Abstract
Plasmodium falciparum is the causative agent of severe human malaria, responsible for over 2 million deaths annually. Of the 5,300 polypeptides predicted to control the parasite life cycle in mosquitoes and humans, 60% are of unknown function. A major challenge of malaria postgenomic biology is to understand how the 5,300 predicted proteins coexist and interact to perform the essential tasks that define the complex life cycle of the parasite. One approach to assign function to these proteins is by identifying their physiological partners. Here we describe the use of tandem affinity purification (TAP) and mass spectrometry for identification of native protein interactions and purification of protein complexes in P. falciparum. Transgenic parasites were generated which express the translation elongation factor PfEF-1β harboring a C-terminal PTP tag which consists of the protein C epitope, a tobacco etch virus protease cleavage site, and two protein A domains. Purification of PfEF-1β-PTP from crude extracts followed by mass spectrometric analysis revealed, in addition to the tagged protein itself, the presence of the native PfEF-1β, the G-protein PfEF-1α, and two new proteins that we named PfEF-1γ and PfEF-1δ based on their homology to other eukaryotic γ and δ translation elongation factor subunits. These data, which constitute the first application of TAP for purification of a protein complex under native conditions in P. falciparum, revealed that the translation elongation complex in this organism contains at least two subunits of PfEF-1β. The success of this approach will set the stage for a systematic analysis of protein interactions in this important human pathogen.
Plasmodium falciparum is the causative agent of the most severe form of human malaria. Over 2 million deaths caused by this parasite are reported annually worldwide (34). The alarming rate of appearance of drug-resistant parasites and mosquitoes, the limited effective antimalarial therapeutic arsenal, and the lack of an effective malaria vaccine have severely hampered international efforts to control the spread of the disease and limit the morbidity and mortality associated with it. The development of new effective malaria eradication strategies requires a better understanding of the parasite cellular biology and advanced knowledge of the metabolic discrepancies that exist between the parasite and the host.
P. falciparum has a nuclear genome size of ∼23 MB distributed among 14 chromosomes varying in size from 0.643 to 3.29 MB (11). This nuclear genome is predicted to encode about 5,300 polypeptides (9). An approximate 60% (3,208 hypothetical proteins) of the predicted open reading frames in the P. falciparum genome encode proteins with no significant homology to proteins in other organisms, and another 5% of the predicted proteins share significant homology to hypothetical proteins of other organisms. The difficulty to genetically manipulate the genome of this parasite has resulted in a very limited analysis of the function of its proteome, with less than 1% of the 5,300 predicted proteins thus far being assessed for their essential function during the parasite life cycle (4, 5, 8, 18, 29-31). The major challenge of malaria postgenomic biology is to define the protein networks that control the parasite life cycle in humans and mosquitoes. Tandem affinity purification (TAP) combined with mass spectrometry has been demonstrated to be an effective and reliable strategy to identify and purify protein complexes under native conditions in different organisms. The original TAP technique was developed in yeast (23, 25) and was based on a translational fusion of the protein of interest to an epitope tag composed of the calmodulin-binding peptide (CBP) and two immunoglobulin G (IgG)-binding domains of the Staphylococcus aureus protein A (ProtA). The CBP and ProtA domains are spaced by a cleavage site of the tobacco etch virus (TEV) protease. The protein complex containing the fusion protein is first bound to an IgG column and, subsequent to washing steps, eluted from the column following TEV protease cleavage. In a second purification step, the fusion protein still harboring the CBP domain is bound to a calmodulin affinity column, a reaction facilitated by addition of Ca2+. Addition of a chelator such as EGTA allows subsequent elution of the protein complex. Although this strategy has been successful for purifying protein complexes from different organisms, it proved to be inefficient in some instances, for example, for the purification of transcription factors in the protistan parasite Trypanosoma brucei (27). It has been proposed that the presence of free calmodulin in cellular extracts may block CBP binding to the calmodulin column, thus reducing the yield of the purified complex (23). Such competition becomes critical when the complex including the tagged protein is present at low levels in the cell. Consequently, new TAP strategies have recently been developed which are based on tags in which CBP is replaced either by a biotinylation tag (3) or the protein C epitope (ProtC) (28). The latter strategy was termed PTP (ProtC-TEV-ProtA) and involves anti-ProtC immunoaffinity chromatography in the second purification step. The PTP method was successfully employed in Trypanosoma brucei for the purification and characterization of a multisubunit transcription factor, the U1 small nuclear RNP and RNA polymerase I (17, 19, 27).
Here we report the use of PTP-based tandem affinity purification for the analysis of protein networks in P. falciparum. We have successfully used this strategy to purify the translation elongation factor complex and determine the identity of its molecular components. Furthermore, we have developed a Gateway-based entry clone for rapid cloning of P. falciparum open reading frames upstream of the PTP tag prior to transfection of the parasite. We present data demonstrating the successful use of this strategy to purify the native phosphoethanolamine methyltransferase Pfpmt of P. falciparum.
MATERIALS AND METHODS
Construction of transfection plasmids.
To make the PfEF-1β-PTP-fused fragment for cloning in the pDCI vector (6), forward primer 5′-CGCTCGAGATGGCTAGTACGCAAGTTTATTAAACGTTA-3′ (with the added XhoI site underlined and the start codon in bold) and reverse primer 5′-ATAGTTTAGCGGCCGCCAATTTGTTAAAGGAAATAATTTCA-3′ (with the added NotI site underlined and the stop codon deleted) were used in combination to amplify the PfEF-1β open reading frame from P. falciparum total cDNA. Forward primer 5′-ATAAGAATGCGGCCGCAGAAGATCAGGTGGATCCTCGTC-3′ (with added NotI site underlined) and reverse primer 5′-CCGCTCGAGTCAGGTTGACTTCCCCGCGGAATTCGCGTCT-3′ (with the added XhoI site underlined and the stop codon in bold) were used to amplify the PTP tag coding sequence cloned in a pN-PURO-PTP vector (28). The PfEF-1β and PTP fragments were digested by NotI and then ligated to create a fusion at the NotI site. The ligation product was rescued by PCR using a combination of PfEF-1β forward and PTP reverse primers and subsequently digested by XhoI and ligated at the XhoI site of the pDCI vector. Directional cloning and the correct sequence of the fused fragment were confirmed by sequencing. To construct the pCHDR-Pfpmt-PTP expression vector, PfPMT cDNA in the pENTR/D-TOPO vector, PTP tag in the pDONRP2R-P3 vector, the HSP86-5′ promoter in pDONRP4-P1R (32), and the pCHDR-3/4 destination vector (32) were combined in an LR reaction. Recombinant plasmids were propagated in Escherichia coli and extracted by Maxi-prep (QIAGEN) columns for transfection of P. falciparum.
