Abstract
The hemiascomycete yeast Dekkera bruxellensis, also known as Brettanomyces bruxellensis, is a major cause of wine spoilage worldwide. Wines infected with D. bruxellensis develop distinctive, unpleasant aromas due to volatile phenols produced by this species, which is highly ethanol tolerant and facultatively anaerobic. Despite its importance, however, D. bruxellensis has been poorly genetically characterized until now. We performed genome survey sequencing of a wine strain of D. bruxellensis to obtain 0.4× coverage of the genome. We identified approximately 3,000 genes, whose products averaged 49% amino acid identity to their Saccharomyces cerevisiae orthologs, with similar intron contents. Maximum likelihood phylogenetic analyses suggest that the relationship between D. bruxellensis, S. cerevisiae, and Candida albicans is close to a trichotomy. The estimated rate of chromosomal rearrangement in D. bruxellensis is slower than that calculated for C. albicans, while its rate of amino acid evolution is somewhat higher. The proteome of D. bruxellensis is enriched for transporters and genes involved in nitrogen and lipid metabolism, among other functions, which may reflect adaptations to its low-nutrient, high-ethanol niche. We also identified an adenyl deaminase gene that has high similarity to a gene in bacteria of the Burkholderia cepacia species complex and appears to be the result of horizontal gene transfer. These data provide a resource for further analyses of the population genetics and evolution of D. bruxellensis and of the genetic bases of its physiological capabilities.
The flavors and aromas of wine are the products of complex interactions between many microorganisms. Saccharomyces cerevisiae is the primary yeast used in wine-making, but other fungi and bacteria, welcome or unwelcome, also contribute to most alcoholic fermentations. Many of these species occur naturally on the skin of grapes and flourish briefly in the initial stages of fermentation before being killed by the rising ethanol concentration (49). Other species appear to make the winery itself a primary habitat, surviving on the walls of the winery, on the interior surfaces of presses and fermentation tanks, or in the wood of the barrels (22), in ideal positions to colonize the fermenting grape must or the maturing wine. Dekkera bruxellensis, a hemiascomycete yeast also known as Brettanomyces bruxellensis, is one such species.
D. bruxellensis is probably the major microbial cause of wine spoilage worldwide and causes substantial economic losses within the wine industry (22). Wines infected by D. bruxellensis are said to have “Brett” character: they may smell mousy or medicinal or of wet wool, burnt plastic, or horse sweat (38). The mousy taint is the result of pyridines synthesized by D. bruxellensis from lysine and ethanol (29), while medicinal or barnyard odors are caused by the volatile phenols 4-ethylguaiacol and 4-ethylphenol, secondary metabolites produced by D. bruxellensis from phenolic acids naturally present in the grape must (14, 30). D. bruxellensis may have been present in wineries for centuries, and a very slight Brett character is traditionally considered desirable in certain styles of wine (22). The frequency and severity of D. bruxellensis contamination have increased in recent decades, however, as winemaking trends have changed to favor the production of wines that contain more residual sugars and that may be unsulfited, unfiltered, or aged on lees (dead yeast cells), all factors that are favorable to D. bruxellensis growth.
D. bruxellensis possesses a number of adaptations that allow it to survive in the physiologically challenging environments of fermenting must and maturing wine. Like S. cerevisiae, it is ethanol tolerant, facultatively anaerobic, and petite positive (it can survive without mitochondrial DNA). Both species are also Crabtree positive and thus ferment preferentially in the presence of high glucose under aerobic conditions (36, 48). This suite of characteristics has evolved independently in each of the two lineages, possibly in response to similar selective pressures. The species differ in a number of other regards, however. For example, D. bruxellensis utilizes glucose less efficiently and grows much more slowly than S. cerevisiae (57) but can assimilate a wider variety of alternative carbon sources (16). Together, these characteristics explain the typical ecological succession observed during the course of D. bruxellensis-contaminated alcoholic fermentations, with S. cerevisiae dominating throughout the primary fermentation phase and then being replaced by D. bruxellensis in the maturation phase, when ethanol concentrations are high and only minute amounts of residual sugars remain (51).
Despite its economic importance and physiological interest, D. bruxellensis has remained largely unstudied at the genomic level. Previous work has focused on sequencing rRNA regions for phylogenetic analysis (67) or to aid in molecular detection of D. bruxellensis contamination (45), and only one nuclear protein-coding gene has been sequenced from this species to date (33). Consequently, the genetic bases of the physiological capabilities of D. bruxellensis remain largely unknown. To investigate these, and to provide a resource for further research on this species, we undertook genome survey sequencing of strain CBS 2499, isolated from wine. We report here a preliminary analysis of the genome organization and gene content of this strain.
MATERIALS AND METHODS
Sequence data.
DNA from Y1031, a petite mutant derived from strain CBS 2499, was used to construct a genomic library of random Sau3AI partial digestion fragments, of average length 5 kb, in the low-copy-number Escherichia coli vector pMCL210. Library construction and sequence data generation were done by Agowa (Berlin, Germany). Sequences were obtained from both ends of the insert for 7,381 clones and from one end only for a further 98 clones. Read sequences were base called by Phred and vector masked using Cross_Match (20). Sequence data were assembled into contigs using Phrap (http://www.phrap.org), and low-quality sequence at the ends of contigs was trimmed using a purpose-written Perl script.
Gene identification.
To identify protein-coding genes in D. bruxellensis, we first compared the contig sequences to a database of S. cerevisiae protein sequences (5,770 protein-coding genes, excluding dubious open reading frames and transposable element genes, downloaded from the Saccharomyces Genome Database in July 2005). Regions of the contigs greater than 100 nucleotides in length were annotated as orthologs to S. cerevisiae genes to which they were reciprocal best BLAST hits (using BLASTX and TBLASTN bit scores) (1).
To identify D. bruxellensis genes that do not have S. cerevisiae orthologs, we repeated this analysis using the nonredundant (NR) protein database (downloaded from the National Center for Biotechnology Information in July 2005). Regions of the D. bruxellensis contigs were considered to be orthologs of characterized genes in the NR database to which they were reciprocal best hits if (i) the contig region had no reciprocal best S. cerevisiae hit or (ii) the BLASTX bit score of the NR reciprocal best hit to the region was at least 100 bits higher than that of the S. cerevisiae hit.
Finally, we used S. cerevisiae rRNA and tRNA gene sequences as BLASTN queries against the contigs, with an upper E-value threshold of 1e-10, to identify D. bruxellensis homologs of these genes.
Phylogenetic position.
We constructed phylogenetic trees using amino acid sequence data from D. bruxellensis, S. cerevisiae, and eight other fungal species for which the whole genome sequence was available (Candida glabrata, Kluyveromyces lactis, Debaryomyces hansenii, Yarrowia lipolytica [19], Candida albicans [34], Ashbya gossypii [18], Aspergillus nidulans [23], and Schizosaccharomyces pombe [66]). We accepted as sets of orthologous genes those genes that were best mutual BLASTP hits, with E values of <1e-40, in all possible pairwise comparisons among all 10 taxa. This yielded 396 sets of orthologous genes.
In those cases where only a partial gene sequence was present in the D. bruxellensis data, we extracted the corresponding segment of the orthologous gene in each of the other species based on BLASTP HSP coordinates; in all other cases, complete gene sequences were used. The amino acid sequences of each set of orthologs were aligned using T-Coffee (46), and regions of uncertain alignment were removed using Gblocks (13). Each of the 396 alignments was separately used to estimate the species phylogeny, using PHYML (26) with the JTT model of amino acid substitution and a gamma distribution of substitution rates with four categories.
We visualized the degree of topological conflict among these trees by constructing a consensus network (31) using SplitsTree4 (32). To test whether one of the three best-supported topologies suggested by this consensus network was a significantly better fit to the total sequence data than the others, we performed the Shimodaira-Hasegawa test (56) as implemented in Tree-Puzzle (54). We concatenated the 396 gene alignments to produce an alignment of 115,036 residues and calculated the log likelihood of each of the three topologies given these sequence data and a JTT model of substitution. To ensure that model misspecification did not affect our results, we repeated the Shimodaira-Hasegawa test using four other substitution models (WAG, VT, BLOSUM 62, and Dayhoff).
Genome architecture.
For each pair of genes adjacent in D. bruxellensis, we determined whether their orthologs were also adjacent (or separated by fewer than five genes) in the S. cerevisiae genome. We also used the same method to calculate the degree of gene order conservation between D. bruxellensis and C. albicans. For this purpose, we considered as orthologous any pair of D. bruxellensis and C. albicans genes that shared a mutual S. cerevisiae ortholog. The list of C. albicans/S.cerevisiae orthologs we used here was obtained from http://www.candidagenome.org.
We used two methods to identify introns in D. bruxellensis. Because many introns occur at conserved genomic locations across the hemiascomycetes (8), we first examined those D. bruxellensis genes orthologous to intron-containing S. cerevisiae genes. We aligned each pair of orthologous genes and determined whether the D. bruxellensis gene contained known hemiascomycete intron splice or branch sites (obtained from reference 27) and/or noncoding, presumably intronic, sequence in the region of the gene corresponding to the S. cerevisiae intron. Secondly, to detect D. bruxellensis-specific introns, we also identified genes that appeared to contain intron length segments of extra sequence when compared to their S. cerevisiae orthologs. To do this, we searched for genes that returned two significant BLASTX HSPs to a single S. cerevisiae protein, where the interval between those HSPs was 50 nucleotides (approximately the minimum known hemiascomycete intron length [8]) or greater in D. bruxellensis but 10 nucleotides or smaller in S. cerevisiae. These genes were then screened for known intronic splice and branch sites, as described above.
