Abstract
Candida albicans expresses specific virulence traits that promote disease establishment and progression. These traits include morphological transitions between yeast and hyphal growth forms that are thought to contribute to dissemination and invasion and cell surface adhesins that promote attachment to the host. Here, we describe the regulation of the adhesin gene ALS3, which is expressed specifically during hyphal development in C. albicans. Using a combination of reporter constructs and regulatory mutants, we show that this regulation is mediated by multiple factors at the transcriptional level. The analysis of ALS3 promoter deletions revealed that this promoter contains two activation regions: one is essential for activation during hyphal development, while the second increases the amplitude of this activation. Further deletion analyses using the Renilla reniformis luciferase reporter delineate the essential activation region between positions −471 and −321 of the promoter. Further 5′ or 3′ deletions block activation. ALS3 transcription is repressed mainly by Nrg1 and Tup1, but Rfg1 contributes to this repression. Efg1, Tec1, and Bcr1 are essential for the transcriptional activation of ALS3, with Tec1 mediating its effects indirectly through Bcr1 rather than through the putative Tec1 sites in the ALS3 promoter. ALS3 transcription is not affected by Cph2, but Cph1 contributes to full ALS3 activation. The data suggest that multiple morphogenetic signaling pathways operate through the promoter of this adhesin gene to mediate its developmental regulation in this major fungal pathogen.
Candida albicans is a major opportunistic pathogen of humans (54). This fungus is a frequent cause of superficial oral and vaginal infections, and in immunocompromised patients, C. albicans can disseminate via the bloodstream to invade internal organs, thereby causing deep-seated, systemic infections that are often fatal (54).
Various factors are thought to contribute to the virulence of C. albicans. These include adhesion to host tissue, the ability to undergo reversible morphogenetic transitions between budding (yeast) and filamentous (hyphae and pseudohyphae) growth forms, the secretion of extracellular hydrolases, and rapid switching between different phenotypic forms (30, 42, 44, 65). The contribution of yeast-hypha morphogenesis to C. albicans virulence has been hotly debated (21, 29, 71). However, it is clear that hyphal development is closely associated with tissue invasion (21, 61, 71, 83).
Adherence plays a key role in fungal colonization (27, 68, 70). C. albicans expresses an array of adhesin genes including HWP1, which encodes a cell surface glycoprotein that acts as a target for mammalian transglutaminases. These enzymes are thought to generate covalent cross-links between Hwp1 on the fungal hyphal surface and proteins on the mammalian cell surface (68, 72). The ALS gene gamily encodes a set of differentially regulated cell surface glycosylphosphatidylinositol-anchored glycoproteins that promote fungal adherence (27, 55). ALS3 was initially identified as a member of this gene family that is expressed specifically during hyphal development (28). A second hypha-specific ALS gene (ALS8) (40) was later identified as an allele of the ALS3 gene (81). C. albicans als3/als3 cells are defective in biofilm formation (53, 82). Furthermore, Als3 is involved in adhesion to endothelial and epithelial cells (55), and als3/als3 cells display an almost total lack of epithelial destruction in a reconstituted buccal human epithelium model (81). ALS3 expression has been detected in clinical vaginal fluid specimens and in a vaginal candidiasis model (13). These observations indicate a role for ALS3 in the pathogenicity of C. albicans.
A complex network of signaling pathways regulates yeast-hypha morphogenesis (10). Following exposure to serum, hyphal development is activated by a cyclic AMP-protein kinase A pathway that regulates the activity of the β-helix-loop-helix transcription factor Efg1 (42, 69). In addition, a mitogen-activated protein kinase pathway, which includes the Ste12-like transcription factor Cph1, activates hyphal development under starvation conditions (38, 41). Additional regulatory factors contribute to the activation of hyphal development, but their relationship to these main signaling pathways remains to be established (11, 16, 39, 57). These include the transcription factors Tec1 and Cph2, the inactivation of which causes defects in hyphal development (37, 62). Tec1 is a TEA (TEF-1, Tec1p, and AbaAp)/ATTS (AbaAp, TEF-1, Tec1p, and Scalloped) motif transcription factor that is also required for C. albicans virulence (62). It was previously suggested that TEC1 is regulated both by Cph2 and Efg1 (36), but their precise roles in gene regulation during hyphal development are not known.
Hyphal development is negatively regulated by the transcriptional repressors Tup1, Nrg1, and Rfg1 (6, 9, 31, 46). In the absence of hypha-inducing signals, the global repressor Tup1 inhibits the transcription of hypha-specific genes. This repression is dependent upon Nrg1, which binds to Nrg1 response elements (NREs) in the promoters of these genes and targets Tup1 to these promoters (9, 18, 46). Current models suggest that Rfg1 is a second DNA-binding protein that targets Tup1 to the promoters of hypha-specific genes, although Nrg1 appears to make the major contribution to the repression of hyphal development (31, 32).
The prevailing view is that these morphogenetic signaling pathways combine to regulate the transcription of hypha-specific genes. Genome-wide and gene-specific studies have revealed only a small number of hypha-specific genes in C. albicans. These include ALS3, ECE1, HGC1, HWP1, HYR1, RBT1, and RBT4 (2, 5, 8, 28, 48, 67, 83). As described above, ALS3 and HWP1 encode adhesins. The inactivation of RBT1 or RBT4 attenuates C. albicans virulence (9). HGC1 encodes a hypha-specific cyclin required for hyphal development and virulence (83). With the exception of HGC1, all known hypha-specific genes appear to encode secreted or cell wall proteins. These observations reinforce the tight link between the formation of hyphae, the cell surface, and C. albicans virulence.
In this paper, we have examined the organization of the ALS3 promoter and determined the relative contributions of key morphogenetic transcription factors to the regulation of this hypha-specific gene. We find that, relative to other C. albicans genes, the promoter regions of ALS3 and other hypha-specific genes are unusually large. We show that the ALS3 promoter is complex, requiring a 150-bp region for hypha-specific activation. This promoter integrates inputs from multiple activators and repressors. Related observations have been made for a second hypha-specific gene (HWP1) by Kim et al. in the accompanying paper (33).
MATERIALS AND METHODS
Strains and growth conditions.
C. albicans strains (Table 1) were grown in YPD at 30°C (63). Hyphal development was induced using 20% bovine calf serum (73).
TABLE 1.
C. albicans strains
| Strain | Genotype or descriptiona | Parent strain | Reference or source |
|---|---|---|---|
| SC5314 | Wild-type clinical isolate | 20 | |
| CAI4 | ura3::λ imm434/ura3::λ imm434 | SC5314 | 17 |
