Abstract
Rhodococcus equi is a facultative intracellular pathogen that causes pneumonia in foals but does not induce disease in adult horses. Virulence of R. equi depends on the presence of a large plasmid, which encodes a family of seven virulence-associated proteins (VapA and VapC to VapH). Eradication of R. equi from the lungs depends on gamma interferon (IFN-γ) production by T lymphocytes. The objectives of the present study were to determine the relative in vivo expression of the vap genes of R. equi in the lungs of infected foals, to determine the recall response of bronchial lymph node (BLN) lymphocytes from foals and adult horses to each of the Vap proteins, and to compare the cytokine profiles of proliferating lymphocytes between foals and adult horses. vapA, vapD, and vapG were preferentially expressed in the lungs of infected foals, and expression of these genes in the lungs was significantly (P < 0.05) higher than that achieved during in vitro growth. VapA and VapC induced the strongest lymphoproliferative responses for foals and adult horses. There was no significant difference in recall lymphoproliferative responses or IFN-γ mRNA expression by bronchial lymph node lymphocytes between foals and adults. In contrast, interleukin 4 (IL-4) expression was significantly higher for adults than for foals for each of the Vap proteins. The ratio of IFN-γ to IL-4 was significantly higher for foals than for adult horses for most Vap proteins. Therefore, foals are immunocompetent and are capable of mounting lymphoproliferative responses of the same magnitude and cytokine phenotype as those of adult horses.
Rhodococcus equi, a gram-positive facultative intracellular pathogen, is one of the most important causes of pneumonia in foals between ages 3 weeks and 5 months. R. equi has also emerged as a significant opportunistic pathogen in immunosuppressed people, especially those infected with the human immunodeficiency virus (3, 9, 14). In foals, the course of the disease is insidious and pathology is often extensive by the time the disease is diagnosed. Unlike environmental R. equi, isolates from pneumonic foals typically contain an 80- to 90-kb plasmid. Plasmid-cured derivatives of virulent R. equi strains lose their ability to replicate and survive in macrophages (12). Plasmid-cured derivatives also fail to induce pneumonia and are completely cleared from the lungs of foals, confirming the absolute necessity of the large plasmid for the virulence of R. equi (12, 37).
A 27.5-kb region of the virulence plasmid bears the hallmark of a pathogenicity island and contains the genes for a family of seven closely related virulence-associated (Vap) proteins, designated VapA and VapC to VapH (35). Although a recent study has proposed the designation vapI for another gene of the pathogenicity island, vapI is not functional (31). In a recent study, an R. equi mutant lacking a 7.9-kb DNA region spanning five vap genes (vapA, -C, -D, -E, and -F) was attenuated for virulence in mice and failed to replicate in macrophages (20). Only complementation with vapA could restore full virulence, whereas complementation with vapC, vapD, or vapE could not (20). All vap genes are expressed during in vitro growth and are upregulated when R. equi is grown in equine macrophage monolayers (32).
Because of the facultative intracellular nature of R. equi, cell-mediated immune mechanisms are thought to be of major importance in resistance. Most knowledge of cell-mediated immunity to R. equi infections comes from studying infection of mice. In mice, functional T lymphocytes are required for the clearance of virulent R. equi. Although both CD4+ and CD8+ T cells contribute to the host defense against R. equi in mice, CD4+ T lymphocytes play the major role and are required for complete pulmonary clearance (21). Studies with mice have also clearly shown that a Th1 response, characterized by gamma interferon (IFN-γ) induction, is sufficient to effect complete pulmonary clearance of R. equi, whereas a Th2 response, characterized by interleukin 4 (IL-4) induction, is detrimental (22, 23). How these findings in mice relate to the foal remains to be determined. As opposed to foals, adult horses completely clear an intrabronchial challenge with virulent R. equi and do not develop clinical signs. Clearance of R. equi in adult horses is associated with a significant increase in bronchoalveolar lavage fluid CD4+ and CD8+ lymphocytes, lymphoproliferative responses to R. equi antigens, including VapA, development of R. equi-specific cytotoxic T lymphocytes, and IFN-γ induction (15-17, 24, 30). A few studies have examined antibody responses of foals and adult horses to the Vap proteins of R. equi (18, 24). However, cell-mediated immune responses and cytokine profiles of foals and adult horses for each of the Vap proteins have never been evaluated. Knowledge of gene products preferentially induced during infection in the natural host would provide important insight into the pathogenesis of the disease and may prove important for vaccine development. Despite the documented importance of some of the vap genes in the virulence of R. equi, relative expression of these genes during active infection in foals has not been studied.
In regard to the above, the objectives of the present study were to determine the relative in vivo expression of the functional vap genes of R. equi in the lungs of infected foals, to determine the recall lymphoproliferative responses of foals to each of the functional Vap proteins and compare them to those of resistant adult horses, and to determine the cytokine profiles of proliferating lymphocytes for both foals and adult horses.
MATERIALS AND METHODS
Animals and intrabronchial challenge.