Transfection of P. falciparum.
Synchronized P. falciparum strain 3D7 parasites cultured in complete medium (6) at 2% hematocrit and 7% parasitemia (predominantly rings) were used for transfection. Prior to electroporation, a 5-ml infected red blood cell (RBC) suspension was washed with cytomix, resuspended in fresh cytomix to a final volume of 400 μl, and kept on ice. About 100 μg of plasmid DNA dissolved in 100 μl of cytomix was mixed with the 400 μl of the cell suspension in a 0.2-cm cuvette. Electroporation was performed using a Gene Pulser II (Bio-Rad) set at 0.31 kV and 950 μF as previously described (6). Transfected parasites were cultured in petri dishes for 48 h without drug selection and, thereafter, the selection drug (WR92210) was introduced in the culture medium at 5 nM final concentration. Drug-resistant parasites were established after 3 weeks of continuous culture.
Tandem affinity purification of PTP-tagged PfEF-1β.
A total of 1.4 liters culture of transgenic and wild-type 3D7 parasites (8% parasitemia, 2% hematocrit) was grown to the trophozoite stage, harvested, and treated with saponin. Pellets containing parasites were resuspended in 10 ml of ice-cold PA-150 buffer (20 mM Tris-HCl, pH 7.7, 150 mM potassium chloride, 3 mM MgCl2, 0.5 mM dithiothreitol, 0.1% Tween 20) containing a tablet of Complete Mini EDTA-free protease inhibitor cocktail (Roche Diagnostics). All subsequent steps were carried out at 4°C. Buffers and solutions were cooled on ice before use. Parasites were disrupted by sonication, and a crude cytoplasmic extract was obtained from the soluble fraction after centrifugation at 16,000 × g for 20 min at 4°C. PfEF1β-PTP was tandem affinity purified from parasite extract essentially as described previously (28), though some modifications were necessary to accommodate the P. falciparum system. About 10.5 ml of the crude extract was applied onto a 0.2-ml resin bed of IgG-Sepharose 6 Fast Flow (Invitrogen) packed into a 0.8- by 4-cm Poly-Prep chromatography column (Bio-Rad) and incubated for 2 h on a rotary mixer at 4°C. The flowthrough fraction was collected and the resin washed three times with 10 ml of PA-150 buffer. Subsequently, 2 ml of TEV buffer (20 mM Tris-HCl, pH 7.7, 150 mM potassium chloride, 3 mM MgCl2, 0.5 mM EDTA, 1 mM dithiothreitol, 0.1% Tween 20) containing 200 U of AcTEV protease (Invitrogen) was added to the settled resin and incubated overnight at 4°C with gentle agitation. The protein complex cleaved off the matrix by AcTEV protease was collected by gravity flow, diluted with 4 ml of PC-150 buffer (PA-150 containing 2 mM CaCl2), and applied to an immobilized HPC4 (anti-ProtC monoclonal antibody) matrix resin bed (0.2 ml) equilibrated with PC-150. After 2 h of incubation at 4°C, the matrix was washed six times with 10 ml of PC-150 buffer and eluted with 1.5 ml of elution buffer (5 mM Tris-HCl, pH 7.7, 10 mM EGTA, 5 mM EDTA, 0.1% Tween 20). The final eluate was concentrated to 0.2 ml using a vacuum concentrator. Subsequently, the proteins were collected with 10 μl of hydrophobic StrataClean resin (Stratagene), released into sodium dodecyl sulfate (SDS) loading buffer at 80°C, separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE), and stained with Sypro ruby (Invitrogen) according to the manufacturer's protocol.
Mass spectrometric analysis.
Individual protein bands were excised from polyacrylamide gels, digested with trypsin, and analyzed by liquid chromatography-tandem mass spectrometry. The resulting micromass files were used to identify P. falciparum proteins through the Sonar and Mascot search engines.
Immunoblot analysis.
Immunoblot analyses were performed as previously described (24, 26) on crude extracts of the wild-type and transgenic parasites as well as on fractions of the tandem affinity purifications using a monoclonal anti-ProtC antibody (Roche Diagnostics) as well as polyclonal anti-PfEF-1β, anti-PfEF-1α, and anti-Pfpmt antibodies (16, 33). PAP (peroxidase-antiperoxidase soluble complex; Sigma) was used to detect the fusion proteins harboring the protein A moiety. Immunoblot signals were measured using the Image J version 1.37v software.
RESULTS
Expression of PTP-tagged PfEF-1β in P. falciparum.
