Abstract
In vivo transcription of the Escherichia coli argO gene, which encodes an arginine (Arg) exporter, requires the LysR-family regulator protein ArgP (previously called IciA) and is induced in the presence of Arg or its naturally occurring antimetabolite analog canavanine. Lysine (Lys) addition, on the other hand, phenocopies an argP mutation to result in the shutoff of argO expression. We now report that the ArgP dimer by itself is able to bind the argO promoter-operator region to form a binary complex, but that the formation of a ternary complex with RNA polymerase is greatly stimulated only in presence of a coeffector. Both Arg and Lys were proficient as coeffectors for ArgP-mediated recruitment of RNA polymerase to, and open complex formation at, the argO promoter, although only Arg (but not Lys) was competent to activate transcription. The two coeffectors competed for binding to ArgP, and the ternary complex that had been assembled on the argO template in the presence of Lys could be chased into a transcriptionally active state upon Arg addition. Our results support a novel mechanism of argO regulation in which Lys-bound ArgP reversibly restrains RNA polymerase at the promoter, at a step (following open complex formation) that precedes, and is common to, both abortive and productive transcription. This represents, therefore, the first example of an environmental signal regulating the final step of promoter clearance by RNA polymerase in bacterial transcription. We propose that, in E. coli cells, the ternary complex remains assembled and poised at the argO promoter at all times to respond, positively or negatively, to instantaneous changes in the ratio of intracellular Arg to Lys concentrations.
Keywords: ArgP, arginine export; bacterial transcription, gene regulation, promoter clearance
All bacteria possess high-affinity transport systems for the active uptake or import of nutrient solutes. A single nutrient may be the substrate for more than one import system, and it is generally accepted that these transporters have provided an evolutionary advantage for bacteria to scavenge often scarce resources from their environments (Lolkema et al. 1998; Broome-Smith et al. 1999).
In contrast to the uptake systems, far fewer nutrient efflux systems or exporters in bacteria have been identified and characterized (Aleshin et al. 1999; Burkovski and Krämer 2002; Eggeling and Sahm 2003). The hypothesis is that the efflux systems serve as safety valves to prevent the accumulation to toxic levels of the nutrients in question, and this hypothesis has been validated by genetic studies in a number of instances (Vrljic et al. 1996; Simic et al. 2001; Kennerknecht et al. 2002; Nandineni and Gowrishankar 2004; Kutukova et al. 2005; Bucarey et al. 2006). One would also expect that the exporters are tightly regulated (Bellmann et al. 2001; Nandineni and Gowrishankar 2004; Trötschel et al. 2005), in order to minimize futile cycling of substrate flux across the cell membrane.
One example of a nutrient exporter is that for arginine (Arg), ArgO (previously designated as YggA), in Escherichia coli. In vivo studies with argO-lac fusions have established that argO is under the strict transcriptional control of ArgP (a member of the LysR family of regulator proteins [Schell 1993]), and that its expression is induced by 1 mM Arg as well as by its toxic analog canavanine (CAN, which is a plant derived naturally occurring antimetabolite) (Nandineni and Gowrishankar 2004). ArgP exists as a dimer (Hwang and Kornberg 1990; Hwang et al. 1992; Azam and Ishihama 1999) at an estimated 100–400 molecules per cell (i.e., ∼0.4–1.6 μM monomers) (Hwang and Kornberg 1992; Azam et al. 1999).
Interestingly, the first identification of ArgP was by Arthur Kornberg and coworkers (Hwang and Kornberg 1990, 1992; Thony et al. 1991; Hwang et al. 1992) ∼15 years ago as a protein that binds to oriC to inhibit chromosomal replication initiation in vitro, which led to it being designated as IciA; however, argP (iciA) mutants are viable and exhibit no replication-associated phenotype, so that the role, if any, of ArgP (IciA) in replication initiation is uncertain (Thony et al. 1991; Lee et al. 1996; Nandineni and Gowrishankar 2004; Nandineni et al. 2004). In the same context, ArgP (IciA) has also been implicated as a transcriptional activator of the dnaA and nrd genes involved in DNA replication, but the equivalent phenotypes for argP-null mutants have not been reported (Lee et al. 1996, 1997; Han et al. 1998). The sites on DNA to which ArgP (IciA) has been shown to bind are all AT rich, but no consensus binding sequence has as yet been discerned (Hwang and Kornberg 1990, 1992; Lee et al. 1996; Wei and Bernander 1996; Han et al. 1998; Azam and Ishihama 1999).
On the other hand, both argO and argP-null mutants are supersensitive to CAN, suggesting both that ArgP-mediated regulation of argO is physiologically relevant and that at least one function of ArgO is to export CAN out of the cells (Nandineni and Gowrishankar 2004). ArgO shares considerable sequence similarity with another exporter, LysE of Corynebacterium glutamicum, which mediates both Arg and lysine (Lys) export, and LysE mutants of C. glutamicum have been shown to be sensitive to excessive intracellular accumulation of Arg or Lys (Vrljic et al. 1996; Bellmann et al. 2001). Both Arg and Lys serve as inducers of lysE transcription, and this regulation is mediated by the protein LsyG, which shares sequence similarity with E. coli ArgP and is another member of the LysR family (Bellmann et al. 2001).
In contrast to the situation with C. glutamicum lysE, transcription of E. coli argO in vivo is completely shut off upon addition of 1 mM Lys (Nandineni and Gowrishankar 2004). Lys supplementation therefore phenocopies an argP mutation (Nandineni and Gowrishankar 2004; Nandineni et al. 2004), and in the presence of both Arg and Lys, it is the Lys effect that dominates. Dominant gain-of-function mutations in argP (argPd) have been identified that confer a CAN-resistant phenotype, associated with constitutively elevated argO transcription and abolition of the inactivating effects of Lys supplementation (Celis 1999; Nandineni and Gowrishankar 2004; Nandineni et al. 2004).
The physiological significance of Lys-mediated transcriptional inactivation of argO is unclear; one suggestion has been that E. coli cells have evolved to control the ratio of intracellular Arg to Lys, rather than the absolute levels of intracellular Arg, but the reason for such regulation is not known (Nandineni and Gowrishankar 2004). Also unknown is the mechanism by which Lys supplementation leads to the shutting down of argO transcription. Genetic evidence suggests that ArgP along with Lys as coeffector also down-regulates transcription of other genes needed for NH4+ assimilation and osmotolerance in E. coli (Nandineni et al. 2004).
In this study, we have investigated the roles of ArgP in argO transcription in vitro. Our results indicate that whereas ArgP activates argO transcription in the presence of Arg through the process of RNA polymerase (RNAP) recruitment (Ptashne and Gann 1997; Browning and Busby 2004), the mechanism by which Lys addition phenocopies an argP mutation is not simply by preventing the binding of ArgP to the argO operator but instead by reversible trapping of the recruited RNAP at the promoter in a novel molecular complex that is proficient for neither productive nor abortive transcription. To our knowledge, this is also the first example of an environmental signal acting at the final step of promoter clearance by RNAP to modulate transcription initiation.
Results
Reconstitution of ArgP-mediated regulation of argO transcription in vitro
Toward reconstitution of argO transcriptional regulation in vitro, the in vivo start site of argO transcription was first mapped (in an argPd strain where such transcription is high and constitutive) by primer extension analysis to an A residue situated 28 bases upstream of the predicted translation start site of the structural gene (see Supplementary Fig. S1). A 402-base-pair (bp) region of argO extending from −293 to +109 (relative to the start site of transcription, taken as +1) (Fig. 1A), cloned upstream of the lacZ reporter gene in a single-copy-number plasmid vector, was shown to confer identical properties of regulated expression of β-galactosidase as that reported earlier for a chromosomal argO-lac transcriptional fusion (see Fig. 1B, plasmid construct designations given in parentheses in the text below; Nandineni and Gowrishankar 2004). Notably, there was a very low level of β-galactosidase expression from the wild-type plasmid in an argP mutant or in the argP+ strain grown in Lys-supplemented medium, whereas Arg supplementation was associated with a substantial induction of transcription (Fig. 1B, top panel). The expression of wild-type argO-lac was also rendered constitutive in the presence of an argPd-S94L mutation (Fig. 1B, bottom panel). Between two alternative overlapping hexamer motifs (underlined and italicized, respectively) in the octamer sequence TATAGTCT upstream of the argO transcription start site (each of the motifs having the consensus invariant residues A at position 2 and T at position 6), the former was shown to be the authentic −10 promoter element since site-directed alterations of the sequence to TCTAGCCT (Pm1.1) or CCTAGTCT (Pm1.2), but not to TATCGTCC (Pm2.1) or TACCGTCT (Pm2.2), were associated with complete loss of argO-lac expression in the argPd strain (Fig. 1B, bottom panel).
Figure 1.
ArgP-mediated regulation by Arg, Lys, and CAN of argO transcription. (A) Sequence of the argO regulatory region used in this study, numbered with respect to the A residue (in bold) representing the start site of transcription (taken as +1). Promoter −35 and −10 motifs and GTG triplet representing start site of translation have lines above them. Inverted-repeat sequence is marked by overhead arrows. The ArgP region from around −85 to −20 (as determined in this study) is underlined and bracketed. The two −10-like motifs in the ITS are italicized. (B) In vivo argO-lac expression, measured as β-galactosidase-specific activities (sp act), in host strain derivatives with plasmid constructs carrying the argO regulatory region, either wild-type (wt) or with different mutations as described in the text. The host strain for top panel data was argP+ (except for bar at extreme left, which was from host with argP-null mutation) and that for bottom panel data was argPd-S94L. (C) Radiolabeled in vitro transcription products obtained with the 427-bp argO template (with sequence from −293 to +109), in reaction mixes with various additives as indicated, and subjected to electrophoresis on denaturing 10% polyacrylamide gel. (RO) Run-off transcript from argO promoter; (EE) end-to-end transcription product; (His) histidine.