Five foals between 7 and 10 days of age and five adult horses between 2 and 12 years of age were used in this study. Adequate transfer of passive immunity was confirmed in foals at 12 to 24 h of age by measurement of the plasma immunoglobulin G concentration using a commercial immunoassay (DVM Stat; Corporation for advanced Applications, Newburg, WI). Foals together with their dams were moved to individual stalls in an isolation facility the day after birth. Adult horses were moved to the isolation facility at least 2 days prior to the beginning of the study. Prior to initiation of the study, all animals were determined to be healthy on the basis of a thorough physical examination, complete blood count, biochemical profile, cytology and bacterial culture of a tracheobronchial aspirate, and thoracic radiographs.
R. equi ATCC 33701, a strain containing an 80-kb virulence plasmid, was used to infect foals (35). Aliquots of R. equi were grown on trypticase soy agar plates for 48 h at 37°C. Bacteria were harvested with 4 ml of sterile phosphate-buffered saline (PBS) per plate. The bacterial concentration was determined from CFU counting. Each animal was infected intrabronchially with an inoculum of 2 × 104 CFU per kg of body weight. This corresponded to a total inoculum of approximately 1 × 106 for each foal and 1 × 107 for each adult horse. The inoculum was diluted in 50 ml of sterile PBS. Prior to infection, animals were sedated with 0.5 mg/kg of xylazine hydrochloride and 0.07 mg/kg of butorphanol tartrate given intravenously. A flexible fiber-optic endoscope was used to deliver 25 ml of the bacterial suspension into each of the main bronchi. Animals were clinically assessed based on daily complete physical examinations as well as twice-daily heart rate, respiratory rate, and temperature recording. Euthanasia was performed 15 days postinfection by intravenous administration of a lethal dose of pentobarbital sodium. Bronchial lymph nodes (BLN) were collected aseptically and placed in sterile PBS for transport to the laboratory. Two lung tissue samples (approximately 1 g each) were collected aseptically from a cranioventral lung lobe. This area corresponded to the most severely affected location in all foals. One sample was immediately frozen in liquid nitrogen for subsequent bacterial RNA isolation, and the other was placed in a sterile bag for quantitative bacterial culture.
Preparation of BLN cells, cell stimulation, and lymphocyte proliferation assay.
Vap proteins (VapA, -C, -D, -E, -F, -G, and -H) for use in proliferation assays were obtained as glutathione S-transferase fusion proteins as described previously (18). Cells used for proliferation assays were collected from BLN. Briefly, BLN were cut into 125-mm3 pieces and the cells were separated in glass tissue grinders. Mononuclear cells were separated by density gradient centrifugation for 30 min at 400 × g using endotoxin-free Ficoll-Paque (Amersham Biosciences, Pittsburgh, PA). Aliquots of 3 × 107 cells were placed in 1 ml of freezing medium containing 90% fetal calf serum and 10% dimethyl sulfoxide. Cells were placed at −70°C for 4 h and then transferred to liquid nitrogen until assayed.
Immediately after thawing, BLN cells were washed twice and placed in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 2 mM glutamine, 25 mM HEPES, and penicillin-streptomycin (100 U and μg per ml, respectively). More than 70% of the cells were viable after thawing. Proliferative responses were assessed using a nonradioactive colorimetric assay. This assay has been shown to correlate closely with conventional radioactive [3H]thymidine incorporation in many species, including the horse (2, 38). Aliquots (100 μl) of cells (1 × 106 cells/ml) were placed in triplicate wells of 96-well black plates with flat, clear-bottom wells (Corning, Inc., Corning, NY). Cells were separately incubated with no antigen (blank), 2 μg/ml of concanavalin A (positive control), 10 μg/ml of Corynebacterium pseudotuberculosis soluble antigens (negative control), or 50 μg/ml of each of the recombinant protein (VapA, -C, -D, -E, -F, -G, and -H). Optimal concentrations of antigens and mitogen were determined based on a dose-response curve with VapA and ConA, respectively. The cells were stimulated at 37°C for 72 h in 6% CO2. Twelve hours before the end of the assay, 20 μl of alamar blue (Accumed International, Inc., Westlake, OH) was added to each of the wells. Plates were read on a fluorometer (Synergy HT; BioTek Instruments, Inc., Winooski, VT) using an excitation wavelength of 530 nm, and emission was measured at 590 nm. The change in fluorescence was calculated as the mean for the stimulated cells minus the mean for the cells without antigen or mitogen (blank).
BLN cells used for quantification of mRNA expression were prepared exactly as described above with the exception that the cells were stimulated with each of the Vap antigens for 12 h. This time selection was based on a time-response curve for IFN-γ and IL-4 mRNA expression.
RNA isolation from BLN cells, DNase treatment of RNA samples, and cDNA synthesis.
Isolation of total RNA from BLN cells was performed using the QIAGEN RNeasy kit (QIAGEN, Inc., Valencia, CA) according to the manufacturer's instructions. The RNA concentration was measured by determining the optical density at 260 nm. All RNA samples were treated with amplification-grade DNase I (Gibco BRL, Rockville, MD) to remove any trace of genomic DNA contamination. Briefly, 1 U of DNase I and 1 μl of 10× DNase I reaction buffer were mixed with 1 μg of total RNA to yield a 10-μl reaction mixture. The mixture was incubated for 10 min at room temperature and then inactivated by addition of 1 μl of 25 mM of EDTA and heating at 65°C for 10 min.
cDNA was synthesized with the Advantage RT-for-PCR kit (Clontech, Palo Alto, CA) by using the protocol of the manufacturer. Briefly, 1 μg of total RNA was mixed with 1 μl of oligo(dT)18 primer (20 μM) and heated at 70°C for 2 min. After cooling to room temperature, the following reagents were added: 4 μl of 5× reaction buffer, 1 μl of deoxynucleoside triphosphates (10 mM each), 0.5 μl of RNase inhibitor (40 U/μl), and 1 μl of Moloney murine leukemia virus reverse transcriptase (200 U/μl). The mixture was incubated at 42°C for 1 h, heated at 94°C for 5 min, diluted to a final volume of 100 μl, and stored at −70°C until used for PCR analysis.
