Abstract
Interdisciplinary work in the life sciences at the boundaries of biology, chemistry and physics is making enormous strides. This progress was showcased at the recent Single Molecule Biophysics conference.
Brilliant blue skies and powdery white slopes set the stage for the 4th biennial Single Molecule Biophysics (SMB) workshop, held February 4–9 at the Aspen Center for Physics in Aspen, Colorado. In addition to capturing 27 medals in a nationally handicapped (NASTAR) ski race, the 98 enthusiastic participants heard over 50 talks and viewed about 40 posters covering both theory and experiment from a balanced mix of graduate students, post-docs and professors. The extraordinary amount of recent (and in many cases unpublished) data presented at SMB 2007 testified to the explosive growth of this new field. This year’s workshop was the largest held so far, yet it managed to maintain an intimate atmosphere in which participants could easily gather to discuss their findings, both on and off the mountain. The SMB conference covers an emerging discipline in which the behaviors of individual macromolecules are directly measured with a variety of biophysical techniques, including optical trapping, atomic force microscopy, fluorescence microscopy and magnetic tweezers. These techniques capture the dynamics of life at the macromolecular scale, elucidating details that are typically lost to ensemble averaging or asynchrony when studied by traditional (‘bulk’) biochemistry. Single-molecule studies seem destined to grow increasingly popular with chemical biologists as the mechanical nature of macromolecular interactions, and their associated stochasticity, takes on a greater explanatory power in describing cellular action. Increasingly, biophysics and chemical biology share scientific goals and stand to gain from each other. Here, we report on a selection of findings reported at the SMB 2007 meeting. Space considerations preclude a balanced treatment of all topics, but these are intended to convey the flavor of the meeting—and the rich possibilities afforded by many of the new approaches.
The central dogma: a broader view
Francis Crick coined the phrase ‘central dogma of biology’ to describe the basic flow of molecular information from DNA to RNA to proteins. In addition to the complex molecular machines that carry out the fundamental reactions underlying gene expression, such as polymerases and ribosomes, a broader view of the central dogma includes the proteins that facilitate the manipulation of DNA, such as topoisomerases and helicases, as well as those that allow for the reverse flow of information, such as reverse transcriptases. Many of these molecules were the subject of new single-molecule studies presented in Aspen.
Gene expression requires modulation of DNA organization and compaction during the cell cycle. Gijs Wuite (Vrije Universiteit) used a four-trap optical tweezers instrument to study H-NS, a protein that compacts DNA by cross-bridging two strands1. The group subjected filaments to an unzipping force and saw a step-like increase in the distance between the traps, with the fundamental step size corresponding to one helical repeat along the DNA. When the two DNA strands were unzipped at 70 base pairs per second (a speed corresponding to the transcription rate of bacterial RNA polymerase), individual H-NS cross-bridges were disrupted by ~7 pN of force. The block associated with an H-NS cross-bridge should therefore not present an insuperable obstacle for RNA polymerase, which is capable of exerting at least 14 pN of force2.
During DNA replication in vivo, torsional strain is induced by unwinding of the DNA ahead of the moving replication complex. This growing strain can be relieved by bacterial topoisomerase IV (Topo IV), an essential ATP-dependent type II topoisomerase that removes positive supercoils by transporting one segment of double-stranded DNA through a transient double-stranded break in a second segment of double-stranded DNA. In vitro, Topo IV relaxes positive supercoils ~20-fold faster than it relaxes negative supercoils. Keir Neuman, in collaboration with Gilles Charvin, Vincent Croquette and David Bensimon (École Normale Supérieure, Paris), presented data collected using magnetic tweezers to study this asymmetric activity (Fig. 1). In experiments in which only a single crossing between strands of double-stranded DNA was relieved, Neuman demonstrated that Topo IV preferentially cuts and relaxes supercoils when the strands cross at an angle of 85°, thereby giving near-equal preference for relaxing positive and negative supercoils. In an assay in which several supercoils were relieved, Neuman showed that whereas a single Topo IV can relax positive supercoils processively, it must unbind and rebind in order to relax multiple negative supercoils, which accounts for the observed asymmetry in bulk.
