Abstract
The car gene cluster of the ascomycete Fusarium fujikuroi encodes two enzymes responsible for torulene biosynthesis (CarRA and CarB), an opsin-like protein (CarO), and a putative carotenoid cleaving enzyme (CarX). It was presumed that CarX catalyzes the formation of the major carotenoid in F. fujikuroi, neurosporaxanthin, a cleavage product of torulene. However, targeted deletion of carX did not impede neurosporaxanthin biosynthesis. On the contrary, ΔcarX mutants showed a significant increase in the total carotenoid content, indicating an involvement of CarX in the regulation of the pathway. In this work, we investigated the enzymatic activity of CarX. The expression of the enzyme in β-carotene-accumulating Escherichia coli cells led to the formation of the opsin chromophore retinal. The identity of the product was proven by high-performance liquid chromatography and gas chromatography-mass spectrometry. Subsequent in vitro assays with heterologously expressed and purified CarX confirmed its β-carotene-cleaving activity and revealed its capability to produce retinal also from other substrates, such as γ-carotene, torulene, and β-apo-8′-carotenal. Our data indicate that the occurrence of at least one β-ionone ring in the substrate is required for the cleavage reaction and that the cleavage site is determined by the distance to the β-ionone ring. CarX represents the first retinal-synthesizing enzyme reported in the fungal kingdom so far. It seems likely that the formed retinal is involved in the regulation of the carotenoid biosynthetic pathway via a negative feedback mechanism.
Carotenoids are widespread lipophilic pigments synthesized by all photosynthetic organisms and some nonphotosynthetic fungi and bacteria. These yellow, orange, and red isoprenoid compounds fulfill diverse functions in all taxa. In addition, carotenoids are the precursors of several physiologically essential compounds, like the ubiquitous chromophore retinal, the phytohormone abscisic acid (ABA), and the fungal sex hormone trisporic acid.
The synthesis of these carotenoid cleavage products, known as apocarotenoids, is catalyzed in general by carotenoid oxygenases, which constitute a new nonheme iron enzyme family common in all taxa (for reviews, see references 1, 11, 15, and 24). Recently, the crystal structure of a member of this family, the Synechocystis apocarotenoid oxygenase, was elucidated at 2.4-Å resolution. The enzyme contains an Fe2+-4-His arrangement at the axis of a seven-bladed β-propeller chain fold covered by a dome formed by six large loops (20). It has also been shown that the 15,15′ BCO I (β-carotene oxygenase I) acts as a monooxygenase (21). However, recent investigations of AtCCD1 (Arabidopsis thaliana carotenoid cleavage dioxygenase 1) suggested a dioxygenase mechanism (31).
VP14 (Viviparous 14), a maize enzyme mediating the oxidative cleavage of 9-cis-violaxanthin and 9′-cis-neoxanthin to form the precursor of ABA, xanthoxin, was the first molecularly identified carotenoid oxygenase (34). Data from sequenced genomes have then revealed the occurrence of VP14 homologs in all taxa, as well as orthologs in several plant species. For instance, the genome of A. thaliana encodes nine members of this oxygenase family (39). The A. thaliana carotenoid oxygenases are involved in ABA biosynthesis (18), the formation of a still unidentified mediator of the apical dominance (10, 32), the synthesis of volatile compounds, e.g., β-ionone (33), and in plastid development (25). Thus, the diversity of biological functions is mirrored in the variety of the substrates, the cleavage sites, and the apocarotenoids formed. In addition to the A. thaliana enzymes, the identification of VP14 has also led to the cloning of further carotenoid oxygenases involved in the formation of apocarotenoid pigments, such as bixin in Bixa orellana (12) and saffron in Crocus sativus (13), and the synthesis of volatile compounds from several plant species (36, 37).