Quantification of cytokine mRNA.
Gene-specific primers and internal oligonucleotide probes for equine glyceraldehyde-3-phosphate dehydrogenase, IL-4, and IFN-γ have been described previously (11). The internal probes were labeled at the 5′ end with the reporter dye 6-carboxyfluorescein and at the 3′ end with the quencher dye 6-carboxytetramethylrhodamine. Amplification of 2 μl of cDNA was performed in a 25-μl PCR mixture containing 900 nM of each primer, 250 nM of TaqMan probe, and 12 μl of TaqMan Universal PCR Mastermix (Applied Biosystems, Foster City, CA). Amplification and detection were performed using the ABI Prism 7700 sequence detection system (Applied Biosystems) with initial incubation steps at 50°C for 2 min and 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Serial dilutions of cDNA from 24-h concanavalin A-stimulated equine blood mononuclear cells were used to generate a standard curve for relative quantification of each gene of interest. Each sample was assayed in triplicate, and the mean value was used for comparison. Samples without cDNA were included in the amplification reactions to determine background fluorescence and check for contamination. To account for variation in the amount and quality of starting material, values obtained for IFN-γ and IL-4 transcripts were normalized by dividing by the glyceraldehyde-3-phosphate dehydrogenase transcript level for the same sample. The sample with the lowest level of gene expression was designated 1, and relative quantification between samples was reported as the n-fold difference relative to cytokine mRNA expression in that sample.
Bacterial RNA isolation, DNase treatment of bacterial RNA samples, and cDNA synthesis.
Bacterial RNA extraction for determination of in vivo expression of the vap genes was performed on lung tissue from the five infected foals. For determination of in vitro expression of the vap genes, one colony of R. equi ATCC 33701 was placed in five separate tubes, each containing 5 ml of brain heart infusion broth with 1% yeast extract. The suspensions were grown in a shaking incubator at 37°C for 18 h. A 125-mm3 section of lung tissue and pellets from in vitro-grown bacteria were independently pulverized in cold Trizol reagents (Invitrogen Corporation, Carlsbad, CA) using glass tissue grinders. Total RNA was extracted using the Trizol reagents by following the instructions from the manufacturer. RNA from each source was further purified independently by passing the resulting sample through RNeasy MinElute spin columns (QIAGEN, Inc.) with on-column DNase treatment according to the manufacturer's instructions. To deplete equine RNA and further enrich bacterial RNA, RNA samples from lung tissue were additionally subjected to the MICROBEnrich kit (Ambion, Inc., Austin, TX). Then, in an attempt to completely eliminate genomic DNA contamination, each RNA sample underwent an additional treatment with amplification-grade DNase I (Gibco BRL) before reverse transcription. Synthesis of cDNA was performed as described above except that random hexamer priming (20 μM) was used. Duplicate RNA samples were subjected to the same reverse transcription protocol minus the reverse transcriptase to confirm the absence of genomic DNA contamination or give a baseline used to subtract traces of remaining genomic DNA contamination.
Quantification of R. equi vap mRNA.
Gene-specific primers and internal oligonucleotide probes for vapA, vapC, vapD, vapE, vapF, vapG, and vapH were selected based on the plasmid DNA sequence of R. equi ATCC 33701 using Primer Express software (Table 1). Primer and probe sequences for 16S rRNA, used as a housekeeping gene in the present study, have been reported previously (29). Serial dilutions of plasmid DNA from strain ATCC 33701 were used to generate a standard curve for relative quantification of the vap genes. Genomic DNA from R. equi strain 33701−, the plasmid-cured derivative of ATCC 33701, was used to generate a standard curve for relative quantification of 16S rRNA. Amplification and detection were performed as described above under “Quantification of cytokine mRNA.” Calculated amplification efficiencies for primer/probe assays ranged between 89.0% and 99.9%. Coefficients of correlation (r) of the standard curves ranged between 0.981 and 0.995. To account for variation in the amount and quality of starting material, values obtained for vap transcripts were normalized by dividing by the 16S rRNA transcript level for the same sample. The sample with the lowest level of gene expression was designated 1, and relative quantification between samples was reported as the n-fold difference relative to mRNA expression in that sample. In rare instances when traces of genomic DNA were detected, the concentration of genomic DNA was subtracted from that for gene expression.
TABLE 1.
