Abstract
Haemophilus influenzae is a major community-acquired pathogen causing significant morbidity and mortality worldwide. Meningitis and bacteremia due to type b strains occur in areas where the protein-conjugated type b vaccine is not in use, whereas nontypeable strains are major causes of otitis media, sinusitis, acute exacerbations of chronic bronchitis, and pneumonia. Antibiotic resistance in this organism is more diverse and widespread than is commonly appreciated. Intrinsic efflux resistance mechanisms limit the activity of the macrolides, azalides, and ketolides. β-Lactamase production is highly prevalent worldwide and is associated with resistance to ampicillin and amoxicillin. Strains with alterations in penicillin binding proteins, particularly PBP3 (β-lactamase negative ampicillin resistant and β-lactamase positive amoxicillin-clavulanate resistant), are increasing in prevalence, particularly in Japan, with increasing resistance to ampicillin, amoxicillin, amoxicillin-clavulanate, and many cephalosporins, limiting the efficacy of expanded-spectrum cephalosporins against meningitis and of many oral cephalosporins against other diseases. Most strains remain susceptible to the carbapenems, which are not affected by penicillin binding protein changes, and the quinolones. The activity of many oral agents is limited by pharmacokinetics achieved with administration by this route, and the susceptibility of isolates based on pharmacokinetic and pharmacodynamic parameters is reviewed.
INTRODUCTION
Haemophilus influenzae includes six encapsulated types, a through f, as well as nonencapsulated or untypeable strains, and these are found as both respiratory tract commensals and respiratory and invasive pathogens. Carriage of H. influenzae type b (Hib) strains is influenced by use of various formulations of type b, protein-conjugated capsular polysaccharide vaccines. The major diseases caused by H. influenzae are childhood pneumonia, meningitis, and bacteremia, which are primarily caused by type b strains, and community-acquired pneumonia (CAP) in adults, acute otitis media (AOM), acute sinusitis, and acute exacerbations of chronic bronchitis (AECB), which are typically caused by untypeable strains.
Prior to introduction of Hib vaccines, Hib was estimated to be responsible worldwide for some three million serious illnesses and an estimated 386,000 deaths per year, chiefly through meningitis and pneumonia, with 95% of cases and 98% of deaths occurring in patients in developing countries (153). Almost all victims are children under the age of 5 years, with those between 4 and 18 months of age especially vulnerable (112). Large, population-based disease burden studies showed annual rates of Hib meningitis at 10 to 60 cases per 100,000 children under 5 years of age in countries where Hib vaccine is not used (153). In developing countries, where the vast majority of Hib deaths occur, pneumonia accounts for a larger number of deaths than meningitis. However, Hib meningitis is also a serious problem in such countries with mortality rates several times higher than seen in developed countries, and 15 to 35% of survivors have permanent disabilities such as mental retardation or deafness. Systematic vaccination has virtually eliminated Hib disease in most industrialized nations, with 89 countries offering infant immunization against Hib by the end of 2004 compared with 38 in 1999. However, Hib vaccine is not available in Japan (56, 57). Ninety-two percent of the populations of developed countries were vaccinated against Hib as of 2003, while vaccination coverage was 42% for developing countries and 8% for least-developed countries.
The current status of resistance mechanisms found in H. influenzae against the antimicrobial agents recommended for empirical and directed treatment of the diseases caused by this pathogen forms the basis of this review.
SUSCEPTIBILITY TESTING OF H. INFLUENZAE
H. influenzae is fastidious, requiring NAD and an iron source in the form of hemoglobin, hematin, or hemin. A variety of susceptibility testing media providing these growth factors have been developed by groups such as Clinical and Laboratory Standards Institute (CLSI), British Society for Antimicrobial Chemotherapy (BSAC), Comité de l′Antibiogramme de la Société Française de Microbiologie, Deutsches Institut für Normung, and Swedish Reference Group of Antibiotics (47). These media include Haemophilus test medium (HTM), Mueller-Hinton broth with 2% lysed horse blood and 15 μg of NAD per ml, IsoSensitest broth with 2% lysed horse blood and 15 μg of NAD per ml, and IsoSensitest broth-based HTM (66). HTM, currently recommended by CLSI, consists of Mueller-Hinton broth base supplemented with 0.5% yeast extract, 15 μg of NAD per ml, and 15 μg of hematin per ml. Considerable variation in the antimicrobial susceptibility of H. influenzae has been reported; the effects of the above test media on the susceptibility of H. influenzae were studied in three laboratories for 21 antimicrobial agents against a panel of 100 selected isolates (66). Overall, all results were very reproducible, with the MIC50, MIC90, and geometric mean MICs being within one doubling dilution by six methods in the three testing centers for 15 of the 21 agents tested. Interlaboratory differences were more marked than intralaboratory differences or differences among media. Cefprozil, cefaclor, and trimethoprim-sulfamethoxazole results differed the most, while results for ampicillin, amoxicillin-clavulanic acid, cefdinir, cefixime, ceftriaxone, and clarithromycin were the most reproducible. Reproducible susceptibility results for a wide range of agents against H. influenzae using a variety of media that support the growth of this fastidious species can therefore be obtained. When testing susceptibility of H influenzae, it is essential to ensure proper quality control of the freshness of the media in every run, as commercial HTM has a rather short shelf-life (66). Additionally, β-lactamase production can be determined by the chromogenic cephalosporin method, using nitrocefin or other suitable substrate (138).
TREATMENT OF H. INFLUENZAE INFECTIONS
Meningitis
Meningitis remains a serious problem in children under 7 years of age in areas where the Hib vaccine is not used (see Introduction), and the Infectious Disease Society of America-recommended empirical antimicrobial treatment of meningitis for this age group is vancomycin plus an expanded-spectrum cephalosporin such as cefotaxime or ceftriaxone (146). Alternate therapies for H. influenzae include chloramphenicol, cefepime and meropenem. For β-lactamase negative, ampicillin susceptible H. influenzae, ampicillin is recommended as standard therapy, with an expanded-spectrum cephalosporin, cefepime, or chloramphenicol as alternate regimens (146). β-Lactamase positive H. influenzae should be treated with an expanded-spectrum cephalosporin, with cefepime or chloramphenicol as alternatives. A fluoroquinolone, either gatifloxacin or moxifloxacin, can be added to the list of recommended alternative agents for adult patients only (146). In Japan, where isolates with non-β-lactamase-mediated β-lactam resistance are now common, meropenem plus an expanded-spectrum cephalosporin such as cefotaxime or ceftriaxone is suggested for treatment of H. influenzae meningitis (57).
Childhood Pneumonia and Bacteremia
In regions where protein-conjugated Hib vaccine is not available, H. influenzae type b is a common bacterial cause of childhood pneumonia and bacteremia between 6 months and 5 years of age. High-dose amoxicillin (90 mg/kg of body weight/day), either alone or with the addition of clavulanic acid, is the first-line drug of choice for empirical treatment of outpatients with childhood pneumonia (15). Recommended empirical therapy for inpatients includes ceftriaxone or cefotaxime to provide coverage for non-penicillin-susceptible Streptococcus pneumoniae and β-lactamase-positive H. influenzae. The addition of azithromycin or erythromycin is recommended to provide coverage for atypical pathogens in older children, and vancomycin should be added for life-threatening pulmonary infections in which virulent, community-acquired, methicillin-resistant Staphylococcus aureus is increasingly being encountered. Directed parenteral therapy for pneumonia and bacteremia due to H. influenzae includes ampicillin for β-lactamase-negative, ampicillin-susceptible strains and ceftriaxone, cefotaxime, or cefuroxime for β-lactamase-positive strains (15).
CAP in Adults
Although S. pneumoniae and Mycoplasma pneumoniae are the most common pathogens for community-acquired pneumonia (CAP), untypeable H. influenzae causes 2 to 12% of cases. Treatment guidelines for management of CAP in immunocompetent adults have been established by the American Thoracic Society and the Infectious Disease Society of America (91, 107). Recommendations for outpatients with no comorbidities include azithromycin, clarithromycin, and doxycycline if no antibiotic therapy had been administered in the past 3 months; if antibiotic therapy had been administered in the past 3 months, recommendations are levofloxacin, gatifloxacin, gemifloxacin, or moxifloxacin as single agents or combination macrolide-β-lactam therapy (azithromycin or clarithromycin with amoxicillin [3 g/day] or amoxicillin-clavulanate [4 g/250 mg/day]). Recommendations for outpatients with comorbidities include azithromycin, clarithromycin, levofloxacin, gatifloxacin, gemifloxacin, or moxifloxacin if no antibiotic therapy had been administered in the past 3 months; if antibiotic therapy had been administered in the past 3 months, recommendations are levofloxacin, gatifloxacin, gemifloxacin, or moxifloxacin as single agents or combination macrolide-β-lactam therapy (azithromycin or clarithromycin with amoxicillin-clavulanate [4 g/250 mg/day]). Amoxicillin-clavulanate or clindamycin is recommended for suspected aspiration pneumonia. High-dose amoxicillin, high-dose amoxicillin-clavulanate, cefpodoxime, cefprozil, cefuroxime axetil, levofloxacin, gatifloxacin, gemifloxacin, or moxifloxacin is recommended for influenza with bacterial superinfection. Recommendations for inpatients in medical wards include levofloxacin, gatifloxacin, gemifloxacin, or moxifloxacin alone or azithromycin or clarithromycin plus cefotaxime, ceftriaxone, ampicillin-sulbactam, or ertapenem. Recommendations for patients requiring intensive care are the same plus inclusion of an antipseudomonal agent if infection with Pseudomonas aeruginosa is a concern. Virulent, community-acquired, methicillin-resistant Staphylococcus aureus (MRSA) is increasingly being encountered, and the addition of vancomycin or other anti-MRSA agents should also be considered.
AOM
AOM occurs most often between 6 months and 3 years of age, especially in children with frequent viral upper respiratory infections, and untypeable H. influenzae is the etiologic agent in 23 to 67% of cases (33). Introduction of the conjugate pneumococcal vaccine in children has resulted in untypeable, β-lactamase-producing H. influenzae becoming more prevalent in patients failing first-line amoxicillin therapy (21). Recent guidelines for empirical treatment of AOM include amoxicillin, 80 to 90 mg/kg/day, as the first-line agent for less severe disease, with azithromycin or clarithromycin as alternatives for patients with type I penicillin allergy and cefdinir, cefuroxime axetil, and cefpodoxime as alternatives for patients with non-type I penicillin allergy (4). Amoxicillin-clavulanate, 90/6.4 mg/kg/day, is recommended as the first-line agent for more severe disease and for patients not responding to treatment after use of amoxicillin for 48 to 72 h. Intramuscular ceftriaxone, 50 mg/kg/day for 1 or 3 days is recommended for initial therapy of less severe disease for patients with penicillin allergy and for 3 days for patients not responding to treatment after use of amoxicillin-clavulanate for 48 to 72 h (4).
Acute Sinusitis
Untypeable H. influenzae is a major sinusitis pathogen in adult and pediatric patients. Recommended empirical therapy for adults with no recent antibiotic use includes amoxicillin (1.5 to 4 g/day), amoxicillin-clavulanate (1.75 g/250 mg/day to 4 g/250 mg/day), cefpodoxime, cefuroxime, and cefdinir, with trimethoprim-sulfamethoxazole, doxycycline, azithromycin, clarithromycin, erythromycin, and telithromycin for patients with type I penicillin allergy (6). Recommendations for patients not responding to therapy after 72 h or that have received antimicrobials in the past 4 to 6 weeks include gatifloxacin, levofloxacin, moxifloxacin, amoxicillin-clavulanate (4g/250 mg/day), intramuscular ceftriaxone, and combination therapy (amoxicillin plus cefixime, clindamycin plus cefixime, amoxicillin plus rifampin, or clindamycin plus rifampin). Recommendations for children are similar to those for adults, with the omission of gatifloxacin, levofloxacin, moxifloxacin, doxycycline, and telithromycin, as they are not approved for pediatric use.
AECB
Untypeable H. influenzae is the most common cause of AECB, and the presence of a new strain of H. influenzae from the sputum of a patient with chronic bronchitis increases the relative risk of an exacerbation twofold (134). Recommendations for treatment of AECB are stratified by the presence of underlying, comorbid conditions and by the severity of the exacerbation. Treatment recommendations for patients without baseline risk factors include azithromycin, clarithromycin, telithromycin, doxycycline, cefuroxime axetil, cefpodoxime, and cefdinir, while those for patients with any of baseline risk factors include amoxicillin-clavulanic acid, levofloxacin, gatifloxacin, gemifloxacin, and moxifloxacin (8, 133).
BASELINE SUSCEPTIBILITY, DEVELOPMENT OF RESISTANCE, AND DEVELOPMENT OF INTERPRETATIVE SUSCEPTIBILITY BREAKPOINTS
H. influenzae, as is the case with most bacterial species, has a baseline, wild-type population with a defined, usually narrow, range of intrinsic activity of antimicrobial agents at the time of introduction of a new antimicrobial drug class (147). This defines the initial spectrum of activity of each antimicrobial agent when susceptibility breakpoints are established based on dosing regimens and sites of infection. Species can then be studied based on baseline activity and susceptibilities of strains with decreased activity, should they be present initially or should they develop. Susceptibility breakpoints between susceptibility of baseline, wild-type populations and those of populations with acquired resistance can be used and are referred to as “microbiological breakpoints” (147). Such breakpoints are very useful but do not necessarily correlate with clinical outcome, and clinically relevant breakpoints are needed for breakpoints to be useful for patient management.
Unfortunately, many breakpoints in common use for H. influenzae are microbiological breakpoints that are of little clinical use rather than clinically relevant breakpoints, and the current CLSI (formerly NCCLS) interpretation guideline for H. influenzae states that results of susceptibility testing using breakpoints provided for the oral β-lactam, macrolide, and ketolide agents “are often not useful for the management of individual patients” but “may be appropriate for surveillance or epidemiologic studies” (27). Clinically relevant susceptibility breakpoints are essential for susceptibility results to be used in patient management, which is typically achieved by setting breakpoints dividing susceptibility measurements into susceptible, intermediate, and resistant categories.
The process of determining clinical susceptibility breakpoints is complex, and is based on multiple factors (27, 47, 49, 73, 99): (i) the pharmacokinetics of the agent, typically reflected by levels achieved in blood and tissues over time with standard dosing regimens (parameters considered include serum protein binding and penetration to various body sites); (ii) susceptibility of relevant species to the agent, typically represented by MIC distributions; (iii) pharmacokinetic/pharmacodynamic (PK/PD) or dose-effect relationships obtained in in vitro studies, animal studies, and human studies; (iv) mathematical modeling processes, such as Monte Carlo simulation, of human pharmacokinetic values and susceptibility values of pathogens may be used to assist the process of breakpoint setting; (v) results of clinical studies relating outcome to susceptibility values; (vi) resulting breakpoints are applied to MIC distributions of each of the major target species and may be adjusted to ensure that the breakpoints do not divide the wild-type distributions of major target species. The ensuing breakpoints may therefore differ somewhat between species. Additional breakpoints may need to be developed for specific sites of infection where drug penetration is poor, such as the central nervous system, or where drugs are concentrated, such as in urine.
Of the above factors used to determine susceptibility breakpoints, clinical studies are very useful in life-threatening diseases such as meningitis but are of limited value in self-limiting diseases such as acute sinusitis and AOM. Bacteriologic outcome studies, such as double tympanocentesis AOM studies, and application of PK/PD parameters from animal models to human infections are currently considered the best parameters available in self-limiting diseases (2, 5, 30, 33, 60).
As PK/PD parameters are of considerable relevance to determining clinical susceptibility breakpoints for H. influenzae, these will be discussed briefly. PK describes the body's absorption, distribution, metabolism, and elimination of drugs, while PD examines the in vivo relationship between the target pathogen and the antimicrobial agent over time, taking into account the effects of variations in drug concentrations on killing and organism growth dynamics (3, 5, 29). These factors interact with the relative susceptibility of the pathogen to the particular agent to affect the rates and extent of bacterial killing in vivo. Based on PK/PD parameters, antimicrobials are divided into time- and concentration-dependent agents. For time-dependent agents, the duration of the non-protein-bound (free or active) drug concentration in plasma over time relative to the established MIC of the agent against the specific pathogen will determine in vivo bacterial killing for time-dependent agents such as β-lactams. The time free (non-protein-bound) plasma concentrations of a β-lactam should be above the MIC of the agent against a pathogen (T > MIC) for a successful outcome varies somewhat between pathogens and β-lactam subclass. For H. influenzae this T > MIC should exceed 30 to 35% of the dosing interval for penicillins, 40 to 50% for cephalosporins and 20 to 25% for carbapenems. Thus, the free concentrations of an agent, based on standard dosing regimens, achieved for the required proportion of the dosing interval can be determined and used as the susceptibility limit or pharmacodynamic breakpoint for time-dependent agents. Most agents other than β-lactams are concentration dependent, with the ratio of non-protein-bound plasma area under the concentration-time curve (AUC) value over 24 h (AUC24) to MICs (AUC24/MIC ratio) correlated with in vivo efficacy. Target AUC24/MIC ratios required for clinical success vary between drug classes, and are around 30 for quinolones, macrolides, and azolides, 20 for tigecycline, and 80 for linezolid (3). Susceptibility breakpoints for concentration-dependent agents can therefore be calculated by dividing AUC24 values by the target applicable to the agent. Susceptibility breakpoints for time- and concentration-dependent agents based on plasma PK are generally applicable to extracellular infections at body sites where extracellular fluid levels are directly related to non-protein-bound plasma levels, such as the respiratory tract and dermis, but not to sites such as the central nervous system, where drug penetration may be limited (5, 30). This approach has been used very successfully for selecting susceptibility breakpoints for pneumococcal infections and for new agents but has not, as yet, been systematically applied to other species or older agents (30, 44-46, 73). As discussed above, breakpoints should be based on PK/PD parameters and appropriate clinical studies and should be the same (or at least similar) for all species associated with each clinical syndrome, e.g., pneumonia, meningitis, cystitis, otitis, etc. Many breakpoints were developed before these principles, particularly PK/PD, were introduced, and some breakpoints in clinical use have been shown not to be appropriate, particularly for oral agents. This is especially the case for H. influenzae, as noted earlier. To overcome this problem, breakpoints based on PK/PD parameters, and, where available, adequate clinical studies have been developed and will be used in this review to enable meaningful use of the terms clinical susceptibility and clinical resistance (4, 65).
