Abstract
Recent progress in genomic sequencing, computational biology, and ontology development has presented an opportunity to investigate biological systems from a unique perspective, that is, examining genomes and transcriptomes through the multiple and hierarchical structure of Gene Ontology (GO). We report here our development of GO Engine, a computational platform for GO annotation, and analysis of the resultant GO annotations of human proteins. Protein annotation was centered on sequence homology with GO-annotated proteins and protein domain analysis. Text information analysis and a multiparameter cellular localization predictive tool were also used to increase the annotation accuracy, and to predict novel annotations. The majority of proteins corresponding to full-length mRNA in GenBank, and the majority of proteins in the NR database (nonredundant database of proteins) were annotated with one or more GO nodes in each of the three GO categories. The annotations of GenBank and SWISS-PROT proteins are available to the public at the GO Consortium web site.
Biomedical research over the last century has made tremendous progress in our understanding of biology and medicine. The recent genomic sequencing of human, mouse, and other organisms, and high-throughput studies, such as those based on microarray technology, have been yielding massive amounts of data. However, the knowledge accumulated so far is mainly fragmented. Full utilization of this data and its integration with existing knowledge can be facilitated by a systematic representation of knowledge, that is, the development of ontology. Ontology is the formalized specification of knowledge in a certain subject. Great potential exists for ontology-based literature retrieval in biomedical research (McGuinness 1999), ontology-based database integration in drug discovery, and ontology-facilitated biomedical research. Recently, the Gene Ontology (GO) Consortium (www.geneontology.org) has developed a systematic and standardized nomenclature for annotating genes in various organisms. Using three main ontologies—molecular function, biological process, and cellular component—a significant number of genes in yeast, Drosophila, mouse, and other model organisms have been annotated, either manually or automatically (Ashburner et al. 2000; The Gene Ontology Consortium 2001).
Association between ontology nodes and proteins, namely, protein annotation through gene ontology, is an integral application of ontology and has many practical uses. For example, designing of microarray probes would be greatly facilitated by a comprehensive understanding of all the genes involved. A microarray aimed to examine a particular process, such as apoptosis, would optimally have probes against all the genes significantly and directly involved in apoptosis. These genes can be chosen using GO annotations.
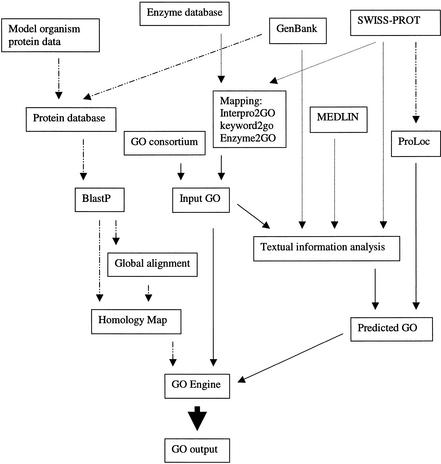
To efficiently annotate proteins, we have developed a software platform, the GO Engine, which combines rigorous sequence homology comparison with text information analysis. During evolution, many new genes arose through mutation, duplication, and recombination of the ancestral genes. When one species evolved into another, the majority of orthologs retained very high levels of homology. The high sequence similarity between orthologs forms one of the foundations of the GO Engine. Text information related to individual genes or proteins is immersed in the vast ocean of biomedical literature. Manual review of the literature to annotate proteins presents a daunting task. Several recent papers described the development of various methods for the automatic extraction of text information (Li et al. 2000; Jenssen et al. 2001). However, the direct applications of these approaches in GO annotation have been minimal. We used simple correlation of text information with specific GO nodes in the training data to predict GO association for unannotated proteins. The GO Engine combines homology information, a unique protein-clustering procedure, and text information analysis to create the best possible annotations, as represented schematically in Figure 1.
Figure 1.
A schematic representation of automatic GO annotation. Solid, dotted, and dot and dash intervened arrows indicate flow of GO information, sequence information, and text information, respectively.
The availability of GO annotations for a significant number of proteins from different organisms presents an opportunity to examine the cellular localization, molecular function, and involvement in a biological process of each of these proteins through the multiple and hierarchical structure of Gene Ontology. We report here our brief analysis of human proteins using the GO Engine annotation system.
RESULTS AND DISCUSSION
Protein Database, Input GO Annotation, and Homology Analysis
As a first step in the annotation process, we collected proteins from different sources to build a database of proteins, some of which have been annotated by the members of the GO Consortium. This database is considered to contain the majority of prototype proteins and served as the main driver of the GO Engine. The rationale for using such a database is that there is a finite number of nonmutated proteins in existence, corresponding to a finite number of cognitively distinguishable functions, locations, or biological processes. If the majority of proteins in this database are accurately annotated with GO nodes, then the annotation for any query protein can be deduced from the annotation of a known protein in this database, which is homologous to the query protein. The National Center for Biotechnology Information (NCBI) nonredundant protein database contains proteins from a diverse array of sources, and thus it served as the major source for our protein database. Proteins from the Saccharomyces genome database (SGD) (Dwight et al. 2002) and the Drosophila genome database (Flybase) (The FlyBase Consortium 2002) were added. The database used in this study comprises 670,130 proteins.
Initial GO annotations of proteins were obtained from several sources. Members of the GO Consortium have annotated a substantial number of proteins. Their annotations were collected and mapped to proteins in our protein database. In addition, various conversion tables that link Enzyme Commission number, InterPro protein motifs, and SWISS-PROT keywords to GO nodes, which are available from the Gene Ontology Consortium web site, are used to annotate additional proteins in the protein database. The combined GO annotations of proteins served as the training data for the text information analysis and also served as input GO annotation for the GO Engine.
