Abstract
Selection of the AUG start codon is a key step in translation initiation requiring hydrolysis of GTP in the eIF2•GTP•Met-tRNAiMet ternary complex (TC) and subsequent Pi release from eIF2•GDP•Pi. It is thought that eIF1 prevents recognition of non-AUGs by promoting scanning and blocking Pi release at non-AUG codons. We show that Sui− mutations in Saccharomyces cerevisiae eIF1, which increase initiation at UUG codons, reduce interaction of eIF1 with 40S subunits in vitro and in vivo, and both defects are diminished in cells by overexpressing the mutant proteins. Remarkably, Sui− mutation ISQLG93–97ASQAA (abbreviated 93–97) accelerates eIF1 dissociation and Pi release from reconstituted preinitiation complexes (PICs), whereas a hyperaccuracy mutation in eIF1A (that suppresses Sui− mutations) decreases the eIF1 off-rate. These findings demonstrate that eIF1 dissociation is a critical step in start codon selection, which is modulated by eIF1A. We also describe Gcd− mutations in eIF1 that impair TC loading on 40S subunits or destabilize the multifactor complex containing eIF1, eIF3, eIF5, and TC, showing that eIF1 promotes PIC assembly in vivo beyond its important functions in AUG selection.
Keywords: AUG selection, Saccharomyces cerevisiae, translation initiation, eIF1
The eukaryotic translation initiation pathway produces an 80S ribosome bound to mRNA with methionyl initiator tRNA (Met-tRNAiMet) base-paired to the AUG start codon in the ribosomal P-site. The Met-tRNAiMet is recruited to the small (40S) ribosomal subunit in a ternary complex (TC) with GTP-bound eIF2, to produce the 43S preinitiation complex (PIC). In budding yeast, this reaction is stimulated by eIF1, eIF1A, eIF3, and eIF5 both in vitro (Danaie et al. 1995; Phan et al. 1998; Asano et al. 2001; Algire et al. 2002) and in vivo (Hinnebusch 2000; Olsen et al. 2003; Fekete et al. 2005; Jivotovskaya et al. 2006). The eIF3, eIF5, eIF1, and TC can be isolated in a multifactor complex (MFC) from yeast (Asano et al. 2000) whose formation promotes binding of all constituent factors to 40S subunits in vivo (Valášek et al. 2002, 2004; Singh et al. 2004; Yamamoto et al. 2005; Jivotovskaya et al. 2006). The 43S PIC interacts with the 5′ end of mRNA, producing the 48S PIC, and scans the leader until the anticodon of Met-tRNAiMet base-pairs with an AUG codon. Scanning is promoted by eIF1, eIF1A, and eIF4F in a reconstituted mammalian system (Pestova et al. 1998; Pestova and Kolupaeva 2002), and genetic data suggest that eIF5, eIF1A, and eIF3 promote scanning in yeast cells (Asano et al. 2001; Nielsen et al. 2004; Fekete et al. 2005). In the 48S PIC, the GTP bound to eIF2 is partially hydrolyzed to GDP and inorganic phosphate (Pi) in a manner stimulated by eIF5, but AUG recognition is required for Pi release from eIF2•GDP•Pi, driving GTP hydrolysis to completion (Algire et al. 2005; Maag et al. 2005b). It is thought that eIF2-GDP releases Met-tRNAiMet into the P-site, allowing subsequent joining of the 60S subunit (Hershey and Merrick 2000; Pestova et al. 2000). The eIF2-GDP is recycled to eIF2-GTP by guanine nucleotide exchange factor eIF2B to allow the reassembly of TC.
Genetic studies in yeast showed that eIF1, subunits of eIF2, and eIF5 regulate AUG selection in vivo, by identifying mutations that increase initiation at UUG codons. Such mutations restore translation of the his4-303 allele lacking the initiation codon, producing the Sui− (Suppressor of initiation codon mutation) phenotype (Donahue 2000). Biochemical analysis of the mammalian system showed that eIF1 antagonizes recognition of non-AUG codons during scanning (Pestova and Kolupaeva 2002). The 40S-binding site of eIF1 was localized near the P-site, and it was proposed that eIF1 promotes an open conformation of the PIC conducive to scanning and restricts base-pairing of Met-tRNAiMet with non-AUG triplets (Lomakin et al. 2003). There is biochemical evidence that eIF1 also restrains the GAP (GTPase activating protein) function of eIF5 at non-AUG codons (Unbehaun et al. 2004; Algire et al. 2005). Consistent with these last results, overexpression of wild-type (WT) eIF1 suppresses the increased initiation at UUG codons conferred by various Sui− mutations in vivo (Valášek et al. 2004).
Our recent experiments with a reconstituted yeast system revealed that AUG recognition stimulates dissociation of eIF1 from the PIC as well as Pi release from eIF2•GDP•Pi (Maag et al. 2005b). The kinetics of eIF1 dissociation and Pi release are similar, and a mutation in eIF1 was found to reduce the rates of both reactions in vitro (Algire et al. 2005). This suggested a model wherein eIF1 blocks non-AUG selection by impeding Pi release, beyond its functions in restraining eIF5 GAP function and promoting scanning. All of these eIF1 functions should be eliminated when AUG base-pairs with Met-tRNAiMet, as this triggers eIF1 dissociation from the PIC (Maag et al. 2005b). However, the importance of eIF1 dissociation in controlling the accuracy of AUG selection in vivo was unclear.
If the foregoing model were valid, then it should be possible to obtain a class of Sui− mutations that evoke more rapid eIF1 dissociation when UUG occupies the P-site. Consistent with this, we found previously that the Sui− eIF1 mutation sui1-D83G reduces eIF1 association with native PICs (Valášek et al. 2004). Here we show that sui1-D83G, sui1-Q84P, and a newly isolated Sui− mutation in eIF1, ISQLG93–97ASQAA (abbreviated 93–97 throughout), all decrease eIF1 affinity for 40S subunits in vitro. The 93–97 and Q84P mutations also decrease the stability of eIF1–40S interaction in vivo, and both the Sui− phenotype and impaired 40S binding of eIF1 are partially corrected by overexpressing the mutant proteins. Importantly, the 93–97 mutation elevates the rates of both eIF1 dissociation and Pi release from eIF2•GDP•Pi in reconstituted PICs. The D83G, Q84P, and 93–97 Sui− mutations also increase selection of non-AUGs in the reconstituted mammalian system independent of GTP hydrolysis. In addition, an eIF1A mutation that suppresses UUG initiation decreases (rather than increases) the rate of eIF1 release from initiation complexes. Together, these results provide strong evidence that release of eIF1 from the 40S subunit is a critical step in AUG selection in vivo, and is coupled to Pi release from eIF2•GDP•Pi and the transition to a closed, scanning-incompatible conformation of the initiation complex.
Results
The FDPF9,12ADPA and G107R mutations impair TC binding to 40S subunits in vivo and in vitro
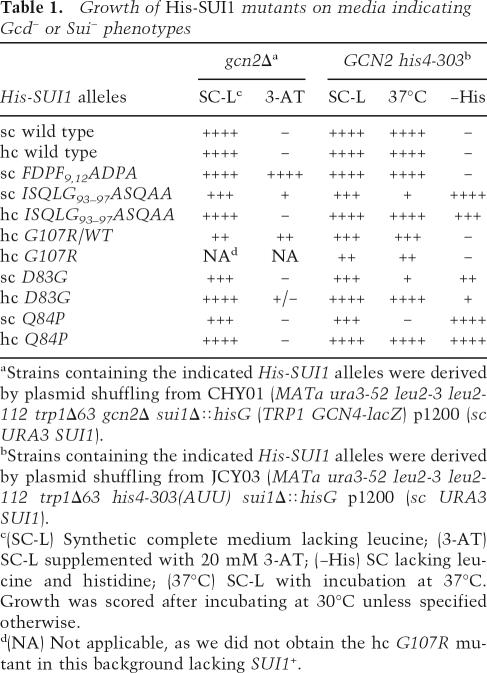
As eIF1 has been implicated in both PIC assembly and AUG selection, we sought to assign these functions to particular residues of the protein through genetic analysis in budding yeast. Whereas the Sui− phenotype signifies reduced stringency of AUG selection, the derepressed translation of GCN4 mRNA (a Gcd− phenotype) is a strong indicator of impaired TC recruitment; that is, 43S PIC assembly. Hence, we constructed alanine substitutions in residues of yeast eIF1 predicted to reside on the surface of the globular domain or unstructured N-terminal tail (NTT) and screened them for Sui− and Gcd− phenotypes. The mutations were made in a plasmid-borne His-SUI1 allele (with hexahistidine encoded at the N terminus), and strains with wild-type SUI1+ replaced by the mutant alleles were produced by plasmid-shuffling. We also constructed His-SUI1 alleles containing the previously described mutations G107R (mof2-1) (Cui et al. 1998), D83G (sui1-1), and Q84P (sui1-17) (Yoon and Donahue 1992). A sui1Δ gcn2Δ strain was used to identify mutations with a Gcd− phenotype, whereas a sui1Δ his4-303 GCN2 strain was used to reveal Sui− phenotypes. The predicted locations in eIF1 of mutations with these phenotypes described in this study are depicted in Figure 1A, and their phenotypes are summarized in Table 1.
Figure 1.
