The mammalian microbiota comprises several hundred different bacterial species, many of which have a beneficial effect on the host. For example, they are involved in preventing colonization of the gut by pathogens and maintaining the gut mucosal immunity (85). The gut microbiota is more abundant in the large intestine of mammals, with densities rising to over 1011 organisms/g intestinal content (84, 86). The number of bacterial cells in the entire gut exceeds the number of eukaryotic cells in the host, but under normal circumstance they coexist without any adverse effect on the host. The influence of the resident microflora on mucosal immune function and gut health has become an area of scientific and clinical importance (22, 26). There is an active dialogue between the commensal microorganisms and the host mucosal immune system (21, 48). This cross talk elicits different host responses to commensal and pathogenic bacteria. Commensal bacteria may even share molecular patterns recognized by toll-like receptors (TLRs), which can recognize patterns associated mainly with pathogens. However, the mucosal immune system of the healthy intestine allows the persistence of this microbiota associated with the intestine and avoids immunological tolerance, maintaining the intestinal homeostasis. Now, there is acceptance of the concept that oral tolerance is not generated by commensal intestinal bacteria; the host would ignore or fail to recognize the presence of indigenous microorganisms (49). The healthy host is able to elicit a good mucosal immune response against luminal antigens and to maintain a “physiological state of inflammation” in the gut, but it is also capable of responding to invading commensal organisms or pathogens. In the healthy host the penetration of the commensal bacteria is usually prevented by the barrier afforded by the intestinal epithelium and the immune cells associated with the mucosa, which are highly adapted to the presence of the normal microbiota (71). The signals sent by these microorganisms prevent their penetration and keep them outside the intestinal tissue. If the commensal microorganisms invade the host tissues, the innate immune mechanisms contribute to their rapid clearance, but when pathogens enter the intestine, innate and adaptive mechanisms are coordinately stimulated to respond to the danger signals (38, 60). Although mucosal epithelial tissues form an efficient barrier that prevents the entrance of the environmental pathogens and the external antigens into the host internal milieu, mucosal tissues represent the main sites of infection by pathogens. Many attempts have been made to understand the gut immunomodulation by pathogenic bacteria but not the mechanisms involved in the modulation of the gut immune system by commensal bacteria and by nonpathogenic microorganisms present in many foods included in the daily diet.
NONPATHOGENIC PROBIOTIC BACTERIA
Interest in the gut microflora has led to numerous investigations to demonstrate that there are beneficial and potentially harmful microorganisms in the intestine and that the one could be used to influence the activities of the other. These findings led to the “probiotic” concept, originally used to describe microbial feed supplements which stimulate the growth of farm animals. Now, the use of live microbes as dietary supplements has been extended to humans. Many definitions of probiotics have been published, starting from Fuller, who defined a probiotic as “a live microbial feed supplement which beneficially affects the host by improving its intestinal microbial balance” (25). A more recent one from FAO/WHO is the following: “live microorganisms that when being administered in appropriate dose, they confer a benefit of health to the receiver.”
Some of the health benefits which have been claimed for probiotics include the following: improvement of the normal microflora (2), prevention of infectious diseases (3, 6, 9, 11, 13, 65, 83) and food allergies (51, 61), reduction of serum cholesterol (23, 77), anticarcinogenic activity (14, 18, 33, 35, 73), stabilization of the gut mucosal barrier (79), immune adjuvant properties (15, 20, 24, 28, 36, 40, 77, 80, 92), alleviation of intestinal bowel disease symptoms (31, 82), and improvement in the digestion of lactose in intolerant hosts (19, 42).
The genera most commonly used in probiotic preparations are Lactobacillus, Bifidobacterium, Streptococcus, and Lactococcus and some fungal strains (58). Foods for human consumption that containing mainly lactic acid bacteria include fermented milks, cheeses, fruit juices, wine, and sausages. Single and mixed cultures of live microorganisms are used in probiotic preparations (4, 88).
The ability of probiotics to prevent or reverse several pathological conditions (27) by stimulating the immune system and all the scientific evidence of immune system activation by probiotics indicate that the ability to generate an immune response should be included in the probiotic definition. Consequently, we suggest probiotics should be defined as follows: “live microorganisms, that when included in foods can influence the composition and activity of the gut microbiota, modulate the inflammatory response, improve the nonspecific intestinal barrier, and reinforce or modulate the mucosal and the systemic immune responses.” This definition ascribes to the probiotic microorganisms in the dietary supplement the potential for the prevention of infections, tumor growth, or other systemic pathologies, including effects in mucosal sites distant from the gut, such as the bronchus (15, 69), mammary glands (16, 17), and the urogenital tract (74, 91). However, for the best use of these microorganisms, the mechanisms by which they work should be understood. We believe that the selection of an appropriate probiotic strain for its inclusion in a probiotic preparation should be made on the basis of its capacity to induce an improved gut immune response without modification of the intestinal homeostasis. To achieve this task, probiotic strains should have the following properties: (i) high cell viability, thus they must be resistant to low pH and bile acids; (ii) ability to persist in the intestine even if the probiotic strain cannot colonize the gut (continuous administration may be necessary); (iii) adhesion to the gut epithelium to cancel the flushing effects of peristalsis. In this last aspect, there are many relevant literature reports of the adhesive property of the probiotic bacteria to epithelial cells in in vitro studies (5, 10, 29, 30, 87). (iv) Also, they should be able to interact or to send signals to the immune cells associated with the gut. There are reports from in vitro assays that show the activation of immune cells after stimulation by probiotics (32, 54, 55, 81).
WHAT ARE THE IMMUNE MECHANISMS INDUCED BY PROBIOTIC BACTERIA?
The functioning of the gut mucosal immune system requires a complex network of signals with multiple interactions between commensal and foreign antigens and the eukaryotic cells. These include epithelial cells, macrophages, dendritic cells, and other cells that belong to the nonspecific barriers, mucus-producing cells such as goblet cells, and Paneth cells, which secrete antimicrobial peptides and produce cryptidins or defensins (76).
The mucosal epithelial cells are crucial in coordinating the defense mechanisms. They respond to environmental signals by releasing chemokines and cytokines that recruit the immune cells from both the innate and adaptive immune responses. These recruited immune cells can in turn act upon the epithelial cells, stimulating the release of cytokines. This response must not be triggered by harmless intestinal commensal bacteria, and the inflammatory response must be controlled. The particular characteristics of soluble, particulate antigens and pathogens will affect the gut immune response in relation to the way that they initiate the interaction with the immune system. At least three different routes exist for the uptake of luminal antigens: dendritic cells, specialized M cells from the Peyer's patches, and individual M cells found in the villous epithelium (39, 43). The anatomical location of the immune cells from the innate response (macrophages and dendritic cells) and the way by which these cells acquire antigens are crucial in determining the nature of the subsequent responses. Thus, the immune response induced can be the result of uptake of antigens by transepithelial sampling involving dendritic cells (75) or by dendritic cells present in the lamina propria of the intestine or by M cells from Peyer's patches or from the intestinal villous.