Oligonucleotide primer and probe sequences for amplification of vap genes from R. equi
| Gene | Primer or probe | Sequence (5′ to 3′) |
|---|---|---|
| vapA | Forward | CATCAACTTCTTCGATAGCTCAGGTA |
| Reverse | GACGCCCACCACAGTACTAACTC | |
| Probe | TCCTCGGCCATATCCAGTCCGGT | |
| vapC | Forward | GGGTCACGCTTGGTGGC |
| Reverse | CCGTATACTGTTCGTCGGCA | |
| Probe | TCGGAGTTCTACGGCCGCACAATAAA | |
| vapD | Forward | CACGAGCCTTTGGGCG |
| Reverse | CATTGAGACCGTTGCGATCA | |
| Probe | TTATTCACTTTCTTGCTCGCGGTGGCT | |
| vapE | Forward | TGAGTACAACGCTGTCGGTCC |
| Reverse | ACGTGCCCCAGCAAACC | |
| Probe | TACTTGAACATCAATCTTTTCGCCGGAGAC | |
| vapF | Forward | CATCAGCTGCTGGCAAAGTACT |
| Reverse | CCCCATGAACCGCATATTG | |
| Probe | CGCCAATCAATAACAATGCCGACGA | |
| vapG | Forward | CACTGCAACCCCGGGA |
| Reverse | GGCGGCGGAAAGACGT | |
| Probe | TCGAAATCCCGCCAGAATCACCA | |
| vapH | Forward | GTCAACTTCTTCGACGGTCACA |
| Reverse | ACGGAGCTCACCCCTCCTA | |
| Probe | CGCCATACTCGGCCATGCACAA |
Statistical analyses.
Statistical analyses were performed by using SigmaStat (version 3.0; SPSS, Inc., Chicago, IL). Differences in lymphocyte proliferation and cytokine expression between foals and adult horses, as well as differences between in vivo versus in vitro expression of each vap gene, were assessed using the Mann-Whitney U test. Differences in lymphocyte proliferation and cytokine expression between Vap proteins for a given group (foals or adults), as well as differences in the relative expression levels of each vap gene within a given sampling site (in vivo or in vitro), were assessed using the Friedman repeated measure analysis of variance on ranks. When appropriate, multiple pairwise comparisons were done using the Student-Newman-Keuls test. Significance was set at a P value of <0.05.
RESULTS
Disease process and macroscopic findings.
All animals maintained normal vital signs during the study and showed no evidence of disease. All five infected foals had macroscopic pulmonary lesions that consisted of mild to moderate areas of consolidation in the ventral lung lobes, along with multiple small nodular lesions up to 1 cm in diameter. The mean number of R. equi bacteria (± standard deviation [SD]) was 9.63 × 106 ± 1.21 × 107 (range, 6.23 × 104 to 2.33 × 107) CFU/g of lung tissue. The lungs of adult horses were free from lesions, and bacterial culture was negative.
In vivo expression of R. equi vap genes.
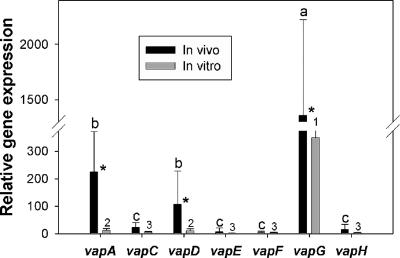
Real-time PCR was used to quantify the relative in vivo expression of the vap genes in the lung tissue of infected foals, and the expression was compared to that of in vitro-grown bacteria. Expression levels of vapA, vapD, and vapG were significantly higher during in vivo growth than during in vitro growth (Fig. 1). Expression of vapG in lung tissue was significantly higher than that of all other vap genes. Expression of vapA and vapD in lung tissue was significantly higher than that of vapC, vapE, vapF, and vapH (Fig. 1). The expression of the vap genes during in vitro growth followed a similar pattern, with significantly greater expression of vapA, vapD, and vapG (Fig. 1).
FIG. 1.
Comparison of in vivo and in vitro expression of the plasmid-borne vap genes of R. equi. In vivo expression was determined in lung tissue from five foals experimentally infected with R. equi. In vitro expression was determined during exponential growth of R. equi in broth. Results are presented as means ± SD. Different letters (a, b, and c) indicate a significant difference in expression in vivo between vap genes. Different numbers (1, 2, and 3) indicate a significant difference in expression in vitro between vap genes. *, significant difference in expression of a given vap gene between in vivo and in vitro growth (P < 0.05).
Vap-specific proliferative responses and cytokine expression.
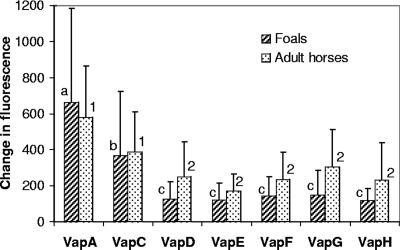
To determine whether differences in lymphoproliferative responses and cytokine profiles may be at the origin of the peculiar susceptibility of foals to infection with R. equi, recall responses of susceptible, infected foals was compared to those of R. equi-resistant adult horses. Proliferation of BLN mononuclear cells following stimulation with each recombinant Vap protein was measured. There were no significant differences in lymphoproliferative responses to each of the Vap proteins between infected foals and adult horses (Fig. 2). For foals, lymphoproliferative responses to VapA were significantly greater than those to all of the other Vap proteins, and lymphoproliferative responses to VapC were significantly greater than those to all Vap proteins except VapA (Fig. 2). In adult horses, lymphoproliferative responses to VapA and VapC were significantly greater than those to the other Vap proteins (Fig. 2).