The current annotation process exploits the transitive nature of protein homology. This homology transitivity has been used previously (Yona et al. 1999; Bolten et al. 2001), and the merits of this approach have been debated. We found that, with additional input data, such as information derived from protein-domain features, text information analysis, and cellular localization prediction, this homology transitivity can be used as the main engine for predicting GO annotations of unknown proteins. Rigorous and detailed homology comparisons among these 670,130 proteins were performed to delineate the degree of homology between protein pairs by using BLASTP in BLAST with default parameters (Altschul et al. 1997). Table 1A lists the distribution of the BLASTP results. Overall, 78.5 million pairs of proteins were found to have E scores lower than 10–2. To accurately calculate the sequence similarity, we performed global alignment for each pair of homologous proteins identified with the BLAST program, using the Needleman-Wunsch algorithm (Needleman and Wunsch 1970). Table 1B shows the distribution of protein pairs in terms of the identity percentage between them. The majority (68.5%) of protein pairs have identity percentages in the range of 10%–50%.
Textmining and Prediction of Cellular Localization
Many earlier GenBank records and all SWISS-PROT records contain text information, which generally describes the functions of gene products. In addition, one or more reference articles were sometimes identified in the respective field of the GenBank and SWISS-PROT records. The reference articles relevant to the proteins in our database were obtained from the MEDLINE database in the National Library of Medicine, National Institutes of Health. Almost all of them have titles, abstracts, and MeSH terms. In total, 115,527 unique proteins from our protein database were linked to 86,599 MEDLINE records. A few of them lack contents in abstracts or medical subject headings (MeSH) terms. Among those proteins, 61,032 were linked with a single paper. Forty-six MEDLINE records have over 100 protein correspondences. Such records tend to be those reporting on high-throughput cDNA sequencing studies. We applied a simple computational linguistics technique to analyze the textual information from titles, abstracts, MeSH terms, and definition lines of gene records. Text contained in the sequence-related papers and definition lines in sequence records were extracted. The extraction process involves elimination of negative sentences, word stemming, and generation of predictive words. Table 2 lists some general statistics of text information from available sequence databases. A simple, yet predictive, probabilistic model was then applied to create possible GO annotations based on the associated text information. Definition lines of sequence records, MeSH term annotations, titles, and abstracts from the sequence-related papers were modeled separately.
Table 2.
Statistics on Textual Information Analyzed by GO Engine
| MeSH term | Title | Abstract | Definition line | |
|---|---|---|---|---|
| Number of proteins | 110608 | 106190 | 113073 | 516952 |
| Number of articles | 71703 | 77314 | 82654 | n/a |
| Number of unique words | 40011 | 18175 | 26630 | 25915 |
| Average number of words per article or per definition line | 19.05 | 2.70 | 11.65 | 6.56 |
Text information was extracted from titles, abstracts, and MeSH terms of articles referenced in GenBank and SWISS-PROT records and from the definition lines of protein records.
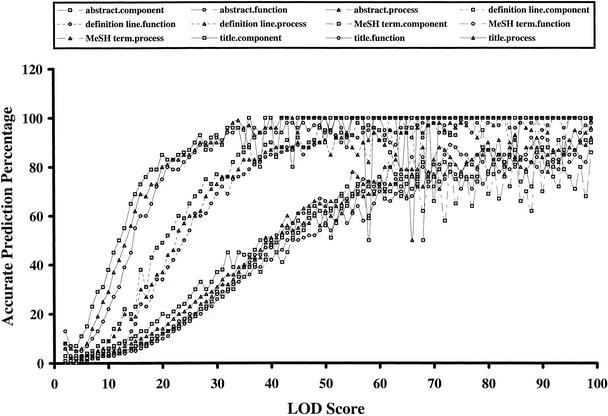
For the text analysis, the frequency of association of a specific term with a specific GO node in the training data was examined. Parameters such as boundaries of the frequency of MeSH terms and other words were optimized through the training process, using self-validation and cross-validation methods. Logarithm of odds (LOD) scores, defined as the logarithm of the ratio between the association frequency of any term–GO pair and the calculated frequency of the random combination of this pair, were used to indicate the relatedness of certain terms with certain GO nodes. These LOD scores were found to be correlative with the accuracy of GO prediction, as shown in Figure 2. Text information from titles of MEDLINE records appears to have more predictive power, in particular at lower LOD scores, than does text information from other categories (Fig. 2). This probably reflects the fact that the title tends to summarize the gist of an article in a straightforward manner. MeSH terms havesimilar predictive capabilities as the abstracts, possibly because the MeSH terms are derived from the abstracts, and thus have similar information content.
Figure 2.
The calculated logarithm of odds (LOD) scores in textual information analysis correlated well with the accuracy of GO predictions. The result is based on self-validation studies. LOD score is calculated as defined in Methods. Only predictions made with LOD scores above 2 were evaluated here and used in the GO Engine. Any LOD scores above 99 are collapsed to 99. GO prediction for any particular protein is considered accurate if the predicted GO node is the same as one of the input GO nodes for this protein or the predicted GO node is a parent or a child of one of the input GO nodes.
Based on text information, a significant number of proteins were predicted to be associated with one or more GO nodes. Table 3 lists the number of proteins with predicted GO nodes from four types of text information in the three categories of GO. These predicted GO annotations were incorporated in GO Engine to increase the accuracy of homology-based GO annotation and to generate de novo annotations. To further enhance the accuracy and coverage of GO Engine, we used a computational platform for predicting cellular localization, ProLoc (A. Novik et al., in prep.), to predict the cellular localization of individual proteins based on their inherent features such as specific localization signatures, protein domains, amino acid composition, isoelectric point (pI), and protein length. Only protein sequences that begin with methionine underwent ProLoc analysis. Thus, 88,997 of 93,110 proteins in SWISS-PROT version 39 were analyzed, and 78,111 proteins have one to three GO predictions in the cellular component category through ProLoc.
Table 3.
