Abstract
The principal capsular polysaccharide of the opportunistic fungal pathogen Cryptococcus neoformans consists of an α-1,3-linked mannose backbone decorated with a repeating pattern of glucuronyl and xylosyl side groups. This structure is critical for virulence, yet little is known about how the polymer, called glucuronoxylomannan (GXM), is faithfully synthesized and assembled. We have generated deletions in two genes encoding predicted parallel β-helix repeat proteins, which we have designated PBX1 and PBX2. Deletion of either gene results in a dry-colony morphology, clumpy cells, and decreased capsule integrity. Two-dimensional nuclear magnetic resonance spectroscopy of purified GXM from the mutants indicated that both the wild-type GXM structure and novel, aberrant linkages were present. Carbohydrate composition and linkage analysis determined that these aberrant structures are correlated with the incorporation of terminal glucose residues that are not found in wild-type capsule polysaccharide. We conclude that Pbx1 and Pbx2 are required for the fidelity of GXM synthesis and may be involved in editing incorrectly added glucose residues. PBX1 and PBX2 knockout mutants showed severely attenuated virulence in a murine inhalation model of cryptococcosis. Unlike acapsular strains, these mutant strains induced delayed symptoms of cryptococcosis, though the infected animals eventually contained the infection and recovered.
Cryptococcus neoformans is the most common systemic fungal infection of individuals infected with human immunodeficiency virus and is estimated to cause 13 to 44% of all AIDS-related deaths in sub-saharan Africa (12, 15, 27). This basidiomycetous yeast exists in four serotypes (A to D). Serotype A strains, in particular, account for >90% of all human cases of cryptococcosis (1, 5, 22). A distinctive feature of this pathogen is the polysaccharide capsule that surrounds the yeast. C. neoformans is the only fungal pathogen of humans that is encapsulated, and this capsule has been shown to be essential for virulence. June Kwon-Chung's group cloned four capsule-associated genes (CAP10, CAP59, CAP60, and CAP64). Deletion of any of these genes resulted in an acapsular phenotype and loss of virulence in animal models of cryptococcosis (6-9). Other studies primarily using cell culture systems have demonstrated antiphagocytic and immunomodulatory properties of the capsule, including inhibition of T-cell activation, modulation of cytokine secretion, and sequestration of complement (4, 32, 34).
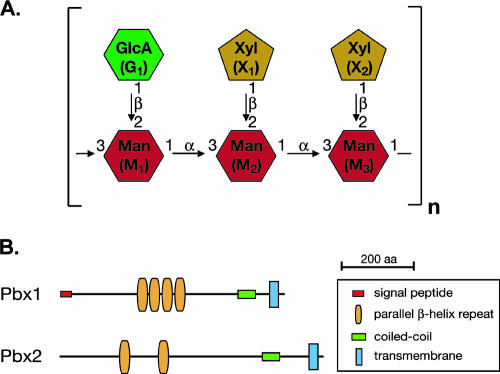
The capsule is composed of two high-molecular-weight polysaccharides, glucuronoxylomannan (GXM) and galactoxylomannan, as well as mannoproteins (3). GXM accounts for most of the capsule mass (∼88%), and differences in the structure of the GXM polymer serve as the antigenic basis for the C. neoformans serotype classification system (21). GXM consists of a backbone of α-1,3-linked mannose residues decorated with xylosyl and glucuronyl side groups. A fraction of the mannose residues are also 6-O-acetylated. Nuclear magnetic resonance (NMR) spectroscopic analysis of GXM from serotypes A to D determined that GXMs from these strains differ in the sites of xylosylation and acetylation. For serotype A, the GXM polymer is composed primarily of a repeating mannose triplet (α-1,3 linked) with glucuronic acid 1→2 linked to the first mannose of the triplet and xylose 1→2 linked to the second and third mannoses (Fig. 1A) (30).
FIG. 1.
(A) Structure of the GXM of serotype A C. neoformans. Not shown are the 6-O-acetyl groups that modify a portion of the mannose residues. (B) Protein domains as predicted by SMART (http://smart.embl-heidelberg.de) for Pbx1 and Pbx2.
Given the capsule's importance in pathogenesis, the proteins involved in the biosynthesis of GXM have been targets of much research. To synthesize GXM, activated nucleotide-sugar precursors (i.e., GDP-mannose, UDP-xylose, and UDP-glucuronic acid) are needed to serve as donors for the GXM polymer. These precursors must then be properly assembled and linked into GXM polymers. By reverse genetic approaches, some of the metabolic enzymes needed to generate these precursors, including the phosphomannose isomerase (Man1), the UDP-glucuronate decarboxylase (Uxs1), and the UDP-glucose dehydrogenase (Ugd1), have been identified and shown to be required for proper GXM synthesis (2, 17, 25, 26, 33). In contrast, the transferases that act directly to catalyze the assembly of GXM have remained elusive.
The assembly of GXM poses an interesting biological problem. Unlike DNA, mRNA, and protein synthesis, polysaccharide synthesis is not templated by the genome. Instead, polysaccharide linkages are determined by the specificity of biosynthetic enzymes. At a minimum, the enzymes involved in the assembly of GXM polymers must accomplish two tasks. First, specific enzymes must create each of the linkages found in the polymer. Second, these enzymes must generate the linkages in the proper order such that the repeating pattern of one glucuronic acid followed by two xyloses linked to three consecutive mannose residues in the backbone is maintained. There is increasing evidence that capsular polysaccharides are synthesized intracellularly (16, 35). These intracellular compartments may contain active glycosyltransferases and noncognate nucleotide sugars such as UDP-glucose that are not needed for capsule polymer synthesis. Thus, it might be predicted that a third function of the enzymes involved in capsule biosynthesis would be to either preclude incorrect sugars from being added to the GXM polymer or cleave them from the polymer as part of a proofreading mechanism.
As mentioned earlier, the sugar transferases that synthesize GXM have yet to be identified. Thus far, an α-1-3-mannosyltransferase, Cmt1, has been purified, but mutants lacking this protein display no defect in capsule synthesis (31). Proteins involved in the second task, targeting of these linkages to the mannose backbone, have been identified in genetic screens for mutants with altered binding to anticapsule antibodies (18, 24, 26). Structural analysis of GXMs from these mutant strains indicated that Cas1 and the Cas3 family of proteins play a role in determining the position and the linkage of the xylose and/or the O-acetyl residues on the mannose backbone. However, whether these proteins are themselves sugar transferases is unknown. Enzymes involved in the third task of preventing the incorporation of incorrect sugars have not been identified.
In this study, we isolated two mutant deletion strains that display a dry-colony morphology, clumpy cells, and defects in capsule integrity. Interestingly, the affected genes, which we have named PBX1 and PBX2, are predicted to encode parallel β-helix domains, which have primarily been found in pectin lyases, polygalactouronases, endorhamnosidases, and other glycosidases (19). Using NMR spectroscopy, carbohydrate composition, and carbohydrate linkage analyses, we have found that a portion of the GXM in these mutants is incorrectly assembled. In particular, novel terminal glucose linkages are present in the mutant capsules, demonstrating a role for Pbx1 and Pbx2 in preventing the incorporation of glucose residues onto the elongating GXM chain and ensuring the fidelity of GXM synthesis. Both mutants were severely attenuated for virulence in a murine inhalation model of cryptococcosis.
