the target nucleotides that pair with nucleotides 10 and 11 of the miRNA (Fig. 3D), which is the characteristic feature of RISC-mediated cleavage (Elbashir et al. 2001; Llave et al. 2002; Rivas et al. 2005).
We tried to confirm the predicted targets of other miRNAs/Candidates using 5′ RACE. For the FAP274 gene (Chlre3_159889), the predicted target of Candidate9, a few of the cleavage sites were mapped to the target regions, albeit more sites were found in the downstream region of the target regions (Fig. 3E). For many other predicted targets, cleavage products were not detectable or cleavage sites were mapped outside the predicted target regions (very often in the downstream region of the predicted target regions). This might attribute to quicker turnover of cleavage products in Chlamydomonas, compared with that in Arabidopsis, or to false-positive prediction of some of the miRNA targets.
Considered together, our results indicate that Chlamydomonas miRNAs can direct the cleavage of their targets.
Expression patterns of miRNAs/Candidates during Chlamydomonas gametogenesis
In multicellular organisms, miRNAs have been shown to play regulatory roles in cellular differentiation, developmental patterning, and timing (Bartel 2004). The discovery of a large number of miRNAs in unicellular Chlamydomonas raises pressing question about their biological functions. The life cycle of Chlamydomonas is rather simple compared with that of plants and animals. Cells of this species are haploid and divide asexually under normal growth conditions. Under stress conditions (e.g., lack of nitrogen), haploid cells of opposite mating types become gametes and conjugate to form diploid zygotes. The zygote loses its flagella and produces a thick cell wall, forming a resting spore. When conditions become normal, the diploid zygote undergoes meiosis and produces four haploid cells that resume asexual reproduction (Harris 1989). As a first step to address the question whether miRNAs are involved in the progression of the Chlamydomonas life cycle, we sought to examine their expression profiles in vegetative cells and cells undergoing gametogenesis.
We probed the expression of eight randomly selected miRNAs/Candidates in vegetative cells and gametes using small RNA Northern blotting. Different expression patterns were observed for these miRNAs (Fig. 4). The expression of miR919-3p.2, Candidate12, and Candidate112 remained similar in both cell types. The expression of miR911 and miR912 was elevated in gametes, whereas that of miR906-3p, Candidate57, and Candidate82 was decreased at different degrees. The changed expression level of these miRNAs in gametes suggests that they might play roles in gametic differentiation.
Figure 4.
Expression patterns of miRNAs in different cell types. The expression of miRNAs in vegetative cells and gametes (CC-621 mt−) was examined by Northern blotting using end-labeled oligonucleotide probes as indicated. Enriched small RNAs prepared from the same number of cells were loaded in each lane. The miRNA signals were quantified and the relative levels in gametes were calculated by comparison with those in vegetative cells (arbitrarily set to 1.0). A synthetic 32P-labeled 21-nt RNA oligo was used as a size marker.
Endogenous siRNAs in Chlamydomonas
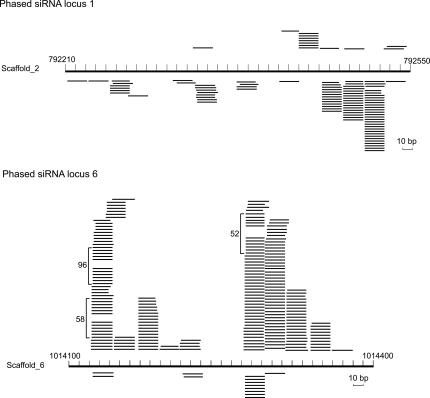
Besides miRNAs/Candidates, we found that many small RNAs were derived from various genomic locations including protein-coding genes and intergenic regions. Interestingly, we identified 11 loci producing small RNAs (Fig. 5; Supplementary Table 4), the majority of which are phased relative to each other, reminiscent of plant ta-siRNAs. It remains to be examined whether these phased siRNAs are produced by a similar mechanism to that used by plant ta-siRNAs, the generation of which requires the actions of miRNAs and RDRs. However, no RDR homologs can be predicted from the current version of the Chlamydomonas genome sequence.
Figure 5.
Phased siRNA loci in Chlamydomonas. Two representative loci produce siRNAs that are phased to each other. The loci are represented by the long bars at the center of each diagram, and the genomic positions of the loci are shown. Short thin lines above the long bars represent small RNAs derived from the antisense strands, and lines below the bars represent small RNAs from the sense strands. The copy numbers are shown for the small RNAs cloned >50 times.
Besides the ta-siRNA-like small RNAs, we also found a large proportion of siRNAs derived from sense or antisense strands of other protein-coding genes or intergenic regions (Table 1). The biogenesis mechanism of these siRNAs remains to be elucidated. It also remains to be examined whether these siRNAs can regulate gene expression in cis and/or in trans.
It has been shown in several organisms that small RNAs can play an important role in silencing of repetitive sequences and formation of heterochromatin (Lippman and Martienssen 2004). We asked whether small RNAs could also act to silence repeats in Chlamydomonas. As a first step, we examined whether there were any siRNAs originating from repeats. By the RepeatMasker program (Kohany et al. 2006), 6.11% of the Chlamydomonas genome could be annotated as transposable elements, low complexity, or simple repeats (Supplementary Table 5). Intriguingly, we only identified 26 unique siRNAs derived from transposable elements, 114 from simple repeat regions, and 46 from low complexity regions. These three groups of unique small RNAs represent 41, 206, and 65 sequences from the redundant data set, respectively (Supplementary Table 5). Therefore, it seems that small RNAs in Chlamydomonas may not be important for silencing of repetitive elements.