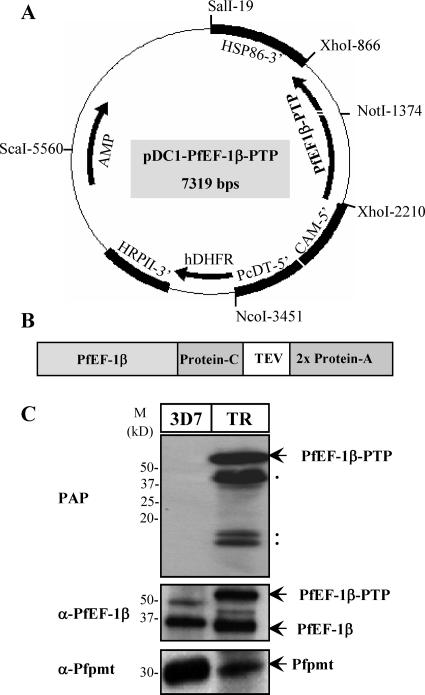
To establish tandem affinity purification as a tool to purify protein complexes from P. falciparum under native conditions and in quantities sufficient for mass spectrometric identification, we chose the translation elongation factor PfEF-1β as a bait. PfEF-1β is a member of a conserved family of translation elongation nucleotide exchange factors that play an essential role in the GTP-dependent elongation step of protein synthesis. This step involves binding of aminoacyl-tRNA to the ribosomal A site, formation of a peptide bond, and translocation of the newly formed peptidyl-tRNA to the P site (15). EF-1β is responsible for the regeneration of a GTP-bound EF-1α necessary for each elongation cycle. A previous study in P. falciparum using radiolabeling and immunoprecipitation demonstrated an interaction between PfEF-1β and PfEF-1α (16). Furthermore, this study revealed that PfEF-1α exists in a protein complex that may play an essential function in protein translation (16). For the purification of PfEF-1β, its coding region was fused to the PTP tag sequence (Fig. 1). The resulting fusion was cloned under the regulatory control of the calmodulin gene promoter into the pDC1 vector (6), which harbors a mutated human dihydrofolate reductase (DHFR) gene that confers resistance to the antimalarial drug WR99210 (Fig. 1A). Following transfection of the P. falciparum 3D7 clone, stable transfectants were selected and analyzed for the expression of the fusion protein by Western blotting. As a control, wild-type 3D7 parasites were also examined. As shown in Fig. 1C, a band of 55 kDa of the expected size of the fusion protein could be detected in 3D7-PfEF-1β-PTP parasites but not in 3D7 using the PAP reagent that recognizes the protein A moiety of the fusion protein. Immunoblotting using anti-PfEF-1β antibodies revealed two bands of 55 and 36 kDa corresponding to the sizes of the PTP-tagged and native PfEF-1β, respectively, whereas only one band of 36 kDa could be detected in the 3D7 clone. As a positive control, antibodies against the P. falciparum phosphoethanolamine methyltransferase, Pfpmt, detected a single band of 30 kDa in both 3D7 and 3D7-PfEF-1β-PTP parasites.
FIG. 1.
(A) Map of the plasmid pDC1-PfEF-1β-PTP. (B) Outline of the PfEF-1β-PTP cassette. The PTP tag consists of protein C, the TEV protease site, and two protein A epitopes. (C) Analysis of expression of native and PTP-tagged PfEF-1β in wild-type (3D7) and transgenic (TR) parasites using the PAP reagent and anti-PfEF-1β antibodies as described in Materials and Methods. Anti-Pfpmt antibodies were used as a control to detect the native Pfpmt enzyme. M, protein marker. The dots indicate degradation products.
Purification of the PfEF-1β protein complex.
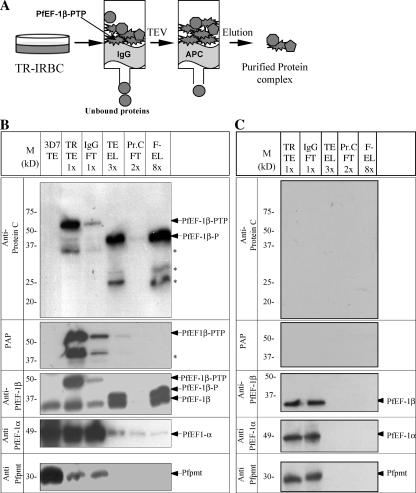
To purify protein complexes that include PfEF-1β, crude extracts were prepared from 3D7-PfEF-1β-PTP transgenic parasites and passed through an IgG column followed by cleavage of the bound material by TEV protease (Fig. 2A). Input extract and released material were analyzed by immunoblotting using the PAP reagent to detect the protein A moiety and anti-ProtC antibodies to detect the protein C moiety (Fig. 2B). The latter antibody detected a band of 55 kDa in the crude extract and a smaller size protein (39 kDa) in the TEV protease-released material consistent with the cleavage of the C-terminal double protein A motif (Fig. 2B). Accordingly, no bands could be detected in the TEV protease-released material using the PAP reagent (Fig. 2B). TEV protease-released proteins were then passed through a column containing an immobilized HPC4 monoclonal antibody that recognizes the ProtC epitope and binds to it with very high affinity in the presence of calcium (Kd, ∼1 nM). The latter property enabled the final elution of the PfEF-1β-ProtC fusion protein and any other proteins that were in complex with it by adding a buffer containing EGTA. The eluted material was separated by SDS-PAGE and further analyzed by immunoblotting using the anti-ProtC monoclonal antibody and the PAP reagent (Fig. 2B). The immunoblot assay demonstrated that both chromatography steps were highly efficient, since the flowthrough fractions from both columns were nearly depleted of tagged PfEF-1β. Densitometric quantification of immunoblot signals indicated ∼13% recovery of the purified proteins in the final eluates relative to their abundance in total extracts. Consistent with previous radiolabeling and immunoprecipitation studies (16), antibodies raised against PfEF-1α detected this protein in the final eluate (Fig. 2B), thus validating the coexistence of PfEF-1β and PfEF-1α in a complex during the P. falciparum intraerythrocytic life cycle. Interestingly, immunoblotting of the purified fractions, including the final eluate using antibodies against PfEF-1β, revealed in addition to the tagged PfEF-1β-ProtC, the presence of the native PfEF-1β protein (Fig. 2B). This suggests that the translation elongation factor complex contains more than one subunit of PfEF-1β. To demonstrate the specificity of the purification, all fractions were tested by immunoblotting using antibodies against Pfpmt, which is involved in parasite phospholipid biosynthesis (20-22, 33). No Pfpmt signal could be detected in the IgG or TEV eluates, whereas this protein was clearly present in the crude extracts (Fig. 2B). As a control for the tandem affinity purification procedure, extracts from wild-type 3D7 parasites were subjected to the same purification strategy as transgenic parasites. As expected, no detectable protein bands could be observed using anti-ProtC antibody or the PAP reagent, and using antibodies against PfEF-1β, PfEF-1α, and Pfpmt, only the endogenous proteins could be detected in the total extract and the flowthrough of the IgG column (Fig. 2C).