MATERIALS AND METHODS
Strains and media.
C. neoformans serotype A strain H99 (a gift from J. K. Lodge) was used as the wild type. Strains were routinely grown in rich medium, YPAD (1% yeast extract, 2% Bacto peptone, 2% glucose, 0.015% l-tryptophan, 0.004% adenine), or minimal medium, YNB (0.45% yeast nitrogen base without amino acids and without ammonium sulfate, 1.5% ammonium sulfate, 2% glucose). For a list of the strains used in this study, see Table S1 in the supplemental material. Doubling times in YNB were calculated by measuring the change in optical density at 600 nm during exponential growth.
Generation of knockout strains.
The wild-type H99 strain was transformed by biolistic techniques as previously described (13). The gene-specific deletion constructs were generated (for the primers used, see Table S2 in the supplemental material) by overlap fusion PCR. Specifically, the nourseothricin resistance (natR) cassette was amplified from plasmids derived from pHL001 with the primers 5′NAT-10 and 3′NAT-10. This cassette was targeted to delete specific genes by fusing the cassette to the genomic sequences flanking the target gene. These flanking sequences were amplified from H99 genomic DNA by PCR with the ***-W1 and ***-W3 primers to amplify an ∼1-kb region 5′ to the target gene and the ***-W4 and ***-W6 primers to amplify an ∼1-kb region 3′ to the target gene (with the asterisks corresponding to the specific gene identifications described below and in Table S2 in the supplemental material). The two flanking regions and the natR cassette were fused into one product by PCR and biolistically transformed. Transformants were selected on YPAD agar plates containing 100 μg/ml nourseothricin. CM104 was generated with the CN966 set of primers and lacks a portion of PBX1 including the parallel β-helices. CM106 and CM109 were generated with the CDS_873 set of primers and lack all of PBX1. CM106 and CM109 were isolated from independent biolistic transformation experiments (i.e., two separate cultures transformed at different times). CM105 was generated with the CN4654 set of primers and lacks a portion of PBX2 including the parallel β-helices. CM107 and CM110 were generated with the CDS_5151 set of primers and lack all of PBX2. CM107 and CM110 were also isolated from independent biolistic transformation experiments. cap59Δ was generated with the CN605 set of primers. The expected recombination events were verified by PCR with the ***-V5 and VER-5-3 primers to detect the 5′ integration event and the ***-V3 and VER3-2 primers to detect the 3′ integration event. The pbx1Δ pbx2Δ double mutant was generated by using a hygromycin resistance (hygR) cassette which was amplified from plasmid pHYG7-KB1 with primers 5′HYG-10 and 3′HYG-10. The hygR cassette was fused to flanks generated with the CDS_5151 set of primers. This knockout construct was then transformed into CM106. Transformants were selected on YPAD agar plates containing 300 μg/ml hygromycin. For the predicted coding sequences of PBX1 and PBX2, see Fig. S2 in the supplemental material.
Capsule induction and India ink assay.
C. neoformans strains were grown in liquid YPAD cultures overnight at 30°C. As previously described (36), the cultures were then diluted 1/100 in either Sabouraud medium and grown for 1 to 2 days at 30°C (noninducing conditions) or 10% Sabouraud medium buffered to pH 7.3 with 50 mM morpholinepropanesulfonic acid (MOPS) and grown at 37°C for 1 to 2 days (capsule-inducing conditions). Equal volumes of culture and India ink were mixed, and the capsule was visualized by bright-field microscopy. For sonication experiments, 1 ml of the induced culture was first placed in a microcentrifuge tube. The cells were then pulsed 10 times with a Sonifier 450 (Branson) equipped with a microtip at a power setting of 6 and a duty cycle of 50%. The capsules were then visualized as described above.
Polysaccharide purification.
Purified exopolysaccharide was obtained by a protocol adapted from previously described methods (11, 30). C. neoformans strains were grown in 100 ml of liquid YNB medium for 5 days. The culture was then autoclaved at 121°C for 15 min, and the cells were removed by centrifugation at 12,000 × g for 15 min. The supernatant was transferred to a new container, and polysaccharide was precipitated by slow addition of 3 volumes of 95% ethanol (∼300 ml). The mixture was stored overnight at 4°C. The precipitate was recovered by centrifugation at 15,000 × g for 1 h. The supernatant was decanted, and the pellet was resuspended in 0.2 M NaCl to a final concentration of 10 mg/ml (∼100 ml). Once dissolved, the solution was sonicated at 4°C with a Sonifier 450 (Branson) equipped with a microtip for 15 min at a power setting of 2 and a duty cycle of 40%. Three milligrams of cetyltrimethylammonium bromide (CTAB) per milligram of precipitate (∼3 g of CTAB) was slowly added with stirring at room temperature. When the CTAB was completely dissolved, 2.5 volumes (∼250 ml) of 0.05% CTAB was slowly added with stirring. The precipitate was recovered by centrifugation at 18,000 × g for 2 h. The supernatant was decanted, and the pellet was washed with 100 ml of 10% ethanol and centrifuged at 18,000 × g for 20 min. The supernatant was decanted, and the pellet was resuspended in 50 ml of 1 M NaCl. The solution was sonicated at 4°C for 2 h at a power setting of 2 and a 40% duty cycle. The solution was then dialyzed (3,500-Da cutoff; Fisher) versus distilled water for 5 to 6 days at 4°C, and the water was replaced multiple times daily. Any precipitate was removed by centrifugation, and the supernatant was lyophilized.
NMR spectroscopy.
Before NMR analysis, polysaccharide samples were exchanged twice in 99.96% D2O (Sigma) and lyophilized. The samples were then dissolved in 0.6 ml of 99.96% D2O and transferred to 5-mm NMR tubes. NMR spectra were acquired at 75°C with a Varian Inova spectrometer operating at 600 MHz and equipped with a 5-mm triple-resonance probe with an actively shielded z-gradient coil. Two-dimensional (2D) gradient-enhanced 13C, 1H heteronuclear single quantum correlation (HSQC) spectra (20) were recorded with 256* t1 and 512* t2 complex points and spectral widths of 8,000 and 3,000 Hz, respectively. The data were acquired with a recycle delay of 1 s and 256 scans per free induction decay. Decoupling of 13C during acquisition was achieved by GARP-1. Data were processed with nmrPipe/nmrDraw software (14). A Lorentzian-to-Gaussian function (lb = −0.5, gb = 0.05) was applied in t2 and sine-bell function shifted by π/2 to the t1 time domain.
Carbohydrate composition and linkage analysis.