Gcd− mutations in eIF1 reduce TC loading on 40S subunits in vitro. (A) Space-filling model (left) or ribbons depiction (right) of human eIF1 (PDB file 2IF1) highlighting residues corresponding to mutations in yeast eIF1 with Gcd− or Sui− phenotypes. (B) Analysis of Gcd− phenotypes in gcn2Δ strains. Serial dilutions of yeast cells were incubated for 2 d at 30°C on SC lacking uracil and leucine (SC-UL) (left) and 4 d on SC-UL lacking histidine and containing 20 mM or 5 mM 3-AT (right). (Rows 2–5) Strains harboring wild-type (JCY103) or 9,12 (JCY115) sc His-SUI1 alleles and either vector or hc TC plasmid p1780-IMT. (Rows 6,7) Strains with hc wild-type (JCY105) or G107R His-SUI1 alleles (JCY211) and sc SUI1+ plasmid p1200. (Rows 8–10) Strains with wildtype (JCY103), 93–97 (JCY137), or D83G (JCY221) His-SUI1 alleles on sc or hc plasmids. (Row 1) Isogenic GCN2 strain H1642. (C) Expression of GCN4-lacZ (with all four uORFs) in the gcn2Δ strains described in B grown in SC-L or SC-UL. β-Galactosidase activities (nanomoles of o-nitrophenyl-β-D-galactopyranoside cleaved per minute per microgram of protein) were measured in WCEs, and the mean and standard error (SE) from three or more measurements on six independent transformants are plotted. (D) GCN2 his4-303 strains with His-SUI1 alleles wild type (JCY145), 93–97 (JCY189), hc wild type (JCY149), or hc G107R (JCY197), and harboring the GCN4-lacZ plasmid (with all four ORFs, from plasmid p180) were grown in SC-UL also lacking Ile/Val (SC-ULIV) for 6 h with or without sulfometuron (0.5 μg/mL), and β-galactosidase activities were measured. (E) eIF1 mutations 9,12 and G107R reduce the rate of TC loading on reconstituted PICs. Mutant or wild-type eIF1 was mixed with preformed TC (eIF2•GDPNP•35S- Met-tRNAiMet), eIF1A, 40S ribosomes, and mRNA, and the fraction of labeled 35S-Met-tRNAiMet bound to 40S subunits was measured over time by native gel electrophoresis. eIF1 was saturating (1 μM) in all cases. (F) Mutation 93–97, but not 9,12 or G107R, significantly reduces eIF1 affinity for 48S PICs. TAMRA-labeled wild-type or mutant eIF1 was mixed with 40S subunits, eIF1A, TC, and mRNA, and the increase in fluorescence anisotropy at equilibrium (yielding the fraction of eIF1 bound to 43S•mRNA complexes) was measured at different concentrations of the complex.
Table 1.
Growth of His-SUI1 mutants on media indicating Gcd− or Sui− phenotypes
aStrains containing the indicated His-SUI1 alleles were derived by plasmid shuffling from CHY01 (MATa ura3-52 leu2-3 leu2-112 trp1Δ63 gcn2Δ sui1Δ∷hisG (TRP1 GCN4-lacZ) p1200 (sc URA3 SUI1).
bStrains containing the indicated His-SUI1 alleles were derived by plasmid shuffling from JCY03 (MATa ura3-52 leu2-3 leu2-112 trp1Δ63 his4-303(AUU) sui1Δ∷hisG p1200 (sc URA3 SUI1).
c(SC-L) Synthetic complete medium lacking leucine; (3-AT) SC-L supplemented with 20 mM 3-AT; (−His) SC lacking leucine and histidine; (37°C) SC-L with incubation at 37°C. Growth was scored after incubating at 30°C unless specified otherwise.
d(NA) Not applicable, as we did not obtain the hc G107R mutant in this background lacking SUI1+.
We failed to evict SUI1+ from the gcn2Δ strain harboring His-SUI1-G107R, suggesting that the G107R mutation is lethal in this strain background. Western analysis with anti-His6 antibodies of whole-cell extracts (WCEs) from the strain harboring both His-SUI1-G107R and untagged SUI1+ showed that G107R evokes somewhat higher than wild-type expression (Supplementary Fig. S1A), thus implying that the mutation disrupts an essential function of eIF1. G107R on a high-copy (hc) plasmid strongly impairs growth but is not lethal in the his4-303 GCN2 strain, allowing us to characterize its effects on initiation as the only source of eIF1. Western analysis of all other His-SUI1 mutants following eviction of SUI1+ showed that they contained eIF1 at wild-type or greater levels, except sui1-D83G (Supplementary Fig. S1A; data not shown).
Translation of GCN4 mRNA is repressed in nutrient-replete cells by short upstream ORFs (uORFs 1–4) in its leader. After translating uORF1, ribosomes resume scanning, rebind TC, and reinitiate at uORFs 2–4, after which they dissociate from the mRNA and leave the GCN4 ORF untranslated. In amino acid starvation, the concentration of TC is reduced by phosphorylation of eIF2α by kinase GCN2. As a result, a fraction of 40S subunits scanning downstream after terminating at uORF1 rebind TC only after bypassing uORFs 2–4 and then reinitiate at GCN4 instead. GCN4 translation can be derepressed in gcn2Δ mutants by Gcd− mutations that reduce the rate of TC loading on 40S subunits (Hinnebusch 2005). gcn2Δ blocks derepression of GCN4 and attendant derepression of histidine biosynthetic enzymes regulated by GCN4, conferring sensitivity to 3-aminotriazole (3-ATS), an inhibitor of histidine biosynthesis. By restoring derepression of GCN4, Gcd− mutations suppress the 3-ATS phenotype of gcn2Δ cells.
The His-SUI1 alleles with the mutations FDPF9,12ADPA (abbreviated throughout as 9,12) or ISQLG93–97ASQAA (93–97) confer, respectively, strong and weak 3-AT-resistance phenotypes (3ATR) in the gcn2Δ background, although the slow-growth (Slg−) of 93–97 probably diminishes its 3ATR phenotype (Table 1; Fig. 1B). The 3-ATR of 9,12 is evident in the presence of SUI1+ (data not shown), indicating its dominance. The hc G107R allele also confers 3ATR despite its strong Slg− phenotype in the SUI1+ strain (Fig. 1B, hc G107R/WT). These dominant phenotypes suggest that 9,12 and G107R mutations impair eIF1 function rather than merely eliminating the factor from PICs. Among the other mutants we tested, only hc D83G exhibited (weak) 3ATR.
Consistent with their 3ATR/Gcd− phenotypes, the 9,12, hc G107R/WT and 93–97 mutations produced 2.5-fold to fourfold derepression of a GCN4-lacZ reporter in nonstarved gcn2Δ cells compared with the single-copy (sc) or hc SUI1+ alleles (Fig. 1C). Similarly, hc G107R and 93–97 evoked approximately sixfold derepression of GCN4-lacZ in nonstarved GCN2+ cells (Fig. 1D). Starvation of the latter GCN2 strains by treatment with sulfometuron (SM), which inhibits an isoleucine/valine biosynthetic enzyme, led to further increases in GCN4-lacZ expression, although the mutants displayed smaller derepression ratios (fourfold or 11-fold) than the corresponding SUI1+ strains (17- or 22-fold) (Fig. 1D). Thus, the hc G107R, 93–97, and 9,12 mutations partially derepress GCN4 translation independent of an increase in eIF2α phosphorylation.
The strong 3-ATR/Gcd− phenotype of the 9,12 mutation was suppressed by overexpressing all three eIF2 subunits and tRNAiMet from a hc plasmid (hc TC) (Fig. 1B). This suggests that 9,12 derepresses GCN4 translation by reducing the rate of TC binding to 40S subunits scanning the GCN4 leader after translating uORF1, allowing them to bypass uORFs 2–4 and reinitiate at GCN4 instead. Increasing the TC concentration with hc TC would restore efficient TC loading by mass action and prevent bypass of uORFs 2–4. (We did not obtain consistent results on suppression of the 3-ATR phenotype of the hc G107R/WT strain by hc TC, possibly because of copy number fluctuations resulting from the presence of two different hc plasmids in the same cells.)
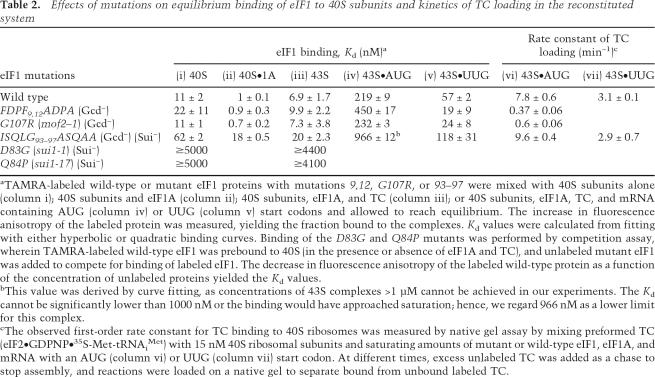
To test our hypothesis that Gcd− mutations in eIF1 impair TC loading on 40S subunits, we measured the kinetics of this reaction in a reconstituted system consisting of eIF1, eIF1A, preassembled TC, 40S subunits, and a model mRNA. As shown in Figure 1E and Table 2 (columns vi–vii), the 9,12 and G107R mutations greatly reduced the rate of TC binding to 40S subunits, whereas saturating 93–97 conferred nearly wild-type loading kinetics. These findings suggest that the Gcd− phenotypes of 9,12 and G107R result directly from impairing eIF1 activity in TC recruitment. As discussed below, we believe that 93–97 affects TC recruitment in vivo by disrupting the MFC or impairing the 40S binding of eIF1 itself.
Table 2.