Discussion
The diversification of small RNA pathways has been well demonstrated in multicellular systems but remains largely unknown in unicellular organisms. Here we have shown that the unicellular green alga Chlamydomonas contains a variety of small RNAs. These include miRNAs and phased siRNAs, as well as other siRNAs originating from intergenic regions, sense and antisense strands of protein-coding genes, and repeat regions. The presence of these small RNAs indicates an unexpected complexity of RNAi-related processes in this unicellular organism. Our discovery of miRNAs in a unicellular alga also supports the concept that the miRNA pathway is a conserved mechanism of gene regulation in eukaryotes that arose prior to the developmental patterning needed for multicellular body plans (Bartel 2004).
We did not find that the Chlamydomonas miRNAs we discovered have any identifiable homologs in higher plants. However, the lack of sequence homology between plant and Chlamydomonas miRNAs does not necessarily reflect the fundamental differences in miRNA silencing pathways. In fact, Chlamydomonas miRNAs share several common features with plant miRNAs. These include being ∼21 nt in length and having a preference for uracil at the 5′ end, as well as being generated from a stem–loop by Dicer and being competent in directing target RNA cleavage. This suggests a conserved machinery for miRNA production and action in Chlamydomonas and in higher plants. It has been proposed that plant miRNAs may have arisen from inverted duplication of their target genes (Allen et al. 2004). Supporting this proposal, several presumably newly evolved Arabidopsis miRNA loci have been shown to share significant sequence similarity to their respective target genes (Allen et al. 2004; Rajagopalan et al. 2006). Moreover, a few small RNA-producing loci that can form relatively long hairpins were identified as transitional forms leading to miRNAs (Allen et al. 2004). Intriguingly, a substantial number of the Chlamydomonas miRNAs/Candidates are located in long hairpins, many of which have nearly perfect base-pairing between the arms and have the potential to produce multiple small RNAs. These long hairpins might represent the evolutionary intermediates of miRNA genes.
It has been well demonstrated in plants and animals that miRNAs play key roles in multicellular development (Bartel 2004). In this study, we identified many miRNAs in the unicellular Chlamydomonas. The biological functions of these miRNAs remain to be understood. It is of great interest to examine whether they play regulatory roles in sexual and/or asexual reproduction, responses to a variety of environmental changes, or metabolic pathways. We found that the expression of some miRNAs/Candidates was changed during gametogenesis (Fig. 4), suggesting the involvement of miRNAs in Chlamydomonas sexual reproduction. We predicted many genes encoding FAPs as miRNAs/Candidates targets, and validated FAP274 as the target of Candidate9, suggesting the involvement of miRNAs in flagellar development. We also have predicted many other targets for Chlamydomonas miRNAs. However, since the Chlamydomonas genome awaits better annotation and functions of many of the annotated genes have yet to be assigned, validation of miRNA targets and elucidation of their functions in Chlamydomonas biology is a major but exciting challenge. Analyzing transgenic lines overexpressing miRNAs and cleavage-resistant targets may reveal which biological process(es) each miRNA is controlling. Additionally, using microarrays to profile the expression of miRNAs and their targets at different steps of the life cycle or under different physiological conditions will help us understand the biological roles of Chlamydomonas miRNAs.
Most animal miRNAs regulate expression of their targets by repressing productive translation with the one exception that mouse miR196 guides the cleavage of its target HoxB8 mRNA (Bartel 2004; Yekta et al. 2004). In contrast, mRNA cleavage is the predominant mechanism used by plant miRNAs to regulate their targets. It remains unclear whether some plant miRNAs can also function as translation repressors (Jones-Rhoades et al. 2006). Through the detection of miRNA cleavage products in vivo by the 5′ RACE method and in vitro by cleavage assay, we showed that Chlamydomonas miRNAs can direct the cleavage of their target mRNAs. However, we do not exclude the possibility that Chlamydomonas miRNAs can also function at the level of translation, in addition to mRNA cleavage. We also point out that the criteria we used for target prediction were adapted from plants; thus, it is possible that we have missed some target genes that are regulated by Chlamydomonas miRNAs through modes similar to those used in animals.