Comparison of CLSI (27), BSAC (87) and PK/PD (30, 63) breakpoints applicable to oral and parenteral agents relevant to H. influenzae are given in Table 1. Susceptibility of worldwide isolates of H. influenzae to 23 antimicrobials is shown in Table 2.
TABLE 1.
Breakpoints used to determine susceptible, intermediate, and resistant categories for H. influenzae based on PK/PD, BSAC, and CLSI interpretative breakpoints
| Antimicrobial | Breakpoint (μg/ml) based ona:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| PK/PD
|
BSAC
|
CLSI
|
||||||
| S | R | S | I | R | S | I | R | |
| Parenteral agents | ||||||||
| Ampicillin | ≤2 | ≥4 | ≤1 | - | ≥2 | ≤1 | 2 | ≥4b |
| Ampicillin-sulbactam | ≤2 | ≥4 | - | - | - | ≤2 | - | ≥2 |
| Piperacillin-tazobactam | ≤8 | ≥16 | - | - | - | ≤1 | - | ≥2 |
| Cefuroxime sodium | ≤4 | ≥4 | ≤1 | - | ≥2 | ≤4 | 8 | ≥16 |
| Cefotaxime | ≤2 | ≥4 | ≤1 | - | ≥2 | ≤2 | - | - |
| Ceftriaxone | ≤2 | ≥4 | ≤1 | - | ≥2 | ≤2 | - | - |
| Cefepime | ≤4 | ≥8 | - | - | - | ≤2 | - | - |
| Ceftazidime | ≤8 | ≥16 | ≤2 | - | ≥4 | ≤2 | - | - |
| Meropenem | ≤4 | ≥8 | ≤4 | - | ≥8 | ≤0.5 | - | - |
| Imipenem | ≤4 | ≥8 | ≤4 | - | ≥8 | ≤4 | - | - |
| Ertapenem | ≤1 | ≥2 | ≤2 | - | ≥4 | ≤0.5 | - | - |
| Parenteral and oral agents | ||||||||
| Erythromycin | ≤0.25 | ≥0.5 | ≤0.5 | 1-8 | ≥16 | - | - | - |
| Clarithromycin | ≤0.25 | ≥0.5 | ≤0.5 | 1-16 | ≥32 | ≤8 | 16 | ≥32 |
| Azithromycin | ≤0.12 | ≥0.25 | ≤0.25 | 0.5-4 | ≥8 | ≤4 | - | - |
| Doxycycline | ≤0.25 | ≥0.5 | - | - | - | - | - | - |
| Trimethoprim-sulfamethoxazolee | ≤0.5/9.5 | ≥1/19 | ≤1.6/30.4 | - | ≥32/60.8 | ≤0.5/9.5 | 1/19-2/38 | ≥4/76 |
| Ciprofloxacin | ≤1 | ≥2 | ≤0.5 | - | ≥1 | ≤1 | - | - |
| Ofloxacin | ≤2 | ≥4 | ≤0.5 | - | ≥1 | ≤2 | - | - |
| Gemifloxacin | ≤0.25 | ≥0.5 | ≤0.25 | - | ≥0.5 | - | - | - |
| Levofloxacin | ≤2 | ≥4 | ≤1 | - | ≥2 | ≤2 | - | - |
| Gatifloxacin | ≤1 | ≥2 | ≤1 | - | ≥2 | ≤1 | - | - |
| Moxifloxacin | ≤1 | ≥2 | ≤0.5 | - | ≥1 | ≤1 | - | - |
| Rifampin | ND | ND | - | - | - | ≤1 | 2 | ≥4 |
| Chloramphenicol | ≤4 | ≥8 | ≤2 | - | ≥4 | ≤2 | 4 | ≥8 |
| Oral agents | ||||||||
| Amoxicillin (1.5 g/day; 45 mg/kg/day) | ≤2 | ≥4 | ≤1 | - | ≥2 | - | - | - |
| Amoxicillin (3-4 g/day; 90 mg/kg/day) | ≤4c | ≥8c | - | - | - | - | - | - |
| Amoxicillin-clavulanate (1.5 g-250 mg/day; 45-6.4 mg/kg/day) | ≤2c | ≥4c | ≤1 | - | ≥2 | ≤4 | - | ≥8d |
| Amoxicillin-clavulanate (4 g-6.4 mg/day; 45 mg/kg/day) | ≤4c | ≥8c | - | - | - | - | - | - |
| Cefaclor | ≤0.5 | ≥1 | ≤1 | - | ≥2 | ≤8 | 16 | ≥32 |
| Cefuroxime axetil | ≤1 | ≥2 | ≤1 | - | ≥2 | ≤4 | 8 | ≥16 |
| Cefixime | ≤1 | ≥2 | - | - | - | ≤1 | - | - |
| Cefprozil | ≤1 | ≥2 | - | - | - | ≤8 | 16 | ≥32 |
| Cefdinir | ≤0.5 | ≥1 | - | - | - | ≤1 | - | - |
| Cefpodoxime | ≤0.5 | ≥1 | - | - | - | ≤2 | - | - |
| Telithromycin | ≤0.5f | ≥1f | ≤0.5 | 1-2 | ≥4 | ≤4 | 8 | ≥16 |
| Tetracycline | ≤2g | ≥4g | ≤1 | - | ≥2 | ≤2 | 4 | ≥8 |
S, susceptible; I, intermediate; R, resistant; ND, not defined; -, no breakpoint available.
CLSI breakpoint used to define BLNAR isolates.
Breakpoints are expressed as amoxicillin component; testing was performed using a 2:1 ratio of amoxicillin/clavulanic acid.
Breakpoint used to defined BLPACR isolates.
Breakpoints are expressed as trimethoprim component; testing was performed using a 1:19 ratio of trimethoprim/sulfamethoxazole.
Limited information is currently available to determine PK/PD breakpoints.
TABLE 2.
Susceptibility of worldwide isolates of H. influenzae (n = 8,523) to 23 antimicrobials and MIC50s and MIC90sa
| Antimicrobial | MIC50 (μg/ml) | MIC90 (μg/ml) | % S by PK/PD | CLSI
|
|
|---|---|---|---|---|---|
| % S | % R | ||||
| Ampicillin | 0.25 | >16 | NA | 81.9 | 17.0 |
| Amoxicillin | 0.5 | >16 | 81.6 | 83.2 | 16.8 |
| Amoxicillin-clavulanate, lower dose | 0.5 | 1 | 98.1 | 99.6 | 0.4 |
| Amoxicillin-clavulanate, higher dose | 0.5 | 1 | 99.6 | NA | NA |
| Cefaclor | 4 | 16 | 1.4 | 89.7 | 3.6 |
| Cefuroxime axetil | 1 | 2 | 83.6 | 98.1 | 0.7 |
| Cefixime | 0.03 | 0.06 | 99.8 | 99.8 | NA |
| Ceftriaxone | ≤0.004 | 0.008 | 100 | 100 | NA |
| Cefprozil | 2 | 8 | 22.3 | 92.5 | 2.6 |
| Cefdinir | 0.25 | 0.5 | 92.0 | 97.6 | NA |
| Erythromycin | 4 | 8 | <0.5 | NA | NA |
| Clarithromycin | 8 | 16 | <0.3 | 79.6 | 0.9 |
| Azithromycin | 1 | 2 | <1.2 | 99.5 | NA |
| Chloramphenicol | 0.5 | 1 | 98.1 | 97.9 | 1.9 |
| Doxycycline | 0.5 | 1 | 28.9 | NA | NA |
| Trimethoprim-sulfamethoxazole | 0.12 | >4 | 78.3 | 78.3 | 17.0 |
| Ciprofloxacin | 0.015 | 0.03 | 99.9 | 99.9 | NA |
| Ofloxacin | 0.03 | 0.06 | 99.9 | 99.9 | NA |
| Gemifloxacin | 0.004 | 0.015 | 99.9 | NA | NA |
| Levofloxacin | 0.015 | 0.015 | 99.9 | 99.9 | NA |
| Gatifloxacin | 0.008 | 0.015 | 99.9 | 99.9 | NA |
| Moxifloxacin | 0.015 | 0.03 | 99.8 | 99.8 | NA |
Data are from the Alexander Project 1998 to 2000 (64). S, susceptible; R, resistant; NA, not available.
MECHANISMS OF RESISTANCE
Relevant genes and gene products associated with selected resistance mechanisms of H. influenzae and discussed in this review are summarized in Table 3.
TABLE 3.
Genes and gene products for selected H. influenzae resistance mechanismsa
| Gene | Gene product and description |
|---|---|
| blaTEM | TEM type β-lactamase; can be specified, as in blaTEM-1, to signify the gene associated with a specific TEM β-lactamase, such as TEM-1 |
| blaROB-1 | ROB-1 β-lactamase (although variants of this gene and enzyme have not been described, it is usual to use the suffix-1 with the gene and enzyme) |
| ftsI | PBP3 (see the text for discussion on PBP3A and PBPB) |
| dacA | PBP5 |
| dacB | PBP4 |
| acrA/acrB | AcrAB efflux pump |
| acrR | AcrR (repressor for AcrAB efflux pump) |
| tet(B) | Tet(B) tetracycline efflux protein |
| tet(M), tet(K) | Tet(M) and Tet(K) ribosomal protection proteins (reported only from non-H. influenzae Haemophilus spp.) |
| cat | Chloramphenicol acetyltransferase |
β-Lactams
In general, the phenotypically expressed susceptibility or resistance to β-lactam antibiotics in an organism is brought about by a complex interaction between the permeability of the bacterial cell to the antibiotic, the presence, spectrum of activity, and level of production of β-lactamases, the presence and efficiency of efflux mechanisms, and the affinity of the antibiotic to the penicillin-binding protein (PBP) target site (67). In contrast to other gram-negative bacilli, particularly the Enterobacteriaceae, the outer membrane of H. influenzae provides very little resistance to the penetration of β-lactams into the cell, so the β-lactam MICs of both susceptible and resistant strains are comparatively lower (28, 95).
In H. influenzae, the resistance to ampicillin and other β-lactam antibiotics is generally limited to either the production of a β-lactamase or, in the case of β-lactamase-negative ampicillin-resistant (BLNAR) strains, the presence of altered PBPs with lowered affinity for β-lactams. A very small proportion of strains possess both mechanisms and are referred to as β-lactamase-positive amoxicillin-clavulanate-resistant (BLPACR) strains (50, 59, 62, 94). Neither lowered cell permeability nor efflux systems have been demonstrated as mechanisms of resistance to β-lactams in H. influenzae (25, 126), although there has been a single report of efflux activity increasing the level of resistance that was due primarily to altered PBPs (72).
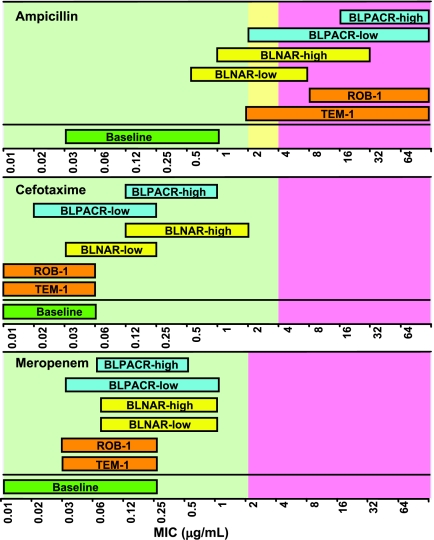
Susceptibility to all β-lactams in H. influenzae is generally predicted by susceptibility to ampicillin as defined by the CLSI MIC breakpoints, which are as follows: ≤1 μg/ml, susceptible; 2 μg/ml, intermediate; and ≥4 μg/ml, resistant (27). However, while some authors use the ampicillin-resistant breakpoint and absence of β-lactamase to define BLNAR strains, others include ampicillin-intermediate strains as BLNAR strains. Additionally, there is no clear definition of BLPACR strains, but most authors use the resistant amoxicillin-clavulanate breakpoint of ≥8 μg/ml (amoxicillin component) to define these strains. A summary of the levels of β-lactam resistance associated with various resistance mechanisms is given in Table 4, and an overview of the degree of overlap of MIC ranges is given in Fig. 1.
TABLE 4.
MIC distributions for H. influenzae and β-lactam resistance mechanismsa
| Drug and strain type | n | MIC (μg/ml)
|
||
|---|---|---|---|---|
| 50% | 90% | Range | ||
| Ampicillin | ||||
| BLNAS | 155 | 0.25 | 0.5 | 0.031-0.5 |
| BLPAR (TEM-1) | 68 | 8 | 16 | 2-64 |
| L-BLNAR | 189 | 1 | 2 | 0.5-2 |
| BLNAR | 138 | 2 | 4 | 1-16 |
| BLPACR-I | 59 | 16 | 32 | 2-64 |
| BLPACR-II | 12 | 32 | 64 | 16-64 |
| Cefotaxime | ||||
| BLNAS | 0.0016 | 0.0031 | 0.004-0.063 | |
| BLPAR (TEM-1) | 0.0016 | 0.0031 | 0.008-0.063 | |
| L-BLNAR | 0.063 | 0.125 | 0.031-0.25 | |
| BLNAR | 0.5 | 1 | 0.125-2 | |
| BLPACR-I | 0.063 | 0.125 | 0.016-0.25 | |
| BLPACR-II | 0.5 | 1 | 0.125-1 | |
| Meropenem | ||||
| BLNAS | 0.031 | 0.063 | 0.008-0.125 | |
| BLPAR (TEM-1) | 0.063 | 0.063 | 0.031-0.063 | |
| L-BLNAR | 0.125 | 0.25 | 0.031-0.5 | |
| BLNAR | 0.125 | 0.25 | 0.031-0.5 | |
| BLPACR-I | 0.125 | 0.25 | 0.031-0.5 | |
| BLPACR-II | 0.125 | 0.25 | 0.063-0.25 | |
Data are from Hasegawa et al. (57). BLNAS, β-lactamase negative ampicillin susceptible; BLPAR, β-lactamase positive ampicillin resistant; L-BLNAR, low-level BLNAR; BLPACR-I, β-lactamase positive amoxicillin-clavulanate resistant (from L-BLNAR); BLPACR-II, β-lactamase positive amoxicillin-clavulanate resistant (from BLNAR).
FIG. 1.
Correlation between β-lactam resistance mechanisms and susceptibility of H. influenzae to ampicillin, cefotaxime, and meropenem. Data were adapted from Hasegawa et al. (57) and Sanbongi et al. (125). The figure was created by M. Jacobs.
Resistance to ampicillin in H. influenzae emerged in the early 1970s and has increased steadily since that time. Resistance rates show enormous variation according to geographical area, and although it is beyond the scope of this work to extensively review this, some general trends are evident. The production of β-lactamase is the most common mechanism of β-lactam resistance in almost all areas, with BLNAR strains being relatively uncommon. One international surveillance study of almost 3,000 strains from 1999 to 2000 showed an overall prevalence of 16.6% β-lactamase-positive strains, ranging from as low as 3% in Germany to as high as 65% in South Korea, and an overall prevalence of only 0.07% for BLNAR strains (59). In some countries, the prevalence of β-lactamase-positive strains has begun to decline, with Canada down from 32% in 1993 to 19% in 2000, Spain down from 28% in 1998 to 16% in 2000, the United States down from 36% in 1994 to 26% in 2002, and Japan down from approximately 25% in 1995 to 3% in 1999 (55, 58, 62, 64).
While the prevalence of BLNAR strains remains low globally, it is probably underestimated due to a range of methodological problems, including a lack of a consensus-defining breakpoint, technical performance (9), and the lack of susceptibility testing beyond a β-lactamase test in some laboratories. However, there are some countries where these strains are relatively common. One study found a prevalence of approximately 10% BLNAR strains in Europe among almost 1,000 isolates from 1997 to 2000, including approximately 20% in Poland and the United Kingdom (50). This study may have overestimated the prevalence, as amoxicillin (MIC ≥ 2 μg/ml) rather than ampicillin MICs were used to categorize strains as BLNAR. Most striking is the situation in Japan, where almost 40% of isolates are reported to be BLNAR strains (55) and over 50% (plus an additional 10% with a combination of altered PBPs and β-lactamase) of Hib strains causing meningitis are BLNAR (56, 57). The incidence of BLNAR strains in France has also been reported to be higher than that reported elsewhere with BLPACR strains also reported with some frequency (31, 32).
