Abstract
The past several years have witnessed an upsurge of genomic data pertaining to the Mycobacterium avium complex (MAC). Despite clear advances, problems with the detection of MAC persist, spanning the tests that can be used, samples required for their validation, and the use of appropriate nomenclature. Additionally, the amount of genomic variability documented to date greatly outstrips the functional understanding of epidemiologically different subsets of the organism. In this review, we discuss how postgenomic insights into the MAC have helped to clarify the relationships between MAC organisms, highlighting the distinction between environmental and pathogenic subsets of M. avium. We discuss the availability of various genetic targets for accurate classification of organisms and how these results provide a framework for future studies of MAC variability. The results of postgenomic M. avium study provide optimism that a functional understanding of these organisms will soon emerge, with genomically defined subsets that are epidemiologically distinct and possess different survival mechanisms for their various niches. Although the status quo has largely been to study different M. avium subsets in isolation, it is expected that attention to the similarities and differences between M. avium organisms will provide greater insight into their fundamental differences, including their propensity to cause disease.
INTRODUCTION
Past reviews on the species Mycobacterium avium have typically focused on two distinct aspects. The first examines organisms classically called M. avium and their role in human disease, such as disseminated disease in AIDS and pulmonary disease (87, 124). This focus has also included other genetically distinct species, such as M. intracellulare and related species that are grouped together as the M. avium complex (MAC). The other focus has been on the Johne's bacillus, previously known as M. paratuberculosis, in the context of veterinary medicine (36, 46, 112). For a number of reasons, spanning from tradition to tools, these two organisms are still usually studied as separate entities, although by genetic criteria they have been classified as subsets of the same species for over a decade (267). As a result, clinical and epidemiologic studies of human exposure, infection, and disease have largely ignored the now-renamed M. avium subsp. paratuberculosis. In parallel, research on M. avium subsp. paratuberculosis often overlooks the existence of other closely related M. avium organisms and the potential impact of these other M. avium organisms on diagnostic and epidemiologic findings.
The advent of genome sequencing projects and comparative genomic tools has provided a renewed opportunity to firmly classify mycobacteria. In the case of the M. tuberculosis complex (MTBC), comparative genomics has provided genomic signatures that define members of the complex (14, 25, 102, 183), and these genomic signatures now serve in diagnostic laboratories to assign identity to clinical isolates (198). Phenotypically ambiguous organisms can now be classified confidently based on their genomic signatures (185), leading to the recognition that certain organisms previously grouped together due to insufficiently discriminatory methods (184, 245) in fact consist of genetically distinct host-associated variants or ecotypes (adapted to a specific habitat), such as the vole bacillus, seal bacillus, dassie bacillus, oryx bacillus, and M. caprae in goats (2, 55, 182). With the recent availability of complete genome sequences for the two principal M. avium subspecies (152) (The Institute for Genomic Research [TIGR] [http://www.tigr.org/]) and results from comparative genomic studies (236, 240, 304), it is now possible to reconsider M. avium in a similar manner. The existence of natural variants of M. avium is expected to initially pose new challenges in taxonomy and diagnostics. However, once the nomenclature is resolved, a postgenomic phylogenetic framework should serve towards improved diagnostics and strain tracking and, additionally, provide a context for studies of disease pathogenesis. The aim of this review is to address current misconceptions and confusion in M. avium taxonomy, to place emphasis on the importance of recognizing the diversity within M. avium strains, and to highlight the opportunities to study M. avium by exploiting the existence of phenotypically variant members of the same species. In addition, we examine how genomic data provide opportunities and challenges for the derivation of novel diagnostic tools, noting in particular the distinction between elements specific by in silico analysis of genome sequence data and those specific by validated laboratory assays. Finally, in the face of accumulating reviews and rebuttals about the potential role of M. avium subsp. paratuberculosis in human Crohn's disease (CD), we consider it especially valuable to reassess the definition of this organism, the methods used for its detection, and the applicability of these methods for epidemiologic investigation of this association.
TAXONOMY AND CLINICAL SIGNIFICANCE
Mycobacteria are defined by their acid-fast properties, cell walls containing mycolic acids, and high (∼61 to 71%) genomic C+G contents (149). There are now over 130 established and validated species and subspecies of mycobacteria (J. P. Euzéby, List of Prokaryotic Names with Standing in Nomenclature [http://www.bacterio.cict.fr]), with the most commonly isolated species in clinical laboratories consisting of members of the MTBC and members of the MAC. Originally described in two separate veterinary settings, MAC organisms have long been recognized as professional pathogens of birds and ruminants. Based on their source of isolation and pathology in animal models, two distinct organisms, namely, the avian tubercle bacillus, the agent of tuberculosis (TB) in birds, and Johne's bacillus, agent of Johne's disease in ruminants, were recognized. With the recognition that the avian tubercle bacillus could occasionally be isolated from human diseases, MAC organisms were also considered opportunistic pathogens of humans. In order to determine the potential sources of human exposure, environmental surveys were undertaken, revealing viable or culturable MAC organisms in a number of sources, including water (reviewed in reference 208). The latter observation led to the concept or belief that MAC organisms are fundamentally environmental mycobacteria. While it appears that some MAC organisms reside primarily in the environment, other subsets are veterinary pathogens with a limited capacity to survive in the environment (143, 300). Therefore, to best appreciate the natural variability among MAC organisms, it is safest to consider the MAC as a microcosm of the mycobacterial genus including both environmental mycobacteria and host-associated pathogens with their own distinct genomic identities.
The definition of MAC varies with the context in which it is discussed (Table 1). Clinicians and health care workers consider MAC to include M. avium, M. intracellulare, and miscellaneous related species. In veterinary medicine, MAC may be recognized the same way but, notably, is distinct from “M. paratuberculosis.” The taxonomist may consider the MAC to contain only the subspecies of M. avium, as the designation implies, including M. avium subsp. paratuberculosis, and recognize that M. intracellulare is a related but clearly distinct species from M. avium. The scientist may or may not adopt any of the definitions described above, depending on the research question being addressed. Confusion sets in when new advances redefine the classic nomenclature. With this being said, we believe that sufficient data now exist to provide clarity in M. avium taxonomy and that a revised taxonomic approach will benefit research into the epidemiology and pathogenesis of diseases due to M. avium.
TABLE 1.
Nomenclature applied to MAC organisms
| Accepted name | Basonyms or synonyms |
|---|---|
| M. avium subsp. avium and/or M. avium subsp. hominissuis | Avian tubercle bacillus, M. tuberculosis avium, M. avium |
| M. avium subsp. silvaticum | Wood pigeon bacillus, M. avium subsp. columbae, M. silvaticum |
| M. avium subsp. paratuberculosis | Johne's bacillus, M. enteritidis, M. johnei, M. paratuberculosis |
| M. intracellulare | Battey bacillus, Nocardia intracellularis, M. battey |
| Collective designation | Battey-avian mycobacteria, M. avium-intracellulare, M. avium- intracellulare-scrofulaceum, M. avium complex |
Classical Definition of MAC Species
A milestone in the characterization and definition of M. avium occurred in 1990 with a publication by Thorel et al. which defined three principal subsets of M. avium, as revealed by prior molecular analyses, such as DNA-DNA hybridization (122, 231, 307), on the basis of growth characteristics and biochemical tests (numerical taxonomy analysis) (267). These three subsets consist of M. avium subsp. avium, M. avium subsp. paratuberculosis, and M. avium subsp. silvaticum.
M. avium subsp. avium.
Before the establishment of the M. avium subsp. avium designation, this organism was simply referred to as M. avium and was recognized to be the cause of avian TB and occasional infections in other animals. The type strain, ATCC 25291, was isolated from a diseased hen. The designation includes the standard M. avium subspecies causing disease in birds but also includes agents of disseminated disease in patients with AIDS, cervical lymphadenitis in children, and chronic lung disease in several settings in adolescents with cystic fibrosis and in older adults. Classically, the designation M. avium subsp. avium has not distinguished avian from human or environmental isolates, and hence, sensitization to M. avium is used as a proxy of exposure to environmental mycobacteria, even though avian purified protein derivative (PPD) was derived from a bird isolate (237). As discussed in greater detail in this review, the failure to distinguish between the environmental and host-associated ecotypes of M. avium is especially problematic for interpreting and comparing data from past studies.
M. avium subsp. paratuberculosis.
M. avium subsp. paratuberculosis refers to the etiologic agent of Johne's disease or paratuberculosis, a chronic granulomatous enteric disease of ruminant livestock and wildlife (112). Difficulties surrounding paratuberculosis control lie primarily in aspects of diagnosis; assays are most accurate when the disease is well established, but detection of subclinical infection is hampered by poor sensitivity (251, 294, 295) and specificity (168). M. avium subsp. paratuberculosis is one of the slowest growing mycobacterial species, such that primary isolation from specimens can take several months (173, 298). The distinguishing phenotype of M. avium subsp. paratuberculosis has classically been an in vitro growth dependency on mycobactin, an iron-chelating agent first obtained from M. phlei (90, 173) which was subsequently replaced by mycobactin J, currently used today, obtained from a strain of M. avium (41, 174). Notably, the type strain of the species, ATCC 19698, isolated from the feces of a cow with paratuberculosis (172), has lost its mycobactin dependency. From phenotypic analysis, the M. avium subsp. paratuberculosis group has been subdivided into two main types, bovine and ovine, that vary in hosts, diseases caused, and growth phenotypes (260, 297, 298).
M. avium subsp. silvaticum.
M. avium subsp. silvaticum applies to the previously named wood pigeon bacillus, an acid-fast organism causing TB-like lesions in these wood pigeons that were not initially successfully cultured in vitro (44, 167). Cultures were obtained for the first time when medium for “M. paratuberculosis” was used for cultivation and were observed for 5 months (249). Subsequently, the organisms were recognized by their mycobactin dependency upon primary isolation, gradually losing this phenotype upon subculture (165). Conflicting experimental data in attempting to classify the organisms led to the performance of DNA-DNA homology studies, ultimately revealing that they belonged to the same species as M. avium and M. avium subsp. paratuberculosis (231, 307). Support for the distinctiveness of M. avium subsp. silvaticum, however, was advanced by distinct patterns obtained with genetic tools such as pulsed-field gel electrophoresis (150), although this type of method is typically used for epidemiological purposes, not to delineate species. Finally, a thorough phenotypic evaluation of the M. avium species revealed that only M. avium subsp. silvaticum was distinct from classical M. avium subsp. avium and M. avium subsp. paratuberculosis based on an inability to grow on egg media and the stimulation of growth at pH 5.5 (267). The type strain, ATCC 48898, represents strain 6409, isolated from the liver and spleen of a wood pigeon and characterized in the numerical taxonomy study (267).
M. intracellulare.
Unlike the M. avium subsets, for which the type strains were isolated from nonhuman hosts, the type strain of M. intracellulare (ATCC 13950) was isolated from a human, specifically a child who died from disseminated disease (63). This organism was initially named Nocardia intracellularis, until Runyon made the link between an atypical mycobacterium called the “battey bacillus” and “N. intracellularis” based on similarities with M. avium and subsequently established the M. intracellulare species (223). Since then, M. intracellulare organisms have been isolated from a variety of animal hosts and environmental sources (225, 266, 269). In general, M. intracellulare has been subject to less study than M. avium, as the latter is more prevalent in clinical and environmental samples, has a wider apparent host range, and contributes almost exclusively to disseminated MAC disease in human immunodeficiency virus patients (276, 305). However, when identification to the species level is performed, M. intracellulare is an important contributor to MAC-associated pulmonary infections in immunocompetent or non-human immunodeficiency virus patients (108, 166, 207, 269, 290). M. intracellulare also appears to have a distinct environmental niche, as it has been found to be more prevalent in biofilms and at significantly higher CFU numbers than M. avium (88). The clinical designation MAC or MAI, used to group M. avium and M. intracellulare, largely reflects the conventional inability of the diagnostic laboratory to distinguish these organisms and the use of the same therapeutic regimens. Sequence-based analysis reveals M. intracellulare as a distinct out-group for resolving subsets of M. avium (277) (for example, see Fig. 1). The implications of blurring the species barrier in clinical, epidemiological, immunological, or bacteriologic studies are unknown but clearly important.
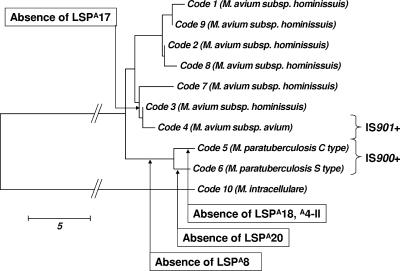
FIG. 1.
hsp65 gene phylogeny based on nucleotide differences (277), superimposed with genetic variation based on LSPs (239). IS901+ strains cluster in one lineage, and all lack LSPA17. IS900+ strains cluster in a lineage identified by codes 5 and 6, and all lack LSPA8. M. intracellulare serves as the outgroup for the M. avium subspecies. M. avium subsp. silvaticum presents with an identical genetic profile to that of code 4. Bar, 5 nucleotides.
New Designations and Related Species
Many strains or groups of strains have been described that share similarity with the MAC, which often results in frustration, confusion, and at times, misleading data and results (137, 147, 250, 292, 293). However, as more sophisticated molecular tools become available, important (or less important) subsets can be identified with greater confidence. Examples of newer and/or less-well-recognized members of M. avium or species associated with MAC include the following.
M. avium subsp. hominissuis.
M. avium subsp. hominissuis was proposed to distinguish organisms found in humans and pigs from those isolated from birds. The hypothesis that there might be host-specific differences within M. avium was suggested when laboratories using genotypic methods noted that M. avium isolates from humans rarely shared the genetic profiles of organisms found in birds (22, 107, 139, 219). To study this further, Mijs et al. undertook a first comprehensive study that encompassed phenotypic assessment as well as several genetic tools (IS1245 restriction fragment length polymorphism (RFLP) analysis, commercial assays, and sequencing) previously used for MAC against a large set of isolates from different hosts and geographical origins (176). This study confirmed that classical avian strains are distinct from human, other mammalian, and environmental MAC isolates. The distinguishing features of M. avium subsp. hominissuis are (i) a multiple copy number of IS1245, (ii) a variable 16S-23S internal transcribed spacer (ITS) sequence (the sequence in avian strains is invariant), and (iii) the ability to grow at a wider temperature range (24 to 45°C) (176). The M. avium strain chosen for genome sequencing, strain 104, is of the M. avium subsp. hominissuis subtype (277). Another important distinguishing feature of M. avium subsp. hominissuis from M. avium subsp. avium is that it does not possess the IS901 insertion sequence (IS) (10, 178, 277), which is occasionally used as a marker of the M. avium species (243, 254). No type strain has been designated to represent M. avium subsp. hominissuis, and consequently, this designation has yet to be formally validated. However, reference strains do exist that represent this subset (Table 2).
TABLE 2.
Commonly used reference M. avium strainsa
| Strain no. | True subspecies designation |
|---|---|
| ATCC 25291T, TMC724T, or DSM 44156T | M. avium subsp. avium |
| “ATCC 12227” or strain 18 | M. avium subsp. avium |
| ATCC 15769 or DSM 44157 | M. avium subsp. avium |
| ATCC 19421, DSM 44158, or NCTC 8559 | M. avium subsp. avium |
| ATCC 35713 or PPD-avian | M. avium subsp. avium |
| ATCC 35718 | M. avium subsp. avium |
| 104 | M. avium subsp. hominissuis |
| ATCC 49601 | M. avium subsp. hominissuis |
| ATCC 700897 | M. avium subsp. hominissuis |
| ATCC 700898 or MAC 101 | M. avium subsp. hominissuis |
| K-10 or ATCC BAA-968 | M. avium subsp. paratuberculosis, C type |
| ATCC 19698T | M. avium subsp. paratuberculosis, C type |
Identities of the strains are based on collective information and results from hsp65 gene sequencing (277), IS901 positivity, IS1245 RFLP profiles, and/or the source of isolation (i.e., diseased bird or AIDS patient).
M. lepraemurium.
M. lepraemurium refers to the agent of rodent leprosy, which was later suspected of causing skin disease in cats and dogs. To date, this organism is generally considered unculturable and can be identified reliably only by sequencing methods (119, 120). The association between M. lepraemurium and MAC stems from a likeness in serological groupings (103) and genetic relatedness by DNA-DNA hybridization methods (3, 123). The organism is characterized by only two single-nucleotide polymorphisms (SNPs) in the 16S rRNA gene from that of M. avium but bears a highly divergent hsp65 sequence (170). While this species is genetically closely related to MAC organisms, it is not typically considered part of the MAC.
Undefined species and novel designations.
Over the years, the MAC has had other taxonyms, including M. avium-intracellulare (MAI), M. avium-intracellulare-scrofulaceum (MAIS), and MAIX, where “X” represents the ambiguous MAC species that could not be assigned as M. avium or M. intracellulare. These isolates typically have similar characteristics to MAC organisms but lack features that typify the species. For example, isolates positive in the MAC AccuProbe (GenProbe) test but not in the species-specific M. avium or M. intracellulare AccuProbe test would fit within this rubric (12, 93, 147, 247, 285). M. scrofulaceum was once grouped with M. avium and M. intracellulare on the basis of phenotypic similarity and its ability to be serotyped, but it has long been accepted as a separate entity from MAC. The recently described species M. palustre (270) and M. saskatchewanense (278) may be confused with MAC due to their positive reactions with the MAC AccuProbe test, but they are otherwise genetically distant from MAC. Other recently described species that are genetically and phenotypically related to MAC, such as M. chimaera (274) and M. colombiense (186), highlight the fact that outliers or MAC-like organisms continue to be isolated and characterized, defying simple classification. While it is tempting to consider the latter to be the same as other MAC organisms, such a simplification risks overlooking potentially informative differences between organisms that may not yet be apparent. Therefore, when faced with such an organism, the clinical or reference laboratory may best report a MAC-like organism.
MAC Terminology Used for This Review
For the remainder of this review, we use the more encompassing term MAC to include the species and subspecies that preceded but focus mostly on the species M. avium. When discussing subsets of M. avium, we use the following terminology. M. avium subsp. avium refers to the avian subtype, including the type strain ATCC 25291 or TMC724. M. avium subsp. hominissuis refers to human, porcine, and environmental isolates, including the strain used for the genome sequence, which we refer to as M. avium 104. M. avium subsp. paratuberculosis refers to bovine and ovine subtypes of Johne's bacillus, including the strain used for the genome sequence, known as M. avium subsp. paratuberculosis K-10. Designations for commonly studied organisms, including type strains, are presented in Table 2.
LABORATORY ASPECTS OF THE MAC
Serotyping and Other Traditional Methods
Prior to the era of molecular diagnostics, identification of mycobacteria to the species level was based on morphology and a set of in vitro biochemical tests (135). These tests were not useful in subidentifying members of the MAC, since MAC organisms are generally nonreactive or produce variable results with most tests used to differentiate between species. Morphologically, the MAC presents with a wide range of colony variability, from smooth to rough and from nonpigmented to cream-colored to bright yellow, and can appear like many other mycobacterial species. High-performance liquid chromatography (HPLC) of mycolic acids became popular as a diagnostic method for species identification of mycobacteria and can distinguish MAC organisms from other mycobacteria (100). HPLC patterns of M. avium and M. intracellulare are very similar, although differentiation may be achieved by using precise interpretive criteria (32). M. avium subsp. paratuberculosis cannot be differentiated from the other subspecies of M. avium by this method (65). Serotyping, one of the earliest typing tools for the MAC (232), was based on differences in the sugar residue compositions of surface glycopeptidolipids (GPLs) and became the preferred method of MAC identification in the premolecular era. More than 30 serovars have been described (reviewed in reference 38). Based on a variety of tests, including DNA probing, serotype numbers could generally be assigned to the following MAC species: serotypes 1 to 6, 8 to 11, and 21 are classical M. avium; serotypes 7 and 12 to 20 are M. intracellulare; and serotypes 26, 27, and 41 to 43 are M. scrofulaceum (225, 293). Interlaboratory reproducibility in serotype numbers, however, was poor, and issues with autoagglutination, failure to react with any serum, or agglutination with two or more antisera were common (276, 293). Also, many serotypes could not be assigned confidently to a MAC species due to a poor consensus or to nonreaction with species-specific antisera, resulting in the ambiguous designation “MAC” for these strains. It was concerning that serotyping in tandem with RFLP methods used for epidemiological purposes was noted to generate different serotypes for isolates with identical RFLP profiles (79, 110). Conversely, the same serotype can be represented across the two subgroups of M. avium subsp. avium and M. avium subsp. hominissuis. Multilocus enzyme electrophoresis could differentiate strains of the MAC into many electrophoretic types on the basis that the enzymes chosen had multiple alleles (291, 306). This technique likely reflected variability at the geographic or ecotype level but did not appear to provide the level of resolution desirable for epidemiological tracking of isolates.
Genetic Methods To Detect IS Elements
When well characterized and used in the proper context, the species-specific IS elements described below can serve as a useful classification tool to distinguish subsets of the MAC (10, 49, 84). However, two problems have consistently hampered their utility for this purpose. First, a number of IS elements have been uncovered in strains considered to be MAC organisms, but without adequate strain characterization, it is difficult to judge which organisms harbor such elements. Second, IS elements are by nature mobile elements, so there is a risk that similar elements are found in unrelated bacteria because of mobility to or from MAC organisms. Therefore, while studies may report on the specificity of these elements across MAC organisms, this degree of specificity is not assured in diagnostic laboratories classifying unknown clinical isolates unless the organisms have first been shown to be MAC organisms by other methods. For instance, a newly discovered element may be found only in M. intracellulare among a panel of MAC strains and therefore appear to be a promising target for PCR-based detection directly from broth culture. However, until it can be ascertained that this element, or something genetically similar, is not found among the over 130 mycobacteria that may present to a reference laboratory, a positive PCR for this element should not on its own be considered sufficient evidence to state that M. intracellulare has been detected.
IS900.
IS900 was the first IS characterized within the Mycobacterium genus (51, 104). It was identified from a pMB22 clone derived from a genomic library from a human M. avium subsp. paratuberculosis isolate from a CD patient and was found to be specific to M. avium subsp. paratuberculosis. Just as IS6110 has been used successfully for genotyping M. tuberculosis strains (280), RFLP analysis of the IS900 element has been used a molecular tool to type M. avium subsp. paratuberculosis isolates. Based on IS900 RFLP patterns, M. avium subsp. paratuberculosis has been divided into two main groups, namely, those isolates represented by a cattle-associated profile (C) and those represented by a sheep-associated profile (S) (11, 52, 57, 202, 297). A third RFLP genotype, called intermediate (I), was also identified from sheep (11, 67). The IS900 element is by far the most widely used target for the molecular detection of M. avium subsp. paratuberculosis and has been used in the form of direct PCR (161, 235, 284), in situ PCR (228), sequence/hybridization capture PCR (109, 159, 161, 177), nested PCR (28, 187, 224), and real-time PCR (89, 214), with the references listed representing only a small portion of what is available in the literature. However, other similar elements found across other mycobacteria, including M. terrae, M. xenopi, M. scrofulaceum and related strains, M. chelonae, and strain 2333 (related to M. cookii), have been shown to cross-react with IS900 primers used for detection of M. avium subsp. paratuberculosis (56, 85, 258). In these cases, the elements were not 100% identical with IS900, with different regions of the elements showing variable sequence identity. Sequencing of the amplified product for IS900 is therefore necessary to confirm that the amplicon is truly IS900. A few studies have reported SNPs in the IS900 element (19, 187), posing a problem for sequence-based verification of IS900-PCR results for molecular detection of M. avium subsp. paratuberculosis. Because sequencing directly from a single-round PCR product (i.e., not from cloned PCR products or nested PCR products) revealed only two specific SNPs, dividing ovine and bovine forms of M. avium subsp. paratuberculosis, the relevance of these other reported polymorphisms is presently unclear (238).