Carbohydrate composition and linkage analysis was performed at the Glycotechnology Core Resource at the University of California, San Diego (http://glycotech.ucsd.edu). Composition analysis was done by high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD) after acid hydrolysis with trifluoroacetic acid. Linkage analysis was done by gas-liquid chromatography-electron ionization mass spectrometry (GLC-EIMS) of partially methylated alditol acetate derivatives.
Virulence studies.
C. neoformans strains were individually grown in liquid YPD cultures overnight at 30°C. Cells were counted with a hemacytometer, washed twice in phosphate-buffered saline, and resuspended in phosphate-buffered saline to a final concentration of 1 × 107 cells/ml. Five- to 6-week-old female A/J (NCI) mice were anesthetized by intraperitoneal injection of ketamine (75 mg/kg) and medetomidine (0.5 to 1.0 mg/kg). The mice were then suspended from a silk thread by their front incisors, and 50 μl of the inoculum (5 × 105 cells) was slowly pipetted into the nares. After 10 min, the mice were lowered and the anesthesia was reversed by intraperitoneal injection of atipamezole (1.0 to 2.5 mg/kg). Eight to 10 mice were infected per inoculum. The concentration of cells in the inoculum was confirmed by plating serial dilutions. Mice were monitored several times a week until onset of symptoms (weight loss, ruffled fur, abnormal gait) and then monitored daily. Mice that displayed signs of severe morbidity (weight loss, abnormal gait, hunched posture, swelling of the cranium) were sacrificed by CO2 inhalation followed by cervical dislocation. This protocol was reviewed and approved by the University of California at San Francisco Institutional Animal Care and Use Committee.
RESULTS
Deletion of genes encoding predicted parallel β-helix proteins causes a defect in capsule stability.
During the generation of knockout strains as part of an ongoing C. neoformans gene deletion project, we identified two mutants, designated CM104 and CM105, that displayed a dry-colony appearance compared to the parental H99 strain when grown on YPAD plates. H99 is a serotype A human clinical isolate. CM104 contains a partial deletion of CNAG_01172.1 (as assigned by the C. neoformans serotype A genome sequencing project at the Broad Institute [http://www.broad.mit.edu/annotation/genome/cryptococcus_neoformans/Home.html]), which we designated PBX1. CM105 contains a partial deletion of CNAG_05562.1, which we designated PBX2. By using gene models from an earlier genome annotation effort (E. D. Chow et al., unpublished data; see Fig. S2 in the supplemental material) and the SMART motif database resource at EMBL (http://smart.embl-heidelberg.de/), we found that both genes are predicted to encode a transmembrane domain, a coiled-coil segment, and parallel β-helix (PbH1) repeats. Pbx1 is also predicted to contain a signal peptide (Fig. 1B). BLAST comparison of Pbx1 against the nonredundant protein database identified only two predicted proteins in other species with significant similarity. The first (E = 5e-48) is a hypothetical protein in Coprinus cinereus (EAU93625) that is predicted by SMART to contain PbH1 repeats and a transmembrane domain. The second (E = 1e-39) is a hypothetical protein in Neurospora crassa (XP_958164) that is predicted by SMART to contain PbH1 repeats, a B lectin domain, and a signal peptide. BLAST comparison of Pbx2 identified the same two proteins.
To rule out the possibility that a second, unknown mutation in these original knockout strains caused the dry-colony phenotype, we constructed new strains containing a complete deletion of either PBX1 (CM106) or PBX2 (CM107). Both of the new knockout strains displayed a dry-colony phenotype, indicating that mutations of these genes were responsible for the dull colony appearance. The growth rates of these complete-knockout strains (CM106 and CM107) in minimal medium were measured at both 30°C and 37°C. At 30°C, the doubling times of the pbx1Δ (2.20 ± 0.04 h) and pbx2Δ (2.28 ± 0.07 h) mutant strains were approximately equal to that of the wild type (2.17 ± 0.05 h). At 37°C, both the pbx1Δ (3.10 ± 0.17 h) and pbx2Δ (3.42 ± 0.15 h) mutant strains displayed a growth defect compared to the wild type (2.64 ± 0.06 h), indicating an increased requirement for these gene products at 37°C.
Analysis of the C. neoformans serotype A genome indicated that there is a third PbH1-containing gene, CNAG_01275.1, which has similarity to Pbx1 (31% amino acid identity) and Pbx2 (28% amino acid identity) but does not appear to encode a transmembrane domain. Deletion of CNAG_01275.1 did not cause a dry-colony phenotype, and the mutant appeared identical to the wild type in the capsule analyses described below (data not shown).
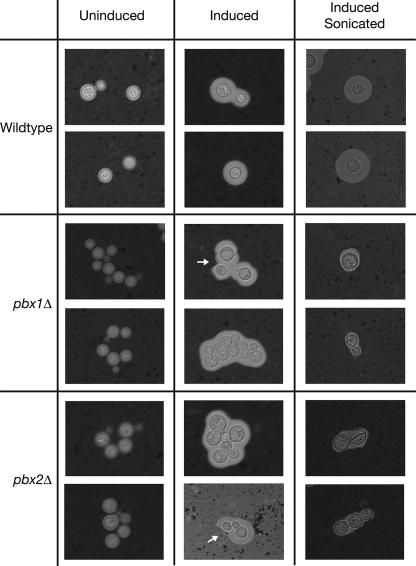
A dry phenotype is characteristic of most known C. neoformans capsule mutants where the polysaccharide capsule is missing or reduced (17, 24, 25, 33). As a result, we tested the mutants for the presence of capsule by India ink staining of strains grown in capsule-inducing medium (36). In wild-type strains, the polysaccharide capsule excludes India ink particles, creating a characteristic halo around an encapsulated cell which can be visualized by bright-field light microscopy.
In these experiments, pbx1Δ and pbx2Δ mutants (CM106 and CM107) were observed to grow in clumps in liquid under both inducing and noninducing conditions, a characteristic common to many hypocapsular and acapsular mutants (17, 24, 25, 33). However, under inducing conditions, sizable halos were still observed around the pbx1Δ and pbx2Δ strains, demonstrating that these strains do produce a capsule (Fig. 2). India ink staining of the original partial deletion strains (CM104 and CM105) yielded similar phenotypes (data not shown). While the clumping of cells makes measurement of capsule size difficult in these strains, the capsules surrounding the pbx1Δ and pbx2Δ strains did not appear significantly smaller than that of the wild type. Interestingly, however, while H99 capsules typically were round and symmetrical, we observed many examples of mutant capsules that were extremely asymmetric (Fig. 2, white arrows). These capsules may have been partially disassembled or become unanchored after synthesis, or portions of these capsules may be more permeable to India ink particles. Thus, while the pbx1Δ and pbx2Δ mutants are able to synthesize and assemble a capsule, the structural integrity and stability of these mutant capsules appear compromised. In support of this idea, we observed that mutant capsules were virtually abolished after a short sonication treatment while H99 capsules remained intact (Fig. 2). A single pbx1Δ pbx2Δ double mutant (CM084) was analyzed and found to be indistinguishable from the single mutants in these experiments (data not shown).
FIG. 2.