The ArgP protein (with a C-terminal hexa-histidine [His6] tag) was overexpressed and purified (see Materials and Methods; Supplementary Fig. S2). Complementation experiments established that the His6 tag did not affect the function of the protein (data not shown). ArgP-mediated regulation of argO transcription was then tested in a defined in vitro run-off transcription system using σ70-bearing RNAP holoenzyme and a linear DNA template that encompasses the argO region from −293 to +109 (Fig. 1C). The data obtained were consistent with those from the in vivo studies in that they demonstrated (1) a basal level of argO transcription in the absence of ArgP (Fig. 1C, lane 1), (2) substantial ArgP-dependent activation occurring in presence of Arg or CAN but not Lys (Fig. 1C, cf. lanes 3,5 and 1,2,4), and (3) reversal of such activation upon addition of Lys but not of a nonspecific amino acid such as histidine (Fig. 1C, cf. lanes 6 and 7). Transcription from a control (phage T7 A1) promoter was unaffected by ArgP or the coeffectors Arg and Lys (see Supplementary Fig. S3). These findings hence established that ArgP, as well as Arg and Lys, act specifically and directly in regulating argO transcription.
ArgP binds to the −85 to −20 region of argO in both the absence and presence of Arg or Lys
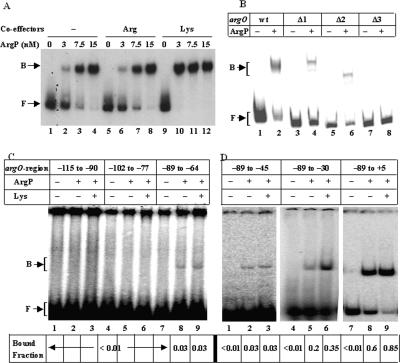
In an electrophoretic mobility shift assay (EMSA), ArgP was shown to specifically bind the radiolabeled argO fragment comprising the sequence from −293 to +109 with a Kd of 6.5 nM (monomer concentration). Binding affinity was unaffected in presence of Arg, whereas it was increased two- to threefold (Kd, 2.5 nM) in presence of Lys (Fig. 2A; see also Supplementary Fig. S4).
Figure 2.
Binary interaction of ArgP with argO regulatory region. In A–C, bands corresponding to free DNA (F) and to DNA in binary complex with ArgP (B) are marked. (A) Electrophoretic mobility of radiolabeled 427-bp argO fragment with indicated monomer concentrations of ArgP in the absence or presence of coeffectors Arg or Lys. (B) Electrophoretic mobility of radiolabeled 427-bp argO fragment (wild type) and its deletion derivatives (described in text) in the absence or presence of 20 nM ArgP (monomer). (C,D) Electrophoretic mobility of radiolabeled fragments corresponding to indicated regions of argO in the absence or presence of ArgP (20 nM monomer) and Lys. Below each lane is given the value, obtained from densitometry, for the fraction of labeled DNA bound by the protein.
Although a 26-bp near-perfect palindromic region from −102 to −77 exists upstream of argO (see Fig. 1A), neither its deletion (designated Δ1) nor that of an overlapping upstream region from −115 to −90 (designated Δ2) led to any significant loss of either ArgP binding to argO in vitro (Fig. 2B) or to Arg- and Lys-modulated transcriptional regulation in vivo (Fig. 1B, top panel). On the other hand, deletion of the overlapping downstream region from −89 to −64 (designated Δ3) substantially reduced both ArgP binding (Fig. 2B) and in vivo argO-lac expression (Fig. 1B, top panel). Likewise, neither the double-stranded 26-mer sequence from −102 to −77 nor that from −115 to −90 exhibited binding to ArgP in EMSA, whereas that from −89 to −64 bound the protein, albeit very weakly, both in the absence and presence of Lys (Fig. 2C).
Since the results above suggested that the argO regulatory region upstream of −89 is not required for ArgP binding, we tested fragments with increasing length of downstream sequence in the EMSA experiments. The affinity of ArgP binding to the sequence from −89 to −45 remained weak, but it was higher for the fragment from −89 to −30 and even more so for that from −89 to +5; in the latter two cases, further increases in ArgP-binding affinity were also observed in the presence of the coeffector Lys (Fig. 2D).
In DNase I footprinting experiments, there was prominent protection by ArgP of the stretch from around −75 to −17 on the top strand (with intervening regions of no protection and hypersensitivity, respectively, around −65 and −47) (Fig. 3A), and from around −85 to −25 on the bottom strand (with a hypersensitive band at −15) (Fig. 3B), both in the absence and presence of Arg or Lys. Small differences in band patterns for the bottom strand (Fig. 3B) were observed between reactions done with ArgP without coeffector (Fig. 3B, lane 3) and those with Arg or Lys (Fig. 3B, lanes 1,2) such as, for example, around positions −18, −50, and −135, but importantly, there were no differences between the patterns obtained on both strands in the presence of Arg and those in the presence of Lys. The footprint of ArgP on argO, as determined above with DNase I digestion, was more or less identical to that inferred from the exonuclease III digestion experiments (see Fig. 6A,B, below).
Figure 3.
Determination of the ArgP-binding site on argO. DNase I digestion patterns were determined for radiolabeled argO fragments that had been uniquely 5′-end-labeled (as described in Materials and Methods) on either the top strand at −115 (A) or the bottom strand toward +109 (B), in the absence or presence of ArgP and coeffectors Arg (A) or Lys (L). The corresponding di-deoxy sequence ladder of argO is represented alongside the numbered lanes in each of the two panels. Vertical lines denote the regions of ArgP footprint, and nucleotide positions of interest are also marked. (C) Comparison of electrophoretic mobility of argO fragment from −85 to −20 with that of the 427-bp fragment (from −293 to +109), in the absence or presence of ArgP (20 nM monomer) and Lys. (F) Free DNA; (B) ArgP–DNA binary complex. Below each lane is given the value, obtained by densitometry, for the fraction of labeled DNA bound by the protein.
Figure 6.
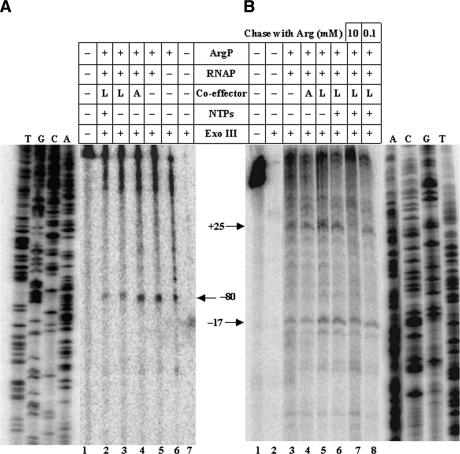
Exonuclease III (Exo III) protection assays to determine upstream and downstream edges of protection on argO by ArgP and RNAP. argO fragments that had been uniquely 5′-end-labeled (as described in Materials and Methods) on either the bottom strand toward +109 (A) or the top strand at −115 (B) were used in reaction mixes with various additives as indicated. (A) Arg; (L) Lys. The corresponding di-deoxy sequence ladder of argO is represented alongside the numbered lanes in each of the two panels. Arrows denote protected bands at the indicated nucleotide positions in the two panels. Lanes 7 and 8 in B represent the reactions involving chase with Arg (0.1 mM and 10 mM, respectively), as described in the text.
Based on these observations, we tested the binding of ArgP in EMSA to an argO fragment from −85 to −20, and the results indicated that this fragment exhibits similar affinities of binding to ArgP in both the absence and presence of Lys as those of the full-length fragment from −293 to +109 (Fig. 3C). The data therefore suggest that most, if not all, of the important sequences or contacts for ArgP lie within the argO region from approximately −85 to −20 (whose sequence is underlined and bracketed in Fig. 1A).
Significant recruitment of RNAP to argO promoter by ArgP occurs only in the presence of either Arg or Lys
EMSA studies were then undertaken with the radiolabeled argO sequence from −293 to +109, ArgP, and RNAP (Fig. 4A). As earlier, with ArgP alone added, all radioactivity was shifted to a sharp band of retarded mobility, indicative of formation of a binary complex that was stable during gel electrophoresis (Fig. 4A, lane 8). On the other hand, with RNAP added in absence of ArgP, only a faint retarded band representing a binary complex of argO with RNAP was observed, but the band corresponding to free probe had also become diffuse, indicating that although a substantial proportion of the binary complex had indeed formed in solution it had then dissociated during gel electrophoresis (Fig. 4A, lane 2); this pattern remained unaffected by addition of Lys to the mixture (Fig. 4A, lane 3).
Figure 4.
Ternary interaction of ArgP and RNAP with argO regulatory region. In A–C, bands corresponding to free DNA (F), DNA in binary complex with ArgP [B(ArgP)] or RNAP [B(RNAP)], and that in ternary complex with ArgP and RNAP (T), are marked. (A) Electrophoretic mobility of radiolabeled argO fragment in the absence or presence (as indicated) of 20 nM ArgP (monomer), 80 nM RNAP, and coeffectors Arg, Lys, or CAN at 0.1 mM. (B) Electrophoretic mobility experiment as in A, in the absence of or following treatment with heparin at 200 μg/mL for the time periods indicated in parentheses. (Note that the samples of free DNA probe preparations used in lane 1 of A and lane 2 of B appear to have been inadvertently contaminated with a small amount of ArgP protein.) (C) Electrophoretic mobility experiment as in B, in the presence of 0.5 mM NTPs in all reactions. (D) In vivo KMnO4 footprinting in cultures of argP mutant (argP−) and argP+ strain grown in 0.2% glucose–minimal A medium without (Nil) or with 1 mM Arg or Lys supplementation as indicated, and then treated with rifampicin. The corresponding di-deoxy sequence ladder of argO is represented alongside, and the −10 region of the argO promoter as well as the HaeIII truncation site at −80 are marked.
In the mixtures containing both ArgP and RNAP, the mobility shift patterns were determined by the presence or absence of coeffectors. In the absence of any coeffector, the radioactivity was distributed in three bands, two of which coincided with the pattern observed with RNAP in absence of ArgP (i.e., corresponding to RNAP–DNA binary complex and to its dissociation to free DNA), while the third (intermediate) band representing the stable ArgP–DNA binary complex displayed reduced intensity (compared with that observed in absence of RNAP) (Fig. 4A, lane 4). These findings indicated that, in the absence of any of the coeffectors, RNAP competes with ArgP for binding to argO, and that the formation of binary complexes of argO with RNAP on the one hand and with ArgP on the other are mutually exclusive with no evidence for occurrence of a ternary complex.