IS901.
Kunze et al. discovered the IS901 element by performing a Southern blot with the pMB22 probe containing the IS900 element across various MAC isolates under low-stringency conditions (142). This element shows ∼60% sequence identity to IS900. Screening across a larger panel of isolates revealed that most isolates from birds and some animals contained the element, whereas isolates obtained from AIDS patients or the environment did not. Furthermore, it was found that most bird isolates had similar IS901 patterns. These isolates were also shown to be strikingly more virulent than AIDS patient isolates in BALB/c mice (142, 204). Evidence that a more pathogenic subset of M. avium exists has been advanced numerous times since, leading some to simply divide M. avium isolates into those that are IS901+ and those that are IS901− (68). In general, isolates from diseased birds and animals with macroscopic lesions are IS901+, while those from humans, swine, or other animals without lesions are IS901− (22, 49, 189, 203). The virulence of IS901+ strains has also been confirmed experimentally (79, 203).
Simultaneous to the publication of the IS901 element, Moss et al., who were also screening for IS900 under low-stringency conditions, observed cross-hybridization with a strain of M. avium subsp. silvaticum and designated the related element IS902 (181). They determined that the element was present in all M. avium subsp. silvaticum isolates they tested, although no other MAC strains were included in the study set. Sequence alignment of the IS901 (X59272) and IS902 (X58030) sequences indicates 99% sequence identity, and upon closer inspection, their differences consist of several sequence gaps and four pairs of GC switches, suggestive of editing errors. IS901 and IS902 are most likely the same element, in which case the existence of an IS902 element specific for M. avium subsp. silvaticum would not be a valid distinction. Consequently, claims that M. avium subsp. silvaticum has been detected in samples based on the presence of IS902 should be interpreted with caution, with a more likely scenario being the detection of a strain containing IS901 or related elements.
IS1311.
IS1311 was first reported as a GenBank entry in 1994 (U16276) and was subsequently used for RFLP analyses (73, 220). The element is present in all members of the M. avium subspecies, including M. avium subsp. avium, M. avium subsp. hominissuis, and M. avium subsp. paratuberculosis (49), and is not present in M. intracellulare (73, 296). The element itself has 85% sequence identity to IS1245 (described below) and therefore results in cross-hybridization with the conventional IS1245 probe (130). With the wide range of M. avium hosts for this element, it is possible that IS1311 represents an “older” IS element which may have been present prior to subspecies divergence. A longer evolutionary time span is consistent with the presence of mutations in some of the IS1311 elements among distinct subsets within the MAC. This was first observed by Whittington et al., who noted one polymorphism specific to the M. avium subsp. paratuberculosis cow or “C” type (a C-to-T change at bp 223 of the U16276 sequence) and other polymorphisms common to both the “C” and “S” types of M. avium subsp. paratuberculosis compared to other M. avium organisms (296). RFLP analysis of IS1311 also revealed distinct pattern types corresponding to cattle and sheep strains of M. avium subsp. paratuberculosis (49). A simple PCR and restriction enzyme analysis (PCR-REA) using the restriction enzyme HinfI was then developed as a rapid diagnostic tool to distinguish bovine M. avium subsp. paratuberculosis isolates from the ovine type (158). Distinct growth characteristics of M. avium subsp. paratuberculosis isolates from bison in Montana prompted investigation using IS1311 PCR-REA and revealed a third IS1311 genotype, “B” (299), and M. avium subsp. paratuberculosis strains obtained from armadillos in Wisconsin were reported to have yet another IS1311 PCR-REA allele (54). In agreement with IS900 RFLP analysis (reviewed in reference 297), cattle and goats have predominantly the C type and sheep have predominantly the S type, while the B type has been found not only in American bison but also in goats and sheep in India (241, 296). Other animals, when tested, generally have the C type (296).
IS1245.
First described in 1995 (107), IS1245 was presented as having a more restricted range than IS1311, being limited to the subspecies of M. avium, i.e., M. avium subsp. avium (that would include M. avium subsp. hominissuis), M. avium subsp. paratuberculosis, and M. avium subsp. silvaticum. By PCR analysis, this element was not found in M. intracellulare or 17 other mycobacterium species. This element, however, has high DNA sequence identity with IS1311, with both belonging to the IS30 family, and it was shown that cross-hybridization of IS1245 probes with IS1311 is widespread; for instance, M. avium subsp. paratuberculosis does not contain the IS1245 element (130). Nonetheless, from this first publication on IS1245, Guerrero et al. observed that human and swine strains contained an elevated number of copies (more than eight; “multicopy”), whereas bird strains, including M. avium subsp. silvaticum, presented a three-band pattern (107). The observation that human and swine strains (now called M. avium subsp. hominissuis) differ from avian strains has since been confirmed numerous times (139, 178, 190, 219, 263), also with the added dimension that environmental strains have similar characteristics to those of the M. avium subset from humans and swine (78, 164). Standardization of IS1245 RFLP analysis was proposed in 1998 as a tool for MAC molecular epidemiology (283). To eliminate cross-hybridization with IS1311, the method was modified, leading to the recognition that the three-band bird type IS1245 RFLP profile in fact consisted of a single IS1245 copy and two copies of IS1311 (130). It remains to be seen if epidemiological value would be added by using an IS1245-specific protocol instead of the standard protocol.
As for any widely tested insertion element, the “presence” of the IS1245 element in species outside its typical host has been documented, although this was not confirmed by sequencing and may have been related elements, such as IS1311 (12, 134). Some M. avium isolates have been documented as being IS1245 negative, but only a few such reports have presented further documentation of strain identity by a sequence-based method (12). In some reports, IS1245-negative isolates have been described that contain an hsp65 sequence identical to that of M. avium but that differ from M. avium in other taxonomic targets, such as the 16S rRNA gene and the ITS sequence (147, 277).
Other insertion elements described for the MAC.
Many other IS elements have been described or detected in various members of the MAC. In most cases, their distribution is either unknown or only partially known. We attempt to put some emphasis on their most likely distribution or lack thereof. When stating that some elements are similar to others, we refer to similarity on the order of 80 to 85% identity at the nucleotide level.
(i) IS elements of rare distribution.
The IS1110 element was identified from a single strain, designated M. avium LR541 (116). It has some similarity to IS900 and IS901 and was found in only a small proportion of M. avium strains. However, which subset of M. avium contains this element is not clear, and screening was done by Southern hybridization, where the signal could have resulted from other related elements. IS1110-like elements have been reported for many species of mycobacteria, but these either have not been sequence confirmed (117) or have been confirmed but do not correspond to IS1110 per se (194, 253). The only information available for the IS1141 element is a GenBank entry dated 1995 (L10239). It was found in a strain identified as M. intracellulare strain Va14. Since then, no new data have been presented on this element or on the strain of M. intracellulare in which it was found. Unfortunately, no IS element has been identified to date that is present in all strains of M. intracellulare or even in any single well-known strain representative of the species. IS1626 was discovered in the same manner as IS901 (and IS902), by Southern blotting of 66 MAC isolates with an IS900 probe (210), and has some similarity to IS900 and to IS1613 (below). Strong hybridization occurred for only one strain, subsequently characterized as M. avium by a variety of molecular tests. It is unclear, however, if this strain or any of the others screened were M. avium subsp. avium or M. avium subsp. hominissuis. This element appears to be uncommon in MAC organisms in general. IS1613 is another element for which very little information is available: a GenBank submission exists (AJ011837), and one publication mentions that it was isolated from an AIDS patient (28), indicating a probable M. avium subsp. hominissuis strain. It is similar to both IS1626 and IS900. None of these elements is present in the genome sequence of strain 104 or K-10.
(ii) IS elements of partially known distribution.
The element IS1612, identified in a strain of M. avium subsp. silvaticum and in M. avium subsp. avium TMC724 (30), corresponds to IS2534 (80), similarly found in strain TMC724. Proper IS nomenclature (244) was eventually assigned to this element, now referred to as ISMav1 (ISFinder [http://www-is.biotoul.fr/]). ISMav1 is present in at least one M. avium subsp. hominissuis strain, the M. avium subsp. avium type strain, and also the M. avium 104 genome sequence. The distribution of this element across a panel of MAC isolates is undetermined. The element IS666 was identified in M. avium isolates from humans (36%), pigs (5%), cattle (12%), and the environment (78%) but not from avian strains (227). Therefore, IS666 is likely present only in some subsets of M. avium subsp. hominissuis, as it was present in 21% of M. avium strains tested. The IS1601 element was identified during a study of the genetic mechanisms behind the variable morphology of M. avium and was implicated in the smooth-to-rough switch in some strains (81). The IS1348 element was uncovered upon further sequencing of the ser2 GPL gene cluster (81). Both IS1601 and IS1348 are present in the M. avium 104 genome but not in M. avium subsp. paratuberculosis K-10. On this basis, these elements appear to be present in at least a subset of M. avium subsp. hominissuis strains and not in M. avium subsp. paratuberculosis strains. ISMav2 is a potentially M. avium subsp. paratuberculosis-specific element, as it was detected in all M. avium subsp. paratuberculosis strains but not in strains of M. avium subsp. avium (243, 254). Unfortunately, IS901-negative strains were not evaluated, and therefore the distribution of ISMav2 in M. avium subsp. hominissuis isolates is unknown. The IS999 element was found in isolates presumed to be M. avium subsp. hominissuis since they were from human clinical samples and was absent from one strain known to be M. avium subsp. avium (144). While its distribution in M. avium subsp. paratuberculosis was not evaluated, it is not present in the K-10 genome sequence. The element ISMpa1, with 80% sequence identity with IS1601, was found in all M. avium subsp. paratuberculosis strains tested, 2 of 13 MAC organisms tested, and no other mycobacterial species (191). The true distribution of this element within the MAC is unknown since only a small panel of isolates was evaluated.
(iii) IS elements newly discovered via genome sequencing projects.
The M. avium subsp. paratuberculosis K-10 genome sequence contains three insertion elements that were previously described, namely, the M. avium subsp. paratuberculosis-specific elements IS900 and ISMav2 and the pan-M. avium element IS1311. Sixteen additional insertion elements were identified and named IS_MAP01 through -16 (152). Of note, IS_MAP12 corresponds to the previously described ISMpa1 (191). The most abundant IS family represented in the K-10 genome is the IS110 family, which includes IS900, ISMpa1, and IS_MAP14 to -16 (152) but also the IS1110, IS901, IS1613, and IS1626 elements described for other MAC strains. A few K-10 IS elements correspond to some found in the M. avium 104 genome, while others have low or no similarity to other bacteria, including mycobacteria, and may potentially serve in the specific diagnosis of M. avium subsp. paratuberculosis. With the 14 IS elements described for the MAC in the pregenomic era, 15 novel IS elements identified in the genome sequence of M. avium subsp. paratuberculosis K-10, and more to be found through the genome sequence of M. avium 104, it is clear that MAC organisms contain a very large number of IS elements, many of which are related to each other. As new genome sequences become available at an increasing pace, so will the number of related insertion elements. For example, IS1110, which was first identified because of its similarity with IS900 and IS901, is now known to have much higher similarity to one of the new insertion elements in M. avium subsp. paratuberculosis K-10 (IS_MAP15) and to also share high similarity with an element in the recently sequenced Mycobacterium sp. strain MCS (GenBank accession no. CP0003841; M. monacense by 16S rRNA gene sequencing). This example illustrates an important limitation of targeting IS for diagnostics, as cross-hybridization with closely related elements has been documented by both PCR and Southern hybridization.
Non-IS-Based PCR Differentiation of MAC
Apart from insertion elements, other genes have been used as diagnostics or to differentiate MAC organisms and can be a more attractive option due to concerns of nonspecificity associated with IS elements. A single-copy sequence named F57 was identified as specific for M. avium subsp. paratuberculosis (206) and later used in a duplex PCR that differentiated the MTBC, M. avium, and M. avium subsp. paratuberculosis (47, 101). More recently, a real-time PCR assay based on the F57 element was developed for the detection of M. avium subsp. paratuberculosis in milk, feces, and tissue (23, 259). Another M. avium subsp. paratuberculosis-specific genetic target showed similarity to the dnaJ family of heat shock protein genes and was designated hspX (83). The specificity of hspX for M. avium subsp. paratuberculosis was subsequently confirmed across a large panel of MAC strains with various genetic and host characteristics (84). To distinguish bovine and ovine M. avium subsp. paratuberculosis strains, a three-primer PCR assay was developed that yields PCR products of different sizes in a single reaction tube (50, 66).
In an attempt to find an M. intracellulare- and M. avium-specific target for use in clinical laboratory diagnostics, Southern hybridization of genomic fragments cloned from the avian type strain revealed two fragments specific for the MAC that were not found in other mycobacterial species (265). Fragment DT1 was specific for all isolates of M. intracellulare and M. avium strains of serotypes 2 and 3, while DT6 was specific for all M. avium isolates and M. avium subsp. paratuberculosis (265). Although it was not considered at the time, most isolates of M. avium tested were from human clinical isolates (276), therefore most likely consisting of M. avium subsp. hominissuis isolates, which could explain why few M. avium strains were positive for DT1. While DT1 appears to be a marker of M. intracellulare (71, 248) and may possibly be a marker of M. avium subsp. avium, the presence of DT1 was also found in several other mycobacterial species closely related to the MAC (72) and was lacking in some M. intracellulare isolates (97). DT1 reveals no similarity to M. avium 104 (the highest match is 61%) and no similarity to M. avium subsp. paratuberculosis K-10 or other sequences available in GenBank to date. Conversely, DT6 is found in both sequenced genomes and the avian type strain and therefore may serve as a marker for subspecies of M. avium.
Sequence-Based Classification
The ribosomal operon.
The 132 established mycobacterial species at last count are known to present almost as many different 16S rRNA gene sequences. Additionally, many other mycobacterial 16S rRNA genotypes are thought to exist for which the organisms are not yet recognized as species (199, 272). Yet the M. avium subspecies (M. avium subsp. avium, M. avium subsp. paratuberculosis, and M. avium subsp. hominissuis) share an identical 16S rRNA gene sequence and hence cannot be differentiated by 16S rRNA gene sequencing. The unvalidated species “M. brunense” (ATCC 23434) also shares 100% identity with the M. avium 16S rRNA gene sequence (279), although it is unclear to which subspecies it corresponds. The closest relatives to M. avium by 16S rRNA gene sequence vary by 6 bp (M. colombiense), 9 bp (M. intracellulare), and 10 bp (M. chimaera) and are considered part of the MAC in a clinical setting. Restricting analysis to the type strains of validated species, the next closest species are M. bohemicum (13 bp) and M. malmoense (17 bp).
Commercial molecular diagnostic assays offer a user-friendly, rapid method of classifying mycobacteria and are typically based on the ribosomal operon. The first such available assay was the AccuProbe test (GenProbe, Inc., San Diego, CA), developed for the most common mycobacteria in human clinical samples, including the MTBC, MAC, M. kansasii, and M. gordonae. In addition to the pan-MAC probe, species-specific probes against M. avium and M. intracellulare are also available. However, cross-reaction of other mycobacterial species with the MAC probe is not uncommon (72, 247), and only one probe can be tested at a time. Two kits based on the technology of reverse hybridization and on line probe assays have recently been developed which can identify several mycobacterial species at once, with the number increasing with new kit versions. The Inno-LiPA Mycobacteria test (Innogenetics, Belgium) is based on the 16S-23S ITS region (146, 256, 273), and the GenoType Mycobacteria test (Hain Lifescience GmbH, Nehren, Germany) is based on the 23S rRNA gene (217, 229). Both of these contain probes that identify M. avium and M. intracellulare, while the Inno-LiPA kit contains additional probes for the “MAIS complex” and M. intracellulare II. Isolates which hybridize to the MAIS complex probe but not the species-specific probes are common and may pose a diagnostic dilemma, but they do emphasize the complexity of strains that resemble MAC organisms and prevent their undue assignment to either species (147). None of these systems was designed to distinguish between the subspecies of M. avium based on the targets chosen.
The 16S-23S ITS is a highly variable genetic region that has been used extensively to study the variability within MAC organisms. To date, 35 MAC sequevars have been identified, including MavA to -H for the species M. avium, MinA to -D for M. intracellulare, and MAC-A to -X for strains which could not be assigned to either species (70, 92, 176, 186) (GenBank no. AY701784 to -86 [unpublished]). Mav and Min sequevars vary by only 1 to 4 bp, while the MAC sequevars present with significant variability and are candidates for new species. To date, three such new species have been described. M. chimaera, characterized by the MAC-A sequevar, and M. colombiense, characterized by the MAC-X sequevar, are genetically related to the MAC and are considered as such in the clinical setting. In contrast, the species M. parascrofulaceum, characterized by the MAC-G sequevar, is a distant species from MAC organisms and should therefore not be considered as such. With that being said, most clinical MAC isolates present with a MavA, MavB, or MinA sequevar (70, 92, 188), and M. avium subsp. avium, M. avium subsp. silvaticum, and M. avium subsp. paratuberculosis belong to the MavA sequevar (277). Therefore, subspecies of M. avium cannot be distinguished from each other by this method, and the majority of the many ITS sequevars are rare and of unknown epidemiological significance.
The hsp65 gene.
Housekeeping genes offer a higher level of sequence variation than do ribosomal genes but are nonetheless useful for taxonomic purposes due to the relative sequence conservation imposed to maintain function. In this category, the stress protein gene hsp65 is a preferred target for mycobacterial identification to the species level, having routinely been used in diagnostics since the development of a rapid PCR-restriction enzyme analysis (PRA) method using a 441-bp section of the ∼1,600-bp gene (262). However, the PRA method, which is dependent on band size interpretation, shows poor interlaboratory correlation in band size designations. Also, since protein-encoding genes generally have higher mutation rates, as little as one SNP in a restriction enzyme site can result in a different PRA pattern, complicating interpretation (145). With more access to sequencing technology, some laboratories perform hsp65 gene sequencing in the same manner as that done for 16S rRNA gene sequencing (170). The use of this target as an epidemiological tool for closely related mycobacteria, including MAC organisms, has been investigated (72, 86, 190, 288, 289, 303). Single SNPs in this region exist among various subsets of M. avium subsp. hominissuis, and great variability can be observed in M. intracellulare (246). However, like the case for the ITS, the fragment targeted cannot distinguish between M. avium subsp. avium (and M. avium subsp. silvaticum) isolates, M. avium subsp. paratuberculosis isolates, and a great proportion of M. avium subsp. hominissuis isolates. Conversely, the hsp65 sequence outside the Telenti fragment offers unique sequence signatures that can help to identify the various subspecies of M. avium. PRA analysis of a 960-bp fragment was shown to differentiate M. avium subsp. avium from M. avium subsp. paratuberculosis (86). Based on this, comparative sequencing of the nearly complete hsp65 gene was performed on a large panel of isolates, revealing that polymorphisms in the 3′ end, beyond the Telenti region, can unambiguously distinguish between M. avium subsp. avium strains, M. avium subsp. paratuberculosis strains of bovine and ovine types, and six sequevars of M. avium subsp. hominissuis (277).
Other housekeeping genes.
Housekeeping genes other than the hsp65 gene have been evaluated, though to a lesser extent, for mycobacterial identification to the species level. Unfortunately, these studies typically do not include all members of the MAC, omitting at least one of the main subtypes from test panels, and therefore the true utility of these sequence-based tools, at least in the detection and epidemiology of MAC organisms, remains unknown. The manganese superoxide dismutase gene (sodA) revealed several distinct sequevars among MAC organisms, including a unique sequevar for M. avium subsp. paratuberculosis (29), compared to human M. avium isolates. However, SNPs present among three sequences submitted to GenBank representing the identical type strain of M. avium subsp. avium (X81384, U11550, and AY544802) make it difficult to establish whether avian strains do or do not have a unique sod sequevar.
A 236-bp fragment of the dnaJ gene has been reported to produce variable sequences across a panel of MAC strains, but the utility of this target requires further evaluation (179). The reference panel of isolates tested included the 28 MAC serotypes and a set of clinical strains but did not include any samples of M. avium subsp. paratuberculosis. Notably, the degree of genetic diversity observed among M. avium isolates was relatively limited compared to that for strains of M. intracellulare (179). Therefore, further study of this gene target across a broader sample, and perhaps including a larger fragment, is required to determine the utility of dnaJ variability for characterization of MAC organisms.
Several other genes have been assessed for their diagnostic potential for mycobacteria, including gyrB (132), recA (21), the 32-kDa protein gene (247), rpoB (98, 136), and a combination of these in a multigene approach (74). However, these studies generally evaluated only a few strains belonging to the MAC, often limited to the type strains of M. avium subsp. avium and M. intracellulare. Therefore, the utility of these genes in distinguishing between epidemiologically important subsets of MAC is largely unknown.
The observed variability in a number of genes across MAC isolates suggests that a multilocus approach may provide greater discrimination than analysis of each target on its own. For other pathogenic bacteria, such as Neisseria meningitidis, Streptococcus pneumoniae, and Staphylococcus aureus, the combination of high-throughput sequencing technologies and recognized variations in housekeeping genes has enabled the emergence of multilocus sequence typing (MLST) (154, 155) as a powerful tool for typing and taxonomic purposes. An important advantage of this method is the existence of more than 30 (and increasing) curated MLST sequence databases freely available on the Internet, permitting direct comparisons with existing data (154). MLST has not formally been initiated in mycobacteriology to date, but given the number of variable genes noted above, this method could easily be implemented and serve as an epidemiologic or phylogenetic tool to characterize MAC organisms on the subspecies, geographic, or ecotype level.
COMPARATIVE GENOMICS OF THE MAC
Genetic Variability in the Pregenomic Era
Prior to the availability of genome sequence data, and hence the capacity to perform microarray studies, genetic methods such as restriction mapping, Southern blotting, suppression subtractive hybridization (SSH), and representational difference analysis (RDA) were employed to identify regions of difference among study strains. These methods and others were applied to MAC organisms in several studies, as described below.