PBX1 and PBX2 mutants produce capsules with decreased integrity. Cells were grown under noninducing conditions (Uninduced), grown under capsule-inducing conditions (Induced), or grown under capsule-inducing conditions and then sonicated (Induced Sonicated). Capsules were visualized by India ink staining and bright-field light microscopy. White arrows point out regions of asymmetry in the mutant capsules. All images were taken at ×160 magnification. Shown are images of the complete-gene deletion strains CM106 (pbx1Δ) and CM107 (pbx2Δ). The original partial-deletion strains CM104 (pbx1Δ) and CM105 (pbx2Δ) displayed similar phenotypes (data not shown). A single double-mutant strain isolate was examined.
NMR analysis of the polysaccharide structure indicated a heterogeneous capsule.
Both PBX1 and PBX2 are predicted to encode parallel β-helix repeats. This structure has been found primarily in enzymes that utilize polysaccharide substrates, such as pectin lyases, polygalactouronases, endorhamnosidases, and other glycosidases (19). The presence of parallel β-helix repeats, as well as transmembrane domains, suggests that these proteins may be directly involved in the synthesis of capsule polymers. We hypothesized then that, without PBX1 or PBX2, the structure of the capsule polysaccharide would be altered in these mutants, resulting in the observed phenotypes. To investigate this possibility, we used 2D nuclear magnetic resonance (NMR) spectroscopy to analyze the structure of the polysaccharide.
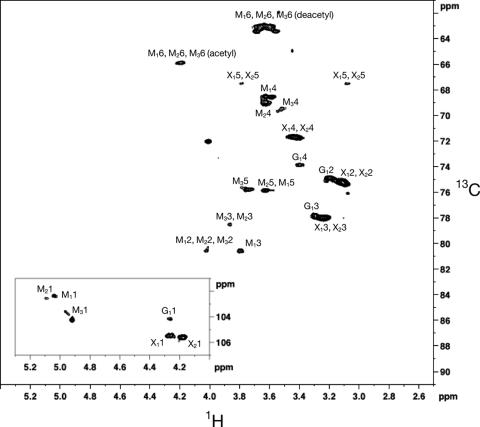
C. neoformans continuously sheds capsular material into the surrounding medium. This material, called the exopolysaccharide, has been purified and analyzed in previous structural studies in order to determine the composition and structure of GXM (24). We purified the exopolysaccharide from H99, pbx1Δ (CM106), pbx2Δ (CM107), and the pbx1Δ pbx2Δ double mutant (CM084) and performed 2D NMR. The 2D 1H, 13C HSQC spectra of H99 were consistent with previous NMR analysis used to determine the structure of GXM from a serotype A strain (Fig. 3). By comparison to the assignments made in this previous study, we were able to assign each cross-peak detected in the H99 spectra to the one glucuronic acid, two xylose, and three mannose residues that form the predominant repeating unit of the C. neoformans serotype A GXM (as shown in Fig. 1A) (30).
FIG. 3.
2D 1H, 13C HSQC spectra of the wild type. The assignments in the spectra are named by the following convention. The first letter and subscript indicate the specific residue as identified in Fig. 1A. The second number is the number of the carbon or hydrogen atom in the respective sugar rings. Assignments were based on comparison to previous work (24).
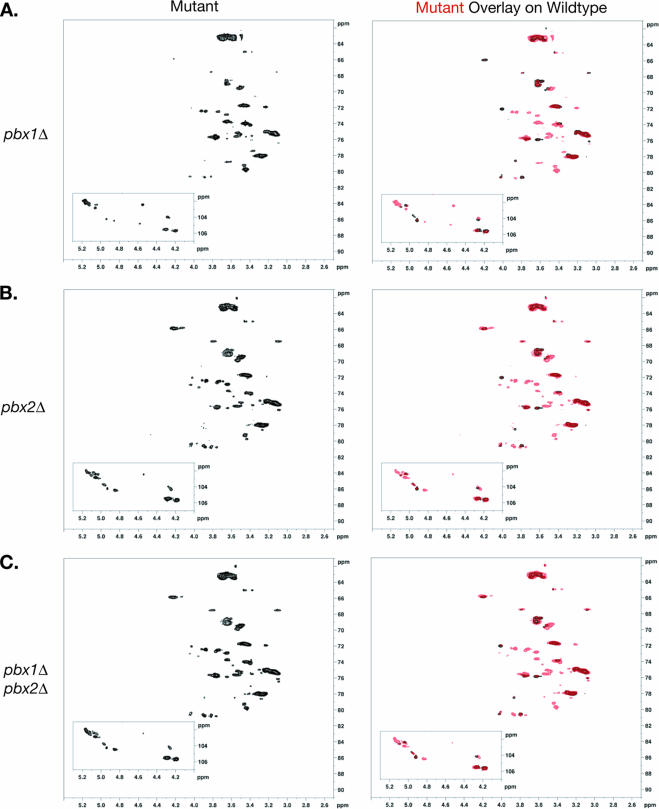
When purified exopolysaccharide from the pbx1Δ strain was analyzed by NMR, two features of the spectra were readily apparent. First, without exception, all of the cross-peaks observed in the H99 spectra were also present in the pbx1Δ strain spectra. Second, more than a dozen additional peaks appeared in the pbx1Δ strain spectra (Fig. 4A). These new signals primarily had 1H and 13C chemical shifts of 3.5 to 4.2 ppm and 72 to 79 ppm, respectively. Novel peaks were also observed in the region of 4.6 to 5 ppm and 98 to 104 ppm. Both of these ranges are consistent with the cross-peaks expected from carbohydrates. Given the presence of all of the cross-peaks observed in the H99 spectra, we concluded that at least a portion of the polysaccharide purified from the pbx1Δ strain maintained the wild-type structure. However, the detection of these additional new signals demonstrated that a significant portion of the mutant polysaccharide consisted of aberrant structures.
FIG. 4.
The pbx1Δ and pbx2Δ strain exopolysaccharides contain both the wild-type structure and aberrant structures. Exopolysaccharides from (A) pbx1Δ strain CM106, (B) pbx2Δ strain CM107, and (C) pbx1Δ pbx2Δ strain CM084 were purified, and 2D [1H,13C] HSQC spectra were acquired (left). Each mutant strain spectrum, in red, was overlaid on that of the wild-type strain, in black, for comparison (right). Similar experiments with exopolysaccharides purified from the original partial-deletion strains CM104 (pbx1Δ) and CM105 (pbx2Δ) resulted in similar patterns of cross-peaks (data not shown). Exopolysaccharide from a single double-mutant strain was analyzed.