Statistics on the Number of Proteins with One or More GO Predictions from MeSH Terms, Titles, and Abstracts of Articles Referenced in the GenBank and SWISS-PROT Records and from Definition Lines of Protein Records
| MeSH term | Title | Abstract | Definition line | Total | |
|---|---|---|---|---|---|
| Cellular component | 57845 | 52094 | 57597 | 514191 | 521396 |
| Molecular function | 57845 | 54152 | 57632 | 516319 | 523384 |
| Biological process | 57845 | 53970 | 57631 | 516402 | 523385 |
GO Annotation
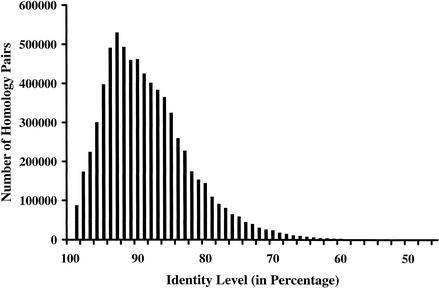
To use the homology transitivity between proteins from different species, we developed a progressive single-linkage clustering process. GO Engine clustered the proteins through single linkage; that is, a protein belongs to a cluster if this protein has sequence homology above a certain threshold with one member of the cluster. The threshold progressed from high homology levels to lower ones, with some defined granularity. The protein clustering and GO annotation were performed at each granularized homology level. The granularity resolution is 1% for global alignment identity; that is, for example, clustering was first performed at 98%, then at 97%, and so on. The granularity is 10-fold for the E score of a BLASTP homology pair; for example, clustering was performed at 10–50, then at 10–49, and so on. To show clustering efficiency and homology transitivity, we examined all homology pairs clustered with at least 90% identity. There were a total of 57,004 clusters containing 263,259 protein members in this level. Among these clusters, 23,321 clusters contained at least three protein members. Figure 3 shows a histogram of identity percentage between proteins with the 90% identity clusters with at least three protein members. The lowest homology pairs had an identity of 46% while being clustered at 90% or higher identity levels.
Figure 3.
The number of protein pairs at different identity levels in clusters identified at greater than 89% identity level through single-linkage clustering. The result indicates that single-linkage clustering efficiently grouped proteins into clusters.
Clusters containing proteins with GO annotations were analyzed along with the GO prediction from the text information, and best annotations for individual proteins of the cluster were selected through an error weight calculation. Table 4A shows the number of the input and output GO annotations through GO Engine. Over 85% of proteins were annotated with one or more GO nodes in each of three GO categories. GO annotations for the majority of proteins with complete coding sequence in GenBank 122 and the majority of SWISS-PROT version 39 were deposited in the www.geneontology.org web site. Table 4B lists the breakdown of the number of proteins annotated at different homology levels. The results indicate that GO annotations were achieved throughout the whole spectrum of homology level.
The accuracy of these annotations by GO Engine was assessed through cross-validation. One-fifth of input GO annotations were withheld during the GO annotation process, and the resultant annotations were compared with these withheld GO nodes. For each protein, the GO node with the lowest error score was examined. Table 4C lists the coverage and accuracy of such a representative test. The coverage ranges from 96% to 99% and the reproducibility is between 65% and 80%. The lower reproducibility of GO annotation in the “cellular component” category, as compared with that in the other two GO categories, is consistent with the notion that a short amino acid segment of a particular protein such as a signal peptide and a nuclear localization signal affects the cellular localization. The presence or absence of short amino acid segments cannot be completely captured through sequence similarity comparisons. Detailed analysis of the validation data indicates that the accuracy of the annotation correlates with the homology level during the annotation (data not shown). Manual validation of GO Engine annotations was performed on a total of over 500 annotations, and about 85%–93% of annotations were found to be correct. The higher percentage of accuracy in the manual examination compared with the automatic cross-validation may result from the incomplete input GO annotations. An additional analysis was performed using manually curated GO annotation from European Bioinformatics Institute (EBI), which was recently available and not yet incorporated in GO Engine. This validation evaluated the overall accuracy of automatic approaches, including various mappings (InterPro2GO, SWISS-PROT keyword2GO, and Enzyme2GO) and GO Engine. In EBI gene association data deposited in the GO consortium web site as of March 6, 2002, there were 9666 human proteins manually annotated with 32,590 GO nodes (with evidence of codes other than ‘IEA’). Among these 9666 human proteins, 5413 have accession numbers present in SWISS-PROT version 39. In GO Engine annotation, there were 5839 human proteins with 27,115 GO annotations from SWISS-PROT version 39 (see Compugen file in the GO Consortium web site). The 5359 overlapping proteins between EBI manual annotations with 18,537 unique GO nodes and GO Engine annotations with 27,115 GO nodes were further used for comparison. Among these 5359 proteins, 3603, with 19,477 GO Engine annotations, had no direct annotations from GO mappings in all three GO categories, indicating the majority of GO annotations in this cohort were from GO Engine. Between the EBI set of data and the GO Engine set of data, 14,695 annotations were exactly matched. Among the rest of the unmatched EBI GO assignments, 47 had no GO Engine assignments in the corresponding GO category, 1329 of them were the parents, 411 of them were the children of one of the GO Engine assignments, and 2055 of them were incompatible with any of the GO Engine assignments for corresponding proteins. A separate analysis was performed to examine the compatibility of GO Engine prediction with the EBI manual curation. GO annotations with the lowest error scores in GO Engine above the homology level of an E score of 10–10 in each of three GO categories for the 5359 overlapping proteins were compared with EBI manual annotation. Among a total of 15,416 GO annotations, 6815 matched exactly with EBI annotation and 1558 were parents of one of the EBI annotations. Such annotations are correct, yet not specific enough. Six hundred thirty six GO Engine annotations were children of one of the EBI annotations. Three thousand two hundred forty three GO Engine annotations are in different paths of GO hierarchy from the corresponding EBI annotations. They are likely to be incorrect, although some of them may indicate novel or rare protein functionalities. In addition, 3164 GO Engine annotations had no corresponding GO annotations from the EBI data set, and thus these annotations may provide some potentially correct annotations. These results suggested that automatic approaches, mainly through GO Engine, could capture the majority of the GO annotations achievable through manual curation, and provide reasonable ground for future curation and experimental verification.