FIG. 2.
Recall lymphoproliferative responses of BLN mononuclear cells from foals and adult horses to recombinant R. equi VapA, VapC, VapD, VapE, VapF, VapG, or VapH. The results are expressed as the fluorescence of stimulated cells minus that of unstimulated cells. The results are displayed as means ± SD. Different letters (a, b, and c) indicate significant differences in lymphoproliferative responses to Vap proteins for foals (P < 0.05). Different numbers (1 and 2) indicate significant differences in lymphoproliferative responses to Vap proteins for adult horses (P < 0.05).
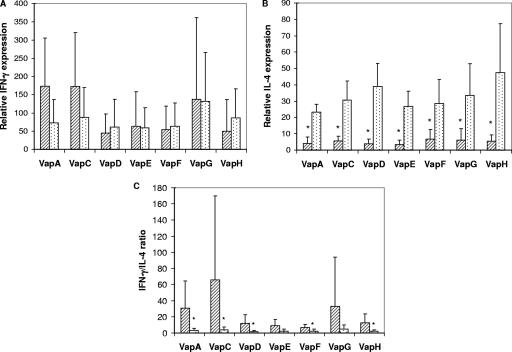
Induction of IFN-γ and IL-4 in proliferating cells was quantified by real-time reverse transcription-PCR. There was no significant difference in IFN-γ expression by BLN cells between foals and adult horses for any of the Vap proteins (Fig. 3A). In contrast, IL-4 mRNA expression for each of the Vap proteins was significantly greater for adult horses than for foals (Fig. 3B). The ratio of IFN-γ to IL-4 was significantly higher with foals than with adult horses for VapA, VapC, VapD, VapF, and VapH (Fig. 3C).
FIG. 3.
Relative IFN-γ (A) or IL-4 (B) mRNA expression or IFN-γ/IL-4 ratio (C) following stimulation of BLN mononuclear cells of foals (striped bars) or adult horses (dotted bars) with recombinant R. equi VapA, VapC, VapD, VapE, VapF, VapG, or VapH. The results are displayed as means ± SD. *, statistically significant difference between results for foals and those for adult horses (P < 0.05).
DISCUSSION
Despite a central role for cell-mediated immune responses in protection against R. equi, most studies have focused on antibody responses against the Vap proteins. This is the first study to examine the expression of vap genes in the lungs of R. equi-infected foals and to measure lymphoproliferative responses and the cytokine profiles of infected foals and adult horses for each of the functional Vap proteins. It complements an earlier study by Hooper-McGrevy et al. (18) that examined the serum immunoglobulin G subisotype responses to each of the Vap proteins for foals and for adult horses, as well as a study evaluating the expression of the pathogenicity island genes in equine macrophages (32).
VapA is expressed on the bacterial surface, and its expression is temperature regulated, occurring between 34 and 41°C (34). In contrast, VapC, -D, and -E are secreted proteins concomitantly regulated by temperature with VapA, being produced at 37°C but not at 30°C (7). The cellular locations of VapF, VapG, and VapH are unknown. In one study, all vap genes were upregulated when R. equi was grown in equine macrophage monolayers compared to expression during in vitro growth at 30°C (32). In the same study, vapA and vapC were the most highly upregulated of the vap genes (32). In the present study, only vapA, vapD, and vapG were expressed at significantly higher levels in vivo than during in vitro growth. The discrepancy between findings of these studies may result from the fact that culture in macrophage monolayers does not accurately reflect all the conditions experienced by the pathogen in the lungs of foals. In addition, relative expression of the vap genes may depend on the stage of infection. The present study evaluated expression in the lungs of foals 2 weeks after infection, whereas the macrophage study evaluated expression 4 h after the monolayers were infected with R. equi. Finally, differences in temperatures for in vitro growth between the two studies may at least partially explain some of the inconsistencies. The macrophage study grew R. equi in vitro at 30°C, a temperature known to decrease expression of the Vap proteins (7, 32). In contrast, the present study used the more physiological temperature of 37°C to maximize vap gene expression during in vitro growth, hence minimizing the likelihood of identifying differences due only to dissimilar growth conditions.
The exact role of each Vap protein remains to be determined, but the proteins are probably involved in preventing acidification and late endosomal maturation of R. equi-containing vacuoles within macrophages (10, 36). Regulation of expression of the vap genes is complex and depends on at least five environmental signals, temperature, pH, oxidative stress, magnesium, and iron (4, 5, 32, 34). The vapA and vapD genes are the major acid-inducible determinants carried by the virulence plasmid (4). In another study, vapA and vapG were predominantly induced by H2O2 treatment (5). These findings have led to the hypothesis that VapA, VapD, and VapG may play a dominant role in protection against macrophage-related stresses (5). That vapA, vapD, and vapG were expressed at significantly higher levels than the other vap genes in the lungs of infected foals in the present study is consistent with this hypothesis. These three genes were also significantly more expressed in vivo than in vitro.