Effects of mutations on equilibrium binding of eIF1 to 40S subunits and kinetics of TC loading in the reconstituted system
aTAMRA-labeled wild-type or mutant eIF1 proteins with mutations 9,12, G107R, or 93–97 were mixed with 40S subunits alone (column i); 40S subunits and eIF1A (column ii); 40S subunits, eIF1A, and TC (column iii); or 40S subunits, eIF1A, TC, and mRNA containing AUG (column iv) or UUG (column v) start codons and allowed to reach equilibrium. The increase in fluorescence anisotropy of the labeled protein was measured, yielding the fraction bound to the complexes. Kd values were calculated from fitting with either hyperbolic or quadratic binding curves. Binding of the D83G and Q84P mutants was performed by competition assay, wherein TAMRA-labeled wild-type eIF1 was prebound to 40S (in the presence or absence of eIF1A and TC), and unlabeled mutant eIF1 was added to compete for binding of labeled eIF1. The decrease in fluorescence anisotropy of the labeled wild-type protein as a function of the concentration of unlabeled proteins yielded the Kd values.
bThis value was derived by curve fitting, as concentrations of 43S complexes >1 μM cannot be achieved in our experiments. The Kd cannot be significantly lower than 1000 nM or the binding would have approached saturation; hence, we regard 966 nM as a lower limit for this complex.
cThe observed first-order rate constant for TC binding to 40S ribosomes was measured by native gel assay by mixing preformed TC (eIF2•GDPNP•35S-Met-tRNAiMet) with 15 nM 40S ribosomal subunits and saturating amounts of mutant or wild-type eIF1, eIF1A, and mRNA with an AUG (column vi) or UUG (column vii) start codon. At different times, excess unlabeled TC was added as a chase to stop assembly, and reactions were loaded on a native gel to separate bound from unbound labeled TC.
To determine if the defects in TC loading conferred by 9,12 and G107R reflect impaired 40S binding of eIF1, we measured the apparent dissociation constants (Kd) in experiments using fluorescent tetramethylrhodamine-tagged eIF1 (eIF1-TAMRA) and (1) 40S subunits alone; (2) 40S and eIF1A (40S•1A complex); (3) 40S, eIF1A, and TC (43S PIC); or (4) 40S, eIF1A, TC, and mRNA (43S•AUG complex). The 9,12 mutant protein exhibits, at most, a twofold decrease in affinity, while the G107R mutant binds with wild-type affinity (Fig. 1F; Table 2, columns i–v). The decreased rate of TC loading by the 9,12 mutant (Fig. 1E) cannot be due to its twofold or less 40S-binding defect because the eIF1 concentration used in the those assays (1 μM) is well above the measured Kd of the mutant protein.
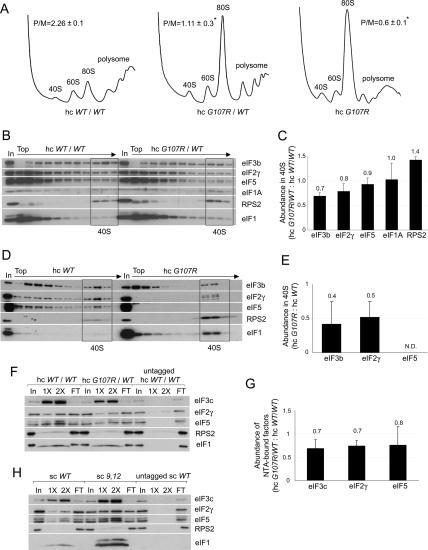
We investigated next whether the 9,12 and G107R mutations decrease TC recruitment in vivo. We began by examining their effects on translation initiation by measuring the polysome:monosome ratios (P/M) in the mutant cells. Consistent with its Slg− phenotype, the hc G107R/WT strain has a diminished P/M, indicating a reduced initiation rate (Fig. 2A, middle), and the P/M is even lower when hc G107R is the only SUI1 allele (Fig. 2A, right). In contrast, the 9,12 mutant shows little decrease in polysome content (data not shown), consistent with its wild-type growth rate on SC medium. We then analyzed the levels of eIF2 and other MFC components associated with native PICs. Treating cells with formaldehyde cross-links factors to ribosomes in vivo, minimizing dissociation of PICs during sedimentation through sucrose gradients (Nielsen et al. 2004). The hc G107R/WT mutant showed moderate reductions in amounts of eIF3 and eIF2 in the 40S fractions, but a higher than wild-type level of free 40S subunits (Fig. 2B,C). Assuming that the excess free 40S subunits in the mutant are competent for PIC assembly, these data indicate that formation or stability of PICs is impaired by hc G107R. In contrast, the 9,12 mutant showed little or no reduction in 40S binding of eIF3 and eIF2 and no accumulation of free 40S subunits (Supplementary Fig. S1B,C).
Figure 2.
Gcd− eIF1 mutation G107R reduces translation initiation and disrupts MFC integrity. (A,B) Effects on translation initiation and PIC levels in vivo. Strains JCY105, JCY211, and JCY197 were grown in SC-UL or SC-L and cross-linked with 1% (v/v) HCHO for 1 h. (A) WCEs were resolved by sedimentation through 4.5%–45% sucrose gradients at 39,000 rpm for 2.5 h and scanned at A254 to determine polysome/monosome ratios (P/M, mean ± SE, n = 3; [*] p < 0.05). (B) Fractions from a 7.5%–30% gradient centrifuged at 41,000 rpm for 5 h and 1% of each WCE (In) were subjected to Western analysis. Fractions containing free 40S subunits are boxed. (C) Initiation factor binding to 40S subunits for the experiment in B and two replicates was quantified by calculating the ratio of eIF signals in 40S fractions of the mutant versus wild-type gradients for each factor. The results plotted are means ± SEs (n = 3). (D) The analysis described in B was performed using strains JCY149 and JCY197, and the 40S fractions were pooled, resolved on a second gradient, and subjected to Western analysis. (E) eIF binding to 40S subunits in the resedimentation experiment in D and two replicates were quantified, plotting the means ± SEs (n = 3). (F) eIF1 mutation G107R impairs MFC integrity in vivo. Nickel chelation chromatography of WCEs from strains JCY105, JCY211, and JCY255. Eluted proteins were subjected to Western analysis. (In) 3% of WCEs input; (1× and 2×) 15% and 30% of eluates; (FT) 1% of flowthrough. (G) For the experiment in F and two replicates, the amounts of eIFs in the eluates were quantified and normalized to the amounts of coeluting His6-eIF1, and the ratio of the normalized values for hc G107R/WT versus hc WT/WT strains is plotted (mean values and SEs; n = 3). (H) WCEs of strains JCY103, JCY115, and JCY253 were analyzed by nickel chelation chromatography as in F.
Additional evidence that G107R reduces the stability of native PICs came from cross-linking analysis of the hc G107R mutant in the GCN2 background, which revealed an obvious decrease in 40S-bound eIF5 and accumulation of free subunits (Supplementary Fig. S1D,E). Discrimination between eIFs cross-linked to 40S subunits and non-cross-linked factors present nonspecifically in 40S fractions can be achieved by resedimentation of the 40S fractions on a second gradient. This is particularly useful for eIF5 and eIF1, which often do not exhibit defined 40S peaks after a single separation (Fekete et al. 2005; Jivotovskaya et al. 2006). Application of the resedimentation protocol confirmed a severe defect in eIF5 binding to 40S subunits and indicated weaker association of eIF2 and eIF3 with native PICs, despite higher than wild-type levels of free subunits and 40S-bound eIF1 (Fig. 2D,E). (The eIFs in the upper fractions after resedimentation likely represent factors not cross-linked to PICs in vivo, although cross-linking could be partially reversed during the in vitro manipulations. Given the latter possibility, the resedimentation protocol could reveal a decrease in stability, rather than diminished formation of mutant PICs in vivo.) We conclude that G107R impairs the ability of 40S-bound eIF1 to promote tight binding of eIF2, eIF3, and especially eIF5 to 40S subunits in vivo.
Further support for this last conclusion came from analysis of MFC integrity in cell extracts. Ni2+ affinity purification of His-tagged eIF1 revealed that G107R reduced copurification of eIF3, eIF2, and eIF5 with eIF1, demonstrating impaired interaction of eIF1 with all MFC components (Fig. 2F,G). In contrast, purification of the 9,12 mutant revealed higher than wild-type levels of copurifying eIFs, commensurate with the higher level of this mutant protein (Fig. 2H). Thus, consistent with its wild-type growth, polysome content, and native 43S PIC levels, the 9,12 mutant showed no defect in association with MFC components.
The nearly wild-type level of 40S-bound eIF2 in the 9,12 mutant extracts (Supplementary Fig. S1B) might seem at odds with the Gcd− phenotype and decreased rate of TC loading in vitro produced by this mutation. However, GCN4 translation is much more sensitive than general translation to defects in TC recruitment (Hinnebusch 2005). Indeed, the strong induction of GCN4 translation in cells starved with SM (e.g., Fig. 1D) occurs with only a small reduction in 40S-bound eIF2 in extracts of cross-linked cells (Supplementary Fig. S1F). Thus, the partial derepression of GCN4 observed in 9,12 cells would not require an observable decrease in 40S-bound eIF2 in vivo. We propose that 9,12 reduces the rate of TC loading by an amount sufficient to partially derepress GCN4 translation but not enough to lower steady-state TC binding to bulk 40S subunits. The G107R mutation, in contrast, impairs MFC integrity and assembly or the stability of bulk PICs in addition to affecting the reinitiation events on GCN4 mRNA.