It has been well established that RNAi plays an important role in the silencing of selfish genetic elements and in the formation of heterochromatin in plants, animals, and fission yeast (Lippman and Martienssen 2004). It has been proposed that siRNAs guide heterochromatic silencing complexes to homologous target loci through RNA/RNA recognition (Motamedi et al. 2004; Sugiyama et al. 2005; Qi et al. 2006). Screening for Chlamydomonas mutants that are defective in silencing of a transgene at either the transcriptional or post-transcriptional level led to the identification of three genes, Mut11 encoding a WD40-repeat protein, Mut9 encoding a Ser//Thr kinase, and Mut6 encoding a DEAH-box RNA helicase (Wu-Scharf et al. 2000; Jeong Br et al. 2002; Zhang et al. 2002). In these mutants, several transposons were reactivated. However, evidence for direct involvement of these proteins in RNAi is still lacking, and the role of RNAi in transposon silencing in Chlamydomonas remains to be evaluated. Among the 4182 unique small RNAs we obtained from Chlamydomonas, <4% of small RNAs were derived from repeats (Supplementary Table 5). This fraction is much less than what would be expected based on a random distribution, considering that the repeats account for 6.11% of the Chlamydomonas genome as defined by RepeatMasker (Kohany et al. 2006). Thus, RNAi might not be a general mechanism used to silence transposons and repetitive elements. Alternatively, RNAi-controlled silencing of repeats may not be dependent on small RNA accumulation in Chlamydomonas under vegetative growth conditions. In Tetrahymena, scnRNAs are only induced during conjugation to direct developmentally programmed DNA elimination (Mochizuki and Gorovsky 2004a; Lee and Collins 2006). It remains possible that RNAi may play a role in silencing repeats when Chlamydomonas undergoes sexual reproduction. Profiling of small RNAs of Chlamydomonas during this process will be necessary to address this possibility.
In plant and animal species studies so far, there are multiple Dicer and Argonaute family members that are genetically and functionally diversified (Carmell et al. 2002; Carmell and Hannon 2004). For instance, in Arabidopsis, there are four Dicer-like and 10 Argonaute proteins. It has been demonstrated that DCL1 has a unique role in miRNA biogenesis, DCL3 produces ∼24-nt hc-siRNAs, and DCL2 and DCL4 are involved in antiviral defense and the production of nat-siRNAs or ta-siRNAs (Vaucheret 2006). AGO1 binds miRNAs to catalyze the cleavage of miRNA target mRNAs (Baumberger and Baulcombe 2005; Qi et al. 2005), while AGO4 recruits hc-siRNAs to direct DNA/histone methylation, sometimes resulting in transcriptional gene silencing (Zilberman et al. 2003; Qi et al. 2006). The Chlamydomonas genome encodes three Dicer-like and at least two Argonaute proteins (Schroda 2006). It will be of great interest to examine whether the different types of small RNAs that we have uncovered are processed by the same or different Dicers, and whether they enter distinct complexes containing different Argonaute proteins. Genetic analysis of RNAi lines or mutants for each Dicer-like and Argonaute gene, in combination with biochemical purification of their complexes, will shed light on the mechanisms of RNAi pathways in Chlamydomonas.
Materials and methods
Chlamydomonas strains and culture conditions
Chlamydomonas strain CC-503 cw92 mt+ was used as the source of small RNA cloning. CC-621 (mt−) was used to produce gametes. Chlamydomonas cells were grown in Tris-acetate-phosphate (TAP) liquid medium (Harris 1989) on a rotary shaker at 23°C, under constant light. Gametes were generated essentially as described (Wegener et al. 1989). Gametic differentiation was induced by transferring vegetative cells in mid-log phase to N-free TAP medium for 18–20 h under the same culture conditions.
Small RNA preparation and cloning
Total RNA was extracted from Chlamydomonas cultures at vegetative growth in TAP medium using Trizol (Invitrogen), and small RNAs were enriched by PEG precipitation method. The isolated small RNAs were separated by 15% denaturing PAGE and visualized by SYBR-gold staining, and small RNAs of 18∼28 nt were gel-purified. Small RNAs were ligated to a 3′ adaptor and a 5′ acceptor sequentially, and then RT–PCR-amplified as described (Lau et al. 2001). PCR products were reamplified using a pair of 454 cloning primers and provided to 454 Life Sciences for sequencing as previously described (Girard et al. 2006).
The genome and transcript sequences
The Chlamydomonas genome (version 3.0) was downloaded from the JGI Chlamydomonas Web site (http://genome.jgi-psf.org/Chlre3/Chlre3.home.html). Since the current Chlamydomonas genome sequences were provided as 1266 scaffolds, to facilitate data analysis, we linked all these scaffolds together following the order of their numbers to build an arbitrary genome. All analyses described in the text were carried out using information extracted from this arbitrary genome. Repeat regions and transposable elements of the Chlamydomonas genome were identified by RepeatMasker (version 3.1.3) according to RepBase (version 10.09) (Jurka et al. 2005).
The sequences of 15,143 annotated Chlamydomonas protein-coding genes (including pseudogenes) were also obtained from the Chlamydomonas Web site (http://genome.jgi-psf.org/Chlre3/Chlre3.home.html). The genomic location and intron–exon structure of each gene were determined by mapping gene sequences to the arbitrary Chlamydomonas genome using the BLAT program (Kent 2002).
The complete genome sequences of O. tauri and O. lucimarinus were downloaded from the O. tauri Web site (http://genome.jgi-psf.org/Ostta4/Ostta4.home.html) and the O. lucimarinus Web site (http://genome.jgi-psf.org/Ost9901_3/Ost9901_3.home.html).
Small RNA analysis
After removing adaptor/acceptor sequences from the raw reads obtained from 454 Life Sciences, the remaining small RNA sequences were mapped to the arbitrary Chlamydomonas genome using Perl scripts. Relationships of small RNAs to annotated genes were determined by comparing the genomic loci of small RNAs with the BLAT results of gene sequences. Small RNAs derived from known noncoding RNAs were identified by comparing small RNAs with the sequences of noncoding RNAs collected in Rfam (http://www.sanger.ac.uk/Software/Rfam) (Griffiths-Jones et al. 2005).