In the gut immune response induced by commensal bacteria, the antigen presentation from the luminal flora leads to the generation of large quantities of local immunoglobulin A (IgA) without induction of systemic immunity (56). The local secretory IgA specific for the pathogen requires the interaction of phagocytic dendritic cells with T and B cells from the Peyer's patches with the antigen-presenting cells in isolated lymphoid follicles or in the mesenteric lymph nodes. The pathway of antigen internalization is crucial for immune cell stimulation and the initiation of mucosal immune responses.
In the complex microenvironment of the gut, how can the transient population of nonpathogenic probiotic bacteria which may be unable to colonize the intestine affect gut mucosal immunity? What kinds of signals do they induce to act as oral adjuvants? Which kind of immune response do they elicit: innate or adaptive? How long do they have to remain in the gut to be effective? What is the quantity of these microorganisms that is needed to achieve the immunomodulatory capacity? Is the viability of the microorganisms a sine qua non condition required to induce such immunomodulation?
In order to survive, probiotic bacteria entering by the mouth must be resistant to pH, bile acid, proteolytic enzymes, antimicrobial peptides, intestinal peristalsis, and luminal secretory IgA blocking. The oral adjuvant capacity of some probiotic bacteria has been well demonstrated in our laboratory (90). How can this particulate antigen, without a virulence factor, evade all the barriers of the host and up- or down-regulate the gut mucosal immune system? It is obvious that these nonpathogenic probiotic bacteria must interact with the epithelial cells and with the immune cells associated with the gut to start the network of immune signals. The increase in the number of IgA-producing cells was the most remarkable property induced by probiotic microorganisms or by fermented milk yogurt (62, 68). The physiological role of IgA in the mucosal surface is unquestionable (34, 45). The IgA+ B cells induced in the Peyer's patches circulate through the mesenteric lymphatic nodes to enter into the blood via the thoracic duct and return to the intestinal mucosa, repopulating distant mucosal sites, such as the bronchus. Similar recirculation also occurs with intestinal T cells (70). Some probiotic microorganisms are also able to increase the IgA cycle, and this effect is dose dependent (15, 67).
T-independent IgA induction was also demonstrated; the cytokines transforming growth factor β (TGF-β), interleukin-4 (IL-4) (50), and IL-2, IL-6, and IL-10 work in a synergistic way from other immune cells different from T cells and can promote the switch from IgM to IgA expression (12, 44).
We have demonstrated that some probiotic bacteria can act as adjuvants of the mucosal and systemic immune response (65, 68). The stimulation with probiotic bacteria induced signals on epithelial and immune cells that evoked different patterns of cytokines in the intestine (53, 64, 89), depending on the dose administered (Table 1), as has also been shown by Massen et al. (47). The quantity of these microorganisms to achieve the adjuvant effect in the mucosal or systemic immune response was 1 × 108 to 1 × 109 CFU/day (68, 90).
TABLE 1.
Effects of administration of lactic acid bacteria on the number of IgA-secreting and cytokine-producing cells in the lamina propria of the small intestinea
| Organism | Feeding period (days) | No. of cells producingb:
|
||||||
|---|---|---|---|---|---|---|---|---|
| Cytokines
|
IgA | |||||||
| TNF-α | IFN-γ | IL-2 | IL-12 | IL-4 | IL-10 | |||
| L. casei CRL 431 | 2 | 90 ± 8* | 124 ± 15* | 24 ± 6 | 13 ± 3 | 86 ± 23* | 40 ± 2* | 99 ± 21 |
| 5 | 74 ± 10* | 116 ± 18* | 28 ± 7 | 19 ± 4* | 38 ± 7 | 27 ± 13 | 100 ± 12 | |
| 7 | 52 ± 7 | 85 ± 19* | 20 ± 9 | 27 ± 8* | 42 ± 6 | 63 ± 8* | 135 ± 17* | |
| L. delbrueckii subsp. bulgaricus CRL 423 | 2 | 79 ± 6* | 59 ± 22* | 40 ± 8 | 23 ± 11 | 67 ± 4* | 85 ± 9* | 135 ± 23* |
| 5 | 59 ± 11 | 72 ± 18* | 42 ± 8 | 17 ± 4 | 51 ± 9* | 68 ± 8* | 92 ± 12 | |
| 7 | 43 ± 12 | 209 ± 34* | 42 ± 15 | 17 ± 4 | 146 ± 8* | 97 ± 23* | 95 ± 18 | |
| L. acidophilus CRL 724 | 2 | 52 ± 7 | 51 ± 25* | 27 ± 8 | 30 ± 16* | 44 ± 11 | 30 ± 7 | 173 ± 24* |
| 5 | 51 ± 9 | 73 ± 11* | 25 ± 7 | 37 ± 11* | 87 ± 19* | 55 ± 6* | 168 ± 25* | |
| 7 | 22 ± 11 | 64 ± 6* | 31 ± 13 | 25 ± 8 | 87 ± 18* | 34 ± 8* | 135 ± 17* | |
| Control | 24 ± 4 | 17 ± 6 | 31 ± 12 | 11 ± 2 | 27 ± 7 | 18 ± 6 | 98 ± 17 | |
The cytokine-producing cells and the IgA-secreting cells were determined on histological slices from the small intestines of BALB/c mice by an immunofluorescence test. The animals were fed in their drinking water lactic acid bacteria (1 × 108 CFU/ml/day) for 2, 5, or 7 consecutive days. L. casei and Lactobacillus acidophilus were isolated from human feces, and Lactobacillus delbrueckii subsp. bulgaricus was from yogurt. The animals received 2.5 or 3 ml/day.
Three measurements were taken, and values are means ± standard deviations. *, significant difference between test and untreated control groups (P < 0.001).
In the analyses of the profiles of cytokines induced by some lactic acid bacteria, we observed the most remarkable effect was the increase in the tumor necrosis factor alpha (TNF-α) and gamma interferon (IFN-γ) and in the regulatory cytokine IL-10 for all the probiotic strains assayed. This effect was obtained without increasing the inflammatory response and only a slight increase in the cellularity was found. However, the induction of TNF-α by the probiotic bacteria would be necessary to initiate the cross talk between the immune cells associated with the lamina propria and the intestinal epithelial cells. IFN-γ would also play a physiological role; it has been demonstrated that this cytokine is necessary for the maturation of some immune cells, such as dendritic cells, and also controls their cellular proliferation at the intestinal level (78).