Figure 1.
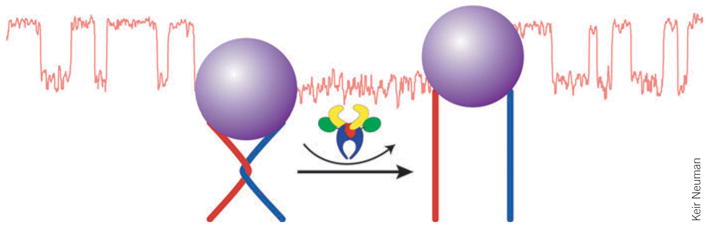
Topoisomerase IV relaxes supercoils in DNA. The cartoon inset shows a single crossing of two duplex DNA molecules (red and blue) that is removed by Topo IV (displayed above reaction arrows). This catalytic activity was monitored by Keir Neuman and co-workers using magnetic tweezers. The data record (red trace) shows the height of a magnetic bead (purple) as a function of time. When the DNA crossing is removed by Topo IV, the bead moves up; the crossing is then reformed by rotating the magnetic bead by a turn, restoring it to the lower position.
For more than a decade, single-molecule techniques have been successfully used to study transcription by prokaryotic RNA polymerase, in part owing to the relative simplicity with which transcription may be initiated in bacteria. To study transcription by eukaryotic RNA polymerase II (Pol II), Eric Galburt and Stephan Grill of Carlos Bustamante’s lab (University of California, Berkeley) finessed the comparative complexity of eukaryotic initiation, which generally requires an entire assortment of initiation factors, by directly assembling a functional transcriptional elongation complex on a preassembled nucleic acid scaffold (a method developed in Mikhail Kashlev’s lab at the National Cancer Institute, US National Institutes of Health). Using a passive, dual-trap optical-tweezers assay, Galburt and Grill observed transcription by single Pol II molecules against steadily increasing loads. Pol II molecules stopped and backtracked at approximately one third of the stall force determined for Escherichia coli RNA polymerase (Nature, in the press). This result is surprising, given that eukaryotic transcriptional elongation routinely proceeds through nucleosomes in vivo, which constitute a structural barrier that must be overcome. The addition of the elongation factor TFIIS, which rescues backtracked transcription complexes by cleaving the extruded RNA, doubled the apparent stall force. This accessory factor–dependent force sensitivity suggests that eukaryotic transcription may be regulated by components that modify the mechanical performance of the polymerase.
Yuhong Wang, working with Yale E. Goldman and Barry S. Cooperman (University of Pennsylvania), studied translation in single E. coli ribosomes using fluorescence resonance energy transfer (FRET) labels located on an incoming tRNA and on the ribosome itself. High levels of FRET occurred when the incoming tRNA was located in the A site, and FRET decreased on subsequent movement of the tRNA. These experiments, and others like them, open the door to probing the kinetics of protein synthesis at the single-ribosome level.
A protein essential to viral replication is the NS3 helicase of hepatitis C. Taekjip Ha and Sua Myong (University of Illinois) reported on experiments in which they directly probed the unwinding and translocation activity of NS3 using single-molecule FRET. They were able to observe discrete changes in FRET levels during unwinding that correspond to advancement of the enzyme in a series of discrete steps, and that are reminiscent of the substeps of 2–5 base pairs observed for this helicase using optical tweezers3. Furthermore, Ha and Myong presented evidence for the existence of even smaller hidden steps, and proposed a ‘slingshot’ model as an unwinding mechanism.
DNA recombination and repair
The eukaryotic genome typically incurs more than 10,000 chemical lesions in a single day, from comparatively benign deaminations to full-blown double-strand breaks. Typically, these lesions are repaired with near-perfect fidelity. One important pathway that is responsible for the repair of double-stranded breaks is homologous recombination. First, the double-stranded breaks are processed to produce 3′ single-stranded DNA overhangs, to which a RecA-like protein binds. Then, the RecA filaments mediate a search for a homologous site within the genome, eventually forming an intermediate known as a joint molecule. The RecA-like protein then catalyzes the exchange of strands to form a Holliday junction, which then moves by branch migration and is resolved to yield the repaired, recombinant products4.