The 93–97, D83G, and Q84P mutations increase UUG selection in vivo and confer scanning defects in vitro
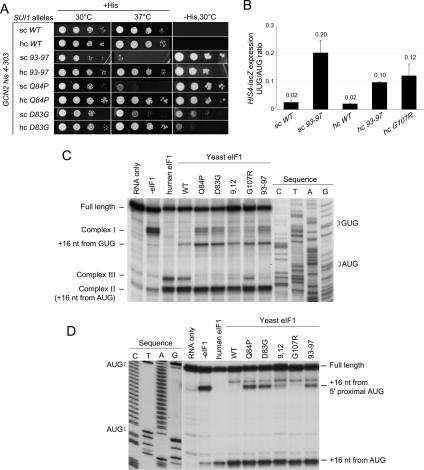
Sui− mutations increase initiation at a UUG codon in the 5′ end of the his4-303 allele, which lacks an AUG start codon, restoring growth on medium lacking histidine (−His). In addition to the previously described mutations D83G and Q84P, the 93–97 mutation produces a strong His+/Sui− phenotype (Fig. 3A; Table 1). Consistent with this, 93–97 confers an 11-fold higher ratio of expression of matched HIS4-lacZ reporters containing UUG versus AUG start codons (Fig. 3B). Using a second set of UUG and AUG luciferase (LUC) reporters, we confirmed our findings for 93–97 and found that D83G and Q84P also produce large increases in the UUG/AUG initiation ratio (Supplementary Fig. S2).
Figure 3.
eIF1 mutations D83G, Q84P, and 93–97 reduce stringent AUG selection in vivo and in vitro. (A) GCN2 his4-303 strains containing His-SUI1 alleles 93–97, D83G, and Q84P were grown on SC-L for 2 d at 30°C or 37°C, and on SC-LH (−His) for 7 d to reveal Sui− phenotypes. (B) Quantification of Sui− phenotypes. Strains from A with HIS4-lacZ reporters with AUG (p367) or UUG (p391), respectively, were grown in SD with His and Trp at 30°C, and β-galactosidase activities were measured in WCEs. The mean ratios of UUG to AUG reporter expression are shown with SEs, from three experiments on six independent transformants. (C) Scanning defects of eIF1 mutants revealed by toeprinting analysis of 48S PICs assembled on native β-globin mRNA in reactions containing mammalian 40S ribosomes, eIF1A, eIF2, eIF3, eIF4A, eIF4B, eIF4F, GTP, and Met-tRNAiMet in the absence or presence of human eIF1 or various yeast wild-type or mutant eIF1 proteins. (D) Toeprinting analysis of 48S complexes formed on a (CAA)nGUS mRNA containing an AUG 1 nt downstream from the 5′ end, as in C.
Compared with the sc 93–97 mutant, the presence of 93–97 on a hc plasmid greatly improved growth on +His medium but decreased growth somewhat on −His medium (Fig. 3A), suggesting that the Sui− phenotype was partially suppressed by overexpressing 93–97. Consistent with this, overexpressing 93–97 decreased the UUG/AUG expression ratio for both HIS4-lacZ (Fig. 3B) and LUC (Supplementary Fig. S2) reporters. Partial suppression of the Sui− phenotypes of Q84P and D83G by overexpression was likewise indicated by the fact that introducing these alleles on hc plasmids improved growth on +His, but reduced or left unaltered growth on −His medium (Fig. 3A), and decreased the UUG/AUG ratios of the LUC reporter (Supplementary Fig. S2). Thus, the 93–97, D83G, and Q84P mutations all increase initiation at UUG in a manner partially reversed by increasing the concentrations of the mutant eIF1 proteins.
Although hc G107R did not produce a His+/Sui− growth phenotype (data not shown), it moderately increased the UUG/AUG ratios for both reporters (Fig. 3B; Supplementary Fig. S2), consistent with previous studies (Cui et al. 1998). Moreover, we found that hc G107R, as well as 93–97, is synthetically lethal with the dominant Sui− mutation in eIF5 encoded by SUI5 (data not shown), as shown previously for D83G (Valášek et al. 2004). These findings suggest that G107R also increases UUG initiation in vivo. Perhaps a His+/Sui− phenotype is masked by the strong Slg− phenotype of this mutant.
We next examined whether the eIF1 mutations alter AUG recognition in a reconstituted mammalian system where eIF1 is needed for efficient scanning to the AUG start codon of β-globin mRNA, using inhibition of primer extension to map the leading edge of the ribosome in the 48S PICs (toeprinting) (Pestova et al. 1998). As eIF5 is omitted, these assays reveal defects in the ability of eIF1 to promote the open, scanning conformation of the PIC independent of GTP hydrolysis and Pi release from eIF2•GDP•Pi. With all human eIFs present except eIF1, most 48S PICs formed aberrantly near the 5′ end (complex I), whereas addition of human or yeast eIF1 allowed the majority to reach the AUG codon (complex II) (Fig. 3C). Smaller proportions of complexes formed 7 nucleotides (nt) upstream of complex II (designated complex III) with both factors, and wild-type yeast eIF1 allowed slight complex formation at the near-cognate GUG upstream of the AUG. (The nature of complex III is currently under investigation.) The previously described Sui− mutations Q84P and D83G, and the 93–97 Sui− mutation isolated here, allowed higher levels of aberrant complex I and GUG complexes, indicating defects in scanning and the ability to suppress recognition of a near-cognate triplet (Fig. 3C). The G107R mutation produced smaller increases in complex I and the GUG complex, and the N-terminal Gcd− mutation 9,12 was even more similar to wild-type yeast eIF1 in both respects. (Note that eIF1 was added at a concentration [17 μM] high enough to compensate for the 40S binding defects [Table 2] of all mutants.)
We analyzed a second mRNA containing an upstream AUG located only 1 nt from the cap. Human and yeast wild-type eIF1 suppressed 48S assembly at the upstream AUG, as expected (Pestova and Kolupaeva 2002), whereas Q84P, D83G, and 93–97 allowed significant recognition of the upstream AUG (Fig. 3D). The 9,12 and G107R mutations conferred little or no complex assembly at the upstream AUG. Thus, 93–97 and the previously isolated Sui− mutations impair the ability of eIF1 to promote scanning past an AUG codon located close to the mRNA cap.
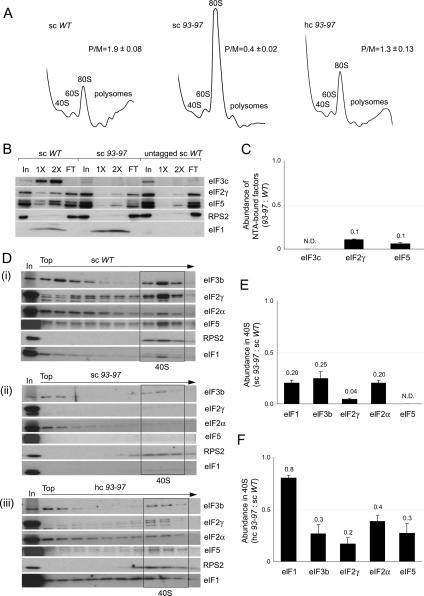
The 93–97 mutation impairs eIF1 association with the MFC and 40S subunits in vivo
We sought next to elucidate the biochemical basis for the Slg− and Sui− phenotypes of the 93–97 mutant. Consistent with its growth defect, the 93–97 mutation greatly decreases the P/M ratio (Fig. 4A) and copurification of other MFC components with His-tagged eIF1 (Fig. 4B,C). Consistent with the latter, 93–97 impairs binding between recombinant eIF1 and the eIF3c N-terminal segment (Supplementary Fig. S3A), the strongest interaction linking eIF1 to the MFC (Asano et al. 2000). 93–97 also reduces the amounts of MFC components that cosediment with 40S subunits in extracts of cross-linked cells (Supplementary Fig. S3B). These defects in eIF binding to native PICs were also observed in the resedimentation protocol, as greatly reduced levels of all MFC constituents, including eIF1 itself, cosedimented with 40S subunits from 93–97 extracts (Fig. 4D, panels i, ii).
Figure 4.
Overexpression of eIF1 mutant 93–97 suppresses its growth defect and partially rescues PIC assembly in vivo. (A) eIF1 mutation 93–97 impairs translation in vivo in a manner suppressed by overexpression. Analysis of polysome profiles, conducted as in Figure 2A, on strains JCY145, JCY189, and JCY193. (B,C) Mutant 93–97 impairs MFC integrity. Nickel chelation chromatography of WCEs from strains JCY145, JCY189, and JCY261 conducted as in Figure 2F,G. (D) 93–97 affects native PIC assembly. Resedimentation analysis of native PICs quantified as described in Figure 2D. (E,F) Quantification of eIF binding to 40S subunits in the experiment shown in D and two replicate experiments, as in Figure 2E.
Interestingly, the 40S-binding defects were partially suppressed by overexpressing the 93–97 allele from a hc plasmid (Fig. 4D, panels ii, panels iii), which correlates with suppression of its Slg− and Sui− phenotypes by overexpression (Fig. 3A,B). Nearly wild-type 40S binding of eIF1 was achieved, whereas 40S binding of other eIFs remained below wild-type levels in cells overexpressing the 93–97 allele (Fig. 4D, panel iii). (As noted above, the eIF1 in the upper fractions after resedimentation likely represents molecules from the 40S fractions of the first gradient that were not cross-linked in vivo or lost their cross-links in vitro; this behavior is exacerbated by eIF1 overexpression.) Importantly, association of all MFC components with polysomes (representing the PICs bound to mRNAs being translated by 80S ribosomes) was substantially recovered in hc 93–97 cells (Supplementary Fig. S3C), in accordance with the nearly complete rescue of polysome content and growth rate. Presumably, 43S PICs remain less stable, while 48S PICs (bound to mRNA) achieve nearly wild-type stability in cells overexpressing the 93–97 mutant. Together, our data indicate that 93–97 impairs translation initiation, MFC integrity, and PIC assembly in a manner partially rescued by increasing the concentration of mutant eIF1. We showed above that overexpression of the Q84P and D83G alleles suppresses their Slg− and Sui− phenotypes (Fig. 3A). Analysis of extracts from cross-linked cells revealed that overexpression likewise rescues 40S binding by these eIF1 mutants (Supplementary Fig. S3D; data not shown).