β-Lactamase mediated.
β-lactamase-mediated ampicillin resistance in H. influenzae was first detected in the early 1970s, and the β-lactamase was identified as a TEM type in 1975 (95). In 1981, Rubin et al. reported the presence of a novel β-lactamase in H. influenzae, later called ROB-1 (122). Both are plasmid-mediated class A serine β-lactamases with similar substrate profiles that confer resistance to ampicillin and are effectively inhibited by β-lactamase inhibitors such as clavulanate (19, 48, 74, 101). In clinical isolates, β-lactamase-mediated ampicillin resistance is usually easily detected, as strains positive for either enzyme exhibit ampicillin MICs well above the resistant breakpoint, with MIC90s of ≥32 μg/ml, and both enzymes are positive for nitrocefin hydrolysis (48).
It is necessary to use molecular methods to determine which of the two β-lactamases is present in a given isolate, and although isoelectric focusing has been used traditionally because of clear differences in isoelectric point (ROB-1 = 8.1, TEM-1 = 5.4) (122), PCR-based methods are now available (48, 55, 74, 131, 144). Some studies have differentiated β-lactamase-positive isolates by enzyme type, with Farrell et al. finding a prevalence of 93.7% TEM-1 and 4.6% ROB-1 in a global survey of over 2,000 β-lactamase-positive strains, which is similar to previous findings (48, 74, 131). Significantly, the prevalence of ROB-1 among β-lactamase-positive isolates varied greatly according to geographical area, with highest being found in Mexico (30%) followed by the United States (13.2%) and Canada (9.4%) (48). Strains positive for both TEM-1 and ROB-1 are rare, with only one reported in that study (48).
(i) TEM β-lactamases.
(a) Background. The plasmid-mediated TEM-1 and TEM-2 β-lactamases are common among gram-negative bacteria, particularly Enterobacteriaceae, and are encoded by blaTEM genes. The enzymes have essentially identical substrate profiles even though TEM-2 has a Gln-to-Lys amino acid substitution at position 37 due to a single base pair difference in the structural region of the blaTEM-2 gene (C317A numbering according to Sutcliffe) (53). Another single base pair difference (C32T) in the regulatory region of the blaTEM-2 gene results in stronger overlapping promoters (Pa/Pb) that produce larger amounts of the enzyme than the P3 promoter of blaTEM-1 (23). The blaTEM genes are known to be transposable, with blaTEM-2 associated with transposon 1 (Tn1) and blaTEM-1 associated with a range of transposons including Tn2 and Tn3 (80). Extensive variation in blaTEM genes in the Enterobacteriaceae has been described over the last 20 years in association with extended-spectrum β-lactamases (ESBLs) or inhibitor-resistant β-lactamases (IRTs) and also in the promoter region (14, 77, 106).
(b) Origin and evolution of TEM in H. influenzae. Extensive investigation of H. influenzae plasmids indicates that the blaTEM in H. influenzae was transposed on Tn2 or Tn3 from Enterobacteriaceae onto cryptic plasmids already present in Haemophilus species (16, 23, 78, 100). Most of the evidence indicates that the TEM β-lactamase in H. influenzae is TEM-1 (131, 145), and early work suggested that it was transcribed exclusively from the Pa/Pb promoter normally associated with TEM-2 (23). It was hypothesized that because of the highly permeable outer membrane of H. influenzae to β-lactam antibiotics, the C32T mutation in blaTEM-1 (P3 to Pa/Pb promoters) was necessary to increase the amount of enzyme produced and confer ampicillin resistance (23). In fact, the need for this mutation following acquisition of the blaTEM-1 gene has been suggested as a contributing factor behind the delayed emergence of TEM-mediated ampicillin resistance in H. influenzae following the earlier emergence in the Enterobacteriaceae (23). Further studies on blaTEM genes in H. influenzae have revealed two other promoter variants (101, 144, 145). Some strains have a 135-bp deletion from bp G23 through to C157 inclusive and a G162T substitution, which together create a theoretically stronger promoter composed of parts of both the Pa/Pb and P3 promoter (101, 145). This has been designated Pdel (for deletion) (145). Other strains have a 54-bp insertion that consists of a repeat of bp 145T to 198A inclusive, inserted immediately after bp 198 to theoretically create an additional promoter to the existing P3. This promoter variant has been designated Prpt (for repeat) (144). There is no direct evidence that the proposed Pdel and Prpt promoters are actually the site of initiation of transcription in H. influenzae, or that they result in the production of different amounts of β-lactamase compared to the Pa/Pb promoter. One of the studies in which the blaTEM-1 with the Pdel promoter was detected found that these strains had higher median and range of MICs to cefaclor and loracarbef than strains with blaTEM-1 without a deletion in the promoter region, which is consistent with increased gene expression (101). The predictions regarding the increased efficiencies of the proposed promoters are based on the known consensus sequences for E. coli (155) and previous evidence that H. influenzae RNA polymerase recognizes and transcribes from similar or identical promoters to the Enterobacteriaceae (23). In Enterobacteriaceae, blaTEM-1 is only very rarely associated with a Pa/Pb promoter (154), Pdel has only occasionally been described in association with TEM type ESBLs (7), and Prpt had not previously been described. In the single published survey of promoter types of blaTEM in H. influenzae, approximately 60% were found to be Pa/Pb, 20% were Pdel, and 20% were Prpt (144). This difference in distribution of promoter types in the blaTEM genes has been interpreted as an indication that selective pressures have influenced the evolution of the genes differently in H. influenzae than in the Enterobacteriaceae (144). Further work is required to clarify the role of the promoter region in the evolution and function of the blaTEM genes in H. influenzae.
(c) TEM-bearing plasmids. There are two distinctly different groups of TEM-specifying plasmids in H. influenzae. The small (<10 kb) nonconjugative plasmids tend to carry blaTEM as the only resistance determinant, whereas the larger (approximately 40 kb) conjugative plasmids often carry other resistance genes such as those for chloramphenicol, tetracycline, or kanamycin resistance (37, 43, 78, 128). The larger plasmids are more common and appear to be more important in the dissemination of blaTEM genes in H. influenzae (79, 128).
Early work on H. influenzae was hampered by a failure to detect plasmid DNA in most ampicillin-resistant isolates. However, the ampicillin resistance was able to be transferred by conjugation to susceptible recipients and plasmid DNA readily detected in transconjugants (103, 137). This indicated that the plasmid was chromosomally integrated in wild strains and excised prior to conjugal transfer, where it replicated extrachromosomally for some time before integration (40, 41, 103, 137).
Further work on these large conjugative plasmids revealed that the core sequences of plasmids from geographically diverse isolates were highly homologous. This indicated that the evolution of these resistance plasmids probably occurred via transposition of resistance elements onto cryptic plasmids already present in H. influenzae (16, 78). This was supported by the observation that the G+C content of these resistance plasmids was the same as the H. influenzae chromosome, suggesting that the core of the plasmid was indigenous (43, 128). In a systematic search for cryptic plasmids, Laufs et al. found plasmid DNA in only 1 of 699 (0.14%) susceptible isolates examined (78).
The major advance in the work on the large H. influenzae plasmids came with the discovery that the integration of the plasmids into the chromosome was site specific. The site of plasmid excision was identified, and PCR primers were designed that could detect and differentiate both integrated and excised plasmids in H. influenzae irrespective of whether or not they carried genes for antibiotic resistance (79).
A survey of antibiotic resistance and plasmid content in a collection comprising nasopharyngeal Haemophilus spp. from volunteers and a separate collection of 541 Hib isolates provided some interesting results (79). In the nasopharyngeal carrier group, 59% were ampicillin-resistant, β-lactamase-producing Haemophilus spp., all of which had plasmid sequences detected by PCR, but of which only 17% were H. influenzae, with the remainder being Haemophilus parainfluenzae. This provides further evidence that H. parainfluenzae carries the same family of large plasmids as H. influenzae (79) and supports the previous suggestion of Dabernat et al. and Scheifele et al. that H. parainfluenzae might be an important reservoir of antibiotic resistance plasmids for H. influenzae (32, 130). In the Hib isolates examined in the study, 10 (2.4%) of 410 fully susceptible β-lactamase-negative strains were PCR positive for large plasmid sequences, indicating the presence of cryptic plasmids (79). This is much higher than Laufs et al. found in the original search for cryptic plasmids because the techniques used initially would only detect extra-chromosomal plasmids, whereas the PCR method also detects chromosomally integrated plasmids (78, 79).
(d) Potential for TEM type ESBL and IRT enzymes. The emergence of BLNAR strains of H. influenzae is testament to the selective pressure of widespread β-lactam use, so it is surprising that TEM type ESBLs have not been detected in H. influenzae (51, 59) This is especially so given the demonstrated capacity of blaTEM to mutate in H. influenzae as evidenced by the degree of promoter variation apparently unique to blaTEM in this organism. There has been some speculation that ESBLs may have emerged in H. influenzae but gone undetected because of problems with methodology and interpretive criteria (143). A study where cloned ESBLs TEM-3, TEM-4, and TEM-5 were expressed in recombinant strains of H. influenzae Rd (Rd is a transformable rough derivative of a capsular type d strain commonly used for recombinant work) revealed that although the cefotaxime MICs were raised significantly (up to 0.5 μg/ml compared to 0.03 μg/ml for a control TEM-1-producing strain), they would not be categorized as resistant based on CLSI criteria (143). Furthermore, the strains would be unlikely to be detected by the adjacent positioning of amoxicillin-clavulanate and cephalosporin disks on a disk diffusion susceptibility test because of significant overlap in inhibition zones that may mask the zone enhancement by clavulanate (143). Further cloning experiments showed that if ESBLs were to emerge in strains of H. influenzae with altered PBPs, then the effect of the ESBLs would be enhanced (13). For example, the cefotaxime MIC of a recombinant strain of H. influenzae Rd expressing a cloned TEM-4 was 0.5 μg/ml, that of a strain expressing the ftsI gene from a BLNAR strain was 1.0 μg/ml, and that of a strain expressing a cloned TEM-1 and the ftsI from a BLNAR strain was 1.0 μg/ml, and when expressing a cloned TEM-4 and the BLNAR ftsI gene, the cefotaxime MIC was increased to 8.0 μg/ml (13).
In 2002, Pitout et al. reported two isolates of H. parainfluenzae from two patients in South Africa that produced a TEM-15 ESBL and had cefotaxime MICs of >16 μg/ml. This is the first report of either an ESBL or cefotaxime resistance in a Haemophilus spp., and the finding by pulsed-field gel typing that the 2 isolates were unique suggests that the ESBL gene was transferred from one strain to the other (116). This is significant because H. parainfluenzae and H. influenzae are known to exchange β-lactamase-containing plasmids (16, 130), and the emergence of ESBLs in H. influenzae would have serious implications for the use of cephalosporins in the treatment of infections caused by H. influenzae.
The question as to why TEM type IRT enzymes have not yet been reported in H. influenzae, especially since amoxicillin-clavulanate is so widely used to treat infections with β-lactamase-positive strains has been posed (22, 106). It is possible that they have emerged and gone undetected, and without access to molecular methods, a routine laboratory could easily interpret the susceptibility profile of an IRT producing H. influenzae as a BLNAR or BLPACR strain.
(ii) ROB β-lactamase.
(a) Origin. The ROB-1 β-lactamase was first described in 1981 in a clinical isolate of Hib causing meningitis (132) and has since been detected in a number of related organisms, including human isolates of Pasteurella multocida and Haemophilus ducreyi, and animal isolates of P. multocida, Pasteurella haemolytica, Pasteurella aerogenes, and Actinobacillus pleuropneumoniae (85, 86, 88, 96, 121, 131). In all but one isolate where the ROB-1 gene was apparently chromosomal, the gene was located on small plasmids ranging from 4.1 to 5.0 kb. The plasmids appeared to be closely related on the basis of sequence homology and indicate a common origin (85, 86, 88, 96, 131). These observations support the original suggestion by Medeiros et al. that animal pathogens might represent a reservoir from which this β-lactamase gene might spread to human pathogens (96).
There is no published data on the nature of the plasmids in the rare strains of H. influenzae isolates that express both TEM-1 and ROB-1, although almost 50 isolates of H. ducreyi from Thailand were found to carry two small plasmids that individually carried the TEM-1 and ROB-1 genes (88).
The nucleotide sequence of the ROB-1 gene and deduced amino acid sequence indicate that it is a class A enzyme but that it is more closely related to the β-lactamases of gram-positive bacteria than gram-negative bacteria (71, 85).
The observation of a chromosomal ROB-1 gene in a P. aerogenes isolate suggests that the gene might be on a transposable element. Moreover, the presence of small cryptic plasmids in many animal strains of P. multocida suggest that the small ROB-1-specifying plasmids might have arisen from insertion of a transposable element into these cryptic plasmids (85). It could be speculated that the transposable element came from a gram-positive source.
(b) ROB-1 and nitrocefin hydrolysis. There appears to be some conflicting information in the literature regarding the sensitivity of nitrocefin hydrolysis in detecting the ROB-1 β-lactamase. When ROB-1 was first described, the presence of a β-lactamase was initially missed because the nitrocefin hydrolysis test was negative. The organism was found to be ampicillin resistant, and the nitrocefin test was positive when repeated with a very heavy inoculum (132). Bell et al. also isolated a strain of ROB-1-positive H. influenzae that was only weakly positive for nitrocefin hydrolysis and determined that the rate of nitrocefin hydrolysis was approximately 13% that of a TEM-1-producing strain (11). Other authors also warn of the difficulty of detecting ROB-1 β-lactamase using nitrocefin hydrolysis (70, 83). Most of the problems with nitrocefin hydrolysis appear to be from earlier work, and more recent studies using both molecular methods and nitrocefin hydrolysis to examine relatively large numbers of strains make no reference of difficulties with nitrocefin hydrolysis (48, 74, 101, 102, 131).
(c) Association of ROB-1 with increased cefaclor MICs. There is a distinct association between the presence of ROB-1 and higher cefaclor MICs in H. influenzae, although the presence of ROB-1 in an individual strain is not predictive of higher cefaclor MICs in the same way that the presence of either ROB-1 or TEM-1 is predictive of ampicillin resistance (Table 5). In a study of 324 β-lactamase-positive isolates from Canada, 80% of the strains producing ROB-1 were cefaclor resistant based on CLSI breakpoints, with the remaining 20% being cefaclor intermediate, and of the 39 strains that were cefaclor resistant, 67% were ROB-1 positive (74). However, in a global survey of 2,225 β-lactamase positive isolates from the PROTEKT study (1999 to 2003), although the ROB-1 positive subpopulation clearly showed increased cefaclor MIC parameters and resistance compared to TEM-1 isolates, 30% of the ROB-1 isolates were still cefaclor susceptible (48).
TABLE 5.
MIC data for β-lactamase-negative and TEM-1- and ROB-1-producing H. influenzae strainsa
| Drug | MIC result (μg/ml) for strain type:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| TEM-1
|
ROB-1
|
β-Lactamase negative
|
|||||||
| 50% | 90% | Range | 50% | 90% | Range | 50% | 90% | Range | |
| Ampicillin | ≥32 | ≥32 | 2-≥32 | ≥32 | ≥32 | 8-≥32 | 0.25 | 0.50 | 0.12-≥32 |
| Amoxicillin-clavulanate | 1 | 2 | 0.12-≥8 | 0.5 | 1.0 | 0.25-2.0 | 0.5 | 1 | 0.12-≥8 |
| Cefaclor | 4 | 16 | 0.5-≥128 | 16 | 64 | 2.0-≥128 | 2 | 8 | 0.5-≥128 |
| Cefpodoxime | 0.06 | 0.12 | ≤0.015-2.0 | 0.06 | 0.06 | ≤0.015-0.25 | 0.06 | 0.12 | ≤0.015-4 |
Data are from Farrell et al. (48).
In Canada, the rates of resistance to cefaclor, based on CLSI breakpoints, have risen from <3% in 1991 to >13% in 1997 to 1998, yet the prevalence of β-lactamase-positive strains and the proportion of TEM-1- to ROB-1-producing strains remained approximately the same (74). Explanations proposed for this rise in cefaclor MICs include mutations in the blaROB-1 or blaTEM-1 genes, producing β-lactamases with enhanced activity against cefaclor, or alterations in PBPs that augment the action of the β-lactamase, especially ROB-1 (74). The possibility of “extended-spectrum” ROB-1 mutants has been explored by Galán et al. The study was based on the premise that high cefaclor use might enrich H. influenzae populations harboring cefaclor-hydrolyzing ROB-1 β-lactamase and lead to an increase in the use of extended-spectrum cephalosporins or β-lactams plus β-lactamase inhibitors. ROB-1 mutants that produced resistance to extended-spectrum cephalosporins or β-lactams plus β-lactamase inhibitors were generated in vitro, but in all cases, the additional spectrum of activity came at a cost of decreased ability to hydrolyze cefaclor. This suggests that the selective pressure of cefaclor use might prevent the evolution of ROB-1 mutants with activity against extended-spectrum β-lactams or β-lactamase inhibitors (51).
The higher prevalence of ROB-1 among β-lactamase-producing strains in the United States and Mexico relative to other countries might be explained by the high levels of consumption of cefaclor in those countries (51). However, the consumption of cephalosporins, including cefaclor, is high in Japan, but the prevalence of ROB-1 strains among β-lactamase-positive strains is low (48, 55), which suggests that there must be other factors involved in high localized ROB-1 prevalence.
