Abstract
Asthma is a complex polygenic disease involving the interaction of many genes. In this study, we investigated the allergic response in experimental asthma. First, we constructed a biological interaction network using the BOND (Biomolecular Object Network Databank) database of literature curated molecular interactions. Second, we mapped differentially expressed genes from microarray data onto the network. Third, we analyzed the topological characteristics of the modulated genes. Fourth, we analyzed the correlation between the topology and biological function using the Gene Ontology classifications. Our results demonstrate that nodes with high connectivity (hubs and superhubs) tend to have low levels of change in gene expression. The significance of our observations was confirmed by permutation testing. Furthermore, our analysis indicates that hubs and superhubs have significantly different biological functions compared with peripheral nodes based on Gene Ontology classification. Our observations have important ramifications for interpreting gene expression data and understanding biological responses. Thus, our analysis suggests that a combination of differential gene expression plus topological characteristics of the interaction network provides enhanced understanding of the biology in our model of experimental asthma.
Keywords: allergic asthma, biological network, Gene Ontology, microarray
Introduction
There is currently an epidemic of allergic disorders including asthma in the US and other developed countries (Braman, 2006). A recent review of the literature identified over 100 genes correlated with asthma in humans by association studies, and over 150 genes in animal models (Ober and Hoffjan, 2006). Thus, both human and animal studies indicate that asthma is a complex polygenic disease (Van Eerdewegh et al, 2002; Xu et al, 2002). In this study, we performed a systems analysis of differential gene expression in an experimental model of asthma to investigate the topological characteristics of modulated genes in a biological interaction network.
We first constructed a biological network from experimentally documented molecular interactions using the Biomolecular Interaction Network Database (BIND), a component database of BOND (Biomolecular Object Network Databank), which is the largest available database of murine molecular interactions. We quantitated differential gene expression with oligonucleotide microarrays in a model of experimental asthma that has been well characterized and shown to develop airway hyperresponsiveness, elevated serum IgE and airway eosinophilia. Next, we mapped the modulated genes onto the interaction network. To investigate the relationship between differentially expressed genes and the interaction network, we performed topological analyses of the network architecture. Our results demonstrated that genes with a high level of change in expression are more likely to be peripheral nodes (low connectivity) in the network, whereas hubs (nodes with higher connectivity) and superhubs (nodes that link hubs) tend to have a lower level of change in expression. The significance of our observations was confirmed by permutation tests. To analyze the biological roles of the modulated genes, we assessed the Gene Ontology (GO) of nodes and hubs. Our analysis identified different annotations of molecular functions based on the topological classifications.
Result and discussion
Differentially expressed genes in murine asthma
To investigate differential expression of genes in the allergic immune response that orchestrate asthma, we used oligonucleotide microarrays to quantitate changes in gene expression. We analyzed a model of asthma previously studied in our laboratory in which wild-type (BALB/c) and recombinase activating gene-deficient (RAG KO) mice were sensitized and challenged with allergen (ovalbumin, OVA) or saline (PBS) control (Krinzman et al, 1996; Velasco et al, 2005). Wild-type mice develop increased serum IgE levels, broncho-alveolar lavage (BAL) eosinophilia, increased BAL IL-13 and IL-4 secretion and airway hyper-responsiveness. The RAG KO mice lack an adaptive immune response and do not generate an allergic asthmatic response (Mombaerts et al, 1992). We analyzed gene expression of whole lung RNA in each experimental group in quadruplicate by Affymetrix Mouse Genome 430 2.0 microarrays. Each array contains over 45 000 probe sets representing approximately 34 000 well-characterized mouse genes. Low expressing and constantly expressing genes were filtered, leaving 11 264 genes subject to analysis. The expression levels of the genes exposed to the OVA allergen versus PBS control were compared by t-statistics. After correcting for false discovery rate (FDR) (Benjamini and Hochberg, 1995), we identified 710 genes that were significantly modulated with FDR adjusted P-value below 0.05 (Supplementary Table I).