Several lines of evidence suggest that immune responses against VapA and potentially other Vap proteins confer a role in protection (17, 24). A recent study with mice has shown that DNA immunization with vapA protects against R. equi infection and that the immunoglobulin G subisotype response is consistent with a Th1-based immune response (13). A similar DNA vaccine containing the vapA gene has been shown to induce strong cell-mediated immune responses in adult horses (25). In foals, production of antibody to VapA and VapC but not of that to other Vap proteins increases following natural exposure to R. equi (18). The present study shows that VapA and VapC are also the proteins inducing the strongest lymphoproliferative recall responses following experimental challenge for both foals and adult horses. A prior study had demonstrated strong lymphoproliferative recall responses to VapA in association with clearance of R. equi in adult horses (24). The present study confirmed this finding and also demonstrated that lymphocytes from foals and from adult horses proliferate similarly in response to each Vap protein.
The documented importance of IFN-γ in protection against infection with R. equi (22) and the recognized Th2 bias in immune responses of neonates from many species (1) have led to the widespread hypothesis that a similar Th2 bias may be at the basis of the peculiar susceptibility of foals to infection by R. equi. The recent finding that young foals are deficient in their ability to produce IFN-γ in response to mitogens led to the conclusion that neonatal foals may also be predisposed to develop a Th2-like immune response (6). In one study, pulmonary lymphocytes from adult horses collected 7 days following challenge with R. equi expressed predominantly IFN-γ, but they also expressed IL-4 mRNA in response to in vitro stimulation with VapA (24). Similar results were obtained following stimulation of BLN lymphocytes with VapA in the present study. In addition, the present study extends these findings to the other Vap proteins of R. equi. The present study also demonstrates that BLN lymphocytes from foals can express IFN-γ in response to stimulation with each Vap protein just as well as those from immune adult horses. In fact, as a result of the significantly lower level of IL-4 expression for foals than for adult horses, foals showed a significantly higher IFN-γ/IL-4 ratio than adult horses in response to stimulation with most Vap proteins. The findings in the present study, therefore, do not support the conclusion that neonatal foals are predisposed to develop a Th2-like immune response. The reason for the peculiar susceptibility of foals to R. equi must be more complex and also must take into account the unique association of the VapA-containing virulence plasmid with disease in foals.
This strong predominating IFN-γ response in a neonate is not unprecedented. Many studies have shown that despite an apparent Th2 bias, neonates can mount strong Th1 responses to some antigens (1, 27). For example, Mycobacterium bovis BCG, a pathogen closely related to R. equi, triggers a Th1 response in human neonates of a magnitude similar to that seen when it is given later in life (26). Virulent R. equi is widespread in the environment of horse breeding farms (28, 33), yet development of clinical disease is the exception rather than the rule. The fact that oral administration of live virulent R. equi to newborn foals confers almost complete protection against subsequent heavy intrabronchial challenge (8, 19) and the fact that most foals on farms where it is endemic do not develop disease or develop subclinical disease and eventually clear the infection are consistent with the results of the present study, which demonstrate that foals develop adequate cell-mediated immune responses to key antigens of R. equi.
In conclusion, the data presented here indicate that vapA, vapD, and vapG are the most biologically relevant vap genes because they are preferentially induced during infection in the natural host. All the Vap proteins are immunogenic, with VapA and VapC providing the strongest lymphoproliferation stimulus. The peculiar susceptibility of foals to infection by R. equi cannot be explained by a failure to mount Th1 immunoreactivity to the Vap proteins. Further work is required to identify the fundamental host basis of the susceptibility of foals to R. equi pneumonia.
Acknowledgments
This project was supported by the Morris Animal Foundation and by the Florida Department of Regulation Pari-Mutuel Wagering Trust Fund. We thank the Florida Thoroughbred Breeders' and Owners' Association for support of the equine research breeding herd.
Footnotes
Published ahead of print on 14 February 2007.