It was previously thought that to have an effect on the immune system, the probiotic strains must remain viable. We demonstrated (52) that this fact is true only for some strains. For Lactobacillus delbrueckii subsp. bulgaricus, viability was not necessary for the induction of positive cells producing cytokines, although the number of positive cells was comparatively lower than the number obtained with viable L. delbrueckii subsp. bulgaricus organisms. The viability was critical for determining the time of residence in the gut with differences between viable and nonviable probiotic bacteria administration; nonviable bacteria were cleared more rapidly. We also demonstrated that the probiotic bacteria must remain in the gut at least 48 to 72 h to be effective; that is the time required for any particulate antigen to induce gut immunostimulation (52, 63). This fact is a very important finding, indicating the importance of daily administration in a dose established for each probiotic bacterium to have an adjuvant effect without the induction of oral tolerance.
We and other workers have demonstrated that probiotic microorganisms are able to induce a gut mucosal immune response (41, 63) which requires the bacteria to interact with the epithelial and immune cells in the gut to induce the network of signals involved in an immune response.
Probiotic bacteria may arrive in the intestine along routes which correspond with the different pathways for the internalization of antigens. These bacteria (as whole cells or as antigenic fragments) must interact with the M cells in the Peyer's patches, with gut epithelial cells, and with the associated immune cells. After contact with these cells, the release of cytokines is induced to up- or down-regulate the immune response.
How do these nonpathogenic bacteria interact with the intestinal epithelial cells? What kind of signals do they induce in the immune cells in order to initiate the gut response?
Mucosal epithelial cells form an efficient barrier which prevents antigens from environmental pathogens from gaining access to the host milieu. Flagellated microorganisms, including commensals, trigger epithelial homeostatic chemokine responses that recruit immune cells of the innate immune system to the epithelium and lamina propria of the intestine to link the innate or/and the adaptive immune response (78). It has also been shown that commensal bacteria can activate TLR signals (37). Although the precise location of these receptors in the intestinal epithelial cells (apical or/and basolateral) is controversial (8), TLR signals are essential, not only for response to pathogens (59) but also to maintain the intestinal barrier function (72).
With in vivo studies in mice, we demonstrated the pathway of internalization of the following probiotic bacteria: Lactobacillus bulgaricus CRL 423, L. casei CRL 431, L. acidophilus CRL 728, and Streptococcus thermophilus CRL 412 (66, 67). We determined the probiotic bacteria in Peyer's patches, in the lamina propria of the villi of the small intestine, and in the nodule of the large intestine. We also demonstrated that Lactobacillus casei CRL 431 interacts with the epithelial cells of the small intestine and that their fragments can internalize and activate the intestinal epithelial cells (52, 63). It was also shown by in vitro and ex vivo studies in a primary culture of intestinal epithelial cells from conventional animals (89) that probiotic bacteria interact with these cells and induce release of IL-6 but not IL-1 and IL-10. This study also demonstrated that TLR2 is involved in this interaction and could be responsible for the signals induced in IL-6 release by epithelial cells (Table 2 and Fig. 1), which would be another source of IL-6 induced by probiotic bacteria than the immune cells associated with the gut.
TABLE 2.
IL-6 production by small intestine epithelial cells isolated from conventional animals and challenged with different concentrations of viable cultures of L. casei CRL 431 and L. helveticus R389
| Expt type, challenge variable, and conditions | IL-6 production (pg/ml) by SIECd
|
|||
|---|---|---|---|---|
| Challenged with:
|
Not challengedf | |||
| L. casei CRL 431 | L. helveticus R389 | LPSe | ||
| In vitro | ||||
| Bacterial concna | ||||
| 108 CFU/ml | 245 ± 10* | 190 ± 20* | ||
| 107 CFU/ml | 405 ± 17* | 500 ± 17* | 878 ± 22* | 310 ± 11 |
| 106 CFU/ml | 414 ± 17* | 368 ± 38 | ||
| Antibody treatment of SIECb | ||||
| Not coated | 531 ± 60* | 641 ± 75* | 863 ± 80* | 399 ± 15 |
| Coated with anti-TLR2 | 376 ± 20 | 514 ± 95* | 755 ± 60* | 410 ± 30 |
| Coated with anti-TLR4 | 510 ± 32* | 481 ± 62* | 740 ± 44* | 390 ± 22 |
| Ex vivoc | ||||
| 2 days | 3,804 ± 82* | 499 ± 85 | ||
| 5 days | 1,109 ± 91* | 452 ± 102 | 494 ± 47 | |
| 7 days | 1,704 ± 83* | 1,740 ± 97* | ||
IL-6 released by small intestine epithelial cells (SIEC) from BALB/c mice when the cells were challenged with different concentrations of the bacterial strains, with lipopolysaccharide (LPS), or were not challenged (control).
IL-6 production by SIEC left untreated or treated with anti-TLR2 and anti-TLR4 antibodies before they were stimulated with the bacterial strains or with LPS.
IL-6 production by SIEC isolated from BALB/c mice that received viable culture (1 × 108 CFU/ml/day) of L. casei isolated from human feces or L. helveticus isolated from cheese for different periods of time (2, 5, or 7 days).
Values are means ± standard deviations. *, significantly different from the corresponding control value (P < 0.05).
LPS was used as a positive control.
Negative control.
FIG. 1.
Scanning electron micrograph showing the interaction of Lactobacillus helveticus R389 with intestinal epithelial cells (IEC) obtained from the small intestine of BALB/c mice. The epithelial cells were recovered from the small intestine after digestion with collagenase in adequate culture medium. A Lactobacillus helveticus suspension (1012 CFU/ml) was added to isolated IEC. After bacterial contact with IEC for 2 h, the samples were processed for scanning electron microscopy. Magnification, ×6,000.
We showed that probiotic bacteria could be also internalized through M cells in the Peyer's patches or villi or may be sampled by dendritic cells as whole cells or their antigenic fragments (52, 63, 66, 67). These may be captured by other dendritic cells or macrophages associated with the lamina propria to increase the signals to the epithelial cells and/or other immune cells. There is scientific evidence that the uptake of nonpathogenic bacteria or their fragments by macrophages or dendritic cells in the lamina propria is possible through direct sampling of luminal antigens for dendritic cells, through TLRs and the CD-206 mannose receptor (1, 46). These bacteria can be cleared or transported to the mesenteric lymph nodes, where they interact with T and B cells to induce specific mucosal IgA or suppress T cells (57).