Sui− mutations in eIF1 elicit more rapid dissociation of eIF1 from reconstituted PICs
The facts that sui1 mutations D83G, Q84P, and 93–97 reduce eIF1 binding to native PICs and that their Sui− phenotypes are diminished by overexpression suggest that increased UUG initiation in these mutants results from weak eIF1 binding to the 40S subunit. This model is consistent with our previous findings using the reconstituted yeast system that AUG stimulates eIF1 dissociation from the PIC and that eIF1 dissociation and Pi release from eIF2•GDP•Pi occur with similar kinetics (Algire et al. 2005; Maag et al. 2005b). Hence, eIF1 mutations that accelerate its dissociation would be predicted to increase selection of non-AUG triplets and confer a Sui− phenotype.
We tested this hypothesis by measuring the effects of the Sui− mutations on the Kd for eIF1 binding to 40S subunits using the binding assays described above. Remarkably, the D83G and Q84P Sui− mutations produce an ∼500-fold increase in the Kd of eIF1 for free 40S subunits and 43S PICs (Table 2, columns i and iii). The 93–97 mutation increases the Kd by fourfold to 15-fold for free 40S, 40S•1A, and 43S•AUG complexes (Fig. 1F; Table 2), suggesting a moderate reduction in eIF1 affinity for the PIC. The Kd of wild-type eIF1 for the 48S PIC is reduced when UUG replaces AUG, reflecting the known higher affinity of eIF1 for 48S PICs with non-AUGs in the P-site (Maag et al. 2005b). The 93–97 mutation decreased eIF1 affinity for the 43S•UUG complex as well (Fig. 5A; Table 2), suggesting that it weakens eIF1 association with the PIC regardless of the P-site triplet.
Figure 5.
Sui− mutation 93–97 increases the rates of eIF1 dissociation and Pi release from eIF2•GDP•Pi in reconstituted 48S PICs. (A) Reduced affinity of the 93–97 mutant for 43S•UUG complexes demonstrated in vitro as described in Table 2. (B) 93–97 accelerates dissociation of eIF1 from 43S•mRNA complexes. 43S complexes reconstituted with TC, eIF1A-fl, and mutant or wild-type eIF1-TAMRA were mixed rapidly with mRNA (AUG) and excess unlabeled eIF1, and the increase in eIF1A-fl fluorescence (caused by decrease in FRET efficiency) was monitored by stopped-flow fluorometry. (C,D) 93–97 accelerates the Pi-release phase of GTP hydrolysis. 43S complexes (with [γ-32P]GTP in the TC) were mixed with eIF5, excess unlabeled GDP, and model mRNA containing AUG (C) or UUG (D) start codons in a rapid quench apparatus, and reactions were quenched at the indicated times with EDTA. The extent of GTP hydrolysis was measured by separating free 32Pi from [γ-32P]GTP by gel electrophoresis followed by PhosphorImager analysis. (E) FL-17–21 mutation in eIF1A decreases the rate of eIF1 dissociation from 43S•mRNA complexes. Kinetic experiments conducted as in B using mutant or wild-type forms of eIF1A-fl and wild-type eIF1-TAMRA in 43S•mRNA (AUG) complexes. For wild-type eIF1A, the rate constants (k) and amplitudes (amp) of the fast (conformational change) and slow (eIF1 release) phases, respectively, are k1 = 9.2 ± 0.3 sec−1, amp1 = 0.16 ± 0.002 and k2 = 0.32 ± 0.003 sec−1, amp2 = 0.83 ± 0.003. The corresponding values for FL-17–21 mutant are k1 = 1.0 ± 0.4 sec−1, amp1 = 0.19 ± 0.03 and k2 = 0.15 ± 0.007 sec−1, amp2 = 0.81 ± 0.09.
To confirm this last conclusion, we examined the effect of 93–97 on eIF1 dissociation kinetics using stopped-flow fluorometry of 43S•AUG PICs assembled with TAMRA-labeled eIF1 and fluorescein-labeled eIF1A (eIF1A-fl). eIF1A-fl acts as the energy donor in fluorescence resonance energy transfer (FRET) between eIF1A-fl and eIF1-TAMRA in these PICs. AUG evokes biphasic loss of FRET and an attendant increase in eIF1A-fl fluorescence, with the slower phase of the reaction corresponding to eIF1 dissociation. The slow phase for wild-type eIF1 has a rate constant of 0.26 sec−1, in accordance with previous results (Maag et al. 2005b), whereas the 93–97 mutation increases the rate constant fivefold to 1.2 sec−1 (Fig. 5B). 93–97 had a similar effect in accelerating eIF1 dissociation from 43S•UUG complexes, in this case by a factor of 2.5 (data not shown). These findings support the idea that 93–97 increases initiation at UUG at least partly by increasing the eIF1 off-rate from the PIC.
We investigated next whether 93–97 accelerates GTP hydrolysis and Pi release from eIF2•GDP•Pi. 43S PICs formed with [γ-32P]GTP in the TC were mixed with saturating eIF5 and mRNA using a rapid quench device. When eIF5 binds, the GTP is hydrolyzed until an internal equilibrium is reached between GDP•Pi and GTP of ∼0.3 (Kint = [GDP•Pi]/[GTP]), which occurs rapidly and is only modestly stimulated by AUG. Dissociation of eIF1 on AUG recognition allows Pi release, driving GTP hydrolysis to completion, and this second phase of the reaction occurs more slowly (∼0.6 sec−1 for wild-type eIF1) and is strongly stimulated by AUG (Algire et al. 2005). In accordance with previous findings, analysis of GTP hydrolysis in the 43S•AUG complex with wild-type eIF1 revealed a fast phase (establishment of the internal equilibrium between GTP and GDP•Pi) of amplitude 0.25 and rate constant of 13 sec−1, and a slow phase (governed by Pi release) of amplitude 0.37 and rate constant 0.8 sec−1. With the 93–97 mutant, the first phase is essentially the same as wild type (amplitude of 0.29 and 12 sec−1), but the second phase is faster (amplitude of 0.33 and 1.6 sec−1) (Fig. 5C). (This twofold increase in rate constant for the 93–97 protein was observed in multiple experiments.) These results agree with the more rapid dissociation of mutant 93–97 from the 43S•AUG complexes observed above.
For the 43S•UUG complex and wild-type eIF1, the slow phase of the reaction (Pi release), but not the fast phase, is reduced for the UUG versus AUG complex (0.45 sec−1 vs. 0.8 sec−1), as expected. Importantly, 93–97 increases the rate of the second phase by sixfold (2.7 sec−1 vs. 0.45 sec−1) (Fig. 5D). (Curiously, the amplitude of the fast phase is decreased by 93–97, suggesting that it perturbs the position of the internal equilibrium between GTP and GDP•Pi.) Thus, 93–97 accelerates the rate of Pi release with AUG or UUG in the P-site. This supports the idea that 93–97 generates a Sui− phenotype at least partly by increasing the rate of GTP hydrolysis and Pi release at UUG codons because of accelerated eIF1 dissociation.
A hyperaccuracy mutation in eIF1A reduces the rate of eIF1 dissociation in vitro
To provide additional evidence that the rate of eIF1 dissociation is a critical determinant of AUG selection, we asked whether mutations in another factor that affect AUG selection in vivo alter the rate of eIF1 dissociation in vitro. We recently described a clustered-alanine substitution in residues 17–21 of the NTT of Flag-tagged eIF1A (FL-17–21) that confers a hyperaccurate phenotype, suppressing the increased UUG initiation in Sui− mutants of eIF5 and eIF2β (Fekete et al. 2007). (Both the N-terminal Flag and Ala substitutions contribute to this phenotype.) We predicted that these mutations would slow down, rather than accelerate, eIF1 dissociation from initiation complexes. After introducing the FL-17–21 mutations into eIF1A-fl, we conducted stopped-flow fluorometry of 43S•AUG PICs assembled with wild-type eIF1-TAMRA. The results indicate that FL-17–21 significantly decreases the rate of eIF1 dissociation (Fig. 5E). Interestingly, it reduces both the rapid phase, which involves a conformational change that separates the C termini of eIF1A and eIF1, and the slow phase of the reaction corresponding to eIF1 dissociation (see legend for Fig. 5). These findings provide evidence that the rapid conformational change and subsequent dissociation of eIF1 are important steps in AUG recognition in vivo, which are modulated by the eIF1A NTT.
Discussion
In this study, we provide several important findings regarding the mechanism of eIF1 function in PIC assembly and AUG selection. First, we identified surface-exposed residues in eIF1 involved in the ability of the factor to stimulate TC recruitment to the 40S subunit, a critical step in 43S PIC assembly. The 9,12, 93–97, and G107R mutations all produce Gcd− phenotypes and partially derepress GCN4 expression in the absence of elevated eIF2α phosphorylation. This phenotype signifies impaired TC loading in vivo, as 40S subunits that resume scanning after translating uORF1 fail to rebind TC quickly enough to ensure reinitiation at uORFs 2–4, allowing a fraction to continue scanning and reinitiate downstream at GCN4. Both 9,12 and G107R greatly reduced the rate of TC binding to reconstituted PICs in vitro. As neither mutation significantly reduces the affinity of eIF1 for 43S or 48S PICs, the 9,12 and G107 residues are required primarily for a function of eIF1 in TC loading rather than 40S binding of eIF1 itself.
