Abstract
A novel subfamily of putative intracellular invertase enzymes (glycoside hydrolase family 32) has previously been identified in fungal genomes. Here, we report phylogenetic, molecular, and biochemical characteristics of SucB, one of two novel intracellular invertases identified in Aspergillus niger. The sucB gene was expressed in Escherichia coli and an invertase-negative strain of Saccharomyces cerevisiae. Enzyme purified from E. coli lysate displayed a molecular mass of 75 kDa, judging from sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis. Its optimum pH and temperature for sucrose hydrolysis were determined to be 5.0 and 37 to 40°C, respectively. In addition to sucrose, the enzyme hydrolyzed 1-kestose, nystose, and raffinose but not inulin and levan. SucB produced 1-kestose and nystose from sucrose and 1-kestose, respectively. With nystose as a substrate, products up to a degree of polymerization of 4 were observed. SucB displayed typical Michaelis-Menten kinetics with substrate inhibition on sucrose (apparent Km, Ki, and Vmax of 2.0 ± 0.2 mM, 268.1 ± 18.1 mM, and 6.6 ± 0.2 μmol min−1 mg−1 of protein [total activity], respectively). At sucrose concentrations up to 400 mM, transfructosylation (FTF) activity contributed approximately 20 to 30% to total activity. At higher sucrose concentrations, FTF activity increased to up to 50% of total activity. Disruption of sucB in A. niger resulted in an earlier onset of sporulation on solid medium containing various carbon sources, whereas no alteration of growth in liquid culture medium was observed. SucB thus does not play an essential role in inulin or sucrose catabolism in A. niger but may be needed for the intracellular conversion of sucrose to fructose, glucose, and small oligosaccharides.
Fructans and fructooligosaccharides (FOS) consist of a chain of fructose molecules linked to a terminal glucose residue. These fructose monomers are linked by either β2,1 (inulin) or β2,6 (levan) glycosidic bonds. Inulin and levan have several favorable properties that make them commercially interesting for applications in pharmaceutical and food industries (36). In the human digestive tract, FOS are almost exclusively fermented by bifidobacteria and lactobacilli, which have beneficial health effects (20, 31, 33). Commercially, FOS are produced by the enzymatic hydrolysis of inulin isolated from plants, primarily chicory and Jerusalem artichoke (36). Alternatively, sucrose can be converted into FOS by using a range of different transfructosylating enzymes originating from plants, bacteria, and fungi (36). Transfructosylation (FTF) and hydrolytic enzymes belong to glycoside hydrolase family 32 (GH32) and GH68 (8) constituting enzyme clan GH-J, based on shared conserved domains (22a). These enzymes have been reported to be present in a variety of plants, bacteria, and fungi (36). FOS synthesis has been reported for the commercially important fungus Aspergillus niger, reflecting a side reaction of an invertase (EC 3.2.1.26) (32) or as the result of the activity of a specific fructosyltransferase (EC. 2.4.1.9) (18). Nguyen et al. (23) previously reported the presence of an intracellular invertase in A. niger IMI303386, grown on sucrose or inulin as the sole carbon source. The purified enzyme produced free glucose and fructose from sucrose hydrolysis as well as 1-kestose and nystose from sucrose FTF. However, the gene encoding this enzyme has not been identified and characterized. Yanai et al. (38) previously reported characteristics of an extracellular β-fructofuranosidase from A. niger 20611. This enzyme displayed increased FTF activity compared to other known Aspergillus invertases. The strain was, however, later reclassified as Aspergillus japonicus ATCC 20611. The true identity, diversity, and characteristics of invertases and FTF enzymes present in A. niger thus remained to be determined.
Recently, the complete genome sequence of A. niger has become available (26) and was analyzed for putative sucrose- and fructan-modifying enzymes (39). In addition to sucA, encoding the previously characterized extracellular invertase (6), two novel putative invertase genes were identified (sucB and sucC) (Fig. 1). The sucB (but not the sucC) gene was (constitutively) expressed at a low level on starch and xylose and up-regulated in the presence of sucrose and inulin (39). Orthologues of these genes have also been identified in other fungal genomes (see below). We have cloned and heterologously expressed the A. niger sucB gene, allowing a biochemical characterization of the purified enzyme. An A. niger sucB gene disruption strain was constructed to determine whether this novel intracellular invertase enzyme plays a significant role in growth on sucrose and inulin. This paper reports the phylogenetic, molecular, and biochemical characterization of SucB. The data show that in addition to invertase activity, SucB displays transfructosylating activity.
FIG. 1.
Neighbor-joining tree of functionally characterized and putative fungal invertases. Bootstrap values are indicated on the nodes of each branch. The tree was created with MEGA 3.1 using default settings for gap and extension penalties. The bar indicates 10% amino acid sequence difference. GenBank accession numbers are in parentheses.
MATERIALS AND METHODS
Phylogenetic analysis.
The complete amino acid sequence of SucB (GenBank accession number DQ233219) (39) was blasted against the protein database at Swissprot (http://www.ncbi.nlm.nih.gov/BLAST/). Identified sequences containing GH32 domains (http://afmb.cnrs-mrs.fr/CAZY/) were aligned with SucB (CLUSTALW interface in MEGA 3.1 [http://www.megasoftware.com]), followed by bootstrap testing of phylogeny (gap opening, 10; extension penalties, 0.2; 1,000 replicates). Sequence logos were created by SequenceLogo (http://bio.cam.ac.uk/seqlogo/).
Strains, plasmids, media, and growth conditions.
A. niger strains N402 (7), NRRL3122 (25), and AB4.1 (34) and Escherichia coli strains XL1-Blue (Stratagene, La Jolla, CA), TOP10, and BL21(DE3) STAR (Invitrogen, Carlsbad, CA) were used in this study. The A. niger genome sequence was derived from strain CBS513.88 (a natural derivative of strain NRRL3122). A. niger strains were grown in Aspergillus minimal medium (MM) (3) or complete medium (MM supplemented with 0.5% [wt/vol] yeast extract and 0.1% [wt/vol] Casamino Acids). Conidiospores were obtained by harvesting spores from a complete medium plate containing 1% (wt/vol) glucose, after 4 to 6 days of growth at 30°C, using a 0.9% (wt/vol) NaCl solution. Transformation of A. niger AB4.1 was performed as described previously (26). Cloning of sucB was performed using Gateway cloning technology (Invitrogen), and the integrity of constructs was verified by DNA sequencing (Baseclear, Leiden, The Netherlands). The Gateway expression vectors pDEST17 and pYES-DEST52 were used for expression in E. coli and the invertase-negative strain of S. cerevisiae (BY4743Δsuc2).