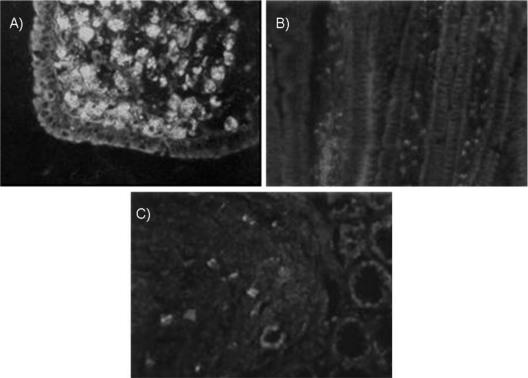
When probiotic bacteria labeled with fluorescein isothiocyanate were administered to mice, we found fluorescent cells at different levels of the intestine in Peyer's patches, lamina propria of the villi, and nodules of the large intestine (Fig. 2A, B, and C) (52, 66). The possible pathway of internalization to the villi of the whole bacteria could be through the M cells present in the villi (39). For the bacterial particles, TLR2 or the CD-206 receptor would be involved, as was demonstrated after L. casei CRL 431 administration (53, 89), with which there was a remarkable increase of both of these receptors in the immune cells associated with the lamina propria or in cells isolated from Peyer's patches. The IgA+ cells in the lamina propria of the small intestine were increased for different lactic acid bacteria, such as L. acidophilus, L. bulgaricus, and S. thermophilus (63, 67, 68, 90). Specific IgA against the probiotic bacteria and modifications in the number of CD4+ population were not found (53, 90). These findings would show that antigenic presentation with production of specific antibodies would not be induced. In previous studies under physiological conditions (healthy animals), we observed that the administration of different probiotic bacteria did not increase the CD4+ or CD8+ population (53, 90). The results obtained for positive cells for cytokine release analyzed in isolated immune cells from Peyer's patches showed that the adherent population (macrophages and dendritic cells) had a more relevant effect on cytokine production (64).
FIG. 2.
Histological slices of intestine from BALB/c mice, showing the pathway of internalization to the gut by lactic acid bacteria. Animals received fluorescein isothiocyanate-labeled lactic acid bacteria (1 × 108 CFU/ml) by intragastric intubation. (A) Fluorescence in Peyer's patches of the small intestine of mice 5 min after administration of labeled Lactobacillus delbrueckii subsp. bulgaricus CRL 423 (isolated from yogurt). (B) Fluorescence in lamina propria of the small intestine of mice that received labeled Lactobacillus casei CRL 431 isolated from human feces. The samples were processed after 5 min of lactobacillus administration. (C) Fluorescence in the nodule and crypts of the large intestine of mice 10 min after administration of labeled Lactobacillus acidophilus CRL 724 isolated from feces. Magnification, ×1,000.
Even though we cannot ignore that other mucosal immune mechanisms, such as the Th1 cell response, can be modulated by probiotic bacteria, this was demonstrated by other authors in pathological processes such as allergy (36), inflammatory bowel disease (7, 8, 18), or colon cancer (14, 16, 17). Our previous scientific evidence under physiological conditions led us to suggest that the probiotic bacteria interact with the epithelial cells and preferentially with the immune cells from the innate immune system, reinforcing this barrier (52, 53, 63, 89). When they interact with cells from Peyer's patches, they can induce an increase of the IgA cycle, as was demonstrated in our laboratory (15, 69). According to these previous studies, where we demonstrated (i) the epithelial interaction of the probiotic bacteria, (ii) the pathway of internalization of probiotics to the gut, (iii) the inducing signals to the immune cells associated with the intestine by an increase in the cytokine production and an increase in the number of IgA-secreting cells; and (iv) the increase of IgA-secreting cells in other distant mucosal sites, such as the bronchus and mammary glands, as a consequence of gut stimulation by probiotic bacteria. We suggested that under physiological conditions, probiotic bacteria can act as mucosal and systemic adjuvants. This last effect would be mediated by the network of cytokines induced after probiotic stimulation. In our opinion, the most important signals induced by probiotic bacteria included in daily food would be mediated through the immune cells involved in the innate immune response. The proposed model for probiotic interaction and gut immune activation in our opinion is shown in Fig. 3 and Fig. 4.
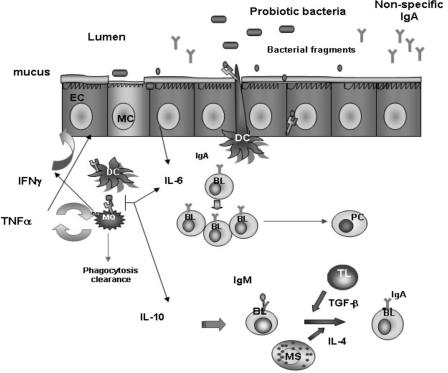
FIG. 3.
The local immune response in the gut induced by the interaction between probiotic bacteria and the epithelial and immune cells associated with the lamina propria of the small intestine. Activation of the innate immune response is shown. There would be different pathways of internalization for the probiotic bacteria present in the lumen of the small intestine: an M cell (MC) is associated with the epithelium, and an epithelial cell (EC) and the interdigitant dendritic cells (DC) are able to sample bacteria. After the interaction with the epithelial cells, probiotic bacteria or their fragments are internalized. The first cells that would interact with them are the antigen-presenting cells (APC), macrophages, and/or dendritic cells associated with the lamina propria of the gut. The interaction with epithelial cells induces IL-6 release. Macrophages and dendritic cells phagocytose the probiotic bacteria or their fragments, and they are induced to produce cytokines such as TNF-α and IFN-γ, which increase epithelial cell stimulation and initiate the cross talk between all the associated immune cells. Mast cells would also be stimulated to produce IL-4. Other cytokines, such as IL-10 and IL-6, are also produced to enhance the cytokine network of signals. The ingested bacteria or their particles could also be eliminated by phagocytosis clearance. IL-6 would favor the clonal expansion of IgA B lymphocytes, increasing the number of IgA-producing cells and the passage of them to plasmatic cells in the lamina propria of the gut. IL-6 together with IL-4 and TGF-β (not determined in our studies) can induce the T-independent switch from IgM to IgA on the surface of B cells and can promote in this way an increase in the number of B cells that are IgA+ in the lamina propria of the gut. EC, intestinal epithelial cells; MQ, macrophages; TL, T lymphocytes; BL, B lymphocytes; MS, mast cells; PC, plasma cells.
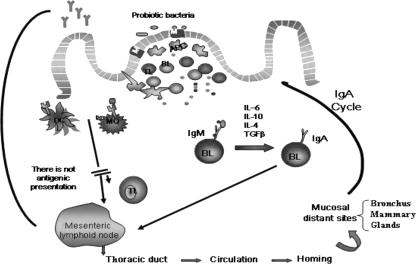
FIG. 4.