Biochemical analysis of native PICs in the 9,12 mutant revealed no obvious defects in MFC integrity, formation or stability of PICs, or polysome assembly. As explained in Results, these findings do not contradict the moderate Gcd− phenotype of this mutant because GCN4 translation is much more sensitive than general initiation to reductions in TC loading. The hc G107R mutant also shows little decrease in 40S-bound eIF2, although a reduction in MFC integrity and less stable 40S association of all MFC components other than eIF1 was detected in hc G107R extracts. Only 40S binding of eIF5 was dramatically decreased in the hc G107R extracts, so the strong initiation defect in this mutant (Slg− phenotype) might result primarily from diminished eIF5 function in promoting MFC binding to 40S subunits (Jivotovskaya et al. 2006) or in stimulating hydrolysis of GTP in the TC.
The 93–97 mutation in eIF1 produces a Gcd− phenotype and decreases binding of eIF2 and other MFC components to native PICs, but does not reduce the rate of TC loading on 40S subunits in vitro. There are several consequences of this mutation in vivo that might account for these results. First, 93–97 reduces 40S binding of eIF1, and this could limit its ability to stimulate TC loading on 40S subunits in vivo. In contrast, its 40S binding defect was overcome in the TC loading assays in vitro by using a saturating concentration of eIF1. Second, 93–97 impairs eIF1 binding to eIF3c and decreases MFC integrity, and optimal 40S binding of eIF2 is enhanced by MFC assembly in vivo (Jivotovskaya et al. 2006). Thus, 93–97 might indirectly impair TC loading in vivo by destabilizing the MFC. As eIF3 was not present in the TC loading assays, the effect of weakened eIF1–eIF3c interaction conferred by 93–97 would not be revealed in these in vitro experiments.
Even though the 93–97 mutation lowers TC occupancy of 40S subunits in vivo, it still does not fully derepress GCN4 translation. This can be explained by noting that 93–97 also impairs the efficiency of scanning, as judged by in vitro toeprint assays. A reduced rate of scanning between uORF1 and uORF4 is expected to compensate for the decreased rate of TC recruitment, restoring reinitiation at uORFs 2–4 and diminishing the Gcd− phenotype (Hinnebusch 2005). The sui1 mutations D83G and Q84P do not produce Gcd− phenotypes despite their pronounced defects in 40S binding and PIC assembly in vitro and in vivo. Presumably, their defects in scanning fully compensate for the defective TC recruitment to maintain repression of GCN4 in nonstarvation conditions. A similar situation was described previously for mutations in eIF1A (Fekete et al. 2005) and eIF5 (Yamamoto et al. 2005) that produce compound defects in PIC assembly and scanning/AUG recognition.
Our findings on the D83G, Q84P, and 93–97 Sui− mutations provide strong in vitro and in vivo evidence supporting the model that eIF1 dissociation from the PIC is an important step in AUG selection. All three mutations dramatically increase initiation at UUG in vivo and confer scanning defects in the reconstituted mammalian system, increasing the frequency at which PICs fail to scan from the 5′ cap, or arrest at an upstream GUG or an AUG located too close to the 5′ end. Accordingly, we propose that these mutations stabilize the scanning-arrested conformation of the PIC at non-AUG triplets and at AUGs lacking optimal 5′ flanking sequences. The D83G and Q84P mutations dramatically impair eIF1 binding to native PICs (Supplementary Fig. S3D; Valášek et al. 2004). While less severe in degree, 93–97 also weakens 40S binding of eIF1 in cell extracts. These in vivo results are commensurate with the increases in Kd for eIF1 binding to 40S subunits or reconstituted PICs of more than two orders of magnitude for D83G and Q84P, and threefold to 18-fold for the 93–97 mutation. Importantly, overexpression of all three mutants diminished their Sui− phenotypes and restored higher than wild-type 40S association in vivo. These findings suggest that the weakened interactions of the mutant eIF1 proteins with 40S subunits (or with eIF3c in the case of 93–97) contribute to their defects in scanning and AUG selection.
The rates of eIF1 dissociation and release of Pi from eIF2•GDP•Pi in reconstituted 48S PICs are higher with AUG versus non-AUG triplets in the P-site. This led us to propose previously that eIF1 impedes Pi release at non-AUG codons, and this activity is eliminated upon AUG recognition by ejection of eIF1 from the 40S subunit. Based on this model, we predicted that one class of Sui− mutations would increase the rate of eIF1 dissociation and Pi release at UUGs. Fulfilling this prediction, the 93–97 mutation increases the rates of both reactions for PICs containing AUG or UUG, helping to explain the Sui− phenotype of 93–97. The Sui− mutations D83G and Q84P impair eIF1 binding to 40S subunits too severely to reconstitute PICs and measure eIF1 dissociation rates, but it seems highly likely that they also accelerate eIF1 dissociation and Pi release with UUG in the P-site. We showed previously that eIF3 reduces the extent of eIF1 dissociation from reconstituted PICs but does not affect the rate constant for eIF1 dissociation, or the rate constant or amplitude of the AUG-dependent conformational change (Maag et al. 2005b). Thus, addition of eIF3 to reconstituted PICs might be expected to modulate the quantitative effects of the eIF1 Sui− mutations but should not alter their overall mechanisms of action.
To explain the partial suppression of the Sui− phenotypes of the 93–97, D83G, and Q84P mutants by overexpression, we propose that increasing the cellular concentrations of the mutant eIF1 proteins allows them to rebind more rapidly after inappropriate dissociation at a UUG codon, preventing Pi release and promoting continued scanning to the next triplet. The same mechanism can be invoked to explain our previous finding that overexpressing wild-type eIF1 partially suppresses the Sui− phenotypes of mutations in eIF3c, eIF5, and eIF2β (Valášek et al. 2004). The fact that overexpressing the Sui− mutants only partially suppresses their Sui− phenotypes, even though they restore wild-type levels of 40S binding, might be explained by proposing that the rate of eIF1 rebinding to a scanning PIC from which eIF1 has dissociated is not rapid enough to completely prevent UUG selection. Another explanation for these genetic characteristics is that more rapid eIF1 dissociation is not the only defect conferred by these mutations. They might also impair eIF1 function in scanning, increasing the dwell time of the PIC and probability of Pi release at non-AUG codons. Such a defect that facilitates UUG selection with eIF1 still bound to the scanning PIC can be invoked to explain the moderate Sui− phenotype of the G107R mutant, as this protein dissociates more slowly than wild-type eIF1 from reconstituted PICs (Algire et al. 2005). Finally, although eIF1 and Pi release are accelerated in vitro by the 93–97 mutation with AUG or UUG in the P-site, initiation at UUG codons is enhanced relative to initiation at AUG in vivo. To explain why increasing the rate of eIF1 release has a smaller effect on initiation at AUG versus UUG, we suggest that other steps in AUG selection—for example, the transition from the open, scanning conformation to the closed complex—become limiting when the rate of eIF1 release exceeds the level achieved by the wild-type factor at AUG.
A model of eIF1 bound to the PIC has been generated by hydroxyl radical mapping of the eIF1-binding site on the 40S subunit (Lomakin et al. 2003). None of the Sui− mutations alter residues in the predicted eIF1–40S interface (Supplementary Fig. S4), suggesting that they impair 40S binding by an indirect mechanism. The fact that overexpressing the mutant eIF1 proteins eliminates their slow-growth phenotypes shows that the mutations do not produce grossly misfolded proteins. As the mutated residues lie on exposed surfaces of eIF1 in the model, they could also affect interactions of eIF1 with TC, other MFC components (including eIF5), or eIF1A, and thereby modulate PIC assembly, scanning, or eIF5 function.
Finally, it is notable that the FL-17–21 mutations in the NTT of eIF1A, which decrease selection of both UUG and AUG triplets in vivo (Fekete et al. 2007), reduce the rates of two reactions involving eIF1 that are triggered by AUG recognition: (1) a conformational change that increases eIF1A–eIF1 separation in the 48S complex and (2) subsequent release of eIF1 from the 40S subunit. This provides strong evidence that both events contribute to AUG selection and are modulated by the eIF1A NTT in vivo. In contrast to eIF1, which binds less tightly to the complex, eIF1A binds more tightly with AUG in the P-site (Maag et al. 2005a). Moreover, the FL-17–21 mutations eliminate eIF1A’s tighter interaction with the PIC upon AUG recognition (Fekete et al. 2007). Thus, tighter binding of eIF1A through its NTT likely promotes the conformational change that separates eIF1A from eIF1 and enhances eIF1 release upon AUG recognition.
Materials and methods
Yeast strain constructions
Deletion of the chromosomal SUI1 gene to construct strains CHY01 and JCY03 is described in the Supplemental Material. The resulting strains were transformed with various His-SUI1 alleles cloned in sc or hc LEU2 plasmids and plated on SC-UL. The resident SUI1+ URA3 plasmid (p1200) was evicted by selection on 5-FOA medium to obtain the relevant mutant strains listed in Supplementary Table S3.
Plasmid constructions and site-directed mutagenesis
Details of the construction of pCFB01 containing the sui1Δ∷hisG-URA3-hisG disruption cassette, and LEU2 plasmids p4389 and pCFB03 containing His-SUI1, are described in the Supplemental Material. Mutations were introduced into the His-SUI1 allele by PCR fusion or the GeneTailor site-directed mutagenesis system (Invitrogen) using the mutagenic primers listed in Supplementary Table S2.
Plasmid pRaugFFuug was derived from a dual luciferase reporter plasmid described previously (Harger and Dinman 2003), modified to express Renilla and firefly luciferase as separate messages, with LUCrenilla under the control of the ADH1 promoter and HIS terminator, LUCfirefly under the GPD promoter and CYC1 terminator, and the start codon of the LUCfirefly ORF altered to TTG.