On the other hand, in the presence of RNAP and ArgP with any one of the coeffectors Arg, Lys, or CAN, the pattern of distribution of radioactivity indicated that the mobility of 100% of the argO molecules had been retarded, with a prominent band that corresponded to the ternary complex of DNA, ArgP, and RNAP leading into a diffuse smear that terminated abruptly at the position corresponding to the ArgP–DNA binary complex (Fig. 4A, lanes 5–7); we interpret the smear as evidence for conversion of a fraction of the ternary complex (RNAP–ArgP–DNA) to the stable binary complex (ArgP–DNA) during electrophoresis.
From densitometric analysis of the gel of Figure 4A (data not shown), we estimate that at the protein concentrations employed (eightfold higher molar RNAP concentration compared with that of ArgP dimer), ∼70% of the DNA forms a largely unstable binary complex with RNAP in either the absence of ArgP or its presence without coeffectors, and that the remaining 30% forms the binary complex with ArgP under the latter conditions. In the presence of a coeffector, all of the DNA is engaged in the ternary complex, which is largely stable, since only ∼25% dissociates during electrophoresis to the ArgP–DNA binary complex. We therefore conclude that the coeffectors promote both the formation and stability of the ternary complex, and that the three coeffectors Arg, Lys, and CAN are more or less similar to each other in this regard.
Open complex formation by RNAP at the argO promoter in the presence of ArgP and Lys
The finding that Lys supplementation was associated with the absence of productive transcription despite evidence for RNAP recruitment to and formation of stable ternary complex on the argO template led us to compare the nature of the argO–ArgP–RNAP complex obtained in the presence of Arg with that in the presence of Lys, and to test the possibility that RNAP is trapped at argO in the presence of ArgP and Lys. Trapping of RNAP to a promoter typically had been described earlier at any one of three different steps (for review, see Hsu 2002), namely those of isomerization from the closed to the open complex, transition from abortive transcription (or a moribund complex) to productive transcription, or pausing of RNAP in the initial transcribed sequence (ITS). These three models are exemplified by, respectively, (one mode of) Lac repressor action at the lac promoter (Straney and Crothers 1987), synthesis of abortive transcripts at a variety of promoters (Hsu 2002), and the ternary transcription complex at the phage λ PR promoter that serves as a substrate for modification by the λ Q protein (Roberts et al. 1998).
With both Arg and with Lys supplementation, the ternary complex of ArgP and RNAP assembled on argO (represented by the supershifted band in EMSA) exhibited roughly similar patterns of heparin sensitivity (dissociating to the binary complex of argO and ArgP) in the absence of nucleoside triphosphates (NTPs) (Fig. 4B, cf. lanes 3–6 and 7–10), and of heparin-resistance in their presence (all four NTPs were added in the case of Lys supplementation, but only three [ATP, UTP, CTP] in case of Arg supplementation, in order to prevent run-off transcription in the latter instance) (Fig. 4C, cf. lanes 2–4 and 7–9). A close examination and densitometric analysis (data not shown) of the autoradiographs depicted in Figure 4, B and C, did indicate a small but significant accentuation of heparin sensitivity for the Lys-associated ternary complex compared with that of the Arg-associated one, both in the absence and presence of NTPs. For example, following heparin treatment in the absence of NTPs (Fig. 4B), a smear extending from the region of the ternary complex band to that of the binary (ArgP–DNA) complex was seen in the lanes for Arg supplementation (Fig. 4B, lanes 3–5) but not in those for Lys supplementation (Fig. 4B, lanes 7–9). These results indicate that, in the absence of NTPs, the Lys-associated ternary complex had completely dissociated in solution itself upon heparin addition whereas a significant fraction (20%) of the Arg-associated ternary complex had persisted in solution and underwent dissociation only during electrophoresis. Likewise, in the experiments involving heparin addition in the presence of the NTPs (Fig. 4C), the band patterns were suggestive of greater instability during electrophoresis of the ternary complex assembled in the presence of Lys (see Fig. 4C, lanes 7–9, in which diffuse bands corresponding to the ArgP–DNA binary complex are visible and the bands representing the ternary complex are less intense) compared with that in the presence of Arg (Fig. 4C, lanes 2–4). We estimate from densitometric analysis (data not shown) that ∼25% and <5%, respectively, of the Lys- and Arg-associated ternary complexes assembled in the presence of NTPs underwent dissociation during gel electrophoresis following treatment with heparin. Nevertheless, these data broadly support the conclusion that the Lys-associated ternary complex is largely resistant to heparin in the presence of NTPs, hence rendering unlikely the possibility that RNAP trapping in the presence of ArgP and Lys is at any step prior to formation of the open complex.
In vivo KMnO4 footprinting studies, undertaken in rifampicin-treated cultures, indicated that melting of the −10 region of the argO promoter is more prominent in the argP+ strain than in the argP mutant (Fig. 4D, cf. lanes 1 and 2–4; see also Supplementary Fig. S7). More significantly, for the argP+ strain, the extent of opening of the −10 region in cells grown with 1 mM Lys supplementation (when argO transcription is inactive) was similar to that in cells grown with 1 mM Arg supplementation (when argO transcription is induced) (Fig. 4D, cf. lanes 3 and 4). (It may be noted that the pattern observed in Fig. 4D, lane 2, for the culture grown without Arg or Lys supplementation, would represent the effects of the endogenous amino acid pools in the cells.) These results provided strong support for the conclusion that RNAP is not impeded for formation of an open complex at argO in the presence of ArgP and Lys.
A novel inactive ternary transcription complex is assembled on the argO promoter with ArgP and RNAP in presence of Lys
Notwithstanding the evidence above for open complex formation at the argO promoter by RNAP in presence of ArgP and Lys, the in vitro transcription experiments revealed that the absence of run-off transcript synthesis is accompanied also by the absence of synthesis of any short paused or abortive products under these conditions, whereas both abortive as well as full-length transcripts were obtained in the presence of ArgP and Arg (Fig. 5, cf. lanes 3 and 4). An inverse pulse labeling experiment, which has been reported to be a more sensitive means to detect the presence of moribund RNAP–promoter complexes synthesizing abortive transcripts and in which the labeled nucleotide is added at different time points to an in vitro transcription mix already containing all four NTPs (Kubori and Shimamoto 1996; Susa et al. 2006), also failed to detect abortive products from the argO template in the presence of ArgP and Lys (data not shown).
Figure 5.

Absence of short products representing paused or abortive transcripts in argO transcription reactions with ArgP and Lys. In vitro transcription products (abortive, run-off from argO promoter [RO], and end-to-end [EE], as marked) from the 427-bp argO template (with sequence from −293 to +109) in reaction mixes with various additives as indicated were subjected to electrophoresis on denaturing 20% polyacrylamide gel.
The data so far had therefore indicated that the inactive transcription ensemble obtained at the argO promoter in the presence of ArgP and Lys is explained neither by a defect in isomerization from closed to open complex nor as a moribund complex that is unable to transit from abortive to productive transcription. The possibility was therefore considered that it represents a paused complex in the ITS, analogous to that characterized in detail for the late gene promoters of the lambdoid phages that are the substrates for action by the cognate antiterminator Q proteins (Ko et al. 1998; Roberts et al. 1998; Marr et al. 2001), as well as for the lacUV5 and additional promoters in E. coli (Brodolin et al. 2004; Nickels et al. 2004). One feature that lent support to this possibility was the presence of two promoter −10-like hexamer motifs starting, respectively, at +5 and +18 in the ITS region of argO (sequence motifs italicized in Fig. 1A), reminiscent of the situation in the lambdoid phage and lacUV5 promoters, in each of which a −10-like element in the ITS binds σ70 in RNAP and leads to pausing of the holoenzyme.
Accordingly, we mutated each of the two −10-like elements of the argO ITS separately, from GATACT to GCTACC (designated ITS-1) and from GAATAT to GCATAC (designated ITS-2), respectively, to test whether Lys-mediated inactivation of argO transcription is abolished in them. The results from both in vitro transcription (see Supplementary Fig. S5) and in vivo argO-lac expression studies (Fig. 1B), however, demonstrated that Lys supplementation in the presence of ArgP leads to a drastic reduction in argO transcription even in the mutants. Also consistent with these data were our findings that (1) no paused transcript was detected in the argO–ArgP–RNAP complex similar to those described in paused complexes at the lambdoid phage promoters (Fig. 5; (2) GreB addition did not relieve the ArgP- and Lys-mediated RNAP trapping at argO (see Supplementary Fig. S6), contrary once again to the situation for the lambdoid and lacUV5 promoters; and (3) KMnO4 footprinting in the absence of rifampicin gave a pattern similar to that with rifampicin addition (see Supplementary Fig. S7), whereas in a model of RNAP pausing at the ITS, the two patterns would have been expected to be different (Ko et al. 1998). We conclude that the mechanism of ArgP-mediated trapping of RNAP at the argO promoter in the presence of Lys is not through pausing mediated by the −10-like motifs in the ITS.
To delineate the position of trapped RNAP in the complex, exonuclease III protection experiments were undertaken with a pair of DNA fragments spanning the argO regulatory region that had been 5′-end-labeled on either the bottom strand toward +109 (Fig. 6A) or the top strand at −115 (Fig. 6B) (to detect, respectively, the upstream and downstream edges of protection by protein binding). With ArgP alone, the upstream and downstream edges of protection of argO were at approximately −80 (Fig. 6A, lane 6) and −17 (data not shown), respectively. These bands were visible also in reaction mixtures comprising ArgP along with other components (Fig. 6A [lanes 2–5], B [lanes 3–8]), suggesting the existence of a subset of molecules in binary complex with ArgP under all the other conditions tested. In addition, another downstream edge of protection at around +25 was observed for reaction mixes with ArgP and RNAP in the absence of coeffectors or with Arg (without NTPs), which represents the footprint of RNAP (Fig. 6B, lanes 3,4). Interestingly, an identical downstream edge of protection at around +25 was also observed for the mixes containing ArgP with RNAP and Lys (both with and without the four NTPs added) (Fig. 6B, lanes 5,6), suggesting that even in the Lys-associated ternary complex, which is inactive for both abortive and productive transcription, RNAP is trapped at the step of the open complex itself or immediately thereafter.