Human Protein GO Annotations and Analysis
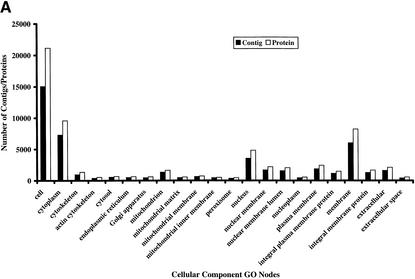
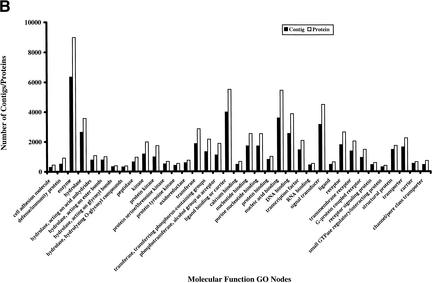
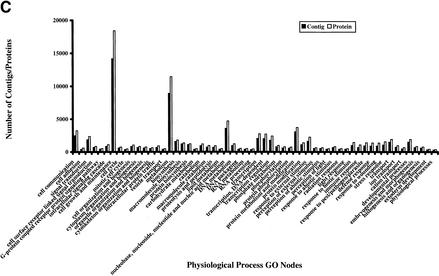
The availability of a large number of GO-annotated proteins from other organisms presents an opportunity to investigate GO features of human proteins in detail. For this purpose, we obtained the Ensembl version 1.0.0 (Hubbard et al. 2002), and annotated proteins through InterPro scanning (Apweiler et al. 2001), InterPro-to-GO node conversion, and GO Engine. Of 27,333 proteins corresponding to 16,913 contigs in version 1.0.0 Ensembl, 23,036 had been annotated in one or more GO categories: cellular component GO for 15,466 contigs, molecular function GO for 15,271 contigs, and biological process GO for 14,939 contigs. Figure 4 indicates the number of proteins and contigs in each major GO node of the three GO categories that contain more than 300 proteins.
Figure 4.
Histograms show the number of proteins and contigs from Ensembl version 1.0.0 in the major nodes in three GO categories: cellular component (A), molecular function (B), and biological process (C). The number of any particular node represents the sum of the number of proteins annotated with this node and that with all children nodes. The sum of all numbers may exceed the total number of proteins or the total numbers of contigs because the annotations of some nodes, which are the children of several higher nodes, are counted multiple times.
The genomic localizations of the majority of Ensembl contigs have been identified in the Ensembl data release. We investigated the distribution of chromosome localization of annotated proteins across the GO hierarchy. Each major GO node in the three GO categories with at least 50 contigs assigned to that GO or its children nodes was analyzed. The actual number of contigs localized in each chromosome was compared with the expected number of contigs for this GO node and its children nodes, and significant differences between these two distributions were identified using a chi-square test. Table 5 lists chromosomal distribution of a few selected GO nodes in biological process, cellular component, and molecular function categories, including the actual number of contigs annotated with the particular GO node and its children nodes, the chi-square test score, and the associated P values. In general agreement with previous reports (International Human Genome Sequencing Consortium 2001; Venter et al. 2001; Wright et al. 2001), current annotation identified 1443 transcriptional factors in molecular function GO and 1771 proteins involved in ‘transcriptional regulation’ in biological process GO. Transcription-related proteins are densely coded in chromosome 19. Although the biological significance of this distribution remains to be determined, it is conceivable that such a clustering of transcriptional factors in specific chromosomal segments, and the more localized clustering in local genomic regions, may be related to particular chromosome structure and, more importantly, this clustering might serve as a regulatory mechanism for the coordinate activation of such genes and the rapid accumulation of transcriptional activity during a period of hyperactivity of gene expression, such as in early development after fertilization, or proliferative activation following a quiescent state. As would be expected, proteins involved in spermatogenesis are clustered on the Y chromosome. Further examination of some of the chromosomal distribution patterns revealed the existence of gene clusters for specific protein families (proteins annotated with a specific GO node or its children GO nodes). For example, we found that in chromosomes 12 and 17, the genes encoding intermediate filament proteins are clustered, and in chromosome 17, at least 10 of those genes also reside in a short stretch of genomic DNA (data not shown). Such observations were not surprising, because protein families such as certain globin family (Ni et al. 2000), protocadherin (Wu et al. 2001), and Hox genes (Ferrier and Holland 2001) among many others, are clustered on a segment of genomic region. The annotation of proteins and the structured hierarchy of GO Engine may allow sophisticated and systematic analyses of human proteins. For instance, a broader definition of ‘gene cluster’ can be used to refer to genes having similar functions, or being involved in a defined process, or being localized in a defined cellular component in a contiguous genomic segment. Such gene clusters may correlate well with specific chromosomal structure, or specific evolutional events. With systematic GO annotation, these clusters can be computationally identified and investigated.
Table 5.