In animals, retinal and its derivatives represent the best known apocarotenoids. This C20 compound is synthesized through central cleavage of β-carotene, a reaction mediated by the enzyme β-carotene oxygenase I (BCO I). The corresponding cDNAs were cloned from several species, i.e., Drosophila melanogaster (42), chickens (44), and mammals (27, 46), allowing the characterization of this enzyme. In addition to BCO I, mammals contain two different members of the carotenoid oxygenase family. The first one, BCO II, mediates the formation of the volatile compound β-ionone from β-carotene through asymmetrical cleavage at the position 9′,10′ (19), while the second member, RPE65 (retinal pigment epithelium 65), acts as a chaperone for retinol and retinylester rather than as a cleavage enzyme (45).
Carotenoid-producing microorganisms have been extensively used as model systems in the investigation of the biochemistry and genetics of this biosynthetic pathway (reviewed in reference 35). Several examples are represented by filamentous fungi, like the zygomycetes Phycomyces blakesleeanus and Mucor circinelloides for β-carotene formation, or the ascomycetes Neurospora crassa and Fusarium fujikuroi for neurosporaxanthin biosynthesis (reviewed in references 4 and 35). The synthesis of neurosporaxanthin, a C35 acidic apocarotenoid, is achieved from geranyl-geranyl-diphosphate by five enzymatic activities: the bifunctional enzyme phytoene synthase/lycopene cyclase (CarRA), the phytoene desaturase CarB (23), the torulene cleaving oxygenase (CarT), and a presumed dehydrogenase. The first two enzymes mediate the reactions needed to produce torulene: the condensation of two geranyl-geranyl-diphosphate molecules, five desaturations, and the introduction of one β-ionone ring. The next step is mediated by CarT, which cleaves torulene to β-apo-4′-carotenal (26a). Finally, the aldehyde is oxidized to the acid neurosporaxanthin by an unknown enzyme. In addition to neurosporaxanthin, F. fujikuroi accumulates minor amounts of torulene and γ- and β-carotene (2), with the later being a final side-product of the CarRA-catalyzed cyclization of γ-carotene.
As deduced from the presence of opsins, the carotenoid pathway of the filamentous fungi is also supposed to be the source of an additional apocarotenoid, retinal. Opsins are a class of seven-transmembrane helices proteins that bind this apocarotenoid via a conserved lysine residue. The light-mediated isomerization of the chromophore enables opsins to function as ion pumps or light sensory receptors in animals, archaea, algae (reviewed in references 16 and 38), and as recently shown, in eubacteria (6, 14), including cyanobacteria (41). In addition, opsin-encoding sequences have been identified in the genomes of the fungi Neurospora crassa (8), Fusarium fujikuroi (26), and Leptosphaeria maculans (17), but more examples are appearing as new fungal genomes become available. Recently, it has been demonstrated that the Leptosphaeria opsin acts as a bacteriorhodopsin-like proton pump (43). It was also shown that the Neurospora opsin NOP-1, when heterologously expressed in the yeast Pichia pastoris, forms an active photoreceptor exhibiting similar characteristics to the archaeal sensory rhodopsin II (7, 9). However, the disruption of nop-1 did not lead to detectable changes in photoregulated processes, like conidation or pigmentation (8). Therefore, the target processes regulated by NOP-1 are still unknown.
Interestingly, the genes responsible for the first steps in the carotenoid biosynthesis (carRA and carB) constitute a gene cluster with two further members, carO and carX, coding for an opsin-like protein and a putative carotenoid oxygenase, respectively. The four genes exhibit the same transcriptional pattern, being expressed at low levels in the dark and induced by illumination to reach a maximum after 1 h of exposure. In addition, all four genes are deregulated in carotenoid-overproducing mutants (23, 26, 40).
As mentioned above, carotenoid biosynthesis in Fusarium fujikuroi is induced by light (3). The occurrence of a gene encoding the opsin-like protein CarO in the car gene cluster suggested a possible role of this photoreceptor in the light regulation of the pathway. However, like in the case of NOP-1 from Neurospora (8), targeted deletion of carO did not lead to a detectable phenotype (26). In contrast to carO, the disruption of carX, which encodes a putative carotenoid oxygenase, resulted in a significant induction of carotenoid biosynthesis (40), indicating that the enzyme is involved in negative feedback regulation of the pathway and not in neurosporaxanthin formation, as presumed before. The car gene cluster resembles the recently described clusters from γ-proteobacteria in that it harbors genes responsible for β-carotene formation and two genes encoding an opsin and a retinal-forming enzyme (29).