Predictions of miRNAs
The prediction of Chlamydomonas miRNAs was carried out using criteria similar to those applied for Arabidopsis miRNA prediction previously (Wang et al. 2004). Basically, the upstream and downstream genomic sequences of each miRNA locus within intergenic and intronic regions were extracted and subjected to RNA secondary structure check using Mfold (Zuker 2003). Genomic sequences with lengths ranging from 70 to 350 nt with 20-nt increments were extracted from each miRNA locus. Since mature miRNAs could be located on either the 5′ arm or the 3′ arm of their hairpin-structured precursors, two genomic sequences with the same length were extracted from each miRNA locus, with the cloned small RNA sequences mapped near the 5′ end or the 3′ end, respectively. If the lowest energy form of any genomic sequence extracted as described above contained a relatively long double-stranded region that included the cloned small RNA sequence, the small RNA was selected as a miRNA candidate and the genomic sequence was considered as a miRNA precursor. All the sequences and structures of the precursors were manually checked. In some cases, the precursor sequences were manually extended up to ∼1 kb to get structures of the lowest free energy. In cases in which two small RNAs were mapped to the same stem–loop and they formed a duplex with a 2-nt overhang, the one with higher cloning frequency was chosen as a miRNA, while the other was considered as miRNA*. When the two small RNAs had the same cloning frequency, then both were selected as miRNAs. In cases in which two small RNAs were mapped to the same stem–loop but they could not form a duplex with a 2-nt overhang, both small RNAs were selected as miRNA candidates.
Prediction of miRNA targets
We modified criteria that were developed for plant miRNA target prediction to predict targets of Chlamydomonas miRNAs (Jones-Rhoades and Bartel 2004; Allen et al. 2005). The putative target sites of all miRNA candidates were identified by aligning miRNA sequences to the annotated gene sequences and ESTs of Chlamydomonas using Perl script. The method enables identification of multiple target sites of a single miRNA on the same target sequence. An initial pool of predicted targets was created, with at most four unpaired nucleotides (including G:U pairs, mismatches, and single-nucleotide bulges) and two single-nucleotide bulges allowed between a miRNA and its targets. A mispair scoring system was then applied to these initial targets. Mismatches and single-nucleotide bulges were each scored as 1, and G:U pairs were each scored as 0.5. The scores were doubled if mismatches, G:U pairs, and bulges were located at positions at 2–13 as counted from the 5′ end of a miRNA. Genes with a mispair score ≤3 were selected as putative miRNA targets. In order to assess the specificity of the prediction method, we generated 10 shuffled cohorts for all the miRNAs/Candidates. The signal/noise ratio was calculated by comparing the total number of targets (the “signal”) predicted for miRNAs/Candidates and that for shuffled cohorts.
Detection of phased siRNAs
Small RNAs with a phased position relationship were identified by checking the distance among all small RNAs within every 1000-nt window, anchored at one end of each small RNA. Small RNAs with interval distances that were a multiple of 21 nt (allowing ±2 nt divergence) were selected as phased small RNAs. Every small RNA within the 1000-nt window was examined to identify most small RNAs with the same phased relationship. The selected small RNAs were removed, and the remaining small RNAs were rescanned to search for other phased relationships until no two small RNAs were within the same phase.
Validation of miRNA target
Target validation was performed using a 5′ RACE assay as described except that a nested PCR reaction was performed (Llave et al. 2002).
Northern blot
Northern blot analysis with enriched small RNAs from total extracts or fractions collected from gel filtration was performed as described (Qi et al. 2005). 32P-end-labeled oligonucleotides complementary to miRNA sequences were used as probes. A synthetic 32P-end-labeled 21-nt RNA oligo was used as a size marker.
Chlamydomonas extract preparation and size chromatography
Chlamydomonas cultures were collected and ground into fine powder under liquid nitrogen, and then homogenized in 1 mL/g extraction buffer (20 mM Tris-HCl, 150 mM NaCl, 4 mM MgCl2 at pH 7.5) containing 5 mM DTT and 1 tablet/10 mL (Roche) protease inhibitor cocktail. Cell debris was removed by centrifugation at 22,000g for 20 min at 4°C. The supernatant was collected and protein concentration was determined by Bio-Rad Protein Assay (Bio-Rad).
For size chromatography, a Sephacryl S-300 HR column was used (Amersham Biosciences) with buffer (20 mM Tris-HCl at pH 7.5, 150 mM NaCl, 4 mM MgCl2, 4% glycerol, 2 mM DTT). Ten milligrams of total protein were loaded onto the column. Fractions were collected for testing RISC activity or for RNA preparation.
Slicer activity assays
Fractions from gel filtration were incubated with 32P-labeled in vitro transcripts of miRNA target genes essentially as described (Qi et al. 2005).
Accession numbers
All genome-matched small RNAs referred to in this study have been deposited in the GenBank database under accession numbers EF495357–EF499538 and in Gene Expression Omnibus Platform GPL5013.