The NMR spectra of purified exopolysaccharide from the pbx2Δ strain were examined. Again, all of the cross-peaks assigned to the wild-type structure were present in the pbx2Δ strain spectra, as were cross-peaks not observed in the H99 spectra (Fig. 4B). Strikingly, these new shifts were almost identical in the pbx1Δ and pbx2Δ strain spectra. The pbx1Δ strain spectra contained one additional signal at 3.6 ppm and 78.5 ppm. The pbx2Δ strain spectra contained two minor cross-peaks around 3.9 to 4.0 ppm and 73 ppm which were not observed in the pbx1Δ strain spectra. Aside from these small differences, all of the new cross-peaks identified in the pbx1Δ strain spectra were also detected in the pbx2Δ strain spectra and vice versa. Exopolysaccharide from the double mutant was also subjected to NMR analysis. The spectra from the pbx1Δ pbx2Δ double mutant were identical to those of the pbx2Δ mutant (Fig. 4C). Similar experiments were performed with the original partial deletion strains, CM104 (pbx1Δ) and CM105 (pbx2Δ), and similar patterns of cross-peaks were detected (data not shown). Exopolysaccharide from only one double-mutant strain was analyzed by NMR.
Taken together, these data demonstrate that Pbx1 and Pbx2 are not absolutely required to properly synthesize the capsule polymers. Much of the exopolysaccharide in the pbx1Δ and pbx2Δ mutants maintained the correct structure, as indicated by the presence of the expected cross-peaks. However, the detection of aberrant structures suggested that the portion of the polysaccharide having the wild-type structure in these mutants was reduced. These results are consistent with a model where Pbx1 and Pbx2 are not required for the synthesis of the capsule polysaccharide per se but, instead, are involved in the fidelity of the process.
Terminal glucose is present in mutant exopolysaccharide.
The novel cross-peaks observed in the NMR spectra of the mutant GXM could result from two possible changes in the capsule structure. First, the known components of GXM, glucuronic acid, xylose, and mannose, may be arranged or linked in unexpected structures. Second, novel residues, such as other sugars, may have been incorrectly added to the capsule polymer. To distinguish between these possibilities, we analyzed the composition of the mutant capsule polysaccharide by HPAEC-PAD.
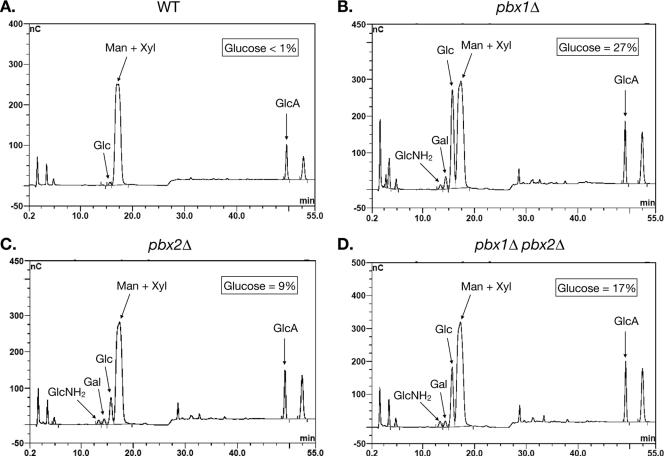
We again purified exopolysaccharide from H99, pbx1Δ strain CM106, pbx2Δ strain CM107, and the pbx1Δ pbx2Δ double mutant CM084. Carbohydrate composition analysis was performed by HPAEC-PAD after acid hydrolysis with trifluoroacetic acid. As expected, the H99 exopolysaccharide was composed almost exclusively of mannose, xylose, and glucuronic acid (Fig. 5A). A trace amount of glucose (<1%) was detected in the sample. Mannose, xylose, and glucuronic acid also formed most of the pbx1Δ strain exopolysaccharide, consistent with the NMR data which indicated that the wild-type GXM structure is present in the mutant exopolysaccharide. However, mannose, xylose, and glucuronic acid were not the only carbohydrates present in the material from the pbx1Δ mutant. Three additional sugars were detectable. Most dramatically, glucose made up almost 27% of the carbohydrate mass in the pbx1Δ strain capsule (Fig. 5B). Galactose (∼3%) and glucosamine (∼1%) were also detectable, though in much smaller amounts. For quantitation of all of the HPAEC-PAD data, see Table S3 in the supplemental material.
FIG. 5.
The pbx1Δ and pbx2Δ strain exopolysaccharides contain glucose. The carbohydrate compositions of the exopolysaccharides from (A) the wild-type strain, (B) pbx1Δ strain CM106, (C) pbx2Δ strain CM107, and (D) pbx1Δ pbx2Δ strain CM084 were determined by HPAEC-PAD after acid hydrolysis with trifluoroacetic acid. Percentages refer to percentages of carbohydrate mass in the sample. Exopolysaccharide purified from a single strain per genotype was analyzed for carbohydrate composition.
Analysis of the pbx2Δ mutant yielded similar results. In the pbx2Δ strain exopolysaccharide, glucose accounted for 9% of the carbohydrate mass in the capsule (Fig. 5C). As in the pbx1Δ strain material, detectable amounts of galactose (∼2%) and glucosamine (∼1%) were measured. In the pbx1Δ pbx2Δ strain exopolysaccharide, glucose accounted for 17% of the carbohydrate mass (Fig. 5D). Galactose (∼2%) and glucosamine (∼2%) were detected as well. Exopolysaccharide from only one strain per genotype was analyzed by HPAEC-PAD.
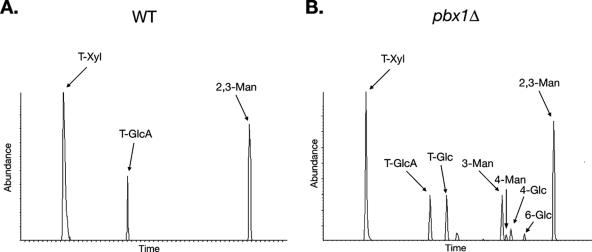
In order to determine how glucose was incorporated into the capsule, we performed carbohydrate linkage analysis by GLC-EIMS of methylated derivatives. Only three types of residues were detected in the H99 exopolysaccharide, 2,3-linked mannose, terminal xylose, and terminal glucuronic acid (Fig. 6A). These linkages are entirely consistent with the known structure of the serotype A GXM structure (Fig. 1A). 6-O-acetylations that were present on the mannose residues were not detectable, since they were lost during the permethylation step. We next examined the pbx1Δ strain exopolysaccharide. The three residue types seen in the H99 exopolysaccharide made up most of the residues found in the pbx1Δ strain CM106 exopolysaccharide, again supporting the idea that most of the mutant capsule retains the normal structure. However, several novel residues were identified. Specifically, most of the glucose in the mutant exopolysaccharide was present as terminal glucose (Fig. 6B). 4-Linked and 6-linked glucose and 3-linked mannose were also detected. Exopolysaccharide from one pbx1Δ strain was analyzed by GLC-EIMS.
FIG. 6.
Most of the glucose in the pbx1Δ strain exopolysaccharide exists as terminal glucose. Residue types found in (A) the wild-type (WT) strain and (B) pbx1Δ strain CM106 were determined by GLC-EIMS carbohydrate linkage analysis. Exopolysaccharide purified from a single pbx1Δ strain was analyzed by GLC-EIMS.
Parallel β-helix genes required for virulence.