FIG. 2.
(A) Schematic representation of the strategy for tandem affinity purification of PTP-tagged proteins from extracts of P. falciparum-infected erythrocytes. Transgenic parasites expressing PTP-tagged proteins (TR-IRBC) were extracted from RBCs by saponin treatment and sonicated, and the soluble fraction was applied to an IgG-Sepharose column. The eluate obtained after TEV protease digestion was applied to an anti-ProtC affinity matrix column (APC), and the bound protein eluted with EDTA-EGTA buffer. IRBC, infected red blood cells. (B) Immunoblot analysis of whole-cell lysates and tandem affinity-purified fractions of wild-type 3D7 and transgenic parasites expressing the PfEF-1β-PTP protein fusion using a monoclonal antibody against protein C, the PAP reagent, and polyclonal antibodies against PfEF-1β, PfEF-1α, and Pfpmt (as a loading control). Identified protein bands are indicated. (C) Extracts from wild-type 3D7 parasites were subjected to the TAP procedure and analyzed by immunoblotting using a monoclonal antibody against protein C, the PAP reagent, and polyclonal antibodies against PfEF-1β, PfEF-1α, and Pfpmt. M, protein marker. Asterisks indicate degradation products. TEV, tobacco etch virus protease; M, protein marker; TE, total extract; TR, transgenic parasites; FT, flowthrough; TE-EL, eluate from IgG column; Pr.C, protein C; F-EL, final eluate from anti-ProtC column. About 0.28% (vol/vol) of the total protein extract (TR-TE), 1.0% of the TEV protease eluate (TE-EL), and 2.5% of the final EGTA eluate (F-EL) were loaded in the respective lanes.
Molecular determination of the components of the P. falciparum translation elongation factor complex.
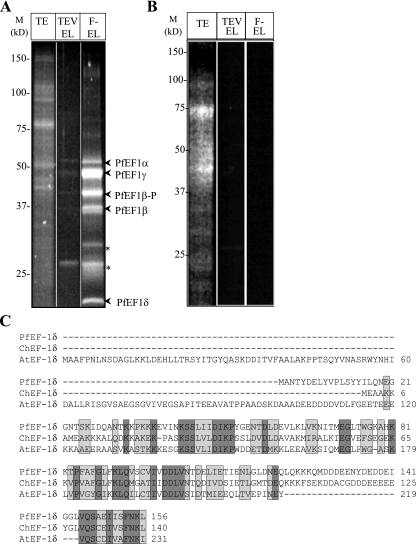
To demonstrate the utility of the method for mass spectrometric identification of proteins that are copurified with PfEF-1β, the proteins of the final eluate were separated by SDS-PAGE and visualized by UV transillumination using Sypro ruby, a highly sensitive fluorescent stain for proteins that is compatible with mass spectrometric analysis. Eight major protein bands could be detected in the final eluate (Fig. 3A). As a control, the same purification conducted with extracts from wild-type 3D7 parasites did not reveal any detectable parasite protein bands (Fig. 3B). For identification of the proteins detected in the Sypro ruby staining of the PfEF-1β purification, bands were excised, digested with trypsin, and subjected to liquid chromatography-tandem mass spectrometry. For each band, several peptides could be detected (Table 1). These were used to identify the corresponding proteins by screening against the available P. falciparum predicted proteins. Consistent with the immunoblotting data obtained using anti-PfEF-1β, anti-ProtC, and anti-PfEF-1α antibodies against the eluate, the tagged and native PfEF-1β (NCBI 23613651) as well as PfEF-1α (NCBI 3410705) were all identified in the purified complex. Interestingly, two additional proteins, PFC0870w and PF13_0214, that we named PfEF-1δ (NCBI 23957766) and PfEF-1γ (NCBI 23619320) were also identified as associated with PfEF-1β. PfEF-1δ is a 156-amino-acid polypeptide highly homologous to the δ subunit of the fungal and plant elongation factor complex (Fig. 3C). PfEF-1δ also shares 55% identity and 72% similarity with PfEF-1β, primarily in the predicted nucleotide exchange domain. PfEF-1γ is a 434-amino-acid polypeptide highly homologous to the γ subunit of the fungal and plant elongation factor complex (not shown). Additional bands detected in the stained gel were found to correspond to degradation forms of native and tagged PfEF-1β as confirmed by mass spectrometry and immunoblotting using anti-PfEF-1β and anti-ProtC antibodies (Fig. 1B and 2B). Together these data provide the first evidence that tandem affinity purification can be used to purify the complete translation elongation factor complex of P. falciparum. Interestingly, further analysis of the data revealed that of the 18 peptides of PfEF-1γ revealed by mass spectrometry, two peptides were found to be phosphorylated on serine 263 (Table 1).
FIG. 3.
(A and B) Sypro ruby staining of SDS-PAGE-resolved protein fractions obtained by tandem affinity purification of protein complexes from transgenic parasites expressing the PfEF-1β-PTP recombinant protein (A) and the wild-type 3D7 parasites (B). About 0.015% (vol/vol) of the total protein extract (TE), 0.5% of the TEV protease eluate (TEV-EL), and 80% of the final EGTA eluate (F-EL) were fractionated on a 12.5% SDS-PAGE gel. All the bands obtained in the F-EL column were excised and submitted for mass spectrometry analysis. (C) Sequence alignment of the polypeptide sequences of EF-1δ from P. falciparum (PfEF-1δ; accession no. NP_473308), Cryptosporidium hominis (ChEF-1δ; accession no. XP_665336), and Arabidopsis thaliana (AtEF-1δ; accession no. NP_174314). Sequence identity is indicated in dark gray, and similarity is indicated in light gray.