(iii) Novel β-lactamases.
In a global survey of β-lactamase types in H. influenzae, Farrell et al. found that just over 1% of the strains that were positive for nitrocefin hydrolysis were negative for both TEM-1 and ROB-1 genes by PCR, which indicates a previously unrecognized β-lactamase. There were no obvious differences between the susceptibility profiles of isolates with this unknown β-lactamase and those isolates with either TEM-1 or ROB-1 (48). A small number of similar strains have also been reported from a Spanish study (101). To date, these β-lactamases have not been identified or characterized.
In a U.S. surveillance study, a single strain out of 406 β-lactamase-positive isolates was found to be ampicillin susceptible with an MIC of 0.5 μg/ml. The isolates were characterized by nitrocefin hydrolysis, and the β-lactamase was not further identified (75). This may represent a β-lactamase not previously described for H. influenzae or a variant of ROB-1 or TEM-1 with decreased activity or level of expression.
BLNAR-altered PBPs. (i) Defining BLNAR strains.
It is difficult to precisely define what constitutes a BLNAR strain. In the strictest sense, the strain must be β-lactamase negative and have an ampicillin MIC above the CLSI resistant breakpoint of >4.0 μg/ml (27). Strains with an MIC of 2.0 μg/ml present difficulties, as they fall between the CLSI susceptible and resistant breakpoints and are interpreted as intermediate, but they are often included in the BLNAR category because they are not susceptible (9). Interpretation of MICs that cluster around the breakpoints is always difficult given the methodological variation of plus or minus one doubling dilution on repeat testing but more so with H. influenzae because of a range of organism-specific methodological problems (9, 66, 131). In addition, there is no international consensus on ampicillin breakpoints: for example, BSAC has a resistance breakpoint of ≥2.0 μg/ml and the Australian Calibrated Dichotomous Sensitivity method has a resistance breakpoint of ≥1.0 μg/ml (10, 87).
As the mechanism of non-β-lactamase-mediated resistance has been clarified, the situation has become more complicated, as it is now possible to categorize strains as BLNAR based on the presence of the resistance mechanisms and not just on the basis of a raised ampicillin MIC. Based on studies by a number of groups and the use of PCR and sequencing to detect specific mutations in the ftsI gene, strains can be categorized as low BLNAR or BLNAR (also referred to as high BLNAR) based on ftsI gene mutations and associated PBP 3 substitutions. Low-BLNAR strains usually have ampicillin MICs in the range from 0.5 to 2.0 μg/ml, and BLNAR strains have ampicillin MICs in the range from 1.0 to 16 μg/ml (Table 4) (55, 56, 148).
The lack of a consensus definition and the broad range of ampicillin MICs associated with BLNAR strains is clearly demonstrated by the range of ampicillin breakpoints used in various surveillance studies to screen for them: Doern et al. (42) used ≥4.0 μg/ml, Ubukata et al. (149) used ≥1.0 μg/ml, Dabernat et al. (31) used ≥1.0 μg/ml, Jacobs et al. (64) used ≥4.0 μg/ml, Fluit et al. (50) used ≥2.0 μg/ml (amoxicillin and not ampicillin in this instance), and Kubota et al. (76) used ≥2.0 μg/ml.
BLNAR strains show reduced susceptibility not only to ampicillin but also to other β-lactam antibiotics, particularly cephalosporins. Livermore et al. suggest that cefaclor resistance is a better indicator of a BLNAR strain than ampicillin resistance (84), and James et al. used cefuroxime resistance (MIC > 4.0 μg/ml) to screen for BLNAR strains (68). The CLSI actually recommends that BLNAR strains also be considered resistant to amoxicillin-clavulanic acid, ampicillin-sulbactam, cefaclor, cefetamet, cefonicid, cefprozil, cefuroxime, and loracarbef despite apparent in vitro susceptibility of some strains to these antibiotics (27).
(ii) PBPs in H. influenzae.
H. influenzae has eight PBPs that were initially named PBP1 to PBP8 in order of decreasing molecular weight (89). These proteins were subsequently referred to as PBPs IA, IB, 2, 3A, 3B, 4, 5 and 6, respectively, based on the observation that the binding capacities of the various proteins for different β-lactams was similar to PBPs in Escherichia coli (97, 103). The use of the original nomenclature in some of the early literature on BLNAR and PBPs, where what are now known as PBP3A and PBP3B were referred to as PBPs 4 and 5 (97, 132), has created some confusion, as has the fact that when cells in the stationary phase of growth are used for PBP binding assays, PBP3A and PBP4 are not always detected (97).
PBP3 is encoded by the ftsI gene and is a relatively large protein that consists of a short cytoplasmic domain, a membrane spanning segment, and a periplasmic domain that has transpeptidase activity and is involved in the synthesis of peptidoglycan in the septum of a dividing cell (septal peptidoglycan synthesis) (90, 152).
Most penicillin binding enzymes, including PBPs, have three highly conserved amino acid motifs which are essential for function (110). In H. influenzae PBP3, these motifs are S327-T-V-K, S379-S-N, and K513-T-G (149).
(iii) Evidence for role of PBP3A and PBP3B.
The first report of ampicillin resistance in β-lactamase-negative strains of H. influenzae was in 1974 when Thornsberry and Kirven serendipitously discovered two strains while evaluating a rapid test for β-lactamase (142). In 1980, Markowitz reported an ampicillin-resistant Hib isolate from a case of endocarditis and meningitis in which the presence of β-lactamase could not be demonstrated despite the use of numerous methods and attempts at induction. The mechanism of ampicillin resistance was not determined, but an alteration to the target site was proposed as an attractive hypothesis (92).
In 1984, Parr and Bryan were able to use genomic DNA from a BLNAR Hib clinical isolate to transform a susceptible H. influenzae Rd strain to a BLNAR phenotype and then demonstrate reduced binding affinity of PBP3A and PBP3B in both the donor and transformed recipient relative to the susceptible parent Rd strain (111). Thus, alterations in PBP3A and PBP3B were established as the mechanism behind BLNAR. These findings were subsequently confirmed by other groups using their own BLNAR strains (25, 90, 97).
The next progression in elucidating the mechanism behind BLNAR was the observation that there appeared to be differences in levels of resistance among different isolates. Clairoux et al. examined a collection of Canadian isolates and placed them into 3 distinct groups based on ampicillin MICs of 0.5 to 1, 2 to 4, and ≥8 μg/ml for groups I, II, and III, respectively (25). When genomic DNA from strains representative of each group was used to transform H. influenzae Rd, transformants showed the same level of resistance to ampicillin and decreased binding affinity of PBP3A and PBP3B as the donors. Furthermore, a thorough examination of outer membrane proteins, porin function, and lipopolysaccharide profiles in the donor clinical strains and the relevant transformants could not explain the differences in levels of ampicillin resistance. The authors concluded that the level of resistance in BLNAR strains might be related to the extent of PBP modification. Extensive cell filamentation was demonstrated in a transformant from one of the group III donors, indicating a high degree of septal dysfunction, which is consistent with alterations in PBP3 (25).
In 2001, Ubukata et al. analyzed the PBP profiles of 25 Japanese BLNAR strains and found that the binding affinity of PBP3A was decreased in all strains and those for PBP3B and PBP4 decreased in some strains (149). They were able to search the published whole-genome sequence of H. influenzae Rd and determine the nucleotide sequences of the ftsI and dacB genes, which encode PBP3 and PBP4, respectively. Using this information, it was possible to sequence the ftsI and dacB genes of the wild strains. The mutations in the ftsI gene, but not the dacB gene, correlated with ampicillin resistance. Straker et al. sequenced the ftsI, dacB, and dacA (encoding PBP 5) genes of a collection of cefuroxime-resistant isolates and found no evidence of association between dacA or dacB mutations and cefuroxime resistance but found evidence supporting the role of ftsI mutations, which were confirmed by transformation studies (136).
When Ubukata et al. used the PCR-amplified 2.2-kb ftsI open reading frame from BLNAR strains to transform H. influenzae Rd, transformants had raised β-lactam MICs consistent with the donors (149). This represented an important step, as previous transformation studies had used genomic DNA, and it was therefore difficult to attribute changes in resistance in transformants solely to PBP3, as other genes may have been transferred during transformation. Notably, both PBP3A and PBP3B affinities were simultaneously decreased in transformants carrying the single ftsI open reading frame. This was interpreted as PBP3A and PBP3B either being different forms of the same protein both transcribed from ftsI or evidence of some posttranslational processing of PBP3A, both of which occur with some of the PBPs of E. coli. (149).
The sequence of the ftsI gene that encodes the transpeptidase region of PBP3 can be used to deduce the amino acid sequences for BLNAR strains and compare them with susceptible control strains. In the strains originally described by Ubukata et al., many of these substitutions appear to be in the vicinity of the conserved PBP3 motifs (149). Using the previously reported three-dimensional crystallographic structure of PBP2X in S. pneumoniae as a guide, it was possible to construct models of PBP3 from BLNAR strains, and it was found that the substitutions were near the active site pocket surrounded by the conserved motifs. This modeling was taken further by Straker et al. in the examination of cefuroxime-resistant strains from the United Kingdom (136). They used computer modeling, again based on the three-dimensional structure of S. pneumoniae PBP2X, to examine how the S357N substitution, common in both their strains and the strains of Ubukata et al. (149), would affect the interaction of cefuroxime with PBP3. It was postulated that the asparagine side chain produced steric hindrance with the adjacent valine at position 362, which would alter the orientation of the V362-P366 loop. Since this loop is adjacent to the active site of the PBP, a change in orientation may limit access to certain β-lactams (136).
(iv) Range of amino acid substitutions.
The amino acid substitutions described in this section and in other studies are summarized in Table 6.
TABLE 6.
Deduced amino acid substitutions in PBP3 for different series of BLNAR strains compared with Rd control
| Strain (n) | Amino acid at positiona:
|
||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 337 | 350 | 352 | 357 | 368 | 377 | 385 | 389 | 437 | 443 | 449 | 490 | 501 | 502 | 511 | 517 | 526 | 528 | 532 | 547 | 562 | 569 | 586 | |
| Rd | A | D | S | S | A | M | S | L | A | T | I | G | R | A | V | R | N | Y | T | V | V | N | A |
| Ubukatab | |||||||||||||||||||||||
| I (9) | N | N | I | T | S | H | S | ||||||||||||||||
| II (12) | N | N | V | K | S | ||||||||||||||||||
| III (4) | N | N | I | T | F | K | L | S | |||||||||||||||
| Straker (14)c | V | N | N | T | I | A | L/H | T/V | A | H | K | H | S | ||||||||||
| Kubota (20)d | N | F | N | I | T | F | A | K | I | L | S | S | |||||||||||
| Dabernate | |||||||||||||||||||||||
| I (7) | N | N | T | H | |||||||||||||||||||
| IIa (5) | K | ||||||||||||||||||||||
| IIb (56) | N | N | L | S | E | V | K | ||||||||||||||||
| IIc (25) | N | T | T | K | |||||||||||||||||||
| IId (15) | V | K | |||||||||||||||||||||
| Hasegawaf | |||||||||||||||||||||||
| I (9) | H | ||||||||||||||||||||||
| II (2) | I | T | F | H | |||||||||||||||||||
| III (7) | T | K | |||||||||||||||||||||
| IV (1) | I | T | K | ||||||||||||||||||||
| V (1) | T | F | K | ||||||||||||||||||||
| VI (35) | I | T | F | K | |||||||||||||||||||
| Fluit (28)g | N | N | T | I | S | V | E | T/V | H | K | |||||||||||||
The BLNAR strains from Ubukata et al. were placed into 3 groups (I, II, and III) based on the presence of different amino acid substitutions in the transpeptidase region of PBP3. Almost all isolates had aspartic acid at position 350 replaced with asparagine (D350N) and S357N; in addition, group I strains had R517H, group II strains had N526K, and group III strains had N526K, M377I, S385T, and L389F. Some group I strains also had M377I and/or S385T (149).
The PBP3 substitutions of a large collection of 108 BLNAR clinical isolates from France were determined, and some significant differences were found compared to the substitutions found by Ubukata et al. (31, 149). None of the French strains had the triple substitution at positions 377, 385, and 389 near the conserved SSN motif that characterized the Japanese group III strains. In addition, all but one of the Japanese strains had a D350N substitution and most had the S357N substitution, whereas only 63 of 108 French strains had D350N, only 3 had S357N, and only a single strain had both substitutions. There were a total of 18 different substitution patterns in the French strains, and they were divided into groups I and II, on the basis of R517H and N526K substitutions, respectively, in an attempt to follow the Ubukata groupings. Other substitutions were also present in both groups, and the group II strains were further divided into subgroups a, b, c, and d to account for the diversity of other substitutions present (31). It was postulated that the different substitution patterns in the strains from the two studies might be related to different antibiotic-prescribing habits in the two countries (31). A large European study found large variation in PBP3 mutations in the BLNAR strains studied but found that certain mutation profiles were country specific (69). Whether this was due to clonal spread or the effect of different prescribing habits was not determined (69).
It is difficult to directly compare the deduced substitutions in the strains analyzed by Straker et al. because these isolates were selected for study on the basis of cefuroxime resistance (MIC ≥ 2 μg/ml, BSAC breakpoint) (87) rather than ampicillin resistance, and this might bias toward substitutions involved in cephalosporin resistance (136). The substitutions A337V, T443A, R501L, V511A, Y528H, and T532S found in these strains were previously unreported. The most common substitutions were S357N, A502T or V, and N526K, all of which are present in the Ubukata strains (149). The substitutions D350N, A502T, and N526K, which are known to be associated with ampicillin resistance, were not associated with cefuroxime resistance, although they may contribute to cefuroxime resistance when present with other substitutions. Interestingly, it might have been expected that the substitutions M377I, S385T, and L389F, which distinguish Ubukata group III BLNAR strains from groups I and II and produce the highest cephalosporin MICs, would be common in the cefuroxime-resistant strains. This was not the case, as only one of the isolates had the M377I substitution and none had an S385T or L389F substitution (136).
Other studies with Japanese isolates have found that the substitutions present do not follow the pattern described by Ubukata. Hasegawa et al. looked at the substitutions around the SSN and KTG motifs in 55 BLNAR strains of H. influenzae type b from cases of meningitis. They found the same substitutions as Ubukata et al. (149), but in different patterns, and assigned them into 6 subgroups (I to VI) (56). In a further study in 2006, Hasegawa et al. again found the same range of substitutions at positions 377, 385, 389, 502, 517, and 526 but divided them into 9 subgroups (57). Kubota et al. found BLNAR strains with substitutions consistent with Ubukata groups II and III, but no strains with the R517H substitution of group I, and single strains each with an individual S352F or V511A not described for the Ubukata strains (76).
In addition to the substitutions mentioned above or in Table 6, substitutions have been found in another 38 positions between position 7 and 603 in Japanese BLNAR strains where more extensive ftsI gene sequencing was undertaken, although no evidence was presented that they were associated with resistance. Of those additional substitutions, A239E was found in 33 of the 36 isolates examined (108).
In summary, substitutions have been described in association with BLNAR for 24 different amino acids in the transpeptidase domain of PBP3. Interestingly, even though the strains described in this review cover the years 1988 to 2004 and are of diverse geographical origin, the substitutions at any given amino acid position are consistent between studies: apparently only specific substitutions at each position are associated with resistance. There is no single substitution that is present in all of the BLNAR strains; however, N526K, R517H, M377I, S357N, and D350N occur frequently. The number of substitutions in a single BLNAR isolate can range from a single substitution (31, 149) to as many as 11 (76).
(v) Correlates of β-lactam resistance.
Although the strains are referred to as BLNAR, it is important to note that decreased susceptibility, and in some cases resistance, to other β-lactam antibiotics, especially cephalosporins, also occurs. In the Ubukata group III strains, the MICs increased approximately 32-fold (0.125 to 4.0 μg/ml) for ampicillin and 16-fold for meropenem (0.03 to 0.5 μg/ml), whereas the cefotaxime MICs increased approximately 250-fold (0.008 to 2.0 μg/ml) compared to wild-type susceptible strains (149). Similar observations have been made in other studies; for example, that by Hasegawa et al., where the MIC50 for BLNAR strains compared to ampicillin-susceptible strains showed only an increase of 4-fold for meropenem, 8-fold for ampicillin, and 32-fold for cefotaxime (55-57).
When the binding affinities for the different H. influenzae PBPs (in a susceptible strain with unmodified PBPs) and various β-lactam antibiotics were examined, cefotaxime and other cephalosporins had the highest affinity for PBP3A and PBP3B, whereas ampicillin had a higher affinity for PBP1A and PBP4 (149). In fact, it is the high affinity of cephalosporins for PBP3 that is central to their good activity against this organism, so under the selective pressure of cephalosporin use, mutations in ftsI are selected (57). It has been proposed that the much higher rates of BLNAR strains found in Japan than in Europe and the United States is a consequence of different prescribing habits (31, 55, 57). In the United States and Europe, amoxicillin-clavulanate has been recommended as a first-line therapy for community-acquired respiratory type infections, whereas in Japan, oral cephalosporins, particularly cefdinir and cefditoren, are widely used (55). Moreover, the latter two drugs are often used in relatively low doses, which promotes partial microbial damage without bactericidal activity and might allow emergence of surviving mutants (55). One study has shown that amoxicillin-clavulanate was unable to select resistant mutants in H. influenzae in vitro, but cefprozil was, and that the cefprozil-resistant mutants had the S357N substitution described for many clinical BLNAR strains (26).