Number of Contigs Annotated with Selected GO Nodes and Their Children Nodes in the Three GO Categories, and Chi-Square Test Results
| GO term | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | X | Y | Contiga | Chib | P valuec |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Physiological process | |||||||||||||||||||||||||||
| Spermatogenesis | 4 | 4 | 2 | 1 | 1 | 7 | 1 | 9 | 3 | 3 | 2 | 3 | 1 | 1 | 1 | 3 | 1 | 0 | 1 | 7 | 1 | 2 | 8 | 11 | 77 | 484 | 0.00E-00 |
| Homophilic cell adhesion | 2 | 0 | 1 | 5 | 24 | 0 | 1 | 1 | 0 | 2 | 3 | 0 | 4 | 1 | 0 | 11 | 0 | 9 | 0 | 3 | 0 | 1 | 3 | 1 | 72 | 235 | L04E-37 |
| Olfaction | 4 | 0 | 0 | 0 | 0 | 12 | 5 | 1 | 4 | 0 | 25 | 1 | 1 | 1 | 0 | 2 | 7 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 68 | 165 | 5-45E-24 |
| DNA-dependent DNA replication | 8 | 5 | 4 | 6 | 2 | 8 | 23 | 1 | 1 | 4 | 1 | 4 | 4 | 2 | 0 | 5 | 8 | 5 | 2 | 5 | 0 | 6 | 1 | 0 | 105 | 103 | L75E-12 |
| Transcription regulation | 143 | 100 | 105 | 49 | 67 | 93 | 93 | 71 | 87 | 52 | 106 | 102 | 41 | 50 | 55 | 76 | 85 | 39 | 171 | 67 | 11 | 34 | 68 | 6 | 1771 | 97 | 2.10E-11 |
| Immune response | 96 | 52 | 43 | 31 | 29 | 44 | 24 | 16 | 20 | 21 | 45 | 29 | 4 | 21 | 22 | 13 | 30 | 8 | 65 | 15 | 8 | 24 | 15 | 0 | 675 | 73 | 1.84E-07 |
| Ribosome biogenesis | 7 | 5 | 4 | 2 | 8 | 3 | 4 | 2 | 4 | 0 | 3 | 5 | 1 | 2 | 3 | 3 | 3 | 1 | 4 | 1 | 0 | 0 | 2 | 0 | 67 | 14 | 0.88 |
| Cell shape and cell size control | 11 | 10 | 5 | 1 | 7 | 6 | 7 | 2 | 7 | 3 | 10 | 8 | 2 | 3 | 5 | 3 | 8 | 2 | 6 | 3 | 1 | 4 | 1 | 0 | 115 | 14 | 0.90 |
| Exocytosis | 20 | 8 | 13 | 7 | 10 | 8 | 13 | 5 | 5 | 8 | 10 | 14 | 2 | 7 | 7 | 5 | 9 | 3 | 7 | 6 | 0 | 5 | 3 | 0 | 175 | 13 | 0.90 |
| Central nervous system development | 9 | 8 | 4 | 5 | 4 | 3 | 5 | 3 | 4 | 2 | 7 | 8 | 2 | 2 | 4 | 5 | 7 | 0 | 7 | 1 | 2 | 1 | 3 | 0 | 96 | 12 | 0.94 |
| Molecular function | |||||||||||||||||||||||||||
| Calcium-dependent cell adhesion molecule | 1 | 0 | 1 | 4 | 23 | 0 | 1 | 1 | 0 | 2 | 2 | 0 | 4 | 1 | 0 | 10 | 0 | 9 | 0 | 3 | 0 | 1 | 2 | 1 | 66 | 238 | 2.59E-38 |
| Olfactory receptor | 6 | 1 | 1 | 0 | 1 | 13 | 5 | 3 | 9 | 0 | 31 | 1 | 1 | 1 | 0 | 3 | 8 | 0 | 9 | 0 | 0 | 0 | 0 | 0 | 93 | 181 | 4338-27 |
| Tumor antigen | 14 | 4 | 0 | 1 | 1 | 2 | 4 | 2 | 3 | 2 | 3 | 5 | 5 | 2 | 0 | 0 | 2 | 3 | 5 | 1 | 1 | 2 | 23 | 0 | 85 | 161 | 3.20B-23 |
| G-protein coupled receptor | 94 | 50 | 59 | 30 | 50 | 84 | 41 | 23 | 30 | 28 | 76 | 33 | 24 | 30 | 9 | 16 | 56 | 11 | 85 | 23 | 2 | 18 | 59 | 1 | 932 | 130 | 2.43E-17 |
| Defense/humanity protein | 75 | 45 | 33 | 16 | 21 | 46 | 20 | 18 | 15 | 9 | 30 | 14 | 6 | 17 | 19 | 7 | 11 | 4 | 55 | 14 | 6 | 22 | 10 | 0 | 513 | 106 | 4.10E-13 |
| Transcription factor | 116 | 78 | 80 | 37 | 58 | 78 | 77 | 55 | 74 | 52 | 95 | 79 | 24 | 41 | 51 | 59 | 60 | 33 | 149 | 51 | 9 | 24 | 59 | 4 | 1443 | 100 | 5.04E-12 |
| Glycopeptide hormone | 6 | 6 | 2 | 0 | 3 | 4 | 3 | 1 | 1 | 1 | 1 | 5 | 2 | 2 | 1 | 1 | 3 | 1 | 5 | 1 | 1 | 1 | 3 | 0 | 54 | 13 | 0.91 |
| Metallopeptidase | 14 | 8 | 11 | 6 | 7 | 6 | 8 | 8 | 4 | 10 | 13 | 13 | 3 | 3 | 5 | 6 | 7 | 3 | 7 | 6 | 2 | 2 | 4 | 0 | 156 | 13 | 0.92 |
| Serotonin receptor | 11 | 9 | 4 | 3 | 7 | 4 | 7 | 4 | 3 | 5 | 11 | 6 | 2 | 1 | 3 | 6 | 6 | 0 | 6 | 4 | 0 | 3 | 3 | 0 | 108 | 13 | 0.92 |
| Protein serine/threonine kinase | 56 | 43 | 32 | 23 | 24 | 29 | 23 | 16 | 23 | 21 | 23 | 31 | 8 | 15 | 18 | 18 | 29 | 5 | 27 | 11 | 5 | 7 | 20 | 1 | 508 | 11 | 0.97 |
| Cellular component | |||||||||||||||||||||||||||
| Secretory vesicle | 5 | 17 | 5 | 1 | 1 | 2 | 3 | 2 | 2 | 5 | 9 | 5 | 1 | 8 | 6 | 4 | 8 | 1 | 1 | 2 | 1 | 12 | 3 | 0 | 104 | 88 | 6.09E-10 |
| Intermediate filament | 6 | 5 | 3 | 2 | 3 | 9 | 2 | 2 | 1 | 2 | 2 | 12 | 0 | 0 | 1 | 3 | 18 | 0 | 0 | 3 | 1 | 2 | 3 | 0 | 80 | 86 | 1.30E-09 |
| 26S proteasome | 11 | 7 | 5 | 1 | 0 | 2 | 3 | 1 | 2 | 3 | 5 | 2 | 1 | 7 | 3 | 5 | 20 | 2 | 2 | 1 | 2 | 1 | 1 | 0 | 87 | 85 | 1.72E-09 |
| Nuclear membrane lumen | 173 | 91 | 87 | 50 | 72 | 79 | 76 | 57 | 64 | 83 | 83 | 97 | 23 | 39 | 63 | 70 | 60 | 48 | 78 | 49 | 12 | 33 | 51 | 22 | 1560 | 82 | 6.79E-09 |
| Collagen | 9 | 11 | 7 | 0 | 1 | 5 | 2 | 5 | 2 | 1 | 5 | 1 | 1 | 7 | 0 | 1 | 2 | 0 | 2 | 2 | 4 | 8 | 3 | 0 | 79 | 78 | 3.12E-08 |
| Respiratory chain complex I | 6 | 4 | 4 | 1 | 5 | 2 | 2 | 1 | 3 | 1 | 4 | 1 | 0 | 2 | 3 | 3 | 4 | 0 | 3 | 0 | 1 | 2 | 2 | 0 | 54 | 12 | 0.94 |
| NADH dehydrogenase (abiquinone) | 6 | 4 | 4 | 1 | 5 | 2 | 2 | 1 | 3 | 1 | 4 | 1 | 0 | 2 | 3 | 3 | 4 | 0 | 3 | 0 | 1 | 2 | 2 | 0 | 54 | 12 | 0.94 |
| Mitochondrial inner membrane | 36 | 31 | 35 | 15 | 25 | 24 | 16 | 17 | 16 | 15 | 28 | 23 | 11 | 13 | 13 | 22 | 23 | 8 | 21 | 12 | 7 | 10 | 14 | 2 | 437 | 12 | 0.95 |
| Small ribosomal subunit | 9 | 6 | 4 | 3 | 4 | 3 | 6 | 2 | 3 | 1 | 4 | 3 | 0 | 3 | 1 | 6 | 4 | 1 | 3 | 3 | 1 | 2 | 2 | 0 | 74 | 11 | 0.96 |
Numbers or x, y in the column headings indicate the chromosome. The results indicate that contigs annotated with some GO nodes or their children nodes such as spermatogenesis and homophilic cell adhesion under “Physiological Process,” or tumor antigen and defense/immunity protein under “Molecular function,” or secretory vesicle and intermediate filament under “Cellular component,” among many others, are distributed unevenly across different chromosomes.