Systemic immune response induced by probiotic bacteria after interaction with the immune cells of the Peyer's patches. In the Peyer's patches, the probiotic bacteria or their fragments are internalized by M cells or in a paracellular way through follicle-associated epithelial cells of the Peyer's patches. After that, the bacteria or their particles interact with the macrophages and dendritic cells, which are activated to produce cytokines. As consequence of the bacterial stimulation to the immune cells in this inductor site of the immune response, cytokine production is enhanced, as well is the switch from IgM to IgA B cells. IL-10, IL-6, IL-4, and TGF-β from immune cells could also promote this T-independent switch. Probiotic stimulation can induce the IgA cycle, increasing the number of IgA+ cells in mucosal sites distant to the intestine. The IgA+ cells migrate to the mesenteric lymphoid node and then via the thoracic duct to the circulation, arriving in the bronchus and mammary glands. The cytokines released by probiotic stimulation in Peyer's patches are the biological messengers of the complex network of signals that activate the systemic immune response. DC, dendritic cells; MQ, macrophages cells; APC, antigen-presenting cells; TL, T lymphocytes; BL, B lymphocytes.
In conclusion, we demonstrated for probiotic microorganisms that the most important mechanisms involved in the gut immune stimulation are the clonal expansion of B-lymphocyte IgA+ and the innate immune response. The magnitude of such stimulation did not enhance the inflammatory immune response. They induced up- or down-regulation of the innate response in order to maintain the intestinal homeostasis. Even though the T-cell population was not modified in the lamina propria of the intestine, we cannot exclude T-cell activation as a source of the cytokines detected.
More studies concerning the different signals induced by probiotic microorganisms involved in the activation of immune cells through distinct receptors are necessary. This research will allow determination of the big difference among the signals induced by pathogens (beside their virulence factors) that use similar receptors, commensal and noncommensal probiotic bacteria, to induce inflammatory immune responses or immunomodulatory effects. The proposed model for the mechanisms induced by probiotic bacteria from our studies shows at least in part the scientific basis of the way in which the probiotics work. This knowledge would also be useful for the influence on the gut immune system under pathological conditions.
Acknowledgments
We thank Roy Fuller for his help in preparation of the manuscript.
This work was financially supported by a grant from Consejo de Investigaciones Universidad Nacional de Tucumán, 26/D231-2001/2003, Argentina, and from PIP 02176 (CONICET), Argentina, and PICT 00/10068.
Footnotes
Published ahead of print on 14 March 2007.
REFERENCES
- 1.Akira, S., K. Takeda, and T. Kaisho. 2001. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat. Immunol. 2:675-680. [DOI] [PubMed] [Google Scholar]
- 2.Alander, M., R. Satokari, R. Korpela, M. Saxelin, T. Vilpponen-Salmela, T. Mattila-Sandholm, and A von. Wright. 1999. Persistence of colonization of human colonic mucosa by a probiotic strain, Lactobacillus rhamnosus GG, after oral consumption. Appl. Env. Microbiol. 65:351-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Axelsson, L. T., T. C. Chung, W. G. Dobrogosz, and S. E. Lindgren. 1989. Production of a broad spectrum antimicrobial substance by Lactobacillus reuteri. Microb. Ecol. Health Dis. 2:131-136. [Google Scholar]
- 4.Berg, R. D. 1998. Probiotic, probiotics or ‘conbiotics’? Trends Microbiol. 6:89-92. [DOI] [PubMed] [Google Scholar]
- 5.Bernet, M. F., D. Brassart, J. R. Neeser, and A. L. Servin. 1993. Adhesion of human bifidobacterial strains to cultured human intestinal epithelial cells and inhibition of enteropathogen-cell interactions. Appl. Environ. Microbiol. 59:4121-4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernet-Camard, M. F., V. Lievin, D. Brassart, J. R. Neeser, A. L. Servin, and S. Hudault. 1997. The human Lactobacillus acidophilus strain LA1 secretes nonbacteriocin antibacterial substances active in vitro and in vivo. Appl. Environ. Microbiol. 63:2747-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bibiloni, R., R. N. Fedorak, J. W. Tannock, K. L. Madsen, P. Gionchetti, M. Campieri, C. De Simone, and R. Balfour Sarpor. 2005. DSL #3 probiotics-misture induces remission in patients with active ulcerative colitis. Am. J. Gastroenterol. 100:1539-1546. [DOI] [PubMed] [Google Scholar]
- 8.Cario, E., G. Gerken, and D. K. Podolsky. 2004. Toll-like receptor-2 enhances ZO-1-associated intestinal epithelial barrier integrity via protein kinase C. Gastroenterology 127:224-238. [DOI] [PubMed] [Google Scholar]
- 9.Coconnier, M. H., M. F. Bernet, S. Kerneis, G. Chauviere, J. Fourniat, and A. L. Servin. 1993. Inhibition of adhesion of enteroinvasive pathogens to human intestinal Caco-2 cells by Lactobacillus acidophilus strain LB decreases bacterial invasion. FEMS Microbiol. Lett. 110:299-305. [DOI] [PubMed] [Google Scholar]
- 10.Coconnier, M. H., T. R. Klaenhammer, S. Kerneis, M. F. Bernet, and A. L. Servin. 1992. Protein-mediated adhesion of Lactobacillus acidophilus BG2FO4 on human enterocyte and mucus-secreting cell lines in culture. Appl. Environ. Microbiol. 58:2034-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coconnier, M. H., V. Levien, E. Hemery, and A. L. Servin. 1998. Antagonistic activity against Helicobacter infection in vitro and in vivo by the human Lactobacillus acidophilus strain LB. Appl. Environ. Microbiol. 64:4573-4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooffman, R. L., D. Lebman, and B. Shrader. 1989. Transforming growth factor β specifically enhances IgA production by lipopolysaccharide-stimulated murine B lymphocytes. J. Exp. Med. 170:1039-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dembele, T., V. Obdrzalek, and M. Votava. 1998. Inhibition of bacterial pathogens by lactobacilli. Zentralbl. Bakteriol. 288:395-401. [DOI] [PubMed] [Google Scholar]
- 14.de Moreno de LeBlanc, A., and G. Perdigón. 2004. Yoghurt feeding inhibits promotion and progression of experimental colorectal cancer. Med. Sci. Monit. 10:96-104. [PubMed] [Google Scholar]
- 15.de Moreno de LeBlanc, A., C. Maldonado Galdeano, S. Chaves, and G. Perdigón. 2005. Oral administration of L. casei CRL 431 increases immunity in bronchus and mammary glands. Eur. J. Inflamm. 3:23-28. [Google Scholar]
- 16.de Moreno de LeBlanc, A., C. Matar, E. Farnworth, and G. Perdigón. 2006. Study of cytokines involved in the prevention of a murine experimental breast cancer by kefir. Cytokine 34:1-8. [DOI] [PubMed] [Google Scholar]
- 17.de Moreno de LeBlanc, A., C. Matar, N. LeBlanc, and G. Perdigón. 2005. Effects of milk fermented by Lactobacillus helveticus R389 on a murine breast cancer model. Breast Cancer Res. 7:477-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Moreno de LeBlanc, A., J. Valdez, and G. Perdigón. 2004. Regulatory effect of yoghurt on intestinal inflammatory immune response. Eur. J. Inflamm. 2:21-61. [Google Scholar]
- 19.de Vrese, M., A. Stegelmann., B. Ritcher, S. Fenselau, C. Laue, and J. Schrezenmeir. 2001. Probiotics: compensation for lactase insufficiency. Am. J. Clin. Nutr. 73:S421-S429. [DOI] [PubMed] [Google Scholar]
- 20.Di Giacinto, C., M. Marinaro, M. Sanchez, W. Strober, and M. Boirivant. 2005. Probiotics ameliorate recurrent Th1-mediated murine colitis by inducing IL-10 and IL-10-dependent TGF-beta-bearing regulatory cells. J. Immunol. 174:3237-3246. [DOI] [PubMed] [Google Scholar]
- 21.Dogi, C. A., and G. Perdigón. 2006. Importance of the host specificity in the selection of probiotic bacteria. J. Dairy Res. 73:357-366. [DOI] [PubMed] [Google Scholar]
- 22.Finegold, M. V., L. Sutter, and G. E. Mathisen. 1983. Normal indigenous intestinal flora, p. 3-31. In D. J. Hentges (ed.), Human intestinal microflora in health and disease. Academic Press, New York, NY.