REFERENCES
- 1.Adkins, B. 2000. Development of neonatal Th1/Th2 function. Int. Rev. Immunol. 19:157-171. [DOI] [PubMed] [Google Scholar]
- 2.Ahmed, S. A., R. M. Gogal, Jr., and J. E. Walsh. 1994. A new rapid and simple non-radioactive assay to monitor and determine the proliferation of lymphocytes: an alternative to [3H]thymidine incorporation assay. J. Immunol. Methods 170:211-224. [DOI] [PubMed] [Google Scholar]
- 3.Arlotti, M., G. Zoboli, G. L. Moscatelli, G. Magnani, R. Maserati, V. Borghi, M. Andreoni, M. Libanore, L. Bonazzi, A. Piscina, and R. Ciammarughi. 1996. Rhodococcus equi infection in HIV-positive subjects: a retrospective analysis of 24 cases. Scand. J. Infect. Dis. 28:463-467. [DOI] [PubMed] [Google Scholar]
- 4.Benoit, S., A. Benachour, S. Taouji, Y. Auffray, and A. Hartke. 2001. Induction of vap genes encoded by the virulence plasmid of Rhodococcus equi during acid tolerance response. Res. Microbiol. 152:439-449. [DOI] [PubMed] [Google Scholar]
- 5.Benoit, S., A. Benachour, S. Taouji, Y. Auffray, and A. Hartke. 2002. H2O2, which causes macrophage-related stress, triggers induction of expression of virulence-associated plasmid determinants in Rhodococcus equi. Infect. Immun. 70:3768-3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breathnach, C. C., T. Sturgill-Wright, J. L. Stiltner, A. A. Adams, D. P. Lunn, and D. W. Horohov. 2006. Foals are interferon gamma-deficient at birth. Vet. Immunol. Immunopathol. 112:199-209. [DOI] [PubMed] [Google Scholar]
- 7.Byrne, B. A., J. F. Prescott, G. H. Palmer, S. Takai, V. M. Nicholson, D. C. Alperin, and S. A. Hines. 2001. Virulence plasmid of Rhodococcus equi contains inducible gene family encoding secreted proteins. Infect. Immun. 69:650-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chirino-Trejo, J. M., J. F. Prescott, and J. A. Yager. 1987. Protection of foals against experimental Rhodococcus equi pneumonia by oral immunization. Can. J. Vet. Res. 51:444-447. [PMC free article] [PubMed] [Google Scholar]
- 9.Donisi, A., M. G. Suardi, S. Casari, M. Longo, G. P. Cadeo, and G. Carosi. 1996. Rhodococcus equi infection in HIV-infected patients. AIDS 10:359-362. [DOI] [PubMed] [Google Scholar]
- 10.Fernandez-Mora, E., M. Polidori, A. Luhrmann, U. E. Schaible, and A. Haas. 2005. Maturation of Rhodococcus equi-containing vacuoles is arrested after completion of the early endosome stage. Traffic 6:635-653. [DOI] [PubMed] [Google Scholar]
- 11.Garton, N. J., M. Gilleron, T. Brando, H. H. Dan, S. Giguère, G. Puzo, J. F. Prescott, and I. C. Sutcliffe. 2002. A novel lipoarabinomannan from the equine pathogen Rhodococcus equi. Structure and effect on macrophage cytokine production. J. Biol. Chem. 277:31722-31733. [DOI] [PubMed] [Google Scholar]
- 12.Giguère, S., M. K. Hondalus, J. A. Yager, P. Darrah, D. M. Mosser, and J. F. Prescott. 1999. Role of the 85-kilobase plasmid and plasmid-encoded virulence-associated protein A in intracellular survival and virulence of Rhodococcus equi. Infect. Immun. 67:3548-3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haghighi, H. R., and J. F. Prescott. 2005. Assessment in mice of vapA-DNA vaccination against Rhodococcus equi infection. Vet. Immunol. Immunopathol. 104:215-225. [DOI] [PubMed] [Google Scholar]
- 14.Harvey, R. L., and J. C. Sunstrum. 1991. Rhodococcus equi infection in patients with and without human immunodeficiency virus infection. Rev. Infect. Dis. 13:139-145. [DOI] [PubMed] [Google Scholar]
- 15.Hines, M. T., K. M. Paasch, D. C. Alperin, G. H. Palmer, N. C. Westhoff, and S. A. Hines. 2001. Immunity to Rhodococcus equi: antigen-specific recall responses in the lungs of adult horses. Vet. Immunol. Immunopathol. 79:101-114. [DOI] [PubMed] [Google Scholar]
- 16.Hines, S. A., D. M. Stone, M. T. Hines, D. C. Alperin, D. P. Knowles, L. K. Norton, M. J. Hamilton, W. C. Davis, and T. C. McGuire. 2003. Clearance of virulent but not avirulent Rhodococcus equi from the lungs of adult horses is associated with intracytoplasmic gamma interferon production by CD4+ and CD8+ T lymphocytes. Clin. Diagn. Lab. Immunol. 10:210-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hooper-McGrevy, K. E., S. Giguère, B. N. Wilkie, and J. F. Prescott. 2001. Evaluation of equine immunoglobulin specific for Rhodococcus equi virulence-associated proteins A and C for use in protecting foals against Rhodococcus equi-induced pneumonia. Am. J. Vet. Res. 62:1307-1313. [DOI] [PubMed] [Google Scholar]
- 18.Hooper-McGrevy, K. E., B. N. Wilkie, and J. F. Prescott. 2003. Immunoglobulin G subisotype responses of pneumonic and healthy, exposed foals and adult horses to Rhodococcus equi virulence-associated proteins. Clin. Diagn. Lab. Immunol. 10:345-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hooper-McGrevy, K. E., B. N. Wilkie, and J. F. Prescott. 2005. Virulence-associated protein-specific serum immunoglobulin G-isotype expression in young foals protected against Rhodococcus equi pneumonia by oral immunization with virulent R. equi. Vaccine 23:5760-5767. [DOI] [PubMed] [Google Scholar]