| CAI8 | ura3::λ imm434/ura3::λ imm434 ade2::hisG/ade2::hisG | CAI4 | 17 |
| RM1000 | ura3::λ imm434/ura3::λ imm434 his1::hisG/his1::hisG | CAI4 | 50 |
| BWP17 | ura3::λ imm434/ura3::λ imm434 arg4::hisG/arg4::hisG his1::hisG/his1::hisG | RM1000 | 79 |
| Ca90 | ura3::λ imm434/ura3::λ imm434 als3::hisG/als3::hisG | CAI4 | This study |
| Ca107 | ura3::λ imm434/ura3::λ imm434 ade2::hisG/ADE2-ALS3696-RrLUC | CAI8 | This study |
| Ca108 | ura3::λ imm434/ura3::λ imm434 ade2::hisG/ADE2-ALS3496-RrLUC | CAI8 | This study |
| Ca109 | ura3::λ imm434/ura3::λ imm434 ade2::hisG/ADE2-ALS3306-RrLUC | CAI8 | This study |
| Ca110 | ura3::λ imm434/ura3::λ imm434 ade2::hisG/ADE2-ALS3129-RrLUC | CAI8 | This study |
| Ca111 | ura3::λ imm434/ura3::λ imm434 ade2::hisG/ADE2-ALS3100-RrLUC | CAI8 | This study |
| Ca176 | ura3::λ imm434/ura3::λ imm434 ade2::hisG/ADE2-ALS3496-307-RrLUC | CAI8 | This study |
| Ca167 | ura3::λ imm434/ura3::λ imm434 ade2::hisG/ADE2-ALS3496-461-RrLUC | CAI8 | This study |
| Ca619 | ura3::λ imm434/ura3::λ imm434 ade2::hisG/ADE2-ALS3483-445-RrLUC | CAI8 | This study |
| Ca168 | ura3::λ imm434/ura3::λ imm434 ade2::hisG/ADE2-ALS3467-421-RrLUC | CAI8 | This study |
| Ca620 | ura3::λ imm434/ura3::λ imm434 ade2::hisG/ADE2-ALS3450-405-RrLUC | CAI8 | This study |
| Ca169 | ura3::λ imm434/ura3::λ imm434 ade2::hisG/ADE2-ALS3429-417-RrLUC | CAI8 | This study |
| Ca170 | ura3::λ imm434/ura3::λ imm434 ade2::hisG/ADE2-ALS3420-386-RrLUC | CAI8 | This study |
| Ca621 | ura3::λ imm434/ura3::λ imm434 ade2::hisG/ADE2-ALS3388-352-RrLUC | CAI8 | This study |
| Ca171 | ura3::λ imm434/ura3::λ imm434 ade2::hisG/ADE2-ALS3374-307-RrLUC | CAI8 | This study |
| Ca622 | ura3::λ imm434/ura3::λ imm434 ade2::hisG/ADE2-ALS3471-321-RrLUC | CAI8 | This study |
| Ca623 | ura3::λ imm434/ura3::λ imm434 ade2::hisG/ADE2-ALS3471-338-RrLUC | CAI8 | This study |
| Ca624 | ura3::λ imm434/ura3::λ imm434 ade2::hisG/ADE2-ALS3448-321-RrLUC | CAI8 | This study |
| Ca625 | ura3::λ imm434/ura3::λ imm434 ade2::hisG/ADE2-ALS3448-338-RrLUC | CAI8 | This study |
| SAC500 | ura3::λ imm434/ura3::λ imm434 his1::hisG/his1::hisG ALS3/ALS3-GFP-URA3 | RM1000 | This study |
| SAC501 | ura3::λ imm434/ura3::λ imm434 his1::hisG/his1::hisG ALS3/HIS1-ALS31449-GFP-URA3 | RM1000 | This study |
| SAC502 | ura3::λ imm434/ura3::λ imm434 his1::hisG/his1::hisG ALS3/HIS1-ALS31449t-GFP-URA3 | RM1000 | This study |
| SAC503 | ura3::λ imm434/ura3::λ imm434 his1::hisG/his1::hisG ALS3/HIS1-ALS31438-GFP-URA3 | RM1000 | This study |
| SAC504 | ura3::λ imm434/ura3::λ imm434 his1::hisG/his1::hisG ALS3/HIS1-ALS31438t-GFP-URA3 | RM1000 | This study |
| SAC505 | ura3::λ imm434/ura3::λ imm434 his1::hisG/his1::hisG ALS3/HIS1-ALS31049-GFP-URA3 | RM1000 | This study |
| SAC506 | ura3::λ imm434/ura3::λ imm434 his1::hisG/his1::hisG ALS3/HIS1-ALS31049t-GFP-URA3 | RM1000 | This study |
| SAC507 | ura3::λ imm434/ura3::λ imm434 his1::hisG/his1::hisG ALS3/HIS1-ALS3885-GFP-URA3 | RM1000 | This study |
| SAC508 | ura3::λ imm434/ura3::λ imm434 his1::hisG/his1::hisG ALS3/HIS1-ALS3885t-GFP-URA3 | RM1000 | This study |
| SAC509 | ura3::λ imm434/ura3::λ imm434 his1::hisG/his1::hisG, ALS3/HIS1-ALS3842-GFP-URA3 | RM1000 | This study |
| SAC510 | ura3::λ imm434/ura3::λ imm434 his1::hisG/his1::hisG ALS3/HIS1-ALS3842t-GFP-URA3 | RM1000 | This study |
| SAC511 | ura3::λ imm434/ura3::λ imm434 his1::hisG/his1::hisG ALS3/HIS1-ALS3696-GFP-URA3 | RM1000 | This study |
| SAC512 | ura3::λ imm434/ura3::λ imm434 his1::hisG/his1::hisG ALS3/HIS1-ALS3471-GFP-URA3 | RM1000 | This study |
| SAC513 | ura3::λ imm434/ura3::λ imm434 his1::hisG/his1::hisG ALS3/HIS1-ALS3306-GFP-URA3 | RM1000 | This study |
| SAC530 | ura3::λ imm434/ura3::λ imm434 ALS3/ALS3-GFP-URA3 | CAI4 | This study |
| MMC4 | ura3::λ imm434/ura3::λ imm434 nrg1::hisG/nrg1::hisG | CAI4 | 46 |
| SAC531 | ura3::λ imm434/ura3::λ imm434 nrg1::hisG/nrg1::hisG ALS3/ALS3-GFP-URA3 | MMC4 | This study |
| DK158 | ura3::λ imm434/ura3::λ imm434 rfg1::hisG/rfg1::hisG | CAI4 | 31 |
| SAC532 | ura3::λ imm434/ura3::λ imm434 rfg1::hisG/rfg1::hisG ALS3/ALS3-GFP-URA3 | DK158 | This study |
| BCa2-10 | ura3::λ imm434/ura3::λ imm434 tup1::hisG/tup1::hisG | CAI4 | 6 |
| SAC533 | ura3::λ imm434/ura3::λ imm434 tup1::hisG/tup1::hisG ALS3/ALS3-GFP-URA3 | BCA2-10 | This study |
| SGC124 | ura3::λ imm434/ura3::λ imm434 ade2::hisG/ade2::hisG ssn6::hisG/ssn6::hisG | CAI8 | 18 |
| SAC534 | ura3::λ imm434/ura3::λ imm434 ade2::hisG/ade2::hisG ssn6::hisG/ssn6::hisG ALS3/ALS3-GFP-URA3 | SGC124 | This study |
| CHY257 | ura3::λ imm434/ura3::λ imm434 a1 a2::hisG | CAI4 | 43 |
| SAC535 | ura3::λ imm434/ura3::λ imm434 a1 a2::hisG ALS3/ALS3-GFP-URA3 | CHY257 | This study |
| JCK18 | ura3::λ imm434/ura3::λ imm434 cph1::hisG/cph1::hisG | CAI4 | 41 |
| SAC536 | ura3::λ imm434/ura3::λ imm434 cph1::hisG/cph1::hisG ALS3/ALS3-GFP-URA3 | This study | |
| HLY1927 | ura3::λ imm434/ura3::λ imm434 his1::hisG/his1::hisG cph2::ARG4/cph2::HIS1 | CAI4 | 37 |
| SAC537 | ura3::λ imm434/ura3::λ imm434 his1::hisG/his1::hisG cph2::ARG4/cph2::HIS1 ALS3/ALS3-GFP-URA3 | This study | |
| HLC67 | ura3::λ imm434/ura3::λ imm434 efg1::hisG/efg1::hisG | CAI4 | 42 |
| SAC538 | ura3::λ imm434/ura3::λ imm434 efg1::hisG/efg1::hisG ALS3/ALS3-GFP-URA3 | HLC67 | This study |
| AS18 | ura3::λ imm434/ura3::λ imm434 tec1::hisG/tec1::hisG (pVEC) | CAI4 | 62 |
| SAC540 | ura3::λ imm434/ura3::λ imm434 tec1::hisG/tec1::hisG (pVEC), post-5-FOA selection | AS18 | This study |
| SAC539 | ura3::λ imm434/ura3::λ imm434 tec1::hisG/tec1::hisG (pVEC), post-5-FOA selection, ALS3/ALS3-GFP-URA3 | AS18 | This study |
| CJN702 | ura3::λ imm434/ura3::λ imm434 arg4::hisG/arg4::hisG his1::hisG/his1::hisG::pHIS1 bcr1::ARG4/bcr1::URA3 | BWP17 | 52 |
| SAC519 | ura3::λ imm434/ura3::λ imm434 arg4::hisG/arg4::hisG his1::hisG/his1::hisG::pHIS1 bcr1::ARG4/bcr1::URA3, post-5-FOA selection | CJN702 | This study |
| SAC528 | ura3::λ imm434/ura3::λ imm434 arg4::hisG/arg4::hisG his1::hisG/his1::hisG::pHIS1 bcr1::ARG4/bcr1::URA3, post-5-FOA selection, ALS3/ALS3-GFP-URA3 | CJN702 | This study |
| CJN688 | ura3::λ imm434/ura3::λ imm434 arg4::hisG/arg4::hisG, his1::hisG/his1::hisG bcr1::ARG4/bcr1::URA3 | BWP17 | 52 |
| SAC518 | ura3::λ imm434/ura3::λ imm434 arg4::hisG/arg4::hisG his1::hisG/his1::hisG bcr1::ARG4/bcr1::URA3, post-5-FOA selection | CJN688 | This study |
| SAC520 | ura3::λ imm434/ura3::λ imm434 arg4::hisG/arg4::hisG his1::hisG/his1::hisG ALS3/ALS3-GFP-HIS1 | BWP17 | This study |
| SAC521 | ura3::λ imm434/ura3::λ imm434 arg4::hisG/arg4::hisG his1::hisG/his1::hisG bcr1::ARG4/bcr1::URA3, post-5-FOA selection, ALS3/ALS3-GFP-HIS1 | BWP17 | This study |
| SAC522 | ura3::λ imm434/ura3::λ imm434 arg4::hisG/arg4::hisG his1::hisG/his1::hisG ALS3/ALS3-GFP-HIS1 RPS1-PYK1-URA3 | BWP17 | This study |
| SAC523 | ura3::λ imm434/ura3::λ imm434 arg4::hisG/arg4::hisG his1::hisG/his1::hisG ALS3/ALS3-GFP-HIS1 RPS1-PYK1-TEC1-URA3 | BWP17 | This study |
| SAC524 | ura3::λ imm434/ura3::λ imm434 arg4::hisG/arg4::hisG his1::hisG/his1::hisG bcr1::ARG4/bcr1::ura3−ALS3/ALS3-GFP-HIS1 RPS1-PYK1-URA3 | BWP17 | This study |
| SAC525 | ura3::λ imm434/ura3::λ imm434 arg4::hisG/arg4::hisG his1::hisG/his1::hisG bcr1::ARG4/bcr1::ura3−ALS3/ALS3-GFP-HIS1 RPS1-PYK1-TEC1-URA3 | BWP17 | This study |
5-FOA, 5-fluoroorotic acid.
Strain construction.