The earliest genetic variability studies of the MAC set out to identify a genetic basis for differences in GPL composition between rough colony variants of M. avium and their smooth counterparts (15, 16, 81). Belisle et al. (15) performed Southern blot analysis against morphological variants of an M. avium serovar 2 strain, using restricted fragments from a plasmid probe containing the complete ser2 gene cluster responsible for biosynthesis of the serovar 2-specific sugar residue characterized earlier (17). Striking differences were observed between rough morphotypes and their parent smooth strains, and it was determined that this was due to genetic deletions, one of which resulted from IS element-mediated recombination causing the loss of the complete ser2 cluster (81). Some genetic differences were also observed between the two serotype 2 strains, i.e., TMC724 from a bird and M. avium 2151, isolated from human sputa (15) and later characterized as having multicopy IS1245 and therefore being M. avium subsp. hominissuis (140). Part of the ser2 gene cluster was also identified by RDA and characterized in an independent study (30, 268), although the association with the ser2 GPL cluster was not confirmed until recently (80). The region was designated “GS,” was described as “genetic island-like” with a lower G+C content, and was found in M. avium subsp. paratuberculosis, M. avium subsp. silvaticum, and some M. avium subsp. avium strains of serotype 2 but not in other strains of M. avium subsp. avium. Although the following was not specifically addressed, MAC strains not containing the GS element could be inferred to be M. avium subsp. hominissuis. In an attempt to identify genetic differences between a virulent M. avium strain (M. avium subsp. avium strain 724) and a less virulent human strain of M. avium (M. avium subsp. hominissuis strain A5), RDA analysis was performed and also revealed genes belonging to the ser2 GPL cluster in strain 724 (141).
SSH of M. avium subsp. paratuberculosis against strains of M. avium subsp. avium identified 42 short genetic regions, 24 of which were deemed specific for M. avium subsp. paratuberculosis on the basis that they also revealed low sequence similarity by BLAST searching of genome sequence data from M. avium strain 104 (138). To identify genetic differences between M. avium subsp. paratuberculosis variants of type I/S/ovine and type II/C/bovine (75), the avian type strain of M. avium subsp. avium (ATCC 25291) served to identify M. avium subsp. paratuberculosis-specific sequences by RDA, and one each of the M. avium subsp. paratuberculosis bovine and ovine types was tested against the other to uncover their unique genetic signatures. Three small genetic regions were identified as unique to M. avium subsp. paratuberculosis versus M. avium subsp. avium. Also, three genetic regions present in type I but not type II strains were also present in M. avium subsp. avium, suggesting that type I M. avium subsp. paratuberculosis is an evolutionary intermediate between M. avium subsp. avium and bovine M. avium subsp. paratuberculosis. No genetic regions unique to type II strains and not present in type I strains were identified in the study. In contrast, Marsh and Whittington did uncover an 11.5-kb region missing from the S/I type and present in the C/II type by RDA (162), although comparison with other M. avium strains was not carried out. RDA analysis also uncovered a 7-kb section which was further characterized as part of a putative 38-kb pathogenicity island specific for M. avium subsp. paratuberculosis (253). Described as an ABC transporter operon (mpt), no similarity was found in sequences available from GenBank or in the sequenced genome of M. avium strain 104 from TIGR. Since the driver used in the RDA experiments was M. avium subsp. avium (ATCC 25291T), it is likely that this region is also not found in avian strains.
While this review is focused on chromosomal genetics, it is important that many strains of MAC organisms are known to harbor a variety of plasmids (58, 60, 163, 171). This aspect of MAC research is significantly understudied at present but has revealed important findings in terms of strain epidemiology (171), virulence (95, 96), adaptability and resistance (91, 94, 205), and other mechanisms attributed to the presence of plasmids (61). While most M. avium strains isolated from AIDS patients in the United States were found to contain plasmids (59, 114, 180), the sequenced strain of M. avium 104 is not known to contain any and was chosen in part due to its rare ability among M. avium isolates to be genetically manipulated (197, 271). Furthermore, to our knowledge, no plasmids have been discovered or identified in strains of M. avium subsp. paratuberculosis.
Genomes of Strains M. avium subsp. paratuberculosis K-10 and M. avium 104
MAC genetics and genomics have advanced exponentially in the past decade, with but one paragraph on this topic in the last comprehensive MAC review (124). The turn of the century brought us the first genome sequences of MAC organisms, for (i) M. avium strain 104 from the blood of an AIDS patient (TIGR), a representative of M. avium subsp. hominissuis; and (ii) M. avium subsp. paratuberculosis strain K-10, isolated from a cow with Johne's disease (152). Several other MAC genome sequencing projects are under way and are expected to generate more data in the coming 3 to 5 years (John Bannantine and Vivek Kapur, personal communication).
The K-10 genome is 4.83 Mb long, has a G+C content of 69.3%, and contains 4,350 open reading frames (ORFs) (152). For comparison, M. avium 104 has an additional ∼700 kb of DNA, for a total genome size of 5.48 Mb, but has a similar G+C content of 69.0%. Comparison of genes orthologous between the two genomes reveals 98 to 99% sequence identity. Approximately 75% of the K-10 genome has homologs in the published M. tuberculosis H37Rv genome sequence (48, 152). Since little is known about the actual virulence mechanisms of M. avium subsp. paratuberculosis, the representation of H37Rv genes associated with virulence was investigated (152). The K-10 genome has significantly fewer PE/PPE genes (i.e., with Pro Glu and Pro Pro Glu motifs) (1% versus 10% of total genes in H37Rv). As well, K-10 has additional mammalian cell entry (mce) gene homologs or operons but also a lack of some of the specific mce genes described for H37Rv. Additionally, K-10 has a larger number of genes possibly involved in lipid metabolism. K-10 is notably lacking two important operons, for polyketide and mycocerosic acid synthesis, that together result in the production of the cell wall component phthiocerol dimycocerosate. However, additional genes were identified which may possibly play a role in phthiocerol dimycocerosate synthesis. Since several of these are also found in M. avium 104, they alone cannot explain the pathogenic nature of M. avium subsp. paratuberculosis.
The raw genome sequence of M. avium 104 has been available from TIGR since about 2003. However, the TIGR annotation was just released in late 2006 (GenBank accession no. CP000479). In the interim, publications on the M. avium 104 genome were based on the in-house annotation efforts of individual groups (240, 304). These predicted the presence of between 4,480 (240) and 4,987 (304) ORFs. The present TIGR annotation includes 5,313 genes, 5,120 of which code for proteins. The differences in gene content reflect differences in annotation methods and improvements in the identification of short genes. A formal published annotation of M. avium 104 will be a useful resource for helping to resolve these differences and opening the door to further postgenomic study of the MAC.
Comparative Genomics of the MAC
Prior to the completion of the two MAC genome sequences, comparative analyses using contig fragments representing the partial genome of K-10 already revealed 27 genes unique to it compared to M. avium strain 104 (6). Some of these were found to be present in other mycobacteria, such as M. intracellulare and other strains of M. avium, when screened by PCR, emphasizing caution in the interpretation of what is truly subset specific and foreshadowing the genetic variability in the complex. Additionally, examination of the origin of replication (oriC) site of M. avium subsp. paratuberculosis compared to that of M. avium subsp. avium as well as random genomic regions beyond the oriC site did not reveal any notable differences that could explain their divergent phenotypes. Nucleotide identity values were no different from those observed for other bacteria belonging to the same species and with identical phenotypes (8).
Based on the availability of genome sequence data, three groups have assembled DNA microarrays, two based on the genome sequence of M. avium 104 (240, 304) and one starting with the M. avium subsp. paratuberculosis K-10 genome as a template (201). These microarrays then served to evaluate genomic variability in the MAC as a whole. The different groups employed similar experimental approaches, beginning with a relatively small panel of MAC strains of different types to identify large sequence polymorphisms (LSPs) or regions of difference by microarray analysis and then confirming the presence/absence of these regions by PCR and sequencing across a larger panel of isolates. In part due to an unawareness of the taxonomic issues detailed above, discrimination between M. avium subsp. hominissuis and “true” M. avium subsp. avium isolates was not taken into consideration, which may have resulted in greater complication and/or simplification in the interpretation of results from these experiments.
The first of these experiments used the first available genome data set, albeit not an annotated set, namely, that of M. avium 104 from TIGR (240), and included 93% of the putative ORFs, representing a first-generation array. Microarray analyses were performed against strains representative of other M. avium subsets, including M. avium subsp. paratuberculosis K-10 (bovine type) and LN20 (ovine type) and the type strain of M. avium subsp. silvaticum. Fourteen LSPs, defined as representing six or more contiguous ORFs missing in test strains, were identified as present in strain 104 but not in the others. These were first labeled LSP1 to LSP14 and subsequently renamed LSPA1 to LSPA14 to distinguish them from the LSPPs described in a subsequent study as present in M. avium subsp. paratuberculosis K-10 but missing from M. avium 104 (236). LSPAs were 21 kb to 197 kb long, for a total of 727 kb (13.5% of the strain 104 genome). Only LSPA11, representing part of the mce2 operon, appeared to be variably present in M. avium subsp. paratuberculosis. Upon further investigation, it was determined to be missing specifically from an ovine strain of a specific IS900 RFLP type (unpublished data), which happened to have been the only ovine strain tested by microarray in this study. Notably, only 3 of the 14 LSPAs were observed as uniformly present in non-M. avium subsp. paratuberculosis MAC strains, indicating that a small minority of the variability observed represented differences between M. avium subsp. hominissuis and M. avium subsp. paratuberculosis. Additionally, no LSPA could serve to distinguish M. avium subsp. silvaticum from strains designated M. avium subsp. avium. Principal observations made from this comparative analysis included a high conservation of PE/PPE genes in all tested strains and variability in the distribution of mce genes. Genes of the mycobactin synthesis operon (mbtA to mbtJ), which is characterized for M. tuberculosis (212), are all present in strains 104 and K-10. However, mbtA, believed to be the initiator of mycobactin synthesis, is truncated in M. avium subsp. paratuberculosis strain K-10 (240), as confirmed by others (152). The hypothesis that this may be the cause of mycobactin dependency in M. avium subsp. paratuberculosis remains to be confirmed formally by functional studies and may be technically hampered by the existence of a number of smaller mutations in other genes of the mbt operon (C. Y. Turenne and M. A. Behr, unpublished).
More recently, another group took a similar approach where smaller regions of deletion, defined as three or more consecutive ORFs, were considered in comparative analyses. Twenty-four LSPs were identified as missing from M. avium subsp. paratuberculosis strains, which included most of those described by Semret et al. plus an additional 96 ORFs distributed among 11 LSPs. Altogether, these ranged from 3 to 196 kb long, totaling 846 kb (17% of the strain 104 genome) (304). In addition to mce operons, genome plasticity was also observed in TetR transcriptional regulators. Finally, three large genetic inversions were described between the 104 and K-10 genomes.
Working in the converse sense, the availability of the M. avium subsp. paratuberculosis K-10 genome has facilitated in silico (236, 304) and microarray-based (201) approaches to determine which genes are present in K-10 but missing from M. avium 104 and other MAC organisms. Semret et al. identified 17 LSPPs spanning 230 kb of sequence in sections of 3 to 66 kb (236). Comparably, Wu et al. identified 18 LSPPs (GI MAP-1 to -18) spanning 240 kb, 16 of which were perfectly shared in both studies, with the only differences being due to short genetic regions spanning a few genes (304). Paustian et al. presented their microarray data according to any individual genes differentially present in MAC strains versus K-10 (201), not restricted to runs of genes. Not surprisingly, many of these consisted of transposase genes specific to M. avium subsp. paratuberculosis. Also, they identified seven large regions of difference that corroborated with the larger LSPPs described in the other two studies and several single genes or genes in small groups that corresponded mostly to the smaller LSPPs.
Studies to date have found that little genomic variability exists among M. avium subsp. paratuberculosis strains. However, the level of variability between M. avium subsp. paratuberculosis and the other MAC organisms is >1 log greater than that observed through nearly a decade of genomic studies on the MTBC. In common with efforts for the MTBC, though different labs find various numbers of elements, LSPs noted across different papers are typically recognizable as the same genomic regions, providing valuable independent confirmation for the findings presented.
Genetic Variability in the Postgenomic Era
Evolutionary events among the MAC organisms can be speculated upon by the use of LSP analysis. LSPs can be the result of (i) horizontal gene transfer (HGT) or genetic insertion events or (ii) deletion events. Which event occurred is not always evident by simple comparative genomics of two strains against each other. Events in intergenic regions reveal no directionality by themselves. One can best assume an insertion event if the flanking regions represent a single gene split in two. Conversely, a deletion event can be inferred if a gene of known function or homology to a closely related species is truncated or if both the stop codon of one gene and the start codon of another are deleted, resulting in a chimeric gene (26). Additionally, different organisms lacking the same element via a deletion should share precisely the same truncation point, as revealed by sequencing across the deletion (183). Another approach, which requires investigation beyond direct comparative genomics, involves determining the distribution of variable genetic elements in closely related strains, in this case restricted to the MAC complex. Ideally, such an approach would serve to inform M. avium genomic studies if the sequence of an M. intracellulare strain were determined. However, since this is not the case, and few nontuberculous mycobacterium genome sequences exist, the tendency is perforce to perform comparative genomics with what is available, mainly the MTBC. HGT in M. avium subsp. paratuberculosis K-10 has been evaluated by this approach (157). However, without taking into consideration other members of the MAC apart from M. avium strain 104, HGT remains difficult to ascertain.
The availability of the genome sequences provides the capacity to build whole-genome arrays of strains 104 and K-10, separately or combined, for screening against other MAC species. Of particular interest is the assessment of genomic variability within M. avium subsp. paratuberculosis as a whole or between the two predominant types, i.e., S and C. Two independent studies identified several different LSPs between the C and S types based on microarray analyses. Firstly, two LSPs spanning clusters of several genes were found to be present in the C type, not the S type. One of these, called LSPA20 (239) or deletion 1 (160), spans from MAP1484c to MAP1491 and contains putative pyruvate dehydrogenase genes. The region is also present in M. avium 104, suggesting that the LSP may represent a true deletion in the S lineage. The second region not present in S-type versus C-type strains, which is 20 kb long and referred to as deletion 2 (160), spans from MAP1728c to MAP1744 and includes mycobacterial membrane protein genes (mmpL5 and mmpS5) previously identified by RDA (162). Comparison with the M. avium 104 genome, however, reveals a complex event. The middle section of deletion 2 is missing from strain 104 and overlaps with the LSP-P9 fragment (236), while the two end clusters of genes found present in 104 are separated by a 203-kb sequence corresponding to the previously designated LSP1 (now LSPA1) (240). Further analysis will be required to establish the sequence of events in that region.
In addition, the use of a combined strain 104 and K-10 genome array uncovered two LSPs that were present in M. avium 104 and M. avium subsp. paratuberculosis S-type strains but deleted from C-type strains (239). The first, LSPA18, is a 16-kb sequence that is replaced by an IS900 element in strain K-10. The second LSP, designated LSPA4-II, is of particular interest because it consists of a 26-kb section of the 197-kb sequence previously designated LSP4 (or LSPA4) that is present in M. avium 104 but not in strain K-10. Interestingly, the mbtA gene is intact in the S type due to the presence of the LSPA4-II sequence, whereas the absence of this element in C-type strains results in the truncation of the mbtA gene in K-10 (239). This finding provides strong evidence that the ovine form is ancestral to bovine M. avium subsp. paratuberculosis and related to the domestication of sheep predating that of cattle (45). Moreover, this genetic finding challenges the mycobactin-dependent phenotype ascribed to M. avium subsp. paratuberculosis, which has been examined more rigorously for C-type strains. Again, functional studies will be required to address this point, and since S-type strains are so fastidious, mycobactin dependence may not be readily quantified. Certainly, other factors are implicated in the painstakingly slow in vitro growth rate of ovine M. avium subsp. paratuberculosis. The evidence obtained from comparative genomics of the C and S types does suggest that the M. avium subsp. paratuberculosis S-type strains are intermediary between M. avium and C-type strains. However, an important limitation of microarray analysis is the inability to uncover an extra genomic region in these strains. Using RDA, fragments of the regions LSPA18 and LSPA4-II had been identified, but no fragments unique to M. avium subsp. paratuberculosis S-type strains were reported (75). Since RDA is only able to detect relatively short genetic sequences and is subject to significant interexperiment variability (162), the ultimate determination of whether ovine strains of M. avium subsp. paratuberculosis contain additional genetic material will come from a complete genomic sequence.
In addition to the study of genetic variability within M. avium subsp. paratuberculosis, microarray analysis of other MAC species has been initiated. A 6-kb region designated LSPA17, spanning from MAP1375c to MAP1381c, was found to be missing from all IS901+ strains tested, i.e., M. avium subsp. avium and M. avium subsp. silvaticum, but was present in all M. avium subsp. paratuberculosis and M. avium subsp. hominissuis strains (with the exception of those represented by a code 3 hsp65 sequence) (239). The significance of the commonality between the avian strains and a specific subset of M. avium subsp. hominissuis strains is unknown but does emphasize the importance of including a representative panel of isolates to avoid bias in the interpretation of comparative genomic data. The directionality of this event, like that for many others, remains to be determined by further analysis.
Work on the ser2 GPL cluster progressed further with the availability of the genome sequence of M. avium 104. A pathway for the biosynthesis of the serovar-specific GPL in MAC strains of serovar 2 was ultimately proposed based on a combination of previous data with sequence comparison with M. avium 104 (serovar 1) and analysis of strain 104 rough morphotype mutants (80). An additional strain, A5 of serotype 4 from an AIDS patient, was evaluated in similar comparative analyses of the GPL cluster (141), providing further insight into the high level of genomic variation, including insertions-deletions and possible rearrangements, within members of M. avium.
Diagnostics and Taxonomy Based on LSPs
There are unique regions of diagnostic interest in M. avium subsp. paratuberculosis which may help to overcome many of the sensitivity and/or specificity hurdles that continue to hinder M. avium subsp. paratuberculosis diagnostics. Comparative genomics of strains 104 and K-10 have allowed for the characterization of genetic regions unique to each, but only in relation to each other. The K-10 genome revealed ∼161 “unique” sequences (152), or approximately 200 kb of sequence data was found in K-10 only, not in strain 104. However, among the 17 LSPs constituting the 200 kb, 10 were also found present in other MAC species (236). The LSPs specific to M. avium subsp. paratuberculosis (designated LSPP2, -4, -11, -12, and -14 to -16) contained, not surprisingly, the previously described hspX gene (in LSPP12; gene MAP2182c), F57 (in LSPP4; gene MAP0865), and the 38-kb pathogenicity island or ABC transporter operon (in LSPP14; MAP3725 to -3764). Whether these regions are present in all M. avium subsp. paratuberculosis strains remains to be clarified. In at least two studies using PCR-based screening, certain elements were reported as variably present (236, 304); however, employing Southern blot analysis, we instead found that these elements are consistently present across M. avium subsp. paratuberculosis strains (Turenne and Behr, unpublished observations). Regarding LSPP14, the surface-exposed MptD protein (encoded by gene MAP3732c) was subsequently used as an M. avium subsp. paratuberculosis-specific target for the development of a peptide-mediated diagnostic assay for detection in bulk milk samples (252). Additionally, M. avium subsp. paratuberculosis-specific DNA sequences previously identified by SSH (138) correspond to sections within LSPP2, -4, -11, -14, and -16.
Strains can also be identified by what they do not contain, as applied to “brand” lineages of the MTBC by their genomic deletion profiles (185, 198, 257). For example, LSPA8 is a region that shares homology with elements of the MTBC and is present in all MAC species except M. avium subsp. paratuberculosis, where it has been deleted (239). Presumably, this deletion involved the loss of genes no longer selected for in the bacterium's host-adapted environment. In the practical sense, this region was used to design a three-primer PCR that could distinguish unambiguously between M. avium subsp. paratuberculosis and non-M. avium subsp. paratuberculosis strains, which is useful not only for rapid identification to the subspecies level but also for the detection of mixed cultures or infections (239).
While there is great genetic variability among MAC organisms, very little genomic variation seems to exist among M. avium subsp. paratuberculosis strains. The principle differences documented to date involve variability between ovine and bovine subtypes. Epidemiological tests also reflect the clonal characteristics of the species, but further comparative genomic studies will be required to determine if unrecognized variability exists within M. avium subsp. paratuberculosis.
Immunodiagnostics of M. avium subsp. paratuberculosis
The search for M. avium subsp. paratuberculosis-specific antigenic proteins is ongoing, as their discovery would have great potential for the design of diagnostic methods for detection of M. avium subsp. paratuberculosis. Not surprisingly, however, most genes identified as coding for M. avium subsp. paratuberculosis antigenic proteins are also present in other mycobacteria or subspecies of the MAC (reviewed in references 112 and 200). Their nonspecificity may therefore translate into false-positive results in serodiagnostic assays, as most hosts for which a diagnosis of M. avium subsp. paratuberculosis infection is being investigated are likely to be exposed to environmental strains of M. avium. For the antigenic proteins known to be M. avium subsp. paratuberculosis specific, their utility in immunodiagnostics is uncertain: the HspX protein can be recognized by a small portion of M. avium subsp. paratuberculosis-infected cattle sera only (7). The differential expression of antigenic proteins in M. avium subsp. paratuberculosis versus other mycobacterial species may offer additional options for immunodiagnostics (192, 193), although much work is needed to investigate this possibility.
With the completion of the M. avium 104 and M. avium subsp. paratuberculosis K-10 genome sequences, Paustian et al. focused on a selection of 13 ORFs deemed specific to M. avium subsp. paratuberculosis based on BLAST searches against public databases and PCR screening against other MAC organisms and mycobacteria (200). Five of these were cloned, expressed in Escherichia coli, purified in adequate amounts, and tested for reactivity against sera from animals experimentally immunized or naturally exposed to M. avium subsp. paratuberculosis. Variable results were observed across the five genes, but the protein from MAP0862 showed great promise, as all serum samples from cattle in the clinical stages of disease but none of the uninfected sera were reactive.
In contrast to efforts in building one protein at a time, an alternative approach recently advanced was to generate protein extracts from the surfaces of the relevant bacteria and then determine whether cross-reactivity was observed. This report suggested the generation of a highly specific preparation for Johne's disease immunodiagnostics (82), although further validation and independent verification are needed.
Comparative Genomics: Current Appreciation and Diagnostic Implications
With the exclusion of M. leprae and the MTBC, both of which are considered strictly host-restricted pathogens, for most other mycobacteria living in the soil or water there should be plenty of opportunity for gene exchange. However, since the most-studied mycobacterial species is the MTBC, documentation of HGT in mycobacteria has been rare. Few genomic studies have been done within the genus outside the MTBC, which is in great part due to the fact that the earliest and most abundant Mycobacterium genome sequences available were for strains of the MTBC.
General observations from comparative genomic analyses of the MAC have been presented in this review and indicate the tremendous degree of genomic variability, the presence of both vertical and horizontal forms of genetic variability, and the potential to exploit these findings for laboratory identification. While further work is clearly indicated to completely catalogue differences between MAC organisms, there are now a number of bacterial targets for identification to the species level and the first examples of protein targets offering the possibility of specific immunodiagnosis. A tabulation of some of these elements and their distribution across MAC organisms for diagnostic purposes is presented in Table 3.