- 23.Fukushima, M., and M. Nakano. 1996. Effects of a mixture of organisms Lactobacillus acidophilus or Streptococcus faecalis on cholesterol metabolism in rats fed on fat-and-cholesterol-enriched diet. Br. J. Nutr. 76:857-867. [DOI] [PubMed] [Google Scholar]
- 24.Fukushima, Y., Y. Kawata, H. Hara, A. Terada, and T. Mitsuoka. 1998. Effect of a probiotic formula on intestinal immunoglobulin A production in healthy children. Int. J. Food Microbiol. 42:39-44. [DOI] [PubMed] [Google Scholar]
- 25.Fuller, R. 1989. Probiotics in man and animals. J. Appl. Bacteriol. 66:365-369. [PubMed] [Google Scholar]
- 26.Fuller, R. 1991. Probiotics in human medicine. Gut 32:439-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Furrie, E., S. Macfarlane, A. Kennedy, J. H. Cummings, S. V. Walsh, D. A. O'Neil, and G. T. Macfarlane. 2005. Synbiotic therapy (Bifidobacterium longum/Sinergy) initiates resolution of inflammation in patients with active ulcerative colitis: a randomized controlled pilot trial. Gut 54:242-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gill, H. S., K. J. Rutherfurd, J. Prasad, and P. K. Gopal. 2000. Enhancement of natural and acquired immunity by Lactobacillus rhamnosus (HN001), Lactobacillus acidophilus (HN017) and Bifidobacterium lactis (HN019). Br. J. Nutr. 83:167-176. [DOI] [PubMed] [Google Scholar]
- 29.Granato, D., F. Perotti, I. Masserey, M. Rouvet, M. Golliard, A. Servin, and D. Brassart. 1999. Cell surface-associated lipoteichoic acid acts as an adhesion factor for attachment of Lactobacillus johnsonii La1to human enterocyte-like Caco-2 cells. Appl. Environ. Microbiol. 65:1071-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greene, J. D., and T. R. Klaenhammer. 1994. Factors involved in adherence of lactobacilli to human Caco-2 cells. Appl. Environ. Microbiol. 60:4487-4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herías, M. V., J. F. J. G. Koninkx, J. G. Vos, J. H. J. Huis in't Veld, and J. E. van Dijk. 2005. Probiotic effects of Lactobacillus casei on DSS-induced ulcerative colitis in mice. Int. J. Food Microbiol. 103:143-155. [DOI] [PubMed] [Google Scholar]
- 32.Hermelijn, H. S., A. Engering, D. van der Kleij, E. C. de Jong, K. Schip perd, T. M. M. van Capela, B. A. J. Zaat, M. Yazdanbakhsh, E. A. Wierenga, Y. van Kooyk, and M. L. Kapsenberg. 2005. Selective probiotic bacteria induce IL-10-producing regulatory T cells in vitro by modulating dendritic cell function through dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin. J. Allergy Clin. Immunol. 115:1260-1267. [DOI] [PubMed] [Google Scholar]
- 33.Hirayama, K., and J. Rafter. 1999. The role of lactic acid bacteria in colon cancer prevention: mechanistic considerations. Antonie Leeuwenhoek 76:391-394. [DOI] [PubMed] [Google Scholar]
- 34.Husband, A. J., and J. L. Cowans. 1978. The origin and antigen dependent distribution of IgA-containing cells in the intestine. J. Exp. Med. 148:1146-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ishikawa, H., I. Akedo, T. Otani, T. Suzuki, T. Nakamura, I. Takeyama, S. Ishiguro, E. Miyaoka, T. Sobue, and T. Kakizoe. 2005. Randomized trial of dietary fiber and Lactobacillus casei administration for prevention of colorectal tumors. Int. J. Cancer 116:762-767. [DOI] [PubMed] [Google Scholar]
- 36.Isolauri, E., Y. Sütas, P. Kankaanpää, H. Arvilommi, and S. Salminen. 2001. Probiotics: effects on immunity. Am. J. Clin. Nutr. 73:444S-450S. [DOI] [PubMed] [Google Scholar]
- 37.Iwasaki, A., and R. Medzhitov. 2004. Toll-like receptor control of the adaptative immune response. Nat. Immunol. 5:987-995. [DOI] [PubMed] [Google Scholar]
- 38.Janeway, C. A., and R. Medzhitov. 2002. Innate immune recognition. Annu. Rev. Immunol. 20:197-216. [DOI] [PubMed] [Google Scholar]
- 39.Jang, M. H., M.-N. Kweon, K. Iwatani, M. Yamamoto, K. Terahara, Ch. Sasakawa, T. Suzuki, T. Nochi, Y. Yokota, P. D. Rennert, T. Hiroi, H. Tamagawa, H. Iijima, J. Kunisawa, Y. Yuki, and H. Kiyono. 2004. Intestinal villous M cells: an antigen entry site in the mucosal epithelium. Proc. Natl. Acad. Sci. USA 101:6110-6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kato, I., K. Tanaka, and T. Yokokura. 1999. Lactic acid bacterium potently induces the production of interleukin-12 and interferon-gamma by mouse splenocytes. Int. J. Immunopharmacol. 21:121-131. [DOI] [PubMed] [Google Scholar]
- 41.Kelly, D., S. Conway, and R. Aminov. 2005. Commensal gut bacteria: mechanisms of immune modulation. Trends Immunol. 26:326-333. [DOI] [PubMed] [Google Scholar]
- 42.Kopp-Hoolihan, L. 2001. Prophylactic and therapeutic uses of probiotics: a review. J. Am. Diet. Assoc. 101:229-238. [DOI] [PubMed] [Google Scholar]
- 43.Krahenbuhl, J. P., and M. R. Neutra. 2000. Epithelial M cells: differentiation and function. Annu. Rev. Cell Dev. Biol. 16:301-332. [DOI] [PubMed] [Google Scholar]
- 44.Kunimoto, D. Y., R. P. Nordan, and W. Strober. 1989. IL-6 is a potent cofactor of IL-1 in IgM synthesis and of IL-5 in IgA synthesis. J. Immunol. 143:2230-2235. [PubMed] [Google Scholar]
- 45.Lamm, M. 2003. IgA function. Mucosal Immunol. Update 11:7-8. [Google Scholar]