To generate the C. albicans strains carrying in situ ALS3-GFP promoter fusions, yeast enhanced green fluorescent protein (GFP) was first integrated immediately downstream of the start codon at the ALS3 locus to create SAC500 (Table 1) (14). This was done by PCR amplifying a GFP-URA3 cassette (19) (primers ALS3-F1 and ALS3-F2) (see Table S1 in the supplemental material) and transforming it into C. albicans RM1000 (77). The correct integration at the ALS3 locus in Ura-positive or -negative transformants was confirmed by whole-cell PCR (60) and Southern blotting (not shown). Promoter deletions were created at the ALS3-GFP allele by PCR amplifying HIS1 (15) with primers designed to target this marker to specific regions of the ALS3 upstream region (see Table S1 in the supplemental material). The correct integration of HIS1 upstream of the ALS3-GFP allele in His-positive or -negative and Ura-positive or -negative transformants was confirmed by PCR and Southern blotting. C. albicans strains with mutated Tec1 sites in the ALS3 promoter were made in the same way by using 5′ PCR primers that contain mutations in these sequences (see Table S1 in the supplemental material). These mutations in the C. albicans genome were confirmed by PCR amplification of the corresponding regions followed by DNA sequence analysis (not shown).
The first set of ALS3-Renilla reniformis luciferase (RrLUC) promoter fusions was constructed by PCR amplifying portions of the ALS3 promoter up to position +4 of the ALS3 coding region (see Table S1 in the supplemental material) and cloning these portions between the ClaI and PstI sites in pCRW3 (66). To create the next sets of RrLUC promoter fusions, new BstEII, NdeI, SpeI, NotI, and MluI sites were introduced into pCRW3 to make pCRW3N (oligonucleotides KpnSal and SalKpn) (see Table S1 in the supplemental material). A basal ALS3 promoter region (positions −306 to +4) was then PCR amplified and inserted between the PstI and MluI sites in pCRW3N, upstream of the RrLUC open reading frame (ORF). Various promoter ALS3 fragments were cloned as oligonucleotides or PCR fragments (see Table S1 in the supplemental material) upstream of this basal ALS3 promoter region. The STRE5-, YRE5-, and GCRE5-RrLUC fusions were made by cloning oligonucleotides with each sequence element upstream of a basal RrLUC reporter containing part of the ADH1 promoter region (51, 74) (see Table S1 in the supplemental material). pCRW3-based plasmids were linearized with HindIII and transformed into C. albicans CAI8 (Table 1) selecting for the ADE2 marker. Single-copy integration at the ade2 locus was confirmed by PCR diagnosis.
To test the roles of Bcr1 and Tec1 in C. albicans, a nonrevertible Ura3− segregant of CJN688 (52) was selected (SAC518) (Table 1). SAC518 was transformed with a GFP-HIS1 cassette (19), as described above, to generate the in situ ALS3-GFP reporter in this bcr1 strain (SAC521). Meanwhile, the TEC1 ORF was PCR amplified (primers TEC1-F4 and TEC1-R) (see Table S1 in the supplemental material) and cloned into pPYK1-GFP (4). This placed TEC1 under the control of the PYK1 promoter. pPYK1-TEC1 and the empty control vector pPYK1 were linearized with StuI and transformed into SAC520 (BCR1) and SAC521 (bcr1) (Table 1). Single-copy integration at RPS1 was confirmed by PCR diagnosis (45).
DNA and RNA analysis.
DNA was prepared and analyzed by Southern blotting as described previously (25, 78). RNA was isolated and Northern analysis was performed as described previously (24, 47). The ALS3-specific probe was PCR amplified using primers ALS3-F and ALS3-R, which were described previously by Hoyer et al. (28). GFP and ACT1 sequences were analyzed using probes corresponding to the PCR-amplified ORFs. Primers are specified in Table S1 in the supplemental material.
Reporter assays.
GFP fluorescence in whole C. albicans cells was quantified in 96-well, black, clear-bottomed microplates (Matrix Technologies, Wilmslow, United Kingdom) using a Tecan Ultra 384 Microplate reader (Tecan Trading AG, Switzerland) running XFluor 4 software. Fluorescence polarization was used to distinguish GFP fluorescence from background autofluorescence (34, 35). The method exploits the high fluorescence anisotropy of GFP compared to other autofluorescing species. The difference between the fluorescence that polarized parallel to the excitation light and that which polarized perpendicular to the excitation light was used as the analytical signal. This measurement is relatively large for GFP and small for autofluorescing molecules. Fluorescence and fluorescence polarization measurements were made at 485-nm excitation and 535-nm emission wavelengths, as described previously (35). Means (in “FP [fluorescence polarization] brightness” units) and standard deviations from two to eight independent transformants are presented. Observations were reproducible in at least two independent experiments. Untransformed C. albicans cells were used as the background control.
Luciferase assays (relative light units/20 μg protein/20 s) were performed using fresh C. albicans protein extracts with a Lumat LB9507 luminometer (EG&G Berthold) as described previously (46). Means and standard deviations from quadruplicate assays are presented, and similar data were obtained in three experiments using independent transformants.
Microscopy.
Cell morphology was monitored using an Olympus BX50 microscope and recorded with an Olympus DP11-P digital video camera. Cell numbers were counted using an Improved Neubauer hemocytometer.
Phase-contrast microscopy and fluorescence microscopy were performed using an Axioplan 2 microscope (Carl Zeiss, United Kingdom) with filter sets XF66 (blue emission), XF67 (red emission), and XF77 (green emission) from Omega Optical Inc. (Brattleboro, VT). Images were generated using a Hamamatsu charge-coupled-device camera and analyzed using Openlab 3.0.9 (Improvision, Coventry, United Kingdom). C. albicans cells were mounted onto polylysine-coated glass slides and covered with Vectashield immunofluorescence mounting medium (Vector Laboratories, Peterborough, United Kingdom) (3).
In silico promoter analysis.
Promoter sequences were analyzed for the presence of putative regulatory elements using MatInspector (12, 56) (http://www.genomatix.de/products/MatInspector/index.html) or Regulatory Sequence Analysis Tools (76) (http://www.flychip.org.uk/rsa-tools/).
RESULTS
ALS3 transcription is activated specifically during hyphal development.
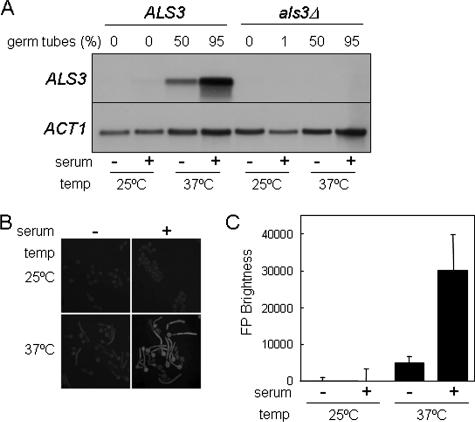
To confirm that the ALS3 gene is expressed specifically during hyphal development in C. albicans, we examined ALS3 mRNA levels by Northern blotting following exposure to three distinct types of morphogenetic signals: serum (Fig. 1A), neutral pH, and N-acetylglucosamine (not shown). The ALS3 mRNA was induced in C. albicans cells growing at 37°C and strongly induced in cells exposed to serum at 37°C (Fig. 1A). This transcript was undetectable in a C. albicans als3/als3 null mutant. ALS3 mRNA levels correlated strongly with the extent of hyphal development in these cultures. The same was true when hyphal development was induced by neutral pH or N-acetylglucosamine (not shown). Our data confirm data from a previous report by Hoyer et al. showing that ALS3 is a hypha-specific gene (28).
FIG. 1.
ALS3 transcription is activated during hyphal development. (A) Northern analysis of ALS3 mRNA levels in C. albicans after 3 h of growth in YPD at 25°C, in YPD containing serum at 25°C, in YPD at 37°C, or in YPD containing serum at 37°C. ALS3, SC5314; als3Δ, Ca90 (Table 1). The proportion of filamentous (as opposed to yeast) cells in each culture is indicated. (B) Fluorescence microscopy of C. albicans SAC500 cells containing the in situ ALS3-GFP reporter under equivalent conditions. (C) Quantification of GFP fluorescence in C. albicans SAC500 cells under the same conditions.
To test whether the developmental expression pattern of ALS3 is mediated at a transcriptional or posttranscriptional level, we generated an ALS3-GFP promoter fusion. This promoter fusion was integrated into the C. albicans genome in situ at the ALS3 locus (SAC530) (Table 1). The expression of this reporter was monitored by assaying fluorescence levels in C. albicans SAC530 cells growing in the presence and absence of serum (Fig. 1B and C). The ALS3-GFP promoter fusion displayed an expression pattern that was similar to that of wild-type ALS3 mRNA, indicating that ALS3 transcription is induced specifically during hyphal development.
Hypha-specific promoters are unusually long.