Construction of a murine interaction network of an allergic response
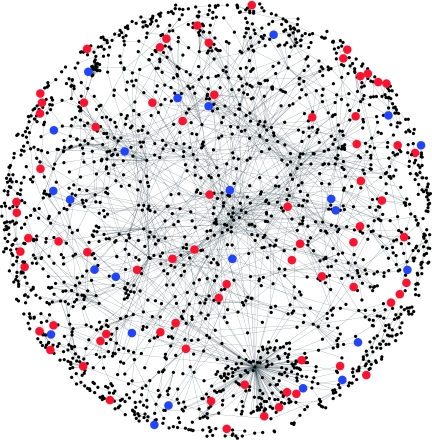
We compared six broadly used databases of known murine molecular interactions. We analyzed the databases for their screening and inclusion criteria of interactions as well as the size of the databases (Supplementary Table II). These analyses indicate that BIND is the largest currently available database of murine interactions. Hence, we constructed a mouse biological interaction network using the BIND database, and visualized it with Cytoscape v2.3 software (Figure 1). Our initial interaction network contained all interactions documented in BIND; however, our preliminary analysis focused on 2054 genes that were present in both the Affymetrix Mouse Genome 430 2.0 microarray and the BIND database. There were 2584 molecular interactions between these 2054 genes. In the mouse gene network, our analysis showed a power law decay of connectivity, which is consistent with a ‘scale free network' reported for most biological networks analyzed to date (Barabasi and Oltvai, 2004) (Supplementary Figure 1). To investigate the relationship between differentially expressed genes and the network, we labeled the up- and downregulated genes by red and blue, respectively (Figure 1).
Figure 1.
Mouse gene network from BIND. Red and blue spots represent genes that were significantly up- or downregulated.
Degree of differential expression is negatively correlated with connectivity
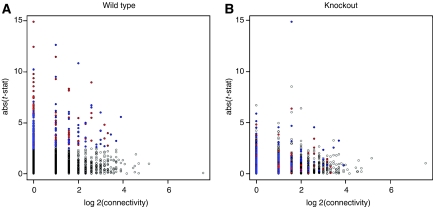
In our network, we identified 106 genes that were significantly modulated. To investigate the topology of the differentially expressed genes, we plotted the scatter plot of the t-statistics versus connectivity (Figure 2A). Interestingly, we found that genes with higher connectivity tend to have a lower dynamic range of t-statistics, and conversely, genes with lower connectivity tend to have higher dynamic range of t-statistics. As expected, no correlation was detected in the RAG KO mice lacking the adaptive immune response (Figure 2B). By integrating gene expression data with the interaction network, our analysis demonstrated that genes exerting major biological roles in general (based on a higher level of connectivity) (Jeong et al, 2001; Haynes et al, 2006) have lower levels of differential expression in the allergic immune response and thus are unlikely to be identified solely by expression measurements.
Figure 2.
Scatter plot of t-statistics versus connectivity for genes modulated in OVA-specific allergic immune response. (A) Wild-type mice. (B) RAG KO mice. X-axis is the log 2 of connectivity (no. of genes directly connected) and Y-axis is the absolute of t-statistics. Red and blue spots represent genes upregulated or downregulated, respectively, in response to OVA sensitization and challenge with individual P-values below 0.05. Empty circles correspond to non-differentially expressed genes.
To determine the significance of our observations, we grouped the genes with the same connectivity, averaged the top 20% of the highest absolute t-statistics in each group and calculated the rank correlation between the average t-statistics and the connectivity. There are 24 groups of genes with connectivity ranging from 1 to 191; however, only gene groups with connectivity below 13 having more than five genes per group were used in the analysis. We then tested the significance of the negative correlation by four permutation tests. In the first permutation test, we randomly shuffled the t-statistics among genes, averaged the top 20% of the highest absolute t-statistics for each connectivity group and then calculated the rank correlation to connectivity. In the second test, we randomly picked eight microarrays from the original 16 including data from both the wild-type and knockout mice, split them into two groups and calculated the t-statistics for each gene. Subsequently, for each group, we averaged the top 20% of the highest absolute t-statistics and calculated the rank correlation to connectivity. In the third test, we randomly picked eight microarrays from the original 16, split them into two groups and calculated the t-statistics for each gene. Subsequently, for each connectivity group, we averaged the t-statistics of the genes that belong to the top 20% in the original data set and then calculated the rank correlation between the average t-statistics and the connectivity. The first three permutations all randomized the differential expression while still using the original gene network topology. In the fourth permutation test, we used the original differential expression data but created a random network by randomly re-assigning edges between nodes while maintaining the same number of nodes and edges as original network, and then calculated the correlation between connectivity and the average of top 20% highest absolute t-statistics in each connectivity group. As the degree of connectivity in a random network has a much smaller dynamic range than a scale free network, we cannot directly compare the rank correlations as performed in the previous three permutation tests. Therefore, we used the corresponding null distribution as a control, that is, we calculated the P-value for rank correlation and then compared the P-value of our interaction network with the P-value of the random network.