At the level of individual bacterial strains, it is difficult to draw conclusions about the relative effects of particular substitutions in PBP3 and the resistance of the isolate because of the range of different substitutions and combinations thereof. The N526K and the R517H appear to be central to phenotypic resistance, and all of the clinical BLNAR strains characterized so far have one of those substitutions, although no strains have both.
Looking primarily at amino acid positions 377, 385, 389, 502, 517, and 526, strains can be characterized as low BLNAR if they only have the R517H or N526K substitution and BLNAR if they also have any of the M377I, S385T, L389F, or A502V substitutions. Low-BLNAR strains tend to have lower levels of resistance than BLNAR strains, with MIC90 of ampicillin of 2.0 μg/ml and 8.0 μg/ml and of cefotaxime of 0.25 μg/ml and 1.0 μg/ml, respectively (55, 57). The three substitutions around the SSN motif seem particularly important for resistance to cephem antibiotics, and the Ubukata strains with substitutions at these three positions are the ones with the highest cefotaxime MICs (149).
An attempt to clarify the relationship between various PBP3 substitutions and resistance was made by Osaki et al. using site-directed mutagenesis to examine single and multiple amino acid substitutions at positions 377, 385, 389, 517, and 526. This study indicated that the addition of S385T or S385T and L389F mutations to a background of either N526K or R517H further increased the level of cephalosporin resistance. The addition of an M377I substitution to various backgrounds did not influence the level of resistance (108).
In a large study of European isolates, Fluit et al. found two BLNAR strains (ampicillin MICs of 8 μg/ml and negative nitrocefin hydrolysis) with no amino acid substitutions in the transpeptidase region of PBP3, which suggests a different mechanism of resistance, perhaps involving PBPs other than PBP3 (50).
Given the diversity of substitutions that have been described, it seems likely that strains will continue to emerge with either new substitutions or new combinations that might produce greater levels of resistance than those currently seen.
(vi) Epidemiology of BLNAR.
With Streptococcus pneumoniae, the clonal spread of penicillin-resistant clones is important, and a very limited number of clones are responsible for a large proportion of disease worldwide. This indicates that the clones are stable and is even more notable given that there is great genetic diversity in penicillin-susceptible S. pneumoniae isolates (39).
In contrast, many studies have shown that BLNAR strains of H. influenzae are genotypically diverse with a general absence of clonal spread (50, 52, 76, 98). Dabernat and coworkers looked at 108 BLNAR strains and found 70 different pulsed-field gel electrophoresis (PFGE) profiles, which is consistent with the general level of heterogeneity of susceptible nonencapsulated strains (31, 123). Given that H. influenzae has an efficient system for uptake of homologous DNA, it may be possible for the mutated ftsI gene to be transferred from BLNAR strains to susceptible strains if patients are colonized and/or infected with more than one strain (25). This issue has not been explored, although Dabernat et al. examined the sequence phylogeny of the ftsI gene in the French isolates and did not find the typical mosaic structure that would typify widespread “clonal” transfer of the gene (31).
There have been some instances of limited spread of specific BLNAR clones, including within cystic fibrosis patients (31), between outpatients attending a single hospital (75), and between parents, children, and siblings (151).
The issue of clonal spread of BLNAR in Hib strains requires some discussion. Hasegawa et al. studied Hib isolates from Japanese patients with meningitis. Of the 51 BLNAR strains subjected to PFGE, 36 belonged to 1 of 2 PFGE profiles (56). The conclusion that this represents the spread of a specific strain should be considered in the light of the limited genetic diversity of Hib isolates in general (76, 135) and the fact that there were 6 different PBP3 substitution patterns within the strains described (56).
BLPACR.
The term BLPACR is a phenotypic description but does not readily portray that the mechanism of resistance is typically a combination of both β-lactamase production and the presence of altered PBP3, as seen in isolation in BLNAR strains. This is because when these strains were first described, and the term BLPACR first used (42), BLNAR strains were very uncommon and the mechanism of resistance not fully characterized or appreciated (42). As a result, when the BLPACR strains were detected during a routine surveillance study of over 1,500 clinical H. influenzae isolates, they were primarily considered β-lactamase-positive isolates that were also resistant to amoxicillin-clavulanate. At that time, it was suggested that the amoxicillin-clavulanate resistance might be due to hyperproduction of TEM-1 or ROB-1 β-lactamase, production of an inhibitor-resistant TEM, ROB, or novel β-lactamase, or elaboration of altered PBPs superimposed on typical TEM-1 or ROB-1 β-lactamase (42). It is now accepted that strains with altered PBP3 that would ordinarily be categorized as BLNAR are categorized as BLPACR if they also express a β-lactamase (31, 149). As contradictory as it might sound, β-lactamase-enhanced BLNAR might be a more useful term for these strains.
A number of studies have shown that when BLPACR strains are compared with BLNAR strains with identical PBP3 amino acid substitutions, the BLPACR strains have much higher ampicillin MICs than the BLNAR strains, typically a range of 2.0 to 64 μg/ml compared to 0.5 to 16 μg/ml, but the MICs of amoxicillin-clavulanate and a range of cephalosporins are essentially the same (31, 57, 149), indicating that the contribution of the β-lactamase to resistance is limited to ampicillin. This was confirmed by Matic et al. when a susceptible strain of H. influenzae was transformed with amplified ftsI genes from 2 BLPACR strains, and the only significant difference in β-lactam susceptibility of the BLPACR strains compared to the transformants was a much higher ampicillin MIC (94).
Problems with detection and different methodologies make comparisons of the incidence of BLPACR strains in different countries difficult to make; indeed, assessing their overall incidence is difficult. Some studies screen isolates with molecular methods to detect mutations in the ftsI gene and the presence of β-lactamase genes; other studies screen using MICs of ampicillin and amoxicillin-clavulanate and nitrocefin hydrolysis. In collections of 395 meningitis isolates between 1999 and 2002 and 621 meningitis isolates between 2000 and 2004 in Japan, Hasegawa et al. found approximately 11% BLPACR strains during both periods in addition to 45% and 56% BLNAR strains in these respective periods (56, 57). Another Japanese study of 150 nasopharyngeal isolates only found 1.3% BLPACR strains (124). The study by Dabernat et al. on 108 French isolates selected on the basis of predetermined reduced susceptibility to β-lactams found 14% BLPACR strains (31) and Karlowsky et al. (75) found only 0.15% of 1,434 isolates from the United States to be BLPACR strains. Only 6 BLPACR strains were found among 8,523 isolates (0.07%) in the Alexander Project during the period 1998 to 2000 (64).
The clinical significance of BLPACR strains is unclear, as many of them show only a slight decrease in amoxicillin-clavulanate susceptibility (0.5 to 2 μg/ml versus 0.25 μg/ml for fully susceptible strains) (31) and are therefore not classified as resistant based on the CLSI breakpoints for amoxicillin-clavulanate of S ≤ 4/2 and R ≥ 8/4 μg/ml (27). In a study of 160 clinical isolates, Kubota et al. found 27 BLNAR strains and 2 BLPACR strains using phenotypic screening, and 9 of 24 β-lactamase-positive isolates had reduced susceptibility but not resistance to amoxicillin-clavulanate (76): some of these 9 latter strains may have been categorized as BLPACR had PBP3 been characterized. On this basis, it seems that BLPACR strains are possibly underreported. This may be a problem because of the CLSI recommendation that a number of other antibiotics should be reported as resistant despite apparent in vitro sensitivity in BLNAR strains (strains with altered PBP3) (27). As is the case with S. pneumoniae, application of PK/PD breakpoints is likely to be useful in predicting the clinical significance of strains of H. influenzae with altered PBPs.
Efflux pump-mediated β-lactam resistance.
Most BLNAR strains have ampicillin MICs of 1 to 4 μg/ml, although some isolates have been reported with MICs in the range 4 to 16 μg/ml. Kaczmarek et al. studied BLNAR strains from North America with such high ampicillin MICs with the aim of determining the mechanism behind the increased resistance. Sequencing of the genes encoding PBP1A, PBP1B, or PBP2 or analysis of outer membrane proteins did not reveal abnormalities that could account for the increased resistance. The mutations in ftsI were consistent with BLNAR strains in Ubukata groups I and II, and when H. influenzae Rd was transformed with PCR-amplified ftsI genes from these clinical strains, the ampicillin MICs were only in the range of 1 to 4 μg/ml, indicating that the ftsI mutations alone could not account for the high ampicillin MICs (72).
The acrR gene, which regulates (represses) the AcrAB efflux pump was sequenced for the strains with the high ampicillin MICs and compared to that of BLNAR strains with ampicillin MICs of <4 μg/ml and with ampicillin-susceptible strains. All of the high-level ampicillin-resistant strains but none of the other strains had single nucleotide insertions in the acrR gene that predicted early termination of the reading frame. The effect that the loss of the AcrR repressor was demonstrated when an acrB knockout was made of one of the high-level resistant clinical isolates and the ampicillin MIC was markedly reduced. Furthermore, when H. influenzae Rd was transformed with the mutant acrR genes from the clinical isolates, the ampicillin MICs of the transformants were significantly increased as a result of disruption of the regulation of the efflux pump (72).
The AcrAB efflux pump is known to play an important role in the efflux of macrolide antibiotics from H. influenzae, but the effect of any efflux of β-lactam antibiotics is thought to be negligible, as it is countered by the rapid influx of these relatively small molecules through the porin channels (127). It is now apparent that, in combination with altered PBPs, the loss of regulation of the efflux pump can contribute to ampicillin resistance (72). It is impossible to say to what extent this loss of regulation contributes to β-lactam resistance, as the acrR gene has not been sequenced in other studies. However, a number of studies have reported H. influenzae Rd transformants with either genomic DNA or PCR amplified ftsI from clinical BLNAR isolates, where the ampicillin MICs of transformants are significantly less than that of the donor strain (25, 72, 108). This could be a result of an undetected acrR gene mutation. Finally, given the importance of AcrAB in producing elevated MICs to the macrolides and ketolides, the use of these antibiotics might select for strains overexpressing AcrAB which would augment any resistance to β-lactam antibiotics (72).
Macrolides, Azalides, and Ketolides
Resistance mechanisms include efflux pumps, either intrinsic or acquired, ribosomal methylase, and alterations in ribosomal proteins and RNA (139, 140). H. influenzae is intrinsically resistant to macrolide-lincosamide-streptogramin B agents and ketolides, which is associated with the presence of an efflux pump homologous (but not identical to) to the acrAB efflux mechanism in E. coli or other efflux pumps and explains the limited activity of these agents against wild-type strains of this pathogen (12, 113, 114, 126, 127). Occasional strains of H. influenzae lack this efflux pump and have lower MICs than wild-type strains, while a few strains have higher MICs associated with L4 and L22 ribosomal protein and 23S rRNA mutations (12, 114). The clinical significance of macrolide efflux in H. influenzae is the subject of much debate, as MICs are relatively low, e.g., ranging from 0.13 to 4 μg/ml for azithromycin, with MIC50 and MIC90 of 1 and 2 μg/ml, respectively. Bacteriologic outcome of otitis media caused by H. influenzae has clearly been shown to be no different from that expected with a placebo (29, 33). A recent study of the clinical significance of macrolide resistance in pneumococcal bacteremia showed that macrolide failures were significantly more common among cases of pneumococcal bacteremia with isolates exhibiting an erythromycin MIC of ≥1 μg/ml than among isolates exhibiting erythromycin MICs of ≤0.25 μg/ml and that the likelihood of macrolide failure was not significantly different between isolates with erythromycin MICs of 1 to 16 μg/ml, associated with efflux-mediated resistance (mefA), and isolates with erythromycin MICs between 1 and >64 μg/ml, associated with ribosomal methylase-mediated resistance (ermB) (34). The macrolides administered to these patients were predominantly clarithromycin and azithromycin, and MICs of these agents against pneumococci are similar to those of erythromycin, allowing extrapolation to H. influenzae, whereas most azithromycin MICs are between 0.5 and 2 μg/ml. While macrolides do accumulate in host cells such as macrophages, the evidence discussed suggests that this does not appear to provide any benefit in infections due to extracellular pathogens such as H. influenzae and macrolide-resistant pneumococci (60, 61).
Tetracyclines
Tetracyclines exert an antimicrobial effect by binding to the 30S subunit of bacterial ribosomes and preventing tRNA from binding to the A or P sites (24). Tetracycline resistance in H. influenzae (and H. parainfluenzae) is associated with an efflux mechanism encoded by the tet(B) gene which is usually located on conjugative plasmids (24). Plasmids carrying ampicillin-chloramphenicol-tetracycline-kanamycin resistance genes have been described for H. influenzae type b isolates in Belgium, Spain, and Cuba (20, 81). The tet(M) and tet(K) genes, which produce tetracycline resistance via production of ribosomal protection proteins rather than efflux, have been found in H. ducreyi and H. aphrophilus, respectively (24, 118). The tet(M) gene in H. ducreyi was plasmid mediated and could be transferred by conjugation in vitro to H. influenzae, leading to the suggestion that tet(M) and possibly other tetracycline resistance determinants might be introduced into H. influenzae (24, 118).
Quinolones
The quinolones have a broad spectrum of activity and exert an antimicrobial effect by interfering with DNA replication and, subsequently, bacterial reproduction. Two enzymes that are important in the replication process are DNA gyrase and topoisomerase IV (150). Resistance to quinolones among strains of H. influenzae, while currently rare, occurs via alterations in the quinolone resistance-determining region of the genes encoding DNA gyrase or topoisomerase IV (35, 105, 120). The newer quinolones are potent against H. influenzae, and the prevalence of resistance among clinical strains is low (62). However, spontaneous quinolone-resistant mutants are readily selected in vitro by exposure to quinolones, and this has resulted in development of considerable resistance to this drug class in other species (35, 36). Quinolones are not currently approved for pediatric use due to concerns about toxicity, and use of quinolones in children is likely to provide the selective pressure needed to create a resistance problem in H. influenzae and S. pneumoniae (129).
Chloramphenicol
Chloramphenicol resistance in H. influenzae is usually associated with plasmid-mediated production of chloramphenicol acetyltransferase (CAT) encoded by the cat gene, with occasional strains having a penetration barrier (18, 119). The cat gene is carried on conjugative plasmids ranging in molecular weight from 34 × 106 to 46 × 106, and these plasmids often carry genes encoding resistance to tetracycline and ampicillin as well. These conjugative plasmids can also be incorporated into the chromosome (117). The CAT enzyme produced resembles the type II CATs produced by Enterobacteriaceae. Resistance associated with a permeability barrier is due to the loss of an outer membrane protein (18).
Folic Acid Metabolism Inhibitors
Trimethoprim and sulfamethoxazole (used alone or in combination) exert an antimicrobial effect by interfering with cellular metabolism and replication by sequentially blocking the production of tetrahydrofolate. During normal cellular metabolism, dihydrofolate is reduced to tetrahydrofolate by the enzyme dihydrofolate reductase (DHFR) (17). Tetrahydrofolate is an important cofactor in many cellular reactions, supplying single carbon moieties for the production of thymidylate, purine nucleotides, methionine, serine, glycine, and other compounds (54). Inhibiting the production of tetrahydrofolate prevents thymine synthesis and, hence, prevents DNA replication (141). Trimethoprim is a substrate analog of dihydrofolate and blocks the reduction of dihydrofolate to tetrahydrofolate by DHFR, whereas sulfamethoxazole is a substrate analog of para-aminobenzoic acid, which is involved in the production of dihydropteroate, a precursor compound of dihydrofolate, blocking the enzyme dihydropteroate synthetase (17). Thus, the use of these compounds in combination limits the production of dihydrofolate and prevents the conversion from dihydrofolate to tetrahydrofolate. Both compounds, trimethoprim and sulfamethoxazole, selectively inhibit bacterial metabolism with little toxicity to humans because humans do not synthesize folic acid; rather, the necessary levels of folic acid are obtained from dietary sources.
Resistance to trimethoprim occurs via alteration in the affinity between trimethoprim and DHFR. The decreased affinity is the result of altered genes that encode DHFR, which often are carried on plasmids or transposons and probably originated from closely related bacteria. Studies have shown that substitutions in the amino acid sequence of DHFR result in resistance to trimethoprim without affecting the affinity of the natural substrates (1, 93, 115). Resistance to trimethoprim-sulfamethoxazole among strains of H. influenzae is common and is caused by an increase in the production of DHFR with altered affinity for trimethoprim (38).
CLINICAL SIGNIFICANCE OF RESISTANCE
Significant advances have recently been made in understanding the relationships between in vitro susceptibility and in vivo response to infection based on PK/PD correlations (3). In the absence of human studies or to complement limited human data, susceptibility breakpoints can be established based on animal models and PK parameters. Clinically relevant susceptibility breakpoints can then be derived based on applying these PK/PD parameters to standard dosing regimens. For nonmeningeal infections, breakpoints can be derived from non-protein-bound plasma drug levels present for 35 to 50% of the dosing interval for time-dependent agents such as β-lactams, and from non-protein-bound AUC/MIC ratios exceeding target values for concentration-dependent agents such as most non-β-lactam agents. These principles have repeatedly been validated in animal models and in bacteriologic outcome human studies of AOM, AECB, and sinusitis (2-4, 29, 30, 63). Breakpoints for agents recommended for use against H. influenzae based on PK/PD parameters, as well as current CLSI and BSAC breakpoints, are shown in Table 1. While PK/PD and BSAC breakpoints are very similar, many CLSI breakpoints are considerably higher and generally represent microbiological rather than clinical breakpoints. Susceptibility of H. influenzae to agents recommended for treatment of diseases due to these pathogens is shown in Table 2.