- 46.Lee, S. J., S. Evers, D. Roeder, A. F. Parlow, J. Risteli, L. Risteli, Y. C. Lee, T. Feizi, H. Langen, and M. C. Nussenzweig. 2002. Mannose receptor-mediated regulation of serum glycoprotein homeostasis. Science 295:1898-1901. [DOI] [PubMed] [Google Scholar]
- 47.Maassen, C., C. van Holten-Neelen, F. Balk, M. Heijne den Bak-Glashouwer, R. Leer, J. Laman, W. Boersma, and E. Claassen. 2000. Strain-dependent induction of cytokine profiles in the gut by orally administered Lactobacillus strains. Vaccine 18:2613-2623. [DOI] [PubMed] [Google Scholar]
- 48.Macpherson, A. J., and N. L. Harris. 2004. Interaction between commensal intestinal bacteria and the immune system. Nat. Rev. Immunol. 4:478-485. [DOI] [PubMed] [Google Scholar]
- 49.Macpherson, A. J., and T. Uhr. 2004. Compartmentalization of the mucosal immune response to commensal intestinal bacteria. Ann. N. Y. Acad. Sci. 1029:36-43. [DOI] [PubMed] [Google Scholar]
- 50.Macpherson, A. J., and T. Uhr. 2004. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science 303:1662-1665. [DOI] [PubMed] [Google Scholar]
- 51.Majamaa, H., and E. Isolauri. 1997. Probiotics: a novel approach in the management of food allergy. J. Allergy Clin. Immunol. 99:179-185. [DOI] [PubMed] [Google Scholar]
- 52.Maldonado Galdeano, C., and G. Perdigón. 2004. Role of viability of probiotic strains in their persistence in the gut and in mucosal immune stimulation. J. Appl. Microbiol. 97:673-681. [DOI] [PubMed] [Google Scholar]
- 53.Maldonado Galdeano, C., and G. Perdigón. 2005. The probiotic bacteria Lactobacillus casei induces activation of the gut mucosal immune system through the innate immunity. Clin. Vaccine Immunol. 13:219-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miettinen, M., S. Matikainen, J. Vuopio-Varkila, and K. Varkila. 1996. Production of tumor necrosis factor alpha, interleukin-6, and interleukin-10 is induced by lactic acid bacteria. Infect. Immun. 64:5403-5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miettinen, M., J. Vuopio-Varkila, J. Pirhonen, K. Varkila, M. Korimoto, and I. Julkunen. 1998. Lactobacilli and streptococci induce interleukin 12 (IL-12), IL-18, and gamma interferon production in human peripheral blood mononuclear cells. Infect. Immun. 66:6058-6062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Milling, S. W. F., L. Cousins, and G. MacPherson. 2005. How do dendritic cells interact with intestinal antigens? Trends Immunol. 26:349-352. [DOI] [PubMed] [Google Scholar]
- 57.Mowat, A. M. 2003. Anatomical basis of tolerance and immunity to intestinal antigens. Nat. Rev. Immunol. 3:331-341. [DOI] [PubMed] [Google Scholar]
- 58.Naidu, A. S., W. R. Bidlack, and R. A. Clemens. 1999. Probiotic spectra of lactic acid bacteria (LAB). Crit. Rev. Food Sci. Nutr. 39:13-126. [DOI] [PubMed] [Google Scholar]
- 59.Netea, M. G., C. van der Graaf, J. W. M. Van der Meer, and B. Jan Kullberg. 2004. Toll-like receptors and the host defense against microbial pathogens: bringing specificity to the innate-immune system. J. Leukoc. Biol. 75:749-755. [DOI] [PubMed] [Google Scholar]
- 60.Neutra, M. R., N. J. Mantis, and J. P. Kraenhenbuhl. 2001. Collaboration of epithelial cells with organized mucosal lymphoid tissues. Nat. Immunol. 2:1004-1009. [DOI] [PubMed] [Google Scholar]
- 61.Pelto, L., E. Isolauri, E. M. Lilius, J. Nuutila, and S. Salminen. 1998. Probiotic bacteria down-regulate the milk-induced inflammatory response in milk-hypersensitive subjects but have an immunostimulatory effect in healthy subjects. Clin. Exp. Allergy 28:1474-1479. [DOI] [PubMed] [Google Scholar]
- 62.Perdigón, G., A. de Moreno de LeBlanc, J. Valdez, and M. Rachid. 2002. Role of yoghurt in the prevention of colon cancer. Eur. J. Clin. Nutr. 56(Suppl. 3):S65-S68. [DOI] [PubMed] [Google Scholar]
- 63.Perdigón, G., C. Maldonado Galdeano, C. G. Vinderola, A. de Moreno, M. Medici, and M. E. Bibas Bonet. 2005. Immunomodulation of mucosal immune response by probiotics. Curr. Trends Immunol. 6:69-85. [Google Scholar]
- 64.Perdigón, G., C. Maldonado Galdeano, J. C. Valdez, and M. Medici. 2002. Interaction of lactic acid bacteria with the gut immune system. Eur. J. Clin. Nutr. 56(Suppl. 4):S21-S26. [DOI] [PubMed] [Google Scholar]
- 65.Perdigón, G., M. E. Nader de Macias, S. Alvarez, G. Oliver, and A. A. Pesce de Ruiz Holgado. 1990. Prevention of gastrointestinal infection using immunobiological methods with milk fermented with Lactobacillus casei and Lactobacillus acidophilus. J. Dairy Res. 57:255-264. [DOI] [PubMed] [Google Scholar]
- 66.Perdigón, G., M. Medina, E. Vintiñi, and J. C. Valdez. 2001. Intestinal pathway of internalization of lactic acid bacteria and gut mucosal immunostimulation. Int. J. Immunol. Pharmacol. 13:141-150. [PubMed] [Google Scholar]
- 67.Perdigón, G., R. Fuller, and M. Medina. 2005. The influence of the lactic acid bacteria and other resident microflora on the immune system of the growing animal, p. 351-375. In W. H. Holzapfel and P. J. Naughton (ed.), Microbial ecology in growing animals. Elsevier Publishing Inc., London, United Kingdom.