- 20.Jain, S., B. R. Bloom, and M. K. Hondalus. 2003. Deletion of vapA encoding virulence associated protein A attenuates the intracellular actinomycete Rhodococcus equi. Mol. Microbiol. 50:115-128. [DOI] [PubMed] [Google Scholar]
- 21.Kanaly, S. T., S. A. Hines, and G. H. Palmer. 1993. Failure of pulmonary clearance of Rhodococcus equi infection in CD4+ T-lymphocyte-deficient transgenic mice. Infect. Immun. 61:4929-4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanaly, S. T., S. A. Hines, and G. H. Palmer. 1995. Cytokine modulation alters pulmonary clearance of Rhodococcus equi and development of granulomatous pneumonia. Infect. Immun. 63:3037-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanaly, S. T., S. A. Hines, and G. H. Palmer. 1996. Transfer of a CD4+ Th1 cell line to nude mice effects clearance of Rhodococcus equi from the lung. Infect. Immun. 64:1126-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lopez, A. M., M. T. Hines, G. H. Palmer, D. C. Alperin, and S. A. Hines. 2002. Identification of pulmonary T-lymphocyte and serum antibody isotype responses associated with protection against Rhodococcus equi. Clin. Diagn. Lab. Immunol. 9:1270-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lopez, A. M., M. T. Hines, G. H. Palmer, D. P. Knowles, D. C. Alperin, and S. A. Hines. 2003. Analysis of anamnestic immune responses in adult horses and priming in neonates induced by a DNA vaccine expressing the vapA gene of Rhodococcus equi. Vaccine 21:3815-3825. [DOI] [PubMed] [Google Scholar]
- 26.Marchant, A., T. Goetghebuer, M. O. Ota, I. Wolfe, S. J. Ceesay, D. De Groote, T. Corrah, S. Bennett, J. Wheeler, K. Huygen, P. Aaby, K. P. McAdam, and M. J. Newport. 1999. Newborns develop a Th1-type immune response to Mycobacterium bovis bacillus Calmette-Guerin vaccination. J. Immunol. 163:2249-2255. [PubMed] [Google Scholar]
- 27.Marchant, A., and M. Goldman. 2005. T cell-mediated immune responses in human newborns: ready to learn? Clin. Exp. Immunol. 141:10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martens, R. J., S. Takai, N. D. Cohen, M. K. Chaffin, H. Liu, K. Sakurai, H. Sugimoto, and S. W. Lingsweiler. 2000. Association of disease with isolation and virulence of Rhodococcus equi from farm soil and foals with pneumonia. J. Am. Vet. Med. Assoc. 217:220-225. [DOI] [PubMed] [Google Scholar]
- 29.Miranda-Casoluengo, R., P. S. Duffy, E. P. O'Connell, B. J. Graham, M. W. Mangan, J. F. Prescott, and W. G. Meijer. 2005. The iron-regulated iupABC operon is required for saprophytic growth of the intracellular pathogen Rhodococcus equi at low iron concentrations. J. Bacteriol. 187:3438-3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patton, K. M., T. C. McGuire, D. G. Fraser, and S. A. Hines. 2004. Rhodococcus equi-infected macrophages are recognized and killed by CD8+ T lymphocytes in a major histocompatibility complex class I-unrestricted fashion. Infect. Immun. 72:7073-7083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Polidori, M., and A. Haas. 2006. VapI, a new member of the Rhodococcus equi Vap family. Antonie Leeuwenhoek 90:299-304. [DOI] [PubMed] [Google Scholar]
- 32.Ren, J., and J. F. Prescott. 2003. Analysis of virulence plasmid gene expression of intra-macrophage and in vitro grown Rhodococcus equi ATCC 33701. Vet. Microbiol. 94:167-182. [DOI] [PubMed] [Google Scholar]
- 33.Takai, S., T. Anzai, K. Yamaguchi, S. Kakizaki, J. Takahagi, Y. Sato, F. Takehara, Y. Tamada, S. Matsukura, A. Tani, M. Kato, N. Seno, Y. Sasaki, S. Tsubaki, and M. Kamada. 1994. Prevalence of virulence plasmids in environmental isolates of Rhodococcus equi from horse-breeding farms in hokkaido. J. Equine Sci. 5:21-25. [Google Scholar]
- 34.Takai, S., N. Fukunaga, K. Kamisawa, Y. Imai, Y. Sasaki, and S. Tsubaki. 1996. Expression of virulence-associated antigens of Rhodococcus equi is regulated by temperature and pH. Microbiol. Immunol. 40:591-594. [DOI] [PubMed] [Google Scholar]
- 35.Takai, S., S. A. Hines, T. Sekizaki, V. M. Nicholson, D. A. Alperin, M. Osaki, D. Takamatsu, M. Nakamura, K. Suzuki, N. Ogino, T. Kakuda, H. Dan, and J. F. Prescott. 2000. DNA sequence and comparison of virulence plasmids from Rhodococcus equi ATCC 33701 and 103. Infect. Immun. 68:6840-6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toyooka, K., S. Takai, and T. Kirikae. 2005. Rhodococcus equi can survive a phagolysosomal environment in macrophages by suppressing acidification of the phagolysosome. J. Med. Microbiol. 54:1007-1015. [DOI] [PubMed] [Google Scholar]
- 37.Wada, R., M. Kamada, T. Anzai, A. Nakanishi, T. Kanemaru, S. Takai, and S. Tsubaki. 1997. Pathogenicity and virulence of Rhodococcus equi in foals following intratracheal challenge. Vet. Microbiol. 56:301-312. [DOI] [PubMed] [Google Scholar]
- 38.Witonsky, S., R. M. Gogal, Jr., V. Buechner-Maxwell, and S. A. Ahmed. 2003. Immunologic analysis of blood samples obtained from horses and stored for twenty-four hours. Am. J. Vet. Res. 64:1003-1009. [DOI] [PubMed] [Google Scholar]