S. cerevisiae strains were grown aerobically at 30°C in S. cerevisiae medium plus glucose (1.7 g liter−1 yeast nitrogen base, 5 g liter−1 ammonium sulfate, 2.5 g liter−1 sodium succinate, 5 g liter−1 Casamino Acids, 0.1 g liter−1 tryptophan, 20 g liter−1 glucose), followed by induction of expression in S. cerevisiae medium plus 20 g liter−1 galactose.
Cloning and purification of SucB.
The coding sequence of sucB was amplified in a two-step procedure. The first two exons were amplified using primers sets SucBGATEF and SucBDNAP1 and SucBDNAP2 and SucBGATER, respectively, followed by the joining of the two exons in a single PCR together with outside primers SucBGATEF and SucBGATER (Table 1). Amplifications were performed in a GeneAmp PCR system 2700 thermal cycler (Applied Biosystems, Foster City, CA) using the Expand High Fidelity PCR system (Roche Diagnostics Corporation, Indianapolis, IN) under the following conditions: an initial denaturation step for 2 min at 94°C, 30 cycles of denaturation at 94°C for 15 s, annealing at 55°C for 30 s, and elongation at 72°C for 90 s, followed by a final elongation step of 7 min at 72°C. Gateway cloning of the fragments was performed according to the manufacturer's instructions (Invitrogen) to create constructs pDEST17-sucB and pYES-DEST52-sucB, respectively.
TABLE 1.
Primers used during this study
| Primer | Sequencea |
|---|---|
| SucBGATEF | GGGGACAAGTTTGTACAAAAAAGCAGGCTTAATAATGGAGCGGCAAACTAGCCCCTCAG |
| SucBGATER | GGGGACCACTTTGTACAAGAAAGCTGGGTCTCACTCGCGCATCGACTTCTTCC |
| SucBcDNAP1 | TGTTGGTGGATGGTGCCATGGAGGACTCTAT |
| SucBcDNAP2 | CATGGCACCATCCACCAACACAAACCAGCCCT |
| SucBP1f | TAGCGGCCGCCATCGCGACTGTCCTCATACA |
| SucBP2r | CAGCGGCTTGGGTCTAGAATCATCCGAACTATTCTCGATT |
| SucBP3f | ATCCTCTAGAGTCGACCTGCAGTCGATGCGCGAGTGAGAT |
| SucBP4r | TGGAATTCCTCATTTACGTCATCGTCGGCGA |
Underlining indicates restriction sites.
Cultures of E. coli BL21(DE3) STAR containing pDEST17-sucB that were grown overnight were inoculated into fresh LB medium containing 100 μg ml−1 ampicillin. Soluble expression of SucB was achieved at 18°C (optical density at 600 nm of approximately 0.4), followed by induction at 18°C for 6 h (1 mM isopropyl-β-d-thiogalactopyranoside [IPTG]). Cells were harvested by centrifugation (10 min at 4°C, 4000 × g), and cell pellets were resuspended in 5 ml 50 mM sodium phosphate buffer (pH 8) containing 250 mM NaCl, 10 mM imidazole, and 5 mM β-mercaptoethanol. Following sonification (seven times for 15 s at 8 μm with 30-s intervals), cell-free lysate was obtained by centrifugation (20 min at 4°C, 10,000 × g). SucB was purified from the cell-free lysates using His tag affinity chromatography according to the manufacturer's instructions (Sigma-Aldrich, St. Louis, MO). Protein concentration, size, and purity were determined using the Bradford reagent (Bio-Rad, Hercules, CA), sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and Biosafe Coomassie staining (Bio-Rad).
Cultures of S. cerevisiae BY4743Δsuc2 cells grown overnight containing the expression vector pYES-DEST52-sucB were used to inoculate 50 ml of fresh medium containing 2% galactose to induce protein expression. After 5 h of growth, cells were pelleted by centrifugation and washed with sterile demineralized water. Y-PER (Pierce Biotechnology, Rockford, IL) was used for cell lysis (according to the manufacturer's recommendations), followed by the preparation of cell-free lysate as described above.
Activity assays.
Enzymatic activity was quantified spectrophotometrically by separate measurements of the released glucose and fructose from sucrose (d-glucose/d-fructose kit; Roche Diagnostics Corporation). The pH and temperature optima were determined by measuring initial rates in 50 mM phosphate-citrate buffer containing 100 mM sucrose using a pH range of 4.0 to 7.5 and a temperature range of 25 to 60°C. The effect of the sucrose concentration on enzyme activity was determined using two independent experiments performed in triplicate by measuring initial rates over a substrate range of 12 sucrose concentrations (2.5 mM to 1 M) in 50 mM acetate buffer at 37°C (pH 5.0). Corrections for background glucose and fructose values at high substrate concentrations were made accordingly. Nonlinear regression curve fitting was done using Sigma Plot (Systat Software, Richmond, CA) by applying the Michaelis-Menten formula for substrate inhibition: (y = Vmax × [S]/Km + [S] + [S]2/Ki), where y equals the specific activity (μmol mg−1 min−1) and [S] is the concentration of substrate.
Substrate specificity and product range.
In order to identify the substrate specificity of SucB and the products made after incubation, 14 μg of purified SucB was incubated overnight in 50 mM acetate buffer (pH 5.0) at 37°C with a range of substrates. The effect of the reducing agents dithiothreitol (DTT) and β-mercaptoethanol (BME) on SucB activity was also tested by incubating the enzyme at optimal conditions overnight in the presence of these agents at concentrations up to 100 mM. Substrate conversion and product formation were analyzed by thin-layer chromatography (TLC) (aluminum sheets, silica gel 60 F 254; Merck and Co., Whitehouse station, NJ) and high-performance anion-exchange chromatography (HPAEC) (Dionex Corporation, Sunnyvale, CA). Sample separation on TLC was performed using a mixture of butanol, ethanol, and water (3.8:3.8:2.4 [vol/vol/vol]) or ethyl acetate, 2-propanol, and water (6:3:1 [vol/vol/vol]). Subsequently, plates were dried and sprayed with developer solution [95% methanol, 5% sulfuric acid, and 3 g liter−1 2-(1-naphthylamino)ethylamine dihydrochloride]. Product formation by SucB was confirmed by HPAEC as described previously (24).