We used Tandem Repeats Finder (6) to identify microsatellites in the assembly. To identify transposable elements in this genome, we used the D. bruxellensis contigs as BLASTX queries against a database of protein sequences of TYA and TYB genes from Ty elements of multiple hemiascomycete species (43) and protein-coding regions of non-long terminal repeat (LTR) retrotransposons and DNA transposons from C. albicans and Y. lipolytica (12, 25, 44). Regions of the contigs that had hits with E values of <1e-10 to any of these proteins were counted as transposable elements. If a region had significant hits to more than one type of element, it was annotated as belonging to the element type of the best hit. Because many of the elements identified were fragmentary and/or substantially diverged from the query sequences used, we identified the longest D. bruxellensis elements from each of the three classes found and repeated the analysis, using these sequences as a TBLASTX database, to identify more diverged elements in our data. To ensure that our estimate of the number of elements was not inflated by double counting 5′ and 3′ ends of a single element on different contigs, we counted only hits that shared a common region of homology to the query as unique elements.
Gene content analyses.
To further examine the differences in the sets of proteins encoded by D. bruxellensis and other yeasts, we searched for three types of loci: individual genes that have undergone recent lineage-specific duplication in D. bruxellensis, gene families that have expanded in D. bruxellensis relative to other species, and genes that have been horizontally transferred. To identify genes in the first two categories, we created a database of “putative genes” in D. bruxellensis. Each such gene was a region of a contig that was over 100 nucleotides long, had a BLASTX hit to an S. cerevisiae protein with E < 1e-10 (but was not necessarily the reciprocal best BLAST hit to that protein), and did not contain a stop codon.
A pair of genes arising from a lineage-specific gene duplication are likely to be one another's best BLAST hits, to the exclusion of orthologs in other lineages. We used the translated putative D. bruxellensis genes as BLASTP queries against a database containing the same putative D. bruxellensis proteins plus the proteomes of S. cerevisiae, C. glabrata, K. lactis, A. gossypii, C. albicans, D. hansenii, and Y. lipolytica. Any D. bruxellensis gene whose top two hits were itself and another D. bruxellensis protein was considered a probable lineage-specific duplication.
To assess whether genes with particular functions are more likely to be duplicated in D. bruxellensis, we tested whether the S. cerevisiae orthologs of the duplicated genes represent a functionally biased subset of the S. cerevisiae proteome. We determined the distribution of the orthologs of the duplicates among gene ontology categories, for each of the three ontologies, using GOToolBox (42). For each ontology, we then compared this distribution to that of the 5,770 S. cerevisiae proteins used in our analyses, using Fisher's exact test. We corrected for multiple tests (5).
We also searched for gene families that have expanded to a greater size in D. bruxellensis than in two closely related species, C. albicans and D. hansenii. The proteomes of these two species were combined with the translated putative gene set of D. bruxellensis into a single database. We queried this database with the protein sequences of S. cerevisiae genes belonging to previously defined gene families with two or more members (a subset, defined by phyletic pattern, of the “consensus families” from http://cbi.labri.fr/Genolevures/fam/index.html). We then calculated the total number of unique hits with an E value of <1e-10 to D. bruxellensis, C. albicans, and D. hansenii proteins across all genes in each family. Families that had more hits to D. bruxellensis than to either C. albicans or D. hansenii were considered to have undergone gene family expansion in D. bruxellensis. This is a conservative threshold, as the D. bruxellensis genome data are far less complete than those of either C. albicans or D. hansenii.
Nucleotide sequence accession numbers.
Read sequences have been deposited in GenBank with accession numbers EI011584 to EI026443, and contig sequences are available on request from the corresponding author. In addition to the genome survey data, we completely sequenced a number of clones containing adenyl deaminase genes and nitrate/nitrite utilization genes (GenBank accession numbers EF364424 to EF364429).
RESULTS AND DISCUSSION
Sequence data and assembly.
Previous analyses using pulsed-field gel electrophoresis have shown that the karyotypes of D. bruxellensis strains vary quite extensively, with estimated total genome sizes ranging from under 20 Mb to over 30 Mb (58). Partial ribosomal DNA (rDNA) sequencing of 30 strains suggests that, nonetheless, most isolates fall into a single major clade (3). We selected strain CBS 2499 for genome analysis because (i) it is a representative of the major D. bruxellensis clade, (ii) its estimated genome size, 19.4 Mb, is towards the lower end of the range observed in this species, (iii) it can be easily manipulated in the laboratory, and (iv) we were able to make a petite derivative from this strain, allowing us to construct a genomic DNA library without mitochondrial contamination. We assembled the sequences of 14,860 random reads from this library (see Materials and Methods) into 5,407 contigs, totalling approximately 7.6 Mb of sequence data. Given the estimated genome size of this strain, and assuming that it is haploid, our data contain approximately 40% of the genome. The principal features of our assembly are listed in Table 1.
TABLE 1.
Genome analysis summary
| Parameter | Value |
|---|---|
| Assembly | |
| No. of contigs | 5,407 |
| Mean contig length (bp) | 1,386 |
| Assembly size (Mb) | 7.6 |
| Estimated genome size (Mb) | 19.4 |
| % of genome sequenced | ∼40 |
| Gene content | |
| Genes with S. cerevisiae orthologs | 2,606 |
| Genes without S. cerevisiae orthologs | 278 |
| Lineage-specific gene duplicates | 89 |
| tRNAs | 24 |
| Retrotransposons, Ty1-like | 3 |
| Retrotransposons, Ty3-like | 16 |
| Retrotransposons, Ty5-like | 13 |
| Estimated total no. of protein-coding genes | ∼7,430 |
| Genome architecture | |
| Mean intergenic length (bp) | 945 |
| Introns, % genes in which present | 2 |
| Introns, mean length (bp) | 195 |
| % GC content (S. cerevisiae value) | |
| Introns, genome | 40.2 (38.3) |
| Introns, coding | 42.9 (39.7) |
| Introns, GC3 | 44.2 (37.0) |
| Introns, intergenic | 39.1 (33.2) |
| Introns, intronic | 34.7 (33.8) |
| Di/trinucleotide microsatellites | 48 |
The sequences appear to be largely single copy. After removing transposable element sequences (see below), we found 270 regions of the assembly longer than 500 bp that were matched by a second region with nucleotide identity of 95% or greater. The duplicate copies of these regions add up to a total of 0.23 Mb, or approximately 3% of our sequence data. This will be an underestimate of the total proportion of the genome that is present in duplicate copy, as our data are likely to have sampled only single copies of many regions that are actually present in duplicate in the genome. As a rough correction, we may assume that the probability of observing a region as duplicated, P(D0), is equal to the probability of the region actually being duplicated, P(DA), times the probability of observing that second region in our sample (in this case, 0.4). As we observe that P(D0) is 0.03, this implies that P(DA) is 0.07, or that approximately 7% of the whole genome is made up of duplicated regions.
Genes and genetic code.
We identified complete or partial sequences of 2,606 D. bruxellensis protein-coding genes with orthologs in S. cerevisiae, listed in Table S1 of the supplemental material. Additionally, 277 genes without S. cerevisiae orthologs, but with orthologs from other Saccharomycetales species, were identified (most of these genes were annotated as hypothetical; the 50 with functional annotations are listed in Table 2), as was a single example of a gene horizontally transferred from a bacterial species (see below). No other genes without fungal homologs were identified. An almost complete rDNA repeat, including 18S, 5.8S, 25S, and 5S rDNAs, is present in the data, as are at least 24 tRNA genes. If we assume that the 40% of the genome represented in our data are a random sample of the whole genome with respect to gene density, the total protein-coding gene complement of D. bruxellensis is likely to be on the order of 7,430 genes, which is comparable to that of other hemiascomycetes.
TABLE 2.