Using magnetic tweezers, Cees Dekker’s group (Technical University of Delft) took the end-to-end distance of DNA strands as a metric of filament formation in the presence of either E. coli RecA or the human RecA-like protein Rad51. They reported that RecA cooperatively forms continuous presynaptic filaments, but that Rad51 binds along DNA less cooperatively, and rather discontinuously, in the form of large clusters of ~35 monomers. Further, strand invasion could also be monitored as a change in the end-to-end distance of supercoiled DNA. Evidence was presented for a triple-helix, joint-molecule intermediate, and strand invasion and filament assembly were monitored in real time following the addition of ATP. Surprisingly, strand invasion and filament disassembly compensate one another so that the displacement loop does not grow beyond 80 nucleotides.
Using a combination of dual-trap optical tweezers and fluorescence techniques, Piero Bianco (State University of New York, Buffalo) directly observed human Rad54, a double-stranded DNA translocase and accessory factor for Rad51, moving at extraordinary rates—up to 2.5 μm per second. Although the speed was highly variable, it did not seem to be sequence dependent. When two Rad54-coated DNA molecules were brought together, the cross-bridging of these structures could be directly observed. The translocase and cross-bridging activities of Rad54 combine to stimulate the synapsis stage of homologous recombination promoted by Rad51.
Although double-stranded breaks are among the most detrimental forms of DNA damage, mismatch mutations are also introduced roughly once every 107 nucleotides, simply as a result of the intrinsic error rate of DNA polymerase. MutS protein in E. coli and Msh2-Msh6 heterodimers in eukaryotes are responsible for the recognition of these mismatch sites. Jason Gorman from Eric Greene’s lab (Columbia University) used single-molecule total internal reflection fluorescence imaging to show that Msh2-Msh6 can diffuse along stretched DNA even in the absence of ATP. The measured diffusion coefficient was consistent with a molecule that both translates and rotates as it tracks along the helical groove of DNA, driven purely by thermal energy.
Nucleic acid folding
Several presentations at SMB 2007 focused on the kinetic and energetic properties of nucleic acid structures. The simplest and most ubiquitous element of secondary structure, the hairpin, was the subject of talks by Michael Woodside (now at National Research Council Canada and the University of Alberta) from Steven Block’s lab (Stanford University) and by Jeffrey Vieregg from Ignacio Tinoco’s lab (University of California, Berkeley). Woodside used dual-trap optical tweezers to study the folding and unfolding of many different types of single DNA hairpins under mechanical loads. He demonstrated that the height and location of the transition state with respect to the fully folded and unfolded states depend on hairpin sequence, and that they can be determined with high precision by making steady state measurements at loads poised near the unfolding force. He further showed that by measuring the extensions of hairpins at constant force for sufficiently long times, the complete energy landscapes for the folding transition can be recovered, and that these landscapes are in close agreement with a simple phenomenological model5. These results were complemented by those of Vieregg, who used a single-trap instrument in passive (open-loop) mode and exerted nonequilibrium forces to unfold RNA hairpins, including the HIV-1 TAR sequence. Vieregg showed that the free energy of unfolding can be obtained robustly by repeatedly extending and releasing the hairpin and applying Crooks’ fluctuation theorem (which relates nonequilibrium work to equilibrium free energy) to the measured work performed on the hairpin by the instrument. He also demonstrated that this free energy is sensitive to monovalent ion concentrations, and to bulges in the hairpin stem.
The analysis of single-molecule data for molecules undergoing transitions among states may soon be aided by the theoretical work of Olga Dudko’s group at the US National Institutes of Health. Extending earlier analyses6,7, Dudko presented a framework for considering force-dependent reactions, such as bond rupture in single-molecule pulling experiments, under regimes of constant force or constant pulling speed. For both regimes, Dudko et al. derived expressions for the force-dependent intrinsic rate coefficient and the height and location of the free energy barrier8. Remarkably, these expressions accounted for so-called Hammond behavior, a phenomenon long understood for reactions of organic molecules as a shift in the apparent location of the transition barrier upon chemical perturbation9, but never explicitly considered for reactions biased by mechanical load. Dudko demonstrated the utility of her framework for extracting the useful kinetic parameters of a reaction by applying it to the unzipping of single DNA hairpins moving through nanopores, obtaining good agreement between theory and experiment.