Construction of the sucB::pyrG gene deletion strain.
A sucB deletion cassette was constructed by PCR amplification of 1.0 kb of 5′ and 3′ DNA flanking regions of the sucB gene using primer pair SucBP1-SucBP4 (Table 1). Both fragments were cloned into pBluescriptII (Stratagene) using appropriate restriction enzymes (Table 1). The A. oryzae pyrG gene from pAO4-13 (11) was isolated as a 2.7-kb XbaI fragment and cloned between the 5′ and 3′ sucB flanking regions to give pΔsucB.
Prior to transformation into AB4.1, pΔsucB was linearized with EcoRI. Uridine prototrophic transformants were purified and screened for sucB deletion by Southern blot analysis (30). Genomic DNA was isolated and digested with XhoI, and the 3′ region flanking the sucB gene was used as a probe. As predicted, a 2.2-kb hybridizing DNA fragment was observed in the wild-type strain, whereas a 4.0-kb DNA fragment was detected in sucB deletion strains (data not shown). Several sucB deletions strains were independently obtained, and strain NC1.1 (ΔsucB) was used throughout this study.
Microtiter plate growth assay.
Growth of A. niger strains N402 and NC1.1 was determined using a HTS7000 BioAssay reader (Perkin-Elmer Life and Analytical Sciences, Inc., Wellesley, MA). Spores (1 × 104) were inoculated into each well of a 96-well microtiter plate (Nalge Nunc International, Rochester, NY) and incubated at 32°C for 56 h. Each well contained 200 μl of MM with 1% (wt/vol) of one of the various carbon sources supplemented with 0.1% (wt/vol) Casamino Acids to stimulate spore germination. Six replicates of each condition were made. Growth was monitored by measuring the optical density at 595 nm every 2 h.
RESULTS
Sequence analysis.
Using the predicted amino acid sequence of SucB in the phylogenetic analysis, we have identified SucB orthologues in various other fungal species. In silico analysis indicated that these putative invertases also lack any recognizable signal peptide sequences, as has been reported previously for SucB (39). Multiple sequence alignment between these new groups of putative intracellular invertases and known fungal invertases indicates that they cluster together in a separate subfamily clearly distinct from known extracellular fungal and yeast invertase proteins (Fig. 1). Table 2 depicts sequence logos (9) constructed from alignments of the SucB subfamily members, revealing the presence of all eight conserved domains characteristic of GH32.
TABLE 2.
Sequence logo depiction of conserved motifs identified in invertases with known function and SucB orthologuesa
Cloning and purification of SucB.
Initial attempts to clone the sucB from a cDNA library constructed from inulin-growing A. niger strain N402 were unsuccessful, probably because of the relatively low level of expression of the sucB gene (39). Following amplification, the full coding region of sucB (1,854 bp) was obtained. The same procedure was followed to clone the second putative intracellular invertase identified in A. niger (sucC); however, we failed to obtain functional expression in either E. coli or S. cerevisiae.
Purification of SucB from E. coli was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis, where the protein size was estimated to 75 kDa (70 kDa, calculated). C-terminally-His-tagged SucB expressed in S. cerevisiae strain BY4743Δsuc2 could not be sufficiently purified using Ni-nitrilotriacetic acid affinity chromatography. Therefore, the cell-free lysate was used for comparative studies. SucB expressed in both E. coli and S. cerevisiae displayed similar characteristics, whereas no activity could be detected in S. cerevisiae BY4743Δsuc2 containing the empty expression vector. SucB in the cell extract of E. coli or S. cerevisiae, as well as the affinity-purified SucB from E. coli, displayed activity only for a maximum storage time of 3 days (4°C or −20°C in 20% glycerol). Thus, for all subsequent analyses, the enzyme was used directly after purification.
Influence of pH and temperature on SucB enzyme activity.
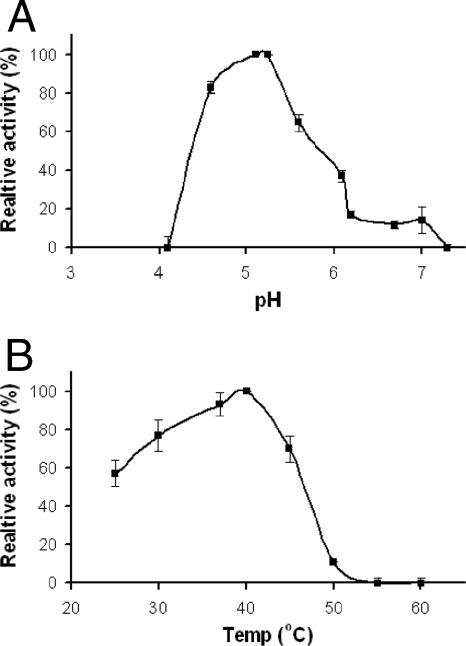
The optimal pH and temperature conditions for SucB activity with sucrose as a substrate were determined by measuring the amount of released glucose enzymatically (total activity). SucB activity could be detected from pH 4.5 to 7, albeit at very low activity levels at pH values above 6.3. SucB displayed maximal activity at pH 5.0 (Fig. 2A). The optimal temperature for SucB total activity is in the range of 37 to 40°C (Fig. 2B). At temperatures of 50°C or higher, no activity could be detected, whereas at lower temperatures, the SucB specific activity remained relatively high, with 50% activity remaining at 25°C.
FIG. 2.
Effect of pH (A) and temperature (B) on SucB activity. The enzymatic activity was determined by measuring the amount of glucose released from the initial reaction of SucB (14 μg) incubated with 200 μl of 100 mM sucrose in citrate-acetate buffer at 37°C. Values depicted are the means of duplicates ± standard errors of the means based on at least two independent experiments.
Kinetic analysis of SucB activity.