Ascomycete proteins that have orthologs in D. bruxellensis but not S. cerevisiae
| Functional class and NCBI accession no. | Species | Gene name | Function |
|---|---|---|---|
| Nitrate metabolism | |||
| CAA11229 | Hansenula polymorpha | YNT1 | Nitrate transporter |
| CAA11232 | Hansenula polymorpha | YNR1 | Nitrate reductase |
| CAA11230 | Hansenula polymorpha | YNI1 | Nitrite reductase |
| CAA11231 | Hansenula polymorpha | YNA1 | Nitrate assimilation pathway-specific Zn(II)2Cys6 transcription factor |
| CAC16081 | Hansenula polymorpha | YNA2 | Nitrate assimilation pathway-specific Zn(II)2Cys6 transcription factor |
| Metabolism of alternative carbon sources | |||
| AAX31178 | Candida albicans | NAG1 | Glucosamine-6-phosphate deaminase |
| CAH02587 | Kluyveromyces lactis | LAC4 | β-Galactosidase |
| CAE47547 | Ambrosiozyma monospora | ALX1 | l-Xylulose reductase |
| CAG88556a | Debaryomyces hansenii | Inositol oxygenase | |
| CAG90181a | Debaryomyces hansenii | Phosphatidylinositol-specific phospholipase C | |
| BAD32688 | Hansenula polymorpha | Glycerol dehydrogenase | |
| BAD32689 | Pichia ofunaensis | Dihydroxyacetone reductase | |
| Transporters | |||
| EAK93787 | Candida albicans | HGT1 | High-affinity glucose transporter |
| AAB17122 | Debaryomyces occidentalis | HAK1 | High-affinity potassium transporter |
| EAL02606 | Candida albicans | NUP | Purine nucleoside permease |
| EAK96574 | Candida albicans | UAP2 | Potential purine permease |
| Lipid metabolism | |||
| BAD11952 | Saccharomyces kluyveri | FAD3 | ω-3 fatty acid desaturase |
| AAU10084 | Pichia pastoris | Δ8-(E)-sphingolipid desaturase | |
| AAU10085 | Pichia pastoris | Δ4-(E)-sphingolipid desaturase | |
| AAZ08581 | Ashbya gossypii | Sphingolipid C9-methyltransferase | |
| AAK73020 | Pichia pastoris | Ceramide glucosyltransferase | |
| Respiratory chain complexes | |||
| CAG86996a | Debaryomyces hansenii | Complex I 14-kDa subunit | |
| CAG88782a | Debaryomyces hansenii | Complex I 20-kDa subunit | |
| CAG84577a | Debaryomyces hansenii | Complex I 24-kDa subunit | |
| CAG89146a | Debaryomyces hansenii | Complex I 49-kDa subunit | |
| CAG89064a | Debaryomyces hansenii | Complex I 51-kDa subunit | |
| CAG90271a | Debaryomyces hansenii | Complex I 75-kDa subunit | |
| CAG85893a | Debaryomyces hansenii | AOX1 | SHAM-sensitive alternative terminal oxidase 1 |
| Peroxisome | |||
| CAA82928 | Hansenula polymorpha | PER1 | Peroxisomal matrix protein |
| AAD52811 | Hansenula polymorpha | PEX1 | Peroxin |
| P78723 | Hansenula polymorpha | PEX14 | Peroxin |
| CAA65646 | Pichia pastoris | PEX2 | Peroxin |
| AAF19606 | Pichia pastoris | PEX17 | Peroxin |
| AAD43507 | Pichia pastoris | PEX19 | Peroxin |
| Other | |||
| CAG60540a | Candida glabrata | PRN1 | Putative pirin |
| CAG85416a | Debaryomyces hansenii | URA9 | Dihydroorotate dehydrogenase 2 |
| CAG86646a | Debaryomyces hansenii | Beta-alanine synthase | |
| CAH00925a | Kluyveromyces lactis | GTP cyclohydrolase II | |
| CAG80587a | Yarrowia lipolytica | FAD-dependent oxidoreductase | |
| BAB12222 | Candida boidinii | DAO1 | d-Amino acid oxidase |
| CAH02585 | Kluyveromyces lactis | Acid phosphatase | |
| AAS51137 | Ashbya gossypii | Calcineurin-like phosphoesterase | |
| EAL00803 | Candida albicans | Potential secreted Cu/Zn superoxide dismutase | |
| EAK95140 | Candida albicans | Possible thiamine biosynthesis enzyme | |
| EAL02572a | Candida albicans | Ras GTPase-activating protein | |
| CAG85348a | Debaryomyces hansenii | Potential histone binding protein | |
| CAG78240a | Yarrowia lipolytica | Translation initiation factor 3 subunit 7 | |
| EAL04857 | Candida albicans | Negative regulator of iron uptake genes | |
| EAK96905 | Candida albicans | Potential fungal zinc cluster transcription factor | |
| AAQ75382 | Hansenula polymorpha | SWI1 | Global transcription activator |
For this gene, the best BLAST hit of the D. bruxellensis sequence in the NR database is an unannotated or hypothetical gene, and the functional annotation given here is taken from the highest annotated hit, where the bit score of the annotated hit was at least 90% that of the best hit and sequence comparison indicates that the genes are homologs.
As a number of hemiascomycete species are known to deviate from the universal genetic code (60), we tested whether there was any evidence of codon reassignment in D. bruxellensis. Each D. bruxellensis gene with an S. cerevisiae ortholog was translated using the universal genetic code, and its product was aligned to the protein sequence encoded by its ortholog. We then returned the D. bruxellensis portions of the alignments to codon format and calculated, across all the gene alignments, how frequently each of the 61 sense codons in the D. bruxellensis data corresponded to each of the 20 amino acids in the S. cerevisiae data. For every codon, the amino acid most frequently observed was that expected under the universal genetic code, suggesting that codon reassignment has not occurred in D. bruxellensis. This method correctly identifies the known CUG reassignment in D. hansenii (60) (data not shown). The universal genetic code was used in all further analyses of D. bruxellensis data.
Phylogenetic position.
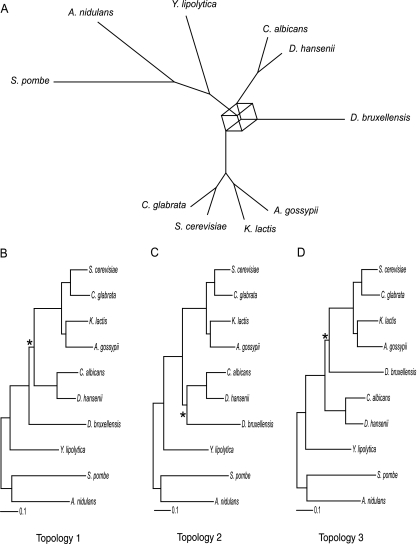
To determine the phylogenetic position of D. bruxellensis among the hemiascomycetes, we constructed 396 trees, based on protein sequences of orthologous genes from D. bruxellensis and nine other fungal species (see Materials and Methods). We then calculated a consensus network to quantify the degree of incongruence between these trees (31). This involves generating a list of splits (bipartitions of the taxa corresponding to branches in the trees) present in the complete set of trees and weighting them by the frequency with which they occur; all splits that occur at a frequency above a certain threshold can then be displayed in a network. Boxes in the network indicate the presence of incompatible splits. Although the major groupings in our network are well supported, there is conflict between the gene trees regarding the position of D. bruxellensis: three incompatible splits are each present in over 25% of trees (Fig. 1A).
FIG. 1.
(A) Consensus network based on 396 protein trees; splits are shown if they are present in 25% or more of the trees. Three incompatible splits involving the placement of D. bruxellensis generate a cube in the network. (B to D) Three likely topologies inferred from the consensus network. Each topology contains one of the incompatible splits shown in panel A; in each case the branch corresponding to the split is marked with an asterisk. Branch lengths for these three trees are estimated by maximum likelihood using concatenated amino acid sequence data.
From this network, we inferred three probable topologies that could explain the sequence data, each of which includes only one of the three incompatible splits identified above (Fig. 1B to D). The marked splits shown in topologies 1, 2, and 3 occur in 34%, 27%, and 26% of gene trees, respectively. A Shimodaira-Hasegawa test showed that topology 2 is the best fit to the concatenated sequence data (log likelihood, −1,344,394), although it is not significantly better than topology 1 (P = 0.27; log likelihood, −1,344,454). Topology 3, however, can be significantly rejected (P = 0.01; log likelihood, −1,344,558). The use of different substitution models made no difference to the outcome of the Shimodaira-Hasegawa test, suggesting that model misspecification is not affecting our results.
Previous estimates of the phylogenetic position of D. bruxellensis reflect the uncertainty we observe in our data: separate analyses based on 18S sequence data have supported both topology 1 (11) and topology 3 (47). Our analysis of almost 400 protein-coding genes significantly rejects topology 3, but we remain unable to determine whether D. bruxellensis diverged from the hemiascomycete lineage shortly before C. albicans and D. hansenii (topology 1) or if these three species briefly formed a shared divergent lineage before the speciation of D. bruxellensis (topology 2). It is likely that very few substitutions were fixed in the short time periods separating these divergence events, making it difficult to accurately reconstruct the true topology. The proteins used in our analysis were well conserved across taxa and thus relatively slowly evolving, and it is possible that more rapidly evolving proteins would provide more substitutions on the branches of interest. Such proteins, however, are more difficult to assign to orthology groups and to reliably align and are more likely to be saturated, possibly leading to decreased phylogenetic accuracy.
Gene sequence statistics.
We calculated the amino acid identity between each D. bruxellensis protein and its S. cerevisiae ortholog by aligning their amino acid sequences and removing gapped sites. D. bruxellensis and S. cerevisiae orthologous proteins have a mean amino acid identity of 48.8% (N = 2,615, standard deviation, 14.6). This is significantly lower than the amino acid identity between C. albicans and S. cerevisiae orthologs for the subset of proteins that are present in all three species (mean C. albicans-S. cerevisiae identity, 52.9%; mean D. bruxellensis-S. cerevisiae identity, 49.9%; N = 2,115; P < 1 × 10−16, Wilcoxon signed-rank test). If we accept that the divergence of D. bruxellensis, S. cerevisiae, and C. albicans is effectively a trichotomy (or that the best-supported tree, topology 2, reflects the real phylogeny), this suggests that the rate of amino acid sequence evolution in D. bruxellensis is higher than that of C. albicans.
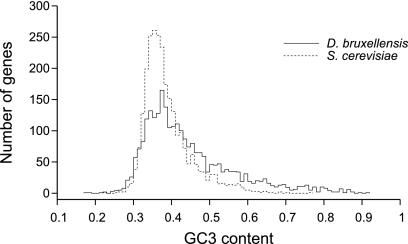
D. bruxellensis genes tend to have higher GC content than their S. cerevisiae orthologs at all classes of sites (Table 1). The difference in GC content across orthologous sites is statistically significant (P < 1 × 10−16 for all genic sites and for codon third position sites; Wilcoxon signed-rank test). GC content also varies among D. bruxellensis genes substantially more than among the orthologous S. cerevisiae genes, a trend that is particularly obvious at third position sites (Fig. 2).
FIG. 2.
Distribution of GC content at third-position codon sites in 2,606 orthologous genes in D. bruxellensis and S. cerevisiae.
Genome architecture.
We examined four aspects of genome organization in D. bruxellensis: the conservation of gene order in D. bruxellensis relative to that in S. cerevisiae and in C. albicans; the lengths of intergenic regions; the positions, lengths, and motif sequences of introns; and the repeat content of the genome.