TABLE 3.
Distributions of genetic elements and gene targets across MAC organisms
| Genetic target | IS familyj | Presence of targetg
|
Comment | Reference(s) for distribution within subspecies | |||||
|---|---|---|---|---|---|---|---|---|---|
| 104a | MAHb,h | K-10c | MAPd,i | MAAe,h | MIf | ||||
| IS900 | IS110 | − | − | + | + | − | − | SNPs present in S type (238) | 10, 49, 51, 84 |
| IS901 | IS110 | − | − | − | − | + | − | 10, 22, 219 | |
| IS1245 | IS256 | + | + | − | − | + | − | 10, 49, 84, 219 | |
| IS1311 | IS256 | + | + | + | + | + | − | SNPs present among MAC subsets (296) | 49, 73 |
| ISMav1 | IS21 | + | +/? | − | ? | +/? | ? | Same as IS1612 and IS2534 | 30, 80 |
| IS666 | IS256 | + | + (21%) | − | − | − | − | 227 | |
| IS1601 | IS256 | + | +/? | − | ? | ? | ? | 81 | |
| ISMav2 | IS481 | − | ? | + | + | − | −/? | 243, 254 | |
| IS999 | IS3 | + | +/? | − | −/? | −/? | ? | 144 | |
| ISMpa1 | IS110 | − | +/? | + | + | −/? | −/? | Same as IS_MAP12 (152) | 191 |
| IS1110 | IS110 | − | +/? | − | ? | ? | ? | 116 | |
| IS1141 | IS3 | − | ? | − | ? | ? | ? | GenBank accession no. L10239 | |
| IS1626 | IS110 | − | −/? | − | ? | ? | ? | 210 | |
| IS1613 | IS110 | − | ? | − | ? | ? | ? | GenBank accession no. AJ011837 | |
| hspX | NA | − | − | + | + | − | − | Present in LSPP12 (236) | 83, 84 |
| F57 | NA | − | − | + | + | − | − | Present in LSPP4 (236) | 206, 258, 282 |
| DT1 | NA | − | −/? | − | − | +/? | + | Found in other species related to the MAC | 72, 264, 265 |
| DT6 | NA | + | + | + | + | + | − | 72, 264, 265 | |
| mptD | NA | − | − | + | + | − | − | Present in LSPP14 (236) | 252 |
Sequenced strain M. avium 104, which is an M. avium subsp. hominissuis strain.
M. avium subsp. hominissuis strains in general.
Sequenced strain M. avium subsp. paratuberculosis K-10.
M. avium subsp. paratuberculosis strains in general.
M. avium subsp. avium and M. avium subsp. silvaticum strains in general.
M. intracellulare.
?, presence unknown; +, present; −, absent; +/? (or −/?), present (or absent) in one or some strains, but distribution throughout is variable or unknown.
While the distinction between the two subsets has not always been taken into consideration, extrapolation could generally be made by the use of known strains or by assumptions.
Presence in both bovine and ovine forms, unless stated otherwise.
NA, not applicable.
UNRESOLVED ISSUES
Does M. avium subsp. silvaticum Really Exist?
The existence of M. avium subsp. silvaticum as a unique subspecies depends upon the interpretation of phenotypic assays, including mycobactin dependence upon primary isolation (267). Results to date of genetic and genomic studies have failed to reveal a distinct molecular profile for M. avium subsp. silvaticum strains compared to that for M. avium subsp. avium strains (79, 239, 277). This discordance raises the issue of whether the phenotypic uniqueness of these organisms is sufficient to merit the designation of a distinct subspecies, and by extension, whether the assays so used have sufficient reliability to serve in this role. The phenotype of mycobactin dependency upon primary isolation is difficult to determine in a controlled manner, particularly if it is dependent on the bacterial burden. Successful culture of these organisms from acid-fast test-positive wood pigeon or roe deer tissue was sparse and required 4 to 5 months for visible growth, even in the presence of mycobactin (131, 249). In one study, wood pigeon mycobacteria grew better in the presence of mycobactin but could also be cultured without mycobactin if a large number of bacilli were present (165). At best, this phenotype might be mycobactin-enhanced growth rather than mycobactin dependence. Barclay and Ratledge found that once mycobactin-dependent M. avium strains lost this phenotype, they were able to produce mycobactin in small amounts, implying a regulatory effect as opposed to a genetic defect (9). Finally, in another study, wood pigeon mycobacteria grew exclusively in 7H11 medium, not on media containing mycobactin, after 6 months (128).
Another important problem in determining the taxonomic status of M. avium subsp. silvaticum is that a small number of strains have been described in the literature, and these isolates have then been passed around from lab to lab for testing by novel methods, contributing to laboratory-generated selection of colonies that replicate best under in vitro conditions. Also, since few M. avium subsp. silvaticum isolations have been reported in the past several decades, it is possible that fastidious nutritional requirements have resulted in underdetection of these organisms. Alternatively, in an era where molecular testing has largely eliminated detailed phenotypic study, organisms previously called M. avium subsp. silvaticum may simply be classified as IS901+ M. avium subsp. avium in the modern era.
At the molecular level, doubt regarding the validity of this species first emerged from a comprehensive study evaluating IS901 and IS1245 RFLP patterns across a large number of M. avium subsp. avium and M. avium subsp. silvaticum isolates from birds, pigs, cattle, humans, and a few odd hosts and reference strains (79). The M. avium subsp. silvaticum strains revealed two IS901 profile types: the first was identical to that from strains of M. avium subsp. avium, and the other profile revealed a single band shift from the pattern for the avian strains. On this basis, the authors emphasized that identification of M. avium subsp. silvaticum could not be made on the basis of mycobactin J dependency and IS901 positivity alone. In other genetic studies, the GS region characterized in M. avium subsp. silvaticum (268) was found to be identical in sequence, including that in the ISMav1 site, to the syntenous region in the avian type strain, TMC724 (80). PCR results for GS-specific genes in other MAC strains of bird origin were also indistinguishable from those for M. avium subsp. silvaticum (30). Finally, in a sequence-based evaluation of the nearly complete hsp65 genes of MAC isolates, M. avium subsp. silvaticum was found to have an identical sequevar to that of M. avium subsp. avium, and all strains in both groups were IS901+. Moreover, no IS901-negative MAC strains presented with that particular sequevar (277). Overall, no genetic data exist to distinguish M. avium subsp. silvaticum from M. avium subsp. avium. Comparative tests have been done, but only against M. avium subsp. paratuberculosis and/or M. avium subsp. hominissuis (under the label of M. avium subsp. avium). With the availability of new genomic tools and the vast amount of information obtained, nothing more has been noted as strikingly distinguishable in M. avium subsp. silvaticum since its description in 1990.
Sequencing the genome of M. avium subsp. silvaticum may provide some answers. However, a genome sequence is not yet available for M. avium subsp. avium or, for that matter, any IS901+ strain. By habit, the M. avium subsp. avium designation is still used by many to denote both the bird pathogen and environmental strains that cause opportunistic infections of humans. Since the sequenced strain M. avium 104 belongs to the M. avium subsp. hominissuis subset, a future genome sequencing project for the MAC should include at minimum the type strain of M. avium subsp. avium (TMC724 or ATCC 25291) or another well-characterized IS901+ strain typical of the pathogenic bird isolates. Sequencing of an M. avium subsp. silvaticum strain, preferably the type strain, ATCC 49884, could be done in tandem or thereafter. Should an M. avium subsp. silvaticum genome sequence be determined, it will be important to not consider unique attributes to be specific to M. avium subsp. silvaticum without first determining if these genetic variants are shared with IS901+ M. avium subsp. avium strains. If there are truly very few differences between the genome sequences of M. avium subsp. avium and M. avium subsp. silvaticum, as suspected, it may provide closure to the confusion and result in the demise of the latter name.
Unique genetic regions in an IS901-positive strain compared to M. avium 104 have already been identified as part of a study characterizing the integration sites of the IS901 element in a cervine strain (126). A gene encoding the p40 protein which is present in the genomes of many mycobacteria, including members of the MAC, was found to be expressed only in IS901+ strains (1, 125). Additional interest in sequencing an avian strain stems from the view that this organism is a professional pathogen, like M. avium subsp. paratuberculosis, that has an unknown capacity to replicate in the environment. Perhaps M. avium subsp. silvaticum does represent a specific host-associated (wood pigeon) subset of the M. avium subsp. avium branch (that can infect a variety of birds), akin to caprine and bovine variants of M. bovis described for the MTBC. With that being said, genetic markers that can make this distinction do not yet exist, and it will be interesting to see what future genomic data can provide.
Is M. avium subsp. hominissuis the Only True Environmental M. avium Subspecies?
While the appropriate designation of a MAC isolate may seem unimportant to the molecular biologist, veterinarian, or physician, this distinction is critical for understanding the epidemiology and capacity to cause disease. For example, where efforts have been applied to distinguish between M. avium subsp. avium and M. avium subsp. hominissuis, studies have consistently revealed that human MAC isolates are generally in the latter category (178, 263), indicating either a reduced exposure or a reduced risk of disease due to the former. Thus, when attempting to determine the source of exposure for a patient with M. avium subsp. hominissuis disease, there is little a priori reason to consider birds or environments that they could contaminate, and consistent with this, when a link between human disease and birds was investigated, it could not be demonstrated (219, 263). A possible explanation is that birds are infected by a specific subset of M. avium strains that are obligate pathogens of birds, in the same fashion that M. avium subsp. paratuberculosis is an obligate pathogen of ruminants. Supporting this notion, avian MAC infection does not seem to affect the bird population on a large scale, as would be expected if this M. avium subset was prevalent in the environment. If this is true, then the vast majority of the biomass of M. avium subsp. avium will be found in birds, not the environment, explaining why humans are generally not infected by avian strains. In this case, the majority of MAC and related species in the environment are simply M. avium subsp. hominissuis and M. intracellulare, and the general statement that M. avium is an environmental organism is misleading.
The highly variable protective efficacy of M. bovis BCG has prompted scientists to evaluate the impact of environmental mycobacteria on mycobacterial vaccination and/or infection (196). Brandt et al. showed that presensitization of mice by environmental mycobacteria resulted in a rapid immune response upon subsequent vaccination which inhibited the multiplication of BCG, thereby diminishing the capacity of BCG to provide protection. This was first tested by presensitization at 2-week intervals with three different ATCC strains and species, including M. avium ATCC 15769, which in actuality is an IS901+ bird strain rarely found in the environment (24). The second experiment evaluated the BCG-blocking capacity of two soil and two sputum isolates of rapidly growing mycobacteria as well as two sputum isolates of M. avium. Of these, only the M. avium strains prevented multiplication of BCG, but the true species or subspecies designation of these strains was not provided. Concurrently, Buddle et al. evaluated the efficacy of BCG and newly attenuated M. bovis strains in calves with strong responses to avian PPD, and hence exposure to “environmental” mycobacteria, and found that the protective effect of BCG in PPD-positive animals was also diminished (27). In a follow-up study, de Lisle et al. performed similar experiments in guinea pigs but methodically compared the effects on vaccine efficacy of presensitization with IS901+ M. avium versus IS901− M. avium strains, based on observations that M. avium strains that caused lesions in farmed deer and cattle were IS901+, while strains that did not cause lesions were IS901− and genetically distinct from the IS901+ strains (68). They found that animals presensitized orally with IS901+ strains received little protection from BCG, whereas presensitization with IS901− strains did not impair BCG-induced protection. These studies show that not all environmental mycobacteria or M. avium strains impair protection by BCG equally. Furthermore, the impact of selecting an IS901+ strain as representative of an environmental isolate for PPD testing is uncertain while IS901+ organisms have not been detected in the environment.
Considerations in Exploring M. avium subsp. paratuberculosis as a Human Pathogen
The hypothesized link between M. avium subsp. paratuberculosis and CD originally dates to similarities noted between the pathology of Johne's disease and that of human cases subsequently classified as CD (64). Unfortunately, this possibility remains a source of debate nearly a century later. One of the critical issues remains the exceptionally slow growth of M. avium subsp. paratuberculosis and its specific growth requirements that are typically not part of routine clinical microbiology protocols. It appears that the best chance at detecting and culturing M. avium subsp. paratuberculosis from human samples, although not assured, is with tremendous patience, experience, and tailored culture media. Despite this, M. avium subsp. paratuberculosis has very seldom been cultured from human specimens (42, 43). One case described the isolation of M. avium subsp. paratuberculosis from 8 of 21 specimens obtained from an AIDS patient over a 3-year period (218). Histology revealed many acid-fast bacteria, but mycobactin was not incorporated into the media at first, and therefore growth was very poor in liquid, and no growth was observed on solid media. Only after 2 years was M. avium subsp. paratuberculosis considered a possibility, and then culture was achieved on Middlebrook medium with mycobactin J. This culture was then determined to be positive for the IS900 element. The authors noted that limited growth of M. avium subsp. paratuberculosis may occur in liquid media if the organism is abundant in clinical specimens, but evidently this is a rare case, because large series of disseminated MAC disease in AIDS patients failed to show the presence of M. avium subsp. paratuberculosis organisms (275, 287, 308). Additionally, this type of disease presentation in a case of AIDS does not reflect the burden of organisms one might expect in CD, where by definition, pathogenic organisms have not been detected. Therefore, if humans with CD have M. avium subsp. paratuberculosis infection, the microbiologic status must be paucibacillary, providing a further technical challenge to its detection amid large numbers of rapidly growing enteric flora.
One hypothesis for the difficulties in isolating M. avium subsp. paratuberculosis from CD tissue is that it may be present in a cell wall-deficient (CWD) form in tissue. M. avium subsp. paratuberculosis in a CWD form was first documented for cultures derived from CD patients after 8 to 15 months of incubation on Herrold's egg yolk medium slants (43). Subsequently, others have cultured CWD cells, or “spheroplasts,” from CD tissue, on the basis that organisms recovered were non-acid-fast or variably acid-fast coccoid or pleomorphic cells and, following a long incubation phase in vitro, eventually adopted the standard acid-fast mycobacterial phenotype (99, 156). In specimens where only the CWD form would exist upon culture, species identification could not be reliably confirmed. CWD forms were also observed from ulcerative colitis patients and control samples (31).
To overcome the noted problems in culturing M. avium subsp. paratuberculosis, and as a means of addressing the possibility of CWD forms, an alternative advanced in the past decade has been to apply molecular methods instead of conventional microbiologic assays. The number of papers existing that incorporate the terms “PCR” and “IS900” currently rivals the number of mycobacterial species (n = 130). In the context of CD, results run the gamut from (i) no evidence of M. avium subsp. paratuberculosis DNA in CD samples (5, 18, 35, 39, 77, 222) to (ii) M. avium subsp. paratuberculosis DNA found primarily in CD samples (4, 28, 69, 224, 226, 233, 235) and (iii) M. avium subsp. paratuberculosis DNA found in CD patient samples, ulcerative colitis patient samples, and healthy controls, in various proportions (53, 187, 255). The controversy will likely continue, and it is not our intention to recapitulate the many arguments presented for and against the hypothesis; 20% of publications containing the terms “Crohn” and “paratuberculosis” are review articles, and the following references address some of these issues (13, 36, 37, 40, 105, 106, 111, 115, 213, 230, 242, 281, 286).
What is certain is that if M. avium subsp. paratuberculosis does infect humans, it is not an easy task to prove that this is the case. A recent study by Jeyanathan et al. set out to evaluate the sensitivities and specificities of in situ detection methods for M. avium subsp. paratuberculosis, employing a murine model where culture-positive paucibacillary M. avium subsp. paratuberculosis infection had been established (129). One simple observation from analysis of these tissues was that diseased tissue may or may not contain visible mycobacteria, but mycobacteria were not seen in histopathologically normal tissue. This observation suggests that the threshold number of M. avium subsp. paratuberculosis organisms needed to elicit a host response is lower than the number required for microscopic detection. Furthermore, to observe acid-fast forms, whether by Ziehl-Neelsen or auramine-rhodamine staining, required searching many fields at a magnification of ×1,000. Considering that the threshold of detection by microscopy is on the order of 10,000 bacilli per ml of sputum or per gram of tissue, it is logical to assume that a low burden of infection (<10 bacilli per mg), akin to that in tuberculoid leprosy or Johne's disease in sheep, will be exceptionally hard to detect and typically reported as “negative” by staining. Another pertinent observation was that M. avium subsp. paratuberculosis cells are significantly smaller than MTBC organisms and present a coccobacillary form, contributing further to the difficulty in their detection (129). Interestingly, methods previously reported as more sensitive, such as in situ hybridization (121, 234) and in situ PCR (228) of the IS900 element, were no more sensitive than cell wall staining methods but were prone to false-positive results in uninfected control animals and M. tuberculosis-infected hosts (129). These results provided no evidence for the CWD hypothesis, as stains targeting nucleic acids did not result in more mycobacterial forms than stains for the cell wall. Notably, while CWD bacteria have been described during the isolation of M. avium subsp. paratuberculosis and other mycobacteria from CD patients (31, 99), to date the existence of M. avium subsp. paratuberculosis in a CWD form has yet to be proven within human tissue. Until the existence of CWD forms of M. avium subsp. paratuberculosis is formally demonstrated, diagnostic strategies based solely on this concept will be difficult to reconcile with discordant results obtained using conventional microbiologic methods.
FUTURE RESEARCH
Experience from MTBC genomics demonstrates the rapid conversion of genomic data into insights about pathogenesis that have subsequently been applied to the clinical laboratory. First, the M. tuberculosis H37Rv genome provided the cornerstone of future research (48). Next, comparative genomic tools permitted the demonstration of variable regions across otherwise closely related organisms (14, 102, 153). The distribution of these regions was then shown to be specific to certain subsets of the MTBC, which was of immediate use in the clinical microbiology laboratory (198, 257). The ability to accurately delineate subspecies and to compare their genomic profiles permitted investigators to derive a phylogenetic scenario for MTBC evolution (25, 183). This scenario then provided a framework for understanding phenotypic variability among MTBC members. An in vitro phenotype that benefited from this framework was the demonstration that M. bovis requires the addition of pyruvate to the culture medium for optimal growth because of a mutated pyruvate kinase gene, pykA, that has resulted in an inactive pyruvate kinase in M. bovis but not M. bovis BCG (133). The most compelling in vivo phenotype spurred on by comparative genomic studies has been the demonstration that the RD1 region, which is absent from BCG strains, in part explains the attenuation of BCG vaccines from between 1908 and 1921 (151, 211). Moreover, the determination that the RD1 region encodes virulence antigens recognized by the infected host has been exploited in the development of immunodiagnostic tests, such as the QuantiFERON-TB Gold test (Cellestis Limited, Carnegie, Australia) and the T-SPOT.TB test (Oxford Immunotec, Abingdon, United Kingdom), which are designed to detect latent tuberculous infection but not sensitization by previous BCG vaccination (195).
The preceding sequence of events can serve as a template for MAC research, using postgenomic findings to both directly bolster diagnostic capacity and guide functional studies of eventual applicability to clinical laboratories. Already, genome sequence data serve as the foundation for comparative sequencing and genomic studies (152, 304). Results of these approaches are already applicable to diagnostic laboratories and serve to brand MAC lineages for further study (239). The concordance of the hsp65-based phylogeny of the MAC with lineage-specific IS and a genomic deletion specific to M. avium subsp. paratuberculosis provides confidence that a phylogenetic framework is now at hand to guide functional studies (Fig. 1). The current challenge, therefore, is to distill the considerable genomic diversity within the MAC into tangible phenotype-genotype associations that can then lend themselves to formal experimental evaluation.
Some outstanding questions about MAC variability include verifying whether all M. avium subsp. paratuberculosis strains are mycobactin dependent, understanding the basis of the fastidious growth of M. avium subsp. paratuberculosis in vitro, and determining why ovine M. avium subsp. paratuberculosis strains are particularly slow-growing in the media currently used. The principal in vivo question is to accurately catalogue virulence, however defined, across MAC organisms in order to determine whether there are particularly robust patterns, such as a greater pathogenicity of bird strains of M. avium or M. avium subsp. paratuberculosis. Since livestock are undoubtedly exposed to environmental mycobacteria, such as M. avium subsp. hominissuis, but only develop classical Johne's disease from M. avium subsp. paratuberculosis, it is intuitive to consider the latter more inherently pathogenic. Despite this, to our knowledge, there is no standardized in vivo or ex vivo model that demonstrates a greater infectivity, persistence, or pathogenicity by M. avium subsp. paratuberculosis than that by M. avium subsp. hominissuis. While comparative genomics of the MTBC identified the absence of RD1 in BCG, it should be remembered that the subsequent experimentation was based on decades-old observations of decreased virulence of BCG strains in mouse, guinea pig, and other higher animal models (33, 76) that could readily be recapitulated in the M. tuberculosis ΔRD1 strain (151). To this end, an important bottleneck in functional genomic study of the MAC may not be the genomic data or even the tools for genetic manipulations (34, 113), but rather a robust virulence phenotype to guide studies trying to understand why certain MAC organisms have profoundly different lifestyles and capacity to cause disease from those of others. As a start, we encourage efforts to generate observational data from macrophage and small-mammal infections on the sequenced strains M. avium 104 and M. avium subsp. paratuberculosis K-10 and also to contrast other pairs of environmental and pathogenic M. avium organisms. If it can be documented that certain MAC organisms have an inherently different biology that impacts their capacity to cause disease (20) or to be detected by the clinical laboratory, then the results of these functional studies may translate into either improved laboratory methodologies (such as media that enhance the growth of ovine M. avium subsp. paratuberculosis) or more tailored diagnostic approaches targeting specific pathogenic subsets.
While we have described, to the best of our ability, what we believe to constitute the different members of the MAC, we accept that additional sequencing or other genetic studies may provide information to further delineate the complex. It is not unreasonable to contemplate that specific MAC types exist that are more pathogenic to specific hosts, with currently available methods unable to formally address this concept. This idea has previously been advanced for humans with different forms of mycobacterial susceptibility by showing differences between isolates from AIDS patients and those from non-AIDS patients (62, 169, 175, 215). Moreover, the latter group of patients, who are often grouped together, comprises distinct age- and sex-specific scenarios, such as lymph node disease in children (302), lung disease in immunocompetent middle-aged males with predisposing lung conditions (148, 221, 290, 301), and lung disease in a particular group of elderly white women without predisposing lung conditions (118, 127, 148, 166, 209, 216, 290). In these cases, disease may therefore involve the interaction of strain-specific properties of the bacterium as well as both genetic and environmentally determined host factors. Since MAC disease in immunocompetent persons is rare overall and disease due to MAC organisms likewise does not affect the majority of birds, pigs, and cattle, factors that determine the outcome of exposure, including whether infection is established and progresses to disease, are in need of further investigation.