Acknowledgments
We thank B. Ding and M. Carmell for critical reading of the manuscript. Y.Q. is supported by Chinese Ministry of Science and Technology, and X.J.W. acknowledges research fundings number 30621001 and number 30570160 from the National Natural Science Foundation of China. G.J.H. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1543507
References
- Allen E., Xie Z., Gustafson A.M., Sung G.H., Spatafora J.W., Carrington J.C., Xie Z., Gustafson A.M., Sung G.H., Spatafora J.W., Carrington J.C., Gustafson A.M., Sung G.H., Spatafora J.W., Carrington J.C., Sung G.H., Spatafora J.W., Carrington J.C., Spatafora J.W., Carrington J.C., Carrington J.C. Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat. Genet. 2004;36:1282–1290. doi: 10.1038/ng1478. [DOI] [PubMed] [Google Scholar]
- Allen E., Xie Z., Gustafson A.M., Carrington J.C., Xie Z., Gustafson A.M., Carrington J.C., Gustafson A.M., Carrington J.C., Carrington J.C. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Aravin A.A., Lagos-Quintana M., Yalcin A., Zavolan M., Marks D., Snyder B., Gaasterland T., Meyer J., Tuschl T., Lagos-Quintana M., Yalcin A., Zavolan M., Marks D., Snyder B., Gaasterland T., Meyer J., Tuschl T., Yalcin A., Zavolan M., Marks D., Snyder B., Gaasterland T., Meyer J., Tuschl T., Zavolan M., Marks D., Snyder B., Gaasterland T., Meyer J., Tuschl T., Marks D., Snyder B., Gaasterland T., Meyer J., Tuschl T., Snyder B., Gaasterland T., Meyer J., Tuschl T., Gaasterland T., Meyer J., Tuschl T., Meyer J., Tuschl T., Tuschl T. The small RNA profile during Drosophila melanogasterdevelopment. Dev. Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- Aravin A., Gaidatzis D., Pfeffer S., Lagos-Quintana M., Landgraf P., Iovino N., Morris P., Brownstein M.J., Kuramochi-Miyagawa S., Nakano T., Gaidatzis D., Pfeffer S., Lagos-Quintana M., Landgraf P., Iovino N., Morris P., Brownstein M.J., Kuramochi-Miyagawa S., Nakano T., Pfeffer S., Lagos-Quintana M., Landgraf P., Iovino N., Morris P., Brownstein M.J., Kuramochi-Miyagawa S., Nakano T., Lagos-Quintana M., Landgraf P., Iovino N., Morris P., Brownstein M.J., Kuramochi-Miyagawa S., Nakano T., Landgraf P., Iovino N., Morris P., Brownstein M.J., Kuramochi-Miyagawa S., Nakano T., Iovino N., Morris P., Brownstein M.J., Kuramochi-Miyagawa S., Nakano T., Morris P., Brownstein M.J., Kuramochi-Miyagawa S., Nakano T., Brownstein M.J., Kuramochi-Miyagawa S., Nakano T., Kuramochi-Miyagawa S., Nakano T., Nakano T., et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- Axtell M.J., Bartel D.P., Bartel D.P. Antiquity of microRNAs and their targets in land plants. Plant Cell. 2005;17:1658–1673. doi: 10.1105/tpc.105.032185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell M.J., Jan C., Rajagopalan R., Bartel D.P., Jan C., Rajagopalan R., Bartel D.P., Rajagopalan R., Bartel D.P., Bartel D.P. A two-hit trigger for siRNA biogenesis in plants. Cell. 2006;127:565–577. doi: 10.1016/j.cell.2006.09.032. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Baumberger N., Baulcombe D.C., Baulcombe D.C. Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. 2005;102:11928–11933. doi: 10.1073/pnas.0505461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani O., Zhu J., Verslues P.E., Sunkar R., Zhu J.K., Zhu J., Verslues P.E., Sunkar R., Zhu J.K., Verslues P.E., Sunkar R., Zhu J.K., Sunkar R., Zhu J.K., Zhu J.K. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell. 2005;123:1279–1291. doi: 10.1016/j.cell.2005.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell M.A., Hannon G.J., Hannon G.J. RNase III enzymes and the initiation of gene silencing. Nat. Struct. Mol. Biol. 2004;11:214–218. doi: 10.1038/nsmb729. [DOI] [PubMed] [Google Scholar]
- Carmell M.A., Xuan Z., Zhang M.Q., Hannon G.J., Xuan Z., Zhang M.Q., Hannon G.J., Zhang M.Q., Hannon G.J., Hannon G.J. The Argonaute family: Tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes & Dev. 2002;16:2733–2742. doi: 10.1101/gad.1026102. [DOI] [PubMed] [Google Scholar]
- Carrington J.C., Ambros V., Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- Elbashir S.M., Lendeckel W., Tuschl T., Lendeckel W., Tuschl T., Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes & Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasciolli V., Mallory A.C., Bartel D.P., Vaucheret H., Mallory A.C., Bartel D.P., Vaucheret H., Bartel D.P., Vaucheret H., Vaucheret H. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr. Biol. 2005;15:1494–1500. doi: 10.1016/j.cub.2005.07.024. [DOI] [PubMed] [Google Scholar]