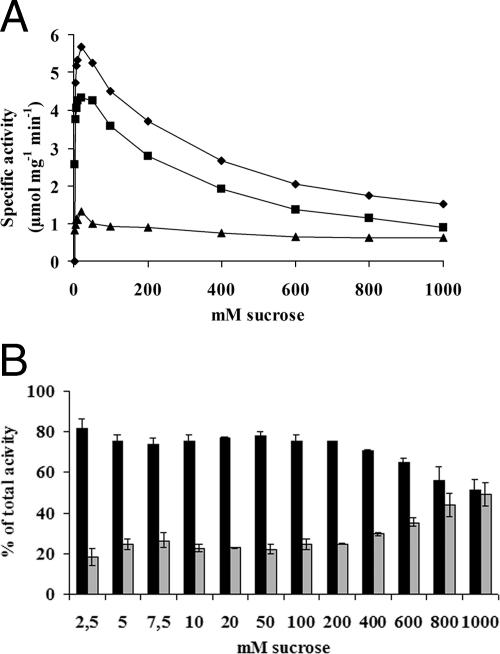
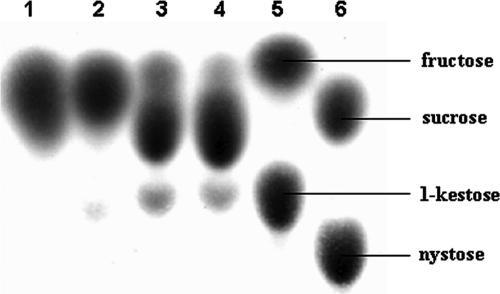
Incubation of SucB with increasing sucrose concentrations led to typical Michaelis-Menten-type kinetics with substrate inhibition (apparent Km, Ki, and Vmax of 2.0 ± 0.2 mM, 268.1 ± 18.1 mM, and 6.6 ± 0.2 μmol mg−1 min−1, respectively). Hydrolysis and FTF reactions displayed similar patterns, with apparent Km values of 2.5 ± 0.2 and 0.9 ± 0.5 mM, apparent Vmax values of 5.5 ± 0.1 and 1.2 ± 0.1 μmol mg−1 min−1, and apparent Ki values of 206.3 ± 12.8 and 797.6 ± 196.5 mM, respectively. Increasing the sucrose concentration from 2.5 mM to 1 M resulted in decreased hydrolysis activity and increased (20% to 50%) FTF activity in relation to total SucB activity (Fig. 3). These observations were confirmed by the TLC product analysis, showing increased 1-kestose and decreased free fructose synthesis at higher sucrose concentrations (Fig. 4). HPAEC analysis showed that apart from the formation of 1-kestose as the major FTF product, minor amounts of nystose were also produced (result not shown).
FIG. 3.
Effect of sucrose concentration on SucB activity. (A) Total activity was determined by measuring the amount of glucose released from the initial reaction of SucB incubated with 12 sucrose concentrations ranging from 2.5 mM to 1 M in 50 mM acetate buffer (pH 5.0) at 37°C. Values depicted were calculated from triplicate measurements. ⧫, total activity; ▪, invertase activity; ▴, FTF activity. (B) Percentage of either hydrolytic (black bars) or FTF (gray bars) activity compared to the total activity of SucB displayed for a range of sucrose concentrations. Measurements are the means of duplicates ± standard errors of the means based on two independent initial measurements. Approximately 14 μg of purified protein was used in each case.
FIG. 4.
TLC analysis of the reaction products after overnight incubation of SucB at 37°C with 50 mM (lane 1) (18), 100 mM (lane 2), 500 mM (lane 3), and 1 M (lane 4) sucrose. Lane 5 shows the standards fructose and kestose, and lane 6 shows sucrose and nystose. Approximately 14 μg of purified protein was used in all incubations. All samples were equilibrated to the same concentration before being spotted onto the plate.
SucB substrate specificity and product formation.
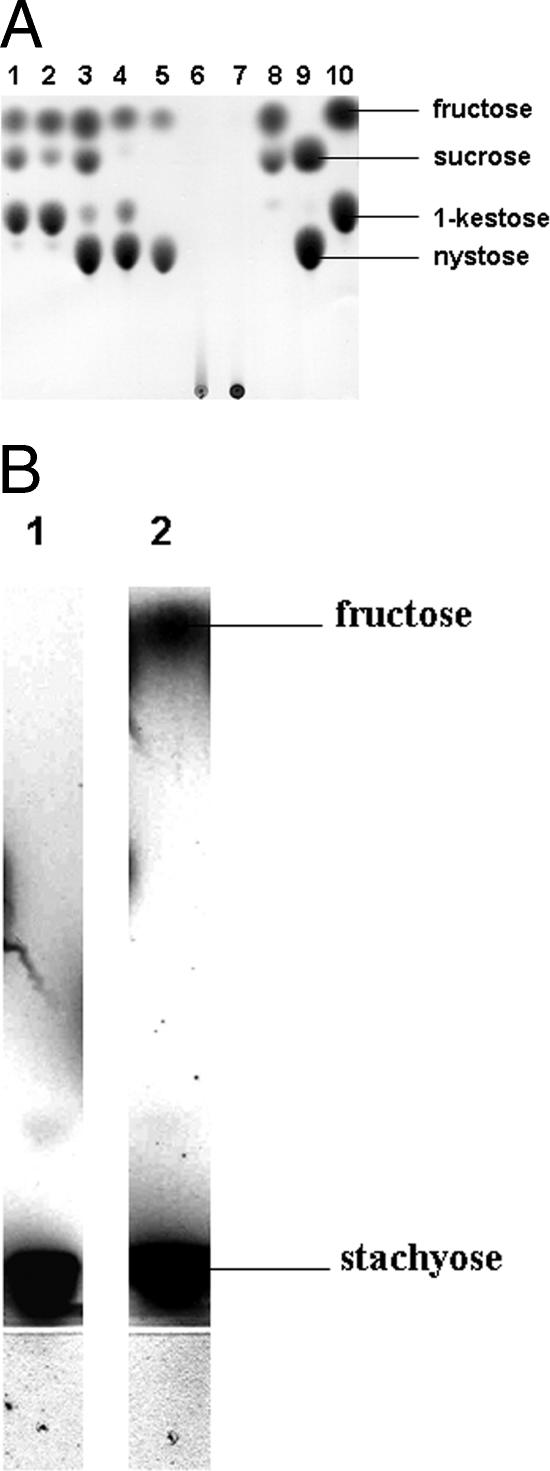
Substrate specificity analysis showed that SucB is able to hydrolyze sucrose, raffinose, and the inulin-type oligosaccharides 1-kestose and nystose (releasing fructose in each case) as well as to perform oligomerization reactions (Fig. 5 A). Incubation of SucB with 100 mM 1-kestose in the presence or absence of 100 mM sucrose produced free fructose, sucrose, and nystose, indicating that 1-kestose could be used as both donor and acceptor substrates. Alternatively, the sucrose formed was subsequently used as a donor substrate for the transfer of free fructose to 1-kestose. Incubations of SucB with nystose alone or nystose plus sucrose yielded only free fructose, sucrose, and 1-kestose (Fig. 5A). HPAEC analysis confirmed that nystose was the largest product produced from the incubation of SucB with kestose and that a minor amount of pentakestose (degree of polymerization of 4) was produced after overnight incubation. SucB incubation in the presence of sucrose plus 1-kestose or nystose did not facilitate product diversification but resulted in an increase in concentration of the observed product only. Furthermore, SucB incubation with galactose and sucrose did not yield any products other than those observed for sucrose alone (Fig. 5A). SucB hydrolyzed the sucrose moieties of the sugars raffinose [α-d-galactose-(1,6)-α-d-glucose-(1,2)-β-d-fructose] (Fig. 5A) and stachyose [α-d-galactose-(1,6)-α-d-galactose-(1,6)-α-d-glucose-(1,2)-β-d-fructose] (Fig. 5B). No hydrolysis of any of the α-glycosidic linkage sugars {trehalose [α-d-glucose-(1,1)-α-d-glucose], turanose [α-d-glucose-(1,3)-β-d-fructose], palatinose [α-d-glucose-(1,6)-β-d-fructose], or melizitose [α-d-glucose-(1,2)-β-d-fructose-(1,3)-α-d-glucose]} was observed. Using trehalose alone or in combination with sucrose (ratios of 5:1 to 1:5) as a substrate, only 1-kestose formation from sucrose could be observed as the FTF product (results not shown). None of these α-glycosidic bond substrates were used as donor/acceptor substrates in FTF reactions with sucrose (ratios 5:1 to 1:5). Hydrolysis of larger polysaccharides such as inulin or levan could not be detected, not even after overnight incubation (Fig. 5A). Similar results were obtained when cell extracts from the recombinant S. cerevisiae strain carrying the construct pYES-DEST52-sucB were used.
FIG. 5.
TLC analysis of substrate specificity of SucB and reaction products after overnight incubation at pH 5.0 and 37°C. (A) A total of 100 mM kestose with sucrose (lane1), kestose (lane 2), nystose with sucrose (lane 3), nystose (lane 4), raffinose (lane 5), 1% inulin (chicory) (see text) (lane 6), and levan (Bacillus subtilis produced) (gift from Cosun Food Technology, The Netherlands) (lane 7) and 100 mM galactose with sucrose (lane 8). Lanes 9 and 10 contain the standards fructose, sucrose, 1-kestose, and nystose. (B) A total of 100 mM stachyose incubated without (lane 1) and with (lane 2) SucB. Released fructose is indicated. Approximately 14 μg of purified protein was used in each case.