We performed each of the four permutation tests 1000 times and compared the rank correlation of these permutations to the rank correlation calculated from the original data of wild-type and knockout mice (Table I). We found that the rank correlation for wild-type mice was significantly lower than the permutations, with all four P-values below 0.05. As expected, the rank correlation for the RAG KO mice was not significant. We also averaged the top 10 or 30% of each connectivity group, or pooled all genes with connectivity greater than 13 as the 14th group in our analysis, and obtained similar results (data not shown).
Table 1. P-value of the rank correlations by permutation tests.
| Permutation method | P-value for wild-type mice | P-value for RAG KO mice |
|---|---|---|
| 1 | 0.025 | 0.424 |
| 2 | 0.048 | 0.382 |
| 3 | 0.003 | 0.125 |
| 4 | 0.013 | 0.261 |
Characterization of hub and superhub genes
To better understand the function of the hubs in our interaction network, we next determined if the topology of our network was consistent with a hierarchical structure. First, we plotted connectivity versus average clustering coefficient, which showed a power law decay (Supplementary Figure 2), which is the signature of a hierarchical network. In addition, the average clustering coefficient was 0.15, which is approximately an order of magnitude greater than for a random network and consistent with a hierarchical network (Ravasz et al, 2002). In our interaction network, we defined ‘hubs' as nodes with connectivity greater than 5 as reported previously (Han et al, 2004; Patil and Nakamura, 2006), and a clustering coefficient below 0.03. When identifying ‘hub' genes, our objective was to select nodes that were candidates to function as signaling centers while simultaneously excluding ‘molecular machines.' Therefore, we used high connectivity and low clustering coefficient as our criterion. We also analyzed our network using a hub definition of connectivity greater than 5 without considering the clustering coefficient; the results were similar (data not shown). In our analysis, we identified 88 hubs of which seven (∼8%) were significantly modulated.
Our next objective was to analyze the interconnections among the hubs. We postulated that the hubs were linked in a hub network that was connected via a core of ‘superhubs.' For this analysis, we found the weighted shortest paths connecting every pair of hub genes to generate a hub network (see Materials and methods), identified nodes with connectivity greater than 5 in the shortest-path hub network and termed these nodes as ‘superhubs'. We identified 16 superhubs (Supplementary Table III). Consistent with our previous observation that genes with high connectivity tend to have low levels of change in expression, none of the superhubs were significantly modulated. A box plot of the t-statistics of superhub, hub and peripheral node genes showed that the superhub genes have a smaller dynamic range than other genes (Supplementary Figure 3). To test the significance, we compared the variation of t-statistics of the three groups of genes by F-statistics. Our analysis showed that the variation of t-statistics of superhub was significantly lower than that of hub genes and non-hub genes, with P-value 0.05 and 0.03, respectively. The difference between hub genes and other non-hub genes was not significant (P=0.38).
After identifying superhubs in the hierarchical architecture of the network based on topological criteria, we next investigated the GO annotations of the superhubs compared to the peripheral nodes using GeneNotes software (see Materials and methods). We determined if specific molecular functions were significantly overrepresented in the superhubs versus the peripheral nodes of the network. Consistent with our hypothesis, our analysis showed that GO molecular functions in the superhubs included evolutionarily ancient processes such as nucleic binding and transcription regulation (Supplementary Table IV). In contrast, overrepresented annotations in the peripheral nodes included more specialized functions (e.g., cytolysis, lipid modification, response to hormone stimulus and induction of apoptosis) (Supplementary Table V). Thus, the relevance of our investigation of the hubs and superhubs is supported by our analysis showing that they have significantly different molecular functions. Furthermore, our results suggest a modular structure to the hierarchical architecture of the network (Ravasz et al, 2002), with the highly connected superhubs performing the most basic biological functions (evolutionarily early), and the more specialized functions (evolutionarily late) performed by the peripheral nodes. Furthermore, changes in gene expression occurred predominantly in the genes (nodes) with low connectivity, but not in the superhubs.
Conclusion
Network analysis has been shown to be a powerful tool to understand biological responses (Sharan and Ideker, 2006). In this paper, we used microarrays to analyze gene expression of allergen-treated mice in experimental asthma. To investigate the regulation of the allergic response, we constructed a murine interaction network from curated data in BIND and mapped the expression profile onto our interaction network. Interestingly, we found an inverse correlation between the level of change of expression and the connectivity of a gene. This observation has both methodological and biological implications.