The relationships between MIC distributions and susceptibility breakpoints are important, as they determine the clinical activity of agents. MICs of clinically useful agents should be below PK/PD breakpoints, and the greater the difference between MICs and breakpoints, the greater the likelihood that the agent will be successful in clinical use. It is therefore important to examine MIC distributions in relation to PK/PD breakpoints, and several patterns are found with H. influenzae (Fig. 2 and 3) (29, 30, 63).
FIG. 2.
MIC distributions of selected β-lactam antimicrobial agents for H. influenzae. Arrows indicate PK/PD breakpoints. The two arrows shown for amoxicillin and amoxicillin-clavulanate indicate breakpoints applicable to lower (left) and higher (right) dosing regimens. The two arrows shown for cefuroxime indicate breakpoints applicable to oral (left) and parenteral (right) dosing regimens. The figure was created by M. Jacobs.
FIG. 3.
MIC distributions of selected non-β-lactam antimicrobial agents for H. influenzae. Arrows indicate PK/PD breakpoints. The figure was created by M. Jacobs.
A unimodal MIC distribution with modal MIC fourfold (i.e., two doubling dilutions) or more below the breakpoint is the case with cefuroxime (parenteral), amoxicillin-clavulanate, cefixime, cefpodoxime, and the quinolones. These agents are therefore highly active against H. influenzae and are most suitable for empirical use.
A unimodal MIC distribution with breakpoint within the MIC distribution is seen with cefuroxime (oral), cefdinir, cefprozil and doxycycline. These are agents with limited clinical activity and their use should be limited to circumstances where other more suitable agents cannot be used.
A unimodal MIC distribution with breakpoint below the MIC distribution is seen with cefaclor, erythromycin, azithromycin, clarithromycin, and telithromycin. These agents have intrinsic resistance due to pharmacokinetic limitations and have essentially no clinically useful activity against H. influenzae.
A bimodal MIC distribution with a clearly defined susceptible population below the breakpoint and a clearly defined resistant population, typically with defined resistance mechanisms, is the case with the ampicillin- and amoxicillin-resistant populations associated with β-lactamase production, tetracycline-resistant population associated with the tet(B) gene, trimethoprim-sulfamethoxazole-resistant population associated with mutations in DHFR, and chloramphenicol-resistant populations associated with the cat gene (MICs of chloramphenicol-susceptible strains are ≤2 μg/ml, while MICs of resistant strains are ≥8 μg/ml). These agents are suitable for directed use against H. influenzae and for empirical use where resistance is low or the consequences of treatment failure are minor.
Comparison of these PK/PD-based susceptibility interpretations with current recommendations for treatment of diseases associated with H. influenzae reveal the following in relation to diseases associated with this species. Current empirical therapy recommendations for meningitis, vancomycin plus an extended-spectrum cephalosporin such as cefotaxime or ceftriaxone or an extended-spectrum cephalosporin alone if presumptive or definitive pathogen identification as H. influenzae are still valid except in areas where high-level BLNAR and BLPACR strains are prevalent and Hib is not in use, such as Japan, where treatment failures with cefotaxime or ceftriaxone monotherapy have been encountered (57). Suggested therapy for areas where high-level BLNAR and BLPACR strains occur is cefotaxime or ceftriaxone plus meropenem based on no additional loss of affinity of meropenem for PBP3 between low- and high-BLNAR strains (Table 4 and Fig. 1) (57, 125). Alternate therapies recommended for H. influenzae, chloramphenicol, cefepime, and meropenem, appear to be valid except in areas where chloramphenicol-resistant strains are prevalent. Gatifloxacin and moxifloxacin are also recommended as alternative agents for adults, and these are also valid options as virtually all H. influenzae strains that are currently susceptible. Although they are not currently approved for pediatric use, these quinolones should also be considered as alternative agents for H. influenzae meningitis in children. Development of susceptibility breakpoints for H. influenzae applicable to meningitis would be very worthwhile, as many current breakpoints are not applicable to meningitis. The only species where meningitis breakpoints are available for a few agents is S. pneumoniae (27).
Current empirical and directed treatment recommendations for childhood pneumonia and bacteremia are valid except for areas where BLNAR and BLPACR strains are prevalent, where the efficacy of oral amoxicillin and amoxicillin-clavulanate against high-level BLNAR strains may be compromised. The efficacy of parenteral cephalosporins and meropenem may also be compromised, but MICs of high-level BLNAR strains are currently below PK/PD breakpoints (Tables 2 and 4 and Fig. 2) (57, 125). The recommendation for CAP in adults of azithromycin, clarithromycin, or doxycycline for outpatients with no comorbidities and of azithromycin or clarithromycin for outpatients with comorbidities if no antibiotic therapy had been administered in the past 3 months are problematic, as these agents have little if any clinical activity against H. influenzae, and the activity of these agents against macrolide-resistant pneumococci as well as the selective pressure of these agents for development of macrolide resistance in the community are also concerns (61). The remaining recommendations for CAP in adults are valid except for areas where BLNAR H. influenzae are found as discussed earlier, in which case respiratory quinolones and meropenem are suitable agents.
Current recommendations for AOM are valid with the exception again of areas where BLNAR H. influenzae strains are found, where MIC90s of amoxicillin, cefaclor, cefpodoxime, and cefdinir against high-level BLNAR strains are above PK/PD breakpoints. The MIC90 of cefditoren against high-level BLNAR strains is 0.25 μg/ml; however, the PK/PD breakpoint for this agent has not been established but is likely to be lower than the MIC90 (82, 104). Cefixime and cefpodoxime may have clinically useful activity against high-BLNAR strains, but additional information is needed and use of quinolones should be considered (104, 129). Similarly, current recommendations for sinusitis are valid, with the exception of areas where BLNAR H. influenzae strains are found.
Recommendations for AECB for patients with baseline risk factors, amoxicillin-clavulanate, and respiratory quinolones are valid, as H. influenzae is the predominant pathogen and these agents are active based on PK/PD breakpoints. However, of the agents recommended for patients without baseline risk factors, azithromycin, clarithromycin, telithromycin, doxycycline, cefuroxime axetil, cefpodoxime, and cefdinir, only cefpodoxime is active against H. influenzae based on PK/PD breakpoints. The rationale for these latter recommendations of agents that will not be effective for the patient group with a high probability of spontaneous resolution is unclear.
CONCLUSIONS
Haemophilus influenzae is a major pathogen causing significant morbidity and mortality in all parts of the world. Meningitis and bacteremia occur in areas where the Hib vaccine is not used, whereas community-acquired respiratory tract infections caused by nontypeable strains occur worldwide. Although it has previously been assumed that little development of drug resistance has occurred in this species, we have reviewed the evidence showing that this is not the case. Apart from β-lactamase production, BLNAR strains occur, especially in Japan and France, and BLPACR strains have also been described. The clinical utility of antimicrobial agents recommended for treating diseases associated with H. influenzae are reviewed, and the limitations of the macrolide, azalide, and ketolide group of agents, tetracyclines, and selected β-lactams against infections caused by this species are noted. Quinolone resistance, though currently rare, may increase in the future. Also, widespread use of the pediatric pneumococcal conjugated vaccine has led to a relative increase in nontypeable H. influenzae compared to pneumococci in community-acquired respiratory tract infections. For all of these reasons, continued epidemiological monitoring of the susceptibility status of these strains is required. New agents are also needed, especially for penicillin-allergic patients in both pediatric patients with otitis media and older patients with AECB.
ADDENDUM IN PROOF
Recent changes have affected the availability of or indications for two of the agents discussed. Gatifloxacin was removed from worldwide markets in 2006 and is no longer being manufactured. In February 2007, the U.S. Food and Drug Administration removed approval of telithromycin for use in the treatment of acute bacterial sinusitis and acute bacterial exacerbations of chronic bronchitis.
A very recent study by Takahata et al. (Antimicrob. Agents Chemother., in press) has presented evidence of in vivo homologous recombination of ftsI genes from H. haemolyticus to H. influenzae to produce a BLNAR phenotype. This study also demonstrated in vitro transfer of the ftsI gene from BLNAR strains of H. influenzae to β-lactamase-negative ampicillin-susceptible strains of H. influenzae.
REFERENCES
- 1.Adrian, P. V., and K. P. Klugman. 1997. Mutations in the dihydrofolate reductase gene of trimethoprim-resistant isolates of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 41:2406-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambrose, P. G., J. B. Anon, J. S. Owen, S. Van Wart, M. E. McPhee, S. M. Bhavnani, M. Piedmonte, and R. N. Jones. 2004. Use of pharmacodynamic end points in the evaluation of gatifloxacin for the treatment of acute maxillary sinusitis. Clin. Infect. Dis. 38:1513-1520. [DOI] [PubMed] [Google Scholar]
- 3.Ambrose, P. G., S. M. Bhavnani, C. M. Rubino, A. Louie, T. Gumbo, A. Forrest, and G. L. Drusano. 2007. Pharmacokinetics-pharmacodynamics of antimicrobial therapy: it's not just for mice anymore. Clin. Infect. Dis. 44:79-86. [DOI] [PubMed] [Google Scholar]
- 4.American Academy of Pediatrics Subcommittee on Management of Acute Otitis Media. 2004. Diagnosis and management of acute otitis media. Pediatrics 113:1451-1465. [DOI] [PubMed] [Google Scholar]
- 5.Andes, D., J. Anon, M. R. Jacobs, and W. A. Craig. 2004. Application of pharmacokinetics and pharmacodynamics to antimicrobial therapy of respiratory tract infections. Clin. Lab. Med. 24:477-502. [DOI] [PubMed] [Google Scholar]
- 6.Anon, J. B., M. R. Jacobs, M. D. Poole, P. G. Ambrose, M. S. Benninger, J. A. Hadley, and W. A. Craig. 2004. Antimicrobial treatment guidelines for acute bacterial rhinosinusitis. Otolaryngol. Head Neck Surg. 130:1-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arlet, G., S. Goussard, P. Courvalin, and A. Philippon. 1999. Sequences of the genes for the TEM-20, TEM-21, TEM-22, and TEM-29 extended-spectrum beta-lactamases. Antimicrob. Agents Chemother. 43:969-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balter, M. S., J. La Forge, D. E. Low, L. Mandell, and R. F. Grossman. 2003. Canadian guidelines for the management of acute exacerbations of chronic bronchitis. Can. Respir. J. 10(Suppl. B):3B-32B. [DOI] [PubMed] [Google Scholar]
- 9.Barry, A. L., P. C. Fuchs, and S. D. Brown. 2001. Identification of beta-lactamase negative, ampicillin-resistant strains of Haemophilus influenzae with four methods and eight media. Antimicrob. Agents Chemother. 45:1585-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bell, S., B. Gatus, J. Pham, and D. Rafferty. 2002. Antibiotic susceptibility testing by the CDS method: a manual for medical and veterinary laboratories. Arthur Productions Pty. Ltd., Sydney, Australia.
- 11.Bell, S. M., M. J. Hardy, J. N. Pham, A. S. Jimenez, and B. J. Gatus. 1993. Resistance of Haemophilus influenzae type b to ampicillin mediated by ROB-1 beta-lactamase. Med. J. Aust. 159:834. [PubMed] [Google Scholar]
- 12.Bogdanovich, T., B. Bozdogan, and P. C. Appelbaum. 2006. Effect of efflux on telithromycin and macrolide susceptibility in Haemophilus influenzae. Antimicrob. Agents Chemother. 50:893-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bozdogan, B., S. Tristram, and P. C. Appelbaum. 2006. Combination of altered PBPs and expression of cloned extended-spectrum beta-lactamases confers cefotaxime resistance in Haemophilus influenzae. J. Antimicrob. Chemother. 57:747-749. [DOI] [PubMed] [Google Scholar]
- 14.Bradford, P. A. 2001. Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 14:933-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bradley, J. S. 2002. Management of community-acquired pediatric pneumonia in an era of increasing antibiotic resistance and conjugate vaccines. Pediatr. Infect. Dis. J. 21:592-598. [DOI] [PubMed] [Google Scholar]
- 16.Brunton, J., D. Clare, and M. A. Meier. 1986. Molecular epidemiology of antibiotic resistance plasmids of Haemophilus species and Neisseria gonorrhoeae. Rev. Infect. Dis. 8:713-724. [DOI] [PubMed] [Google Scholar]
- 17.Burchall, J. J., and G. H. Hitchings. 1965. Inhibitor binding analysis of dihydrofolate reductases from various species. Mol. Pharmacol. 1:126-136. [PubMed] [Google Scholar]
- 18.Burns, J. L., P. M. Mendelman, J. Levy, T. L. Stull, and A. L. Smith. 1985. A permeability barrier as a mechanism of chloramphenicol resistance in Haemophilus influenzae. Antimicrob. Agents Chemother. 27:46-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bush, K., G. A. Jacoby, and A. A. Medeiros. 1995. A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Campos, J., M. Chanyangam, R. deGroot, A. L. Smith, F. C. Tenover, and R. Reig. 1989. Genetic relatedness of antibiotic resistance determinants in multiply resistant Hemophilus influenzae. J. Infect. Dis. 160:810-817. [DOI] [PubMed] [Google Scholar]
- 21.Casey, J. R., and M. E. Pichichero. 2004. Changes in frequency and pathogens causing acute otitis media in 1995-2003. Pediatr. Infect. Dis. J. 23:824-828. [DOI] [PubMed] [Google Scholar]
- 22.Chaibi, E. B., D. Sirot, G. Paul, and R. Labia. 1999. Inhibitor-resistant TEM beta-lactamases: phenotypic, genetic and biochemical characteristics. J. Antimicrob. Chemother. 43:447-458. [DOI] [PubMed] [Google Scholar]
- 23.Chen, S. T., and R. C. Clowes. 1987. Nucleotide sequence comparisons of plasmids pHD131, pJB1, pFA3, and pFA7 and beta-lactamase expression in Escherichia coli, Haemophilus influenzae, and Neisseria gonorrhoeae. J. Bacteriol. 169:3124-3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chopra, I., and M. Roberts. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology and epidemiology of bacterial resistance. Clin. Microbiol. Rev. 65:232-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clairoux, N., M. Picard, A. Brochu, N. Rousseau, P. Gourde, D. Beauchamp, T. R. Parr, Jr., M. G. Bergeron, and F. Malouin. 1992. Molecular basis of the non-beta-lactamase-mediated resistance to beta-lactam antibiotics in strains of Haemophilus influenzae isolated in Canada. Antimicrob. Agents Chemother. 36:1504-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clark, C., B. Bozdogan, M. Peric, B. Dewasse, M. R. Jacobs, and P. C. Appelbaum. 2002. In vitro selection of resistance in Haemophilus influenzae by amoxicillin-clavulanate, cefpodoxime, cefprozil, azithromycin, and clarithromycin. Antimicrob. Agents Chemother. 46:2956-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial susceptibility testing; 16th informational supplement. M100-S16. CLSI, Wayne, PA.