The structures of larger nucleic acids, such as ribozymes, are often more amenable to study using FRET. David Rueda (Wayne State University) introduced fluorophores into the U2–U6 spliceosomal RNA complex and demonstrated that folding is Mg2+-dependent and occurs through an obligatory intermediate. A common theme emerging from FRET-based studies of nucleic acids is the observation of population heterogeneity in both folding and unfolding rates. David Nesbitt (JILA and University of Colorado, Boulder) reported such heterogeneity in the real-time, reversible docking and undocking of the GAAA tetraloop10, a component of the Tetrahymena thermophila intron, and David Lilley (University of Dundee) presented data showing that the heterogeneity observed during the folding of the adenine riboswitch is significantly reduced in the presence of adenine11.
Advances in single-molecule methods
Single-molecule methods are now probing the biological world at ever greater levels of detail. Much of the recent work has focused on improving the resolution limit of fluorescence microscopy to allow near-nanometer precision, on developing methods of manipulating freely diffusing single molecules and measuring their interactions, and on improving the precision of optical trapping techniques to the subnanometer scale. Xiaowei Zhuang (Harvard University) presented a technique for achieving subdiffraction-limited resolution in images based on stochastic optical reconstruction microscopy (STORM)12. In this method, multiple photoswitchable fluorescent dye molecules are attached to biological samples of interest. A fraction of the dye tags can then be switched on at random, resolved separately from the remainder using centroid analysis, and then switched off. By sequentially turning on small random subsets of the fluorescent molecules, an overall imaging resolution below 20 nm can be achieved. Mark Greene reported on work at the US National Institute of Standards and Technology demonstrating successful optical trapping of small aqueous droplets suspended in immiscible hydrophobic solvent (“hydrosomes”)13 and the simultaneous measurement of freely diffusing green fluorescent protein molecules inside these. Hydrosomes function as convenient nano test tubes that can be readily produced by micropipets, manipulated by optical methods, and even fused to measure interactions among single molecules using fluorescence approaches such as FRET. Laser-based optical trapping technology continues to establish new records for spatial and temporal sensitivity, and in the past year or so, it has broken the ‘nanometer barrier’ to reach true atomic-level resolution. Tom Perkins (JILA, and University of Colorado, Boulder) reported a new method for stabilizing surface-tethered molecules in optical trapping systems to within one angstrom in all three dimensions using two independent optical measures of surface drift, thereby affording unprecedented positional resolution14. This and related optical technologies that can surpass nanometer resolution hold enormous promise in the years to come for biophysical measurements at the level of individual macromolecules.
F1Fo-type ATP synthase
The F1Fo-ATPase complex is a membrane-based, reversible rotary machine that provides electrochemical coupling of the transmembrane proton potential and ATP synthesis, thereby furnishing respiring cells with their ATP energy currency. The water-soluble portion of this motor, F1, can hydrolyze ATP to drive rotation of the γ ‘driveshaft’ subunit within its α3β3 subunits in 120° steps, each coinciding with the hydrolysis of a single ATP molecule15. However, the nature of the cooperativity between the three β subunits is not fully understood. Takayuki Ariga (Osaka University) presented experiments in which one, two or all three β subunits of F1 were mutated to slow catalysis in individual subunits. With one mutant β subunit, at least two of the three 120° steps were slowed significantly. This effect was linear as additional β subunits were mutated, which suggests that each subunit undertakes a full round of catalysis during one revolution of the motor, separated from the others by 120° phase lags.