First, a major challenge in the analysis of microarray data is interpreting the biological relevance of changes in expression. Common approaches rely on fold change or statistics (e.g., t-statistic). As both approaches preferentially select genes with large changes in expression, our analysis suggests that many genes with important biological functions would not be detected. Specifically, hub and superhub genes, which have high connectivity and putatively high biological importance, may not be detected. Thus, our study indicates that biological understanding is enhanced by combining information including levels of change in gene expression plus topological criteria from the analysis of interaction networks.
Second, the mechanisms of regulation of biological responses by webs of molecular interactions remain poorly understood. Our study indicates that at least some biological responses, for example, an allergic immune response, are mediated by larger changes in nodes with low connectivity and smaller changes in the hubs and superhubs. Our observations suggest the hypothesis that ‘fine-tuning' or regulating an immune response is facilitated by modulating genes with low connectivity. This notion is indirectly supported by previous studies showing that the deletion of ‘hub' genes, which tend to encode proteins with greater intrinsic disorder in yeast, produced a higher frequency of synthetic sick phenotype or lethal effects (Jeong et al, 2001; Haynes et al, 2006) than the deletion of genes with low connectivity (Barabasi and Oltvai, 2004). Therefore, we postulate that the negative correlation we observed in this allergic immune response analysis may be a general characteristic of gene regulation in other biological responses.
Our study was restrained to the correlation between the regulation of mRNA levels and molecular interactions obtained from the BIND database, with observations obtained from a murine experimental model of asthma. Thus, there are several potential caveats to our observations. For example, additional important regulatory events might not be detected due to experimental conditions, experimental noise or the limited power of the study. In addition, although BIND remains the largest currently available database of murine interactions, this knowledge is incomplete and potentially biased by experimental techniques or research interests.
Future studies combining more information into our experimental approach should be able to provide a more complete view of the correlation between gene regulation and network topology. For example, it will be important to determine if other biological responses, in addition to the allergic immune response, are similarly regulated. Interestingly, our study shows the limitations of interpreting levels of change in gene expression in isolation, and that concomitant analysis of gene expression data and topologic interaction networks may provide critical insights into biological processes. In conclusion, we demonstrate that hubs, and to a greater degree superhubs, exhibit low levels of change in gene expression during an allergic immune response, but based on topological analysis of interaction networks, are likely to play an important role in regulating the biological response.
Materials and methods
Protocol for murine experimental asthma
Six- to eight-week-old BALB/c and RAG KO murine strain (which generates only an innate immune response owing to the absence of functional T and B lymphocytes) mice were purchased from Jackson Laboratory (Bar Harbor, ME, USA). The mice were maintained according to the guidelines of the Committee on Animals and the Committee on the Care and Use of Laboratory Animals of the Institute of Laboratory Animal Resources National Research Council. Mice were sensitized and challenged with the allergen chicken OVA. Briefly, mice were sensitized via intraperitoneal injection with 10 μg OVA (Sigma, St Louis, MO, USA) and 1 mg Al(OH)s (alum) (Sigma, St Louis, MO, USA) in 0.2 ml of PBS, followed by a boosting injection on day 7 with the identical reagents. Control mice received 1 mg alum in 0.2 ml of PBS without OVA. On days 14–20, mice received aerosolized challenges with 6% OVA or PBS for 20 min/day via an ultrasonic nebulizer (Model 5000; DeVilbiss, Somerset, PA). All groups were killed at day 21. Each experiment was repeated in quadruplicate on different mice independently.
RNA preparation
Total RNA was isolated using TRIZOL reagent (Gibco-BRL Life Technologies) according to the manufacturer's protocol. RNA purity was initially determined by spectroscopy with a 260/280 ratio=1.85–2.01 and then by scanning with an Agilent 2100 Bioanalyzer using the RNA 6000 Nano LabChip®. Samples not meeting these basic parameters of quality were excluded from microarray analysis.