Biochemical assays with yeast extracts
Assays of β-galactosidase activity in WCEs were performed as described previously (Moehle and Hinnebusch 1991). Measurements of luminescence in WCEs were conducted essentially as described (Dyer et al. 2000). Analyses of polysome profiles and fractionation of native PICs in WCEs from HCHO cross-linked cells, including resedimentation analysis, were conducted essentially as already described (Nielsen et al. 2004; Fekete et al. 2005; Jivotovskaya et al. 2006). For Western analysis of WCEs, extracts were prepared by breaking the cells in 1:1 (v/v) cracking buffer (20 mM Tris-HCl at pH 7.5, 100 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 5 mM NaF with 7 mM β-mercaptoethanol, 1 mM PMSF, Complete protease inhibitor cocktail tablets freshly added) and vortexing with glass beads. WCEs were also prepared by trichloroacetic acid extraction as previously described (Reid and Schatz 1982). Ni2+ chelation chromatography with WCEs was conducted as previously described (Valášek et al. 2001). WCEs were incubated with 4 μL of 50% Ni2+-NTA-silica resin (Qiagen) suspended in 200 μL of buffer A for 2 h at 4°C, followed by washing and elution. In vitro GST pull-down assays were conducted as already described (Asano et al. 1998).
Biochemical assays in the reconstituted yeast system
Reagent preparation is described in the Supplemental Material. Fluorescence anisotropy measurements of equilibrium binding constants were performed as previously described using C-terminally TAMRA-labeled eIF1 or mutants of eIF1 (Maag and Lorsch 2003; Maag et al. 2005b). For all experiments, buffer conditions were 30 mM HEPES (pH 7.4), 100 mM potassium acetate (pH 7.4), 3 mM MgCl2, and 2 mM dithiothreitol. 43S complex formation was measured by gel mobility shift assays as described previously (Algire et al. 2002) and in the Supplemental Material.
FRET experiments were carried out as previously described (Maag et al. 2005b). GTPase assays were conducted by preforming 43S complexes with [γ32P]GTP and initiating the reaction by addition of saturating eIF5 and mRNA using a rapid quench apparatus (KinTek), as previously described (Algire et al. 2005). Component concentrations were 810 nM Met-tRNAi, 800 nM eIF2, 830 nM eIF1, 800 nM eIF1A, 300 nM 40S subunits, 10 μM mRNA(AUG) or 25 μM mRNA(UUG), and 1 μM eIF5.
Toeprinting analysis in the reconstituted mammalian system
48S complexes were assembled in 40-μL reactions containing 3 pmol 40S subunits, 9 pmol eIF2, 9 pmol eIF3, 5 pmol eIF4F, 10 pmol eIF4A, 10 pmol eIF4B, 9 pmol eIF1A, 680 pmol eIF1 (recombinant human or yeast mutants), 5 pmol Met-tRNAiMet, 2 pmol β-globin or CAUG(CAA)nGUS mRNA, 20 mM Tris-HCl (pH 7.5), 100 mM potassium acetate, 2.5 mM magnesium chloride, 1 mM DTT, 0.25 mM spermidine, 1 mM ATP, 0.2 mM GTP, and 2 U/mL ribonuclease inhibitor (RNAseOUT, Invitrogen). Primer extension and polyacrylamide gel electrophoresis of cDNA products were performed as described (Pestova et al. 1998; Pestova and Kolupaeva 2002).
Acknowledgments
We thank Azadeh Esmaeli for technical assistance; Tom Donahue, Ernie Hannig, and Jon Warner for strains and antibodies; Assan Marintchev for help with the eIF1-30S model; and Tom Dever for many helpful suggestions. This work was supported in part by the Intramural Program of the NIH. T.V.P. and J.R.L. acknowledge support from NIH Grants GM59660 and GM62128, respectively.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1528307
References
- Algire M.A., Maag D., Savio P., Acker M.G., Tarun S.Z., Jr., Sachs A.B., Asano K., Nielsen K.H., Olsen D.S., Phan L., Maag D., Savio P., Acker M.G., Tarun S.Z., Jr., Sachs A.B., Asano K., Nielsen K.H., Olsen D.S., Phan L., Savio P., Acker M.G., Tarun S.Z., Jr., Sachs A.B., Asano K., Nielsen K.H., Olsen D.S., Phan L., Acker M.G., Tarun S.Z., Jr., Sachs A.B., Asano K., Nielsen K.H., Olsen D.S., Phan L., Tarun S.Z., Jr., Sachs A.B., Asano K., Nielsen K.H., Olsen D.S., Phan L., Sachs A.B., Asano K., Nielsen K.H., Olsen D.S., Phan L., Asano K., Nielsen K.H., Olsen D.S., Phan L., Nielsen K.H., Olsen D.S., Phan L., Olsen D.S., Phan L., Phan L., et al. Development and characterization of a reconstituted yeast translation initiation system. RNA. 2002;8:382–397. doi: 10.1017/s1355838202029527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Algire M.A., Maag D., Lorsch J.R., Maag D., Lorsch J.R., Lorsch J.R. π release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol. Cell. 2005;20:251–262. doi: 10.1016/j.molcel.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Asano K., Phan L., Anderson J., Hinnebusch A.G., Phan L., Anderson J., Hinnebusch A.G., Anderson J., Hinnebusch A.G., Hinnebusch A.G. Complex formation by all five homologues of mammalian translation initiation factor 3 subunits from yeast Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:18573–18585. doi: 10.1074/jbc.273.29.18573. [DOI] [PubMed] [Google Scholar]
- Asano K., Clayton J., Shalev A., Hinnebusch A.G., Clayton J., Shalev A., Hinnebusch A.G., Shalev A., Hinnebusch A.G., Hinnebusch A.G. A multifactor complex of eukaryotic initiation factors eIF1, eIF2, eIF3, eIF5, and initiator tRNAMet is an important translation initiation intermediate in vivo. Genes & Dev. 2000;14:2534–2546. doi: 10.1101/gad.831800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K., Shalev A., Phan L., Nielsen K., Clayton J., Valášek L., Donahue T.F., Hinnebusch A.G., Shalev A., Phan L., Nielsen K., Clayton J., Valášek L., Donahue T.F., Hinnebusch A.G., Phan L., Nielsen K., Clayton J., Valášek L., Donahue T.F., Hinnebusch A.G., Nielsen K., Clayton J., Valášek L., Donahue T.F., Hinnebusch A.G., Clayton J., Valášek L., Donahue T.F., Hinnebusch A.G., Valášek L., Donahue T.F., Hinnebusch A.G., Donahue T.F., Hinnebusch A.G., Hinnebusch A.G. Multiple roles for the carboxyl terminal domain of eIF5 in translation initiation complex assembly and GTPase activation. EMBO J. 2001;20:2326–2337. doi: 10.1093/emboj/20.9.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Dinman J.D., Kinzy T.G., Peltz S.W., Dinman J.D., Kinzy T.G., Peltz S.W., Kinzy T.G., Peltz S.W., Peltz S.W. The Mof2/Sui1 protein is a general monitor of translational accuracy. Mol. Cell. Biol. 1998;18:1506–1516. doi: 10.1128/mcb.18.3.1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danaie P., Wittmer B., Altmann M., Trachsel H., Wittmer B., Altmann M., Trachsel H., Altmann M., Trachsel H., Trachsel H. Isolation of a protein complex containing translation initiation factor Prt1 from Saccharomyces cerevisiae. J. Biol. Chem. 1995;270:4288–4292. doi: 10.1074/jbc.270.9.4288. [DOI] [PubMed] [Google Scholar]
- Donahue T. Genetic approaches to translation initiation in Saccharomyces cerevisiae. In: Sonenberg N., et al., editors. Translational control of gene expression. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. pp. 487–502. [Google Scholar]
- Dyer B.W., Ferrer F.A., Klinedinst D.K., Rodriguez R., Ferrer F.A., Klinedinst D.K., Rodriguez R., Klinedinst D.K., Rodriguez R., Rodriguez R. A noncommercial dual luciferase enzyme assay system for reporter gene analysis. Anal. Biochem. 2000;282:158–161. doi: 10.1006/abio.2000.4605. [DOI] [PubMed] [Google Scholar]
- Fekete C.A., Applefield D.J., Blakely S.A., Shirokikh N., Pestova T., Lorsch J.R., Hinnebusch A.G., Applefield D.J., Blakely S.A., Shirokikh N., Pestova T., Lorsch J.R., Hinnebusch A.G., Blakely S.A., Shirokikh N., Pestova T., Lorsch J.R., Hinnebusch A.G., Shirokikh N., Pestova T., Lorsch J.R., Hinnebusch A.G., Pestova T., Lorsch J.R., Hinnebusch A.G., Lorsch J.R., Hinnebusch A.G., Hinnebusch A.G. The eIF1A C-terminal domain promotes initiation complex assembly, scanning and AUG selection in vivo. EMBO J. 2005;24:3588–3601. doi: 10.1038/sj.emboj.7600821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekete C., Mitchell S., Cherkasova V., Applefield D., Algire M.A., Maag D., Saini A., Lorsch J.R., Hinnebusch A.G., Mitchell S., Cherkasova V., Applefield D., Algire M.A., Maag D., Saini A., Lorsch J.R., Hinnebusch A.G., Cherkasova V., Applefield D., Algire M.A., Maag D., Saini A., Lorsch J.R., Hinnebusch A.G., Applefield D., Algire M.A., Maag D., Saini A., Lorsch J.R., Hinnebusch A.G., Algire M.A., Maag D., Saini A., Lorsch J.R., Hinnebusch A.G., Maag D., Saini A., Lorsch J.R., Hinnebusch A.G., Saini A., Lorsch J.R., Hinnebusch A.G., Lorsch J.R., Hinnebusch A.G., Hinnebusch A.G. N- and C-terminal residues of eIF1A have opposing effects on the fidelity of start codon selection. EMBO J. 2007;26:1602–1614. doi: 10.1038/sj.emboj.7601613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harger J.W., Dinman J.D., Dinman J.D. An in vivo dual-luciferase assay system for studying translational recoding in the yeast Saccharomyces cerevisiae. RNA. 2003;9:1019–1024. doi: 10.1261/rna.5930803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey J.W.B., Merrick W.C., Merrick W.C. Pathway and mechanism of initiation of protein synthesis. In: Sonenberg N., et al., editors. Translational control of gene expression. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. pp. 33–88. [Google Scholar]