A challenge to studies of this important molecular motor arises in understanding the interactions between the membrane-bound Fo component and the soluble F1 component. Michael Börsch (University of Stuttgart) presented evidence showing individual, membrane-bound F1Fo-ATP complexes rotating in opposite directions during either hydrolysis or synthesis of ATP. The result was deduced by labeling the rotary ε subunit of F1 and the static b subunits of Fo with fluorescent dyes and observing the resultant FRET signals16. A similar approach was used to monitor c-ring rotation in Fo. These single-molecule studies are the first of their kind on a membrane-bound complex, and should aid our understanding of the mechanical interplay between coupled motors in the full F1Fo-type ATP synthase.
Modulating the cytoskeleton with force
The growth and shrinkage of microtubules (produced by the polymerization and depolymerization of tubulin subunits) are likely regulated by force during cellular processes such as mitosis and chromosome segregation. Using a new in vitro assay, Charles Asbury (University of Washington) showed that the Dam1 ring-like complex found in yeast kinetochores is sufficient to couple the effect of external load to microtubule dynamics in the absence of other accessory factors. Exerting tension on the microtubule through the Dam1 complex slowed microtubule depolymerization and decreased the incidence of catastrophes but increased that of rescues. Marileen Dogterom (Institute for Atomic and Molecular Physics, Dutch Foundation for Fundamental Research on Matter) reported on a technique she developed for applying compressive forces to the ends of microtubules or actin bundles using multiple optical traps and microstructures engineered on a coverslip surface17. Addition of the microtubule-binding protein XMAP215 allowed microtubules to grow in steps of 60 nm—increments much larger than the size subtended by individual tubulin heterodimers, which suggests the possibility of a templating mechanism for assisted microtubule growth (Fig. 2). Furthermore, she found that microtubule bundles are capable of bearing loads in proportion to the number of microtubules within the bundle. In contrast, results on large actin bundles showed that these are unable to grow against loads much larger than those sufficient to stall an individual filament18. The results from these two laboratories constitute major breakthroughs in our understanding of how cytoskeletal filament length is regulated—a crucial topic for chemical biologists interested in developing novel agents that influence cell division.
Figure 2.
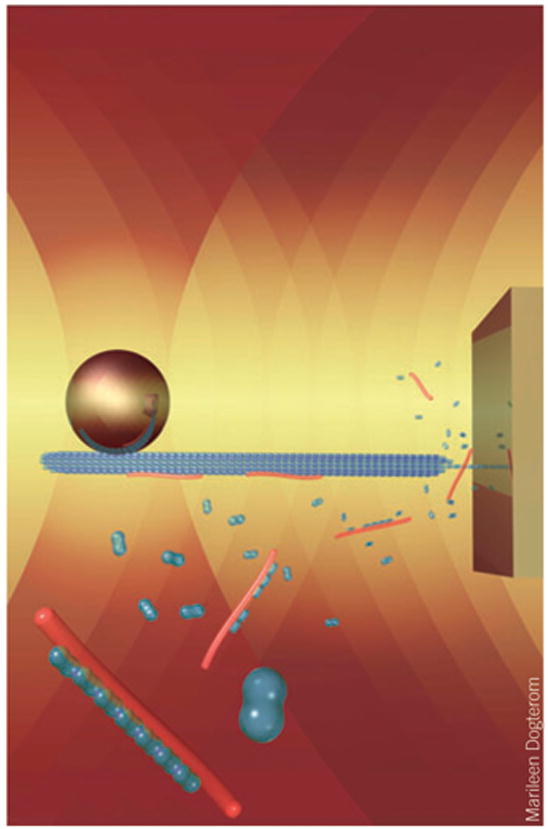
Cartoon showing the experimental geometry used by Dogterom and co-workers to apply compressive loads to a microtubule. A microtubule is attached to a bead held in an optical trap (left) while a time-shared optical trap orients the microtubule perpendicular to a microfabricated barrier (right). Microtubule growth through the addition of tubulin heterodimers (blue) is templated by XMAP215 proteins (red), according to their model.