Microarray methods
The DNA microarray studies were performed in collaboration with the Partners Genetics & Genomics Core Facility of the Harvard Medical School and Partners Healthcare Center for Genetics and Genomics. All protocols were performed according to the Affymetrix GeneChip® Manual. The cDNA synthesis was performed using 100 pM of the T7 dT primer 5′-GGCCAGTGAATTGTAATACGACTCACTATAGGGAGGCGG (dT)24. The cDNA and double-stranded product were processed using the GeneChip Cleanup Module kit. In vitro transcription (IVT) and preparation of labeled RNA was performed using the ENZO BioArray High Yield RNA Transcription Labeling Kit. The IVT sample was quantified on a Bio-Tek UV Plate Reader. Twenty grams of the IVT material was hybridized to the Affymetrix Mouse Genome 430 2.0 array (Affymetrix, Santa Clara, CA). The arrays were incubated in a model 320 hybridization oven at a constant temperature of 45°C overnight. The microarray was washed on a Model 450 Fluidics station and scanned on an Affymetrix Model 3000 scanner with autoloader.
Microarray data analysis
Raw microarray data were normalized and gene expression levels were calculated by the GCRMA algorithm (Wu and Irizarry, 2004) using R language (http://cran.r-project.org/) and Bioconductor project (http://www.bioconductor.org/). The expression data were deposited in GEO database (accession number GSE6858). The expression levels were log 2 transformed before any analysis. To eliminate genes with low levels of expression or constant expression, a filtering process was applied to the whole data set of 45 000 probe sets. For each gene, we calculated the average expression levels of the four replicates in OVA treatment and control groups, and included one with the maximum across conditions. We eliminated genes with low expression (maximum conditional mean of gene expression below 20), and genes with coefficient of variation across samples below 0.05. The remaining subset of genes was subjected to later analysis.
The mean of log-transformed expression level of each gene under different conditions (control and OVA stimulated) was compared by t-statistics and raw P-values were calculated. In order to correct for multiple hypothesis tests, these raw P-values were adjusted to control FDR (Benjamini and Hochberg, 1995). We selected a set of differentially expressed genes with the criterion of FDR adjusted P-values below 0.05.
Gene interaction network analysis
We compared six broadly used databases of known murine molecular interactions. We analyzed the databases for their screening and inclusion criteria of interactions as well as the size of the database (Supplementary Table II). Molecular interaction data were downloaded from the BIND (http://bond.unleashedinformatics.com/), analyzed by R language and visualized by Cytoscape v2.3 software (http://www.cytoscape.org/). (The Cytoscape file of interaction network and annotations of gene symbol as well as up- or downregulated genes are in Supplementary information.) In order to map the mRNA expression data onto gene interaction network, we used Entrez Gene ID as the unique identifier for genes. When there are multiple probe sets corresponding to the same gene, we used the one with the maximum t-statistic as a representative.
Finding a unique shortest path between a pair of genes
A shortest path represents the minimum requirement for the transduction of a response from one molecule to another. Also, the molecules involved in multiple shortest paths may be the important upstream regulators that control many downstream effectors. Because the BIND database is an assembly of information from various sources, simply counting the number of intermediate genes as the length of the shortest path will often generate multiple shortest paths with the same length. Also, the total number of shortest paths or the number of molecules involved in these shortest paths will grow exponentially with the increase in the number of starting nodes. We addressed this issue by weighting the edges based on the clustering coefficient of nodes. We calculated clustering coefficient of each node, and assigned weight to an edge based on the sum of the clustering coefficient of the two genes being connected by the edge. Therefore, the length of the paths became the sum of edge weights along the path instead of simply counting the number of genes.
Gene ontology analysis
GO provides three structured, controlled vocabulary (ontology) to describe gene and gene product attributes in any organism, in terms of their associated biological processes, cellular components and molecular functions. After selecting differentially expressed genes, we employed GeneNotes software (http://combio.cs.brandeis.edu/GeneNotes/index.htm) to identify the enriched GO terms associated with subsets of nodes. The GeneNotes software uses a hyper-geometric test to compare the number of genes in the experimental group within each GO term with the total number of genes in that term, and reports a P-value for each GO term. In the GO reports listed in Supplementary Tables IV and V, we reported P-values for each GO term. Because GO annotation has a hierarchical structure and the GO terms are correlated with P-values but not independent, we did not adjust P-values for multiple hypothesis testing.
Supplementary Material
Supplementary Figures
Supplementary Table I
Supplementary Table II
Supplementary Table III
Supplementary Table IVa
Supplementary Table IVb
Supplementary Table IVc
Supplementary Table Va
Supplementary Table Vb
Supplementary Table Vc
Supplementary Data 1
Supplementary Data 2
Supplementary Data 3
Acknowledgments
This work is supported by NIH grants R01HL081663 and R01AI053878.