- 28.Coulton, J. W., P. Mason, and D. Dorrance. 1983. The permeability barrier of Haemophilus influenzae type b against beta-lactam antibiotics. J. Antimicrob. Chemother. 12:436-449. [DOI] [PubMed] [Google Scholar]
- 29.Craig, W. A. 1998. Pharmacokinetic/pharmacodynamic parameters: rationale for antibacterial dosing of mice and men. Clin. Infect. Dis. 26:1-10. [DOI] [PubMed] [Google Scholar]
- 30.Craig, W. A. 2003. Basic pharmacodynamics of antibacterials with clinical applications to the use of beta-lactams, glycopeptides, and linezolid. Infect. Dis. Clin. N. Am. 17:479-501. [DOI] [PubMed] [Google Scholar]
- 31.Dabernat, H., C. Delmas, M. Seguy, R. Pelissier, G. Faucon, S. Bennamani, and C. Pasquier. 2002. Diversity of beta-lactam resistance-conferring amino acid substitutions in penicillin-binding protein 3 of Haemophilus influenzae. Antimicrob. Agents Chemother. 46:2208-2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dabernat, H., R. Bauriaud, C. Delmas, and M. B. Lareng. 1983. Haemophilus colonization in children: serotypes, biotypes and antibiotic sensitivity. Pathol. Biol. (Paris) 31:103-106. [PubMed] [Google Scholar]
- 33.Dagan, R., and E. Leibovitz. 2002. Bacterial eradication in the treatment of otitis media. Lancet Infect. Dis. 2:593-604. [DOI] [PubMed] [Google Scholar]
- 34.Daneman, N., A. McGeer, K. Green, and D. E. Low. 2006. Macrolide resistance in bacteremic pneumococcal disease: implications for patient management. Clin. Infect. Dis. 43:432-438. [DOI] [PubMed] [Google Scholar]
- 35.Davies, T. A., L. M. Kelly, D. B. Hoellman, L. M. Ednie, C. L. Clark, S. Bajaksouzian, M. R. Jacobs, and P. C. Appelbaum. 2000. Activities and postantibiotic effects of gemifloxacin compared to those of 11 other agents against Haemophilus influenzae and Moraxella catarrhalis. Antimicrob. Agents Chemother. 44:633-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davies, T. A., L. M. Kelly, G. A. Pankuch, K. L. Credito, M. R. Jacobs, and P. C. Appelbaum. 2000. Antipneumococcal activities of gemifloxacin compared to those of nine other agents. Antimicrob. Agents Chemother. 44:304-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Graaff, J., L. P. Elwell, and S. Falkow. 1976. Molecular nature of two beta-lactamase-specifying plasmids isolated from Haemophilus influenzae type b. J. Bacteriol. 126:439-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Groot, R., D. O. Chaffin, M. Kuehn, and A. L. Smith. 1991. Trimethoprim resistance in Haemophilus influenzae is due to altered dihydrofolate reductase(s). Biochem. J. 274(Pt 3):657-662. [PMC free article] [PubMed] [Google Scholar]
- 39.De Lencastre, H., and A. Tomasz. 2002. From ecological reservoir to disease: the nasopharynx, day-care centres and drug-resistant clones of Streptococcus pneumoniae. J. Antimicrob. Chemother. 50(Suppl. S2):75-81. [DOI] [PubMed] [Google Scholar]
- 40.Dimopoulou, I. D., J. E. Russell, Z. Mohd-Zain, R. Herbert, and D. W. Crook. 2002. Site-specific recombination with the chromosomal tRNA(Leu) gene by the large conjugative Haemophilus resistance plasmid. Antimicrob. Agents Chemother. 46:1602-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dimopoulou, I. D., W. A. Kraak, E. C. Anderson, W. W. Nichols, M. P. Slack, and D. W. Crook. 1992. Molecular epidemiology of unrelated clusters of multiresistant strains of Haemophilus influenzae. J. Infect. Dis. 165:1069-1075. [DOI] [PubMed] [Google Scholar]
- 42.Doern, G. V., A. B. Brueggemann, G. Pierce, H. P. Holley, Jr., and A. Rauch. 1997. Antibiotic resistance among clinical isolates of Haemophilus influenzae in the United States in 1994 and 1995 and detection of beta-lactamase-positive strains resistant to amoxicillin-clavulanate: results of a national multicenter surveillance study. Antimicrob. Agents Chemother. 41:292-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elwell, L. P., J. De Graaff, D. Seibert, and S. Falkow. 1975. Plasmid-linked ampicillin resistance in Haempohilus influenzae type b. Infect. Immun. 12:404-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.European Committee on Antimicrobial Susceptibility Testing. 2006. EUCAST Technical Note on daptomycin. Clin. Microbiol. Infect 12:599-601. [DOI] [PubMed] [Google Scholar]
- 45.European Committee on Antimicrobial Susceptibility Testing. 2006. EUCAST Technical Note on linezolid. Clin. Microbiol. Infect. 12:1243-1245. [DOI] [PubMed] [Google Scholar]
- 46.European Committee on Antimicrobial Susceptibility Testing. 2006. EUCAST technical note on tigecycline. Clin. Microbiol. Infect 12:1147-1149. [DOI] [PubMed] [Google Scholar]
- 47.European Committee on Antimicrobial Susceptibility Testing. 22 December 2006, accession date. EUCAST procedure for harmonising and defining breakpoints. http://www.srga.org/Eucastwt/bpsetting.htm.
- 48.Farrell, D. J., I. Morrissey, S. Bakker, S. Buckridge, and D. Felmingham. 2005. Global distribution of TEM-1 and ROB-1 {beta}-lactamases in Haemophilus influenzae. J. Antimicrob. Chemother. 56:773-776. [DOI] [PubMed] [Google Scholar]
- 49.Ferraro, M. J. 2001. Should we reevaluate antibiotic breakpoints? Clin. Infect. Dis. 33(Suppl. 3):S227-S229. [DOI] [PubMed] [Google Scholar]
- 50.Fluit, A. C., A. Florijn, J. Verhoef, and D. Milatovic. 2005. Susceptibility of European beta-lactamase-positive and -negative Haemophilus influenzae isolates from the periods 1997/1998 and 2002/2003. J. Antimicrob. Chemother. 56:133-138. [DOI] [PubMed] [Google Scholar]
- 51.Galán, J. C., M. I. Morosini, M. R. Baquero, M. Reig, F. Baquero, J. A. Karlowsky, G. Verma, G. G. Zhanel, and D. J. Hoban. 2003. Haemophilus influenzae bla(ROB-1) mutations in hypermutagenic delta ampC Escherichia coli conferring resistance to cefotaxime and beta-lactamase inhibitors and increased susceptibility to cefaclor. Antimicrob. Agents Chemother. 47:2551-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gazagne, L., C. Delmas, E. Bingen, and H. Dabernat. 1998. Molecular epidemiology of ampicillin-resistant non-beta-lactamase-producing Haemophilus influenzae. J. Clin. Microbiol. 36:3629-3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goussard, S., and P. Courvalin. 1991. Sequences of the genes blat-1B and blat-2. Gene 102:71-73. [DOI] [PubMed] [Google Scholar]
- 54.Hartman, P. G. 1993. Molecular aspects and mechanism of action of dihydrofolate reductase inhibitors. J. Chemother. 5:369-376. [PubMed] [Google Scholar]
- 55.Hasegawa, K., K. Yamamoto, N. Chiba, R. Kobayashi, K. Nagai, M. R. Jacobs, P. C. Appelbaum, K. Sunakawa, and K. Ubukata. 2003. Diversity of ampicillin-resistance genes in Haemophilus influenzae in Japan and the United States. Microb. Drug Resist. 9:39-46. [DOI] [PubMed] [Google Scholar]
- 56.Hasegawa, K., N. Chiba, R. Kobayashi, S. Y. Murayama, S. Iwata, K. Sunakawa, and K. Ubukata. 2004. Rapidly increasing prevalence of beta-lactamase-nonproducing, ampicillin-resistant Haemophilus influenzae type b in patients with meningitis. Antimicrob. Agents Chemother. 48:1509-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hasegawa, K., R. Kobayashi, E. Takada, A. Ono, N. Chiba, M. Morozumi, S. Iwata, K. Sunakawa, and K. Ubukata. 2006. High prevalence of type b {beta}-lactamase-non-producing ampicillin-resistant Haemophilus influenzae in meningitis: the situation in Japan where Hib vaccine has not been introduced. J. Antimicrob. Chemother. 57:1077-1082. [DOI] [PubMed] [Google Scholar]
- 58.Heilmann, K. P., C. L. Rice, A. L. Miller, N. J. Miller, S. E. Beekmann, M. A. Pfaller, S. S. Richter, and G. V. Doern. 2005. Decreasing prevalence of beta-lactamase production among respiratory tract isolates of Haemophilus influenzae in the United States. Antimicrob. Agents Chemother. 49:2561-2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hoban, D., and D. Felmingham. 2002. The PROTEKT surveillance study: antimicrobial susceptibility of Haemophilus influenzae and Moraxella catarrhalis from community-acquired respiratory tract infections. J. Antimicrob. Chemother. 50(Suppl. S1):49-59. [DOI] [PubMed] [Google Scholar]
- 60.Jacobs, M. 2001. Optimisation of antimicrobial therapy using pharmacokinetic and pharmacodynamic parameters. Clin. Microbiol. Infect. 7:589-596. [DOI] [PubMed] [Google Scholar]
- 61.Jacobs, M. R. 2002. In vivo veritas: in vitro macrolide resistance in systemic Streptococcus pneumoniae infections does result in clinical failure. Clin. Infect. Dis. 36:565-569. [DOI] [PubMed] [Google Scholar]
- 62.Jacobs, M. R. 2003. Worldwide trends in antimicrobial resistance among common respiratory tract pathogens in children. Pediatr. Infect. Dis. J. 22:S109-S119. [DOI] [PubMed] [Google Scholar]
- 63.Jacobs, M. R. 2004. Anti-infective pharmacodynamics-maximizing efficacy, minimizing toxicity. Drug Discov. Today 1:505-512. [Google Scholar]
- 64.Jacobs, M. R., D. Felmingham, P. C. Appelbaum, and R. N. Gruneberg. 2003. The Alexander Project 1998-2000: susceptibility of pathogens isolated from community-acquired respiratory tract infection to commonly used antimicrobial agents. J. Antimicrob. Chemother. 52:229-246. [DOI] [PubMed] [Google Scholar]
- 65.Jacobs, M. R., S. Bajaksouzian, A. Windau, C. E. Good, G. Lin, G. A. Pankuch, and P. C. Appelbaum. 2004. Susceptibility of Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis to 17 oral antimicrobial agents based on pharmacodynamic parameters: 1998-2001 U. S. Surveillance Study. Clin. Lab. Med. 24:503-530. [DOI] [PubMed] [Google Scholar]
- 66.Jacobs, M. R., S. Bajaksouzian, A. Windau, P. C. Appelbaum, G. Lin, D. Felmingham, C. Dencer, L. Koeth, M. E. Singer, and C. E. Good. 2002. Effects of various test media on the activities of 21 antimicrobial agents against Haemophilus influenzae. J. Clin. Microbiol. 40:3269-3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.James, P. A., and D. S. Reeves. 1996. Bacterial resistance to cephalosporins as a function of outer membrane permeability and access to their target. J. Chemother. 8(Suppl. 2):37-47. [PubMed] [Google Scholar]
- 68.James, P. A., D. A. Lewis, J. Z. Jordens, J. Cribb, S. J. Dawson, and S. A. Murray. 1996. The incidence and epidemiology of beta-lactam resistance in Haemophilus influenzae. J. Antimicrob. Chemother. 37:737-746. [DOI] [PubMed] [Google Scholar]
- 69.Jansen, W. T., A. Verel, M. Beitsma, J. Verhoef, and D. Milatovic. 2006. Longitudinal European surveillance study of antibiotic resistance of Haemophilus influenzae. J. Antimicrob. Chemother. 58:873-877. [DOI] [PubMed] [Google Scholar]
- 70.Jorgensen, J. H. 1992. Update on mechanisms and prevalence of antimicrobial resistance in Haemophilus influenzae. Clin. Infect. Dis. 14:1119-1123. [DOI] [PubMed] [Google Scholar]
- 71.Juteau, J. M., and R. C. Levesque. 1990. Sequence analysis and evolutionary perspectives of ROB-1 beta-lactamase. Antimicrob. Agents Chemother. 34:1364-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kaczmarek, F. S., T. D. Gootz, F. Dib-Hajj, W. Shang, S. Hallowell, and M. Cronan. 2004. Genetic and molecular characterization of beta-lactamase-negative ampicillin-resistant Haemophilus influenzae with unusually high resistance to ampicillin. Antimicrob. Agents Chemother. 48:1630-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kahlmeter, G., D. F. Brown, F. W. Goldstein, A. P. MacGowan, J. W. Mouton, I. Odenholt, A. Rodloff, C. J. Soussy, M. Steinbakk, F. Soriano, and O. Stetsiouk. 2006. European Committee on Antimicrobial Susceptibility Testing (EUCAST) technical notes on antimicrobial susceptibility testing. Clin. Microbiol. Infect. 12:501-503. [DOI] [PubMed] [Google Scholar]
- 74.Karlowsky, J. A., G. Verma, G. G. Zhanel, and D. J. Hoban. 2000. Presence of ROB-1 beta-lactamase correlates with cefaclor resistance among recent isolates of Haemophilus influenzae. J. Antimicrob. Chemother. 45:871-875. [DOI] [PubMed] [Google Scholar]
- 75.Karlowsky, J. A., I. A. Critchley, R. S. Blosser-Middleton, E. A. Karginova, M. E. Jones, C. Thornsberry, and D. F. Sahm. 2002. Antimicrobial surveillance of Haemophilus influenzae in the United States during 2000-2001 leads to detection of clonal dissemination of a beta-lactamase-negative and ampicillin-resistant strain. J. Clin. Microbiol. 40:1063-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kubota, T., F. Higa, N. Kusano, I. Nakasone, S. Haranage, M. Tateyama, N. Yamane, and J. Fujita. 2006. Genetic analyses of beta-lactamase negative ampicillin-resistant strains of Haemophilus influenzae isolated in Okinawa, Japan. Jpn. J. Infect. Dis. 59:36-41. [PubMed] [Google Scholar]
- 77.Lartigue, M. F., V. Leflon-Guibout, L. Poirel, P. Nordmann, and M. H. Nicolas-Chanoine. 2002. Promoters P3, Pa/Pb, P4, and P5 upstream from blaTEM genes and their relationship to β-lactam resistance. Antimicrob. Agents Chemother. 46:4035-4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Laufs, R., F. C. Riess, G. Jahn, R. Fock, and P. M. Kaulfers. 1981. Origin of Haemophilus influenzae R factors. J. Bacteriol. 147:563-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Leaves, N. I., I. Dimopoulou, I. Hayes, S. Kerridge, T. Falla, O. Secka, R. A. Adegbola, M. P. Slack, T. E. Peto, and D. W. Crook. 2000. Epidemiological studies of large resistance plasmids in Haemophilus. J. Antimicrob. Chemother. 45:599-604. [DOI] [PubMed] [Google Scholar]
- 80.Levesque, R. C., and G. A. Jacoby. 1988. Molecular structure and interrelationships of multiresistance beta-lactamase transposons. Plasmid 19:21-29. [DOI] [PubMed] [Google Scholar]
- 81.Levy, J., G. Verhaegen, P. De Mol, M. Couturier, D. Dekegel, and J. P. Butzler. 1993. Molecular characterization of resistance plasmids in epidemiologically unrelated strains of multiresistant Haemophilus influenzae. J. Infect. Dis. 168:177-187. [DOI] [PubMed] [Google Scholar]
- 82.Liu, P., K. H. Rand, B. Obermann, and H. Derendorf. 2005. Pharmacokinetic-pharmacodynamic modelling of antibacterial activity of cefpodoxime and cefixime in in vitro kinetic models. Int. J. Antimicrob. Agents 25:120-129. [DOI] [PubMed] [Google Scholar]
- 83.Livermore, D. M., and D. F. Brown. 2001. Detection of beta-lactamase-mediated resistance. J. Antimicrob. Chemother. 48(Suppl. 1):59-64. [DOI] [PubMed] [Google Scholar]
- 84.Livermore, D. M., T. G. Winstanley, and K. P. Shannon. 2001. Interpretative reading: recognizing the unusual and inferring resistance mechanisms from resistance phenotypes. J. Antimicrob. Chemother. 48(Suppl. 1):87-102. [DOI] [PubMed] [Google Scholar]
- 85.Livrelli, V., J. Peduzzi, and B. Joly. 1991. Sequence and molecular characterization of the ROB-1 beta-lactamase gene from Pasteurella haemolytica. Antimicrob. Agents Chemother. 35:242-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Livrelli, V. O., A. Darfeuille-Richaud, C. D. Rich, B. H. Joly, and J. L. Martel. 1988. Genetic determinant of the ROB-1 beta-lactamase in bovine and porcine Pasteurella strains. Antimicrob. Agents Chemother. 32:1282-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.MacGowan, A. P., and R. Wise. 2001. Establishing MIC breakpoints and the interpretation of in vitro susceptibility tests. J. Antimicrob. Chemother. 48(Suppl. 1):17-28. [DOI] [PubMed] [Google Scholar]
- 88.Maclean, I. W., L. Slaney, J. M. Juteau, R. C. Levesque, W. L. Albritton, and A. R. Ronald. 1992. Identification of a ROB-1 beta-lactamase in Haemophilus ducreyi. Antimicrob. Agents Chemother. 36:467-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Makover, S. D., R. Wright, and E. Telep. 1981. Penicillin-binding proteins in Haemophilus influenzae. Antimicrob. Agents Chemother. 19:584-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Malouin, F., T. R. Parr, Jr., and L. E. Bryan. 1990. Identification of a group of Haemophilus influenzae penicillin-binding proteins that may have complementary physiological roles. Antimicrob. Agents Chemother. 34:363-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mandell, L. A., J. G. Bartlett, S. F. Dowell, T. M. File, Jr., D. M. Musher, and C. Whitney. 2003. Update of practice guidelines for the management of community-acquired pneumonia in immunocompetent adults. Clin. Infect. Dis. 37:1405-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Markowitz, S. M. 1980. Isolation of an ampicillin-resistant, non-beta-lactamase-producing strain of Haemophilus influenzae. Antimicrob. Agents Chemother. 17:80-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Maskell, J. P., A. M. Sefton, and L. M. Hall. 2001. Multiple mutations modulate the function of dihydrofolate reductase in trimethoprim-resistant Streptococcus pneumoniae. Antimicrob. Agents Chemother. 45:1104-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Matic, V., B. Bozdogan, M. R. Jacobs, K. Ubukata, and P. C. Appelbaum. 2003. Contribution of beta-lactamase and PBP amino acid substitutions to amoxicillin/clavulanate resistance in beta-lactamase-positive, amoxicillin/clavulanate-resistant Haemophilus influenzae. J. Antimicrob. Chemother. 52:1018-1021. [DOI] [PubMed] [Google Scholar]
- 95.Medeiros, A. A., and T. F. O'Brien. 1975. Ampicillin-resistant Haemophilus influenzae type B possessing a TEM-type beta-lactamase but little permeability barrier to ampicillin. Lancet i:716-719. [DOI] [PubMed] [Google Scholar]
- 96.Medeiros, A. A., R. Levesque, and G. A. Jacoby. 1986. An animal source for the ROB-1 beta-lactamase of Haemophilus influenzae type b. Antimicrob. Agents Chemother. 29:212-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mendelman, P. M., D. O. Chaffin, and G. Kalaitzoglou. 1990. Penicillin-binding proteins and ampicillin resistance in Haemophilus influenzae. J. Antimicrob. Chemother. 25:525-534. [DOI] [PubMed] [Google Scholar]
- 98.Mendelman, P. M., D. O. Chaffin, J. M. Musser, R. De Groot, D. A. Serfass, and R. K. Selander. 1987. Genetic and phenotypic diversity among ampicillin-resistant, non-beta-lactamase-producing, nontypeable Haemophilus influenzae isolates. Infect. Immun. 55:2585-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Metlay, J. P., J. H. Powers, M. N. Dudley, K. Christiansen, and R. G. Finch. 2006. Antimicrobial drug resistance, regulation, and research. Emerg. Infect. Dis. 12:183-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mohd-Zain, Z., S. L. Turner, A. M. Cerdeno-Tarraga, A. K. Lilley, T. J. Inzana, A. J. Duncan, R. M. Harding, D. W. Hood, T. E. Peto, and D. W. Crook. 2004. Transferable antibiotic resistance elements in Haemophilus influenzae share a common evolutionary origin with a diverse family of syntenic genomic islands. J. Bacteriol. 186:8114-8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Molina, J. M., J. Cordoba, A. Monsoliu, N. Diosdado, and M. Gobernado. 2003. Haemophilus influenzae and betalactam resistance: description of bla TEM gene deletion. Rev. Esp. Quimioter. 16:195-203. [PubMed] [Google Scholar]
- 102.Molina, J. M., J. Cordoba, R. Esteban, B. Lainez, A. Monsoliu, V. Gregori, A. Hernandez, N. Diosdado, and M. Gobernado. 2002. Study of the betalactam resistance of Haemophilus influenzae conferred by the bla(ROB-1) gene. Rev. Esp. Quimioter. 15:148-151. [PubMed] [Google Scholar]
- 103.Murphey-Corb, M., M. Nolan-Willard, and R. S. Daum. 1984. Integration of plasmid DNA coding for beta-lactamase production in the Haemophilus influenzae chromosome. J. Bacteriol. 160:815-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nakamura, T., and H. Takahashi. 2004. Antibacterial activity of oral cephems against various clinically isolated strains and evaluation of efficacy based on the pharmacokinetics/pharmacodynamics theory. Jpn. J. Antibiot. 57:465-474. [PubMed] [Google Scholar]
- 105.Nazir, J., C. Urban, N. Mariano, J. Burns, B. Tommasulo, C. Rosenberg, S. Segal-Maurer, and J. J. Rahal. 2004. Quinolone-resistant Haemophilus influenzae in a long-term care facility: clinical and molecular epidemiology. Clin. Infect. Dis. 38:1564-1569. [DOI] [PubMed] [Google Scholar]
- 106.Nicolas-Chanoine, M. H. 1997. Inhibitor-resistant beta-lactamases. J. Antimicrob. Chemother. 40:1-3. [DOI] [PubMed] [Google Scholar]
- 107.Niederman, M. S., L. A. Mandell, A. Anzueto, J. B. Bass, W. A. Broughton, G. D. Campbell, N. Dean, T. File, M. J. Fine, P. A. Gross, F. Martinez, T. J. Marrie, J. F. Plouffe, J. Ramirez, G. A. Sarosi, A. Torres, R. Wilson, and V. L. Yu. 2001. Guidelines for the management of adults with community-acquired pneumonia. Diagnosis, assessment of severity, antimicrobial therapy, and prevention. Am. J. Respir. Crit. Care Med. 163:1730-1754. [DOI] [PubMed] [Google Scholar]
- 108.Osaki, Y., Y. Sanbongi, M. Ishikawa, H. Kataoka, T. Suzuki, K. Maeda, and T. Ida. 2005. Genetic approach to study the relationship between penicillin-binding protein 3 mutations and Haemophilus influenzae beta-lactam resistance by using site-directed mutagenesis and gene recombinants. Antimicrob. Agents Chemother. 49:2834-2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Reference deleted.