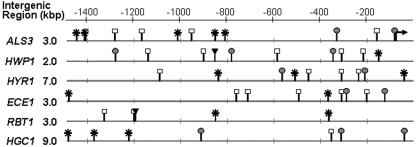
Having established that the developmental regulation of ALS3 is mediated at the transcriptional level, we performed an in silico comparison of the ALS3 promoter region and other hypha-specific promoters (ECE1, HGC1, HWP1, HYR1, RBT1, and RBT4). Our aim was to identify common sequence elements that might contribute to the coordinate regulation of these genes during hyphal development. To achieve this, we analyzed the intergenic regions that lie upstream of these genes (Fig. 2). Two main observations were made. First, the 5′-intergenic regions for hypha-specific genes are unusually long compared to C. albicans genes in general. The estimated average length of intergenic regions for divergently transcribed C. albicans genes is 1,088 bp, that for convergently transcribed genes is 521 bp, and that for tandemly transcribed genes is 770 bp (26). In contrast, the average length of the upstream intergenic regions for these seven hypha-specific genes is 4.5 kbp (based on the latest genome assembly available in the Candida Genome Database) (http://www.candidagenome.org/ [accessed October 2006]). The ALS3 intergenic region is 3.0 kbp, and HCG1 has the longest region at 9.0 kbp. This provided our first clue that morphogenetically regulated promoters in C. albicans might be relatively complex. This view is consistent with observations of budding yeast. For example, the developmentally regulated FLO11 and HO genes in Saccharomyces cerevisiae both have unusually long and complex promoters (49, 58).
FIG. 2.
In silico analysis of hypha-specific promoters. The lengths of the intergenic regions of hypha-specific genes and the organization of specific sequence elements in their 5′ regions are presented. Asterisks, Tec1 sites (CATTCY); open squares, E box (CANNTG); gray circles, Nrg1 sites (MVCCCT); closed triangles, Rfg1 sites (YYYATTGTTCTC). The lengths of the intergenic regions were calculated from assembly 20 of the C. albicans genome sequence (see the CGD website at www.candidagenome.org/ [accessed September 2006]).
Our second observation was that hypha-specific promoters contain putative binding sites for many known transcription factors in C. albicans. These include putative sites for Efg1, Tec1, Nrg1, Rfg1, Cph1, Cph2, Rim101, Cap1, and Gcn4. (Cap1 and Gcn4 are transcription factors that play key roles in responses to oxidative stress and amino acid starvation, respectively [1, 74].) However, only a small number of these sites are conserved in all the hypha-specific genes analyzed (Fig. 2). These include putative Efg1, Tec1, and Nrg1 sites. It should be noted, however, that the Efg1 consensus site (E box) is likely to occur by chance in sequences of this length.
Contribution of morphogenetic transcriptional factors to ALS3 regulation.
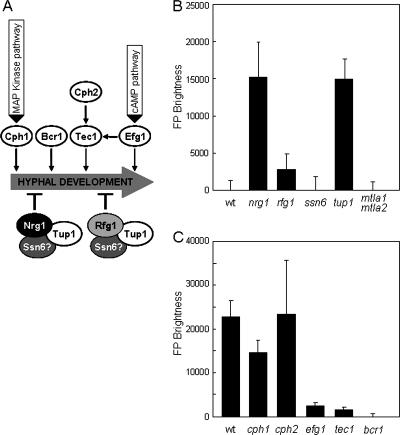
The ALS3 promoter region contains Nrg1 sites but no obvious Rfg1 sites on the basis of the Rfg1 consensus sequence (Fig. 2). However, genome-wide transcriptional profiling studies have suggested that ALS3 is transcriptionally repressed by Rfg1 as well as by Nrg1 and Tup1 (32, 46). Therefore, we compared the influence of these transcription factors upon ALS3 directly by using the in situ ALS3-GFP reporter (Fig. 3). In S. cerevisiae, Tup1 acts in concert with Ssn6, forming a Tup1-Ssn6 corepressor complex that represses the expression of many target genes (64). However, in C. albicans, Ssn6 is not thought to play a role in the Tup1-mediated repression of hypha-specific genes largely on the basis of transcript profiling (18). Therefore, we tested this further by examining the role of Ssn6 in ALS3 gene regulation. The ALS3 promoter also contains a putative site for the a1/α2 repressor, which involved in the repression of “haploid-specific” genes in C. albicans (75). Therefore, we included the a1/α2 repressor in this analysis.
FIG. 3.
Contribution of transcriptional regulators to the regulation of ALS3. (A) Cartoon illustrating the putative impact of transcriptional activators and repressors upon hyphal development. MAP, mitogen-activated protein; cAMP, cyclic AMP. (B) Effect of repressor mutations upon the expression of the ALS3-GFP reporter after growth for 2 h in YPD at 25°C. (C) Effect of inactivating transcriptional activators on the ALS3-GFP reporter after 90 min of growth in YPD containing serum at 37°C. wt, wild type.
The ALS3-GFP-URA3 cassette was transformed into wild-type, nrg1, rfg1, tup1, ssn6, and mtla1 mtla2 cells. GFP fluorescence levels were measured in these C. albicans strains during growth in the yeast form (Fig. 3B). As expected, the ALS3-GFP reporter was repressed in wild-type yeast cells. Nrg1 acts through two NREs in the ALS3 promoter at positions −330 and −80 (46). Hence, the derepression of the ALS3-GFP reporter in nrg1 and tup1 cells was also expected (Fig. 3B). However, this reporter was only partially derepressed in rfg1 cells and was not derepressed in ssn6 or mtla1 mtla2 cells. These data reinforce the idea that ALS3 is repressed mainly by Nrg1 and Tup1 in an Ssn6-independent fashion and that Rfg1 plays a minor role in the regulation of ALS3 (18, 32). The data also suggest that although the ALS3 promoter contains a putative a1/α2 site, this repressor is not required for ALS3 regulation under these conditions.
The ALS3 promoter also contains putative sites for several transcription factors that are known to contribute to the activation of hyphal development: Efg1, Cph1, Cph2, and Tec1. Therefore, we examined the contributions of these factors to the activation of ALS3 expression during hyphal development (Fig. 3). The activity of the in situ ALS3-GFP reporter was compared in wild-type, efg1, cph1, cph2, and tec1 cells following serum induction (Fig. 3C). Both Efg1 and Tec1 were required for the full activation of the ALS3-GFP reporter. In contrast, Cph2 was not essential for activation, although Cph2 has been reported to regulate TEC1 (36). We did observe considerable variation in ALS3-GFP expression levels in the cph2 mutant, and this is reflected in relatively large error bars even though this experiment was performed five times with up to eight independent transformants (Fig. 3C). Decreased ALS3-GFP expression was observed in cph1 cells, suggesting that this mitogen-activated protein kinase pathway does contribute to ALS3 activation following serum stimulation, although this pathway is not required for hyphal development under these conditions (10, 42). We also examined the impact of Bcr1 upon ALS3-GFP; this is discussed below. Taken together, the data indicate that the transcription factors Efg1, Tec1, Nrg1, and Tup1 play important roles in regulating ALS3 expression and that Rfg1 and Cph1 contribute to ALS3 regulation.
The ALS3 promoter contains two main activation regions.
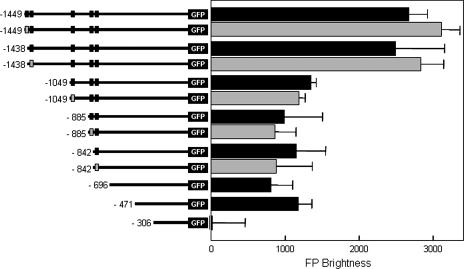
A set of mutations was generated at the ALS3 locus to examine the organization of its promoter. These mutations were generated by inserting a HIS1 cassette at a range of positions in the 5′ intergenic region of the ALS3-GFP allele in C. albicans strain SAC500 (Table 1). Essentially, this created a set of promoter mutations in situ at the ALS3 locus, the activities of which were monitored during hyphal development by measuring GFP fluorescence following serum stimulation.
The removal of sequences between positions −1438 and −1049 (with respect to the first base of the coding region) from the promoter caused a twofold decrease in the activity of the ALS3-GFP allele (Fig. 4). The further removal of sequences between positions −1049 and −471 had no significant effect upon expression. However, the removal of sequences between positions −471 and −306 blocked ALS3-GFP activation completely. We conclude that the full activation of ALS3 depends upon two promoter regions. One region (A1 [positions −471 to −306]) is essential for activation, while a second region (A2 [positions −1438 to −1049]) enhances this activation.
FIG. 4.
Effect of in situ promoter mutations upon ALS3-GFP expression. GFP fluorescence was quantified in each C. albicans SAC strain (Table 1) after 90 min of growth in YPD containing serum at 37°C. The coordinate of each promoter deletion endpoint is provided. Wild-type Tec1 sites are indicated by black boxes, and mutated Tec1 sites are indicated by gray boxes.
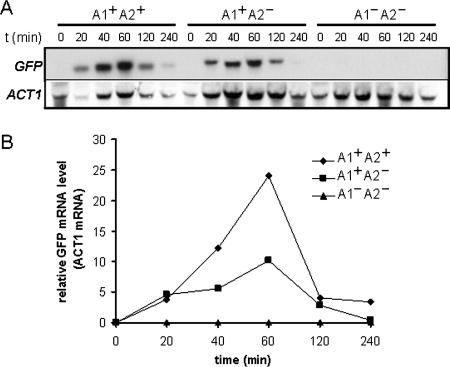
We examined the kinetics of induction of ALS3-GFP transcripts to further investigate the contributions of the A1 and A2 activation regions to the hypha-specific induction of ALS3. Northern analysis was performed on C. albicans SAC501, SAC505, and SAC513 cells following serum stimulation. SAC501 contains both the A1 and A2 activation regions (ALS31499-GFP), and SAC505 lacks A2 but contains A1 (ALS31049-GFP), whereas SAC513 lacks both A1 and A2 (ALS3306-GFP). All three strains developed hyphae at similar rates following serum stimulation, as expected. However, no induction of GFP mRNA was observed for the negative control containing the ALS3306-GFP fusion (Fig. 5). In contrast, GFP mRNA was strongly induced from the positive control containing both activation regions, reaching a maximum at 60 min. Similar kinetics of GFP mRNA induction were observed for the ALS3-GFP construct that contains only the A1 region. However, the GFP mRNA levels reached only about one-third of those in the positive control (Fig. 5), which correlates well with GFP fluorescence levels from these and related constructs (Fig. 4). This reproducible observation was consistent with the idea that the A1 region is essential for transcriptional activation during hyphal development, while the A2 region increases the amplitude of this activation.