Total number of contigs the annotated with the GO node listed under GO term column or its children nodes.
CHi-square test score.
Significance level from the chi-square test.
METHODS
Data Collection
Gene Ontology and gene association files were obtained from the Gene Ontology Consortium http://www.geneontology.org/, InterPro from http://www.ebi.ac.uk/interpro/ and the enzyme database from http://expasy.proteome.org.au/enzyme/. The following databases and versions were used: GenBank release 122.0; SWISS-PROT release 39.0 (Bairoch and Apweiler 2000); Enzyme database release 26.0; InterPro database as of April 6, 2001; NCBI LocusLink data as of March 6, 2001; MEDLINE databases as of April 6, 2001; and the following files from the Gene Ontology Consortium: gene_association.fb (version 1.26, 2001/02/19), gene_association.mgi (version 1.19, 2001/03/01), gene_association.sgd (version 1.251,2001/03/13), gene_association.pombase (version 1.2, 2000/07/22), ec2go (version 1.2, 2000/10/23), and swp2go (version 1.4, 2000/11/15). Fifty-eight thousand one hundred eighteen SWISS-PROT proteins have been assigned with at least one GO node by the following sources: 15,534 proteins were assigned with at least a functional GO node by conversion of enzyme nomenclature (EC) to GO nodes. Mouse Genome Informatics (MGI) has assigned 5984 SWISS-PROT proteins with GO nodes (http://www.informatics.jax.org). Thirty-one thousand eight hundred sixty-nine SWISS-PROT proteins were assigned at least one GO node using SWISS-PROT keyword correspondence and 33,048 SWISS-PROT proteins were assigned at least one GO node by InterPro scanning. The nonredundant protein database is constructed from GenPep file from NCBI, along with proteins collected from Saccharomyces Genome Database (SGD) and Flybase, with a total of 670,130 proteins.
Sequence Similarity Analysis
A two-stage strategy was used to build a detailed homology map between all proteins in our protein database. In the first stage, all protein pairs with an E score lower than 0.01 using BLASTP with default parameters were cataloged. In the second stage, all of these homologous protein pairs were aligned through the Needlman-Wunsch algorithm with a global alignment to obtain the percentage of identical amino acids between the two proteins. BLOSUM62 was used as the substitution matrix. The percentage of identity is defined as the number of amino acids aligned with nonnegative scores divided by the number of amino acids in both aligned and unaligned length of two proteins in the global alignment. This two-stage homology searching was the most computation-intensive part of GO annotation.
Text Information Extraction
Both GenBank and European Molecular Biology Laboratory (EMBL) databases contain references to the bibliographic information. NCBI staff also add specific MeSH terms to MEDLINE records. In addition, almost all of these records contain abstracts. Efforts were made to obtain the correlations between the presence of specific MeSH terms, or specific English words, in the referred papers and GO assignments in the training data. The correlations were then used to predict GO nodes for unassigned genes.
Noncharacters in titles and abstracts and in the definition line of gene records were eliminated and words were stemmed through the Lingua::stem module from www.cpan.org. Because of the standardized and curated nature of MeSH terms, MeSH terms were not parsed or stemmed. The frequency of each word in all the available text information was calculated. Words that occur at least five times over the whole text information space are retained for further studies. This cutoff threshold is used to eliminate rare words, wrong spellings, and sometimes even the base-pair sequence present in either the definition lines or abstracts. In addition, an upper limit of word frequency (common words such as ‘and,’ ‘gene,’ and ‘protein’ have very high frequencies) and a lower limit of word frequency are defined through a repeated training process and manual review. The words within the upper and the lower limits are considered as predictive. Because the correlation between the GO nodes and specific words is in a positive nature, negative sentences with words such as ‘not’ and its variants, such as ‘unlikely’ or ‘unresponsive,’ were excluded from consideration. Proteins with GO annotation from other sources such as GO Consortium, InterPro scanning, or keyword mappings were used as training data to obtain the correlation between specific words with specific GO nodes. The following formula was used: S = log(P(m,g)/P(m)P(g)), where S is the LOD score for the word m–GO g combination, P(m,g) is the frequency of the term m and GO node g co-occurrence among all word and GO combinations, P(m) is the frequency of occurrence of the term m among all word occurrences, and P(g) is the frequency of occurrence of GO node g among all GO occurrences. To predict GO nodes for any specific protein that is linked to one to a few dozen words, we calculate and sort the sums of LOD scores from all of these words for each possible GO node, and we use them for further GO annotation. Multiple MeSH terms–GO correlations were tested and were found to be no more informative than the single MeSH term–GO correlation, and therefore they were not used.