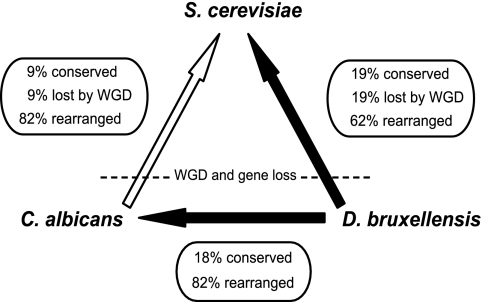
We compared the order of genes along D. bruxellensis contigs with that of their orthologs on the S. cerevisiae genome to determine the degree of synteny conservation between these two species. Most of our D. bruxellensis contigs contain only a single gene, but 286 contain two or more genes, allowing us to analyze 318 pairs of adjacent genes. Of these gene pairs, 19% are also immediate neighbors in S. cerevisiae and a further 4% are close neighbors separated by one to five genes. Because we expect gene order relationships between D. bruxellensis and S. cerevisiae to have been affected by the whole genome duplication that occurred on the S. cerevisiae lineage (65), we also assessed the degree of gene order conservation between D. bruxellensis and C. albicans, a species that has not undergone whole genome duplication. In this case, of 233 pairs of adjacent D. bruxellensis genes with C. albicans orthologs, 18% have adjacent C. albicans orthologs and an additional 8% are separated by one to five genes. Only 17 pairs of adjacent genes in D. bruxellensis are also adjacent in both C. albicans and S. cerevisiae (see Table S2 in the supplemental material); these genes show no obvious functional bias.
These results suggest that D. bruxellensis has a somewhat lower rate of small-scale rearrangements than C. albicans (Fig. 3). An earlier analysis using the same method determined that only 9% of gene pairs adjacent in C. albicans were also adjacent in S. cerevisiae (55). As we expect approximately half of the links between gene pairs in pre-whole-genome duplication species to be broken due to random gene loss in the S. cerevisiae lineage subsequent to duplication (65), we can estimate that approximately 62% of D. bruxellensis and 82% of C. albicans neighboring gene pairs have been disrupted in S. cerevisiae due to chromosomal rearrangement. The lower value for D. bruxellensis is not unusual: it has been shown that the rate of small chromosomal inversions differs by over an order of magnitude among six completely sequenced yeast genomes (21), and our estimate of the percentage of adjacent gene pairs in D. bruxellensis conserved in S. cerevisiae (19%) is consistent with those for D. hansenii (16%) and C. tropicalis (18%) (39).
FIG. 3.
Estimated rates of chromosomal rearrangements between D. bruxellensis, C. albicans, and S. cerevisiae. Black arrows indicate analyses performed in this study, while the white arrow refers to a previous study (55). The percentage of adjacent gene pairs conserved between species was calculated from the data, and the percentage of gene pair adjacencies disrupted by gene loss following whole genome duplication (WGD) in S. cerevisiae is estimated to be approximately equal to the percentage conserved. The remaining gene pairs have been disrupted by chromosomal rearrangements.
The mean intergenic distance for the set of 318 adjacent D. bruxellensis genes is 700 nucleotides. This may be an unrealistically low estimate of the average genomic intergenic distance, however, as the relatively short lengths of the contigs mean that long intergenic regions (complete with flanking genes) are unlikely to be detected. Approximately 1,300 contigs have no significant BLASTX hit to any protein in the NR database, nor do they contain rRNAs or tRNAs. If we assume that these contigs consist of intergenic sequence and include their lengths in our calculation of mean intergenic distance, this value becomes 945 nucleotides. This may represent a substantial underestimate (if many intergenic regions are much longer than individual contigs) or a slight overestimate (if fragments of genes at the ends of contigs are not detected by BLAST analysis) of the true average intergenic length in this species. Although higher than the mean intergenic distance in S. cerevisiae, this value is well within the range observed in other hemiascomycete species (10).
We identified introns in 40 D. bruxellensis genes whose S. cerevisiae orthologs also contain introns (Table 3). Of these, two (DYN2 and RPL7A and their D. bruxellensis orthologs) each contain two introns. We found an additional eight introns that appear to be specific to D. bruxellensis (Table 3). This may be an underestimate of the true number of novel introns, as our detection method requires that exons both upstream and downstream of the intron be identified as HSPs by BLAST. Hemiascomycete introns (including those we observe in our data) have a strong bias towards the 5′ end of genes, and in many cases the first exon encodes five or fewer amino acids; such first exons may not be detected as an HSP.
TABLE 3.
Introns in D. bruxellensis genes
|
S. cerevisiae ortholog
|
5′ Motifa | Branch Motif | 3′ Motif | Intron lengthb (bp)
|
||
|---|---|---|---|---|---|---|
| Systematic name | Standard name | D. bruxellensis | S. cerevisiae | |||
| YBL001C | ECM15 | GTAAGT | TACTAAC | CAG | 65 | NA |
| YBL050W | SEC17 | — | TATTAAC | TAG | ||
| YBR159W | YBR159W | GTATGT | TACTAAC | CAG | 79 | NA |
| YBR189W | RPS9B | GTAAGT | TACTAAC | TAG | 344 | 413 |
| YDL082W | RPL13A | — | TACTAAC | TAG | ||
| YDL108W | KIN28 | — | TAATAAT | AAG | ||
| YDL125C | HNT1 | GTAAGT | TACTAAC | AAG | 65 | 111 |
| YDR025W | RPS11A | GTATGC | TACTAAC | TAG | 403 | 339 |
| YDR064W | RPS13 | — | TACTAAC | TAG | ||
| YDR092W | UBC13 | GTATGT | TACTAAC | AAG | 61 | 268 |
| YDR367W | YDR367W | — | GACTAAC | TAG | ||
| YDR424C | DYN2 | — | TACTAAC | TAG | ||
| YDR424C | DYN2 | GTAAGT | TACTAAC | CAG | 125 | 80 |
| YDR471W | RPL27B | — | TACTAAC | TAG | ||
| YER009W | NTF2 | GTAAGT | TACTAAC | TAG | 260 | NA |
| YGL076C | RPL7A | — | TACTAAC | TAG | ||
| YGL076C | RPL7A | GTAAGT | TACTAAC | CAG | 276 | 468 |
| YGL103W | RPL28 | GTAAGT | TACTAAC | TAG | 419 | 511 |
| YGR029W | ERV1 | GTAAGT | TACTAAC | TAG | 143 | 83 |
| YGR243W | YGR243W | GTAAGT | TACTAAC | AAG | 118 | NA |
| YHL001W | RPL14B | — | TACTAAC | TAG | ||
| YHR001W-A | QCR10 | GTAAGT | TACTAAC | CAG | 67 | 63 |
| YHR016C | YSC84 | GTATGT | TACTAAC | CAG | 113 | 168 |
| YHR021C | RPS27B | — | AACTAAT | TAG | ||
| YIL018W | RPL2B | — | TACTAAC | CAG | ||
| YIL069C | RPS24B | — | TACTAAC | TAG | ||
| YIL133C | RPL16A | — | TACTAAC | CAG | ||
| YJL191W | RPS14B | — | TACTAAC | CAG | ||
| YKR057W | RPS21A | — | TACTAAC | TAG | ||
| YLR185W | RPL37A | — | TACTAAC | CAG | ||
| YLR306W | UBC12 | GTAAGT | TACTAAC | CAG | 65 | 134 |
| YLR344W | RPL26A | GTATGT | TACTAAC | TAG | 357 | 447 |
| YLR448W | RPL6B | — | TACTAAC | TAG | ||
| YML067C | ERV41 | GTAAGT | TACTAAC | CAG | 92 | 93 |
| YML094W | GIM5 | GTAAGT | TACTAAC | CAG | 75 | 83 |
| YMR116C | ASC1 | — | TACTAAC | TAG | ||
| YMR143W | RPS16A | GTATGT | TACTAAC | TAG | 359 | 544 |
| YMR194W | RPL36A | GTATGT | AACTAAC | CAG | 472 | 463 |
| YNL310C | YNL310C | GTAAGT | TACTAAC | TAG | 77 | NA |
| YNR053C | NOG2 | — | TACTAAC | AAG | ||
| YOL120C | RPL18A | GTAAGT | TACTAAC | TAG | 328 | 447 |
| YOL127W | RPL25 | — | TACTAAC | TAG | ||
| YOL132W | GAS4 | GTAGGT | TACTAAC | CAG | 62 | NA |
| YOL146W | PSF3 | GTAAGT | TACTAAC | TAG | 65 | NA |
| YOR122C | PFY1 | GTAAGT | TACTAAC | CAG | 161 | 209 |
| YOR182C | RPS30B | — | TACTAAC | CAG | ||
| YPL090C | RPS6A | GTATGT | TACTAAC | CAG | 357 | 394 |
| YPL143W | RPL33A | GTAAGT | TACTAAC | TAG | 359 | 525 |
| YPR028W | YOP1 | — | TACTAAC | TAG | ||
| YPR062W | FCY1 | GTAAGT | TACTAAC | CAG | 100 | NA |
—, the start of the intron is not present in our sequence data; for these cases, the intron length cannot be given.
Where the intron length is known in D. bruxellensis, the length of the corresponding intron in S. cerevisiae is given for comparative purposes. NA, the orthologous gene in S. cerevisiae does not contain an intron.
In total, approximately 2% of the D. bruxellensis genes we identified contain introns, and the mean length of the introns is 195 nucleotides. These values are consistent with those found in other hemiascomycetes: the proportion of genes that contain introns varies from 1% in D. hansenii to 4% in S. cerevisiae, with a concomitant increase in average intron length from around 100 to over 250 nucleotides (8). The D. bruxellensis introns have 5′, 3′, and branch site motifs that are very similar to the consensus in S. cerevisiae.
We examined two kinds of repeated sequences in the D. bruxellensis genome: microsatellites and transposable elements. The assembly contains 48 microsatellites with between 8 and 34 dinucleotide or trinucleotide repeat units. On the basis of results from other hemiascomycete species (37), it is likely that at least some of these are polymorphic and may thus be of use in future analyses of population diversity.
The great majority of transposable elements so far described in hemiascomycete yeasts are LTR retrotransposons, although non-LTR retrotransposons and DNA transposons have been found in C. albicans and Y. lipolytica (12, 25, 44). The LTR retrotransposons fall into two broad categories: Ty1/copia (including Ty1, Ty2, T4, and Ty5 elements) and Ty3/gypsy (43). The D. bruxellensis data contain at least 42 transposable elements, all of which are LTR retrotransposons: three Ty1-like, 16 Ty3-like, and 13 Ty5-like. This number includes only those elements that retain at least fragments of the TYA or TYB protein-coding genes and is thus a minimum estimate of the number of elements present; many solo LTR sequences are likely to be present but too degenerate to identify by comparison with LTR sequences from other species.