FIG. 5.
Kinetic analysis of ALS3-GFP transcript levels during serum-induced hyphal development. (A) Northern analysis of ALS3-GFP transcripts at various times (minutes) after the serum induction of C. albicans strains carrying different promoter deletions (Table 1). A1+ A2+, SAC501 cells in which the ALS3-GFP fusion contains both activation regions; A1+ A2−, SAC505 cells in which the ALS3-GFP fusion contains only the A1 activation region; A1− A2−, SAC513 cells in which the ALS3-GFP fusion lacks both activation regions. PCR-amplified ALS3 and ACT1 probes were used (see Materials and Methods). (B) Quantification of ALS3-GFP transcript levels relative to the internal ACT1 mRNA control. Similar results were obtained when quantifying relative to 26S rRNA. Also, similar results were obtained in a second independent experiment.
The ALS3 promoter is complex.
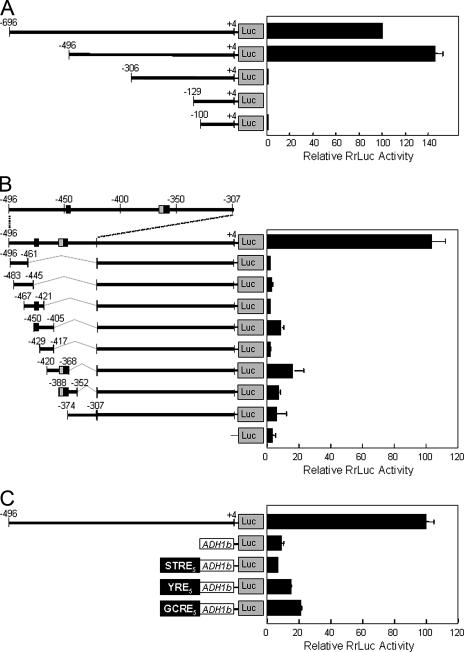
To examine the essential activation region (A1) in more detail, we turned to the sensitive RrLUC reporter (66). First, we tested the robustness of this approach for the dissection of the ALS3 promoter. A set of ALS3-RrLUC promoter fusions containing or lacking the A1 region were integrated into the genome of C. albicans CAI8, and their expression was examined in yeast and hyphal cells. As expected, all of these constructs were inactive in yeast cells (not shown), and only those containing the A1 region (positions −496 to −306) were induced in hyphal cells (Fig. 6A). This indicated that the ALS3-RrLUC fusions accurately reflected the behavior of in situ ALS3-GFP fusions and confirmed the presence of an essential activation region in this part of the promoter.
FIG. 6.
Analysis of the A1 activation region in the ALS3 promoter. Various RrLUC promoter-RrLUC fusions were constructed and transformed into C. albicans CAI8 (Table 1). The expression levels of these luciferase fusions were assayed after 3 h of growth in YPD containing serum at 37°C. (A) The expression of ALS3 promoter deletions that target the A1 activation region was assayed. (B) Fragments of the A1 activation region were cloned upstream of the ALS3306-RrLUC fusion, and the expression of these constructs was assayed. Black boxes, putative YRE; gray boxes, putative GCRE. (C) Oligonucleotides containing multiple STREs, YREs, or GCREs were cloned upstream of a basal RrLUC reporter, and the luciferase levels generated by these constructs were assayed.
Additional ALS3-RrLUC constructs were generated to further define the 5′ and 3′ ends of the A1 region. Hypha-specific activation was lost if 5′ sequences between positions −471 and −448 were deleted (not shown). Activation was retained if 3′ sequences between positions −321 and −307 were removed, but further 3′ deletions to position −331 resulted in reduced levels of expression in hyphal cells and the derepression of RrLUC expression in yeast cells. This was consistent with the disruption of activating sequences and the loss of Nrg1-mediated repression through the deletion of the NRE at position −330. We concluded that the A1 activation region lies between positions −471 and −321. This activation region does not correlate well with an in silico analysis of putative regulatory elements in the ALS3 promoter (Fig. 2), reinforcing the view that in isolation, in silico analyses of promoter elements are a poor predictor of regulatory function.
In an attempt to define the A1 region more precisely, we generated a further set of RrLUC constructs containing short overlapping fragments from the A1 region. None of these constructs displayed expression levels equivalent to those of the control (Fig. 6B), indicating that no single enhancer element within the A1 region was sufficient to confer hypha-specific activation. Weak activation (<20% of the control) was observed for some fragments. This might have suggested that multiple copies of a weak element could combine to provide strong activation. However, none of these fragments shared any obvious sequence elements.
Putative binding sites for the transcription factors Msn4/Msn2 (STRE [C4T]), Cap1 (YRE [TTA[G/C]TAA]), and Gcn4 (GCRE [TGACTC]) do exist in the promoters of hypha-specific genes, and these elements are present in ALS3 promoter fragments that provide weak transcriptional activation. Therefore, we tested whether STRE, YRE, or GCRE elements can activate transcription in response to serum induction (Fig. 6C). The YRE- and GCRE-RrLUC reporters displayed weak activation compared with the ALS3-RrLUC control, suggesting that these elements might contribute to the weak activation seen for the short ALS3 promoter fusions examined in Fig. 6B. However, the YRE element mediates transcriptional activation in C. albicans yeast cells in response to oxidative stress (51), and the GCRE activates transcription in yeast cells in response to amino acid starvation (74). Neither Cap1 nor Gcn4 is required for serum-induced morphogenesis. Hence, these elements cannot account for the hypha specificity of the A1 promoter region. Nevertheless, it is conceivable that YRE and GCRE elements might contribute to the transcriptional activation of hypha-specific genes in the context of the natural promoters.
Taken together, the data suggest that the A1 promoter region is complex. Sequence elements close to the 5′ and 3′ ends of this region are required for the transcriptional activation of ALS3 during hyphal development. These elements appear to function in combination to mediate hypha-specific activation.
Tec1 acts indirectly through Bcr1 to regulate ALS3 transcription.
Putative Tec1 sites exist in all hypha-specific promoter regions (Fig. 2). Five such sites are present in the ALS3 promoter at positions −1499, −1438, −1049, −885, and −842. Furthermore, Tec1 is required for the morphogenetic activation of ALS3 (Fig. 3C). Therefore, we reasoned that Tec1 might act directly upon the ALS3 promoter via (some of) the putative Tec1 sites. To test this, we generated a set of in situ ALS3 promoter mutants in which the Tec1 sites were sequentially inactivated and compared them to a parallel set of control mutations containing the Tec1 sites (Fig. 4 and Table 1). No significant difference in expression level was observed between each Tec1 site mutation (Fig. 4, gray bars) and its corresponding control (black bars). This indicated that the putative Tec1 sites are not required for the hypha-specific activation of ALS3 and hence that Tec1 might act indirectly upon this gene.
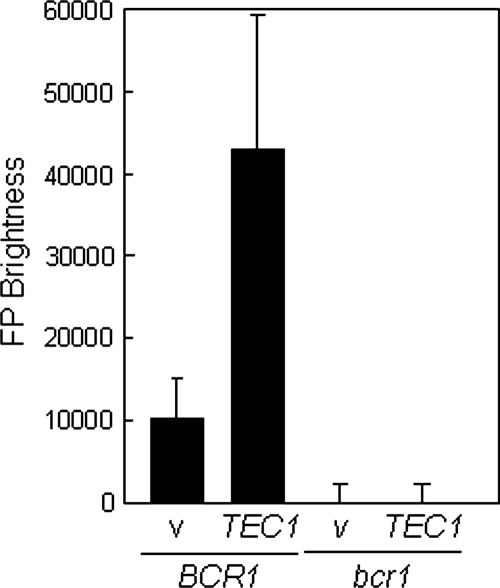
Recently, Nobile and Mitchell (52) identified BCR1 as being a regulator of biofilm formation in C. albicans. During the course of that work, they showed that ALS3 mRNA levels are reduced in bcr1 cells and that BCR1 expression is reduced in a tec1 mutant. This raised the possibility that Tec1 might regulate ALS3 indirectly via Bcr1. We tested this idea by first asking whether BCR1 is required for the transcriptional activation of the ALS3-GFP reporter. ALS3-GFP expression was lost in bcr1 cells, indicating that Bcr1 is essential for the transcriptional activation of ALS3 during hyphal development (Fig. 3C). We then tested whether TEC1 overexpression enhances ALS3 expression and whether this effect is dependent upon BCR1. TEC1 overexpression was engineered by transforming a PYK1-TEC1 fusion into C. albicans SAC520 cells and growing them on glucose-containing medium to activate the PYK1 promoter (4). This led to the significant overexpression of ALS3-GFP (Fig. 7). This overexpression was blocked in a bcr1 mutant background, confirming that Tec1 acts indirectly upon ALS3 transcription via Bcr1.