CONCLUSION
In an editorial comment published in 1998, Telenti noted that while lab resources may be best directed towards TB research, publications on nontuberculous mycobacteria are abundant: “If we are going to do the work anyway, let's at least do it properly by calling things by a useful name” (261). This issue remains at large in MAC research nearly a decade later, as certain species names carry little information, while other terms, such as MAC and M. avium, obscure the existence of distinct subsets of organisms. Several examples have been provided in this review to underscore how flawed interpretation can follow when not all members of the MAC are considered in the experimental design. Most notably, this issue is likely to be critical in pursuing the functional genomics of MAC organisms and understanding the relative capacities of different organisms to cause infection and disease based on their natural reservoirs and their pathogenicities. Advances in evolutionary studies and taxonomy of the MTBC have changed the definition of the species in the last few years. Similarly, we now have the tools to confidently type MAC organisms and to define precisely which organisms are the subjects of study, providing valuable strain information to other laboratories aiming to replicate and build upon published findings. M. avium is genomically very heterogenous (more like E. coli than M. tuberculosis), and it is important to be cognizant of the various subsets to study them appropriately. Therefore, providing only the host type or source of M. avium subspecies is unlikely to adequately brand an organism, given that the distribution of various subtypes is not uniform or, more commonly, is unknown. At the other extreme, the degree of genetic similarity between phenotypically different organisms is often not recognized and used to focus study towards specific differences. For instance, to understand why M. avium subsp. paratuberculosis is a pathogen, one should consider parallel control experiments with M. avium subsp. hominissuis to increase the chance of determining which specific features of M. avium subsp. paratuberculosis translate into the host phenotype (disease). If an M. avium subsp. paratuberculosis virulence factor is also found in M. avium subsp. hominissuis, it may be true that the absence of the gene confers an attenuated phenotype, but it cannot readily be advanced that the presence of the gene explains why M. avium subsp. paratuberculosis is a pathogen. With this in mind, we encourage a convergence of studies related to M. avium subsp. paratuberculosis and of those related to the other subspecies of M. avium. Mycobacteria, whether saprophytic or pathogenic, have many similar characteristics that characterize the genus, but it is setting apart what makes one different from another that will help us to truly understand the biological differences between different species and subspecies of the MAC.
Acknowledgments
We thank David Alexander and Makeda Semret for input and discussions on MAC genomic variability.
C.Y.T. has a Lloyd-Carr Harris McGill Majors Fellowship and an F. C. Harrison Scholarship. M.A.B. is a Chercheur-Boursier Senior of the Fonds de la Recherche en Santé du Québec (FRSQ). Work on MAC genomics has been funded by an operating grant from the Natural Sciences and Engineering Research Council of Canada (NSERC).
REFERENCES
- 1.Ahrens, P., S. B. Giese, J. Klausen, and N. F. Inglis. 1995. Two markers, IS901-IS902 and p40, identified by PCR and by using monoclonal antibodies in Mycobacterium avium strains. J. Clin. Microbiol. 33:1049-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aranaz, A., D. Cousins, A. Mateos, and L. Dominguez. 2003. Elevation of Mycobacterium tuberculosis subsp. caprae Aranaz et al. 1999 to species rank as Mycobacterium caprae comb. nov., sp. nov. Int. J. Syst. Evol. Microbiol. 53:1785-1789. [DOI] [PubMed] [Google Scholar]
- 3.Athwal, R. S., S. S. Deo, and T. Imaeda. 1984. Deoxyribonucleic acid relatedness among Mycobacterium leprae, Mycobacterium lepraemurium, and selected bacteria by dot blot and spectrophotometric deoxyribonucleic acid hybridization assays. Int. J. Syst. Bacteriol. 34:371-375. [Google Scholar]
- 4.Autschbach, F., S. Eisold, U. Hinz, S. Zinser, M. Linnebacher, T. Giese, T. Loffler, M. W. Buchler, and J. Schmidt. 2005. High prevalence of Mycobacterium avium subspecies paratuberculosis IS900 DNA in gut tissues from individuals with Crohn's disease. Gut 54:944-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baksh, F. K., S. D. Finkelstein, S. M. Ariyanayagam-Baksh, P. A. Swalsky, E. C. Klein, and J. C. Dunn. 2004. Absence of Mycobacterium avium subsp. paratuberculosis in the microdissected granulomas of Crohn's disease. Mod. Pathol. 17:1289-1294. [DOI] [PubMed] [Google Scholar]
- 6.Bannantine, J. P., E. Baechler, Q. Zhang, L. Li, and V. Kapur. 2002. Genome scale comparison of Mycobacterium avium subsp. paratuberculosis with Mycobacterium avium subsp. avium reveals potential diagnostic sequences. J. Clin. Microbiol. 40:1303-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bannantine, J. P., and J. R. Stabel. 2000. HspX is present within Mycobacterium paratuberculosis-infected macrophages and is recognized by sera from some infected cattle. Vet. Microbiol. 76:343-358. [DOI] [PubMed] [Google Scholar]
- 8.Bannantine, J. P., Q. Zhang, L. L. Li, and V. Kapur. 2003. Genomic homogeneity between Mycobacterium avium subsp. avium and Mycobacterium avium subsp. paratuberculosis belies their divergent growth rates. BMC Microbiol. 3:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barclay, R., and C. Ratledge. 1983. Iron-binding compounds of Mycobacterium avium, M. intracellulare, M. scrofulaceum, and mycobactin-dependent M. paratuberculosis and M. avium. J. Bacteriol. 153:1138-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartos, M., P. Hlozek, P. Svastova, L. Dvorska, T. Bull, L. Matlova, I. Parmova, I. Kuhn, J. Stubbs, M. Moravkova, J. Kintr, V. Beran, I. Melicharek, M. Ocepek, and I. Pavlik. 2006. Identification of members of Mycobacterium avium species by Accu-Probes, serotyping, and single IS900, IS901, IS1245 and IS901-flanking region PCR with internal standards. J. Microbiol. Methods 64:333-345. [DOI] [PubMed] [Google Scholar]
- 11.Bauerfeind, R., S. Benazzi, R. Weiss, T. Schliesser, H. Willems, and G. Baljer. 1996. Molecular characterization of Mycobacterium paratuberculosis isolates from sheep, goats, and cattle by hybridization with a DNA probe to insertion element IS900. J. Clin. Microbiol. 34:1617-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beggs, M. L., R. Stevanova, and K. D. Eisenach. 2000. Species identification of Mycobacterium avium complex isolates by a variety of molecular techniques. J. Clin. Microbiol. 38:508-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Behr, M. A., M. Semret, A. Poon, and E. Schurr. 2004. Crohn's disease, mycobacteria, and NOD2. Lancet Infect. Dis. 4:136-137. [DOI] [PubMed] [Google Scholar]
- 14.Behr, M. A., M. A. Wilson, W. P. Gill, H. Salamon, G. K. Schoolnik, S. Rane, and P. M. Small. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520-1523. [DOI] [PubMed] [Google Scholar]
- 15.Belisle, J. T., K. Klaczkiewicz, P. J. Brennan, W. R. Jacobs, Jr., and J. M. Inamine. 1993. Rough morphological variants of Mycobacterium avium. Characterization of genomic deletions resulting in the loss of glycopeptidolipid expression. J. Biol. Chem. 268:10517-10523. [PubMed] [Google Scholar]
- 16.Belisle, J. T., M. R. McNeil, D. Chatterjee, J. M. Inamine, and P. J. Brennan. 1993. Expression of the core lipopeptide of the glycopeptidolipid surface antigens in rough mutants of Mycobacterium avium. J. Biol. Chem. 268:10510-10516. [PubMed] [Google Scholar]
- 17.Belisle, J. T., L. Pascopella, J. M. Inamine, P. J. Brennan, and W. R. Jacobs, Jr. 1991. Isolation and expression of a gene cluster responsible for biosynthesis of the glycopeptidolipid antigens of Mycobacterium avium. J. Bacteriol. 173:6991-6997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernstein, C. N., G. Nayar, A. Hamel, and J. F. Blanchard. 2003. Study of animal-borne infections in the mucosas of patients with inflammatory bowel disease and population-based controls. J. Clin. Microbiol. 41:4986-4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhide, M., E. Chakurkar, L. Tkacikova, S. Barbuddhe, M. Novak, and I. Mikula. 2006. IS900-PCR-based detection and characterization of Mycobacterium avium subsp. paratuberculosis from buffy coat of cattle and sheep. Vet. Microbiol. 112:33-41. [DOI] [PubMed] [Google Scholar]
- 20.Birkness, K. A., W. E. Swords, P. H. Huang, E. H. White, C. S. Dezzutti, R. B. Lal, and F. D. Quinn. 1999. Observed differences in virulence-associated phenotypes between a human clinical isolate and a veterinary isolate of Mycobacterium avium. Infect. Immun. 67:4895-4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blackwood, K. S., C. He, J. Gunton, C. Y. Turenne, J. Wolfe, and A. M. Kabani. 2000. Evaluation of recA sequences for identification of Mycobacterium species. J. Clin. Microbiol. 38:2846-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bono, M., T. Jemmi, C. Bernasconi, D. Burki, A. Telenti, and T. Bodmer. 1995. Genotypic characterization of Mycobacterium avium strains recovered from animals and their comparison to human strains. Appl. Environ. Microbiol. 61:371-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bosshard, C., R. Stephan, and T. Tasara. 2006. Application of an F57 sequence-based real-time PCR assay for Mycobacterium paratuberculosis detection in bulk tank raw milk and slaughtered healthy dairy cows. J. Food Prot. 69:1662-1667. [DOI] [PubMed] [Google Scholar]
- 24.Brandt, L., C. J. Feino, O. A. Weinreich, B. Chilima, P. Hirsch, R. Appelberg, and P. Andersen. 2002. Failure of the Mycobacterium bovis BCG vaccine: some species of environmental mycobacteria block multiplication of BCG and induction of protective immunity to tuberculosis. Infect. Immun. 70:672-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brosch, R., S. V. Gordon, M. Marmiesse, P. Brodin, C. Buchrieser, K. Eiglmeier, T. Garnier, C. Gutierrez, G. Hewinson, K. Kremer, L. M. Parsons, A. S. Pym, S. Samper, D. van Soolingen, and S. T. Cole. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. USA 99:3684-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brosch, R., A. S. Pym, S. V. Gordon, and S. T. Cole. 2001. The evolution of mycobacterial pathogenicity: clues from comparative genomics. Trends Microbiol. 9:452-458. [DOI] [PubMed] [Google Scholar]
- 27.Buddle, B. M., B. J. Wards, F. E. Aldwell, D. M. Collins, and G. W. de Lisle. 2002. Influence of sensitisation to environmental mycobacteria on subsequent vaccination against bovine tuberculosis. Vaccine 20:1126-1133. [DOI] [PubMed] [Google Scholar]
- 28.Bull, T. J., E. J. McMinn, K. Sidi-Boumedine, A. Skull, D. Durkin, P. Neild, G. Rhodes, R. Pickup, and J. Hermon-Taylor. 2003. Detection and verification of Mycobacterium avium subsp. paratuberculosis in fresh ileocolonic mucosal biopsy specimens from individuals with and without Crohn's disease. J. Clin. Microbiol. 41:2915-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bull, T. J., D. C. Shanson, and L. C. Archard. 1995. Rapid identification of mycobacteria from AIDS patients by capillary electrophoretic profiling of amplified SOD gene. Clin. Mol. Pathol. 48:M124-M132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bull, T. J., J. M. Sheridan, H. Martin, N. Sumar, M. Tizard, and J. Hermon-Taylor. 2000. Further studies on the GS element. A novel mycobacterial insertion sequence (IS1612), inserted into an acetylase gene (mpa) in Mycobacterium avium subsp. silvaticum but not in Mycobacterium avium subsp. paratuberculosis. Vet. Microbiol. 77:453-463. [DOI] [PubMed] [Google Scholar]
- 31.Burnham, W. R., J. E. Lennard-Jones, J. L. Stanford, and R. G. Bird. 1978. Mycobacteria as a possible cause of inflammatory bowel disease. Lancet ii:693-696. [DOI] [PubMed] [Google Scholar]
- 32.Butler, W. R., L. Thibert, and J. O. Kilburn. 1992. Identification of Mycobacterium avium complex strains and some similar species by high-performance liquid chromatography. J. Clin. Microbiol. 30:2698-2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Calmette, A. 1927. La vaccination préventive contre la tuberculose par le BCG. Masson et Cie, Paris, France.
- 34.Cavaignac, S. M., S. J. White, G. W. de Lisle, and D. M. Collins. 2000. Construction and screening of Mycobacterium paratuberculosis insertional mutant libraries. Arch. Microbiol. 173:229-231. [DOI] [PubMed] [Google Scholar]
- 35.Cellier, C., H. De Beenhouwer, A. Berger, C. Penna, F. Carbonnel, R. Parc, P. H. Cugnenc, Y. Le Quintrec, J. P. Gendre, J. P. Barbier, and F. Portaels. 1998. Mycobacterium paratuberculosis and Mycobacterium avium subsp. silvaticum DNA cannot be detected by PCR in Crohn's disease tissue. Gastroenterol. Clin. Biol. 22:675-678. [PubMed] [Google Scholar]
- 36.Chacon, O., L. E. Bermudez, and R. G. Barletta. 2004. Johne's disease, inflammatory bowel disease, and Mycobacterium paratuberculosis. Annu. Rev. Microbiol. 58:329-363. [DOI] [PubMed] [Google Scholar]
- 37.Chamberlin, W., D. Y. Graham, K. Hulten, H. M. El Zimaity, M. R. Schwartz, S. Naser, I. Shafran, and F. A. El Zaatari. 2001. Review article: Mycobacterium avium subsp. paratuberculosis as one cause of Crohn's disease. Aliment. Pharmacol. Ther. 15:337-346. [DOI] [PubMed] [Google Scholar]
- 38.Chatterjee, D., and K. H. Khoo. 2001. The surface glycopeptidolipids of mycobacteria: structures and biological properties. Cell. Mol. Life Sci. 58:2018-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chiba, M., T. Fukushima, Y. Horie, M. Iizuka, and O. Masamune. 1998. No Mycobacterium paratuberculosis detected in intestinal tissue, including Peyer's patches and lymph follicles, of Crohn's disease. J. Gastroenterol. 33:482-487. [DOI] [PubMed] [Google Scholar]
- 40.Chiodini, R. J. 1989. Crohn's disease and the mycobacterioses: a review and comparison of two disease entities. Clin. Microbiol. Rev. 2:90-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chiodini, R. J. 1993. Abolish Mycobacterium paratuberculosis strain 18. J. Clin. Microbiol. 31:1956-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chiodini, R. J., H. J. Van Kruiningen, R. S. Merkal, W. J. Thayer, and J. A. Coutu. 1984. Characteristics of an unclassified Mycobacterium species isolated from patients with Crohn's disease. J. Clin. Microbiol. 20:966-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chiodini, R. J., H. J. Van Kruiningen, W. R. Thayer, and J. A. Coutu. 1986. Spheroplastic phase of mycobacteria isolated from patients with Crohn's disease. J. Clin. Microbiol. 24:357-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Christiansen, M., H. E. Ottosen, and N. Plum. 1946. A peculiar infection with acid-fast bacteria in wood pigeons (Columba palumbus L.). Skand. Vet.-Tidsskr. 36:352-369. [PubMed] [Google Scholar]
- 45.Clutton-Brock, J. 1999. A natural history of domesticated mammals. Cambridge University Press, Cambridge, United Kingdom.
- 46.Cocito, C., P. Gilot, M. Coene, M. de Kesel, P. Poupart, and P. Vannuffel. 1994. Paratuberculosis. Clin. Microbiol. Rev. 7:328-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coetsier, C., P. Vannuffel, N. Blondeel, J. F. Denef, C. Cocito, and J. L. Gala. 2000. Duplex PCR for differential identification of Mycobacterium bovis, M. avium, and M. avium subsp. paratuberculosis in formalin-fixed paraffin-embedded tissues from cattle. J. Clin. Microbiol. 38:3048-3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 396:190. [DOI] [PubMed] [Google Scholar]
- 49.Collins, D. M., S. Cavaignac, and G. W. de Lisle. 1997. Use of four DNA insertion sequences to characterize strains of the Mycobacterium avium complex isolated from animals. Mol. Cell. Probes 11:373-380. [DOI] [PubMed] [Google Scholar]
- 50.Collins, D. M., M. De Zoete, and S. M. Cavaignac. 2002. Mycobacterium avium subsp. paratuberculosis strains from cattle and sheep can be distinguished by a PCR test based on a novel DNA sequence difference. J. Clin. Microbiol. 40:4760-4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Collins, D. M., D. M. Gabric, and G. W. de Lisle. 1989. Identification of a repetitive DNA sequence specific to Mycobacterium paratuberculosis. FEMS Microbiol. Lett. 51:175-178. [DOI] [PubMed] [Google Scholar]
- 52.Collins, D. M., D. M. Gabric, and G. W. de Lisle. 1990. Identification of two groups of Mycobacterium paratuberculosis strains by restriction endonuclease analysis and DNA hybridization. J. Clin. Microbiol. 28:1591-1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Collins, M. T., G. Lisby, C. Moser, D. Chicks, S. Christensen, M. Reichelderfer, N. Hoiby, B. A. Harms, O. O. Thomsen, U. Skibsted, and V. Binder. 2000. Results of multiple diagnostic tests for Mycobacterium avium subsp. paratuberculosis in patients with inflammatory bowel disease and in controls. J. Clin. Microbiol. 38:4373-4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Corn, J. L., E. J. Manning, S. Sreevatsan, and J. R. Fischer. 2005. Isolation of Mycobacterium avium subsp. paratuberculosis from free-ranging birds and mammals on livestock premises. Appl. Environ. Microbiol. 71:6963-6967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cousins, D. V., R. Bastida, A. Cataldi, V. Quse, S. Redrobe, S. Dow, P. Duignan, A. Murray, C. Dupont, N. Ahmed, D. M. Collins, W. R. Butler, D. Dawson, D. Rodriguez, J. Loureiro, M. I. Romano, A. Alito, M. Zumarraga, and A. Bernardelli. 2003. Tuberculosis in seals caused by a novel member of the Mycobacterium tuberculosis complex: Mycobacterium pinnipedii sp. nov. Int. J. Syst. Evol. Microbiol. 53:1305-1314. [DOI] [PubMed] [Google Scholar]
- 56.Cousins, D. V., R. Whittington, I. Marsh, A. Masters, R. J. Evans, and P. Kluver. 1999. Mycobacteria distinct from Mycobacterium avium subsp. paratuberculosis isolated from the faeces of ruminants possess IS900-like sequences detectable by IS900 polymerase chain reaction: implications for diagnosis. Mol. Cell. Probes 13:431-442. [DOI] [PubMed] [Google Scholar]
- 57.Cousins, D. V., S. N. Williams, A. Hope, and G. J. Eamens. 2000. DNA fingerprinting of Australian isolates of Mycobacterium avium subsp. paratuberculosis using IS900 RFLP. Aust. Vet. J. 78:184-190. [DOI] [PubMed] [Google Scholar]
- 58.Crawford, J. T., and J. H. Bates. 1979. Isolation of plasmids from mycobacteria. Infect. Immun. 24:979-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Crawford, J. T., and J. H. Bates. 1986. Analysis of plasmids in Mycobacterium avium-intracellulare isolates from persons with acquired immunodeficiency syndrome. Am. Rev. Respir. Dis. 134:659-661. [DOI] [PubMed] [Google Scholar]
- 60.Crawford, J. T., M. D. Cave, and J. H. Bates. 1981. Characterization of plasmids from strains of Mycobacterium avium-intracellulare. Rev. Infect. Dis. 3:949-952. [DOI] [PubMed] [Google Scholar]
- 61.Crawford, J. T., M. D. Cave, and J. H. Bates. 1981. Evidence for plasmid-mediated restriction-modification in Mycobacterium avium intracellulare. J. Gen. Microbiol. 127:333-338. [DOI] [PubMed] [Google Scholar]