- Girard A., Sachidanandam R., Hannon G.J., Carmell M.A., Sachidanandam R., Hannon G.J., Carmell M.A., Hannon G.J., Carmell M.A., Carmell M.A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S., Moxon S., Marshall M., Khanna A., Eddy S.R., Bateman A., Moxon S., Marshall M., Khanna A., Eddy S.R., Bateman A., Marshall M., Khanna A., Eddy S.R., Bateman A., Khanna A., Eddy S.R., Bateman A., Eddy S.R., Bateman A., Bateman A. Rfam: Annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33:D121–D124. doi: 10.1093/nar/gki081. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivna S.T., Beyret E., Wang Z., Lin H., Beyret E., Wang Z., Lin H., Wang Z., Lin H., Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes & Dev. 2006;20:1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman A.R., Harris E.E., Hauser C., Lefebvre P.A., Martinez D., Rokhsar D., Shrager J., Silflow C.D., Stern D., Vallon O., Harris E.E., Hauser C., Lefebvre P.A., Martinez D., Rokhsar D., Shrager J., Silflow C.D., Stern D., Vallon O., Hauser C., Lefebvre P.A., Martinez D., Rokhsar D., Shrager J., Silflow C.D., Stern D., Vallon O., Lefebvre P.A., Martinez D., Rokhsar D., Shrager J., Silflow C.D., Stern D., Vallon O., Martinez D., Rokhsar D., Shrager J., Silflow C.D., Stern D., Vallon O., Rokhsar D., Shrager J., Silflow C.D., Stern D., Vallon O., Shrager J., Silflow C.D., Stern D., Vallon O., Silflow C.D., Stern D., Vallon O., Stern D., Vallon O., Vallon O., et al. Chlamydomonas reinhardtii at the crossroads of genomics. Eukaryot. Cell. 2003;2:1137–1150. doi: 10.1128/EC.2.6.1137-1150.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton A., Voinnet O., Chappell L., Baulcombe D., Voinnet O., Chappell L., Baulcombe D., Chappell L., Baulcombe D., Baulcombe D. Two classes of short interfering RNA in RNA silencing. EMBO J. 2002;21:4671–4679. doi: 10.1093/emboj/cdf464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon G.J. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- Harris E.H. The Chlamydomonas sourcebook: A comprehensive guide to biology and laboratory use. Academic Press; New York: 1989. [DOI] [PubMed] [Google Scholar]
- Jeong Br B.R., Wu-Scharf D., Zhang C., Cerutti H., Wu-Scharf D., Zhang C., Cerutti H., Zhang C., Cerutti H., Cerutti H. Suppressors of transcriptional transgenic silencing in Chlamydomonas are sensitive to DNA-damaging agents and reactivate transposable elements. Proc. Natl. Acad. Sci. 2002;99:1076–1081. doi: 10.1073/pnas.022392999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades M.W., Bartel D.P., Bartel D.P. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades M.W., Bartel D.P., Bartel B., Bartel D.P., Bartel B., Bartel B. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- Jurka J., Kapitonov V.V., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J., Kapitonov V.V., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J., Klonowski P., Kohany O., Walichiewicz J., Kohany O., Walichiewicz J., Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Kent W.J. BLAT—The BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohany O., Gentles A.J., Hankus L., Jurka J., Gentles A.J., Hankus L., Jurka J., Hankus L., Jurka J., Jurka J. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinformatics. 2006;7:474. doi: 10.1186/1471-2105-7-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau N.C., Lim L.P., Weinstein E.G., Bartel D.P., Lim L.P., Weinstein E.G., Bartel D.P., Weinstein E.G., Bartel D.P., Bartel D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lau N.C., Seto A.G., Kim J., Kuramochi-Miyagawa S., Nakano T., Bartel D.P., Kingston R.E., Seto A.G., Kim J., Kuramochi-Miyagawa S., Nakano T., Bartel D.P., Kingston R.E., Kim J., Kuramochi-Miyagawa S., Nakano T., Bartel D.P., Kingston R.E., Kuramochi-Miyagawa S., Nakano T., Bartel D.P., Kingston R.E., Nakano T., Bartel D.P., Kingston R.E., Bartel D.P., Kingston R.E., Kingston R.E. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- Lee S.R., Collins K., Collins K. Two classes of endogenous small RNAs in Tetrahymena thermophila. Genes & Dev. 2006;20:28–33. doi: 10.1101/gad.1377006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre P.A., Silflow C.D., Silflow C.D. Chlamydomonas: The cell and its genomes. Genetics. 1999;151:9–14. doi: 10.1093/genetics/151.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis L.A., McCourt R.M., McCourt R.M. Green algae and the origin of land plants. Am. J. Bot. 2004;91:1535–1556. doi: 10.3732/ajb.91.10.1535. [DOI] [PubMed] [Google Scholar]
- Lippman Z., Martienssen R., Martienssen R. The role of RNA interference in heterochromatic silencing. Nature. 2004;431:364–370. doi: 10.1038/nature02875. [DOI] [PubMed] [Google Scholar]