TABLE 1.
Liquid chromatography-tandem mass spectrometry-identified peptides of proteins purified by TAP of PfEF-1β
| Protein | Peptidea |
|---|---|
| PfEF-1α | QIVVGVNK |
| IGGIGTVPVGR | |
| STTTGHIIYK | |
| SGDSALVSLEPK | |
| GYVASDTKNEPAK | |
| YFFTVIDAPGHK | |
| QIVVGVNKMDTVK | |
| FTAQVIILNHPGEIK | |
| AGMVLNFAPSAVVSECK | |
| EVLEEARPGDNIGFNVK | |
| VDFIPISGFEGDNLIEK | |
| SVEMHKEVLEEARPGDNIGFNVK | |
| VGYQADKVDFIPISGFEGDNLIEK | |
| PfEF-1β-P | IYHQIK |
| IVDENIK | |
| SIDTDIAK | |
| DTYPHLFR | |
| QKIVDENIK | |
| DSASNLLLEK | |
| KLPVAFGLYK | |
| IENIDLDNEEDK | |
| IVDENIKWGEEVK | |
| AAGGDDDDDNDIDLFGDDDDNTK | |
| LHMSCIIYDDFVNTNELIEK | |
| AAGGDDDDDNDIDLFGDDDDNTKDSASNLLLEK | |
| PfEF-1β | IVDENIK |
| SIDTDIAK | |
| DTYPHLFR | |
| QKIVDENIK | |
| DSASNLLLEK | |
| SIDTDIAKIPK | |
| DSASNLLLEKK | |
| IENIDLDNEEDK | |
| IENIDLDNEEDKK | |
| AAGGDDDDDNDIDLFGDDDDNTKDSASNLLLEK | |
| PfEF-1γ | LENNFSK |
| HIEDTLK | |
| YLCSIHR | |
| LLAPKNDVR | |
| GTDIPFEMK | |
| LDINNTQDK | |
| LYDTISNQK | |
| YVFACDQNK | |
| LDINNTQDKK | |
| VQTVASFCNVK | |
| YVFACDQNKK | |
| ETVDNRPLVDR | |
| ETVDNRPLVDRK | |
| LNMPNFELGKDNK | |
| TYDPNGFSLYYMK | |
| GTDIPFEMKDHPSFEYHIFK | |
| DDNNNNNNNDADNQHADLLSDDLAEK | |
| DDNNNNNNNDADNQHADLLSDDLAEKK | |
| PfEF-1δ | TPFAFGLFK |
| SSLIIDIKPYGENTDLDEVLK |
The phosphorylated serine residues are underlined.
Gateway-based PTP tagging of P. falciparum proteins.
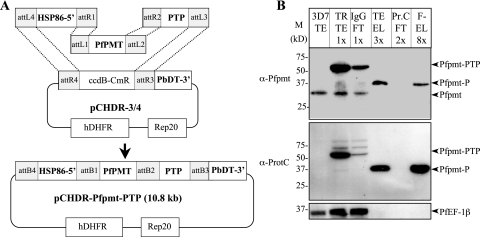
The results described above show that the PTP-based strategy can be used for purification and identification of protein complexes in P. falciparum. For rapid construction of targeting vectors harboring various P. falciparum open reading frames fused to PTP and to facilitate the large-scale purification of proteins under native conditions or the determination of protein networks in this parasite, we have developed a PTP entry vector compatible with the Gateway-based expression vectors reported by van Dooren and colleagues (32). As a proof of principle we used this strategy to construct a targeting vector harboring a fusion between the phosphoethanolamine methyltransferase open reading frame, PfPMT, and the PTP sequence (Fig. 4A) and used it to transfect P. falciparum 3D7 clone parasites. Transgenic parasites harboring this vector expressed the fusion protein, as was demonstrated using anti-ProtC and anti-Pfpmt antibodies (Fig. 4B). As shown by immunoblotting, the tandem affinity purification of Pfpmt was as efficient as the PfEF-1β purification (Fig. 4B). Only one protein was purified this time and it was unambiguously identified by mass spectrometry as Pfpmt (data not shown). Unlike PfEF-1β, the native Pfpmt did not copurify with the tagged Pfpmt (Fig. 4B), suggesting that under native conditions Pfpmt does not dimerize. This strategy would thus make it possible to biochemically characterize the native Pfpmt enzyme. Together, the combination of Gateway-based cloning and PTP tagging would ease and expedite the whole procedure of purification of protein complexes as well as proteins that otherwise cannot be expressed in heterologous systems for subsequent biochemical or cell biological characterization under their native states.
FIG. 4.
(A) Schematic representation of gene recombination strategy using the MultiSite Gateway system to clone the PfPMT open reading frame fused to PTP tag under the regulatory control of the P. falciparum HSP-86 promoter (HSP86-5′) and the P. chabaudi DHFR-TS terminator (PbDT-3′). (B) Immunoblot analysis of whole-cell lysates and tandem affinity-purified fractions of wild-type 3D7 and transgenic parasites expressing the Pfpmt-PTP protein fusion using a monoclonal antibody against protein C and polyclonal antibodies against Pfpmt and PfEF-1β (as control). Identified protein bands are indicated. TEV, tobacco etch virus protease; M, protein marker; TE, total extract; TR, transgenic parasites; FT, flowthrough; TE-EL, eluate from IgG column; Pr.C, protein C; F-EL, final eluate from anti-ProtC column. Asterisks indicate native protein degradation products. About 0.28% (vol/vol) of the total protein extract (TR-TE), 1.0% of the TEV protease eluate (TE-EL), and 2.5% of the final EGTA eluate (F-EL) were loaded in the respective lanes.