Conversion of the Lys-associated inactive ternary transcription complex at argO to a productive form upon Arg addition
Finally, we tested whether the inactive transcription complex formed in the presence of ArgP and Lys is reversible and can be converted to a productive form upon Arg addition. In one approach where the chase was undertaken in solution, to the preincubated mixture of argO template (from −293 to +109) with ArgP, RNAP, and Lys, was added the labeled substrate–heparin mix to generate a primary reaction mix that was inactive for transcription (Fig. 7A, lane 3). An aliquot taken from this mix was then chased with an excess (10 mM) of Arg; the presence of heparin in the mix ensured that new binding of RNAP to the template did not occur following secondary Arg addition, and yet productive transcription was obtained (Fig. 7A, lane 4); in the control experiment in which Lys was omitted from the primary reaction mix, the secondary addition of Arg failed to elicit productive transcription (Fig. 7A, lane 2).
Figure 7.
Arg (or CAN) chase of ArgP–RNAP–argO complex assembled in the presence of Lys and NTPs, either in solution (A) or on streptavidin-coated beads (B). See text for details. Unless otherwise indicated, ArgP was added to all reaction mixes. Other additions were as indicated above the lanes: The primary additives were those that were present in the initial reactions, and the secondary additives were those that were added during the chase step. (A) Arg; (L) Lys; (C) CAN; (RO) run-off transcript from argO promoter; (EE) end-to-end transcription product. In B, the fractions from the primary reaction mixes (pellet [P]; supernatant [S]; unseparated primary reaction mix [P + S]) that were used for preparation of samples for loading on the corresponding lanes of the gel are indicated. Transcription products were resolved on denaturing 6% (A) or 10% (B) polyacrylamide gels.
We also determined the exonuclease III protection pattern that was obtained following addition of either 0.1 mM or 10 mM Arg to a solution mix of argO template DNA (5′-end-labeled on the top strand at −115) with ArgP, Lys (0.1 mM), RNAP, the four NTPs, and heparin. Although the band at +25 (representing the downstream edge of RNAP at the argO promoter) was still visible upon chase with 0.1 mM Arg, it was abolished upon addition of excess (10 mM) Arg (Fig. 6B, cf. lanes 7 and 8). These data are consistent with the notion that under the latter conditions, RNAP molecules from the entire population of ArgP- and Lys-associated inactive ternary complexes had been released to undergo productive transcription.
In the second approach to test the effect of an Arg chase, a biotinylated argO template (from −293 to +109) was initially incubated with ArgP and RNAP in the absence or presence of Arg or Lys, immobilized on streptavidin-coated magnetic beads, and treated with heparin and the four NTPs. An aliquot of this primary reaction mix was saved for analysis; from the remainder, the beads and supernatant were separated, and then the beads were washed (with Arg-supplemented transcription buffer) and incubated (without fresh ArgP or RNAP addition) with Arg and the four NTPs.
As expected, both run-off (argO promoter-encoded) and end-to-end transcription products were obtained in the total primary reaction mix (i.e., of beads and supernatant together) containing Arg (Fig. 7B, lane 1; Supplementary Fig. S8, lane 1); however, little if any of these products was released into the supernatant under the conditions of our experiment (Fig. 7B, lane 2), which is consistent with data from other groups that long RNA products are preferentially retained in the pellet after in vitro transcription on immobilized templates (Kubori and Shimamoto 1997; R. Sen, pers. comm.). Again as expected, in the case of each of the total primary reaction mixes with either Lys or no amino acid added, only the end-to-end transcription product (and no promoter-specific run-off transcripts) was obtained (Supplementary Fig. S8, lanes 2,3), which too was not released into the supernatant (Fig. 7B, lanes 4,6,8). On the other hand, both run-off and end-to-end transcripts were observed following secondary addition of Arg and NTPs to the pellet fractions obtained from the primary mixes that had contained Arg or Lys (Fig. 7B, lanes 3,7), but the same did not occur for similarly treated pellet fractions from the primary mix that had neither amino acid added (Fig. 7B, lane 9). It is possible that, in the case of the sample from the Arg-containing primary mix, at least some of the transcripts observed were those that had been synthesized in the primary reaction itself and retained in the pellet; however, those observed for beads from the Lys-containing primary mix must represent new synthesis following Arg addition in the secondary reaction. Such activation by an Arg chase could be demonstrated even for complexes that had been incubated with Lys for up to 2 h in the primary mix (data not shown). CAN addition was also moderately effective in eliciting run-off transcription from the argO promoter on immobilized template first treated with Lys and ArgP (Fig. 7B, lane 5). We conclude that the transcription complex formed in the presence of ArgP and Lys is in an inactive yet reversible state and that it can be chased into the productive form with Arg (or CAN) addition.
Arg and Lys compete with each other for binding to dimeric ArgP
Gel filtration and glutaraldehyde cross-linking experiments established that ArgP exists as a dimer in solution as had been shown in earlier studies (Hwang and Kornberg 1990; Hwang et al. 1992; Azam et al. 1999), and that its oligomer status was unaffected in the presence of Arg or Lys (Supplementary Fig. S9). Fluorescence spectral changes obtained upon addition of either Arg or Lys to ArgP indicated that the protein binds both ligands, with Kds of ∼150 μM and 70 μM, respectively (Supplementary Fig. S10A,B). Circular dichroism spectroscopy measurements suggested that conformational change(s) are elicited upon Lys binding that result in reduced content of α-helix in the protein (Supplementary Fig. S10C).
That Arg and Lys compete for binding to ArgP was demonstrated in reciprocal competition experiments using 14C-labeled Arg (or Lys) and an excess of cold Lys (or Arg), as follows. In gel filtration experiments done with ArgP mixed with either labeled amino acid alone in the absence of competitor, elution of the label occurred in two well-separated peaks, one corresponding to the protein eluate fraction (in the void volume), which represents the bound state of the ligand, and the other corresponding to the free amino acid (Fig. 8, cf. positions of twin peaks in D,E and that of each of the single peaks of A–C). In presence of the heterologous cold competitor in 100-fold excess, the labeled bound fraction was markedly reduced or absent and almost all of the label was eluted in the fraction representing free amino acid (for both 14C-Arg and 14C-Lys) (Fig. 8F,G). Binding of the labeled amino acid to ArgP was competed also by 100-fold excess of unlabeled CAN (Fig. 8I), but not by a nonspecific amino acid such as alanine (Fig. 8H).
Figure 8.
Binding studies of coeffectors to Arg. Indicated in each chromatograph panel is the material (protein, labeled amino acid, and/or cold competitor amino acid in 100-fold excess) that was loaded on the column. See the text for details. A represents the plot of fluorescence intensity (in arbitrary units, AU) in the various fractions, and B–I represent plots of radioactivity measurements (in counts per min, cpm) in the fractions. (Ala) Alanine.
Discussion
A model for ArgP-mediated argO transcriptional regulation
The model of ArgP-mediated regulation of argO transcription that emerges from the results described above is the following. In the absence of ArgP, RNAP engages with the argO promoter to form a relatively unstable binary complex, resulting in just a low basal level of productive argO transcription. The ArgP dimer has a common binding site(s) for its ligands Arg and Lys (and probably also CAN), and is able to bind the sequence between approximately −85 and −20 of argO to form a stable binary complex in both its liganded and unliganded states. However, the binding of unliganded ArgP and of RNAP to the argO regulatory region appear to be mutually exclusive, and it is only ArgP in its ligand-bound form that is able to cooperatively bind along with RNAP at argO to establish formation of a stable open complex. (It had earlier been speculated that ArgP [IciA] contacts the α-subunit of RNAP in the process of recruiting the latter to different promoters [Lee and Hwang 1997].) Whereas the open complex established by RNAP through the mediation of Arg (or CAN)-bound ArgP is proficient for productive transcription, that mediated by Lys-bound ArgP is trapped in a paused and inactive state. The latter, however, can be chased into the productive state by competitive replacement of the Lys ligand, which is bound to ArgP by Arg. That this model is valid for the situation in vivo is suggested by the evidence obtained for ArgP-dependent open complex formation at the argO promoter in cells grown in Lys-supplemented medium (Fig. 4D; Supplementary Fig. S7), despite the absence of productive argO transcription under these conditions (Fig. 1B).
Environmental regulation at the final step of promoter escape
The argO case is thus the first known instance of an environmental signal regulating transcription initiation at the final stage of promoter escape (i.e., after open complex formation) by RNAP. In related examples, Smith and Sauer (1996) have described enhancement of promoter clearance by a phage-encoded regulator protein that, however, is not ostensibly modulated by any environmental signal. The H-NS protein mediates RNAP trapping following open complex formation at promoters such as rrnB P1 (Schroder and Wagner 2000; Dame et al. 2002) or hdeAB (Shin et al. 2005), but without any apparent environmental regulation thereof. On the other hand, Tagami and Aiba (1998, 1999) have shown that RNAP can form either nonproductive or productive binary open complexes at several promoters depending on the absence or presence of cAMP-CRP (whose in vivo concentration is determined by environmental cues), but they have argued that the CRP–RNAP interaction is needed only transiently prior to open complex formation and that CRP has no role in promoter clearance. Likewise, GalR-mediated repression of the gal operon P1 promoter (in the absence of galactose) is postulated to occur by a mechanism involving trapped RNAP in a complex that is an intermediate between the closed and the open complexes (Choy et al. 1995).