FIG. 7.
Effect of ectopic TEC1 expression and BCR1 inactivation on ALS3-GFP expression. The in situ ALS3-GFP reporter was introduced into wild-type (BCR1) and bcr1 cells, and these strains were transformed with the empty PYK1 expression vector (v) or the PYK1-TEC1 plasmid (TEC1) to generate strains SAC522 (v) (BCR1), SAC523 (TEC1) (BCR1), SAC524 (v) (bcr1), and SAC525 (TEC1) (bcr1) (Table 1). GFP fluorescence levels were assayed in these strains after 90 min of growth in YPD-containing serum at 37°C.
DISCUSSION
Yeast-hypha morphogenesis has been studied intensively in C. albicans because of its likely contribution to the pathogenicity of this fungus (21, 22, 29, 61, 71, 83). A complex network of signaling pathways has been shown to control hyphal development, but the mechanistic relationships between these pathways remain obscure (10). These signaling pathways are thought to converge on the promoters of those genes that respond specifically during hyphal development (7, 10). ALS3 is one of a small set of hypha-specific genes in C. albicans that includes ALS3, ECE1, HGC1, HWP1, HYR1, RBT1, and RBT4 (2, 5, 8, 28, 40, 48, 67, 83). In this study, we have confirmed that the hypha-specific activation of ALS3 is mediated at the transcriptional level (Fig. 1). Clearly, a complete understanding of morphogenetic signaling depends upon the dissection of hypha-specific promoters and the mechanisms by which these pathways regulate these promoters.
We have shown that ALS3 is regulated by a complex array of transcription factors: Efg1, Cph1, Tec1, Bcr1, Nrg1, Rfg1, and Tup1 (Fig. 3). When C. albicans cells grow in the yeast form, ALS3 transcription is repressed mainly by Nrg1, which binds to NREs located at positions −330 and −80 in the promoter (46). Rfg1 also contributes to ALS3 repression (Fig. 3) (32), but the promoter element(s) through which Rfg1 operates in C. albicans has not been experimentally defined. Both Nrg1 and Rfg1 are thought to act by interacting with the global repressor Tup1, which mediates transcription through direct interactions with the transcription complex, by positioning nucleosomes on the promoter, or by a combination of both mechanisms (23, 80). In S. cerevisiae, interactions between Tup1 and its cognate DNA binding proteins often depend on Ssn6 (64). However, this does not appear to be the case for Nrg1 in the context of hypha-specific genes. It has been suggested that the repression of hypha-specific genes by Nrg1 and Tup1 does not depend upon Ssn6 (18), and we have confirmed this for ALS3 in this study (Fig. 3).
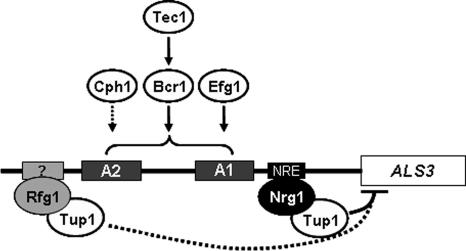
Cph1 and the A2 region of the promoter are required only for full ALS3 activation. This might suggest that Cph1 enhances ALS3 transcription via the A2 region. However, there are no obvious occurrences of the putative Cph1 consensus site in the ALS3 promoter, and therefore, Cph1 might act indirectly to regulate ALS3 transcription (Fig. 8).
FIG. 8.
Working model illustrating the effects of morphogenetic regulators on the transcriptional regulation of ALS3. As described in the text, the ALS3 promoter has two activation regions (A1 and A2), with A1 being essential for hypha-specific activation (Fig. 4, 5, and 6). ALS3 activation is dependent upon Efg1, Bcr1, and Tec1 (Fig. 3), with the latter acting through Bcr1 (Fig. 7) (53). These factors might act through the A1 region, but no direct interaction with this region has been demonstrated. Like the A2 promoter region, Cph1 contributes to ALS3 activation but is not essential for this activation (Fig. 3 and 5). It is not known whether Cph1 acts directly or indirectly upon the ALS3 promoter (dotted line). Nrg1 represses transcription in a Tup1-dependent fashion (9) by binding to NREs in the ALS3 promoter (46). Rfg1 contributes to this repression, but the ALS3 promoter contains no obvious Rfg1 sites (Fig. 2), and it is not known whether Rfg1 acts directly upon the ALS3 promoter (dotted line).
In contrast, Efg1 is essential for the transcriptional activation of ALS3 during hyphal development (Fig. 3C). Efg1 has been shown to bind an E box in vitro (40), and hypha-specific promoters do contain this type of sequence element (Fig. 2). However, the degenerate E-box consensus is likely to occur frequently by chance (1/256), and to date, there are no reports confirming that Efg1 regulates transcription via the E box in C. albicans.
Although Tec1 is essential for the activation of ALS3 (Fig. 4) and putative Tec1 sites exist in the ALS3 promoter (Fig. 2), these sites do not contribute significantly to ALS3 activation (Fig. 4). Instead, Tec1 regulates ALS3 transcription indirectly through Bcr1 (Fig. 7), which is also essential for ALS3 activation (Fig. 3). These observations are entirely consistent with recent data from Nobile et al. They showed that Tec1 and Bcr1 are required for the formation of biofilms in C. albicans and that Bcr1 acts downstream of Tec1 to regulate the expression of adhesin genes required for biofilm formation, such as ALS3 and HWP1 (52, 53).
The transcriptional activation of ALS3 is dependent upon the A1 promoter region (Fig. 4) as well as upon Efg1, Tec1, and Bcr1 (Fig. 3). The A1 promoter region is complex: no single sequence element within this 150-bp region was capable of driving hypha-specific expression, and the trimming of sequences at either the 5′ or 3′ end of this A1 region blocked hypha-specific activation (Fig. 6). This is consistent with the idea that several different regulatory factors converge upon the A1 region to cooperate in ALS3 activation. Hence, Tec1-Bcr1 and Efg1 might regulate ALS3 cooperatively via the A1 promoter region (Fig. 8). An NRE lies at the 3′ border of the A1 region at position −330. It has been reported that Nrg1 might act as a transcriptional activator under some circumstances (47, 59). Hence, it is conceivable that Nrg1 might also contribute to the hyphal activation of ALS3.
In parallel studies, Kim and coworkers (33) made similar observations about the regulation of a second hypha-specific gene, HWP1. The HWP1 promoter also contains two activation regions. One region, which binds an array of chromatin remodeling proteins, is essential for HWP1 activation, whereas the second distal region increases the amplitude of this activation (33). Hence, this class of developmentally regulated genes appears to be controlled by complex interactions between several critical transcription factors at the level of their promoters. It has long been recognized that C. albicans responds to an extremely disparate range of environmental conditions by forming hyphae (54). The unusual length of promoters of hypha-specific genes and the complexity and diversity of factors regulating their transcription not only are compatible with the diversity of conditions known to favor hypha formation but also suggest that morphogenetic changes in C. albicans may be affected by events in several regulatory pathways whose stimulation may not always be specifically or directly related to cell shape.
Supplementary Material
Acknowledgments
We thank Susan Budge for excellent technical assistance. We also thank Paula Sundstrom for making data available to us prior to publication.
This work was supported by funding from the Wellcome Trust (063204 and 068143), the BBSRC (1/CEL 4563 and 1/P17124), and the EC (QLK2CT-2000-00795 and MRTN-CT-2003-504148).
Footnotes
Published ahead of print on 2 February 2007.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Alarco, A. M., and M. Raymond. 1999. The bZip transcription factor Cap1p is involved in multidrug resistance and oxidative stress response in Candida albicans. J. Bacteriol. 181:700-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey, D. A., P. J. F. Feldmann, M. Bovey, N. A. R. Gow, and A. J. P. Brown. 1996. The Candida albicans HYR1 gene, which is activated in response to hyphal development, belongs to a gene family encoding yeast cell wall proteins. J. Bacteriol. 178:5353-5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barelle, C. J., C. L. Manson, D. M. MacCallum, F. C. Odds, N. A. R. Gow, and A. J. P. Brown. 2004. GFP as a quantitative reporter of gene regulation in Candida albicans. Yeast 21:333-340. [DOI] [PubMed] [Google Scholar]
- 4.Barelle, C. J., C. L. Priest, D. M. MacCallum, N. A. R. Gow, F. C. Odds, and A. J. P. Brown. 2006. Niche-specific regulation of central metabolic pathways in a fungal pathogen. Cell. Microbiol. 8:961-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birse, C. E., M. Y. Irwin, W. A. Fonzi, and P. S. Sypherd. 1993. Cloning and characterization of ECE1, a gene expressed in association with cell elongation of the dimorphic pathogen Candida albicans. Infect. Immun. 61:3648-3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun, B. R., and A. D. Johnson. 1997. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science 277:105-109. [DOI] [PubMed] [Google Scholar]
- 7.Braun, B. R., and A. D. Johnson. 2000. TUP1, CPH1 and EFG1 make independent contributions to filamentation in Candida albicans. Genetics 155:57-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braun, B. R., W. S. Head, M. X. Wang, and A. D. Johnson. 2000. Identification and characterization of TUP1-regulated genes in Candida albicans. Genetics 156:31-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braun, B. R., D. Kadosh, and A. D. Johnson. 2001. NRG1, a repressor of filamentous growth in Candida albicans, is down-regulated during filament induction. EMBO. J. 20:4753-4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown, A. J. P. 2002. Morphogenetic signalling pathways in Candida albicans, p 95-106. In R. Calderone (ed.), Candida and candidiasis. ASM Press, Washington, DC.