In silico translation of the SucB reading frame revealed the presence of 13 cysteine residues, which can potentially form disulfide bridges. The addition of up to 100 mM of the reducing agents DTT or BME did not influence the sucrose hydrolysis or FTF activities of SucB, indicating the absence of any structurally important disulfide bridges.
Disruption of the sucB gene.
Disruption of sucB did not result in a significant change in growth rates and yields of A. niger in liquid medium containing sucrose, inulin, glucose, fructose, xylose, maltose, or starch (data not shown). Interestingly, growth of the sucB deletion mutant strains (e.g., strain NC1.1) on solid medium containing these substrates resulted in an earlier onset (approximately 1 day) of sporulation compared to that of the wild-type A. niger. The inclusion of additional uridine in the culture medium to exclude suboptimal complementation by PyrG did not rescue the NC1.1 strain from this sporulation effect. No difference in colony diameter was observed between the NC1.1 and wild-type strains on the various carbon sources, indicating that the growth of the ΔsucB strain was not affected.
DISCUSSION
The recent availability of the complete genome sequence of A. niger (25) enabled the identification of two novel putative intracellular invertases (sucB and sucC) (39). Although these genes share conserved amino acid residues with other GH32 members, phylogenetically, they cluster together with other putative intracellular invertases of fungal origin in a new distinct group (Fig. 1). These new putative invertases also contain all the conserved catalytic residues, as depicted in the sequence logos (Table 2). The only exception is Sir-1, which is missing the catalytic aspartate in domain A. This invertase was isolated from A. niger strain IBT10sb and displayed increased FTF properties (32).
We have previously shown that SucB expression was upregulated by sucrose and inulin, whereas the enzyme was constitutively expressed at a low level with all other substrates used (39). The sucB gene also appears to be under catabolite repression control, which was evident from expression profiling in a creA deletion strain (39). With regard to sucC, no expression could be detected in mycelial mRNA under the same conditions. The use of chromosomal DNA to construct the predicted open reading frame of SucC also failed to produce a functional protein in both E. coli and S. cerevisiae.
In silico analysis of the SucB sequence revealed the absence of any recognizable signal peptide sequences for protein secretion, indicating that it may play a role in A. niger intracellular metabolism. In view of the low levels of expression observed for SucB in A. niger, and to avoid simultaneous separation with other invertases/fructosyltransferases present in A. niger (18), we overproduced the SucB enzyme in E. coli as well as in an invertase-negative strain of S. cerevisiae and subsequently determined its biochemical properties.
SucB clearly acts as an invertase, which is able to hydrolyze the glucose-fructose glycosidic linkage in the smaller fructose-containing oligosaccharides sucrose, kestose, and nystose. Oligosaccharides larger than nystose, including polymeric inulin and levan, could not be hydrolyzed. The enzyme was unable to hydrolyze α-glycosidic bonds in substrates or to use these compounds as donor/acceptor substrates in FTF reactions. Weak hydrolysis of the sucrose moiety of stachyose, but not melizitose, indicates that the fructose of the sucrose moiety should thus be positioned terminally to enable correct orientation and binding in the active site. A similar observation was made for the β-fructosidase from Thermotoga maritima, where it was shown that hydrolysis occurs in a typical exo fashion (19). Taking into account the diversity of the substrates hydrolyzed, one could assume that the −1 subsite in the active-site cleft (for numbering, see reference 10) can accommodate fructose (1, 2, 22) and that the enzyme most probably does not contain multiple binding sites for fructose (as in the case of the exoinulinase of Aspergillus awamori) (17). In the three-dimensional structure of the Cichorium intybus fructan 1-exohydrolase, Verhaest et al. (35) previously observed the presence of multiple glycerol molecules bound in the cavity between the N- and C-terminal domains of the protein. This forms an open cleft, which is emerging from the active site and is believed to be responsible for the binding of inulin or higher-molecular-weight fructans. Obstruction of this cleft could possibly influence the binding of high-molecular-weight inulins, limiting the enzyme to the hydrolysis of small oligosaccharides only (2, 35). In view of the low amino acid similarity between SucB and other characterized invertases and the absence of structural data, we can only speculate that the same obstructing feature is present in SucB. This feature could explain the inability of the enzyme to bind and hydrolyze larger oligo- and polymeric fructans.