When one considers RNAP holoenzyme bearing σ subunits other than σ70, activation of σ54-RNAP transcription during nitrogen limitation is achieved by increased rates of conversion of closed to open complexes at cognate promoters (Buck et al. 2000), and Lee and Gralla (2004) have proposed that osmoresponsivity of osmY transcription may be explained by the stimulatory effect of potassium glutamate (which is believed to accumulate as a cytoplasmic osmolyte in cells grown at high osmolarity) on the escape of σS-RNAP from the osmY promoter, but this conclusion was based on findings only from in vitro studies. Finally, environmental regulation at the level of promoter escape has been described also in eukaryotic RNAP II transcription, although the mechanisms involved are different from that at argO (Saunders et al. 2006).
Mechanism of ArgP- and Lys-mediated RNAP trapping at argO promoter
The step after open complex formation at which RNAP is trapped at the argO promoter by Lys-bound ArgP is also different from other examples of RNAP trapping at bacterial promoters described earlier (for review, see Hsu 2002), even as a recent report suggests that productive transcription fails to occur from nearly one-quarter of all RNAP-bound promoters in vivo (Reppas et al. 2006). One mechanism of RNAP trapping previously characterized is that involving promoter-proximal pausing of the holoenzyme, which is mediated by the binding of σ70 to −10-like hexamer motifs present in the ITS region (Ko et al. 1998; Roberts et al. 1998; Marr et al. 2001; Brodolin et al. 2004; Nickels et al. 2004), and we have shown above that the Lys-induced transcriptional inactivation at argO does not fall into this category. All the other known examples of RNAP trapping (many of which are caused by increased affinity between promoter and RNAP) are associated with increased synthesis of abortive transcripts, and it has been shown in several cases that the GreA/GreB factors are able to revert the paused complex to the productive form (Ellinger et al. 1994; Hsu et al. 1995, 2006; Monsalve et al. 1997; Schroder and Wagner 2000; Sen et al. 2001; Hsu 2002; Liu et al. 2004; Kulbachinskiy and Mustaev 2006; Susa et al. 2006).
On the other hand, no abortive products were observed for the argO promoter in the presence of ArgP and Lys, and the GreB factor was not able to restore productive transcription. We suggest that, under these conditions, RNAP exists in a dormant Rip Van Winkle state after melting of the promoter duplex at the −10 region, at a step that precedes, and is common to, both abortive and productive transcription. For example, deletion of the tip of the σ70 region 3.2 loop has been shown to result in defective promoter clearance of the cognate holoenzyme associated with reduced synthesis of abortive transcripts (Kulbachinskiy and Mustaev 2006), and one possibility is that Lys-bound ArgP triggers a similar, but reversible, conformational change in wild-type RNAP in the open complex at argO as that observed in binary open complexes with the mutant holoenzyme. Since the role of the σ70 3.2 loop region in transcription initiation appears to be to reduce the Km for binding of the second templated nucleotide to the i + 1 site of RNAP (Kulbachinskiy and Mustaev 2006), this model may also serve to explain our findings (Fig. 4B,C) that the Lys-associated ternary initiation complexes assembled on argO are more heparin resistant in the presence of NTPs than in their absence.
A second model, not mutually exclusive, is that the RNAP complex at argO in the presence of ArgP and Lys is blocked in the step of DNA scrunching, which has recently been shown to be an obligatory prerequisite both for abortive initiation as well as for promoter clearance during productive transcription (Kapanidis et al. 2006; Revyakin et al. 2006). Additional mechanistic insights into this phenomenon may come from characterization of the different argPd mutants described earlier (Nandineni and Gowrishankar 2004), as well as by identification of mutations in argO and in the RNAP subunit genes that affect Arg- or Lys-mediated argO regulation.
Physiological relevance of the argO regulatory mechanism
Finally, what is the likely physiological importance of this mode of argO transcriptional regulation? The answer is at present unclear, but we had earlier proposed that the ArgO exporter performs two functions in E. coli—namely, that of exporting CAN (which is a plant-derived naturally occurring antimetabolite), and that of excreting Arg so as to maintain an optimal ratio of the intracellular Arg to Lys pools (rather than simply to keep the cytoplasmic Arg level below an arbitrary absolute value) (Nandineni and Gowrishankar 2004). Both functions are expected to be self-limited by feedback regulation, since the reduction in Arg or CAN concentrations consequent to their export through ArgO would in turn lead to the shutoff of argO transcription.
One could speculate that, with the particular mechanism of Lys-mediated regulation of argO transcription that has been identified in this study, the cells are poised to switch rapidly between transcription-activating and -inactivating complexes at the argO promoter depending upon instantaneous changes in the intracellular Arg to Lys ratios. If, in such a situation, the ArgO exporter were also to exhibit a rapid turnover, then the transcription switch would result in instantaneously tunable rates of Arg export out of the cells. Our finding that the Kd for Arg binding to ArgP is relatively high (∼150 μM) is consistent with the notion of avoidance of futile cycling of Arg uptake and export across the cytoplasmic membrane, so that Arg export would occur only when the twin conditions of high intracellular Arg and low intracellular Lys concentrations are simultaneously satisfied.
Materials and methods
In vivo argO-lac expression studies
In vivo argO-lac expression was monitored with plasmids derived from the single-copy-number plasmid vector pMU575 bearing the promoter-less lacZ gene (Andrews et al. 1991), upstream of which the 402-bp argO fragment from −293 to +109 or its mutant derivatives had been cloned. Mutations in argO were generated as follows: (1) in the case of each of the base substitutions, by site-directed mutagenesis using the QuikChange kit (Stratagene) with oligonucleotide primer pairs incorporating the mutation(s); and (2) in the case of each of the deletions, by a plasmid-based PCR strategy as described (Rajkumari and Gowrishankar 2001), with a pair of outwardly reading primers designed to amplify the entire plasmid except for the region to be deleted, and with EcoRI sites incorporated in the primers to facilitate recircularization of the deletant plasmids.
The wild-type and argP mutant host strains used were MC4100 recA and GJ4892, respectively (Nandineni and Gowrishankar 2004). Wherever required, the latter strain was rendered argPd by introduction of plasmid pHYD926, which carries the argPd-S94L allele (Nandineni and Gowrishankar 2004). Measurements of β-galactosidase-specific activity were performed as described (Miller 1992), in cultures grown to mid-exponential phase in 0.2% glucose–minimal A medium supplemented with Arg or Lys at 1 mM.
Radioactive chemicals and proteins for transcription
14C-labeled amino acids and 32P-labeled nucleotides were obtained from BRIT or Amersham Biosciences. σ70-Bearing RNAP holoenzyme was purchased from USB Corporation, and GreB protein was a gift from Ranjan Sen (Centre for DNA Fingerprinting and Diagnostics, Hyderabad, India).
To prepare the ArgP protein, a derivative of the plasmid vector pET21b (EMD Biosciences) was constructed that carries the PCR-amplified argP+ fragment downstream from the phage T7 promoter, such that the encoded ArgP protein bears a C-terminal His6 tag provided by the vector DNA sequence. The resultant plasmid was transformed into a derivative of strain GJ1158 (Bhandari and Gowrishankar 1997) that expresses phage T7 RNAP from the salt-inducible proU promoter. His6-ArgP overproduction was achieved by incubation for 3 h following addition of 0.3 M NaCl and 1 mM isopropyl thio-β-D-galactoside to a 500-mL volume of culture of the transformant grown in low-osmolarity medium, as described (Bhandari and Gowrishankar 1997). The protein was then purified from the cell lysate by chromatography through a Ni-NTA column (Qiagen), using buffer with 250 mM imidazole for elution of the bound protein (Supplementary Fig. S2). The protein preparation was concentrated to ∼1 mg/mL and stored at −20°C in buffer containing 20 mM Tris-Cl (pH 8), 150 mM NaCl, 10 mM dithiothreitol, and 20% glycerol.
argO DNA fragments
The argO DNA regions that were used in the in vitro experiments included the following: (1) a 427-bp fragment comprising the argO sequence from −293 to +109 obtained by PCR with the forward and reverse argO primers 5′-GTGCGCCTGCAGGA ACTTGGTG-3′ and 5′-GTATGCCCGGATCCATCACAAAA-3′, respectively (residues in italics represent base substitutions in native argO sequence, and those in bold in the two primers correspond, respectively, to positions −293 and +109 of argO); (2) deletion and substitution variants of the 427-bp argO fragment obtained by PCR using the forward and reverse argO primers above on plasmid templates carrying the corresponding mutations; (3) a 234-bp fragment comprising the argO region from −115 to +109 obtained by PCR using the reverse primer as above along with a 26-base primer whose sequence matches that on the top strand of argO from −115 to −90; (4) a 94-bp fragment comprising the argO sequence from −89 to +5 obtained by PCR using a pair of primers corresponding to the sequences from −89 to −64 of the top strand and from +5 to −22 of the bottom strand of argO; (5) a 45-bp fragment comprising the argO region from −89 to −45 obtained following digestion of the 94-bp fragment above with SspI (which cuts argO at −45); and (6) other fragments such as those from −115 to −90, −102 to −77, −89 to −64, −89 to −30, and −85 to −20, each of which was obtained by annealing the corresponding top- and bottom-strand synthetic oligonucleotide sequences. All but the 45-bp fragment were 5′-end-labeled, as required, with γ-32P-ATP and polynucleotide kinase. For labeling of the 45-bp fragment from −89 to −45, SspI digestion was done after PCR amplification of the 94-bp fragment with the 5′-end-labeled top-strand primer.
EMSA
EMSA reactions were performed each in 20 μL of EMSA binding buffer (10 mM Tris-Cl at pH 7.5, 1 mM EDTA, 50 mM MgCl2, 1 mM dithiothreitol, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride) containing (1) 5′-end-labeled argO DNA fragment at 0.5 nM, (2) 1 μg each of bovine serum albumin and poly(dI)· poly(dC), (3) ArgP at indicated monomer concentrations, and (4) when required, coeffectors at 0.1 mM. After incubation for 30 min at room temperature, the complexes were resolved by electrophoresis on a nondenaturing 4% polyacrylamide gel (39:1 acrylamide:bisacrylamide). In the reactions involving both ArgP and RNAP, the labeled argO fragment was first incubated with ArgP (at 20 nM) and indicated coeffector for 5 min at room temperature followed by further incubations with RNAP (at 80 nM) for 30 min before being analyzed by gel electrophoresis. In heparin challenge experiments (in the presence or absence of NTPs at 0.5 mM, as indicated), argO–ArgP–RNAP complexes obtained as described above were treated with heparin at 200 μg/mL and incubations continued for varying times at room temperature prior to gel electrophoresis.