DISCUSSION
The major challenge of malaria biology following the decoding of several Plasmodium genomes has been to understand how a complex life cycle spent between two hosts, within different host cells, in different cellular locations, and under different morphological, physiological, and metabolic states is controlled by less than 6,000 proteins. Large-scale microarray and mass spectrometry analyses during different life cycles of P. falciparum have revealed a highly coordinated expression pattern in this parasite (1, 2, 7, 13, 14, 35). It remains, however, unknown how, at any given time during the parasite's life cycle, different proteins interact to accomplish their essential tasks. A large-scale two-hybrid screen using P. falciparum cDNA derived from mixed stages of the parasite intraerythrocytic life cycle has been reported (10). The pair-wise interactions reported have not been validated in P. falciparum, and their physiological relevance remains to be established, for most of the interactions were performed in a heterologous system (i.e., yeast) and involved fragments rather than full-length cDNAs (10). Furthermore, due to the mixed population of RNAs from different intraerythrocytic stages, some interactions may occur between different proteins in vitro, whereas in vivo these proteins do not coexist at the same time.
The strategy described herein provides a better way to study protein networks in the parasite and at any given time during any stage of the parasite life cycle. We have applied this strategy to the purification of the protein complex that includes the translation elongation factor PfEF-1β. Evidence for the presence of such a complex in P. falciparum was first demonstrated by radiolabeling of P. falciparum-infected erythrocytes followed by immunoprecipitation using anti-PfEF-1β and anti-PfEF-1α antibodies (16). PfEF-1β was found to coimmunoprecipitate with PfEF-1α and vice versa. Interestingly, additional proteins also coimmunoprecipitated with PfEF-1β and PfEF-1α; however, the identity of those proteins remained unknown (16). Employing tandem affinity purification and mass spectrometry, we were able to identify in addition to the tagged PfEF-1β, the native PfEF-1β, and three copurified proteins, PfEF-1α, PfEF-1γ, and PfEF-1δ, all homologs of known translation elongation factor subunits (15). These data suggest that the PTP-based strategy has resulted in the purification of the translation elongation factor EF-1 complex of the parasite. Furthermore, detailed characterization of the purified proteins revealed that serine 263 of PfEF-1γ is phosphorylated. This finding is consistent with a previous study using orthophosphate labeling that showed that some components of the EF-1 complex of P. falciparum are phosphorylated (16). The finding that the native PfEF-1β copurifies with the tagged PfEF-1β suggests that at least two PfEF-1β subunits are part of the translation elongation factor complex in P. falciparum. To the best of our knowledge, our study is the first of its nature to provide direct evidence that more than one subunit of EF-1β exist in the EF-1 complex.
Unlike human EF-1δ, none of the components of the malarial translation elongation complex contains leucine zipper domains. These domains have been proposed to play a role in the protein-protein interactions that define the ordered quaternary structure of the translation elongation complex (15). PfEF-1δ shares significant homology with the C-terminal domain of PfEF-1β, which is predicted to be responsible for the nucleotide exchange activity of the protein. Whereas PfEF-1β has been demonstrated to catalyze the GTP/GDP exchange on PfEF-1α, the function of PfEF-1δ as an exchange factor remains to be elucidated. Interestingly, while tagged and native PfEF-1β proteins as well as PfEF-1γ and PfEF-1δ concentrated in the final purified complex, only a fraction of PfEF-1α was found to associate with the translation elongation complex. This finding is consistent with previous studies that showed PfEF-1α as a monomeric protein as well as in association with high-molecular-weight protein aggregates (26) and with its well-defined cycle of association and dissociation and transient interactions with the other components of the translation elongation complex. Furthermore, besides its essential role in protein translation, EF-1α has also been shown to interact with tubulin and actin and to be involved in different cellular processes, such as organization of the cytoskeleton, cell differentiation, and apoptosis (for a review, see reference 12).
Unlike PfEF-1β, TAP analysis of Pfpmt did not reveal detectable copurified proteins. This might reflect an inherent biological property of this enzyme or might be due to low expression of Pfpmt-PTP, which was under the control of the HSP86 promoter. The latter hypothesis could be tested in future studies using the endogenous PfPMT promoter or a stronger promoter, such as the CAM promoter.
The highly efficient purification of P. falciparum proteins demonstrated in this study will set the stage for the purification of other important protein complexes and the identification of new proteins involved in important physiological functions during the parasite's life cycle. TAP tagging and purification may, for example, help better understand the mechanism of parasite invasion by TAP tagging components of the micronemes and rhoptries or the molecular determinants of protein trafficking to the red blood cell membrane, the food vacuole, or the apicoplast by tagging components of the Mauer's cleft, the food vacuole membrane, or the apicoplast membrane. This strategy may also be useful in the identification of components of the transcription complexes of the parasite as well as new regulators that specifically bind to specific promoters and regulate their expression during the parasite's life cycle. Our proposed strategy to combine the Gateway-based cloning and PTP tagging will make it possible to achieve these goals and will facilitate the large-scale functional analysis of the malarial proteome.
Acknowledgments
We are grateful to Harriett Zawistowski (General Clinical Research Center, University of Connecticut Health Center) for technical help and Mary Ann Gawinowicz (Protein Core Facility, Columbia University) for excellent mass spectometry analyses.
This research was supported by NIH and DOD grants AI51507, AI58962, PR033005, and BWF award 1006267 to C.B.M. and NIH grant AI059377 to A.G. UCHC General Clinical Research Center is supported by NIH grant M01RR06192. C.B.M. is a recipient of the Burroughs Wellcome Award, Investigators of Pathogenesis of Infectious Disease.
Footnotes
Published ahead of print on 16 February 2007.