Apart from the hydrolytic activity observed for SucB, the enzyme was also able to perform FTF reactions. This activity was already detected at sucrose concentrations as low as 2.5 mM, with 1-kestose as the major oligomerization product (20 to 30% of total activity). At 1 M, SucB displayed approximately 50% FTF activity, due largely to a decrease in hydrolytic activity. Nystose was also produced as a minor product in the FTF reaction when 1-kestose was used as a substrate. Also, the presence of a minor amount of pentakestose was observed after overnight incubation with nystose. Larger SucB products have never been observed. The data thus indicate that SucB is responsible for the intracellular production of small inulin-type oligosaccharides. In 1995, Muramatsu and Nakakuki (21) previously described the purification and characterization of an intracellular beta-fructofuranosidase from Aspergillus sydowi that could transfer fructose from sucrose to trehalose, thus creating novel oligofructosyl trehaloses. However, when SucB was incubated with trehalose, no novel oligosaccharides were observed.
The SucB characteristics differ from the previously published data on the extracellular A. niger invertase Suc1/SucA/INV enzyme (6, 18, 37), and other invertases, in a number of aspects. SucB displayed an apparent Km of 2.0 ± 0.2 mM for sucrose, which is substantially lower than that reported previously (30 and 160 mM for Suc1 and 35.67 mM for INV) but comparable to that of the extracellular acid invertase of Fusarium solani (3.57 mM) (5). Extracellular invertases of both fungal as well as bacterial origin generally display lower affinities for sucrose than was observed for SucB (13, 19, 28, 37). However, Rubio and Maldonado (29) previously described the purification and characterization of an invertase from an A. niger strain isolated from lemons. The invertase was purified from the mycelial lysate and displayed a substrate affinity of 0.0625 mM for sucrose and a temperature optimum of 60°C. These characteristics clearly deviate from what has been observed for SucB, indicating that this protein might be either another intracellular invertase or an isoform of SucB produced intracellularly in A. niger and not during recombinant expression in E. coli or yeast.
Using sucrose as a substrate, SucB displayed an apparent Vmax (total activity) of 6.6 ± 0.2 μmol mg−1 min−1 (this study). This figure is more than a thousandfold lower than the Vmax for the extracellular invertase in A. niger AS0023 (7,758.3 μmol mg−1 min−1) (18). The purified extracellular A. niger invertase also did not display any detectable FTF activity, not even at a sucrose concentration as high as 2.2 M (18), suggesting that the FTF described previously in literature could have derived from contaminating fructosyltransferases (16).
Compared to the extracellular invertase of A. niger (Suc1), SucB displayed a lower optimum temperature value (37 to 40°C versus 50°C, respectively), whereas a comparable pH optimum was determined (5, 37). However, SucB was active only in a narrow pH range (above pH 4 and below pH 7) (Fig. 2A) compared to that observed for Suc1 (above pH 3 and below pH 10) (5, 37). In the extracellular environment, Suc1 should be able to function in fluctuating pH conditions, where the extracellular pH could vary between 1.5 and 7.0 (14). In the intracellular environment, however, the cytoplasmic and vacuolar pHs of A. niger are kept constant at 7.6 and 6.2, respectively. This balance is maintained in order to control pH-sensitive processes such as DNA transcription and protein synthesis (14). Taking these facts into consideration, and in the absence of any detectable sequence for protein export, we conclude that SucB functions suboptimally in the intracellular environment.
Although intracellular invertases have been identified in fungi previously (12, 21, 23), little is known about the role that they play in the intracellular environment. To determine whether SucB plays a crucial role in the metabolism of A. niger, a SucB disruption mutant strain was constructed. No difference in growth rate, yield, and morphology was observed between the sucB disruptant and wild-type A. niger N402 using liquid minimal medium with sucrose or inulin as a carbon source. When the NC1.1 sucB disruptant strain was grown on solid minimal medium containing various substrates, an earlier onset of sporulation was observed compared to that for the wild type. However, as observed in liquid medium, no difference in growth was observed, since the colony diameter was equal to that of wild-type A. niger. Supplementing the culture medium with uridine to minimize suboptimal complementation by the inserted pyrG gene did not alleviate the observed effect, suggesting that SucB (in)directly plays a role in the sporulation of A. niger.
Intracellular proteins usually do not contain disulfide bridges, which play a crucial role in the structure and function of extracellular proteins (27). These bridges could be disrupted by the addition of reducing agents, e.g., DTT and BME, which in turn may cause a loss of activity or decreased enzyme stability. The inability of high concentrations of either DTT or BME to disrupt SucB activity gives a further indication that no disulfide bridges crucial to activity or structural integrity are present. This further supports the view that SucB is functioning in the intracellular environment in A. niger.
Considering the high affinity for sucrose, the narrow functional pH range, and the absence of an export signal sequence and functionally important disulfide bridges, we speculate that SucB plays an intracellular role in salvaging low concentrations of sucrose, kestose, or nystose into fructose and glucose as energy sources. SucB may also function in the transfer of fructose units from sucrose to fructan or unknown acceptor molecules. These molecules may be responsible for energy storage or the regulation of osmolarity or may play a role in the induction of other proteins involved in the modification of fructans.
Further analysis and complementation studies should be conducted to determine the effect of sucB gene disruption on the expression of other fructan-modifying enzymes and the possible role it could play in initiating the earlier onset of sporulation.