It has previously been suggested that Ty1 elements first appeared in the genomes of hemiascomycetes at the base of the S. cerevisiae/K. lactis clade (43), after the divergence of D. bruxellensis from the main hemiascomycete lineage. The presence of three putative Ty1 elements in our data may indicate that elements of this class arose at an earlier stage in hemiascomycete evolution and have subsequently been lost from other species in the C. albicans/D. hansenii clade (as they have been from Saccharomyces bayanus, for example). The Ty1-like elements present in D. bruxellensis are substantially diverged from one another and from their closest homologs in other species (the most similar has 26% amino acid identity to a Tkm1 element from Kluyveromyces marxianus), indicating that they are unlikely to be the result of a recent horizontal gene transfer event.
Differences in gene content.
The variations in physiological capabilities between D. bruxellensis and other yeasts are caused by differences in gene complement and regulation in these species. To investigate the extent of gene content differences, we identified genes in D. bruxellensis that are absent from S. cerevisiae (Table 2), and we also sought evidence for increases in gene number that could underlie various traits. We identified 89 pairs of lineage-specific duplicated genes (see Table S3 in the supplemental material). Enzymes and transporters were notably overrepresented in this set of genes, among other significantly enriched GO categories (see Tables S4a to c in the supplemental material). Notably, we found duplicated copies of three genes in the allantoin catabolism pathway (DAL1, DCG1, and MLS1), all with 97% DNA sequence identity between copies. We also identified 18 gene families that are expanded in D. bruxellensis relative to both C. albicans and D. hansenii, including several families functioning in sterol, phosphocholine, and phospholipid synthesis (Table 4).
TABLE 4.
Gene families that are expanded in D. bruxellensis relative to both C. albicans and D. hansenii
| Gene familya | S. cerevisiae genes in family | No. of unique BLASTP hitsb
|
||
|---|---|---|---|---|
| D. bruxellensis | C. albicans | D. hansenii | ||
| GLC.1379 | BNI1 BNR1 | 4 | 2 | 2 |
| GLC.1390 | ITC1 YPL216W | 2 | 1 | 1 |
| GLC.1609 | DIN7 EXO1 RAD2 RAD27 YEN1 | 5 | 4 | 4 |
| GLC.1623 | SEC1 SLY1 VPS33 VPS45 | 5 | 4 | 4 |
| GLC.1700 | CDC46 CDC47 CDC54 MCM3 MCM6 | 9 | 8 | 6 |
| GLC.1702 | CDC16 CDC27 | 4 | 3 | 3 |
| GLC.1711 | RDS2 YBR239C YJL103C | 4 | 3 | 3 |
| GLC.1749 | CCC2 DNF1 DNF2 DNF3 DRS2 ENA1 ENA2 ENA5 NEO1 PCA1 PMA1 PMA2 PMC1 PMR1 SPF1 YOR291W | 19 | 18 | 18 |
| GLC.1765 | NCL1 NOP2 YNL022C | 4 | 3 | 3 |
| GLC.1826 | SHC1 SKT5 | 3 | 1 | 1 |
| GLC.1900 | ECM22 UPC2 | 2 | 1 | 1 |
| GLC.1967 | HSP26 HSP42 | 2 | 0 | 0 |
| GLC.2074 | GDE1 PHO81 | 4 | 2 | 2 |
| GLC.2091 | APL1 APL2 APL3 APL4 APL5 APL6 SEC26 | 8 | 7 | 7 |
| GLC.2291 | HMG1 HMG2 NCR1 | 5 | 2 | 2 |
| GLC.2339 | GEA1 GEA2 SEC7 | 7 | 2 | 2 |
| GLC.2379 | DHR2 ECM16 PRP16 PRP2 PRP22 PRP43 YLR419W | 11 | 9 | 8 |
| GLC.2424 | ARX1 MAP1 MAP2 | 5 | 3 | 3 |
| GLC.2529 | SCS3 YDR319C | 2 | 1 | 0 |
| GLC.2545 | ACC1 HFA1 | 6 | 4 | 3 |
Gene families are from reference 19.
Total number of unique BLASTP hits (with E values of <1e-10) of S. cerevisiae proteins in that family to each of the three species, respectively. Note that as the D. bruxellensis genome is not completely sequenced, the numbers of hits given for this species will be a substantial underestimate of the true figure.
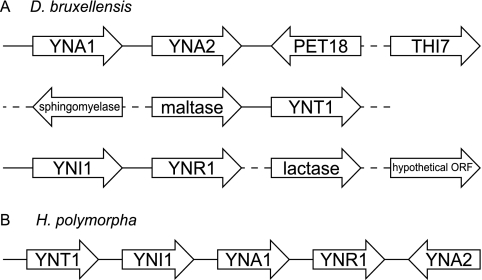
A number of the genes present in our data that do not have S. cerevisiae orthologs (Table 2) probably underpin known species-specific differences in metabolic capabilities between S. cerevisiae and D. bruxellensis (36). For example, a minority of D. bruxellensis strains have recently been shown to grow on lactose or arabinose (16), and the presence in our data of genes coding for β-glucosidase (lactase) and l-xylulose reductase (required for l-arabinose catabolism [64]) suggests that, unlike S. cerevisiae, strain CBS 2499 may be able to utilize both these carbon sources. S. cerevisiae cannot utilize nitrate as a nitrogen source, while D. bruxellensis can; five genes required for nitrate assimilation are present in the D. bruxellensis sequence data. These five genes (YNT1, YNR1, YNI1, YNA1, and YNA2) are clustered in the genome of the yeast Hansenula polymorpha (4), as are subsets of these genes in a number of other more distantly related fungi (2, 17). Due to the fragmented nature of the D. bruxellensis sequence data, we cannot currently determine the exact degree of clustering of these genes in this species. It is clear, however, that all five genes do not occur in a single uninterrupted cluster and that the relative positions and orientations of the genes differ between D. bruxellensis and H. polymorpha (Fig. 4).
FIG. 4.
(A) Gene order relationships (not drawn to scale) of five nitrate assimilation genes and their neighboring genes in the D. bruxellensis data. See Table 2 for full gene names. The arrowheads indicate the direction of transcription. Solid lines between genes indicate sequenced intergenic regions; dotted lines indicate that the gene order is inferred from clone end pair information. (B) Order and orientation of the cluster of orthologous genes in Hansenula polymorpha (based on information provided in reference 4).
Genes involved in lipid metabolism appear to be substantially enriched in the D. bruxellensis genome: it possesses genes in these pathways that are not found in S. cerevisiae (Table 2), and known families of lipid metabolism genes that are present in S. cerevisiae have undergone expansion in D. bruxellensis (Table 4). Some of these genes may contribute to the high ethanol tolerance of this species (35). Numerous others have orthologs in C. albicans whose precise role in the cell is currently unknown (e.g., the sphingolipid desaturase and methyltransferase genes) (9).
D. bruxellensis also possesses genes coding for several subunits of the respiratory chain complex I and for a SHAM-sensitive alternative oxidase (AOX1) (Table 2), suggesting that this species is capable of alternative respiration. This pathway bypasses complexes III and IV of the respiratory chain; instead, electrons from the ubiquinone pool are used by the alternative oxidase to reduce oxygen to water. Alternative respiration and aerobic fermentation have previously been hypothesized to represent two alternative responses to glycolytic overflow (63). Under this theory, when high levels of glucose are available to the yeast cell, rapid glycolysis may lead to saturation of the respiratory pathway and subsequent accumulation of pyruvate and NADH. To prevent glycolysis from stalling, this NADH may then be reoxidized to NAD+ either via alternative respiration (in Crabtree-negative yeasts) or by the reduction of pyruvate to ethanol (in Crabree-positive yeasts). The presence of complex I and AOX1 in D. bruxellensis, a Crabtree-positive yeast capable of aerobic fermentation, is therefore somewhat unexpected (62), although alternative respiration has previously been demonstrated in some strains of Dekkera and Brettanomyces yeasts (7). It has been suggested that this alternative respiratory pathway may be implicated in the Custers effect (anaerobic inhibition of fermentation) shown by these yeasts (7) or NADH reoxidation through glycerol production, but little is known of the mechanisms of these processes.
Finally, seven D. bruxellensis genes coding for peroxisome structural and regulatory proteins were also identified as having ascomycete but no S. cerevisiae orthologs. Peroxisomes are found in almost all eukaryotes, and some peroxins (including that encoded by PEX19) are conserved across this taxonomic spectrum (53), suggesting that S. cerevisiae orthologs to at least some of these genes are likely to exist. Many of the ascomycete peroxisome genes in Table 2 have in fact been previously annotated as homologous to S. cerevisiae genes (59), although their products often have low amino acid identity. Phylogenetic analyses suggest that S. cerevisiae peroxin genes are indeed orthologous to the D. bruxellensis genes but are too divergent to be detected using our method (data not shown).
Horizontal gene transfer.
While comparing our contig sequences to the entire NCBI database, we identified a gene fragment in D. bruxellensis that was the reciprocal best BLAST hit to a Burkholderia cenocepacia (α-Proteobacteria) gene annotated as encoding an adenosine deaminase (ADA). We sequenced three additional plasmid clones to obtain the full sequence of this gene, the surrounding intergenic regions, and portions of the neighboring genes. The gene is 1,053 nucleotides long and is located downstream of a URA4 homolog and upstream of a transporter of the DAL5 family. Both of the neighboring genes show clear evidence of fungal origin, which rules out the possibility that the ADA gene-like gene arose from bacterial contamination of our D. bruxellensis library. The complete gene sequence identifies the gene as a member of the adenyl deaminase gene subfamily, which includes both adenosine deaminase and adenine deaminase (ADE) genes (41, 52).