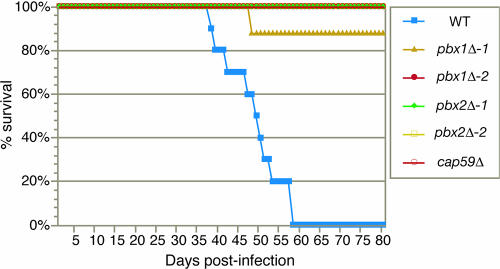
The effect of the pbx1Δ and pbx2Δ mutations on virulence was examined with a murine inhalation model of cryptococcosis. H99, two independently generated pbx1Δ full-deletion strains generated from different transformation experiments (CM106 and CM109), two independently generated pbx2Δ full-deletion strains (CM107 and CM110), and a mutant strain harboring a deletion of the CAP59 gene (cap59Δ) were inoculated intranasally (5 × 105 cells) into 5-week old female A/J mice. Eight to 10 mice were used per strain (Fig. 7). Half of the mice infected with H99 died by day 49 postinfection, and 100% died by day 58 postinfection. In contrast, only one mouse infected with either the pbx1Δ or pbx2Δ strains died during the course of the experiment (all mice were sacrificed 80 days postinfection). The two independently generated pbx1Δ strains and the two independently generated pbx2Δ strains displayed the same phenotype, indicating that the attenuation of virulence was due to the gene deletion. However, unlike mice infected with the cap59Δ mutant, 9 of 16 mice infected with the pbx1Δ mutants and 2 of 16 mice infected with the pbx2Δ mutants displayed transient symptoms of illness, including weight loss (see Fig. S1 in the supplemental material). Other symptoms included ruffled fur, shallow breathing, and lethargy. The onset of symptoms varied from approximately 38 to 73 days postinfection. Symptoms lasted approximately 4 to 8 days. In all cases, the infected mice recovered, and once they recovered, they were indistinguishable from uninfected mice or mice infected with the cap59Δ mutant strain. Independent experiments examining the virulence of the original partial-deletion strains (CM104 and CM105) with the same murine inhalation model of cryptococcosis yielded similar results (data not shown). In these experiments, none of the mice infected with CM104 (partial deletion of PBX1) or CM105 (partial deletion of PBX2) died during the course of the experiment. Five of 13 mice infected with CM104 and 8 of 12 mice infected with CM105 displayed transient symptoms of illness.
FIG. 7.
The pbx1Δ and pbx2Δ strain are hypovirulent. Eight to 10 mice (A/J) were infected intranasally with 5 × 105 cells of the indicated strain, and progression to severe morbidity was monitored. CM106 (pbx1Δ-1) and CM109 (pbx1Δ-2) were generated in separate, independent biolistic transformations. CM107 (pbx2Δ-1) and CM110 (pbx2Δ-2) were generated in separate, independent biolistic transformations. Infections with the partial-deletion strains CM104 (pbx1Δ) and CM105 (pbx2Δ) yielded similar results (data not shown). WT, wild type.
DISCUSSION
As part of an ongoing C. neoformans gene deletion project, we discovered that deletion of either CNAG_01172.1, which we have designated PBX1, or CNAG_05562.1, which we have designated PBX2, results in a dry-colony morphology, clumpy cells, and decreased capsule integrity. NMR spectroscopy demonstrated that while much of the GXM polymers were correctly assembled in these mutants, a portion of the exopolysaccharide material purified from these deletion strains contained aberrant structures not seen in that of the wild type. Carbohydrate composition analysis determined that these aberrant structures were due to the presence of a significant amount of glucose in the purified mutant exopolysaccharide, a sugar not found in the wild-type capsule. Carbohydrate linkage analysis showed that most of this glucose was present as terminal glucose, suggesting that these aberrant glucose residues may be linked directly to the main mannose backbone of the GXM polymers, taking the place of either glucuronic acid or xylose. Thus, we conclude that Pbx1 and Pbx2 play a role in the fidelity of polysaccharide capsule synthesis. Specifically, these two proteins are needed directly or indirectly to remove glucose and perhaps other sugars that appear to be incorrectly added to the GXM polymer. Without either of these two proteins, these erroneous residues cannot be removed, resulting in the aberrant structures detected in the NMR spectra.
Both PBX1 and PBX2 are predicted to encode proteins that contain parallel β-helices. Further analysis with the PHYRE algorithm (http://www.sbg.bio.ic.ac.uk/phyre/) predicts specifically single-stranded right-handed β-helices. Right-handed parallel β-helices are highly unusual in proteins. They have been found primarily in enzymes that cleave linkages in polysaccharide substrates, such as pectate lyases, rhamnogalactouronases, chondroitinases, and pectin methylesterases, though examples of proteins that contain right-handed parallel β-helices but lack any known enzymatic activity have been found (19). Given this significant correlation between right-handed parallel β-helices and glycosidic activity, we hypothesize that Pbx1 and Pbx2 may be directly responsible for cleaving these aberrant glucose linkages. Biochemical reconstitution is needed to test this prediction.
Alternatively, we cannot rule out the possibility that Pbx1 and Pbx2 act indirectly to inhibit addition of glucose to the GXM polymers. For example, they may act as negative regulators of glucosyltransferase expression or glucosyltransferase activity. In this model, deletion of PBX1 or PBX2 results in aberrant glucosyltransferase expression or activity that results in the addition of glucose to the capsule polysaccharide. Such negative regulators have been identified for other glycosyltransferases (10, 37). Another possibility is that Pbx1 and Pbx2 are glucosyltransferases themselves and can compete for UDP-glucose substrates. Deletion of PBX1 or PBX2 results in an increase in available UDP-glucose and addition of glucose to the GXM polymers. Further experimentation is needed to rule out these models.
The sensitivity of the pbx1Δ and pbx2Δ strain capsules to sonication treatment indicates that these capsules have decreased structural integrity. It seems likely that the abnormal GXM structures detected in the NMR spectra and the decrease in capsule stability are related. Electron microscopy of both whole cells and purified GXM has revealed that the capsule is composed of a highly entangled meshwork of GXM polymers that can self-associate (23, 28). The presence of aberrant, glucose-containing capsule polysaccharides in the mutants may compromise interpolymer interactions in these capsules and result in decreased capsule stability. Experiments testing the density and antibody-binding properties of these capsules, as well as the associative properties of the mutant GXM polymers, may help elucidate the architectural defects in these mutants. It is also a possibility that deletion of PBX1 or PBX2 results in a GXM anchoring defect. GXM is known to be attached to the cell wall in an α-1-3-glucan-dependent manner (29), and this interaction may be disrupted in a PBX1 or PBX2 mutant. This model, however, seems less likely since the mutant capsule is stably attached to the cell prior to sonication.