Acknowledgments
We thank N. Carvalho for her assistance in generating the A. niger sucB gene deletion strain and P. van Heusden for providing us with the S. cerevisiae suc1 deletion strain. We thankfully acknowledge COSUN Food Technology Centre (CFTC, Roosendaal, The Netherlands) for providing substrates, HPAEC analysis, and stimulating discussions.
The national IOP program (The Netherlands) is acknowledged for funding this project (project code IGE 1021). This project is part of the CarbNet program (Carbohydrate Modifying Enzyme Network of Aspergillus niger).
Footnotes
Published ahead of print on 9 February 2007.
REFERENCES
- 1.Alberto, F., C. Bignon, G. Sulzenbacher, B. Henrissat, and M. Czjzek. 2004. The three-dimensional structure of invertase (beta-fructosidase) from Thermotoga maritima reveals a bimodular arrangement and an evolutionary relationship between retaining and inverting glycosidases. J. Biol. Chem. 279:18903-18910. [DOI] [PubMed] [Google Scholar]
- 2.Alberto, F., E. Jordi, B. Henrissat, and M. Czjzek. 2006. Crystal structure of inactivated Thermotoga maritima invertase in complex with the trisaccharide substrate raffinose. Biochem. J. 395:457-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennet, J. W., and L. L. Lasure. 1991. Growth media, p. 441-457. In J. W. Bennet and L. L. Lasure (ed.), More gene manipulation in fungi. Academic Press, New York, NY.
- 4.Reference deleted.
- 5.Bhatti, H. N., M. Asgher, A. Abbas, R. Nawaz, and M. A. Sheikh. 2006. Studies on kinetics and thermostability of a novel acid invertase from Fusarium solani. J. Agric. Food Chem. 54:4617-4623. [DOI] [PubMed] [Google Scholar]
- 6.Boddy, L. M., T. Berges, C. Barreau, M. H. Vainstein, M. J. Dobson, D. J. Ballance, and J. F. Peberdy. 1993. Purification and characterisation of an Aspergillus niger invertase and its DNA sequence. Curr. Genet. 24:60-66. [DOI] [PubMed] [Google Scholar]
- 7.Bos, C. J., A. J. Debets, K. Swart, A. Huybers, G. Kobus, and S. M. Slakhorst. 1988. Genetic analysis and the construction of master strains for assignment of genes to six linkage groups in Aspergillus niger. Curr. Genet. 14:437-443. [DOI] [PubMed] [Google Scholar]
- 8.Coutinho, P. M., and B. Henrissat. 1999. Carbohydrate-active enzymes: an integrated database approach, p. 3-12. In H. J. Gilbert, G. Davies, B. Henrissat, and B. Svensson (ed.), Recent advances in carbohydrate bioengineering. The Royal Society of Chemistry, Cambridge, United Kingdom.
- 9.Crooks, G. E., G. Hon, J. M. Chandonia, and S. E. Brenner. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies, G. J., K. S. Wilson, and B. Henrissat. 1997. Nomenclature for sugar-binding subsites in glycosyl hydrolases. Biochem. J. 321:557-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Ruiter-Jacobs, Y. M., M. Broekhuijsen, S. E. Unkles, E. I. Campbell, J. R. Kinghorn, R. Contreras, P. H. Pouwels, and C. A. van den Hondel. 1989. A gene transfer system based on the homologous pyrG gene and efficient expression of bacterial genes in Aspergillus oryzae. Curr. Genet. 16:159-163. [DOI] [PubMed] [Google Scholar]
- 12.Gascon, S., and J. O. Lampen. 1968. Purification of the internal invertase of yeast. J. Biol. Chem. 243:1567-1572. [PubMed] [Google Scholar]
- 13.Gascon, S., N. P. Neumann, and J. O. Lampen. 1968. Comparative study of the properties of the purified internal and external invertases from yeast. J. Biol. Chem. 243:1573-1577. [PubMed] [Google Scholar]
- 14.Hesse, S. J., G. J. Ruijter, C. Dijkema, and J. Visser. 2002. Intracellular pH homeostasis in the filamentous fungus Aspergillus niger. Eur. J. Biochem. 269:3485-3494. [DOI] [PubMed] [Google Scholar]
- 15.Reference deleted.
- 16.Hirayama, M., N. Sumi, and H. Hidaka. 2006. Purification and characterization of a fructooligosaccharide-producing beta-fructofuranosidase from Aspergillus niger ATCC 20611. Agric. Biol. Chem. 53:667-673. [Google Scholar]
- 17.Kulminskaya, A. A., M. Arand, E. V. Eneyskaya, D. R. Ivanen, K. A. Shabalin, S. M. Shishlyannikov, A. N. Saveliev, O. S. Korneeva, and K. N. Neustroev. 2003. Biochemical characterization of Aspergillus awamori exoinulinase: substrate binding characteristics and regioselectivity of hydrolysis. Biochim. Biophys. Acta 1650:22-29. [DOI] [PubMed] [Google Scholar]
- 18.L'Hocine, L., Z. Wang, B. Jiang, and S. Xu. 2000. Purification and partial characterization of fructosyltransferase and invertase from Aspergillus niger AS0023. J. Biotechnol. 81:73-84. [DOI] [PubMed] [Google Scholar]
- 19.Liebl, W., D. Brem, and A. Gotschlich. 1998. Analysis of the gene for beta-fructosidase (invertase, inulinase) of the hyperthermophilic bacterium Thermotoga maritima, and characterisation of the enzyme expressed in Escherichia coli. Appl. Microbiol. Biotechnol. 50:55-64. [DOI] [PubMed] [Google Scholar]
- 20.May, T., R. I. Mackie, G. C. Fahey, Jr., J. C. Cremin, and K. A. Garleb. 1994. Effect of fiber source on short-chain fatty acid production and on the growth and toxin production by Clostridium difficile. Scand. J. Gastroenterol. 29:916-922. [DOI] [PubMed] [Google Scholar]
- 21.Muramatsu, M., and T. Nakakuki. 1995. Enzymatic synthesis of novel fructosyl and oligofructosyl trehaloses by Aspergillus sydowi beta-fructofuranosidase. Biosci. Biotechnol. Biochem. 59:208-212. [DOI] [PubMed] [Google Scholar]