GO Assignment–GO Engine
GO Engine uses the existing GO annotations as inputs. A substantial number of proteins have been annotated by different groups in the GO Consortium. Their association files and LocusLink GO association were obtained. Additional protein–GO associations were built by using translation files between Enzyme nomenclature and GO nodes, between InterPro entries and GO nodes, and between SWISS-PROT keywords and GO nodes. All of these translation files were available in www.geneontology.org.
Progressive single-linkage clustering was used to assign GO node to proteins. The assignment started with clustering proteins at the highest homology—99% identity from the global alignment. In any cluster with GO-assigned proteins, other proteins are assigned GO nodes based on both the cluster GO nodes and the GO predictions based on text information analysis and ProLoc for any individual proteins and for other proteins in the cluster. For any cluster with a single GO node, all members of this cluster are assigned this GO node. For any cluster with multiple GO inputs, an error weight scheme was applied to determine the final GO nodes.
The error weight scheme works as follows. For each prediction method, the error score matrix is obtained from the validation studies. For example, during text information analysis of titles, a LOD score of 29 has 90% accurate GO prediction in the cellular component category from validation studies (see Fig. 2); then any GO cellular component prediction made at a LOD score of 29 has an error score of 0.10. During GO Engine annotation, in any cluster, for any input GO node, the product of all error scores associated with this GO node is the final error score of that GO node. The final GO outputs are sorted according to low final error score for each GO node. The error score of any GO assignment is inherited throughout the GO Engine process.
Precedence was also given to an individual protein with its own GO textual information predictions and then the combined GO textual information predictions for the whole cluster. A specified number of GO nodes (for example, from 1 to 5) were predicted for each protein in the cluster along with the calculated error score. After the assignment of a cluster, the homology linkage with the cluster was broken. The newly assigned protein with GO nodes and associated error scores was then clustered with other proteins at a lower homology level, and proteins in the new cluster could then be assigned with GO inputs from the original GO input from other proteins, if any, and the input GO nodes from these just-annotated proteins, using the same error weight scheme. The clustering and GO annotation reiterated from highest homology 99% to 35% (with the granularity of 1%) and shifted to the BLASTP-based homology scheme (E score with the granularity of E-1). At all homology levels, any clusters with only GO inputs from text information analysis were not analyzed, and the clusters were not broken until at least one protein member had original GO annotation, or had GO annotation through the GO Engine. By then, GO Engine assigned GO nodes to each member protein of the cluster, and the cluster was then broken. In the final step, any cluster containing proteins with only one or more textual information GO predictions were analyzed, and proteins within the cluster were assigned GO nodes accordingly. ProLoc predictions were treated the same as the prediction from textual information analysis with its own error score consideration.
Statistical Analysis and Quality Assurance
The chi-square test was used for investigating the GO distribution across chromosomes. After each production, a manual check was performed for at least 100 GO assignments. The manual check involved homology searches, literature review, and expert evaluation.
WEB SITE REFERENCES
http://www.cpan.org; Peri modules.
http://www.ebi.ac.uk/interpro/; Source for InterPro files.
http://www.ensembl.org; Ensembl project.
http://expasy.proteome.org.au/enzyme/; The enzyme database.
http://www.geneontology.org/; Gene Ontology Consortium web site.
http://www.informatics.jax.org; MGI web site.
http://www.ncbi.nlm.nih.gov; MEDLINE.
Table 1A.
Distribution of the Homology Levels among Pairs of Proteins in Our Protein Database
| E score | Percentage |
|---|---|
| 10−10–10−2 | 17.58 |
| 10−20–10−10 | 13.81 |
| 10−30–10−20 | 11.02 |
| 10−40–10−30 | 12.91 |
| 10−50–10−40 | 10.24 |
| 10−60–10−50 | 5.81 |
| 10−70–10−60 | 3.64 |
| 10−80–10−70 | 2.65 |
| 10−90–10−80 | 2.86 |
| 10−100–10−90 | 2.53 |
| 10−110–10−100 | 2.18 |
| 10−120–10−110 | 1.58 |
| 10−130–10−120 | 1.50 |
| 10−140–10−130 | 1.13 |
| 10−150–10−140 | 1.01 |
| 10−160–10−150 | 1.01 |
| 10−170–10−160 | 0.92 |
| 10−178–10−170 | 0.90 |
| 0.00 | 6.72 |
Results were obtained using BLASTP and expressed as percentage of total homology pairs with E score below 10−2 in various E-score homology ranges.
Table 1B.
Statistics of Identity Levels of Homologous Protein Pairs Identified by Blastp in Table 1A
| Identity level % | Percentage |
|---|---|
| 0–10 | 5.67 |
| 10–20 | 24.66 |
| 20–30 | 19.94 |
| 30–40 | 10.94 |
| 40–50 | 7.31 |
| 50–60 | 7.09 |
| 60–70 | 7.24 |
| 70–80 | 6.70 |
| 80–90 | 5.98 |
| 90–100 | 4.47 |
Protein pairs were aligned globally using BLOSUM62 as the scoring matrix, and results are expressed as the percentage of all homology pairs in various identity ranges.
Table 4A.