REFERENCES
- 1.Ben Mamoun, C., I. Y. Gluzman, C. Hott, S. K. MacMillan, A. S. Amarakone, D. L. Anderson, J. M. Carlton, J. B. Dame, D. Chakrabarti, R. K. Martin, B. H. Brownstein, and D. E. Goldberg. 2001. Co-ordinated programme of gene expression during asexual intraerythrocytic development of the human malaria parasite Plasmodium falciparum revealed by microarray analysis. Mol. Microbiol. 39:26-36. [DOI] [PubMed] [Google Scholar]
- 2.Bozdech, Z., J. Zhu, M. P. Joachimiak, F. E. Cohen, B. Pulliam, and J. L. DeRisi. 2003. Expression profiling of the schizont and trophozoite stages of Plasmodium falciparum with a long-oligonucleotide microarray. Genome Biol. 4:R9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drakas, R., M. Prisco, and R. Baserga. 2005. A modified tandem affinity purification tag technique for the purification of protein complexes in mammalian cells. Proteomics 5:132-137. [DOI] [PubMed] [Google Scholar]
- 4.Duraisingh, M. T., T. S. Voss, A. J. Marty, M. F. Duffy, R. T. Good, J. K. Thompson, L. H. Freitas-Junior, A. Scherf, B. S. Crabb, and A. F. Cowman. 2005. Heterochromatin silencing and locus repositioning linked to regulation of virulence genes in Plasmodium falciparum. Cell 121:13-24. [DOI] [PubMed] [Google Scholar]
- 5.El Bissati, K., R. Zufferey, W. H. Witola, N. S. Carter, B. Ullman, and C. Ben Mamoun. 2006. The plasma membrane permease PfNT1 is essential for purine salvage in the human malaria parasite Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 103:9286-9291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fidock, D. A., T. Nomura, and T. E. Wellems. 1998. Cycloguanil and its parent compound proguanil demonstrate distinct activities against Plasmodium falciparum malaria parasites transformed with human dihydrofolate reductase. Mol. Pharmacol. 54:1140-1147. [DOI] [PubMed] [Google Scholar]
- 7.Florens, L., M. P. Washburn, J. D. Raine, R. M. Anthony, M. Grainger, J. D. Haynes, J. K. Moch, N. Muster, J. B. Sacci, D. L. Tabb, A. A. Witney, D. Wolters, Y. Wu, M. J. Gardner, A. A. Holder, R. E. Sinden, J. R. Yates, and D. J. Carucci. 2002. A proteomic view of the Plasmodium falciparum life cycle. Nature 419:520-526. [DOI] [PubMed] [Google Scholar]
- 8.Furuya, T., J. Mu, K. Hayton, A. Liu, J. Duan, L. Nkrumah, D. A. Joy, D. A. Fidock, H. Fujioka, A. B. Vaidya, T. E. Wellems, and X. Z. Su. 2005. Disruption of a Plasmodium falciparum gene linked to male sexual development causes early arrest in gametocytogenesis. Proc. Natl. Acad. Sci. USA 102:16813-16818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gardner, M. J., N. Hall, E. Fung, O. White, M. Berriman, R. W. Hyman, J. M. Carlton, A. Pain, K. E. Nelson, S. Bowman, I. T. Paulsen, K. James, J. A. Eisen, K. Rutherford, S. L. Salzberg, A. Craig, S. Kyes, M. S. Chan, V. Nene, S. J. Shallom, B. Suh, J. Peterson, S. Angiuoli, M. Pertea, J. Allen, J. Selengut, D. Haft, M. W. Mather, A. B. Vaidya, D. M. Martin, A. H. Fairlamb, M. J. Fraunholz, D. S. Roos, S. A. Ralph, G. I. McFadden, L. M. Cummings, G. M. Subramanian, C. Mungall, J. C. Venter, D. J. Carucci, S. L. Hoffman, C. Newbold, R. W. Davis, C. M. Fraser, and B. Barrell. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419:498-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.LaCount, D. J., M. Vignali, R. Chettier, A. Phansalkar, R. Bell, J. R. Hesselberth, L. W. Schoenfeld, I. Ota, S. Sahasrabudhe, C. Kurschner, S. Fields, and R. E. Hughes. 2005. A protein interaction network of the malaria parasite Plasmodium falciparum. Nature 438:103-107. [DOI] [PubMed] [Google Scholar]
- 11.Lai, Z., J. Jing, C. Aston, V. Clarke, J. Apodaca, E. T. Dimalanta, D. J. Carucci, M. J. Gardner, B. Mishra, T. S. Anantharaman, S. Paxia, S. L. Hoffman, J. Craig Venter, E. J. Huff, and D. C. Schwartz. 1999. A shotgun optical map of the entire Plasmodium falciparum genome. Nat. Genet. 23:309-313. [DOI] [PubMed] [Google Scholar]
- 12.Lamberti, A., M. Caraglia, O. Longo, M. Marra, A. Abbruzzese, and P. Arcari. 2004. The translation elongation factor 1A in tumorigenesis, signal transduction and apoptosis. Amino Acids 26:443-448. [DOI] [PubMed] [Google Scholar]
- 13.Le Roch, K. G., J. R. Johnson, L. Florens, Y. Zhou, A. Santrosyan, M. Grainger, S. F. Yan, K. C. Williamson, A. A. Holder, D. J. Carucci, J. R. Yates III, and E. A. Winzeler. 2004. Global analysis of transcript and protein levels across the Plasmodium falciparum life cycle. Genome Res. 14:2308-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le Roch, K. G., Y. Zhou, P. L. Blair, M. Grainger, J. K. Moch, J. D. Haynes, P. De La Vega, A. A. Holder, S. Batalov, D. J. Carucci, and E. A. Winzeler. 2003. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science 301:1503-1508. [DOI] [PubMed] [Google Scholar]
- 15.Le Sourd, F., S. Boulben, R. Le Bouffant, P. Cormier, J. Morales, R. Belle, and O. Mulner-Lorillon. 2006. eEF1B: at the dawn of the 21st century. Biochim. Biophys. Acta 1759:13-31. [DOI] [PubMed] [Google Scholar]