- 22.Nagem, R. A., A. L. Rojas, A. M. Golubev, O. S. Korneeva, E. V. Eneyskaya, A. A. Kulminskaya, K. N. Neustroev, and I. Polikarpov. 2004. Crystal structure of exo-inulinase from Aspergillus awamori: the enzyme fold and structural determinants of substrate recognition. J. Mol. Biol. 344:471-480. [DOI] [PubMed] [Google Scholar]
- 22a.Naumoff, D. G. 2001. Beta-fructosidase superfamily: homology with some alpha-L-arabinases and beta-D-xylosidases. Proteins 42:66-76. [DOI] [PubMed] [Google Scholar]
- 23.Nguyen, Q. D., F. Mattes, Ã. Hoschke, J. Rezessy-Szabó, and M. K. Bhat. 1999. Production, purification and identification of fructooligosaccharides produced by beta-fructofuranosidase from Aspergillus niger IMI 303386. Biotechnol. Lett. 21:183-186. [Google Scholar]
- 24.Ozimek, L. K., S. Kralj, M. J. van der Maarel, and L. Dijkhuizen. 2006. The levansucrase and inulosucrase enzymes of Lactobacillus reuteri 121 catalyse processive and non-processive transglycosylation reactions. Microbiology 152:1187-1196. [DOI] [PubMed] [Google Scholar]
- 25.Pel, H. J., J. H. de Winde, D. B. Archer, P. S. Dyer, G. Hofmann, P. J. Schaap, G. Turner, R. P. de Vries, R. Albang, K. Albermann, M. R. Andersen, J. D. Bendtsen, J. A. E. Benen, M. van den Berg, S. Breestraat, M. X. Caddick, R. Contreras, M. Cornell, P. M. Coutinho, E. G. J. Danchin, A. J. M. Debets, P. Dekker, P. W. M. van Dijck, A. van Dijk, L. Dijkhuizen, A. J. M. Driessen, C. d'Enfert, S. Geysens, C. Goosen, G. S. P. Groot, P. W. J. de Groot, T. Guillemette, B. Henrissat, M. Herweijer, J. P. T. W. van den Hombergh, C. A. M. J. J. van den Hondel, R. T. J. M. van der Heijden, R. M. van der Kaaij, F. M. Klis, H. J. Kools, C. P. Kubicek, P. A. van Kuyk, J. Lauber, X. Lu, M. J. E. C. van der Maarel, R. Meulenberg, H. Menke, A. M. Mortimer, J. Nielsen, S. G. Oliver, M. Olsthoorn, K. Pal, N. N. M. E. van Peij, A. F. J. Ram, U. Rinas, J. A. Roubos, C. M. J. Sagt, M. Schmoll, J. Sun, D. Ussery, J. Varga, W. Vervecken, P. J. I. van de Vondervoort, H. Wedler, H. A. B. Wösten, A. Zeng, A. J. J. van Ooyen, J. Visser, and H. Stam. 2007. Genome sequence of Aspergillus niger strain CBS 513.88: a versatile cell factory. Nat. Biotechnol. 25:221-231. [DOI] [PubMed] [Google Scholar]
- 26.Punt, P. J., and C. A. van den Hondel. 1992. Transformation of filamentous fungi based on hygromycin B and phleomycin resistance markers. Methods Enzymol. 216:447-457. [DOI] [PubMed] [Google Scholar]
- 27.Raina, S., and D. Missiakas. 1997. Making and breaking disulfide bonds. Annu. Rev. Microbiol. 51:179-202. [DOI] [PubMed] [Google Scholar]
- 28.Reddy, A., and F. Maley. 1996. Studies on identifying the catalytic role of Glu-204 in the active site of yeast invertase. J. Biol. Chem. 271:13953-13957. [DOI] [PubMed] [Google Scholar]
- 29.Rubio, M. C., and M. C. Maldonado. 1995. Purification and characterization of invertase from Aspergillus niger. Curr. Microbiol. 31:80-83. [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 31.Sghir, A., J. M. Chow, and R. I. Mackie. 1998. Continuous culture selection of bifidobacteria and lactobacilli from human faecal samples using fructooligosaccharide as selective substrate. J. Appl. Microbiol. 85:769-777. [DOI] [PubMed] [Google Scholar]
- 32.Somiari, R., H. Brzeski, R. Tate, S. Bieleck, and J. Polak. 1997. Cloning and sequencing of an Aspergillus niger gene coding for [beta]-fructofuranosidase. Biotechnol. Lett. 19:1243-1247. [Google Scholar]
- 33.Tannock, G. W. 1997. Probiotic properties of lactic-acid bacteria: plenty of scope for fundamental R & D. Trends Biotechnol. 15:270-274. [DOI] [PubMed] [Google Scholar]
- 34.van Hartingsveldt, W., I. E. Mattern, C. M. van Zeijl, P. H. Pouwels, and C. A. van den Hondel. 1987. Development of a homologous transformation system for Aspergillus niger based on the pyrG gene. Mol. Gen. Genet. 206:71-75. [DOI] [PubMed] [Google Scholar]
- 35.Verhaest, M., W. V. Ende, K. L. Roy, C. J. De Ranter, A. V. Laere, and A. Rabijns. 2005. X-ray diffraction structure of a plant glycosyl hydrolase family 32 protein: fructan 1-exohydrolase IIa of Cichorium intybus. Plant J. 41:400-411. [DOI] [PubMed] [Google Scholar]
- 36.Vijn, I., and S. Smeekens. 1999. Fructan: more than a reserve carbohydrate. Plant Physiol. 120:351-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wallis, G. L., F. W. Hemming, and J. F. Peberdy. 1997. Secretion of two beta-fructofuranosidases by Aspergillus niger growing in sucrose. Arch. Biochem. Biophys. 345:214-222. [DOI] [PubMed] [Google Scholar]
- 38.Yanai, K., A. Nakane, A. Kawate, and M. Hirayama. 2001. Molecular cloning and characterization of the fructooligosaccharide-producing beta-fructofuranosidase gene from Aspergillus niger ATCC 20611. Biosci. Biotechnol. Biochem. 65:766-773. [DOI] [PubMed] [Google Scholar]
- 39.Yuan, X.-L., C. Goosen, H. Kools, M. J. E. C. van der Maarel, C. A. M. J. J. van den Hondel, L. Dijkhuizen, and A. F. J. Ram. 2006. Database mining and transcriptional analysis of genes encoding inulin-modifying enzymes of Aspergillus niger. Microbiology 152:3061-3073. [DOI] [PubMed] [Google Scholar]