- 62.Crowle, A. J., E. R. Ross, D. L. Cohn, J. Gilden, and M. H. May. 1992. Comparison of the abilities of Mycobacterium avium and Mycobacterium intracellulare to infect and multiply in cultured human macrophages from normal and human immunodeficiency virus-infected subjects. Infect. Immun. 60:3697-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cuttino, J. T., and A. M. McCabe. 1949. Pure granulomatous nocardiosis: a new fungus disease distinguished by intracellular parasitism. Am. J. Clin. Pathol. 25:1-34. [PMC free article] [PubMed] [Google Scholar]
- 64.Dalziel, T. K. 1913. Chronic intestinal enteritis. Br. Med. J. 2:1068-1070. [Google Scholar]
- 65.Dei, R., E. Tortoli, A. Bartoloni, M. T. Simonetti, and E. Lillini. 1999. HPLC does not differentiate Mycobacterium paratuberculosis from Mycobacterium avium. Vet. Microbiol. 65:209-213. [DOI] [PubMed] [Google Scholar]
- 66.de Lisle, G. W., M. C. Cannon, G. F. Yates, and D. M. Collins. 2006. Use of a polymerase chain reaction to subtype Mycobacterium avium subspecies paratuberculosis, an increasingly important pathogen from farmed deer in New Zealand. N. Z. Vet. J. 54:195-197. [DOI] [PubMed] [Google Scholar]
- 67.de Lisle, G. W., D. M. Collins, and H. F. Huchzermeyer. 1992. Characterization of ovine strains of Mycobacterium paratuberculosis by restriction endonuclease analysis and DNA hybridization. Onderstepoort J. Vet. Res. 59:163-165. [PubMed] [Google Scholar]
- 68.de Lisle, G. W., B. J. Wards, B. M. Buddle, and D. M. Collins. 2005. The efficacy of live tuberculosis vaccines after presensitization with Mycobacterium avium. Tuberculosis 85:73-79. [DOI] [PubMed] [Google Scholar]
- 69.Dell'Isola, B., C. Poyart, O. Goulet, J. F. Mougenot, E. Sadoun-Journo, N. Brousse, J. Schmitz, C. Ricour, and P. Berche. 1994. Detection of Mycobacterium paratuberculosis by polymerase chain reaction in children with Crohn's disease. J. Infect. Dis. 169:449-451. [DOI] [PubMed] [Google Scholar]
- 70.De Smet, K. A., I. N. Brown, M. Yates, and J. Ivanyi. 1995. Ribosomal internal transcribed spacer sequences are identical among Mycobacterium avium-intracellulare complex isolates from AIDS patients, but vary among isolates from elderly pulmonary disease patients. Microbiology 141:2739-2747. [DOI] [PubMed] [Google Scholar]
- 71.Devallois, A., M. Picardeau, K. S. Goh, C. Sola, V. Vincent, and N. Rastogi. 1996. Comparative evaluation of PCR and commercial DNA probes for detection and identification to species level of Mycobacterium avium and Mycobacterium intracellulare. J. Clin. Microbiol. 34:2756-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Devallois, A., M. Picardeau, C. N. Paramasivan, V. Vincent, and N. Rastogi. 1997. Molecular characterization of Mycobacterium avium complex isolates giving discordant results in AccuProbe tests by PCR-restriction enzyme analysis, 16S rRNA gene sequencing, and DT1-DT6 PCR. J. Clin. Microbiol. 35:2767-2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Devallois, A., and N. Rastogi. 1997. Computer-assisted analysis of Mycobacterium avium fingerprints using insertion elements IS1245 and IS1311 in a Caribbean setting. Res. Microbiol. 148:703-713. [DOI] [PubMed] [Google Scholar]
- 74.Devulder, G., D. M. Perouse, and J. P. Flandrois. 2005. A multigene approach to phylogenetic analysis using the genus Mycobacterium as a model. Int. J. Syst. Evol. Microbiol. 55:293-302. [DOI] [PubMed] [Google Scholar]
- 75.Dohmann, K., B. Strommenger, K. Stevenson, L. de Juan, J. Stratmann, V. Kapur, T. J. Bull, and G. F. Gerlach. 2003. Characterization of genetic differences between Mycobacterium avium subsp. paratuberculosis type I and type II isolates. J. Clin. Microbiol. 41:5215-5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dubos, R. J., and C. H. Pierce. 1956. Differential characteristics in vitro and in vivo of several substrains of BCG. Am. Rev. Tuberc. 74:655-717. [DOI] [PubMed] [Google Scholar]
- 77.Dumonceau, J. M., A. Van Gossum, M. Adler, P. A. Fonteyne, J. P. Van Vooren, J. Deviere, and F. Portaels. 1996. No Mycobacterium paratuberculosis found in Crohn's disease using polymerase chain reaction. Dig. Dis. Sci. 41:421-426. [DOI] [PubMed] [Google Scholar]
- 78.Dvorska, L., M. Bartos, O. Ostadal, J. Kaustova, L. Matlova, and I. Pavlik. 2002. IS1311 and IS1245 restriction fragment length polymorphism analyses, serotypes, and drug susceptibilities of Mycobacterium avium complex isolates obtained from a human immunodeficiency virus-negative patient. J. Clin. Microbiol. 40:3712-3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dvorska, L., T. J. Bull, M. Bartos, L. Matlova, P. Svastova, R. T. Weston, J. Kintr, I. Parmova, D. van Soolingen, and I. Pavlik. 2003. A standardised restriction fragment length polymorphism (RFLP) method for typing Mycobacterium avium isolates links IS901 with virulence for birds. J. Microbiol. Methods 55:11-27. [DOI] [PubMed] [Google Scholar]
- 80.Eckstein, T. M., J. T. Belisle, and J. M. Inamine. 2003. Proposed pathway for the biosynthesis of serovar-specific glycopeptidolipids in Mycobacterium avium serovar 2. Microbiology 149:2797-2807. [DOI] [PubMed] [Google Scholar]
- 81.Eckstein, T. M., J. M. Inamine, M. L. Lambert, and J. T. Belisle. 2000. A genetic mechanism for deletion of the ser2 gene cluster and formation of rough morphological variants of Mycobacterium avium. J. Bacteriol. 182:6177-6182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Eda, S., J. P. Bannantine, W. R. Waters, Y. Mori, R. H. Whitlock, M. C. Scott, and C. A. Speer. 2006. A highly sensitive and subspecies-specific surface antigen enzyme-linked immunosorbent assay for diagnosis of Johne's disease. Clin. Vaccine Immunol. 13:837-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ellingson, J. L., C. A. Bolin, and J. R. Stabel. 1998. Identification of a gene unique to Mycobacterium avium subspecies paratuberculosis and application to diagnosis of paratuberculosis. Mol. Cell. Probes 12:133-142. [DOI] [PubMed] [Google Scholar]
- 84.Ellingson, J. L., J. R. Stabel, W. R. Bishai, R. Frothingham, and J. M. Miller. 2000. Evaluation of the accuracy and reproducibility of a practical PCR panel assay for rapid detection and differentiation of Mycobacterium avium subspecies. Mol. Cell. Probes 14:153-161. [DOI] [PubMed] [Google Scholar]
- 85.Englund, S., G. Bolske, and K. E. Johansson. 2002. An IS900-like sequence found in a Mycobacterium sp. other than Mycobacterium avium subsp. paratuberculosis. FEMS Microbiol. Lett. 209:267-271. [DOI] [PubMed] [Google Scholar]
- 86.Eriks, I. S., K. T. Munck, T. E. Besser, G. H. Cantor, and V. Kapur. 1996. Rapid differentiation of Mycobacterium avium and M. paratuberculosis by PCR and restriction enzyme analysis. J. Clin. Microbiol. 34:734-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Falkinham, J. O., III. 1994. Epidemiology of Mycobacterium avium infections in the pre- and post-HIV era. Res. Microbiol. 145:169-172. [DOI] [PubMed] [Google Scholar]
- 88.Falkinham, J. O., III, C. D. Norton, and M. W. LeChevallier. 2001. Factors influencing numbers of Mycobacterium avium, Mycobacterium intracellulare, and other mycobacteria in drinking water distribution systems. Appl. Environ. Microbiol. 67:1225-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fang, Y., W. H. Wu, J. L. Pepper, J. L. Larsen, S. A. Marras, E. A. Nelson, W. B. Epperson, and J. Christopher-Hennings. 2002. Comparison of real-time, quantitative PCR with molecular beacons to nested PCR and culture methods for detection of Mycobacterium avium subsp. paratuberculosis in bovine fecal samples. J. Clin. Microbiol. 40:287-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Francis, J., H. Macturk, J. Madinaveitia, and G. Snow. 1953. Mycobactin, a growth factor for Mycobacterium johnei. I. Isolation from Mycobacterium phlei. Biochem. J. 55:596-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Franzblau, S. G., T. Takeda, and M. Nakamura. 1986. Mycobacterial plasmids: screening and possible relationship to antibiotic resistance in Mycobacterium avium/Mycobacterium intracellulare. Microbiol. Immunol. 30:903-907. [DOI] [PubMed] [Google Scholar]
- 92.Frothingham, R., and K. H. Wilson. 1993. Sequence-based differentiation of strains in the Mycobacterium avium complex. J. Bacteriol. 175:2818-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Frothingham, R., and K. H. Wilson. 1994. Molecular phylogeny of the Mycobacterium avium complex demonstrates clinically meaningful divisions. J. Infect. Dis. 169:305-312. [DOI] [PubMed] [Google Scholar]
- 94.Fry, K. L., P. S. Meissner, and J. O. Falkinham III. 1986. Epidemiology of infection by nontuberculous mycobacteria. VI. Identification and use of epidemiologic markers for studies of Mycobacterium avium, M. intracellulare, and M. scrofulaceum. Am. Rev. Respir. Dis. 134:39-43. [DOI] [PubMed] [Google Scholar]
- 95.Gangadharam, P. R., V. K. Perumal, J. T. Crawford, and J. H. Bates. 1988. Association of plasmids and virulence of Mycobacterium avium complex. Am. Rev. Respir. Dis. 137:212-214. [DOI] [PubMed] [Google Scholar]
- 96.Gangadharam, P. R., V. K. Perumal, B. T. Jairam, N. R. Podapati, R. B. Taylor, and J. F. LaBrecque. 1989. Virulence of Mycobacterium avium complex strains from acquired immune deficiency syndrome patients: relationship with characteristics of the parasite and host. Microb. Pathog. 7:263-278. [DOI] [PubMed] [Google Scholar]
- 97.Garriga, X., P. Cortes, F. March, P. Rodriguez, M. Garrigo, C. Moreno, E. Garcia, P. Coll, and G. Prats. 1999. Characterization to species level of clinical isolates of the Mycobacterium avium complex by DNA probes, DT1-DT6 PCR and PCR-restriction enzyme analysis. Clin. Microbiol. Infect. 5:379-382. [DOI] [PubMed] [Google Scholar]
- 98.Gingeras, T. R., G. Ghandour, E. Wang, A. Berno, P. M. Small, F. Drobniewski, D. Alland, E. Desmond, M. Holodniy, and J. Drenkow. 1998. Simultaneous genotyping and species identification using hybridization pattern recognition analysis of generic Mycobacterium DNA arrays. Genome Res. 8:435-448. [DOI] [PubMed] [Google Scholar]
- 99.Gitnick, G., J. Collins, B. Beaman, D. Brooks, M. Arthur, T. Imaeda, and M. Palieschesky. 1989. Preliminary report on isolation of mycobacteria from patients with Crohn's disease. Dig. Dis. Sci. 34:925-932. [DOI] [PubMed] [Google Scholar]
- 100.Glickman, S. E., J. O. Kilburn, W. R. Butler, and L. S. Ramos. 1994. Rapid identification of mycolic acid patterns of mycobacteria by high-performance liquid chromatography using pattern recognition software and a Mycobacterium library. J. Clin. Microbiol. 32:740-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Godfroid, J., C. Delcorps, L. M. Irenge, K. Walravens, S. Marche, and J. L. Gala. 2005. Definitive differentiation between single and mixed mycobacterial infections in red deer (Cervus elaphus) by a combination of duplex amplification of p34 and f57 sequences and Hpy188I enzymatic restriction of duplex amplicons. J. Clin. Microbiol. 43:4640-4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gordon, S. V., R. Brosch, A. Billault, T. Garnier, K. Eiglmeier, and S. T. Cole. 1999. Identification of variable regions in the genomes of tubercle bacilli using bacterial artificial chromosome arrays. Mol. Microbiol. 32:643-655. [DOI] [PubMed] [Google Scholar]
- 103.Grange, J. M., M. D. Yates, and E. Boughton. 1990. The avian tubercle bacillus and its relatives. J. Appl. Bacteriol. 68:411-431. [DOI] [PubMed] [Google Scholar]
- 104.Green, E. P., M. L. Tizard, M. T. Moss, J. Thompson, D. J. Winterbourne, J. J. Mcfadden, and J. Hermon-Taylor. 1989. Sequence and characteristics of IS900, an insertion element identified in a human Crohn's disease isolate of Mycobacterium paratuberculosis. Nucleic Acids Res. 17:9063-9073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Greenstein, R. J. 2003. Is Crohn's disease caused by a mycobacterium? Comparisons with leprosy, tuberculosis, and Johne's disease. Lancet Infect. Dis. 3:507-514. [DOI] [PubMed] [Google Scholar]
- 106.Grimes, D. S. 2003. Mycobacterium avium subspecies paratuberculosis as a cause of Crohn's disease. Gut 52:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Guerrero, C., C. Bernasconi, D. Burki, T. Bodmer, and A. Telenti. 1995. A novel insertion element from Mycobacterium avium, IS1245, is a specific target for analysis of strain relatedness. J. Clin. Microbiol. 33:304-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Guthertz, L. S., B. Damsker, E. J. Bottone, E. G. Ford, T. F. Midura, and J. M. Janda. 1989. Mycobacterium avium and Mycobacterium intracellulare infections in patients with and without AIDS. J. Infect. Dis. 160:1037-1041. [DOI] [PubMed] [Google Scholar]
- 109.Halldorsdottir, S., S. Englund, S. F. Nilsen, and I. Olsaker. 2002. Detection of Mycobacterium avium subsp. paratuberculosis by buoyant density centrifugation, sequence capture PCR and dot blot hybridisation. Vet. Microbiol. 87:327-340. [DOI] [PubMed] [Google Scholar]
- 110.Hampson, S. J., F. Portaels, J. Thompson, E. P. Green, M. T. Moss, T. J. Hermon, and J. J. McFadden. 1989. DNA probes demonstrate a single highly conserved strain of Mycobacterium avium infecting AIDS patients. Lancet i:65-68. [DOI] [PubMed] [Google Scholar]
- 111.Harris, J. E., and A. M. Lammerding. 2001. Crohn's disease and Mycobacterium avium subsp. paratuberculosis: current issues. J. Food Prot. 64:2103-2110. [DOI] [PubMed] [Google Scholar]
- 112.Harris, N. B., and R. G. Barletta. 2001. Mycobacterium avium subsp. paratuberculosis in veterinary medicine. Clin. Microbiol. Rev. 14:489-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Harris, N. B., Z. Feng, X. Liu, S. L. Cirillo, J. D. Cirillo, and R. G. Barletta. 1999. Development of a transposon mutagenesis system for Mycobacterium avium subsp. paratuberculosis. FEMS Microbiol. Lett. 175:21-26. [DOI] [PubMed] [Google Scholar]
- 114.Hellyer, T. J., I. N. Brown, J. W. Dale, and C. S. Easmon. 1991. Plasmid analysis of Mycobacterium avium-intracellulare (MAI) isolated in the United Kingdom from patients with and without AIDS. J. Med. Microbiol. 34:225-231. [DOI] [PubMed] [Google Scholar]
- 115.Hermon-Taylor, J. 2001. Protagonist: Mycobacterium avium subspecies paratuberculosis is a cause of Crohn's disease. Gut 49:755-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hernandez Perez, M., N. G. Fomukong, T. Hellyer, I. N. Brown, and J. W. Dale. 1994. Characterization of IS1110, a highly mobile genetic element from Mycobacterium avium. Mol. Microbiol. 12:717-724. [DOI] [PubMed] [Google Scholar]
- 117.Hernandez Perez, M., Z. M. Kunze, S. Brown, M. A. Yakrus, J. J. Mcfadden, and J. W. Dale. 1997. Strain variation in Mycobacterium avium: polymorphism of IS1110-related sequences. Int. J. Infect. Dis. 1:192-198. [Google Scholar]
- 118.Huang, J. H., P. N. Kao, V. Adi, and S. J. Ruoss. 1999. Mycobacterium avium-intracellulare pulmonary infection in HIV-negative patients without preexisting lung disease: diagnostic and management limitations. Chest 115:1033-1040. [DOI] [PubMed] [Google Scholar]
- 119.Hughes, M. S., N. W. Ball, L. A. Beck, G. W. de Lisle, R. A. Skuce, and S. D. Neill. 1997. Determination of the etiology of presumptive feline leprosy by 16S rRNA gene analysis. J. Clin. Microbiol. 35:2464-2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hughes, M. S., G. James, M. J. Taylor, J. McCarroll, S. D. Neill, S. C. Chen, D. H. Mitchell, D. N. Love, and R. Malik. 2004. PCR studies of feline leprosy cases. J. Feline Med. Surg. 6:235-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hulten, K., H. M. El Zimaity, T. J. Karttunen, A. Almashhrawi, M. R. Schwartz, D. Y. Graham, and F. A. El Zaatari. 2001. Detection of Mycobacterium avium subspecies paratuberculosis in Crohn's diseased tissues by in situ hybridization. Am. J. Gastroenterol. 96:1529-1535. [DOI] [PubMed] [Google Scholar]
- 122.Hurley, S. S., G. A. Splitter, and R. A. Welch. 1988. Deoxyribonucleic acid relatedness of Mycobacterium paratuberculosis to other members of the family Mycobacteriaceae. Int. J. Syst. Bacteriol. 38:143-146. [Google Scholar]
- 123.Imaeda, T., L. Barksdale, and W. F. Kirchheimer. 1982. Deoxyribonucleic acid of Mycobacterium lepraemurium: its genome size, base ratio and homology with those of other mycobacteria. Int. J. Syst. Bacteriol. 32:456-458. [Google Scholar]
- 124.Inderlied, C. B., C. A. Kemper, and L. E. Bermudez. 1993. The Mycobacterium avium complex. Clin. Microbiol. Rev. 6:266-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Inglis, N. F., K. Stevenson, R. C. Davies, D. G. Heaslip, and J. M. Sharp. 2001. Unique expression of a highly conserved mycobacterial gene in IS901(+) Mycobacterium avium. Microbiology 147:1557-1564. [DOI] [PubMed] [Google Scholar]
- 126.Inglis, N. F., K. Stevenson, D. G. Heaslip, and J. M. Sharp. 2003. Characterisation of IS901 integration sites in the Mycobacterium avium genome. FEMS Microbiol. Lett. 221:39-47. [DOI] [PubMed] [Google Scholar]
- 127.Iseman, M. D. 1989. Mycobacterium avium complex and the normal host: the other side of the coin. N. Engl. J. Med. 321:896-898. [DOI] [PubMed] [Google Scholar]
- 128.Jahans, K. L. 1978. The isolation of an atypical Mycobacterium from three wood pigeons with tuberculosis-like lesions. Res. Vet. Sci. 24:378-379. [PubMed] [Google Scholar]
- 129.Jeyanathan, M., D. C. Alexander, C. Y. Turenne, C. Girard, and M. A. Behr. 2006. Evaluation of in situ methods used to detect Mycobacterium avium subsp. paratuberculosis in samples from patients with Crohn's disease. J. Clin. Microbiol. 44:2942-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Johansen, T. B., B. Djonne, M. R. Jensen, and I. Olsen. 2005. Distribution of IS1311 and IS1245 in Mycobacterium avium subspecies revisited. J. Clin. Microbiol. 43:2500-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Jorgensen, J. B., and B. Clausen. 1976. Mycobacteriosis in a roe-deer caused by wood-pigeon mycobacteria. Nord. Vet. Med. 28:539-546. [PubMed] [Google Scholar]
- 132.Kasai, H., T. Ezaki, and S. Harayama. 2000. Differentiation of phylogenetically related slowly growing mycobacteria by their gyrB sequences. J. Clin. Microbiol. 38:301-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Keating, L. A., P. R. Wheeler, H. Mansoor, J. K. Inwald, J. Dale, R. G. Hewinson, and S. V. Gordon. 2005. The pyruvate requirement of some members of the Mycobacterium tuberculosis complex is due to an inactive pyruvate kinase: implications for in vivo growth. Mol. Microbiol. 56:163-174. [DOI] [PubMed] [Google Scholar]
- 134.Keller, A. P., M. L. Beggs, B. Amthor, F. Bruns, P. Meissner, and W. H. Haas. 2002. Evidence of the presence of IS1245 and IS1311 or closely related insertion elements in nontuberculous mycobacteria outside of the Mycobacterium avium complex. J. Clin. Microbiol. 40:1869-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kent, P. T., and G. P. Kubica. 1985. Public health mycobacteriology: a guide for the level III laboratory. U.S. Department of Health and Human Services, Centers for Disease Control, Atlanta, Ga.