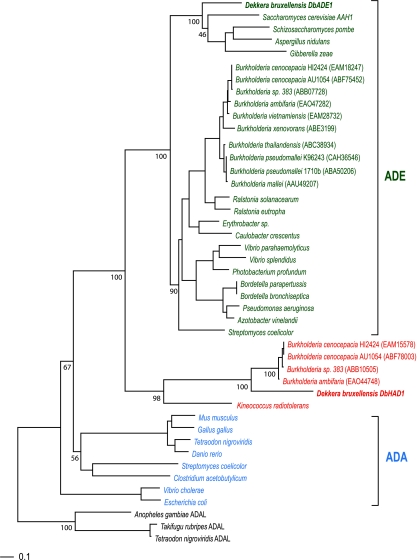
We constructed a phylogenetic tree using sequences of the D. bruxellensis gene, its close BLAST hits, and a number of known ADA and ADE genes (Fig. 5). The D. bruxellensis gene clusters with neither the ADA nor the ADE gene clade but forms a separate clade with sequences from four species of the Burkholderia cepacia complex (40) and one sequence from Kineococcus radiotolerans. The clear separation of this clade from the fungal ADE genes, together with the existence of a canonical ADE gene in D. bruxellensis (DbADE1) (Fig. 5), suggests that horizontal gene transfer from a bacterial species is the most likely origin for the Burkholderia-like gene, which we named DbHAD1 for D. bruxellensis horizontally transferred adenyl deaminase gene.
FIG. 5.
Maximum likelihood phylogenetic tree, based on amino acid sequences, showing the relationships between adenine deaminases (ADEs, green), adenosine deaminases (ADAs, blue), and the cluster of horizontally transferred genes (red). Bootstrap values (1,000 replicates) are given for branches of interest. GenBank accession numbers are given in parentheses for Burkholderia genes.
The four B. cepacia complex species that have HAD genes also have canonical ADE genes present elsewhere in their genomes (Fig. 5). We could not identify orthologs of the HAD gene in any of the other Burkholderia species for which whole genome sequences are available, although canonical ADE genes are present in those species. This distribution of the HAD gene among Burkholderia species suggests that it was acquired by a relatively recent common ancestor of these four species. Inspection of the genomic region surrounding the HAD gene in the four B. cepacia complex species suggests that it is part of an operon specific to these species (e.g., genes Bcep18194_B0386 to Bcep18194_B0389 in the Burkholderia sp. strain 383 genome; GenBank accession number NC_007511.1) (data not shown), but none of the other genes in the operon can be detected in the D. bruxellensis sequence data or among publicly available K. radiotolerans sequences.
It seems likely that both D. bruxellensis and the four B. cepacia complex species received this gene as a horizontal transfer, either independently or sequentially, from some other species not yet represented in the public sequence databases. The four B. cepacia complex species, K. radiotolerans, and D. anomala (the sister species of D. bruxellensis) have all been isolated from soil (15, 50, 61), a common habitat that provides the opportunity for genetic transfers, and B. cepacia complex species are known to have genomic characteristics that facilitate the receipt and transfer of nonendogenous DNA (15). Comparative analysis of conserved amino acid residues gave no indication of the substrate specificity of DbHAD1, although the residues required for catalytic function are conserved (see Table S5 in the supplemental material). It therefore seems likely that this gene remains functional in D. bruxellensis; it is difficult to speculate on precisely what role it plays, however.
While the genomes of some other hemiascomycete species have been shown to contain a number of horizontally transferred genes (19, 28), DbADE1 was the only example found in our data. We found no evidence of an ortholog of URA1, the horizontally transferred cytosolic dihydroorotate dehydrogenase (DHODase) gene present in S. cerevisiae and closely related species (24). Most basal hemiascomycetes, like other eukaryotes, have a mitochondrial DHODase, encoded by URA9; the activity of this enzyme is coupled to the mitochondrial respiratory chain because a quinone is the terminal electron acceptor. As URA1 uses fumarate as an electron acceptor, the presence of this gene effectively decouples uracil biosynthesis from respiration, and it has been proposed that the transfer of URA1 from an anaerobic bacterium to an ancestor of S. cerevisiae and K. lactis was an essential step in the evolution of facultative anaerobiosis in this group of yeasts (24, 28, 48).
Some species in the URA1-possessing clade, such as K. lactis, retain both URA1 and URA9, while in others, like Saccharomyces cerevisiae, URA9 has been lost and only URA1 has been retained (24, 28). Our sequence data contain a complete URA9 gene (Table 2), indicating that a parallel gene displacement event has not occurred in D. bruxellensis. It is not possible to determine from our data, however, whether D. bruxellensis has an undetected URA1 gene that allows it to live anaerobically or whether it has found an alternative evolutionary solution to this challenge.
Conclusions.
The genome survey sequence data analyzed here show that D. bruxellensis strain CBS 2499 has genome characteristics typical of other hemiascomycetes in many respects, including estimated gene number, intron size and number, intergenic length, and gene content. Its rate of sequence evolution is faster than that of its relative C. albicans, though it has a lower rate of genomic rearrangement; neither rate, however, is extreme. Although aneuploidy may contribute to the variation in karyotypes observed between strains of D. bruxellensis (58), the relatively low proportion of duplicated sequence we see in our data, together with the small number of lineage-specific duplicate genes identified, suggests that this strain is haploid; this is consistent with its relatively small estimated genome size when compared with other strains.
These data should additionally provide a resource for future genetic study of this species. For example, although our sequences do not include the gene coding for the cinnamate decarboxylase required for the production of ethylphenols and ethylguaiacols (30), they contain numerous uncharacterized enzymes, some of which may be involved in the generation of the other volatile compounds that contribute to “Brett” character in wine (29, 38). These data may also facilitate analyses of population structure and variation in D. bruxellensis. Such analyses are essential if we are to develop optimal strategies to control contamination and to determine whether strains of D. bruxellensis that are neutral or even beneficial to wine character exist and may be harnessed by the viticultural industry (22).
Supplementary Material
Acknowledgments
We thank Kevin Byrne, Gavin Conant, Brian Cusack, Carolin Frank, Jonathan Gordon, Linda Hellborg, Nora Khaldi, David Lynn, Stephanie Maier, Ted Phelps, Devin Scannell, Marie Semon, Matthew Webster, and Simon Wong for helpful discussions and/or assistance during this study.
This work was supported by Science Foundation Ireland, the E. P. Sörensens Foundation, and the Nilsson-Ehle Foundation.
Footnotes
Published ahead of print on 2 February 2007.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. H. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amaar, Y. G., and M. M. Moore. 1998. Mapping of the nitrate assimilation gene cluster (crnA-niiA-niaD) and characterization of the nitrite reductase gene (niiA) in the opportunistic fungal pathogen Aspergillus fumigatus. Curr. Genet. 33:206-215. [DOI] [PubMed] [Google Scholar]
- 3.Andersen, M. R. 2005. Preliminary molecular and genetic analysis of Brettanomyces/Dekkera yeasts. M.Sc. thesis. Technical University of Denmark, Lyngby, Denmark.