- 68.Perdigón, G., R. Fuller, and R. Raya. 2001. Lactic acid bacteria and their effect on the immune system. Curr. Iss. Intest. Microbiol. 2:27-42. [PubMed] [Google Scholar]
- 69.Perdigón, G., S. Alvarez, M. Medina, E. Vintiñi, and E. Roux. 1999. Influence of the oral administration of lactic acid bacteria on IgA producing cells associated to bronchus. Int. J. Immunol. Pharmacol. 12:97-102. [PubMed] [Google Scholar]
- 70.Phillips-Quagliata, J. M., M. E. Roux, M. Arny, P. Kelly-Hatfield, M. McWilliams, and M. E. Lamm. 1983. Migration and regulation of B cells in the mucosal immune system, p. 194-202. In J. R. McGhee and J. Mestecky (ed.), The secretory immune system. The New York Academy of Science, New York, NY. [DOI] [PubMed]
- 71.Raibaud, P. 1992. Bacterial interactions in the gut, p. 9-28. In R. Fuller (ed.), Probiotics. Chapman & Hall, London, England.
- 72.Rakoff-Nahoum, S., J. Paglino, F. Eslami-Varzaneh, S. Edberg, and R. Medzhitov. 2004. Recognition of commensal microflora by Toll-like receptors is required for intestinal homeostasis. Cell 118:229-241. [DOI] [PubMed] [Google Scholar]
- 73.Reddy, B. S. 1999. Possible mechanisms by which pro-and prebiotics influence colon carcinogenesis and tumor growth. J. Nutr. 129:1478S-1482S. [DOI] [PubMed] [Google Scholar]
- 74.Reid, G., and A. W. Bruce. 2006. Probiotics to prevent urinary tract infections: the rationale and evidence. World J. Urol. 24:28-32. [DOI] [PubMed] [Google Scholar]
- 75.Rescigno, M., M. Urbano, B. Valzasina, M. Francolini, G. Rotta, R. Bonasio, F. Granucci, J. P. Kraehenbuhl, and P. Ricciardi-Castagnoli. 2001. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat. Immunol. 2:361-367. [DOI] [PubMed] [Google Scholar]
- 76.Rook, G. A., and L. R. Brunet. 2005. Microbes, immunoregulation and the gut. Gut 54:317-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Roos, N. M., and M. B. Katan. 2000. Effects of probiotic bacteria on diarrhea, lipid metabolism and carcinogenesis: a review of papers published between 1988 and 1998. Am. J. Clin. Nutr. 71:405-411. [DOI] [PubMed] [Google Scholar]
- 78.Rumbo, M., P. Anderle, A. Didierlaurent, F. Sierra, N. Debard, J. C. Sirard, D. Finke, and J. P. Kraenhenbuhl. 2004. How the gut link innate and adaptative immunity. Ann. N. Y. Acad. Sci. 1029:16-21. [DOI] [PubMed] [Google Scholar]
- 79.Salminen, S., E. Isolauri, and E. Salminen. 1996. Clinical uses of probiotics for stabilizing the gut mucosal barrier: successful strains and future challenges. Antonie Leeuwenhoek 70:347-358. [DOI] [PubMed] [Google Scholar]
- 80.Schiffrin, E. J., F. Rochat, H. Link-Amster, J. M. Aeschlimann, and A. Donnet-Hughes. 1995. Immunomodulation of human blood cells following the ingestion of lactic acid bacteria. J. Dairy Sci. 78:491-497. [DOI] [PubMed] [Google Scholar]
- 81.Schiffrin, E. J., D. Brassart, A. L. Servin, F. Rochat, and A. Donnet-Hughes. 1997. Immune modulation of blood leukocytes in humans by lactic acid bacteria: criteria for strain selection. Am. J. Clin. Nutr. 66:515S-520S. [DOI] [PubMed] [Google Scholar]
- 82.Schultz, M. et al. 2000. Probiotics and inflammatory bowel disease. Am. J. Gastroenterol. 1(Suppl.):S19-S21. [DOI] [PubMed] [Google Scholar]
- 83.Silva, A. M., E. A. Bambirra, A. L. Oliveira, P. P. Souza, D. A. Gomes, E. C. Vieira, and J. R. Nicoli. 1999. Protective effect of bifidus milk on the experimental infection with Salmonella enteritidis subsp. Typhimurium in conventional and gnotobiotic mice. J. Appl. Microbiol. 86:331-336. [DOI] [PubMed] [Google Scholar]
- 84.Simmering, R., B. Kleessen, and M. Blaut. 1999. Quantification of the flavonoid-degrading bacterium Eubacterium ramulus in human fecal samples with a species-specific oligonucleotide hybridization probe. Appl. Environ. Microbiol. 65:3705-3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tancrede, C. 1992. Role of human microflora in health and disease. Eur. J. Microbiol. Infect. Dis. 11:1012-1015. [DOI] [PubMed] [Google Scholar]
- 86.Tani, K., K. Kurokawa, and M. Nasu. 1998. Development of a direct in situ PCR method for detection of specific bacteria in natural environments. Appl. Environ. Microbiol. 64:1536-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tuomola, E. M., and S. J. Salminen. 1998. Adhesion of some probiotic and dairy Lactobacillus strains to Caco-2 cells cultures. Int. J. Food Microbiol. 41:45-51. [DOI] [PubMed] [Google Scholar]
- 88.Vaughan, E. E., B. Mollet, and W. M. Vos. 1999. Functionality of probiotics and intestinal lactobacilli: light in the intestinal tract tunnel. Curr. Opin. Biotechnol. 10:505-510. [DOI] [PubMed] [Google Scholar]
- 89.Vinderola, C. G., C. Matar, and G. Perdigón. 2005. Role of the epithelial cells in the immune effects mediated by gram-positive probiotic bacteria. Involvement of Toll-like receptors. Clin. Diagn. Lab. Immunol. 12:1075-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vintiñi, E., S. Alvarez, M. Medina, M. Medici, M. V. de Budeguer, and G. Perdigón. 2000. Gut mucosal immunostimulation by lactic acid bacteria. Biocell 24:223-232. [PubMed] [Google Scholar]
- 91.Vintiñi, E., V. Ocaña, and M. E. Nader de Macías. 2004. Effect of lactobacilli administration in the vaginal tract of mice: evaluation of side effects and local immune response by local administration of selected strains. Methods Mol. Biol. 268:401-410. [DOI] [PubMed] [Google Scholar]
- 92.Yasui, H., K. Shida, T. Matsuzaki, and T. Yokokura. 1999. Immunomodulatory function of lactic acid bacteria. Antonie Leeuwenhoek 76:383-389. [PubMed] [Google Scholar]