Our linkage analysis of the mutant exopolysaccharide detected the presence of 4-linked and 6-linked glucose along with terminal glucose, suggesting that some of the glucose in the capsule was involved in more complex linkages and perhaps more elaborate side chains off of the mannose backbone. Alternatively, the 4-linked and 6-linked glucose may result from some cell wall contamination in the exopolysaccharide sample. This possibility, however, cannot account for most of the glucose seen in the exopolysaccharide given that most of it is terminal glucose. More elaborate side chains may also explain the 3-linked mannose that was detected in the mutant capsule. These 3-linked mannose residues could be the result of mannose residues in the backbone that are not decorated with any side chain. The presence of glucose residues and/or larger glucose side chains on an adjacent mannose and/or the incorporation of glucose into the backbone itself may preclude the correct glucuronic acid or xylose side chains from being added to these mannose residues.
The incorporation of glucose into these mutant capsules indicates that GXM may be normally assembled in the presence of UDP-glucose and glucosyltransferases. This is consistent with recent evidence demonstrating that capsule polysaccharide synthesis occurs intracellularly (16, 35). It is unclear, however, whether the addition of glucose to the elongating GXM polymer is a normal step in GXM synthesis. It is possible that glucose residues serve as placeholders and/or protecting groups that are used to mark the mannose backbone. In this scenario, these marks would need to be removed to allow for the addition of xylose and/or glucuronic acid at the appropriate moieties. Only then would glucose residues be removed by the direct or indirect action of Pbx1 and Pbx2. On the other hand, the addition of glucose to the growing polysaccharide could reflect an error in synthesis. Given the potential abundance of UDP-glucose, which is a precursor of UDP-xylose and UDP-glucuronic acid, in the membrane-bound compartments where GXM may be synthesized, it could be that glucose is added erroneously to the GXM polymer at some rate. These incorrectly added residues would need to be removed by the action of Pbx1 and Pbx2 in order for the correct polysaccharide structure to be maintained throughout the polymer. This second model, where incorporation of glucose into the capsule reflects errors in the biosynthetic process, is supported by the fact that much of the GXM is assembled correctly in the pbx1Δ and pbx2Δ mutant exopolysaccharides.
Interestingly, the pbx1Δ and pbx2Δ mutant exopolysaccharides have almost identical NMR spectra and the pbx1Δ pbx2Δ double mutant does not appear to have a stronger defect than the single mutants. This suggests that Pbx1 and Pbx2 could function together in a complex that trims aberrant glucose residues from the mannose backbone. Interestingly, both Pbx1 and Pbx2 are predicted to contain coiled-coil domains which may mediate protein-protein interactions. Again, biochemical reconstitution is necessary to test this idea. It is possible that epigenetic changes affecting the structure of the capsule may occur in some portion of the population during in vitro culturing, explaining the appearance or disappearance of minor peaks in the NMR spectra.
The pbx1Δ and pbx2Δ strains are unlike many of the capsule-defective mutants that have previously been tested for virulence in that pbx1Δ and pbx2Δ cells build a sizable capsule, most of which is correctly structured. Interestingly, while the pbx1Δ and pbx2Δ strains are both strongly attenuated for virulence, they induced delayed symptoms of cryptococcosis as late as 9 weeks postinfection. Although significant disease progression was observed, all of the mice except one recovered despite being given a dose that is uniformly lethal for the wild-type strain. In contrast, mice infected with the cap59Δ mutant, which is acapsular, never became ill. We hypothesize that the pbx1Δ and pbx2Δ strain capsules retain a portion of their biological properties. A portion of this virulence defect may be attributable to the modest growth defect observed in these mutant strains at 37°C. However, the onset of disease symptoms at 6 to 9 weeks postinfection suggests that the pbx1Δ and pbx2Δ mutant strains can at the least survive or evade the initial innate immune response. Previous studies have concluded that the wild-type C. neoformans polysaccharide capsule does not induce a strong antibody response. It is possible that the aberrant structures found in the mutant capsules were more immunogenic than the wild-type structure, allowing for a more successful adaptive immune response. The overall stability of the mutant capsules appeared compromised as well, which may also contribute to its defect in pathogenesis, perhaps by making yeast cells more susceptible to certain types of immune attack. Further study of these mutants may allow uncoupling of the various roles the capsule has been hypothesized to play during infection.
Supplementary Material
Acknowledgments
We thank J. Lodge for strain H99, natR and hygR cassettes, and instruction on knockout methodology; G. Cox for instruction on animal infections; A. Datta for help with carbohydrate composition and linkage analysis; and L. O'Campo for assistance with capsule images and microscopy.
This work was supported by an Opportunity Grant from the Herb and Marion Sandler Foundation and a grant from the National Institute of Allergy and Infectious Diseases (R01AI065519) to H.D.M. O.W.L. is supported by a Howard Hughes Medical Institute predoctoral fellowship.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 2 March 2007.
REFERENCES
- 1.Banerjee, U., K. Datta, and A. Casadevall. 2004. Serotype distribution of Cryptococcus neoformans in patients in a tertiary care center in India. Med. Mycol. 42:181-186. [DOI] [PubMed] [Google Scholar]
- 2.Bar-Peled, M., C. L. Griffith, and T. L. Doering. 2001. Functional cloning and characterization of a UDP-glucuronic acid decarboxylase: the pathogenic fungus Cryptococcus neoformans elucidates UDP-xylose synthesis. Proc. Natl. Acad. Sci. USA 98:12003-12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bose, I., A. J. Reese, J. J. Ory, G. Janbon, and T. L. Doering. 2003. A yeast under cover: the capsule of Cryptococcus neoformans. Eukaryot. Cell 2:655-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buchanan, K. L., and J. W. Murphy. 1998. What makes Cryptococcus neoformans a pathogen? Emerg. Infect. Dis. 4:71-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Canteros, C. E., M. Brudny, L. Rodero, D. Perrotta, and G. Davel. 2002. Distribution of Cryptococcus neoformans serotypes associated with human infections in Argentina. Rev. Argent. Microbiol. 34:213-218. [PubMed] [Google Scholar]