- 4.Avila, J., C. Gonzalez, N. Brito, F. Machin, M. D. Perez, and J. M. Siverio. 2002. A second Zn(II)2Cys6 transcriptional factor encoded by the YNA2 gene is indispensable for the transcriptional activation of the genes involved in nitrate assimilation in the yeast Hansenula polymorpha. Yeast 19:537-544. [DOI] [PubMed] [Google Scholar]
- 5.Benjamini, Y., and Y. Hochberg. 1995. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57:289-300. [Google Scholar]
- 6.Benson, G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27:573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blondin, B., P. Gonde, R. Ratomahenina, A. Arnaud, and P. Galzy. 1984. A study of cyanide-insensitive respiration in the genus Dekkera and Brettanomyces. Microbiol. Immunol. 28:637-644. [DOI] [PubMed] [Google Scholar]
- 8.Bon, E., S. Casaregola, G. Blandin, B. Llorente, C. Neuveglise, M. Munsterkotter, U. Guldener, H. W. Mewes, J. Van Helden, B. Dujon, and C. Gaillardin. 2003. Molecular evolution of eukaryotic genomes: hemiascomycetous yeast spliceosomal introns. Nucleic Acids Res. 31:1121-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braun, B. R., M. V. Hoog, C. d'Enfert, M. Martchenko, J. Dungan, A. Kuo, D. O. Inglis, M. A. Uhl, H. Hogues, M. Berriman, M. Lorenz, A. Levitin, U. Oberholzer, C. Bachewich, D. Harcus, A. Marcil, D. Dignard, T. Iouk, R. Zito, L. Frangeul, F. Tekaia, K. Rutherford, E. Wang, C. A. Munro, S. Bates, N. A. Gow, L. L. Hoyer, G. Kohler, J. Morschhauser, G. Newport, S. Znaidi, M. Raymond, B. Turcotte, G. Sherlock, M. Costanzo, J. Ihmels, J. Berman, D. Sanglard, N. Agabian, A. P. Mitchell, A. D. Johnson, M. Whiteway, and A. Nantel. 2005. A human-curated annotation of the Candida albicans genome. PLoS Genet. 1:36-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Byrnes, J. K., G. P. Morris, and W. H. Li. 2006. Reorganization of adjacent gene relationships in yeast genomes by whole-genome duplication and gene deletion. Mol. Biol. Evol. 23:1136-1143. [DOI] [PubMed] [Google Scholar]
- 11.Cai, J. P., I. N. Roberts, and M. D. Collins. 1996. Phylogenetic relationships among members of the ascomycetous yeast genera Brettanomyces, Debaryomyces, Dekkera, and Kluyveromyces deduced by small-subunit rRNA gene sequences. Int. J. Syst. Bacteriol. 46:542-549. [DOI] [PubMed] [Google Scholar]
- 12.Casaregola, S., C. Neuveglise, E. Bon, and C. Gaillardin. 2002. Ylli, a non-LTR retrotransposon L1 family in the dimorphic yeast Yarrowia lipolytica. Mol. Biol. Evol. 19:664-677. [DOI] [PubMed] [Google Scholar]
- 13.Castresana, J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17:540-552. [DOI] [PubMed] [Google Scholar]
- 14.Chatonnet, P., D. Dubourdieu, J. N. Boidron, and M. Pons. 1992. The origin of ethylphenols in wines. J. Sci. Food Agric. 60:165-178. [Google Scholar]
- 15.Chiarini, L., A. Bevivino, C. Dalmastri, S. Tabacchioni, and P. Visca. 2006. Burkholderia cepacia complex species: health hazards and biotechnological potential. Trends Microbiol. 14:277-286. [DOI] [PubMed] [Google Scholar]
- 16.Conterno, L., C. M. L. Joseph, T. J. Arvik, T. Henick-Kling, and L. F. Bisson. 2006. Genetic and physiological characterization of Brettanomyces bruxellensis strains isolated from wines. Am. J. Enol. Vitic. 57:139-147. [Google Scholar]
- 17.Cutler, S. B., and C. E. Caten. 1999. Characterisation of the nitrite reductase gene (NII1) and the nitrate-assimilation gene cluster of Stagonospora (Septoria) nodorum. Curr. Genet. 36:282-289. [DOI] [PubMed] [Google Scholar]
- 18.Dietrich, F. S., S. Voegeli, S. Brachat, A. Lerch, K. Gates, S. Steiner, C. Mohr, R. Pohlmann, P. Luedi, S. D. Choi, R. A. Wing, A. Flavier, T. D. Gaffney, and P. Phillippsen. 2004. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science 304:304-307. [DOI] [PubMed] [Google Scholar]
- 19.Dujon, B., D. Sherman, G. Fischer, P. Durrens, S. Casaregola, I. Lafontaine, J. de Montigny, C. Marck, C. Neuveglise, E. Talla, N. Goffard, L. Frangeul, M. Aigle, V. Anthouard, A. Babour, V. Barbe, S. Barnay, S. Blanchin, J. M. Beckerich, E. Beyne, C. Bleykasten, A. Boisrame, J. Boyer, L. Cattolico, F. Confanioleri, A. de Daruvar, L. Despons, E. Fabre, C. Fairhead, H. Ferry-Dumazet, A. Groppi, F. Hantraye, C. Hennequin, N. Jauniaux, P. Joyet, R. Kachouri, A. Kerrest, R. Koszul, M. Lemaire, I. Lesur, L. Ma, H. Muller, J. M. Nicaud, M. Nikolski, S. Oztas, O. Ozier-Kalogeropoulos, S. Pellenz, S. Potier, G. F. Richard, M. L. Straub, A. Suleau, D. Swennen, F. Tekaia, M. Wesolowski-Louvel, E. Westhof, B. Wirth, M. Zeniou-Meyer, I. Zivanovic, M. Bolotin-Fukuhara, A. Thierry, C. Bouchier, B. Caudron, C. Scarpelli, C. Gaillardin, J. Weissenbach, P. Wincker, and J. L. Souciet. 2004. Genome evolution in yeasts. Nature 430:35-44. [DOI] [PubMed] [Google Scholar]
- 20.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 21.Fischer, G., E. P. C. Rocha, F. G. Brunet, M. Vergassola, and B. Dujon. 2006. Highly variable rates of genome rearrangements between hemiascomycetous yeast lineages. PLoS Genet. 2:e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fugelsang, K. C. 1997. Wine microbiology. Chapman and Hall, New York, NY.
- 23.Galagan, J. E., S. E. Calvo, C. Cuomo, L. J. Ma, J. R. Wortman, S. Batzoglou, S. I. Lee, M. Basturkmen, C. C. Spevak, J. Clutterbuck, V. Kapitonov, J. Jurka, C. Scazzocchio, M. Farman, J. Butler, S. Purcell, S. Harris, G. H. Braus, O. Draht, S. Busch, C. D'Enfert, C. Bouchier, G. H. Goldman, D. Bell-Pedersen, S. Griffiths-Jones, J. H. Doonan, J. Yu, K. Vienken, A. Pain, M. Freitag, E. U. Selker, D. B. Archer, M. A. Penalva, B. R. Oakley, M. Momany, T. Tanaka, T. Kumagai, K. Asai, M. Machida, W. C. Nierman, D. W. Denning, M. Caddick, M. Hynes, M. Paoletti, R. Fischer, B. Miller, P. Dyer, M. S. Sachs, S. A. Osmani, and B. W. Birren. 2005. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438:1105-1115. [DOI] [PubMed] [Google Scholar]
- 24.Gojkovic, Z., W. Knecht, E. Zameitat, J. Warneboldt, J. B. Coutelis, Y. Pynyaha, C. Neuveglise, K. Moller, M. Loffler, and J. Piškur. 2004. Horizontal gene transfer promoted evolution of the ability to propagate under anaerobic conditions in yeasts. Mol. Genet. Genomics 271:387-393. [DOI] [PubMed] [Google Scholar]
- 25.Goodwin, T. J. D., J. E. Ormandy, and R. T. M. Poulter. 2001. L1-like non-LTR retrotransposons in the yeast Candida albicans. Curr. Genet. 39:83-91. [DOI] [PubMed] [Google Scholar]
- 26.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 27.Guldener, U., M. Munsterkotter, G. Kastenmuller, N. Strack, J. van Helden, C. Lemer, J. Richelles, S. J. Wodak, J. Garcia-Martinez, J. E. Perez-Ortin, H. Michael, A. Kaps, E. Talla, B. Dujon, B. Andre, J. L. Souciet, J. De Montigny, E. Bon, C. Gaillardin, and H. W. Mewes. 2005. CYGD: the Comprehensive Yeast Genome Database. Nucleic Acids Res. 33:D364-D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hall, C., S. Brachat, and F. S. Dietrich. 2005. Contribution of horizontal gene transfer to the evolution of Saccharomyces cerevisiae. Eukaryot. Cell 4:1102-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heresztyn, T. 1986. Formation of substituted tetrahydropyridines by species of Brettanomyces and Lactobacillus isolated from mousy wines. Am. J. Enol. Vitic. 37:127-132. [Google Scholar]
- 30.Heresztyn, T. 1986. Metabolism of volatile phenolic compounds from hydroxycinnamic acids by Brettanomyces yeast. Arch. Microbiol. 146:96-98. [Google Scholar]
- 31.Holland, B. R., K. T. Huber, V. Moulton, and P. J. Lockhart. 2004. Using consensus networks to visualize contradictory evidence for species phylogeny. Mol. Biol. Evol. 21:1459-1461. [DOI] [PubMed] [Google Scholar]
- 32.Huson, D. H., and D. Bryant. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23:254-267. [DOI] [PubMed] [Google Scholar]
- 33.Ibeas, J. I., I. Lozano, F. Perdigones, and J. Jimenez. 1996. Detection of Dekkera-Brettanomyces strains in sherry by a nested PCR method. Appl. Environ. Microbiol. 62:998-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones, T., N. A. Federspiel, H. Chibana, J. Dungan, S. Kalman, B. B. Magee, G. Newport, Y. R. Thorstenson, N. Agabian, P. T. Magee, R. W. Davis, and S. Scherer. 2004. The diploid genome sequence of Candida albicans. Proc. Natl. Acad. Sci. USA 101:7329-7334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kajiwara, S., A. Shirai, T. Fujii, T. Toguri, K. Nakamura, and K. Ohtaguchi. 1996. Polyunsaturated fatty acid biosynthesis in Saccharomyces cerevisiae: expression of ethanol tolerance and the FAD2 gene from Arabidopsis thaliana. Appl. Environ. Microbiol. 62:4309-4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kurtzman, C. P., and J. W. Fell. 1998. The yeasts, a taxonomic study, 4th ed. Elsevier Science B.V., Amsterdam, The Netherlands.
- 37.Lasker, B. A., G. Butler, and T. J. Lott. 2006. Molecular genotyping of Candida parapsilosis group I clinical isolates by analysis of polymorphic microsatellite markers. J. Clin. Microbiol. 44:750-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Licker, J. L., T. E. Acree, and T. Henick-Kling. 1999. What is “Brett” (Brettanomyces) flavour? A preliminary investigation, p. 96-115. In A. L. Waterhouse and S. E. Ebeler (ed.), Chemistry of wine flavour. American Chemical Society, Washington, DC.