- 136.Kim, B. J., S. H. Lee, M. A. Lyu, S. J. Kim, G. H. Bai, G. T. Chae, E. C. Kim, C. Y. Cha, and Y. H. Kook. 1999. Identification of mycobacterial species by comparative sequence analysis of the RNA polymerase gene (rpoB). J. Clin. Microbiol. 37:1714-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kirschner, P., and E. C. Bottger. 1992. Headaches for taxonomists: the M. avium-M. intracellulare complex. Int. J. Syst. Bacteriol. 42:335-336. [DOI] [PubMed] [Google Scholar]
- 138.Klitgaard Nielsen, K., and P. Ahrens. 2002. Putative in vitro expressed gene fragments unique to Mycobacterium avium subspecies paratuberculosis. FEMS Microbiol. Lett. 214:199-203. [DOI] [PubMed] [Google Scholar]
- 139.Komijn, R. E., P. E. de Haas, M. M. Schneider, T. Eger, J. H. Nieuwenhuijs, R. J. van den Hoek, D. Bakker, F. G. van Zijd Erveld, and D. van Soolingen. 1999. Prevalence of Mycobacterium avium in slaughter pigs in The Netherlands and comparison of IS1245 restriction fragment length polymorphism patterns of porcine and human isolates. J. Clin. Microbiol. 37:1254-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Krzywinska, E., J. Krzywinski, and J. S. Schorey. 2004. Phylogeny of Mycobacterium avium strains inferred from glycopeptidolipid biosynthesis pathway genes. Microbiology 150:1699-1706. [DOI] [PubMed] [Google Scholar]
- 141.Krzywinska, E., and J. S. Schorey. 2003. Characterization of genetic differences between Mycobacterium avium subsp. avium strains of diverse virulence with a focus on the glycopeptidolipid biosynthesis cluster. Vet. Microbiol. 91:249-264. [DOI] [PubMed] [Google Scholar]
- 142.Kunze, Z. M., S. Wall, R. Appelberg, M. T. Silva, F. Portaels, and J. J. Mcfadden. 1991. IS901, a new member of a widespread class of atypical insertion sequences, is associated with pathogenicity in Mycobacterium avium. Mol. Microbiol. 5:2265-2272. [DOI] [PubMed] [Google Scholar]
- 143.Larsen, A. B., R. S. Merkal, and T. H. Vardaman. 1956. Survival time of Mycobacterium paratuberculosis. Am. J. Vet. Res. 17:549-551. [PubMed] [Google Scholar]
- 144.Laurent, J. P., S. Faske, and G. A. Cangelosi. 2002. Characterization of IS999, an unstable genetic element in Mycobacterium avium. Gene 294:249-257. [DOI] [PubMed] [Google Scholar]
- 145.Leao, S. C., M. R. Briones, M. P. Sircili, S. C. Balian, N. Mores, and J. S. Ferreira-Neto. 1999. Identification of two novel Mycobacterium avium allelic variants in pig and human isolates from Brazil by PCR-restriction enzyme analysis. J. Clin. Microbiol. 37:2592-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lebrun, L., N. Gonullu, N. Boutros, A. Davoust, M. Guibert, D. Ingrand, J. C. Ghnassia, V. Vincent, and F. Doucet-Populaire. 2003. Use of INNO-LIPA assay for rapid identification of mycobacteria. Diagn. Microbiol. Infect. Dis. 46:151-153. [DOI] [PubMed] [Google Scholar]
- 147.Lebrun, L., F. X. Weill, L. Lafendi, F. Houriez, F. Casanova, M. C. Gutierrez, D. Ingrand, P. Lagrange, V. Vincent, and J. L. Herrmann. 2005. Use of the INNO-LiPA-MYCOBACTERIA assay (version 2) for identification of Mycobacterium avium-Mycobacterium intracellulare-Mycobacterium scrofulaceum complex isolates. J. Clin. Microbiol. 43:2567-2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Levin, D. L. 2002. Radiology of pulmonary Mycobacterium avium-intracellulare complex. Clin. Chest Med. 23:603-612. [DOI] [PubMed] [Google Scholar]
- 149.Levy-Frebault, V. V., and F. Portaels. 1992. Proposed minimal standards for the genus Mycobacterium and for description of new slowly growing Mycobacterium species. Int. J. Syst. Bacteriol. 42:315-323. [DOI] [PubMed] [Google Scholar]
- 150.Levy-Frebault, V. V., M. F. Thorel, A. Varnerot, and B. Gicquel. 1989. DNA polymorphism in Mycobacterium paratuberculosis, “wood pigeon mycobacteria,” and related mycobacteria analyzed by field inversion gel electrophoresis. J. Clin. Microbiol. 27:2823-2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lewis, K. N., R. Liao, K. M. Guinn, M. J. Hickey, S. Smith, M. A. Behr, and D. R. Sherman. 2003. Deletion of RD1 from Mycobacterium tuberculosis mimics Bacille Calmette-Guerin attenuation. J. Infect. Dis. 187:117-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Li, L., J. P. Bannantine, Q. Zhang, A. Amonsin, B. J. May, D. Alt, N. Banerji, S. Kanjilal, and V. Kapur. 2005. The complete genome sequence of Mycobacterium avium subspecies paratuberculosis. Proc. Natl. Acad. Sci. USA 102:12344-12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Mahairas, G. G., P. J. Sabo, M. J. Hickey, D. C. Singh, and C. K. Stover. 1996. Molecular analysis of genetic differences between Mycobacterium bovis BCG and virulent M. bovis. J. Bacteriol. 178:1274-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Maiden, M. C. 2006. Multilocus sequence typing of bacteria. Annu. Rev. Microbiol. 60:561-588. [DOI] [PubMed] [Google Scholar]
- 155.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Markesich, D. C., D. Y. Graham, and H. H. Yoshimura. 1988. Progress in culture and subculture of spheroplasts and fastidious acid-fast bacilli isolated from intestinal tissues. J. Clin. Microbiol. 26:1600-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Marri, P. R., J. P. Bannantine, M. L. Paustian, and G. B. Golding. 2006. Lateral gene transfer in Mycobacterium avium subspecies paratuberculosis. Can. J. Microbiol. 52:560-569. [DOI] [PubMed] [Google Scholar]
- 158.Marsh, I., R. Whittington, and D. Cousins. 1999. PCR-restriction endonuclease analysis for identification and strain typing of Mycobacterium avium subsp. paratuberculosis and Mycobacterium avium subsp. avium based on polymorphisms in IS1311. Mol. Cell. Probes 13:115-126. [DOI] [PubMed] [Google Scholar]
- 159.Marsh, I., R. Whittington, and D. Millar. 2000. Quality control and optimized procedure of hybridization capture-PCR for the identification of Mycobacterium avium subsp. paratuberculosis in faeces. Mol. Cell. Probes 14:219-232. [DOI] [PubMed] [Google Scholar]
- 160.Marsh, I. B., J. P. Bannantine, M. L. Paustian, M. L. Tizard, V. Kapur, and R. J. Whittington. 2006. Genomic comparison of Mycobacterium avium subsp. paratuberculosis sheep and cattle strains by microarray hybridization. J. Bacteriol. 188:2290-2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Marsh, I. B., and R. J. Whittington. 2001. Progress towards a rapid polymerase chain reaction diagnostic test for the identification of Mycobacterium avium subsp. paratuberculosis in faeces. Mol. Cell. Probes 15:105-118. [DOI] [PubMed] [Google Scholar]
- 162.Marsh, I. B., and R. J. Whittington. 2005. Deletion of an mmpL gene and multiple associated genes from the genome of the S strain of Mycobacterium avium subsp. paratuberculosis identified by representational difference analysis and in silico analysis. Mol. Cell. Probes 19:371-384. [DOI] [PubMed] [Google Scholar]
- 163.Masaki, S., T. Konishi, G. Sugimori, A. Okamoto, Y. Hayashi, and F. Kuze. 1989. Plasmid profiles of Mycobacterium avium complex isolated from swine. Microbiol. Immunol. 33:429-433. [DOI] [PubMed] [Google Scholar]
- 164.Matlova, L., L. Dvorska, K. Palecek, L. Maurenc, M. Bartos, and I. Pavlik. 2004. Impact of sawdust and wood shavings in bedding on pig tuberculous lesions in lymph nodes, and IS1245 RFLP analysis of Mycobacterium avium subsp. hominissuis of serotypes 6 and 8 isolated from pigs and environment. Vet. Microbiol. 102:227-236. [DOI] [PubMed] [Google Scholar]
- 165.Matthews, P. R., A. McDiarmid, P. Collins, and A. Brown. 1978. The dependence of some strains of Mycobacterium avium on mycobactin for initial and subsequent growth. J. Med. Microbiol. 11:53-57. [DOI] [PubMed] [Google Scholar]
- 166.Maugein, J., M. Dailloux, B. Carbonnelle, V. Vincent, and J. Grosset. 2005. Sentinel-site surveillance of Mycobacterium avium complex pulmonary disease. Eur. Respir. J. 26:1092-1096. [DOI] [PubMed] [Google Scholar]
- 167.McDiarmid, A. 1948. The occurence of tuberculosis in the wild wood-pigeon. J. Comp. Pathol. 58:128-133. [DOI] [PubMed] [Google Scholar]
- 168.McDonald, W. L., S. E. Ridge, A. F. Hope, and R. J. Condron. 1999. Evaluation of diagnostic tests for Johne's disease in young cattle. Aust. Vet. J. 77:113-119. [DOI] [PubMed] [Google Scholar]
- 169.McGarvey, J. A., and L. E. Bermudez. 2001. Phenotypic and genomic analyses of the Mycobacterium avium complex reveal differences in gastrointestinal invasion and genomic composition. Infect. Immun. 69:7242-7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.McNabb, A., D. Eisler, K. Adie, M. Amos, M. Rodrigues, G. Stephens, W. A. Black, and J. Isaac-Renton. 2004. Assessment of partial sequencing of the 65-kilodalton heat shock protein gene (hsp65) for routine identification of Mycobacterium species isolated from clinical sources. J. Clin. Microbiol. 42:3000-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Meissner, P. S., and J. O. Falkinham III. 1986. Plasmid DNA profiles as epidemiological markers for clinical and environmental isolates of Mycobacterium avium, Mycobacterium intracellulare, and Mycobacterium scrofulaceum. J. Infect. Dis. 153:325-331. [DOI] [PubMed] [Google Scholar]
- 172.Merkal, R. S. 1979. Proposal of ATCC 19698 as the neotype strain of Mycobacterium paratuberculosis Bergey et al. 1923. Int. J. Syst. Bacteriol. 29:263-264. [Google Scholar]
- 173.Merkal, R. S., and B. J. Curran. 1974. Growth and metabolic characteristics of Mycobacterium paratuberculosis. Appl. Microbiol. 28:276-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Merkal, R. S., and W. G. McCullough. 1982. A new mycobactin, mycobactin J, from Mycobacterium paratuberculosis. Curr. Microbiol. 7:333-335. [Google Scholar]
- 175.Meyer, M., P. W. von Grunberg, T. Knoop, P. Hartmann, and G. Plum. 1998. The macrophage-induced gene mig as a marker for clinical pathogenicity and in vitro virulence of Mycobacterium avium complex strains. Infect. Immun. 66:4549-4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Mijs, W., P. de Haas, R. Rossau, T. Van der Laan, L. Rigouts, F. Portaels, and D. van Soolingen. 2002. Molecular evidence to support a proposal to reserve the designation Mycobacterium avium subsp. avium for bird-type isolates and ‘M. avium subsp. hominissuis’ for the human/porcine type of M. avium. Int. J. Syst. Evol. Microbiol. 52:1505-1518. [DOI] [PubMed] [Google Scholar]
- 177.Millar, D. S., S. J. Withey, M. L. Tizard, J. G. Ford, and T. J. Hermon. 1995. Solid-phase hybridization capture of low-abundance target DNA sequences: application to the polymerase chain reaction detection of Mycobacterium paratuberculosis and Mycobacterium avium subsp. silvaticum. Anal. Biochem. 226:325-330. [DOI] [PubMed] [Google Scholar]
- 178.Mobius, P., P. Lentzsch, I. Moser, L. Naumann, G. Martin, and H. Kohler. 2006. Comparative macrorestriction and RFLP analysis of Mycobacterium avium subsp. avium and Mycobacterium avium subsp. hominissuis isolates from man, pig, and cattle. Vet. Microbiol. 117:284-291. [DOI] [PubMed] [Google Scholar]
- 179.Morita, Y., S. Maruyama, H. Kabeya, A. Nagai, K. Kozawa, M. Kato, T. Nakajima, T. Mikami, Y. Katsube, and H. Kimura. 2004. Genetic diversity of the dnaJ gene in the Mycobacterium avium complex. J. Med. Microbiol. 53:813-817. [DOI] [PubMed] [Google Scholar]
- 180.Morris, S. L., D. A. Rouse, A. Malik, S. D. Chaparas, and F. G. Witebsky. 1990. Characterisation of plasmids extracted from AIDS-associated Mycobacterium avium isolates. Tubercle 71:181-185. [DOI] [PubMed] [Google Scholar]
- 181.Moss, M. T., Z. P. Malik, M. L. Tizard, E. P. Green, J. D. Sanderson, and T. J. Hermon. 1992. IS902, an insertion element of the chronic-enteritis-causing Mycobacterium avium subsp. silvaticum. J. Gen. Microbiol. 138:139-145. [DOI] [PubMed] [Google Scholar]
- 182.Mostowy, S., D. Cousins, and M. A. Behr. 2004. Genomic interrogation of the dassie bacillus reveals it as a unique RD1 mutant within the Mycobacterium tuberculosis complex. J. Bacteriol. 186:104-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Mostowy, S., D. Cousins, J. Brinkman, A. Aranaz, and M. A. Behr. 2002. Genomic deletions suggest a phylogeny for the Mycobacterium tuberculosis complex. J. Infect. Dis. 186:74-80. [DOI] [PubMed] [Google Scholar]
- 184.Mostowy, S., J. Inwald, S. Gordon, C. Martin, R. Warren, K. Kremer, D. Cousins, and M. A. Behr. 2005. Revisiting the evolution of Mycobacterium bovis. J. Bacteriol. 187:6386-6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Mostowy, S., A. Onipede, S. Gagneux, S. Niemann, K. Kremer, E. P. Desmond, M. Kato-Maeda, and M. Behr. 2004. Genomic analysis distinguishes Mycobacterium africanum. J. Clin. Microbiol. 42:3594-3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Murcia, M. I., E. Tortoli, M. C. Menendez, E. Palenque, and M. J. Garcia. 2006. Mycobacterium colombiense sp. nov., a novel member of the Mycobacterium avium complex and description of MAC-X as a new ITS genetic variant. Int. J. Syst. Evol. Microbiol. 56:2049-2054. [DOI] [PubMed] [Google Scholar]
- 187.Naser, S. A., G. Ghobrial, C. Romero, and J. F. Valentine. 2004. Culture of Mycobacterium avium subspecies paratuberculosis from the blood of patients with Crohn's disease. Lancet 364:1039-1044. [DOI] [PubMed] [Google Scholar]
- 188.Novi, C., L. Rindi, N. Lari, and C. Garzelli. 2000. Molecular typing of Mycobacterium avium isolates by sequencing of the 16S-23S rDNA internal transcribed spacer and comparison with IS 1245-based fingerprinting. J. Med. Microbiol. 49:1091-1095. [DOI] [PubMed] [Google Scholar]
- 189.O'Grady, D., O. Flynn, E. Costello, F. Quigley, A. Gogarty, J. McGuirk, J. O'Rourke, and N. Gibbons. 2000. Restriction fragment length polymorphism analysis of Mycobacterium avium isolates from animal and human sources. Int. J. Tuberc. Lung Dis. 4:278-281. [PubMed] [Google Scholar]
- 190.Oliveira, R. S., M. P. Sircili, E. M. Oliveira, S. C. Balian, J. S. Ferreira-Neto, and S. C. Leao. 2003. Identification of Mycobacterium avium genotypes with distinctive traits by combination of IS1245-based restriction fragment length polymorphism and restriction analysis of hsp65. J. Clin. Microbiol. 41:44-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Olsen, I., T. B. Johansen, H. Billman-Jacobe, S. F. Nilsen, and B. Djonne. 2004. A novel IS element, ISMpa1, in Mycobacterium avium subsp. paratuberculosis. Vet. Microbiol. 98:297-306. [DOI] [PubMed] [Google Scholar]
- 192.Olsen, I., L. J. Reitan, G. Holstad, and H. G. Wiker. 2000. Alkyl hydroperoxide reductases C and D are major antigens constitutively expressed by Mycobacterium avium subsp. paratuberculosis. Infect. Immun. 68:801-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Olsen, I., M. Tryland, H. G. Wiker, and L. J. Reitan. 2001. AhpC, AhpD, and a secreted 14-kilodalton antigen from Mycobacterium avium subsp. paratuberculosis distinguish between paratuberculosis and bovine tuberculosis in an enzyme-linked immunosorbent assay. Clin. Diagn. Lab. Immunol. 8:797-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Pagnout, C., J. F. Ferard, and P. Poupin. 2006. Characterization of IS1110-like sequences found in Mycobacterium species other than Mycobacterium avium. Res. Microbiol. 157:650-658. [DOI] [PubMed] [Google Scholar]
- 195.Pai, M., L. W. Riley, and J. M. Colford, Jr. 2004. Interferon-gamma assays in the immunodiagnosis of tuberculosis: a systematic review. Lancet Infect. Dis. 4:761-776. [DOI] [PubMed] [Google Scholar]
- 196.Palmer, C. E., and M. W. Long. 1966. Effects of infection with atypical mycobacteria on BCG vaccination and tuberculosis. Am. Rev. Respir. Dis. 94:553-568. [DOI] [PubMed] [Google Scholar]
- 197.Parker, A. E., and L. E. Bermudez. 1997. Expression of the green fluorescent protein (GFP) in Mycobacterium avium as a tool to study the interaction between mycobacteria and host cells. Microb. Pathog. 22:193-198. [DOI] [PubMed] [Google Scholar]
- 198.Parsons, L. M., R. Brosch, S. T. Cole, A. Somoskovi, A. Loder, G. Bretzel, D. van Soolingen, Y. M. Hale, and M. Salfinger. 2002. Rapid and simple approach for identification of Mycobacterium tuberculosis complex isolates by PCR-based genomic deletion analysis. J. Clin. Microbiol. 40:2339-2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Pauls, R. J., C. Y. Turenne, J. N. Wolfe, and A. Kabani. 2003. A high proportion of novel mycobacteria species identified by 16S rDNA analysis among slowly growing AccuProbe-negative strains in a clinical setting. Am. J. Clin. Pathol. 120:560-566. [DOI] [PubMed] [Google Scholar]
- 200.Paustian, M. L., A. Amonsin, V. Kapur, and J. P. Bannantine. 2004. Characterization of novel coding sequences specific to Mycobacterium avium subsp. paratuberculosis: implications for diagnosis of Johne's disease. J. Clin. Microbiol. 42:2675-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Paustian, M. L., V. Kapur, and J. P. Bannantine. 2005. Comparative genomic hybridizations reveal genetic regions within the Mycobacterium avium complex that are divergent from Mycobacterium avium subsp. paratuberculosis isolates. J. Bacteriol. 187:2406-2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Pavlik, I., A. Horvathova, L. Dvorska, J. Bartl, P. Svastova, R. du Maine, and I. Rychlik. 1999. Standardisation of restriction fragment length polymorphism analysis for Mycobacterium avium subspecies paratuberculosis. J. Microbiol. Methods 38:155-167. [DOI] [PubMed] [Google Scholar]
- 203.Pavlik, I., P. Svastova, J. Bartl, L. Dvorska, and I. Rychlik. 2000. Relationship between IS901 in Mycobacterium avium complex strains isolated from birds, animals, humans, and the environment and virulence for poultry. Clin. Diagn. Lab. Immunol. 7:212-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Pedrosa, J., M. Florido, Z. M. Kunze, A. G. Castro, F. Portaels, J. McFadden, M. T. Silva, and R. Appelberg. 1994. Characterization of the virulence of Mycobacterium avium complex (MAC) isolates in mice. Clin. Exp. Immunol. 98:210-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Pethel, M. L., and J. O. Falkinham III. 1989. Plasmid-influenced changes in Mycobacterium avium catalase activity. Infect. Immun. 57:1714-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Poupart, P., M. Coene, H. Van Heuverswyn, and C. Cocito. 1993. Preparation of a specific RNA probe for detection of Mycobacterium paratuberculosis and diagnosis of Johne's disease. J. Clin. Microbiol. 31:1601-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Prammananan, T., S. Phunpruch, N. Tingtoy, S. Srimuang, and A. Chaiprasert. 2006. Distribution of hsp65 PCR-restriction enzyme analysis patterns among Mycobacterium avium complex isolates in Thailand. J. Clin. Microbiol. 44:3819-3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Primm, T. P., C. A. Lucero, and J. O. Falkinham III. 2004. Health impacts of environmental mycobacteria. Clin. Microbiol. Rev. 17:98-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Prince, D. S., D. D. Peterson, R. M. Steiner, J. E. Gottlieb, R. Scott, H. L. Israel, W. G. Figueroa, and J. E. Fish. 1989. Infection with Mycobacterium avium complex in patients without predisposing conditions. N. Engl. J. Med. 321:863-868. [DOI] [PubMed] [Google Scholar]
- 210.Puyang, X., K. Lee, C. Pawlichuk, and D. Y. Kunimoto. 1999. IS1626, a new IS900-related Mycobacterium avium insertion sequence. Microbiology 145:3163-3168. [DOI] [PubMed] [Google Scholar]
- 211.Pym, A. S., P. Brodin, R. Brosch, M. Huerre, and S. T. Cole. 2002. Loss of RD1 contributed to the attenuation of the live tuberculosis vaccines Mycobacterium bovis BCG and Mycobacterium microti. Mol. Microbiol. 46:709-717. [DOI] [PubMed] [Google Scholar]
- 212.Quadri, L. E., J. Sello, T. A. Keating, P. H. Weinreb, and C. T. Walsh. 1998. Identification of a Mycobacterium tuberculosis gene cluster encoding the biosynthetic enzymes for assembly of the virulence-conferring siderophore mycobactin. Chem. Biol. 5:631-645. [DOI] [PubMed] [Google Scholar]
- 213.Quirke, P. 2001. Antagonist: Mycobacterium avium subspecies paratuberculosis is a cause of Crohn's disease. Gut 49:757-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Rajeev, S., Y. Zhang, S. Sreevatsan, A. S. Motiwala, and B. Byrum. 2005. Evaluation of multiple genomic targets for identification and confirmation of Mycobacterium avium subsp. paratuberculosis isolates using real-time PCR. Vet. Microbiol. 105:215-221. [DOI] [PubMed] [Google Scholar]
- 215.Reddy, V. M., K. Parikh, J. Luna-Herrera, J. O. Falkinham III, S. Brown, and P. R. Gangadharam. 1994. Comparison of virulence of Mycobacterium avium complex (MAC) strains isolated from AIDS and non-AIDS patients. Microb. Pathog. 16:121-130. [DOI] [PubMed] [Google Scholar]
- 216.Reich, J. M., and R. E. Johnson. 1992. Mycobacterium avium complex pulmonary disease presenting as an isolated lingular or middle lobe pattern. The Lady Windermere syndrome. Chest 101:1605-1609. [DOI] [PubMed] [Google Scholar]
- 217.Richter, E., S. Rusch-Gerdes, and D. Hillemann. 2006. Evaluation of the GenoType Mycobacterium assay for identification of mycobacterial species from cultures. J. Clin. Microbiol. 44:1769-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Richter, E., J. Wessling, N. Lugering, W. Domschke, and S. Rusch-Gerdes. 2002. Mycobacterium avium subsp. paratuberculosis infection in a patient with HIV, Germany. Emerg. Infect. Dis. 8:729-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Ritacco, V., K. Kremer, T. Van der Laan, J. E. Pijnenburg, P. E. de Haas, and D. van Soolingen. 1998. Use of IS901 and IS1245 in RFLP typing of Mycobacterium avium complex: relatedness among serovar reference strains, human and animal isolates. Int. J. Tuberc. Lung Dis. 2:242-251. [PubMed] [Google Scholar]
- 220.Roiz, M. P., E. Palenque, C. Guerrero, and M. J. Garcia. 1995. Use of restriction fragment length polymorphism as a genetic marker for typing Mycobacterium avium strains. J. Clin. Microbiol. 33:1389-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Rosenzweig, D. Y. 1979. Pulmonary mycobacterial infections due to Mycobacterium intracellulare-avium complex. Clinical features and course in 100 consecutive cases. Chest 75:115-119. [DOI] [PubMed] [Google Scholar]
- 222.Rowbotham, D. S., N. P. Mapstone, L. K. Trejdosiewicz, P. D. Howdle, and P. Quirke. 1995. Mycobacterium paratuberculosis DNA not detected in Crohn's disease tissue by fluorescent polymerase chain reaction. Gut 37:660-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Runyon, E. H. 2004. Mycobacterium intracellulare. Am. Rev. Respir. Dis. 95:861-865. [DOI] [PubMed] [Google Scholar]