References
- Barabasi AL, Oltvai ZN (2004) Network biology: understanding the cell's functional organization. Nat Rev Genet 5: 101–113 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B 57: 289–300 [Google Scholar]
- Braman SS (2006) The global burden of asthma. Chest 130: 4S–12S [DOI] [PubMed] [Google Scholar]
- Han JJD, Bertin N, Hao T, Goldberg DS, Berriz GF, Zhang LV, Dupuy D, Walhout AJM, Cusick ME, Roth FP, Vidal M. (2004) Evidence for dynamically organized modularity in the yeast protein–protein interaction network. Nature 430: 88–93 [DOI] [PubMed] [Google Scholar]
- Haynes C, Oldfield CJ, Ji F, Klitgord N, Cusick ME, Radivojac P, Uversky VN, Vidal M, Iakoucheva LM (2006) Intrinsic disorder is a common feature of hub proteins from four eukaryotic interactomes. PLoS Comput Biol 2: e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H, Mason SP, Barabasi AL, Oltvai ZN (2001) Lethality and centrality in protein networks. Nature 411: 41–42 [DOI] [PubMed] [Google Scholar]
- Krinzman SJ, De Sanctis GT, Cernadas M, Kobzik L, Listman JA, Christiani DC, Perkins DL, Finn PW (1996) T cell activation in a murine model of asthma. Am J Physiol 271: L476–L483 [DOI] [PubMed] [Google Scholar]
- Mombaerts P, Iacomini J, Johnson RS, Herrup K, Tonegawa S, Papaionnou VE. (1992) RAG-1 deficient mice have no mature B and T lymphocytes. Cell 68: 869. [DOI] [PubMed] [Google Scholar]
- Ober C, Hoffjan S (2006) Asthma genetics 2006: the long and winding road to gene discovery. Genes Immun 7: 95–100 [DOI] [PubMed] [Google Scholar]
- Patil A, Nakamura H. (2006) Disordered domains and high surface charge confer hubs with the ability to interact with multiple proteins in interaction networks. FEBS Lett 580: 2041–2045 [DOI] [PubMed] [Google Scholar]
- Ravasz E, Somera AL, Mongru DA, Oltvai ZN, Barabasi AL (2002) Hierarchical organization of modularity in metabolic networks. Science 297: 1551–1555 [DOI] [PubMed] [Google Scholar]
- Sharan R, Ideker T (2006) Modeling cellular machinery through biological network comparison. Nat Biotech 24: 427–433 [DOI] [PubMed] [Google Scholar]
- van Eerdewegh P, Little RD, Dupuis J, Del Mastro RG, Falls K, Simon J, Torrey D, Pandit S, McKenny J, Braunschweiger K, Walsh A, Liu Z, Hayward B, Folz C, Manning SP, Bawa A, Saracino L, Thackston M, Benchekroun Y, Capparell N, Wang M, Adair R, Feng Y, Dubois J, FitzGerald MG, Huang H, Gibson R, Allen KM, Pedan A, Danzig MR, Umland SP, Egan RW, Cuss FM, Rorke S, Clough JB, Holloway JW, Holgate ST, Keith TP (2002) Association of the ADAM33 gene with asthma and bronchial hyperresponsiveness. Nature 418: 426–430 [DOI] [PubMed] [Google Scholar]
- Velasco G, Campo M, Manrique OJ, Bellou A, He H, Arestides RS, Schaub B, Perkins DL, Finn PW. (2005) Toll-like receptor 4 or 2 agonists decrease allergic inflammation. Am J Respir Cell Mol Biol 32: 218–224 [DOI] [PubMed] [Google Scholar]
- Wu Z, Irizarry RA (2004) Preprocessing of oligonucleotide array data. Nat Biotechnol 22: 656. [DOI] [PubMed] [Google Scholar]
- Xu J, Bleecker ER, Jongepier H, Howard TD, Koppelman GH, Postma DS, Meyers DA (2002) Major recessive gene(s) with considerable residual polygenic effect regulating adult height: confirmation of genome wide scan results for chromosomes 6, 9, and 12. Am J Hum Genet 71: 646–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures
Supplementary Table I
Supplementary Table II
Supplementary Table III
Supplementary Table IVa
Supplementary Table IVb
Supplementary Table IVc
Supplementary Table Va
Supplementary Table Vb
Supplementary Table Vc
Supplementary Data 1
Supplementary Data 2
Supplementary Data 3