In vitro transcription
Unless otherwise indicated, single-round in vitro transcription reactions were carried out in transcription buffer (40 mM Tris-glutamate at pH 8, 0.1 mM bovine serum albumin, 1 mM dithiothreitol) as previously described (Rajkumari et al. 1996), with some modifications. Typically, to a 5-μL volume of mix containing the linear DNA template (10 nM), ArgP (20 nM), σ70-bearing RNAP holoenzyme (40 nM), and coeffector(s) where indicated (0.1 mM) that had been preincubated at 37°C for 10 min was added an equal volume of labeled substrate–heparin mix to give final concentrations of 500 μM each of UTP, GTP, and ATP, 50 μM CTP, 200 μg/mL heparin, and 10 μCi of 32P-α-CTP (>3000 Ci/mmol), and incubation was continued for 15 min. The reactions were stopped and the products were processed and analyzed as described (Rajkumari et al. 1996) on denaturing polyacrylamide gels (i.e., with 7 M urea).
For the Arg-chase experiments on immobilized template, the argO template fragment from −293 to +109 was prepared by PCR as described above, with the modification that the forward argO primer used was biotinylated at its 5′ end. After preincubation of template with RNAP (and other additives as appropriate) as above, labeled substrate–heparin mix was added together with streptavidin-coated magnetic beads (Promega Corp.) and the primary reaction mixtures were incubated for 15 min to allow for immobilization of the biotinylated template on the beads. Where indicated, the beads were then separated from the supernatant, washed with transcription buffer containing 0.1 mM Arg (or CAN, as appropriate), and then incubated in the chase step with freshly labeled substrate–heparin mix supplemented with 0.1 mM Arg or CAN for 15 min. Each of the various preparations (i.e., the primary reaction mix before separation of beads from supernatant, the separated supernatant, and the mix obtained following chase reaction on the pellet) was analyzed for labeled transcription products by denaturing gel electrophoresis as described above.
Exonuclease III protection and footprinting assays
For the exonuclease III protection assay, the 5′-end-labeled 427-bp PCR-amplified argO fragment (comprising the sequence from −293 to +109) was digested with PstI (whose recognition site is present in the forward primer used in the PCR) to obtain an argO sequence uniquely labeled at the end corresponding to +109 (i.e., the 5′ end of the bottom strand); likewise, the 5′-end-labeled 234-bp argO fragment (with sequence from −115 to +109) was digested with BamHI (whose recognition site is present in the argO reverse primer used in PCR) to obtain a fragment uniquely labeled at the end corresponding to −115 (i.e., the 5′ end of the top strand). Exonuclease III (50 U) was added to a 10-μL volume of transcription buffer containing either of the labeled DNA fragments (3 nM), 20 nM ArgP (monomer), and, where indicated, 80 nM RNAP, NTPs at 0.5 mM each, and Arg or Lys at 0.1 mM. After incubation for 20 min at room temperature, the DNA was phenol-extracted and analyzed by electrophoresis on a 10% polyacrylamide gel with 7 M urea, along with argO di-deoxy sequence ladders generated with the aid of the argO forward or reverse primers, as appropriate.
For DNase I footprinting, the 427-bp and 234-bp argO fragments uniquely labeled at the 5′ ends of the bottom strand and top strand, respectively, as described above, were each dissolved at 0.6 nM in 50 μL of EMSA binding buffer and, where indicated, incubated with ArgP at 20 nM (monomer) in the presence or absence of Arg or Lys at 0.1 mM. After 30 min at room temperature, each mixture was treated with 0.2 U of DNase I for 60 sec prior to addition of 50 μL of stop solution (50 mM EDTA at pH 8, 2% sodium dodecyl sulfate). The DNA was then phenol-extracted and electrophoresed on a 10% denaturing polyacylamide gel along with argO sequence ladders generated as described above.
For in vivo KMnO4 footprinting, derivatives of the argP+ and argP mutant strains carrying a multicopy plasmid bearing the argO fragment from −293 to +109 were grown to mid-exponential phase in 0.2% glucose–minimal A medium (supplemented with 1 mM Arg or Lys, where indicated), and treated with KMnO4 at 5 mM for 4 min. Where required, rifampicin was added to 100 μg/mL to the culture prior to the KMnO4 treatment. Plasmid DNA prepared from each culture was first digested with HaeIII (which cuts argO at −80) and then used as template for 45 cycles of primer extension with AccuTaq DNA polymerase (Sigma-Aldrich) in a thermal cycler, using the 5′-end-labeled argO reverse primer described above. The extension products were detected by phosphorimaging following electrophoresis on a 10% denaturing polyacrylamide gel alongside an argO sequence ladder obtained as described above, and their image intensities were normalized (for each lane) to that of the extension product corresponding to the HaeIII site in the template.
Coeffector binding studies
In a 50-μL volume of running buffer containing 20 mM Tris-Cl (pH 8) and 100 mM NaCl, ArgP protein at 10 μM (monomer) was incubated with 14C-Arg or 14C-Lys (250 mCi/mmol) at 0.1 mM for 30 min at 4°C before the mixture was loaded on a 4-mL Sephadex G-25 gravity column (Amersham Biosciences) and subjected to gel filtration chromatography in running buffer. In each of the 0.2-mL fractions that were collected, protein was quantitated by fluorimetry (with excitation and emission wavelengths set at 295 nm and 330 nm, respectively) and radioactivity by liquid scintillation counting. In the competition experiments, ArgP was first incubated with 10 mM of the indicated cold competitor amino acid prior to addition of 14C-Arg or 14C-Lys.
Other methods
Bacterial genetic procedures were as described by Miller (1992), and those involving recombinant DNA and gel electrophoreses were as described by Sambrook et al. (1989). Band intensities in gel autoradiographs were determined by densitometry (after correction for background), with the aid of the Amersham Biosciences Image Quant system.
Acknowledgments
We thank Ranjan Sen for the gifts of GreB protein and template with phage T7 A1 promoter, advice on transcription and footprinting assays, and discussions. This work was supported by a CSIR Research Fellowship to R.S.L. and a research grant (BT/PR 4924/MED/12/191/2004) to J.G. from the Department of Biotechnology, Government of India. J.G. is a recipient of the J.C. Bose National Fellowship.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1520507
References
- Aleshin V.V., Zakataeva N.P., Livshits V.A., Zakataeva N.P., Livshits V.A., Livshits V.A. A new family of amino-acid-efflux proteins. Trends Biochem. Sci. 1999;24:133–135. doi: 10.1016/s0968-0004(99)01367-5. [DOI] [PubMed] [Google Scholar]
- Andrews A.E., Lawley B., Pittard A.J., Lawley B., Pittard A.J., Pittard A.J. Mutational analysis of repression and activation of the tyrP gene in Escherichia coli. J. Bacteriol. 1991;173:5068–5078. doi: 10.1128/jb.173.16.5068-5078.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azam T.A., Ishihama A., Ishihama A. Twelve species of the nucleoid-associated protein from Escherichia coli. Sequence recognition specificity and DNA binding affinity. J. Biol. Chem. 1999;274:33105–33113. doi: 10.1074/jbc.274.46.33105. [DOI] [PubMed] [Google Scholar]
- Azam T.A., Iwata A., Nishimura A., Ueda S., Ishihama A., Iwata A., Nishimura A., Ueda S., Ishihama A., Nishimura A., Ueda S., Ishihama A., Ueda S., Ishihama A., Ishihama A. Growth phase-dependent variation in protein composition of the Escherichia coli nucleotid. J. Bacteriol. 1999;181:6361–6370. doi: 10.1128/jb.181.20.6361-6370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellmann A., Vrljić M., Patek M., Sahm H., Krämer R., Eggeling L., Vrljić M., Patek M., Sahm H., Krämer R., Eggeling L., Patek M., Sahm H., Krämer R., Eggeling L., Sahm H., Krämer R., Eggeling L., Krämer R., Eggeling L., Eggeling L. Expression control and specificity of the basic amino acid exporter LysE of Corynebacterium glutamicum. Microbiol. 2001;147:1765–1774. doi: 10.1099/00221287-147-7-1765. [DOI] [PubMed] [Google Scholar]
- Bhandari P., Gowrishankar J., Gowrishankar J. An Escherichia coli host strain useful for efficient overproduction of cloned gene products with NaCl as the inducer. J. Bacteriol. 1997;179:4403–4406. doi: 10.1128/jb.179.13.4403-4406.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodolin K., Zenkin N., Mustaev A., Mamaeva D., Heumann H., Zenkin N., Mustaev A., Mamaeva D., Heumann H., Mustaev A., Mamaeva D., Heumann H., Mamaeva D., Heumann H., Heumann H. The σ70 subunit of RNA polymerase induces lacUV5 promoter-proximal pausing of transcription. Nat. Struct. Mol. Biol. 2004;11:551–557. doi: 10.1038/nsmb768. [DOI] [PubMed] [Google Scholar]