- 11.Brown, D. H., A. D. Giusani, X. Chen, and C. A. Kumamoto. 1999. Filamentous growth of Candida albicans in response to physical environmental cues and its regulation by the unique CZF1 gene. Mol. Microbiol. 34:651-662. [DOI] [PubMed] [Google Scholar]
- 12.Cartharius, K., K. Frech, K. Grote, B. Klocke, M. Haltmeier, A. Klingenhoff, M. Frisch, M. Bayerlein, and T. Werner. 2005. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21:2933-2942. [DOI] [PubMed] [Google Scholar]
- 13.Cheng, G., K. Wozniak, M. A. Wallig, P. L. Fidel, S. R. Trupin, and L. L. Hoyer. 2005. Comparison between Candida albicans agglutinin-like sequence gene expression patterns in human clinical specimens and models of vaginal candidiasis. Infect. Immun. 73:1656-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cormack, B., G. Bertram, M. Egerton, N. A. R. Gow, S. Falkow, and A. J. P. Brown. 1997. Yeast-enhanced green fluorescent protein (yEGFP): a reporter of gene expression in Candida albicans. Microbiology 143:303-311. [DOI] [PubMed] [Google Scholar]
- 15.Dennison, P. M. J., M. Ramsdale, C. L. Manson, and A. J. P. Brown. 2005. Gene disruption in Candida albicans using a synthetic codon-optimised Cre-loxP system. Fungal Genet. Biol. 42:737-748. [DOI] [PubMed] [Google Scholar]
- 16.Doedt, T., S. Krishnamurthy, D. P. Bockmühl, B. Tebarth, C. Stempel, C. L. Russell, A. J. P. Brown, and J. F. Ernst. 2004. APSES proteins regulate morphogenesis and metabolism in Candida albicans. Mol. Biol. Cell 15:3167-3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.García-Sánchez, S., A. Mavor, C. L. Russell, S. Argimón, P. Dennison, B. Enjalbert, and A. J. P. Brown. 2005. Global roles of Ssn6 in Tup1- and Nrg1-dependent gene regulation in the fungal pathogen, Candida albicans. Mol. Biol. Cell 16:2913-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerami-Nejad, M., J. Berman, and C. A. Gale. 2001. Cassettes for PCR-mediated construction of green, yellow, and cyan fluorescent protein fusions in Candida albicans. Yeast 18:859-864. [DOI] [PubMed] [Google Scholar]
- 20.Gillum, A. M., E. Y. Tsay, and D. R. Kirsch. 1984. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 198:179-182. [DOI] [PubMed] [Google Scholar]
- 21.Gow, N. A. R., A. J. P. Brown, and F. C. Odds. 2002. Fungal morphogenesis and host invasion. Curr. Opin. Microbiol. 5:366-371. [DOI] [PubMed] [Google Scholar]
- 22.Gow, N. A. R., Y. Knox, C. A. Munro, and W. D. Thompson. 2003. Infection of chick chorioallantoic membrane (CAM) as a model for invasive hyphal growth and pathogenesis of Candida albicans. Med. Mycol. 41:331-338. [DOI] [PubMed] [Google Scholar]
- 23.Green, S. R., and A. D. Johnson. 2004. Promoter-dependent roles for the Srb10 cyclin-dependent kinase and the Hda1 deacetylase in Tup1-mediated repression in Saccharomyces cerevisiae. Mol. Biol. Cell 15:4191-4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hauser, N. C., M. Vingron, M. Scheideler, B. Krems, K. Hellmuth, K.-D. Entian, and J. D. Hoheisel. 1998. Transcriptional profiling on all open reading frames of Saccharomyces cerevisiae. Yeast 14:1209-1221. [DOI] [PubMed] [Google Scholar]
- 25.Hoffman, C. S., and F. Winston. 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of E. coli. Gene 57:267-272. [DOI] [PubMed] [Google Scholar]
- 26.Holton, N. J., T. J. D. Goodwin, M. I. Butler, and R. T. M. Poulter. 2001. An active retrotrasposon in Candida albicans. Nucleic Acids Res. 29:635-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoyer, L. L. 2001. The ALS gene family of Candida albicans. Trends Microbiol. 9:176-180. [DOI] [PubMed] [Google Scholar]
- 28.Hoyer, L. L., T. L. Payne, M. Bell, A. M. Myers, and S. Scherer. 1998. Candida albicans ALS3 and insights into the nature of the ALS gene family. Curr. Genet. 33:451-459. [DOI] [PubMed] [Google Scholar]
- 29.Hube, B. 2004. From commensal to pathogen: stage- and tissue-specific gene expression of Candida albicans. Curr. Opin. Microbiol. 7:336-341. [DOI] [PubMed] [Google Scholar]
- 30.Hube, B., and J. Naglik. 2002. Extracellular hydrolases, p. 107-122. In R. Calderone (ed.), Candida and candidiasis. ASM Press, Washington, DC.
- 31.Kadosh, D., and A. D. Johnson. 2001. Rfg1, a protein related to the S. cerevisiae hypoxic regulator Rox1, controls filamentous growth and virulence in C. albicans. Mol. Cell. Biol. 21:2496-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kadosh, D., and A. D. Johnson. 2005. Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: a genome-wide analysis. Mol. Biol. Cell 16:2903-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim, S., M. J. Wolyniak, J. F. Staab, and P. Sundstrom. 2007. A 368-base-pair cis-acting HWP1 promoter region, HCR, of Candida albicans confers hypha-specific gene regulation and binds architectural transcription factors Nhp6 and Gcf1p. Eukaryot. Cell 6:693-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knight, A. W., N. J. Goddard, P. R. Fielden, M. G. Barker, N. Billinton, and R. M. Walmsley. 1999. Fluorescence polarisation of green fluorescent protein (GFP). A strategy for improved wavelength discrimination for GFP determinations. Analyt. Commun. 36:113-117. [Google Scholar]
- 35.Knight, A. W., N. J. Goddard, N. Billinton, P. A. Cahill, and R. M. Walmsley. 2002. Fluorescence polarization discriminates green fluorescent protein from interfering autofluorescence in a microplate assay for genotoxicity. J. Biochem. Biophys. Methods 51:165-177. [DOI] [PubMed] [Google Scholar]
- 36.Lane, S., C. Birse, S. Zhou, R. Matson, and H. P. Liu. 2001. DNA array studies demonstrate convergent regulation of virulence factors by Cph1, Cph2, and Efg1 in Candida albicans. J. Biol. Chem. 276:48988-48996. [DOI] [PubMed] [Google Scholar]
- 37.Lane, S., S. Zhou, T. Pan, Q. Dai, and H. P. Liu. 2001. The basic helix-loop-helix transcription factor Cph2 regulates hyphal development in Candida albicans partly via Tec1. Mol. Cell. Biol. 21:6418-6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leberer, E., D. Harcus, I. D. Broadbent, K. L. Clark, D. Dignard, K. Ziegelbauer, A. Schmidt, N. A. R. Gow, A. J. P. Brown, and D. Y. Thomas. 1996. Homologs of the Ste20p and Ste7p protein kinases are involved in hyphal formation of Candida albicans. Proc. Natl. Acad. Sci. USA 93:13217-13222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leng, P., P. E. Sudbery, and A. J. P. Brown. 2000. Rad6p represses yeast-hypha morphogenesis in the human fungal pathogen, Candida albicans. Mol. Microbiol. 35:1264-1275. [DOI] [PubMed] [Google Scholar]
- 40.Leng, P., P. Lee, H. Wu, and A. J. P. Brown. 2001. Efg1, a morphogenetic regulator in Candida albicans, is a sequence-specific DNA binding protein. J. Bacteriol. 183:4090-4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu, H., J. R. Kohler, and G. R. Fink. 1994. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 266:1723-1726. [DOI] [PubMed] [Google Scholar]
- 42.Lo, H. J., J. R. Kohler, B. DiDomenico, D. Loebenberg, A. Cacciapuoti, and G. R. Fink. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90:939-949. [DOI] [PubMed] [Google Scholar]
- 43.Miller, M. G., and A. D. Johnson. 2002. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell 110:293-302. [DOI] [PubMed] [Google Scholar]
- 44.Mitchell, A. P. 1998. Dimorphism and virulence in Candida albicans. Curr. Opin. Microbiol. 1:687-692. [DOI] [PubMed] [Google Scholar]
- 45.Murad, A. M. A., P. R. Lee, I. D. Broadbent, C. J. Barelle, and A. J. P. Brown. 2000. CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast 16:325-327. [DOI] [PubMed] [Google Scholar]
- 46.Murad, A. M. A., P. Leng, M. Straffon, J. Wishart, S. Macaskill, D. MacCallum, N. Schnell, D. Talibi, D. Marechal, F. Tekaia, C. d'Enfert, C. Gaillardin, F. C. Odds, and A. J. P. Brown. 2001. NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 20:4742-4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murad, A. M. A., C. d'Enfert, C. Gaillardin, H. Tournu, F. Tekaia, D. Talibi, D. Marechal, V. Marchais, J. Cottin, and A. J. P. Brown. 2001. Transcript profiling in Candida albicans reveals new cellular functions for the transcriptional repressors, CaTup1, CaMig1 and CaNrg1. Mol. Microbiol. 42:981-993. [DOI] [PubMed] [Google Scholar]
- 48.Nantel, A., D. Dignard, C. Bachewich, D. Harcus, A. Marcil, A.-P. Bouin, C. W. Sensen, H. Hogues, M. van het Hoog, P. Gordon, T. Rigby, F. Benoit, D. C. Tessier, D. Y. Thomas, and M. Whiteway. 2002. Transcript profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol. Biol. Cell 13:3452-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nasmyth, K. 1985. At least 1400 base pairs of 5′-flanking DNA is required for the correct expression of the HO gene in yeast. Cell 42:213-223. [DOI] [PubMed] [Google Scholar]
- 50.Negredo, A., L. Monteoliva, C. Gil, J. Pla, and C. Nombela. 1997. Cloning analysis and one-step disruption of the ARG5,6 gene of Candida albicans. Microbiology 143:297-302. [DOI] [PubMed] [Google Scholar]
- 51.Nicholls, S., M. Straffon, B. Enjalbert, A. Nantel, S. Macaskill, M. Whiteway, and A. J. P. Brown. 2004. Msn2/4-like transcription factors play no obvious roles in the stress responses of the fungal pathogen Candida albicans. Eukaryot. Cell 3:1111-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nobile, C. J., and A. P. Mitchell. 2005. Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor Bcr1p. Curr. Biol. 15:1150-1155. [DOI] [PubMed] [Google Scholar]
- 53.Nobile, C. J., D. R. Andes, J. E. Nett, F. J. Smith, F. Yue, Q.-T. Phan, J. E. Edwards, S. G. Filler, and A. P. Mitchell. 2006. Critical role of Bcr1-dependent adhesins in C. albicans biofilm formation in vitro and in vivo. PLoS Pathog. 2:636-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Odds, F. C. 1988. Candida and candidosis, 2nd ed. Bailliere Tindall, London, United Kingdom.