- 6.Chang, Y. C., and K. J. Kwon-Chung. 1994. Complementation of a capsule-deficient mutation of Cryptococcus neoformans restores its virulence. Mol. Cell. Biol. 14:4912-4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, Y. C., and K. J. Kwon-Chung. 1998. Isolation of the third capsule-associated gene, CAP60, required for virulence in Cryptococcus neoformans. Infect. Immun. 66:2230-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, Y. C., and K. J. Kwon-Chung. 1999. Isolation, characterization, and localization of a capsule-associated gene, CAP10, of Cryptococcus neoformans. J. Bacteriol. 181:5636-5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, Y. C., L. A. Penoyer, and K. J. Kwon-Chung. 1996. The second capsule gene of Cryptococcus neoformans, CAP64, is essential for virulence. Infect. Immun. 64:1977-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, W., J. Tang, and P. Stanley. 2005. Suppressors of α(1,3)fucosylation identified by expression cloning in the LEC11B gain-of-function CHO mutant. Glycobiology 15:259-269. [DOI] [PubMed] [Google Scholar]
- 11.Cherniak, R., L. C. Morris, B. C. Anderson, and S. A. Meyer. 1991. Facilitated isolation, purification, and analysis of glucuronoxylomannan of Cryptococcus neoformans. Infect. Immun. 59:59-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corbett, E. L., G. J. Churchyard, S. Charalambos, B. Samb, V. Moloi, T. C. Clayton, A. D. Grant, J. Murray, R. J. Hayes, and K. M. De Cock. 2002. Morbidity and mortality in South African gold miners: impact of untreated disease due to human immunodeficiency virus. Clin. Infect. Dis. 34:1251-1258. [DOI] [PubMed] [Google Scholar]
- 13.Davidson, R. C., M. C. Cruz, R. A. Sia, B. Allen, J. A. Alspaugh, and J. Heitman. 2000. Gene disruption by biolistic transformation in serotype D strains of Cryptococcus neoformans. Fungal Genet. Biol. 29:38-48. [DOI] [PubMed] [Google Scholar]
- 14.Delaglio, F., S. Grzesiek, G. W. Vuister, G. Zhu, J. Pfeifer, and A. Bax. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6:277-293. [DOI] [PubMed] [Google Scholar]
- 15.French, N., K. Gray, C. Watera, J. Nakiyingi, E. Lugada, M. Moore, D. Lalloo, J. A. Whitworth, and C. F. Gilks. 2002. Cryptococcal infection in a cohort of HIV-1-infected Ugandan adults. AIDS 16:1031-1038. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Rivera, J., Y. C. Chang, K. J. Kwon-Chung, and A. Casadevall. 2004. Cryptococcus neoformans CAP59 (or Cap59p) is involved in the extracellular trafficking of capsular glucuronoxylomannan. Eukaryot. Cell 3:385-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Griffith, C. L., J. S. Klutts, L. Zhang, S. B. Levery, and T. L. Doering. 2004. UDP-glucose dehydrogenase plays multiple roles in the biology of the pathogenic fungus Cryptococcus neoformans. J. Biol. Chem. 279:51669-51676. [DOI] [PubMed] [Google Scholar]
- 18.Janbon, G., U. Himmelreich, F. Moyrand, L. Improvisi, and F. Dromer. 2001. Cas1p is a membrane protein necessary for the O-acetylation of the Cryptococcus neoformans capsular polysaccharide. Mol. Microbiol. 42:453-467. [DOI] [PubMed] [Google Scholar]
- 19.Jenkins, J., and R. Pickersgill. 2001. The architecture of parallel beta-helices and related folds. Prog. Biophys. Mol. Biol. 77:111-175. [DOI] [PubMed] [Google Scholar]
- 20.Kay, L., E. P. Keifer, and T. Saarinen. 1992. Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J Am. Chem. Soc. 114:10663-10665. [Google Scholar]
- 21.Kwon-Chung, K. J., and A. Varma. 2006. Do major species concepts support one, two or more species within Cryptococcus neoformans? FEMS Yeast Res. 6:574-587. [DOI] [PubMed] [Google Scholar]
- 22.Litvintseva, A. P., R. Thakur, L. B. Reller, and T. G. Mitchell. 2005. Prevalence of clinical isolates of Cryptococcus gattii serotype C among patients with AIDS in sub-saharan Africa. J. Infect. Dis. 192:888-892. [DOI] [PubMed] [Google Scholar]
- 23.McFadden, D. C., M. De Jesus, and A. Casadevall. 2006. The physical properties of the capsular polysaccharides from Cryptococcus neoformans suggest features for capsule construction. J. Biol. Chem. 281:1868-1875. [DOI] [PubMed] [Google Scholar]
- 24.Moyrand, F., Y. C. Chang, U. Himmelreich, K. J. Kwon-Chung, and G. Janbon. 2004. Cas3p belongs to a seven-member family of capsule structure designer proteins. Eukaryot. Cell 3:1513-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moyrand, F., and G. Janbon. 2004. UGD1, encoding the Cryptococcus neoformans UDP-glucose dehydrogenase, is essential for growth at 37°C and for capsule biosynthesis. Eukaryot. Cell 3:1601-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moyrand, F., B. Klaproth, U. Himmelreich, F. Dromer, and G. Janbon. 2002. Isolation and characterization of capsule structure mutant strains of Cryptococcus neoformans. Mol. Microbiol. 45:837-849. [DOI] [PubMed] [Google Scholar]
- 27.Okongo, M., D. Morgan, B. Mayanja, A. Ross, and J. Whitworth. 1998. Causes of death in a rural, population-based human immunodeficiency virus type 1 (HIV-1) natural history cohort in Uganda. Int. J. Epidemiol. 27:698-702. [DOI] [PubMed] [Google Scholar]
- 28.Pierini, L. M., and T. L. Doering. 2001. Spatial and temporal sequence of capsule construction in Cryptococcus neoformans. Mol. Microbiol. 41:105-115. [DOI] [PubMed] [Google Scholar]
- 29.Reese, A. J., and T. L. Doering. 2003. Cell wall α-1,3-glucan is required to anchor the Cryptococcus neoformans capsule. Mol. Microbiol. 50:1401-1409. [DOI] [PubMed] [Google Scholar]
- 30.Sheng, S., and R. Cherniak. 1997. Structure of the 13C-enriched O-deacetylated glucuronoxylomannan of Cryptococcus neoformans serotype A determined by NMR spectroscopy. Carbohydr. Res. 301:33-40. [DOI] [PubMed] [Google Scholar]
- 31.Sommer, U., H. Liu, and T. L. Doering. 2003. An α-1,3-mannosyltransferase of Cryptococcus neoformans. J. Biol. Chem. 278:47724-47730. [DOI] [PubMed] [Google Scholar]
- 32.Vecchiarelli, A. 2000. Immunoregulation by capsular components of Cryptococcus neoformans. Med. Mycol. 38:407-417. [DOI] [PubMed] [Google Scholar]
- 33.Wills, E. A., I. S. Roberts, M. Del Poeta, J. Rivera, A. Casadevall, G. M. Cox, and J. R. Perfect. 2001. Identification and characterization of the Cryptococcus neoformans phosphomannose isomerase-encoding gene, MAN1, and its impact on pathogenicity. Mol. Microbiol. 40:610-620. [DOI] [PubMed] [Google Scholar]
- 34.Yauch, L. E., J. S. Lam, and S. M. Levitz. 2006. Direct inhibition of T-cell responses by the Cryptococcus capsular polysaccharide glucuronoxylomannan. PLoS Pathogens 2(11):e120. doi: 10.1371/journal.ppat.0020120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoneda, A., and T. L. Doering. 2006. A eukaryotic capsular polysaccharide is synthesized intracellularly and secreted via exocytosis. Mol. Biol. Cell. 17:5131-5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zaragoza, O., and A. Casadevall. 2004. Experimental modulation of capsule size in Cryptococcus neoformans. Biol. Proced. Online 6:10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang, A., B. Potvin, A. Zaiman, W. Chen, R. Kumar, L. Phillips, and P. Stanley. 1999. The gain-of-function Chinese hamster ovary mutant LEC11B expresses one of two Chinese hamster FUT6 genes due to the loss of a negative regulatory factor. J. Biol. Chem. 274:10439-10450. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.