- Llave C., Xie Z., Kasschau K.D., Carrington J.C., Xie Z., Kasschau K.D., Carrington J.C., Kasschau K.D., Carrington J.C., Carrington J.C. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- Lu C., Tej S.S., Luo S., Haudenschild C.D., Meyers B.C., Green P.J., Tej S.S., Luo S., Haudenschild C.D., Meyers B.C., Green P.J., Luo S., Haudenschild C.D., Meyers B.C., Green P.J., Haudenschild C.D., Meyers B.C., Green P.J., Meyers B.C., Green P.J., Green P.J. Elucidation of the small RNA component of the transcriptome. Science. 2005;309:1567–1569. doi: 10.1126/science.1114112. [DOI] [PubMed] [Google Scholar]
- Margulies M., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Berka J., Braverman M.S., Chen Y.J., Chen Z., Braverman M.S., Chen Y.J., Chen Z., Chen Y.J., Chen Z., Chen Z., et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G., Tuschl T., Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- Mochizuki K., Gorovsky M.A., Gorovsky M.A. Conjugation-specific small RNAs in Tetrahymena have predicted properties of scan (scn) RNAs involved in genome rearrangement. Genes & Dev. 2004a;18:2068–2073. doi: 10.1101/gad.1219904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K., Gorovsky M.A., Gorovsky M.A. Small RNAs in genome rearrangement in Tetrahymena. Curr. Opin. Genet. Dev. 2004b;14:181–187. doi: 10.1016/j.gde.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Motamedi M.R., Verdel A., Colmenares S.U., Gerber S.A., Gygi S.P., Moazed D., Verdel A., Colmenares S.U., Gerber S.A., Gygi S.P., Moazed D., Colmenares S.U., Gerber S.A., Gygi S.P., Moazed D., Gerber S.A., Gygi S.P., Moazed D., Gygi S.P., Moazed D., Moazed D. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell. 2004;119:789–802. doi: 10.1016/j.cell.2004.11.034. [DOI] [PubMed] [Google Scholar]
- Palmer J.D., Soltis D.E., Chase M.W., Soltis D.E., Chase M.W., Chase M.W. The plant tree of life: An overview and some points of view. Am. J. Bot. 2004;91:1437–1445. doi: 10.3732/ajb.91.10.1437. [DOI] [PubMed] [Google Scholar]
- Peragine A., Yoshikawa M., Wu G., Albrecht H.L., Poethig R.S., Yoshikawa M., Wu G., Albrecht H.L., Poethig R.S., Wu G., Albrecht H.L., Poethig R.S., Albrecht H.L., Poethig R.S., Poethig R.S. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes & Dev. 2004;18:2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y., Denli A.M., Hannon G.J., Denli A.M., Hannon G.J., Hannon G.J. Biochemical specialization within Arabidopsis RNA silencing pathways. Mol. Cell. 2005;19:421–428. doi: 10.1016/j.molcel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- Qi Y., He X., Wang X.J., Kohany O., Jurka J., Hannon G.J., He X., Wang X.J., Kohany O., Jurka J., Hannon G.J., Wang X.J., Kohany O., Jurka J., Hannon G.J., Kohany O., Jurka J., Hannon G.J., Jurka J., Hannon G.J., Hannon G.J. Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature. 2006;443:1008–1012. doi: 10.1038/nature05198. [DOI] [PubMed] [Google Scholar]
- Rajagopalan R., Vaucheret H., Trejo J., Bartel D.P., Vaucheret H., Trejo J., Bartel D.P., Trejo J., Bartel D.P., Bartel D.P. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes & Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas F.V., Tolia N.H., Song J.J., Aragon J.P., Liu J., Hannon G.J., Joshua-Tor L., Tolia N.H., Song J.J., Aragon J.P., Liu J., Hannon G.J., Joshua-Tor L., Song J.J., Aragon J.P., Liu J., Hannon G.J., Joshua-Tor L., Aragon J.P., Liu J., Hannon G.J., Joshua-Tor L., Liu J., Hannon G.J., Joshua-Tor L., Hannon G.J., Joshua-Tor L., Joshua-Tor L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 2005;12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- Schroda M. RNA silencing in Chlamydomonas: Mechanisms and tools. Curr. Genet. 2006;49:69–84. doi: 10.1007/s00294-005-0042-1. [DOI] [PubMed] [Google Scholar]
- Shrager J., Hauser C., Chang C.W., Harris E.H., Davies J., McDermott J., Tamse R., Zhang Z., Grossman A.R., Hauser C., Chang C.W., Harris E.H., Davies J., McDermott J., Tamse R., Zhang Z., Grossman A.R., Chang C.W., Harris E.H., Davies J., McDermott J., Tamse R., Zhang Z., Grossman A.R., Harris E.H., Davies J., McDermott J., Tamse R., Zhang Z., Grossman A.R., Davies J., McDermott J., Tamse R., Zhang Z., Grossman A.R., McDermott J., Tamse R., Zhang Z., Grossman A.R., Tamse R., Zhang Z., Grossman A.R., Zhang Z., Grossman A.R., Grossman A.R. Chlamydomonas reinhardtii genome project. A guide to the generation and use of the cDNA information. Plant Physiol. 2003;131:401–408. doi: 10.1104/pp.016899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T., Cam H., Verdel A., Moazed D., Grewal S.I., Cam H., Verdel A., Moazed D., Grewal S.I., Verdel A., Moazed D., Grewal S.I., Moazed D., Grewal S.I., Grewal S.I. RNA-dependent RNA polymerase is an essential component of a self-enforcing loop coupling heterochromatin assembly to siRNA production. Proc. Natl. Acad. Sci. 2005;102:152–157. doi: 10.1073/pnas.0407641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talmor-Neiman M., Stav R., Klipcan L., Buxdorf K., Baulcombe D.C., Arazi T., Stav R., Klipcan L., Buxdorf K., Baulcombe D.C., Arazi T., Klipcan L., Buxdorf K., Baulcombe D.C., Arazi T., Buxdorf K., Baulcombe D.C., Arazi T., Baulcombe D.C., Arazi T., Arazi T. Identification of trans-acting siRNAs in moss and an RNA-dependent RNA polymerase required for their biogenesis. Plant J. 2006;48:511–521. doi: 10.1111/j.1365-313X.2006.02895.x. [DOI] [PubMed] [Google Scholar]