- Hinnebusch A.G. Mechanism and regulation of initiator methionyl-tRNA binding to ribosomes. In: Sonenberg N., et al., editors. Translational control of gene expression. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. pp. 185–243. [Google Scholar]
- Hinnebusch A.G. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- Jivotovskaya A.V., Valasek L., Hinnebusch A.G., Nielsen K.H., Valasek L., Hinnebusch A.G., Nielsen K.H., Hinnebusch A.G., Nielsen K.H., Nielsen K.H. Eukaryotic translation initiation factor 3 (eIF3) and eIF2 can promote mRNA binding to 40S subunits independently of eIF4G in yeast. Mol. Cell. Biol. 2006;26:1355–1372. doi: 10.1128/MCB.26.4.1355-1372.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin I.B., Kolupaeva V.G., Marintchev A., Wagner G., Pestova T.V., Kolupaeva V.G., Marintchev A., Wagner G., Pestova T.V., Marintchev A., Wagner G., Pestova T.V., Wagner G., Pestova T.V., Pestova T.V. Position of eukaryotic initiation factor eIF1 on the 40S ribosomal subunit determined by directed hydroxyl radical probing. Genes & Dev. 2003;17:2786–2797. doi: 10.1101/gad.1141803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maag D., Lorsch J.R., Lorsch J.R. Communication between eukaryotic translation initiation factors 1 and 1A on the yeast small ribosomal subunit. J. Mol. Biol. 2003;330:917–924. doi: 10.1016/s0022-2836(03)00665-x. [DOI] [PubMed] [Google Scholar]
- Maag D., Algire M.A., Lorsch J.R., Algire M.A., Lorsch J.R., Lorsch J.R. Communication between eukaryotic translation initiation factors 5 and 1A within the ribosomal pre-initiation complex plays a role in start site selection. J. Mol. Biol. 2005a;356:724–737. doi: 10.1016/j.jmb.2005.11.083. [DOI] [PubMed] [Google Scholar]
- Maag D., Fekete C.A., Gryczynski Z., Lorsch J.R., Fekete C.A., Gryczynski Z., Lorsch J.R., Gryczynski Z., Lorsch J.R., Lorsch J.R. A conformational change in the eukaryotic translation preinitiation complex and release of eIF1 signal recognition of the start codon. Mol. Cell. 2005b;17:265–275. doi: 10.1016/j.molcel.2004.11.051. [DOI] [PubMed] [Google Scholar]
- Moehle C.M., Hinnebusch A.G., Hinnebusch A.G. Association of RAP1 binding sites with stringent control of ribosomal protein gene transcription in Saccharomyces cerevisiae. Mol. Cell Biol. 1991;11:2723–2735. doi: 10.1128/mcb.11.5.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen K.H., Szamecz B., Valasek L., Jivotovskaya A., Shin B.S., Hinnebusch A.G., Szamecz B., Valasek L., Jivotovskaya A., Shin B.S., Hinnebusch A.G., Valasek L., Jivotovskaya A., Shin B.S., Hinnebusch A.G., Jivotovskaya A., Shin B.S., Hinnebusch A.G., Shin B.S., Hinnebusch A.G., Hinnebusch A.G. Functions of eIF3 downstream of 48S assembly impact AUG recognition and GCN4 translational control. EMBO J. 2004;23:1166–1177. doi: 10.1038/sj.emboj.7600116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen D.S., Savner E.M., Mathew A., Zhang F., Krishnamoorthy T., Phan L., Hinnebusch A.G., Savner E.M., Mathew A., Zhang F., Krishnamoorthy T., Phan L., Hinnebusch A.G., Mathew A., Zhang F., Krishnamoorthy T., Phan L., Hinnebusch A.G., Zhang F., Krishnamoorthy T., Phan L., Hinnebusch A.G., Krishnamoorthy T., Phan L., Hinnebusch A.G., Phan L., Hinnebusch A.G., Hinnebusch A.G. Domains of eIF1A that mediate binding to eIF2, eIF3 and eIF5B and promote ternary complex recruitment in vivo. EMBO J. 2003;22:193–204. doi: 10.1093/emboj/cdg030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova T.V., Kolupaeva V.G., Kolupaeva V.G. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes & Dev. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova T.V., Borukhov S.I., Hellen C.U.T., Borukhov S.I., Hellen C.U.T., Hellen C.U.T. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature. 1998;394:854–859. doi: 10.1038/29703. [DOI] [PubMed] [Google Scholar]
- Pestova T.V., Lomakin I.B., Lee J.H., Choi S.K., Dever T.E., Hellen C.U.T., Lomakin I.B., Lee J.H., Choi S.K., Dever T.E., Hellen C.U.T., Lee J.H., Choi S.K., Dever T.E., Hellen C.U.T., Choi S.K., Dever T.E., Hellen C.U.T., Dever T.E., Hellen C.U.T., Hellen C.U.T. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature. 2000;403:332–335. doi: 10.1038/35002118. [DOI] [PubMed] [Google Scholar]
- Phan L., Zhang X., Asano K., Anderson J., Vornlocher H.P., Greenberg J.R., Qin J., Hinnebusch A.G., Zhang X., Asano K., Anderson J., Vornlocher H.P., Greenberg J.R., Qin J., Hinnebusch A.G., Asano K., Anderson J., Vornlocher H.P., Greenberg J.R., Qin J., Hinnebusch A.G., Anderson J., Vornlocher H.P., Greenberg J.R., Qin J., Hinnebusch A.G., Vornlocher H.P., Greenberg J.R., Qin J., Hinnebusch A.G., Greenberg J.R., Qin J., Hinnebusch A.G., Qin J., Hinnebusch A.G., Hinnebusch A.G. Identification of a translation initiation factor 3 (eIF3) core complex, conserved in yeast and mammals, that interacts with eIF5. Mol. Cell. Biol. 1998;18:4935–4946. doi: 10.1128/mcb.18.8.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid G.A., Schatz G., Schatz G. Import of proteins into mitochondria. J. Biol. Chem. 1982;257:13062–13067. [PubMed] [Google Scholar]
- Singh C.R., He H M., Ii, Yamamoto Y., Asano K., He H M., Ii, Yamamoto Y., Asano K., Yamamoto Y., Asano K., Asano K. Efficient incorporation of eukaryotic initiation factor 1 into the multifactor complex is critical for formation of functional ribosomal preinitiation complexes in vivo. J. Biol. Chem. 2004;279:31910–31920. doi: 10.1074/jbc.M313940200. [DOI] [PubMed] [Google Scholar]
- Unbehaun A., Borukhov S.I., Hellen C.U., Pestova T.V., Borukhov S.I., Hellen C.U., Pestova T.V., Hellen C.U., Pestova T.V., Pestova T.V. Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon–anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes & Dev. 2004;18:3078–3093. doi: 10.1101/gad.1255704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valášek L., Phan L., Schoenfeld L.W., Valásková V., Hinnebusch A.G., Phan L., Schoenfeld L.W., Valásková V., Hinnebusch A.G., Schoenfeld L.W., Valásková V., Hinnebusch A.G., Valásková V., Hinnebusch A.G., Hinnebusch A.G. Related eIF3 subunits TIF32 and HCR1 interact with an RNA recoginition motif in PRT1 required for eIF3 integrity and ribosome binding. EMBO J. 2001;20:891–904. doi: 10.1093/emboj/20.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valášek L., Nielsen K.H., Hinnebusch A.G., Nielsen K.H., Hinnebusch A.G., Hinnebusch A.G. Direct eIF2–eIF3 contact in the multifactor complex is important for translation initiation in vivo. EMBO J. 2002;21:5886–5898. doi: 10.1093/emboj/cdf563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valášek L., Nielsen K.H., Zhang F., Fekete C.A., Hinnebusch A.G., Nielsen K.H., Zhang F., Fekete C.A., Hinnebusch A.G., Zhang F., Fekete C.A., Hinnebusch A.G., Fekete C.A., Hinnebusch A.G., Hinnebusch A.G. Interactions of eukaryotic translation initiation factor 3 (eIF3) subunit NIP1/c with eIF1 and eIF5 promote preinitiation complex assembly and regulate start codon selection. Mol. Cell. Biol. 2004;24:9437–9455. doi: 10.1128/MCB.24.21.9437-9455.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y., Singh C.R., Marintchev A., Hall N.S., Hannig E.M., Wagner G., Asano K., Singh C.R., Marintchev A., Hall N.S., Hannig E.M., Wagner G., Asano K., Marintchev A., Hall N.S., Hannig E.M., Wagner G., Asano K., Hall N.S., Hannig E.M., Wagner G., Asano K., Hannig E.M., Wagner G., Asano K., Wagner G., Asano K., Asano K. The eukaryotic initiation factor (eIF) 5 HEAT domain mediates multifactor assembly and scanning with distinct interfaces to eIF1, eIF2, eIF3, and eIF4G. Proc. Natl. Acad. Sci. 2005;102:16164–16169. doi: 10.1073/pnas.0507960102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon H.J., Donahue T.F., Donahue T.F. The sui1 suppressor locus in Saccharomyces cerevisiae encodes a translation factor that functions during tRNAiMet recognition of the start codon. Mol. Cell. Biol. 1992;12:248–260. doi: 10.1128/mcb.12.1.248. [DOI] [PMC free article] [PubMed] [Google Scholar]