- 39.Llorente, B., A. Malpertuy, C. Neuveglise, J. de Montigny, M. Aigle, F. Artiguenave, G. Blandin, M. Bolotin-Fukuhara, E. Bon, P. Brottier, S. Casaregola, P. Durrens, C. Gaillardin, A. Lepingle, O. Ozier-Kalogeropoulos, S. Potier, W. Saurin, F. Tekaia, C. Toffano-Nioche, M. Wesolowski-Louvel, P. Wincker, J. Weissenbach, J. L. Souciet, and B. Dujon. 2000. Genomic exploration of the hemiascomycetous yeasts. 18. Comparative analysis of chromosome maps and synteny with Saccharomyces cerevisiae. FEBS Lett. 487:101-112. [DOI] [PubMed] [Google Scholar]
- 40.Mahenthiralingam, E., T. A. Urban, and J. B. Goldberg. 2005. The multifarious, multireplicon Burkholderia cepacia complex. Nat. Rev. Microbiol. 3:144-156. [DOI] [PubMed] [Google Scholar]
- 41.Maier, S. A., J. R. Galellis, and H. E. McDermid. 2005. Phylogenetic analysis reveals a novel protein family closely related to adenosine deaminase. J. Mol. Evol. 61:776-794. [DOI] [PubMed] [Google Scholar]
- 42.Martin, D., C. Brun, E. Remy, P. Mouren, D. Thieffry, and B. Jacq. 2004. GOToolBox: functional analysis of gene datasets based on gene ontology. Genome Biol. 5:R101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neuveglise, C., H. Feldmann, E. Bon, C. Gaillardin, and S. Casaregola. 2002. Genomic evolution of the long terminal repeat retrotransposons in hemiascomycetous yeasts. Genome Res. 12:930-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neuveglise, C., F. Chalvet, P. Wincker, C. Gaillardin, and S. Casaregola. 2005. Mutator-like element in the yeast Yarrowia lipolytica displays multiple alternative splicings. Eukaryot. Cell 4:615-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nisiotou, A. A., and G. R. Gibson. 2005. Isolation of culturable yeasts from market wines and evaluation of the 5.8S-ITS rDNA sequence analysis for identification purposes. Lett. Appl. Microbiol. 41:454-463. [DOI] [PubMed] [Google Scholar]
- 46.Notredame, C., D. G. Higgins, and J. Heringa. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302:205-217. [DOI] [PubMed] [Google Scholar]
- 47.Padovan, A. C. B., G. F. O. Sanson, A. Brunstein, and M. R. S. Briones. 2005. Fungi evolution revisited: application of the penalized likelihood method to a Bayesian fungal phylogeny provides a new perspective on phylogenetic relationships and divergence dates of Ascomycota groups. J. Mol. Evol. 60:726-735. [DOI] [PubMed] [Google Scholar]
- 48.Piškur, J., E. Rożpędowska, S. Polakova, A. Merico, and C. Compagno. 2006. How did Saccharomyces evolve to become a good brewer? Trends Genet. 22:183-186. [DOI] [PubMed] [Google Scholar]
- 49.Pretorius, I. S. 2000. Tailoring wine yeast for the new millennium: novel approaches to the ancient art of winemaking. Yeast 16:675-729. [DOI] [PubMed] [Google Scholar]
- 50.Rainey, F. A., K. Ray, M. Ferreira, B. Z. Gatz, M. F. Nobre, D. Bagaley, B. A. Rash, M. J. Park, A. M. Earl, N. C. Shank, A. M. Small, M. C. Henk, J. R. Battista, P. Kampfer, and M. S. da Costa. 2005. Extensive diversity of ionizing-radiation-resistant bacteria recovered from Sonoran Desert soil and description of nine new species of the genus Deinococcus obtained from a single soil sample. Appl. Environ. Microbiol. 71:5225-5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Renouf, V., M. Falcou, C. Miot-Sertier, M. C. Perello, G. De Revel, and A. Lonvaud-Funel. 2006. Interactions between Brettanomyces bruxellensis and other yeast species during the initial stages of winemaking. J. Appl. Microbiol. 100:1208-1219. [DOI] [PubMed] [Google Scholar]
- 52.Ribard, C., M. Rochet, B. Labedan, B. Daignan-Fornier, P. Alzari, C. Scazzocchio, and N. Oestreicher. 2003. Sub-families of α/β barrel enzymes: a new adenine deaminase family. J. Mol. Biol. 334:1117-1131. [DOI] [PubMed] [Google Scholar]
- 53.Schluter, A., S. Fourcade, R. Ripp, J. L. Mandel, O. Poch, and A. Pujol. 2006. The evolutionary origin of peroxisomes: an ER-peroxisome connection. Mol. Biol. Evol. 23:838-845. [DOI] [PubMed] [Google Scholar]
- 54.Schmidt, H. A., K. Strimmer, M. Vingron, and A. von Haeseler. 2002. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18:502-504. [DOI] [PubMed] [Google Scholar]
- 55.Seoighe, C., N. Federspiel, T. Jones, N. Hansen, V. Bivolarovic, R. Surzycki, R. Tamse, C. Komp, L. Hulzar, R. W. Davis, S. Scherer, E. Tait, D. J. Shaw, D. Harris, L. Murphy, K. Oliver, K. Taylor, M. A. Rajandream, B. G. Barrell, and K. H. Wolfe. 2000. Prevalence of small inversions in yeast gene order evolution. Proc. Natl. Acad. Sci. USA 97:14433-14437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shimodaira, H., and M. Hasegawa. 1999. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol. Biol. Evol. 16:1114-1116. [Google Scholar]
- 57.Silva, P., H. Cardoso, and H. Geros. 2004. Studies on the wine spoilage capacity of Brettanomyces/Dekkera spp. Am. J. Enol. Vitic. 55:65-72. [Google Scholar]
- 58.Siurkus, J. 2004. Preliminary molecular biology studies of Dekkera bruxellensis yeast. M.Sc. thesis. Technical University of Denmark, Lyngby, Denmark.
- 59.Snyder, W. B., K. N. Faber, T. J. Wenzel, A. Koller, G. H. Luers, L. Rangell, G. A. Keller, and S. Subramani. 1999. Pex19p interacts with Pex3p and Pex10p and is essential for peroxisome biogenesis in Pichia pastoris. Mol. Biol. Cell 10:1745-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tekaia, F., G. Blandin, A. Malpertuy, B. Llorente, P. Durrens, C. Toffano-Nioche, O. Ozier-Kalogeropoulos, E. Bon, C. Gaillardin, M. Aigle, M. Bolotin-Fukuhara, S. Casaregola, J. de Montigny, A. Lepingle, C. Neuveglise, S. Potier, J. L. Souciet, M. Wesolowski-Louvel, and B. Dujon. 2000. Genomic exploration of the hemiascomycetous yeasts. 3. Methods and strategies used for sequence analysis and annotation. FEBS Lett. 487:17-30. [DOI] [PubMed] [Google Scholar]
- 61.Toro, M. E., N. P. Oro, A. D. Vega, Y. P. Maturano, M. C. Nally, E. Fernandez, E. Pucheta, and F. Vazquez. 2005. Diversidad de levaduras en canopias y suelos asociados con Bulnesia retama y Larrea divaricata. Rev. Argentina Microbiol. 37:209-213. [PubMed] [Google Scholar]
- 62.Veiga, A., J. D. Arrabaca, and M. C. Loureiro-Dias. 2000. Cyanide-resistant respiration is frequent, but confined to yeasts incapable of aerobic fermentation. FEMS Microbiol. Lett. 190:93-97. [DOI] [PubMed] [Google Scholar]
- 63.Veiga, A., J. D. Arrabaca, and M. C. Loureiro-Dias. 2003. Cyanide-resistant respiration, a very frequent metabolic pathway in yeasts. FEMS Yeast Res. 3:239-245. [DOI] [PubMed] [Google Scholar]
- 64.Verho, R., M. Putkonen, J. Londesborough, M. Penttila, and P. Richard. 2004. A novel NADH-linked L-xylulose reductase in the L-arabinose catabolic pathway of yeast. J. Biol. Chem. 279:14746-14751. [DOI] [PubMed] [Google Scholar]
- 65.Wolfe, K. H., and D. C. Shields. 1997. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387:708-713. [DOI] [PubMed] [Google Scholar]
- 66.Wood, V., R. Gwilliam, M. A. Rajandream, M. Lyne, R. Lyne, A. Stewart, J. Sgouros, N. Peat, J. Hayles, S. Baker, D. Basham, S. Bowman, K. Brooks, D. Brown, S. Brown, T. Chillingworth, C. Churcher, M. Collins, R. Connor, A. Cronin, P. Davis, T. Feltwell, A. Fraser, S. Gentles, A. Goble, N. Hamlin, D. Harris, J. Hidalgo, G. Hodgson, S. Holroyd, T. Hornsby, S. Howarth, E. J. Huckle, S. Hunt, K. Jagels, K. James, L. Jones, M. Jones, S. Leather, S. McDonald, J. McLean, P. Mooney, S. Moule, K. Mungall, L. Murphy, D. Niblett, C. Odell, K. Oliver, S. O'Neil, D. Pearson, M. A. Quail, E. Rabbinowitsch, K. Rutherford, S. Rutter, D. Saunders, K. Seeger, S. Sharp, J. Skelton, M. Simmonds, R. Squares, S. Squares, K. Stevens, K. Taylor, R. G. Taylor, A. Tivey, S. Walsh, T. Warren, S. Whitehead, J. Woodward, G. Volckaert, R. Aert, J. Robben, B. Grymonprez, I. Weltjens, E. Vanstreels, M. Rieger, M. Schafer, S. Muller-Auer, C. Gabel, M. Fuchs, C. Fritzc, E. Holzer, D. Moestl, H. Hilbert, K. Borzym, I. Langer, A. Beck, H. Lehrach, R. Reinhardt, T. M. Pohl, P. Eger, W. Zimmermann, H. Wedler, R. Wambutt, B. Purnelle, A. Goffeau, E. Cadieu, S. Dreano, S. Gloux, V. Lelaure, S. Mottier, F. Galibert, S. J. Aves, Z. Xiang, C. Hunt, K. Moore, S. M. Hurst, M. Lucas, M. Rochet, C. Gaillardin, V. A. Tallada, A. Garzon, G. Thode, R. R. Daga, L. Cruzado, J. Jimenez, M. Sanchez, F. del Rey, J. Benito, A. Dominguez, J. L. Revuelta, S. Moreno, J. Armstrong, S. L. Forsburg, L. Cerrutti, T. Lowe, W. R. McCombie, I. Paulsen, J. Potashkin, G. V. Shpakovski, D. Ussery, B. G. Barrell, and P. Nurse. 2002. The genome sequence of Schizosaccharomyces pombe. Nature 415:871-880. [DOI] [PubMed] [Google Scholar]
- 67.Yamada, Y., M. Matsuda, K. Maeda, and K. Mikata. 1994. The phylogenetic relationships of species of the genus Dekkera Van Der Walt based on the partial sequences of 18S and 26S ribosomal RNAs (Saccharomycetaceae). Biosci. Biotechnol. Biochem. 58:1803-1808. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.