- 55.Oh, S. H., G. Cheng, J. A. Nuessen, R. Jajko, K. M. Yeater, X. Zhao, C. Pujol, D. R. Soll, and L. L. Hoyer. 2005. Functional specificity of Candida albicans Als3p proteins and clade specificity of ALS3 alleles discriminated by the number of copies of the tandem repeat sequence in the central domain. Microbiology 151:673-681. [DOI] [PubMed] [Google Scholar]
- 56.Quandt, K., K. Frech, H. Karas, E. Wingender, and T. Werner. 1995. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 23:4878-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ramon, A. M., A. Porta, and W. A. Fonzi. 1999. Effect of environmental pH on morphological development of Candida albicans is mediated via the PacC-regulated transcription factor encoded by PRR2. J. Bacteriol. 181:7524-7530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rupp, S., E. Summers, H.-J. Lo, H. Madhani, and G. R. Fink. 1999. MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene. EMBO. J. 18:1257-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Russell, C. L., and A. J. P. Brown. 2005. Expression of one-hybrid fusions with Staphylococcus aureus lexA in Candida albicans confirms that Nrg1 is a transcriptional repressor and that Gcn4 is a transcriptional activator. Fungal Genet. Biol. 42:676-683. [DOI] [PubMed] [Google Scholar]
- 60.Sathe, G. M., S. O'Brien, M. M. McLaughlin, F. Watson, and G. P. Livi. 1991. Use of polymerase chain reaction for rapid detection of gene insertions in whole yeast cells. Nucleic Acids Res. 19:4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saville, S. P., A. L. Lazzell, C. Monteagudo, and J. L. Lopez-Ribot. 2003. Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot. Cell 2:1053-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schweizer, A., S. Rupp, B. N. Taylor, M. Rollinghoff, and K. Schröppel. 2001. The TEA/ATTS transcription factor CaTec1p regulates hyphal development and virulence in Candida albicans. Mol. Microbiol. 38:435-445. [DOI] [PubMed] [Google Scholar]
- 63.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3-21. [DOI] [PubMed] [Google Scholar]
- 64.Smith, R. L., and A. D. Johnson. 2000. Turning off genes by Ssn6-Tup1: a conserved system of transcriptional repression in eukaryotes. Trends Biochem. Sci. 25:325-330. [DOI] [PubMed] [Google Scholar]
- 65.Soll, D. R. 2002. Phenotypic switching, p. 123-142. In R. Calderone (ed.), Candida and candidiasis. ASM Press, Washington, DC.
- 66.Srikantha, T., A. Klapach, W. W. Lorenz, L. K. Tsai, L. A. Laughlin, J. A. Gorman, and D. R. Soll. 1996. The sea pansy Renilla reniformis luciferase serves as a sensitive bioluminescent reporter for differential gene expression in Candida albicans. J. Bacteriol. 178:121-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Staab, J. F., C. A. Ferrer, and P. Sundstrom. 1996. Developmental expression of a tandemly repeated, proline and glutamine-rich amino acid motif on hyphal surfaces of Candida albicans. J. Biol. Chem. 271:6298-6305. [DOI] [PubMed] [Google Scholar]
- 68.Staab, J. F., S. D. Bradway, P. L. Fidel, and P. Sundstrom. 1999. Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science 283:1535-1538. [DOI] [PubMed] [Google Scholar]
- 69.Stoldt, V. R., A. Sonneborn, C. E. Leuker, and J. F. Ernst. 1997. Efg1p, an essential regulator of morphogenesis of the human fungal pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 16:1982-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sundstrom, P. 2002. Adhesion in Candida spp. Cell. Microbiol. 4:461-469. [DOI] [PubMed] [Google Scholar]
- 71.Sundstrom, P. 2006. Candida albicans hyphal formation and virulence, p. 45-47. In J. Heitman, S. G. Filler, J. E. Edwards, Jr., and A. P. Mitchell (ed.), Molecular principles of fungal pathogenesis. ASM Press, Washington, DC.
- 72.Sundstrom, P., E. Balish, and C. M. Allen. 2002. Essential role of the Candida albicans transglutaminase substrate, hyphal wall protein 1, in lethal oroesophageal candidiasis in immunodeficient mice. J. Infect. Dis. 185:521-530. [DOI] [PubMed] [Google Scholar]
- 73.Swoboda, R. K., G. Bertram, S. Delbruck, J. F. Ernst, N. A. R. Gow, G. W. Gooday, and A. J. P. Brown. 1994. Fluctuations in glycolytic mRNA levels during the yeast-to-hyphal transition in Candida albicans reflect underlying changes in growth rather than a response to cellular dimorphism. Mol. Microbiol. 13:663-672. [DOI] [PubMed] [Google Scholar]
- 74.Tripathi, G., C. Wiltshire, S. Macaskill, H. Tournu, S. Budge, and A. J. P. Brown. 2002. CaGcn4 co-ordinates morphogenetic and metabolic responses to amino acid starvation in Candida albicans. EMBO J. 21:5448-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tsong, A. E., M. G. Miller, R. M. Raisner, and A. D. Johnson. 2003. Evolution of a combinatorial transcriptional circuit: a case study in yeasts. Cell 115:389-399. [DOI] [PubMed] [Google Scholar]
- 76.van Helden, J., B. André, and J. Collado-Vides. 2000. A website for the computational analysis of yeast regulatory sequences. Yeast 16:177-187. [DOI] [PubMed] [Google Scholar]
- 77.Walther, A., and J. Wendland. 2003. An improved transformation protocol for the human fungal pathogen Candida albicans. Curr. Genet. 42:339-343. [DOI] [PubMed] [Google Scholar]
- 78.Wicksteed, B. L., I. Collins, A. Dershowitz, L. I. Stateva, R. P. Green, S. G. Oliver, A. J. P. Brown, and C. S. Newlon. 1994. A physical comparison of chromosome III in six strains of Saccharomyces cerevisiae. Yeast 10:39-57. [DOI] [PubMed] [Google Scholar]
- 79.Wilson, R. B., D. Davis, and A. P. Mitchell. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181:1868-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang, Z., and J. C. Reese. 2004. Redundant mechanisms are used by Ssn6-Tup1 in repressing chromosomal gene transcription in Saccharomyces cerevisiae. J. Biol. Chem. 279:39240-39250. [DOI] [PubMed] [Google Scholar]
- 81.Zhao, X., S.-H. Oh, G. Cheng, C. B. Green, J. A. Nuessen, K. Yeater, R. P. Leng, A. J. P. Brown, and L. L. Hoyer. 2004. ALS3 and ALS8 represent a single locus that encodes a Candida albicans adhesion; functional comparisons between Als3p and Als1p. Microbiology 150:2415-2428. [DOI] [PubMed] [Google Scholar]
- 82.Zhao, X., K. J. Daniels, S.-H. Oh, C. B. Green, K. M. Yeater, D. R. Soll, and L. L. Hoyer. 2006. Candida albicans Als3p is required for wild-type biofilm formation on silicone elastomer surfaces. Microbiology 152:2287-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zheng, X., Y. Wang, and Y. Wang. 2004. Hgc1, a novel hypha-specific G1 cyclin-related protein regulates Candida albicans hyphal morphogenesis. EMBO J. 23:1845-1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.