- 110.Pares, S., N. Mouz, Y. Petillot, R. Hakenbeck, and O. Dideberg. 1996. X-ray structure of Streptococcus pneumoniae PBP2x, a primary penicillin target enzyme. Nat. Struct. Biol. 3:284-289. [DOI] [PubMed] [Google Scholar]
- 111.Parr, T. R., Jr., and L. E. Bryan. 1984. Mechanism of resistance of an ampicillin-resistant, beta-lactamase-negative clinical isolate of Haemophilus influenzae type b to beta-lactam antibiotics. Antimicrob. Agents Chemother. 25:747-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Peltola, H. 2000. Worldwide Haemophilus influenzae type b disease at the beginning of the 21st century: global analysis of the disease burden 25 years after the use of polysaccharide vaccine and a decade after the advent of conjugates. Clin. Microbiol. Rev. 13:302-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Peric, M., B. Bozdogan, M. R. Jacobs, and P. C. Appelbaum. 2003. Effects of an efflux mechanism and ribosomal mutations on macrolide susceptibility of Haemophilus influenzae clinical isolates. Antimicrob. Agents Chemother. 47:1017-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Peric, M., B. Bozdogan, C. Galderisi, D. Krissinger, T. Rager, and P. C. Appelbaum. 2004. Inability of L22 ribosomal protein alteration to increase macrolide MICs in the absence of an efflux mechanism in Haemophilus influenzae. J. Antimicrob. Chemother. 54:393-400. [DOI] [PubMed] [Google Scholar]
- 115.Pikis, A., J. A. Donkersloot, W. J. Rodriguez, and J. M. Keith. 1998. A conservative amino acid mutation in the chromosome-encoded dihydrofolate reductase confers trimethoprim resistance in Streptococcus pneumoniae. J. Infect. Dis. 178:700-706. [DOI] [PubMed] [Google Scholar]
- 116.Pitout, M., K. MacDonald, H. Musgrave, C. Lindique, K. Forward, M. Hiltz, and R. Davidson. 2002. Characterization of extended spectrum β-lactamase (ESBL) activity in Haemophilus influenzae, abstr. C2-645, p. 96. Abstr. 42nd Intersci. Conf. Antimicrob. Chemother., San Diego, CA, 2002. American Society for Microbiology, Washington, DC.
- 117.Powell, M., and D. M. Livermore. 1988. Mechanisms of chloramphenicol resistance in Haemophilus influenzae in the United Kingdom. J. Med. Microbiol. 27:89-93. [DOI] [PubMed] [Google Scholar]
- 118.Roberts, M. C. 1989. Plasmid mediated Tet M in Haemophilus ducreyi. Antimicrob. Agents Chemother. 33:1611-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Roberts, M. C., C. D. Swenson, L. M. Owens, and A. L. Smith. 1980. Characterization of chloramphenicol-resistant Haemophilus influenzae. Antimicrob. Agents Chemother. 18:610-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rodríguez-Martínez, J. M., L. López, I. García, and A. Pascual. 2006. Characterization of a clinical isolate of Haemophilus influenzae with a high level of fluoroquinolone resistance. J. Antimicrob. Chemother. 57:577-578. [DOI] [PubMed] [Google Scholar]
- 121.Rosenau, A., A. Labigne, F. Escande, P. Courcoux, and A. Philippon. 1991. Plasmid-mediated ROB-1 beta-lactamase in Pasteurella multocida from a human specimen. Antimicrob. Agents Chemother. 36:2419-2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rubin, L. G., A. A. Medeiros, R. H. Yolken, and E. R. Moxon. 1981. Ampicillin treatment failure of apparently beta-lactamase-negative Haemophilus influenzae type b meningitis due to novel beta-lactamase. Lancet ii:1008-1010. [DOI] [PubMed] [Google Scholar]
- 123.Saito, M., A. Umeda, and S. Yoshida. 1999. Subtyping of Haemophilus influenzae strains by pulsed-field gel electrophoresis. J. Clin. Microbiol. 37:2142-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sakai, A., M. Hotomi, D. S. Billal, K. Yamauchi, J. Shimada, S. Tamura, K. Fujihara, and N. Yamanaka. 2005. Evaluation of mutations in penicillin binding protein-3 gene of non-typeable Haemophilus influenzae isolated from the nasopharynx of children with acute otitis media. Acta Otolaryngol. 125:180-183. [DOI] [PubMed] [Google Scholar]
- 125.Sanbongi, Y., T. Suzuki, Y. Osaki, N. Senju, T. Ida, and K. Ubukata. 2006. Molecular evolution of beta-lactam-resistant Haemophilus influenzae: 9-year surveillance of penicillin-binding protein 3 mutations in isolates from Japan. Antimicrob. Agents Chemother. 50:2487-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sanchez, L., S. Leranoz, M. Puig, J. G. Loren, H. Nikaido, and M. Vinas. 1997. Molecular basis of antimicrobial resistance in non-typeable Haemophilus influenzae. Microbiologia 13:309-314. [PubMed] [Google Scholar]
- 127.Sanchez, L., W. Pan, M. Vinas, and H. Nikaido. 1997. The acrAB homolog of Haemophilus influenzae codes for a functional multidrug efflux pump. J. Bacteriol. 179:6855-6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Saunders, J. R., L. P. Elwell, S. Falkow, R. B. Sykes, and M. H. Richmond. 1978. Beta-lactamases and R-plasmids of Haemophilus influenzae. Scand. J. Infect. Dis. Suppl. 1978:16-22. [PubMed] [Google Scholar]
- 129.Schaad, U. B. 2005. Fluoroquinolone antibiotics in infants and children. Infect. Dis. Clin. N. Am. 19:617-628. [DOI] [PubMed] [Google Scholar]
- 130.Scheifele, D. W., S. J. Fussell, and M. C. Roberts. 1982. Characterization of ampicillin-resistant Haemophilus parainfluenzae. Antimicrob. Agents Chemother. 21:734-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Scriver, S. R., S. L. Walmsley, C. L. Kau, D. J. Hoban, J. Brunton, A. McGeer, T. C. Moore, E. Witwicki, Canadian Haemophilus Study Group, and D. E. Low. 1994. Determination of antimicrobial susceptibilities of Canadian isolates of Haemophilus influenzae and characterization of their beta-lactamases. Antimicrob. Agents Chemother. 38:1678-1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Serfass, D. A., P. M. Mendelman, D. O. Chaffin, and C. A. Needham. 1986. Ampicillin resistance and penicillin-binding proteins of Haemophilus influenzae. J. Gen. Microbiol. 132:2855-2861. [DOI] [PubMed] [Google Scholar]
- 133.Sethi, S., and T. F. Murphy. 2004. Acute exacerbations of chronic bronchitis: new developments concerning microbiology and pathophysiology-impact on approaches to risk stratification and therapy. Infect. Dis. Clin. N. Am. 18:861-882, ix. [DOI] [PubMed] [Google Scholar]
- 134.Sethi, S., N. Evans, B. J. Grant, and T. F. Murphy. 2002. New strains of bacteria and exacerbations of chronic obstructive pulmonary disease. N. Engl. J. Med. 347:465-471. [DOI] [PubMed] [Google Scholar]
- 135.Smith-Vaughan, H. C., K. S. Sriprakash, A. J. Leach, J. D. Mathews, and D. J. Kemp. 1998. Low genetic diversity of Haemophilus influenzae type b compared to nonencapsulated H. influenzae in a population in which H. influenzae is highly endemic. Infect. Immun. 66:3403-3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Straker, K., M. Wootton, A. M. Simm, P. M. Bennett, A. P. MacGowan, and T. R. Walsh. 2003. Cefuroxime resistance in non-beta-lactamase Haemophilus influenzae is linked to mutations in ftsI. J. Antimicrob. Chemother. 51:523-530. [DOI] [PubMed] [Google Scholar]
- 137.Stuy, J. H. 1980. Chromosomally integrated conjugative plasmids are common in antibiotic-resistant Haemophilus influenzae. J. Bacteriol. 142:925-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sutton, L. D., D. J. Biedenbach, A. Yen, and R. N. Jones. 1995. Development, characterization, and initial evaluations of S1. A new chromogenic cephalosporin for beta-lactamase detection. Diagn. Microbiol. Infect. Dis. 21:1-8. [DOI] [PubMed] [Google Scholar]
- 139.Tait-Kamradt, A., T. Davies, M. Cronan, M. R. Jacobs, P. C. Appelbaum, and J. Sutcliffe. 2000. Mutations in 23S rRNA and ribosomal protein L4 account for resistance in pneumococcal strains selected in vitro by macrolide passage. Antimicrob. Agents Chemother. 44:2118-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Tait-Kamradt, A., T. Davies, P. C. Appelbaum, F. Depardieu, P. Courvalin, J. Petitpas, L. Wondrack, A. Walker, M. R. Jacobs, and J. Sutcliffe. 2000. Two new mechanisms of macrolide resistance in clinical strains of Streptococcus pneumoniae from Eastern Europe and North America. Antimicrob. Agents Chemother. 44:3395-3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Then, R., and P. Angehrn. 1973. Nature of the bacterial action of sulfonamides and trimethoprim, alone and in combination. J. Infect. Dis. 128(Suppl.):498-501. [DOI] [PubMed] [Google Scholar]
- 142.Thornsberry, C., and L. A. Kirven. 1974. Ampicillin resistance in Haemophilus influenzae as determined by a rapid test for beta-lactamase production. Antimicrob. Agents Chemother. 6:653-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tristram, S. G. 2003. Effect of extended-spectrum beta-lactamases on the susceptibility of Haemophilus influenzae to cephalosporins. J. Antimicrob. Chemother. 51:39-43. [DOI] [PubMed] [Google Scholar]
- 144.Tristram, S. G., and S. Nichols. 2006. A multiplex PCR for {beta}-lactamase genes of Haemophilus influenzae and description of a new blaTEM promoter variant. J. Antimicrob. Chemother. 58:183-185. [DOI] [PubMed] [Google Scholar]
- 145.Tristram, S. G., R. Hawes, and J. Souprounov. 2005. Variation in selected regions of blaTEM genes and promoters in Haemophilus influenzae. J. Antimicrob. Chemother. 56:481-484. [DOI] [PubMed] [Google Scholar]
- 146.Tunkel, A. R., B. J. Hartman, S. L. Kaplan, B. A. Kaufman, K. L. Roos, W. M. Scheld, and R. J. Whitley. 2004. Practice guidelines for the management of bacterial meningitis. Clin. Infect. Dis. 39:1267-1284. [DOI] [PubMed] [Google Scholar]
- 147.Turnidge, J., G. Kahlmeter, and G. Kronvall. 2006. Statistical characterisation of bacterial wild-type MIC value distributions and the determination of epidemiological cut-off values. Clin. Microbiol. Infect. 12:418-425. [DOI] [PubMed] [Google Scholar]
- 148.Ubukata, K., N. Chiba, K. Hasegawa, Y. Shibasaki, K. Sunakawa, M. Nonoyama, S. Iwata, and M. Konno. 2002. Differentiation of beta-lactamase-negative ampicillin-resistant Haemophilus influenzae from other H. influenzae strains by a disc method. J. Infect. Chemother. 8:50-58. [DOI] [PubMed] [Google Scholar]
- 149.Ubukata, K., Y. Shibasaki, K. Yamamoto, N. Chiba, K. Hasegawa, Y. Takeuchi, K. Sunakawa, M. Inoue, and M. Konno. 2001. Association of amino acid substitutions in penicillin-binding protein 3 with beta-lactam resistance in beta-lactamase-negative ampicillin-resistant Haemophilus influenzae. Antimicrob. Agents Chemother. 45:1693-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wang, J. C. 1985. DNA topoisomerases. Annu. Rev. Biochem. 54:665-697. [DOI] [PubMed] [Google Scholar]
- 151.Watanabe, H., K. Hoshino, R. Sugita, N. Asoh, K. Watanabe, K. Oishi, and T. Nagatake. 2004. Possible high rate of transmission of nontypeable Haemophilus influenzae, including beta-lactamase-negative ampicillin-resistant strains, between children and their parents. J. Clin. Microbiol. 42:362-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Weiss, D. S., K. Pogliano, M. Carson, L. M. Guzman, C. Fraipont, M. Nguyen Disteche, R. Losick, and J. Beckwith. 1997. Localization of the Escherichia coli cell division protein Ftsl (PBP3) to the division site and cell pole. Mol. Microbiol. 25:671-681. [DOI] [PubMed] [Google Scholar]
- 153.World Health Organization. 2005. Haemophilus influenzae type B (HiB). WHO fact sheet no. 294, December 2005. http://www/who.int/mediacentre/factsheets/fs294/en/index.html.
- 154.Wu, P. J., K. Shannon, and I. Phillips. 1995. Mechanisms of hyperproduction of TEM-1 beta-lactamase by clinical isolates of Escherichia coli. J. Antimicrob. Chemother. 36:927-939. [DOI] [PubMed] [Google Scholar]
- 155.Xu, J., B. C. McCabe, and G. B. Koudelka. 2001. Function-based selection and characterization of base-pair polymorphisms in a promoter of Escherichia coli RNA polymerase-sigma(70). J. Bacteriol. 183:2866-2873. [DOI] [PMC free article] [PubMed] [Google Scholar]