- Broome-Smith J.K., Baumberg S., Stirling C.J., Ward F.B., Baumberg S., Stirling C.J., Ward F.B., Stirling C.J., Ward F.B., Ward F.B. Transport of molecules across microbial membranes, Society for General Microbiology. Symposium, No. 58. Cambridge University Press; New York: 1999. [Google Scholar]
- Browning D.F., Busby S.W.J., Busby S.W.J. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2004;2:1–9. doi: 10.1038/nrmicro787. [DOI] [PubMed] [Google Scholar]
- Bucarey S.A., Villagra N.A., Fuentes J.A., Mora G.C., Villagra N.A., Fuentes J.A., Mora G.C., Fuentes J.A., Mora G.C., Mora G.C. The cotranscribed Salmonella enterica sv. Typhi tsx and impX genes encode opposing nucleoside-specific import and export proteins. Genetics. 2006;173:25–34. doi: 10.1534/genetics.105.054700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck M., Gallegos M.-T., Studholme D.J., Guo Y., Gralla J.D., Gallegos M.-T., Studholme D.J., Guo Y., Gralla J.D., Studholme D.J., Guo Y., Gralla J.D., Guo Y., Gralla J.D., Gralla J.D. The bacterial enhancer-dependent σ54 (σN) transcription factor. J. Bacteriol. 2000;182:4129–4136. doi: 10.1128/jb.182.15.4129-4136.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkovski A., Krämer R., Krämer R. Bacterial amino acid transport proteins: Occurrence, functions, and significance for biotechnological applications. Appl. Microbiol. Biotechnol. 2002;58:265–274. doi: 10.1007/s00253-001-0869-4. [DOI] [PubMed] [Google Scholar]
- Celis R.T.F. Repression and activation of arginine transport genes in Escherichia coli K 12 by the ArgP protein. J. Mol. Biol. 1999;294:1087–1095. doi: 10.1006/jmbi.1999.3308. [DOI] [PubMed] [Google Scholar]
- Choy H.E., Park S.W., Aki T., Parrack P., Fujita N., Ishihama A., Adhya S., Park S.W., Aki T., Parrack P., Fujita N., Ishihama A., Adhya S., Aki T., Parrack P., Fujita N., Ishihama A., Adhya S., Parrack P., Fujita N., Ishihama A., Adhya S., Fujita N., Ishihama A., Adhya S., Ishihama A., Adhya S., Adhya S. Repression and activation of transcription by Gal and Lac repressors: Involvement of α subunit of RNA polymerase. EMBO J. 1995;14:4523–4529. doi: 10.1002/j.1460-2075.1995.tb00131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dame R.T., Wyman C., Wurm R., Wagner R., Goosen N., Wyman C., Wurm R., Wagner R., Goosen N., Wurm R., Wagner R., Goosen N., Wagner R., Goosen N., Goosen N. Structural basis for H-NS-mediated trapping of RNA polymerase in the open initiation complex at the rrnB P1. J. Biol. Chem. 2002;277:2146–2150. doi: 10.1074/jbc.C100603200. [DOI] [PubMed] [Google Scholar]
- Eggeling L., Sahm H., Sahm H. New ubiquitous translocators: Amino acid export by Corynebacterium glutamicum and Escherichia coli. Arch. Microbiol. 2003;180:155–160. doi: 10.1007/s00203-003-0581-0. [DOI] [PubMed] [Google Scholar]
- Ellinger T., Behnke D., Bujard H., Gralla J.D., Behnke D., Bujard H., Gralla J.D., Bujard H., Gralla J.D., Gralla J.D. Stalling of Escherichia coli RNA polymerase in the +6 to +12 region in vivo is associated with tight binding to consensus promoter elements. J. Mol. Biol. 1994;239:455–465. doi: 10.1006/jmbi.1994.1388. [DOI] [PubMed] [Google Scholar]
- Han J.S., Kwon H.S., Yim J.-B., Hwang D.S., Kwon H.S., Yim J.-B., Hwang D.S., Yim J.-B., Hwang D.S., Hwang D.S. Effect of IciA protein on the expression of the nrd gene encoding ribonucleoside diphosphate reductase in E. coli. Mol. Gen. Genet. 1998;259:610–614. doi: 10.1007/s004380050854. [DOI] [PubMed] [Google Scholar]
- Hsu L.M. Promoter clearance and escape in prokaryotes. Biochim. Biophys. Acta. 2002;1577:191–207. doi: 10.1016/s0167-4781(02)00452-9. [DOI] [PubMed] [Google Scholar]
- Hsu L.M., Vo N.V., Chamberlin M.J., Vo N.V., Chamberlin M.J., Chamberlin M.J. Escherichia coli transcript cleavage factors GreA and GreB stimulate promoter escape and gene expression in vivo and in vitro. Proc. Natl. Acad. Sci. 1995;92:11588–11592. doi: 10.1073/pnas.92.25.11588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu L.M., Cobb I.M., Ozmore J.R., Khoo M., Nahm G., Xia L., Bao Y., Ahn C., Cobb I.M., Ozmore J.R., Khoo M., Nahm G., Xia L., Bao Y., Ahn C., Ozmore J.R., Khoo M., Nahm G., Xia L., Bao Y., Ahn C., Khoo M., Nahm G., Xia L., Bao Y., Ahn C., Nahm G., Xia L., Bao Y., Ahn C., Xia L., Bao Y., Ahn C., Bao Y., Ahn C., Ahn C. Initial transcribed sequence mutations specifically affect promoter escape properties. Biochemistry. 2006;45:8841–8854. doi: 10.1021/bi060247u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang D.S., Kornberg A., Kornberg A. A novel protein binds a key origin sequence to block replication of an E. coli minichromosome. Cell. 1990;63:325–331. doi: 10.1016/0092-8674(90)90165-b. [DOI] [PubMed] [Google Scholar]
- Hwang D.S., Kornberg A., Kornberg A. Opposed actions of regulatory proteins, DnaA and IciA, in opening the replication origin of Escherichia coli. J. Biol. Chem. 1992;267:23087–23091. [PubMed] [Google Scholar]
- Hwang D.S., Thony B., Kornberg A., Thony B., Kornberg A., Kornberg A. IciA protein, a specific inhibitor of initiation of Escherichia coli chromosomal replication. J. Biol. Chem. 1992;267:2209–2213. [PubMed] [Google Scholar]
- Kapanidis A.N., Margeat E., Ho S.O., Kortkhonjia E., Weiss S., Ebright R.H., Margeat E., Ho S.O., Kortkhonjia E., Weiss S., Ebright R.H., Ho S.O., Kortkhonjia E., Weiss S., Ebright R.H., Kortkhonjia E., Weiss S., Ebright R.H., Weiss S., Ebright R.H., Ebright R.H. Initial transcription by RNA polymerase proceeds through a DNA-scrunching mechanism. Science. 2006;314:1144–1147. doi: 10.1126/science.1131399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennerknecht N., Sahm H., Yen M.R., Patek M., Saier M.H., Jr., Eggeling L., Sahm H., Yen M.R., Patek M., Saier M.H., Jr., Eggeling L., Yen M.R., Patek M., Saier M.H., Jr., Eggeling L., Patek M., Saier M.H., Jr., Eggeling L., Saier M.H., Jr., Eggeling L., Eggeling L. Export of L-isoleucine from Corynebacterium glutamicum: A two-gene-encoded member of a new translocator family. J. Bacteriol. 2002;184:3947–3956. doi: 10.1128/JB.184.14.3947-3956.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko D.C., Marr M.T., Guo J., Roberts J.W., Marr M.T., Guo J., Roberts J.W., Guo J., Roberts J.W., Roberts J.W. A surface of Escherichia coli σ70 required for promoter function and antitermination by phage λ Q protein. Genes & Dev. 1998;12:3276–3285. doi: 10.1101/gad.12.20.3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubori T., Shimamoto N., Shimamoto N. A branched pathway in the early stage of transcription by Escherichia coli RNA polymerase. J. Mol. Biol. 1996;256:449–457. doi: 10.1006/jmbi.1996.0100. [DOI] [PubMed] [Google Scholar]
- Kubori T., Shimamoto N., Shimamoto N. Physical interference between Escherichia coli RNA polymerase molecules transcribing in tandem enhances abortive synthesis and misincorporation. Nucleic Acids Res. 1997;25:2640–2647. doi: 10.1093/nar/25.13.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulbachinskiy A., Mustaev A., Mustaev A. Region 3.2 of the σ subunit contributes to the binding of the 3′-initiating nucleotide in the RNA polymerase active center and facilitates promoter clearance during initiation. J. Biol. Chem. 2006;281:18273–18276. doi: 10.1074/jbc.C600060200. [DOI] [PubMed] [Google Scholar]
- Kutukova E.A., Livshits V.A., Altman I.P., Ptitsyn L.R., Zyiatdinov M.H., Tokmakova I.L., Zakataeva N.P., Livshits V.A., Altman I.P., Ptitsyn L.R., Zyiatdinov M.H., Tokmakova I.L., Zakataeva N.P., Altman I.P., Ptitsyn L.R., Zyiatdinov M.H., Tokmakova I.L., Zakataeva N.P., Ptitsyn L.R., Zyiatdinov M.H., Tokmakova I.L., Zakataeva N.P., Zyiatdinov M.H., Tokmakova I.L., Zakataeva N.P., Tokmakova I.L., Zakataeva N.P., Zakataeva N.P. The yeaS (leuE) gene of Escherichia coli encodes an exporter of leucine, and the Lrp protein regulates its expression. FEBS Lett. 2005;579:4629–4634. doi: 10.1016/j.febslet.2005.07.031. [DOI] [PubMed] [Google Scholar]
- Lee S.J., Gralla J.D., Gralla J.D. Osmo-regulation of bacterial transcription via poised RNA polymerase. Mol. Cell. 2004;14:153–162. doi: 10.1016/s1097-2765(04)00202-3. [DOI] [PubMed] [Google Scholar]
- Lee Y.S., Hwang D.S., Hwang D.S. Occlusion of RNA polymerase by oligomerization of DnaA protein over the dnaA promoter of Escherichia coli. J. Biol. Chem. 1997;272:83–88. [PubMed] [Google Scholar]
- Lee Y.S., Kim H., Hwang D.S., Kim H., Hwang D.S., Hwang D.S. Transcriptional activation of the dnaA gene encoding the initiator for oriC replication by IciA protein, an inhibitor of in vitro oriC replication in Escherichia coli. Mol. Microbiol. 1996;19:389–396. doi: 10.1046/j.1365-2958.1996.485902.x. [DOI] [PubMed] [Google Scholar]
- Lee Y., Lee H., Yim J., Hwang D., Lee H., Yim J., Hwang D., Yim J., Hwang D., Hwang D. The binding of two dimers of IciA protein to the dnaA promoter 1P element enhances the binding of RNA polymerase to the dnaA promoter 1P. Nucleic Acids Res. 1997;25:3486–3489. doi: 10.1093/nar/25.17.3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Garges S., Adhya S., Garges S., Adhya S., Adhya S. lacP1 promoter with an extended −10 motif. Pleiotropic effects of cyclic AMP protein at different steps of transcription initiation. J. Biol. Chem. 2004;279:54552–54557. doi: 10.1074/jbc.M408609200. [DOI] [PubMed] [Google Scholar]