Mechanisms for mechanochemical coupling in classical motors
Questions about the molecular mechanisms responsible for powering cellular transport motors such as kinesin, myosin and dynein have driven technical developments and analytical methods in single-molecule biophysics for over two decades. During this time, biophysicists have resolved the fundamental step sizes for many of these motor proteins, measured their stall forces, and teased apart some of the mechanochemistry responsible for converting ATP hydrolysis into mechanical motion. All this progress notwithstanding, issues regarding ‘classical motors’ abound, including the size and variability of the dynein step, the effects of various microtubule-associated proteins on motor processivity, and the origins of head cooperativity in kinesin. Arne Gennerich of Ron Vale’s lab (University of California, San Francisco) presented an in-depth biophysical characterization of yeast cytoplasmic dynein. The apparent step size was reported to be load dependent, with larger steps in integral multiples of the 8-nm tubulin repeat spacing taking place only at the lowest loads (<3 pN), whereas at high loads (~6 pN), 8-nm steps alone dominated. Near the stall force (>7 pN), dynein motors remained attached to the microtubule substrate for long periods of time. These unusual characteristics contrast strongly with the more extensively characterized microtubule motor kinesin, which invariably takes 8-nm steps and remains associated only briefly with the microtubule under forces near stall. The load-dependent behavior of dynein has been suggested to arise from separate conformations of the AAA ATPase domains of the motor. Ram Dixit in Erika Holzbaur’s lab (University of Pennsylvania) reported on the effects of microtubule-associated proteins τ23 and τ40 on the processivity of both kinesin and dynein. Dynein processivity seemed to be comparatively less sensitive than that of conventional kinesin, which became completely nonprocessive at high concentrations of τ, which implies a possible mechanism for the selective transport of dynein over kinesin along coated microtubules. Technically impressive results from Johnson Chung and Jeff Gelles (Brandeis University) answered a long-standing question of head coordination in kinesin. They found that fluorescent ADP produced by the hydrolysis of fluorescently labeled ATP nucleotides is not immediately released from the enzyme active site. Instead, the fluorescent ADP seemed to remain in the active site of one head until its partner head subsequently bound a new ATP molecule. Pre-steady state measurements at the ensemble level had previously implied the existence of such a ‘head-gating’ mechanism19, but these single-molecule experiments show that such a mechanism continues to coordinate the two heads of kinesin during steady state turnover.
Biophysics in vivo
Several presentations pointed to a new frontier for the single-molecule field: living cells. Giovanni Cappello (Institut Curie), who also won the alpine ski race, presented a technique for introducing kinesin-1 and myosin V labeled with fluorescent quantum dots into HeLa cells via pinocytocysis induced by osmotic shock20. Both types of motors were found to move with velocities and processivities consistent with values previously observed in vitro. However, the motion inside cells was much less uniform, with the molecules of kinesin-1 temporarily confined to particular regions before breaking free for longer runs. Using a somewhat different approach, Sunney Xie and Xiaolin Nan (Harvard University) showed that gold nanoparticles encapsulated in endosomes and transported by endogenous kinesin and dynein can be tracked in vivo with a precision of 1.5 nm in 25 μs via darkfield microscopy. This remarkable time resolution is afforded by the strong light-scattering signal generated by gold nanoparticles and has allowed clear visualization of individual kinesin and dynein steps over the full range of cargo velocities (0–8 μm per sec). They reported that whereas kinesin takes 8-nm steps in vivo, dynein has a variable step size whose value depends on the dimension of the particle to which the motor is attached (and therefore, by implication, to the load). This finding is consistent with previous results on dynein stepping in vitro21. The new experiments avoid the tedious biochemistry involved in setting up in vitro assays and underscore the physiological relevance for the large steps of dynein. Work by George Shubeita from Steve Gross’s lab (University of California, Irvine) went even further by optically trapping lipid droplets inside developing Drosophila melanogaster embryos to apply controlled loads to the kinesin and dynein motors responsible for moving these organelles. The Gross group has reported bidirectional motion of such droplets, and they have obtained evidence suggesting that motors with opposite polarities are controlled cooperatively to move cargo. Shubeita observed that the apparent stall-force values were quantized, which is consistent with the notion that nearly all organelles were transported by fewer than five motors, but that ~60% of cargoes carried more than one motor.