- 224.Ryan, P., M. W. Bennett, S. Aarons, G. Lee, J. K. Collins, G. C. O'Sullivan, J. O'Connell, and F. Shanahan. 2002. PCR detection of Mycobacterium paratuberculosis in Crohn's disease granulomas isolated by laser capture microdissection. Gut 51:665-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Saito, H., H. Tomioka, K. Sato, H. Tasaka, and D. J. Dawson. 1990. Identification of various serovar strains of Mycobacterium avium complex by using DNA probes specific for Mycobacterium avium and Mycobacterium intracellulare. J. Clin. Microbiol. 28:1694-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Sanderson, J. D., M. T. Moss, M. L. Tizard, and J. Hermon-Taylor. 1992. Mycobacterium paratuberculosis DNA in Crohn's disease tissue. Gut 33:890-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Sangari, F. J., M. Bachli, L. E. Bermudez, and T. Bodmer. 2000. Characterization of IS666, a newly described insertion element of Mycobacterium avium. Microb. Comp. Genomics 5:181-188. [DOI] [PubMed] [Google Scholar]
- 228.Sanna, E., C. J. Woodall, N. J. Watt, C. J. Clarke, M. Pittau, A. Leoni, and A. M. Nieddu. 2000. In situ-PCR for the detection of Mycobacterium paratuberculosis DNA in paraffin-embedded tissues. Eur. J. Histochem. 44:179-184. [PubMed] [Google Scholar]
- 229.Sarkola, A., J. Makinen, M. Marjamaki, H. J. Marttila, M. K. Viljanen, and H. Soini. 2004. Prospective evaluation of the GenoType assay for routine identification of mycobacteria. Eur. J. Clin. Microbiol. Infect. Dis. 23:642-645. [DOI] [PubMed] [Google Scholar]
- 230.Sartor, R. B. 2005. Does Mycobacterium avium subspecies paratuberculosis cause Crohn's disease. Gut 54:896-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Saxegaard, F., and I. Baess. 1988. Relationship between Mycobacterium avium, Mycobacterium paratuberculosis and “wood pigeon mycobacteria.” Determinations by DNA-DNA hybridization. APMIS 96:37-42. [PubMed] [Google Scholar]
- 232.Schaefer, W. B. 1965. Serologic identification and classification of the atypical mycobacteria by their agglutination. Am. Rev. Respir. Dis. 92:85-93. [DOI] [PubMed] [Google Scholar]
- 233.Schwartz, D., I. Shafran, C. Romero, C. Piromalli, J. Biggerstaff, N. Naser, W. Chamberlin, and S. A. Naser. 2000. Use of short-term culture for identification of Mycobacterium avium subsp. paratuberculosis in tissue from Crohn's disease patients. Clin. Microbiol. Infect. 6:303-307. [DOI] [PubMed] [Google Scholar]
- 234.Sechi, L. A., M. Mura, F. Tanda, A. Lissia, A. Solinas, G. Fadda, and S. Zanetti. 2001. Identification of Mycobacterium avium subsp. paratuberculosis in biopsy specimens from patients with Crohn's disease identified by in situ hybridization. J. Clin. Microbiol. 39:4514-4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Sechi, L. A., A. M. Scanu, P. Molicotti, S. Cannas, M. Mura, G. Dettori, G. Fadda, and S. Zanetti. 2005. Detection and isolation of Mycobacterium avium subspecies paratuberculosis from intestinal mucosal biopsies of patients with and without Crohn's disease in Sardinia. Am. J. Gastroenterol. 100:1529-1536. [DOI] [PubMed] [Google Scholar]
- 236.Semret, M., D. C. Alexander, C. Y. Turenne, P. de Haas, P. Overduin, D. van Soolingen, D. Cousins, and M. A. Behr. 2005. Genomic polymorphisms for Mycobacterium avium subsp. paratuberculosis diagnostics. J. Clin. Microbiol. 43:3704-3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Semret, M., D. Bakker, N. Smart, I. Olsen, K. Haslov, and M. A. Behr. 2006. Genetic analysis of Mycobacterium avium complex strains used for producing purified protein derivatives. Clin. Vaccine Immunol. 13:991-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Semret, M., C. Y. Turenne, and M. A. Behr. 2006. Insertion sequence IS900 revisited. J. Clin. Microbiol. 44:1081-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Semret, M., C. Y. Turenne, P. de Haas, D. M. Collins, and M. A. Behr. 2006. Differentiating host-associated variants of Mycobacterium avium by PCR for detection of large sequence polymorphisms. J. Clin. Microbiol. 44:881-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Semret, M., G. Zhai, S. Mostowy, C. Cleto, D. Alexander, G. Cangelosi, D. Cousins, D. M. Collins, D. van Soolingen, and M. A. Behr. 2004. Extensive genomic polymorphism within Mycobacterium avium. J. Bacteriol. 186:6332-6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Sevilla, I., S. V. Singh, J. M. Garrido, G. Aduriz, S. Rodriguez, M. V. Geijo, R. J. Whittington, V. Saunders, R. H. Whitlock, and R. A. Juste. 2005. Molecular typing of Mycobacterium avium subspecies paratuberculosis strains from different hosts and regions. Rev. Sci. Tech. 24:1061-1066. [PubMed] [Google Scholar]
- 242.Shanahan, F., and J. O'Mahony. 2005. The mycobacteria story in Crohn's disease. Am. J. Gastroenterol. 100:1537-1538. [DOI] [PubMed] [Google Scholar]
- 243.Shin, S. J., Y. F. Chang, C. Huang, J. Zhu, L. Huang, H. S. Yoo, K. S. Shin, S. Stehman, S. J. Shin, and A. Torres. 2004. Development of a polymerase chain reaction test to confirm Mycobacterium avium subsp. paratuberculosis in culture. J. Vet. Diagn. Investig. 16:116-120. [DOI] [PubMed] [Google Scholar]
- 244.Siguier, P., J. Perochon, L. Lestrade, J. Mahillon, and M. Chandler. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34:D32-D36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Smith, N. H., K. Kremer, J. Inwald, J. Dale, J. R. Driscoll, S. V. Gordon, D. van Soolingen, R. G. Hewinson, and J. M. Smith. 2006. Ecotypes of the Mycobacterium tuberculosis complex. J. Theor. Biol. 239:220-225. [DOI] [PubMed] [Google Scholar]
- 246.Smole, S. C., F. McAleese, J. Ngampasutadol, C. F. von Reyn, and R. D. Arbeit. 2002. Clinical and epidemiological correlates of genotypes within the Mycobacterium avium complex defined by restriction and sequence analysis of hsp65. J. Clin. Microbiol. 40:3374-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Soini, H., E. Eerola, and M. K. Viljanen. 1996. Genetic diversity among Mycobacterium avium complex AccuProbe-positive isolates. J. Clin. Microbiol. 34:55-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Sola, C., A. Devallois, K. S. Goh, E. Legrand, and N. Rastogi. 1996. Molecular characterization of Mycobacterium avium complex isolates from Caribbean patients by DT1/DT6-PCR, nonradioactive Southern hybridization, and the Accuprobe system. Curr. Microbiol. 33:352-358. [DOI] [PubMed] [Google Scholar]
- 249.Soltys, M. A., and D. R. Wise. 1967. Atypical mycobacterium in tuberculosis-like lesions in wood pigeons. J. Pathol. Bacteriol. 93:351-352. [DOI] [PubMed] [Google Scholar]
- 250.Springer, B., L. Stockman, K. Teschner, G. D. Roberts, and E. C. Bottger. 1996. Two-laboratory collaborative study on identification of mycobacteria: molecular versus phenotypic methods. J. Clin. Microbiol. 34:296-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Stabel, J. R. 1998. Johne's disease: a hidden threat. J. Dairy Sci. 81:283-288. [DOI] [PubMed] [Google Scholar]
- 252.Stratmann, J., K. Dohmann, J. Heinzmann, and G. F. Gerlach. 2006. Peptide aMptD-mediated capture PCR for detection of Mycobacterium avium subsp. paratuberculosis in bulk milk samples. Appl. Environ. Microbiol. 72:5150-5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Stratmann, J., B. Strommenger, R. Goethe, K. Dohmann, G. F. Gerlach, K. Stevenson, L. L. Li, Q. Zhang, V. Kapur, and T. J. Bull. 2004. A 38-kilobase pathogenicity island specific for Mycobacterium avium subsp. paratuberculosis encodes cell surface proteins expressed in the host. Infect. Immun. 72:1265-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Strommenger, B., K. Stevenson, and G. F. Gerlach. 2001. Isolation and diagnostic potential of ISMav2, a novel insertion sequence-like element from Mycobacterium avium subspecies paratuberculosis. FEMS Microbiol. Lett. 196:31-37. [DOI] [PubMed] [Google Scholar]
- 255.Suenaga, K., Y. Yokoyama, K. Okazaki, and Y. Yamamoto. 1995. Mycobacteria in the intestine of Japanese patients with inflammatory bowel disease. Am. J. Gastroenterol. 90:76-80. [PubMed] [Google Scholar]
- 256.Suffys, P. N., R. A. da Silva, M. de Oliveira, C. E. Dias Campos, A. M. Werneck Barreto, F. Portaels, L. Rigouts, G. Wouters, G. Jannes, G. van Reybroeck, W. Mijs, and B. Vanderborght. 2001. Rapid identification of mycobacteria to the species level using INNO-LiPA Mycobacteria, a reverse hybridization assay. J. Clin. Microbiol. 39:4477-4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Talbot, E. A., D. L. Williams, and R. Frothingham. 1997. PCR identification of Mycobacterium bovis BCG. J. Clin. Microbiol. 35:566-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Tasara, T., L. E. Hoelzle, and R. Stephan. 2005. Development and evaluation of a Mycobacterium avium subspecies paratuberculosis (MAP) specific multiplex PCR assay. Int. J. Food Microbiol. 104:279-287. [DOI] [PubMed] [Google Scholar]
- 259.Tasara, T., and R. Stephan. 2005. Development of an F57 sequence-based real-time PCR assay for detection of Mycobacterium avium subsp. paratuberculosis in milk. Appl. Environ. Microbiol. 71:5957-5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Taylor, A. W. 1951. Varieties of Mycobacterium johnei isolated from sheep. J. Pathol. Bacteriol. 63:333-336. [DOI] [PubMed] [Google Scholar]
- 261.Telenti, A. 1998. More on “what's in a name…”—pragmatism in mycobacterial taxonomy. Int. J. Tuberc. Lung Dis. 2:182-183. [PubMed] [Google Scholar]
- 262.Telenti, A., F. Marchesi, M. Balz, F. Bally, E. C. Bottger, and T. Bodmer. 1993. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J. Clin. Microbiol. 31:175-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Thegerstrom, J., B. I. Marklund, S. Hoffner, D. Axelsson-Olsson, J. Kauppinen, and B. Olsen. 2005. Mycobacterium avium with the bird type IS1245 RFLP profile is commonly found in wild and domestic animals, but rarely in humans. Scand. J. Infect. Dis. 37:15-20. [DOI] [PubMed] [Google Scholar]
- 264.Thierry, D., S. Bauge, J. D. Poveda, V. Vincent, and J. L. Guesdon. 1993. Rapid identification of Mycobacterium avium-intracellulare complex strains: clinical practice evaluation of DT6 and DT1 probes. J. Infect. Dis. 168:1337-1338. [DOI] [PubMed] [Google Scholar]
- 265.Thierry, D., V. Vincent, F. Clement, and J. L. Guesdon. 1993. Isolation of specific DNA fragments of Mycobacterium avium and their possible use in diagnosis. J. Clin. Microbiol. 31:1048-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Thorel, M. F., H. F. Huchzermeyer, and A. L. Michel. 2001. Mycobacterium avium and Mycobacterium intracellulare infection in mammals. Rev. Sci. Tech. 20:204-218. [DOI] [PubMed] [Google Scholar]
- 267.Thorel, M. F., M. Krichevsky, and V. V. Levy-Frebault. 1990. Numerical taxonomy of mycobactin-dependent mycobacteria, emended description of Mycobacterium avium, and description of Mycobacterium avium subsp. avium subsp. nov., Mycobacterium avium subsp. paratuberculosis subsp. nov., and Mycobacterium avium subsp. silvaticum subsp. nov. Int. J. Syst. Bacteriol. 40:254-260. [DOI] [PubMed] [Google Scholar]
- 268.Tizard, M., T. Bull, D. Millar, T. Doran, H. Martin, N. Sumar, J. Ford, and J. Hermon-Taylor. 1998. A low G+C content genetic island in Mycobacterium avium subsp. paratuberculosis and M. avium subsp. silvaticum with homologous genes in Mycobacterium tuberculosis. Microbiology 144:3413-3423. [DOI] [PubMed] [Google Scholar]
- 269.Tomioka, H., K. Sato, H. Saito, and Y. Yamada. 1989. Susceptibility of Mycobacterium avium and Mycobacterium intracellulare to various antibacterial drugs. Microbiol. Immunol. 33:509-514. [DOI] [PubMed] [Google Scholar]
- 270.Torkko, P., S. Suomalainen, E. Iivanainen, E. Tortoli, M. Suutari, J. Seppanen, L. Paulin, and M. L. Katila. 2002. Mycobacterium palustre sp. nov., a potentially pathogenic, slowly growing mycobacterium isolated from clinical and veterinary specimens and from Finnish stream waters. Int. J. Syst. Evol. Microbiol. 52:1519-1525. [DOI] [PubMed] [Google Scholar]
- 271.Torrelles, J. B., D. Ellis, T. Osborne, A. Hoefer, I. M. Orme, D. Chatterjee, P. J. Brennan, and A. M. Cooper. 2002. Characterization of virulence, colony morphotype and the glycopeptidolipid of Mycobacterium avium strain 104. Tuberculosis (Edinburgh) 82:293-300. [DOI] [PubMed] [Google Scholar]
- 272.Tortoli, E., A. Bartoloni, E. C. Bottger, S. Emler, C. Garzelli, E. Magliano, A. Mantella, N. Rastogi, L. Rindi, C. Scarparo, and P. Urbano. 2001. Burden of unidentifiable mycobacteria in a reference laboratory. J. Clin. Microbiol. 39:4058-4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Tortoli, E., A. Mariottini, and G. Mazzarelli. 2003. Evaluation of INNO-LiPA MYCOBACTERIA v2: improved reverse hybridization multiple DNA probe assay for mycobacterial identification. J. Clin. Microbiol. 41:4418-4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Tortoli, E., L. Rindi, M. J. Garcia, P. Chiaradonna, R. Dei, C. Garzelli, R. M. Kroppenstedt, N. Lari, R. Mattei, A. Mariottini, G. Mazzarelli, M. I. Murcia, A. Nanetti, P. Piccoli, and C. Scarparo. 2004. Proposal to elevate the genetic variant MAC-A, included in the Mycobacterium avium complex, to species rank as Mycobacterium chimaera sp. nov. Int. J. Syst. Evol. Microbiol. 54:1277-1285. [DOI] [PubMed] [Google Scholar]
- 275.Truffot-Pernot, C., H. F. Lecoeur, L. Maury, B. Dautzenberg, and J. Grosset. 1989. Results of blood cultures for detection of mycobacteria in AIDS patients. Tubercle 70:187-191. [DOI] [PubMed] [Google Scholar]
- 276.Tsang, A. Y., J. C. Denner, P. J. Brennan, and J. K. McClatchy. 1992. Clinical and epidemiological importance of typing of Mycobacterium avium complex isolates. J. Clin. Microbiol. 30:479-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Turenne, C. Y., M. Semret, D. V. Cousins, D. M. Collins, and M. A. Behr. 2006. Sequencing of hsp65 distinguishes among subsets of the Mycobacterium avium complex. J. Clin. Microbiol. 44:433-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Turenne, C. Y., L. Thibert, K. Williams, T. V. Burdz, V. J. Cook, J. N. Wolfe, D. W. Cockcroft, and A. Kabani. 2004. Mycobacterium saskatchewanense sp. nov., a novel slowly growing scotochromogenic species from human clinical isolates related to Mycobacterium interjectum and AccuProbe-positive for Mycobacterium avium complex. Int. J. Syst. Evol. Microbiol. 54:659-667. [DOI] [PubMed] [Google Scholar]
- 279.Turenne, C. Y., L. Tschetter, J. Wolfe, and A. Kabani. 2001. Necessity of quality-controlled 16S rRNA gene sequence databases: identifying nontuberculous Mycobacterium species. J. Clin. Microbiol. 39:3637-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.van Embden, J. D., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, T. M. Shinnick, et al. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Van Kruiningen, H. J. 1999. Lack of support for a common etiology in Johne's disease of animals and Crohn's disease in humans. Inflamm. Bowel Dis. 5:183-191. [DOI] [PubMed] [Google Scholar]
- 282.Vansnick, E., P. De Rijk, F. Vercammen, D. Geysen, L. Rigouts, and F. Portaels. 2004. Newly developed primers for the detection of Mycobacterium avium subspecies paratuberculosis. Vet. Microbiol. 100:197-204. [DOI] [PubMed] [Google Scholar]
- 283.van Soolingen, D., J. Bauer, V. Ritacco, S. C. Leao, I. Pavlik, V. Vincent, N. Rastogi, A. Gori, T. Bodmer, C. Garzelli, and M. J. Garcia. 1998. IS1245 restriction fragment length polymorphism typing of Mycobacterium avium isolates: proposal for standardization. J. Clin. Microbiol. 36:3051-3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Vary, P. H., P. R. Andersen, E. Green, J. Hermon-Taylor, and J. J. McFadden. 1990. Use of highly specific DNA probes and the polymerase chain reaction to detect Mycobacterium paratuberculosis in Johne's disease. J. Clin. Microbiol. 28:933-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Viljanen, M. K., L. Olkkonen, and M. L. Katila. 1993. Conventional identification characteristics, mycolate and fatty acid composition, and clinical significance of MAIX AccuProbe-positive isolates of Mycobacterium avium complex. J. Clin. Microbiol. 31:1376-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Vinh, D. C., and C. N. Bernstein. 2005. Crohn's disease and M. paratuberculosis: where's the beef. Inflamm. Bowel Dis. 11:1025-1027. [DOI] [PubMed] [Google Scholar]
- 287.von Reyn, C. F., R. D. Arbeit, C. R. Horsburgh, M. A. Ristola, R. D. Waddell, S. M. Tvaroha, M. Samore, L. R. Hirschhorn, J. Lumio, A. D. Lein, M. R. Grove, and A. N. Tosteson. 2002. Sources of disseminated Mycobacterium avium infection in AIDS. J. Infect. 44:166-170. [DOI] [PubMed] [Google Scholar]
- 288.Wallace, R. J., Jr., B. A. Brown-Elliott, J. Brown, A. G. Steigerwalt, L. Hall, G. Woods, J. Cloud, L. Mann, R. Wilson, C. Crist, K. C. Jost, Jr., D. E. Byrer, J. Tang, J. Cooper, E. Stamenova, B. Campbell, J. Wolfe, and C. Turenne. 2005. Polyphasic characterization reveals that the human pathogen Mycobacterium peregrinum type II belongs to the bovine pathogen species Mycobacterium senegalense. J. Clin. Microbiol. 43:5925-5935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Wallace, R. J., Jr., B. A. Brown-Elliott, R. W. Wilson, L. Mann, L. Hall, Y. Zhang, K. C. Jost, Jr., J. M. Brown, A. Kabani, M. F. Schinsky, A. G. Steigerwalt, C. J. Crist, G. D. Roberts, Z. Blacklock, M. Tsukamura, V. Silcox, and C. Turenne. 2004. Clinical and laboratory features of Mycobacterium porcinum. J. Clin. Microbiol. 42:5689-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Wallace, R. J., Jr., Y. Zhang, B. A. Brown, D. Dawson, D. T. Murphy, R. Wilson, and D. E. Griffith. 1998. Polyclonal Mycobacterium avium complex infections in patients with nodular bronchiectasis. Am. J. Respir. Crit. Care Med. 158:1235-1244. [DOI] [PubMed] [Google Scholar]
- 291.Wasem, C. F., C. M. McCarthy, and L. W. Murray. 1991. Multilocus enzyme electrophoresis analysis of the Mycobacterium avium complex and other mycobacteria. J. Clin. Microbiol. 29:264-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Wayne, L. G., R. C. Good, E. C. Bottger, R. Butler, M. Dorsch, T. Ezaki, W. Gross, V. Jonas, J. Kilburn, P. Kirschner, M. I. Krichevsky, M. Ridell, T. M. Shinnick, B. Springer, E. Stackebrandt, I. Tarnok, Z. Tarnok, H. Tasaka, V. Vincent, N. G. Warren, C. A. Knott, and R. Johnson. 1996. Semantide- and chemotaxonomy-based analyses of some problematic phenotypic clusters of slowly growing mycobacteria, a cooperative study of the International Working Group on Mycobacterial Taxonomy. Int. J. Syst. Bacteriol. 46:280-297. [DOI] [PubMed] [Google Scholar]
- 293.Wayne, L. G., R. C. Good, A. Tsang, R. Butler, D. Dawson, D. Groothuis, W. Gross, J. Hawkins, J. Kilburn, M. Kubin, K. H. Schröder, V. A. Silcox, C. Smith, M. F. Thorel, C. Woodley, and M. A. Yakrus. 1993. Serovar determination and molecular taxonomic correlation in Mycobacterium avium, Mycobacterium intracellulare, and Mycobacterium scrofulaceum: a cooperative study of the International Working Group on Mycobacterial Taxonomy. Int. J. Syst. Bacteriol. 43:482-489. [DOI] [PubMed] [Google Scholar]
- 294.Whitlock, R. H., and C. Buergelt. 1996. Preclinical and clinical manifestations of paratuberculosis (including pathology). Vet. Clin. N. Am. Food Anim. Pract. 12:345-356. [DOI] [PubMed] [Google Scholar]
- 295.Whitlock, R. H., S. J. Wells, R. W. Sweeney, and J. Van Tiem. 2000. ELISA and fecal culture for paratuberculosis (Johne's disease): sensitivity and specificity of each method. Vet. Microbiol. 77:387-398. [DOI] [PubMed] [Google Scholar]
- 296.Whittington, R., I. Marsh, E. Choy, and D. Cousins. 1998. Polymorphisms in IS1311, an insertion sequence common to Mycobacterium avium and M. avium subsp. paratuberculosis, can be used to distinguish between and within these species. Mol. Cell. Probes 12:349-358. [DOI] [PubMed] [Google Scholar]
- 297.Whittington, R. J., A. F. Hope, D. J. Marshall, C. A. Taragel, and I. Marsh. 2000. Molecular epidemiology of Mycobacterium avium subsp. paratuberculosis: IS900 restriction fragment length polymorphism and IS1311 polymorphism analyses of isolates from animals and a human in Australia. J. Clin. Microbiol. 38:3240-3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Whittington, R. J., I. Marsh, S. McAllister, M. J. Turner, D. J. Marshall, and C. A. Fraser. 1999. Evaluation of modified BACTEC 12B radiometric medium and solid media for culture of Mycobacterium avium subsp. paratuberculosis from sheep. J. Clin. Microbiol. 37:1077-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Whittington, R. J., I. B. Marsh, and R. H. Whitlock. 2001. Typing of IS 1311 polymorphisms confirms that bison (Bison bison) with paratuberculosis in Montana are infected with a strain of Mycobacterium avium subsp. paratuberculosis distinct from that occurring in cattle and other domesticated livestock. Mol. Cell. Probes 15:139-145. [DOI] [PubMed] [Google Scholar]
- 300.Whittington, R. J., D. J. Marshall, P. J. Nicholls, I. B. Marsh, and L. A. Reddacliff. 2004. Survival and dormancy of Mycobacterium avium subsp. paratuberculosis in the environment. Appl. Environ. Microbiol. 70:2989-3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Wolinsky, E. 1979. Nontuberculous mycobacteria and associated diseases. Am. Rev. Respir. Dis. 119:107-159. [DOI] [PubMed] [Google Scholar]
- 302.Wolinsky, E. 1995. Mycobacterial lymphadenitis in children: a prospective study of 105 nontuberculous cases with long-term follow-up. Clin. Infect. Dis. 20:954-963. [DOI] [PubMed] [Google Scholar]
- 303.Wong, D. A., P. C. Yip, D. T. Cheung, and K. M. Kam. 2001. Simple and rational approach to the identification of Mycobacterium tuberculosis, Mycobacterium avium complex species, and other commonly isolated mycobacteria. J. Clin. Microbiol. 39:3768-3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Wu, C. W., J. Glasner, M. Collins, S. Naser, and A. M. Talaat. 2006. Whole-genome plasticity among Mycobacterium avium subspecies: insights from comparative genomic hybridizations. J. Bacteriol. 188:711-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Yakrus, M. A., and R. C. Good. 1990. Geographic distribution, frequency, and specimen source of Mycobacterium avium complex serotypes isolated from patients with acquired immunodeficiency syndrome. J. Clin. Microbiol. 28:926-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Yakrus, M. A., M. W. Reeves, and S. B. Hunter. 1992. Characterization of isolates of Mycobacterium avium serotypes 4 and 8 from patients with AIDS by multilocus enzyme electrophoresis. J. Clin. Microbiol. 30:1474-1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Yoshimura, H. H., and D. Y. Graham. 1988. Nucleic acid hybridization studies of mycobactin-dependent mycobacteria. J. Clin. Microbiol. 26:1309-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Young, L. S., C. B. Inderlied, O. G. Berlin, and M. S. Gottlieb. 1986. Mycobacterial infections in AIDS patients, with an emphasis on the Mycobacterium avium complex. Rev. Infect. Dis. 8:1024-1033. [DOI] [PubMed] [Google Scholar]