- Lolkema J.S., Poolman B., Konings W.N., Poolman B., Konings W.N., Konings W.N. Bacterial solute uptake and efflux systems. Curr. Opin. Microbiol. 1998;1:248–253. doi: 10.1016/s1369-5274(98)80018-0. [DOI] [PubMed] [Google Scholar]
- Marr M.T., Datwyler S.A., Meares C.F., Roberts J.W., Datwyler S.A., Meares C.F., Roberts J.W., Meares C.F., Roberts J.W., Roberts J.W. Restructuring of an RNA polymerase holoenzyme elongation complex by lambdoid phage Q proteins. Proc. Natl. Acad. Sci. 2001;98:8972–8978. doi: 10.1073/pnas.161253298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.H. A short course in bacterial genetics: A laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1992. [Google Scholar]
- Monsalve M., Calles B., Mencía M., Salas M., Rojo F., Calles B., Mencía M., Salas M., Rojo F., Mencía M., Salas M., Rojo F., Salas M., Rojo F., Rojo F. Transcription activation or repression by phage Φ29 protein p4 depends on the strength of the RNA polymerase–promoter interactions. Mol. Cell. 1997;1:99–107. doi: 10.1016/s1097-2765(00)80011-8. [DOI] [PubMed] [Google Scholar]
- Nandineni M.R., Gowrishankar J., Gowrishankar J. Evidence for an arginine exporter encoded by yggA (argO) that is regulated by the LysR-type transcriptional regulator ArgP of Escherichia coli. J. Bacteriol. 2004;186:3539–3546. doi: 10.1128/JB.186.11.3539-3546.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandineni M.R., Laishram R.S., Gowrishankar J., Laishram R.S., Gowrishankar J., Gowrishankar J. Osmosensitivity associated with insertions in argP (iciA) or glnE in glutamate synthase (GOGAT)-deficient mutants of Escherichia coli. J. Bacteriol. 2004;186:6391–6399. doi: 10.1128/JB.186.19.6391-6399.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickels B.E., Mukhopadhyay J., Garrity S.J., Ebright R.H., Hochschild A., Mukhopadhyay J., Garrity S.J., Ebright R.H., Hochschild A., Garrity S.J., Ebright R.H., Hochschild A., Ebright R.H., Hochschild A., Hochschild A. The σ70 subunit of RNA polymerase mediates a promoter-proximal pause at the lac promoter. Nat. Struct. Mol. Biol. 2004;11:544–550. doi: 10.1038/nsmb757. [DOI] [PubMed] [Google Scholar]
- Ptashne M., Gann A., Gann A. Transcription activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- Rajkumari K., Gowrishankar J., Gowrishankar J. In vivo expression from the RpoS-dependent P1 promoter of the osmotically regulated proU operon in Escherichia coli and Salmonella enterica serovar Typhimurium: Activation by rho and hns mutations and by cold stress. J. Bacteriol. 2001;183:6543–6550. doi: 10.1128/JB.183.22.6543-6550.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkumari K., Kusano S., Ishihama A., Mizuno T., Gowrishankar J., Kusano S., Ishihama A., Mizuno T., Gowrishankar J., Ishihama A., Mizuno T., Gowrishankar J., Mizuno T., Gowrishankar J., Gowrishankar J. Effects of H-NS and potassium glutamate on σS- and σ70-directed transcription in vitro from osmotically regulated P1 and P2 promoters of proU in Escherichia coli. J. Bacteriol. 1996;178:4176–4181. doi: 10.1128/jb.178.14.4176-4181.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reppas N.B., Wade J.T., Church G.M., Struhl K., Wade J.T., Church G.M., Struhl K., Church G.M., Struhl K., Struhl K. The transition between transcriptional initiation and elongation in E. coli is highly variable and often rate limiting. Mol. Cell. 2006;24:747–757. doi: 10.1016/j.molcel.2006.10.030. [DOI] [PubMed] [Google Scholar]
- Revyakin A., Liu C., Ebright R.H., Strick T.R., Liu C., Ebright R.H., Strick T.R., Ebright R.H., Strick T.R., Strick T.R. Abortive initiation and productive initiation by RNA polymerase involve DNA scrunching. Science. 2006;314:1139–1143. doi: 10.1126/science.1131398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts J.W., Yarnell W., Bartlett E., Guo J., Marr M., Ko D.C., Sun H., Roberts C.W., Yarnell W., Bartlett E., Guo J., Marr M., Ko D.C., Sun H., Roberts C.W., Bartlett E., Guo J., Marr M., Ko D.C., Sun H., Roberts C.W., Guo J., Marr M., Ko D.C., Sun H., Roberts C.W., Marr M., Ko D.C., Sun H., Roberts C.W., Ko D.C., Sun H., Roberts C.W., Sun H., Roberts C.W., Roberts C.W. Antitermination of bacteriophage λ Q protein. Cold Spring Harb. Symp. Quant. Biol. 1998;63:319–325. doi: 10.1101/sqb.1998.63.319. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T., Fritsch E.F., Maniatis T., Maniatis T. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Saunders A., Core L.J., Lis J.T., Core L.J., Lis J.T., Lis J.T. Breaking barriers to transcription elongation. Nat. Rev. Mol. Cell Biol. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- Schell M.A. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 1993;47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- Schroder O., Wagner R., Wagner R. The bacterial DNA-binding protein H-NS represses ribosomal RNA transcription by trapping RNA polymerase in the initiation complex. J. Mol. Biol. 2000;298:737–748. doi: 10.1006/jmbi.2000.3708. [DOI] [PubMed] [Google Scholar]
- Sen R., Nagai H., Shimamoto N., Nagai H., Shimamoto N., Shimamoto N. Conformational switching of Escherichia coli RNA polymerase-promoter binary complex is facilitated by elongation factor GreA and GreB. Genes Cells. 2001;6:389–401. doi: 10.1046/j.1365-2443.2001.00436.x. [DOI] [PubMed] [Google Scholar]
- Shin M., Song M., Rhee J.H., Hong Y., Kim Y.J., Seok Y.J., Ha K.S., Jung S.H., Choy H.E., Song M., Rhee J.H., Hong Y., Kim Y.J., Seok Y.J., Ha K.S., Jung S.H., Choy H.E., Rhee J.H., Hong Y., Kim Y.J., Seok Y.J., Ha K.S., Jung S.H., Choy H.E., Hong Y., Kim Y.J., Seok Y.J., Ha K.S., Jung S.H., Choy H.E., Kim Y.J., Seok Y.J., Ha K.S., Jung S.H., Choy H.E., Seok Y.J., Ha K.S., Jung S.H., Choy H.E., Ha K.S., Jung S.H., Choy H.E., Jung S.H., Choy H.E., Choy H.E. DNA looping-mediated repression by histone-like protein H-NS: Specific requirement of E.σ70 as a cofactor for looping. Genes & Dev. 2005;19:2388–2398. doi: 10.1101/gad.1316305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simic P., Sahm H., Eggeling L., Sahm H., Eggeling L., Eggeling L. L-threonine export: Use of peptides to identify a new translocator from Corynebacterium glutamicum. J. Bacteriol. 2001;183:5317–5324. doi: 10.1128/JB.183.18.5317-5324.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T.L., Sauer R.T., Sauer R.T. Dual regulation of open-complex formation and promoter clearance by Arc explains a novel repressor to activator switch. Proc. Natl. Acad. Sci. 1996;93:8868–8872. doi: 10.1073/pnas.93.17.8868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straney S.B., Crothers D.M., Crothers D.M. Lac repressor is a transient gene-activating protein. Cell. 1987;51:699–707. doi: 10.1016/0092-8674(87)90093-6. [DOI] [PubMed] [Google Scholar]
- Susa M., Kubori T., Shimamoto N., Kubori T., Shimamoto N., Shimamoto N. A pathway branching in transcription initiation in Escherichia coli. Mol. Microbiol. 2006;59:1807–1817. doi: 10.1111/j.1365-2958.2006.05058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami H., Aiba H., Aiba H. A common role of CRP in transcription activation: CRP acts transiently to stimulate events leading to open complex formation at a diverse set of promoters. EMBO J. 1998;17:1759–1767. doi: 10.1093/emboj/17.6.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami H., Aiba H., Aiba H. An inactive open complex mediated by an UP element at Escherichia coli promoters. Proc. Natl. Acad. Sci. 1999;96:7202–7207. doi: 10.1073/pnas.96.13.7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thony B., Hwang D.S., Fradkin L., Kornberg A., Hwang D.S., Fradkin L., Kornberg A., Fradkin L., Kornberg A., Kornberg A. iciA, an Escherichia coli gene encoding a specific inhibitor of chromosomal initiation of replication in vitro. Proc. Natl. Acad. Sci. 1991;88:4066–4070. doi: 10.1073/pnas.88.10.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trötschel C., Deutenberg D., Bathe B., Burkovski A., Krämer R., Deutenberg D., Bathe B., Burkovski A., Krämer R., Bathe B., Burkovski A., Krämer R., Burkovski A., Krämer R., Krämer R. Characterization of methionine export in Corynebacterium glutamicum. J. Bacteriol. 2005;11:3786–3794. doi: 10.1128/JB.187.11.3786-3794.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrljic M., Sahm H., Eggeling L., Sahm H., Eggeling L., Eggeling L. A new type of transporter with a new type of cellular function: L-lysine export from Corynebacterium glutamicum. Mol. Microbiol. 1996;22:815–826. doi: 10.1046/j.1365-2958.1996.01527.x. [DOI] [PubMed] [Google Scholar]
- Wei T., Bernander R., Bernander R. Interaction of the IciA protein with AT-rich regions in plasmid replication origins. Nucleic Acids Res. 1996;24:1865–1872. doi: 10.1093/nar/24.10.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]