Outlook
The interdisciplinary field of single-molecule biophysics is currently enjoying growth and success. The evident breadth and vigor of the scientific discourse at SMB 2007 demonstrates that few aspects of molecular or cell biology remain off-limits to biophysics. Chemical biologists and biophysicists share the common goal of understanding biology in full molecular detail. We can anticipate that the future will bring ever more powerful techniques and collaborations that promise to unravel life’s persistent questions.

The SMB 2007 conference was held at the Aspen Center for Physics in the historic West End of Aspen, Colorado, minutes from the chairlifts.
Acknowledgments
Financial support for SMB 2007 was generously provided by the Martin & Beate Block Scholarship Endowment, the US National Science Foundation, the Biophysical Society, Academia Sinica (Taiwan), Nikon Instruments Inc., Chroma Inc., Visigen Inc., Olympus America Inc., Hamamatsu Inc., and Zeiss Inc. Logistical details at the Aspen Center for Physics were ably coordinated by its administrative director, J. Kelly.
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare no competing financial interests.
References
- 1.Dame RT, Noom MC, Wuite GJ. Nature. 2006;444:387–390. doi: 10.1038/nature05283. [DOI] [PubMed] [Google Scholar]
- 2.Yin H, et al. Science. 1995;270:1653–1657. doi: 10.1126/science.270.5242.1653. [DOI] [PubMed] [Google Scholar]
- 3.Dumont S, et al. Nature. 2006;439:105–108. doi: 10.1038/nature04331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bianco PR, Tracy RB, Kowalczykowski SC. Front Biosci. 1998;3:D570–D603. doi: 10.2741/a304. [DOI] [PubMed] [Google Scholar]
- 5.Woodside MT, et al. Science. 2006;314:1001–1004. doi: 10.1126/science.1133601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kramers HA. Physica. 1940;7:284–304. [Google Scholar]
- 7.Bell GI. Science. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 8.Dudko OK, Hummer G, Szabo A. Phys Rev Lett. 2006;96:108101. doi: 10.1103/PhysRevLett.96.108101. [DOI] [PubMed] [Google Scholar]
- 9.Hammond GS. J Am Chem Soc. 1955;77:334–338. [Google Scholar]
- 10.Hodak JH, Downey CD, Fiore JL, Pardi A, Nesbitt DJ. Proc Natl Acad Sci USA. 2005;102:10505–10510. doi: 10.1073/pnas.0408645102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lemay JF, Penedo JC, Tremblay R, Lilley DM, Lafontaine DA. Chem Biol. 2006;13:857–868. doi: 10.1016/j.chembiol.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 12.Rust MJ, Bates M, Zhuang X. Nat Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reiner JE, et al. Appl Phys Lett. 2006;89:013904. [Google Scholar]
- 14.Carter AR, et al. Appl Opt. 2007;46:421–427. doi: 10.1364/ao.46.000421. [DOI] [PubMed] [Google Scholar]
- 15.Yasuda R, Noji H, Kinosita K, Jr, Yoshida M. Cell. 1998;93:1117–1124. doi: 10.1016/s0092-8674(00)81456-7. [DOI] [PubMed] [Google Scholar]
- 16.Zimmermann B, Diez M, Zarrabi N, Graber P, Borsch M. EMBO J. 2005;24:2053–2063. doi: 10.1038/sj.emboj.7600682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kerssemakers JW, et al. Nature. 2006;442:709–712. doi: 10.1038/nature04928. [DOI] [PubMed] [Google Scholar]
- 18.Footer MJ, Kerssemakers JW, Theriot JA, Dogterom M. Proc Natl Acad Sci USA. 2007;104:2181–2186. doi: 10.1073/pnas.0607052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hackney DD. Proc Natl Acad Sci USA. 1994;91:6865–6869. doi: 10.1073/pnas.91.15.6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Courty S, Luccardini C, Bellaiche Y, Cappello G, Dahan M. Nano Lett. 2006;6:1491–1495. doi: 10.1021/nl060921t. [DOI] [PubMed] [Google Scholar]
- 21.Mallik R, Carter BC, Lex SA, King SJ, Gross SP. Nature. 2004;427:649–652. doi: 10.1038/nature02293. [DOI] [PubMed] [Google Scholar]