Statistics on the Number of Proteins in Each of the Three GO Categories with Original GO Input (from GO Consortium, Enzyme Conversion, SWISS-PROT Keyword Mapping, InterPro Mapping) and the Number of Proteins in GO Engine Output
| Input | Output | ||
|---|---|---|---|
| GO consortium annotation, enzyme conversion, interpro mapping, etc | Textmining proloc | ||
| Cellular component | 44702 | 522179 | 574607 |
| Molecular function | 85626 | 526083 | 580767 |
| Biological process | 69726 | 525842 | 578636 |
The number of proteins with GO Engine annotations (output) includes the proteins with original GO inputs.
Table 4B.
Statistics on the Number of Proteins Annotated at Different Homology Ranges in Each of the Three GO Categories
| Cellular component | Molecular function | Biological process | |
|---|---|---|---|
| Texta | 32257 | 34137 | 30149 |
| 10−2–10−10 | 87967 | 71717 | 74277 |
| 10−10–10−50 | 122992 | 70088 | 79318 |
| 10−50–0.0 | 98059 | 55132 | 59051 |
| 35%–75% | 111130 | 97209 | 108334 |
| 75%–90% | 38509 | 68282 | 67429 |
| 90%–99% | 38991 | 98576 | 90352 |
| Input GOb | 44702 | 85626 | 69726 |
Text indicates the number of proteins annotated with only the GO predictions from the text information analysis and without any from the annotated proteins through homology comparison.
Input GO indicates the number of proteins with original GO inputs.
Table 4C.
Results of One of the Cross-Validation Tests of GO Engine Annotation
| Total | Predicted GO | Accurate GO | |
|---|---|---|---|
| Cellular component | 7431 | 7186 | 4642 |
| Molecular function | 12999 | 12864 | 10138 |
| Biological process | 10811 | 10690 | 8080 |
Cross-validation was performed by withholding one-fifth of the original GO inputs, and using the GO Engine to predict the GO annotation for these withheld proteins. The predicted GO nodes were compared with the withheld GO nodes. For each protein, only the GO node with the lowest error score was examined. GO prediction was considered accurate if the predicted GO node was the same as, or a child or parent of, the withheld GO node.
Acknowledgments
The authors thank the Gene Ontology Consortium, Enzyme, and InterPro databases for making their data available through the web. Thanks are also due to Compugen LEADS production, ProLoc production, Gencarta group, and NJ R & D group for various productions and help. The authors appreciate greatly the review and critique of Dr. Galit Rotman in Compugen Ltd.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-mail: han@cgen.com; E-mail: liat@cgen.com; FAX: (609) 655-5114.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.86902.
REFERENCES
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler R, Attwood TK, Bairoch A, Bateman A, Birney E, Biswas M, Bucher P, Cerutti L, Corpet F, Croning MDR, et al. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 2001;29:37–40. doi: 10.1093/nar/29.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A, Apweiler R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 2000;28:45–48. doi: 10.1093/nar/28.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolten E, Schliep A, Schneckener S, Schomburg D, Schrader R. Clustering protein sequences-structure prediction by transitive homology. Bioinformatics. 2001;17:935–941. doi: 10.1093/bioinformatics/17.10.935. [DOI] [PubMed] [Google Scholar]
- Dwight SS, Harris MA, Dolinski K, Ball CA, Binkley G, Christie KR, Fisk DG, Issel-Tarver L, Schroeder M, Sherlock G, et al. Saccharomyces Genome Database (SGD) provides secondary gene annotation using the Gene Ontology (GO) Nucleic Acids Res. 2002;30:69–72. doi: 10.1093/nar/30.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrier DE, Holland PW. Ancient origin of the Hox gene cluster. Nat Rev Genet. 2001;2:33–38. doi: 10.1038/35047605. [DOI] [PubMed] [Google Scholar]
- The FlyBase Consortium. The FlyBase database of the Drosophila genome projects and community literature. Nucleic Acids Res. 2002;30:106–108. doi: 10.1093/nar/30.1.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium. Creating the gene ontology resource: Design and implementation. Genome Res. 2001;11:1425–1433. doi: 10.1101/gr.180801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard T, Barker D, Birney E, Cameron G, Chen Y, Clark L, Cox T, Cuff J, Curwen V, Down T, et al. The Ensembl genome database project. Nucleic Acids Res. 2002;30:38–41. doi: 10.1093/nar/30.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Jenssen TK, Laegreid A, Komorowski J, Hovig E. A literature network of human genes for high-throughput analysis of gene expression. Nat Genet. 2001;28:21–28. doi: 10.1038/ng0501-21. [DOI] [PubMed] [Google Scholar]
- Li, Q., Shilane, P., Noy, N.F., and Musen, M.A. 2000. Ontology acquisition from on-line knowledge sources. Proc. AMIA Symp. 497–501. [PMC free article] [PubMed]
- McGuinness DL. Proceedings of the International Medical Informatics Association Working Group 6–Medical Concept Representation and Natural Language Processing Conference. Phoenix, AZ. 1999. Ontology-enhanced search for primary care medical literature. [Google Scholar]
- Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Ni J, Kalff-Suske M, Gentz R, Schageman J, Beato M, Klug J. All human genes of the uteroglobin family are localized on chromosome 11q12.2 and form a dense cluster. Ann NY Acad Sci. 2000;923:25–42. doi: 10.1111/j.1749-6632.2000.tb05517.x. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Wright FA, Lemon WJ, Zhao WD, Sears R, Zhuo D, Wang JP, Yang HY, Baer T, Stredney D, Spitzner J, et al. A draft annotation and overview of the human genome. Genome Biol. 2001;2:RESEARCH0025. doi: 10.1186/gb-2001-2-7-research0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Zhang T, Cheng JF, Kim Y, Grimwood J, Schmutz J, Dickson M, Noonan JP, Zhang MQ, Myers RM, et al. Comparative DNA sequence analysis of mouse and human protocadherin gene clusters. Genome Res. 2001;11:389–404. doi: 10.1101/gr.167301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yona G, Linial N, Linial M. ProtoMap: Automatic classification of protein sequences, a hierarchy of protein families, and local maps of the protein space. Proteins. 1999;37:360–378. [PubMed] [Google Scholar]