- Vagin V.V., Sigova A., Li C., Seitz H., Gvozdev V., Zamore P.D., Sigova A., Li C., Seitz H., Gvozdev V., Zamore P.D., Li C., Seitz H., Gvozdev V., Zamore P.D., Seitz H., Gvozdev V., Zamore P.D., Gvozdev V., Zamore P.D., Zamore P.D. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- Vaucheret H. Post-transcriptional small RNA pathways in plants: Mechanisms and regulations. Genes & Dev. 2006;20:759–771. doi: 10.1101/gad.1410506. [DOI] [PubMed] [Google Scholar]
- Vazquez F., Vaucheret H., Rajagopalan R., Lepers C., Gasciolli V., Mallory A.C., Hilbert J.L., Bartel D.P., Crete P., Vaucheret H., Rajagopalan R., Lepers C., Gasciolli V., Mallory A.C., Hilbert J.L., Bartel D.P., Crete P., Rajagopalan R., Lepers C., Gasciolli V., Mallory A.C., Hilbert J.L., Bartel D.P., Crete P., Lepers C., Gasciolli V., Mallory A.C., Hilbert J.L., Bartel D.P., Crete P., Gasciolli V., Mallory A.C., Hilbert J.L., Bartel D.P., Crete P., Mallory A.C., Hilbert J.L., Bartel D.P., Crete P., Hilbert J.L., Bartel D.P., Crete P., Bartel D.P., Crete P., Crete P. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell. 2004;16:69–79. doi: 10.1016/j.molcel.2004.09.028. [DOI] [PubMed] [Google Scholar]
- Wang X.J., Reyes J.L., Chua N.H., Gaasterland T., Reyes J.L., Chua N.H., Gaasterland T., Chua N.H., Gaasterland T., Gaasterland T. Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol. 2004;5:R65. doi: 10.1186/gb-2004-5-9-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegener D., Treier U., Beck C.F., Treier U., Beck C.F., Beck C.F. Procedures for the generation of mature Chlamydomonas reinhardtii zygotes for molecular and biochemical analyses. Plant Physiol. 1989;90:512–515. doi: 10.1104/pp.90.2.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu-Scharf D., Jeong B., Zhang C., Cerutti H., Jeong B., Zhang C., Cerutti H., Zhang C., Cerutti H., Cerutti H. Transgene and transposon silencing in Chlamydomonas reinhardtii by a DEAH-box RNA helicase. Science. 2000;290:1159–1162. doi: 10.1126/science.290.5494.1159. [DOI] [PubMed] [Google Scholar]
- Xie Z., Johansen L.K., Gustafson A.M., Kasschau K.D., Lellis A.D., Zilberman D., Jacobsen S.E., Carrington J.C., Johansen L.K., Gustafson A.M., Kasschau K.D., Lellis A.D., Zilberman D., Jacobsen S.E., Carrington J.C., Gustafson A.M., Kasschau K.D., Lellis A.D., Zilberman D., Jacobsen S.E., Carrington J.C., Kasschau K.D., Lellis A.D., Zilberman D., Jacobsen S.E., Carrington J.C., Lellis A.D., Zilberman D., Jacobsen S.E., Carrington J.C., Zilberman D., Jacobsen S.E., Carrington J.C., Jacobsen S.E., Carrington J.C., Carrington J.C. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Allen E., Wilken A., Carrington J.C., Allen E., Wilken A., Carrington J.C., Wilken A., Carrington J.C., Carrington J.C. DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc. Natl. Acad. Sci. 2005;102:12984–12989. doi: 10.1073/pnas.0506426102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M.C., Chao J.L., Chao J.L. RNA-guided DNA deletion in Tetrahymena: An RNAi-based mechanism for programmed genome rearrangements. Annu. Rev. Genet. 2005;39:537–559. doi: 10.1146/annurev.genet.39.073003.095906. [DOI] [PubMed] [Google Scholar]
- Yekta S., Shih I.H., Bartel D.P., Shih I.H., Bartel D.P., Bartel D.P. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- Yoshikawa M., Peragine A., Park M.Y., Poethig R.S., Peragine A., Park M.Y., Poethig R.S., Park M.Y., Poethig R.S., Poethig R.S. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes & Dev. 2005;19:2164–2175. doi: 10.1101/gad.1352605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Wu-Scharf D., Jeong B.R., Cerutti H., Wu-Scharf D., Jeong B.R., Cerutti H., Jeong B.R., Cerutti H., Cerutti H. A WD40-repeat containing protein, similar to a fungal co-repressor, is required for transcriptional gene silencing in Chlamydomonas. Plant J. 2002;31:25–36. doi: 10.1046/j.1365-313x.2002.01331.x. [DOI] [PubMed] [Google Scholar]
- Zilberman D., Cao X., Jacobsen S.E., Cao X., Jacobsen S.E., Jacobsen S.E. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299:716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold Web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]