- 16.Mamoun, C. B., and D. E. Goldberg. 2001. Plasmodium protein phosphatase 2C dephosphorylates translation elongation factor 1β and inhibits its PKC-mediated nucleotide exchange activity in vitro. Mol. Microbiol. 39:973-981. [DOI] [PubMed] [Google Scholar]
- 17.Nguyen, T. N., B. Schimanski, A. Zahn, B. Klumpp, and A. Günzl. 2006. Purification of an eight subunit RNA polymerase I complex in Trypanosoma brucei. Mol. Biochem. Parasitol. 149:27-37. [DOI] [PubMed] [Google Scholar]
- 18.Omara-Opyene, A. L., P. A. Moura, C. R. Sulsona, J. A. Bonilla, C. A. Yowell, H. Fujioka, D. A. Fidock, and J. B. Dame. 2004. Genetic disruption of the Plasmodium falciparum digestive vacuole plasmepsins demonstrates their functional redundancy. J. Biol. Chem. 279:54088-54096. [DOI] [PubMed] [Google Scholar]
- 19.Palfi, Z., B. Schimanski, A. Günzl, S. Lucke, and A. Bindereif. 2005. U1 small nuclear RNP from Trypanosoma brucei: a minimal U1 snRNA with unusual protein components. Nucleic Acids Res. 33:2493-2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pessi, G., and C. Ben Mamoun. 2006. Pathways for phosphatidylcholine biosynthesis: targets and strategies for antimalarial drugs. Future Med. Future Lipidol. 1:173-180. [Google Scholar]
- 21.Pessi, G., J. Y. Choi, J. M. Reynolds, D. R. Voelker, and C. B. Mamoun. 2005. In vivo evidence for the specificity of Plasmodium falciparum phosphoethanolamine methyltransferase and its coupling to the Kennedy pathway. J. Biol. Chem. 280:12461-12466. [DOI] [PubMed] [Google Scholar]
- 22.Pessi, G., G. Kociubinski, and C. B. Mamoun. 2004. A pathway for phosphatidylcholine biosynthesis in Plasmodium falciparum involving phosphoethanolamine methylation. Proc. Natl. Acad. Sci. USA 101:6206-6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Puig, O., F. Caspary, G. Rigaut, B. Rutz, E. Bouveret, E. Bragado-Nilsson, M. Wilm, and B. Seraphin. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24:218-229. [DOI] [PubMed] [Google Scholar]
- 24.Rager, N., C. B. Mamoun, N. S. Carter, D. E. Goldberg, and B. Ullman. 2001. Localization of the Plasmodium falciparum PfNT1 nucleoside transporter to the parasite plasma membrane. J. Biol. Chem. 276:41095-41099. [DOI] [PubMed] [Google Scholar]
- 25.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 26.Santiago, T. C., R. Zufferey, R. S. Mehra, R. A. Coleman, and C. Ben Mamoun. 2004. The Plasmodium falciparum PfGatp is an endoplasmic reticulum membrane protein important for the initial step of malarial glycerolipid synthesis. J. Biol. Chem. 279:9222-9232. [DOI] [PubMed] [Google Scholar]
- 27.Schimanski, B., T. N. Nguyen, and A. Günzl. 2005. Characterization of a multisubunit transcription factor complex essential for spliced-leader RNA gene transcription in Trypanosoma brucei. Mol. Cell. Biol. 25:7303-7313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schimanski, B., T. N. Nguyen, and A. Günzl. 2005. Highly efficient tandem affinity purification of trypanosome protein complexes based on a novel epitope combination. Eukaryot. Cell 4:1942-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sijwali, P. S., K. Kato, K. B. Seydel, J. Gut, J. Lehman, M. Klemba, D. E. Goldberg, L. H. Miller, and P. J. Rosenthal. 2004. Plasmodium falciparum cysteine protease falcipain-1 is not essential in erythrocytic stage malaria parasites. Proc. Natl. Acad. Sci. USA 101:8721-8726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sijwali, P. S., J. Koo, N. Singh, and P. J. Rosenthal. 2006. Gene disruptions demonstrate independent roles for the four falcipain cysteine proteases of Plasmodium falciparum. Mol. Biochem. Parasitol. 150:96-106. [DOI] [PubMed] [Google Scholar]
- 31.Sijwali, P. S., and P. J. Rosenthal. 2004. Gene disruption confirms a critical role for the cysteine protease falcipain-2 in hemoglobin hydrolysis by Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 101:4384-4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Dooren, G. G., M. Marti, C. J. Tonkin, L. M. Stimmler, A. F. Cowman, and G. I. McFadden. 2005. Development of the endoplasmic reticulum, mitochondrion and apicoplast during the asexual life cycle of Plasmodium falciparum. Mol. Microbiol. 57:405-419. [DOI] [PubMed] [Google Scholar]
- 33.Witola, W. H., G. Pessi, K. El Bissati, J. M. Reynolds, and C. B. Mamoun. 2006. Localization of the phosphoethanolamine methyltransferase of the human malaria parasite Plasmodium falciparum to the Golgi apparatus. J. Biol. Chem. 281:21305-21311. [DOI] [PubMed] [Google Scholar]
- 34.World Health Organization. 2000. Report of the WHO Expert Committee on Malaria. WHO Tech. Rep. Ser. 892:1-74. [PubMed] [Google Scholar]
- 35.Young, J. A., Q. L. Fivelman, P. L. Blair, P. de la Vega, K. G. Le Roch, Y. Zhou, D. J. Carucci, D. A. Baker, and E. A. Winzeler. 2005. The Plasmodium falciparum sexual development transcriptome: a microarray analysis using ontology-based pattern identification. Mol. Biochem. Parasitol. 143:67-79. [DOI] [PubMed] [Google Scholar]