In the light of these findings, we investigated the ability of CarX to cleave carotenes. Here we report on CarX as the first retinal-forming enzyme from fungi as deduced from in vivo and in vitro studies.
MATERIALS AND METHODS
Cloning of carX.
Five micrograms of total RNA, isolated from mycelia grown for 1 h under white light, was used for cDNA synthesis using SuperScript RnaseH− reverse transcriptase (Invitrogen, Paisley, United Kingdom) according to the instructions of the manufacturer. Two microliters of cDNA was then applied for the amplification of carX using the following primers: CarX-1, 5′-ATGAAGTTTCTGCAACAAAATTCC-3′; CarX-2, 5′-TCATCCAACAGCTTTCTCCAACTTC-3′. The PCR was performed using 100 ng of each primer, 200 μM concentrations of deoxynucleoside triphosphates, and 1 μl Advantage cDNA polymerase mix (BD Biosciences) in the buffer provided, as follows: 2 min of initial denaturation at 94°C, 32 cycles (30 s at 94°C, 30 s at 54°C, 2 min at 68°C), and 10 min of final polymerization at 68°C. The obtained PCR product was purified using GFX PCR DNA and Gel Band Purification kit (Amersham Biosciences, NJ) and cloned into the pCR2.1-TOPO and pBAD/TOPO (Invitrogen, Paisley, United Kingdom) vectors to yield pCR-CarX and pBAD-CarX, respectively. The nature of the product was verified by sequencing.
In vivo test using β-carotene-accumulating Escherichia coli cells.
β-Carotene- and lycopene-accumulating E. coli XL1-Blue cells were generated by introduction of the plasmids pBeta and pLyc, derivatives of pACYC177 harboring the required biosynthetic genes (crtE, crtB, and crtI in pLyc; crtE, crtB, crtI, and crtY in pBeta) from Erwinia herbicola, allowing the formation of lycopene and β-carotene, respectively. Cells were then transformed with pBAD-CarX. Overnight cultures of the obtained strain β-CarX and the corresponding control β-Con, containing CarX in the antisense orientation, were inoculated into LB medium, grown at 28°C to an optical density at 600 nm of 0.5, and induced with 0.08% arabinose. Cells were then harvested after 4 h. Carotenoids and retinoids were extracted using formaldehyde according to the method of von Lintig and Vogt (42).
Protein expression and purification.
To express CarX as a glutathione S-transferase (GST) fusion, the corresponding cDNA was excised as a blunt-end fragment from pCR-CarX and then ligated into SmaI-digested and alkaline phosphatase-treated pGEX-4T-1 (Amersham Biosciences, NJ) to yield pGEX-CarX. The blunt-end CarX fragment was obtained through NotI digestion, followed by a T4 DNA polymerase treatment and Ecl136II digestion. Subsequently, E. coli BL21 cells were transformed with pGEX-CarX, grown at 28°C in 2× YT medium, and induced at an optical density at 600 nm of 0.5 with 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG). After overnight incubation at 18°C, cells were harvested by centrifugation. The fusion protein was then purified using glutathione-Sepharose 4B (Amersham Biosciences, NJ) according to the instructions of the manufacturer. The fusion protein was eluted twice with elution buffer (50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 10 mM glutathione, 0.2% Triton X-100 [vol/vol]) for 20 min at room temperature. Purification steps and protein expression were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The control strain carried an antisense plasmid expressing only GST.
Preparation of the substrates.
β-Carotene and lycopene were obtained from Sigma-Aldrich (Deisenhofen, Germany) and Roth (Karlsruhe, Germany). β-Apo-8′-carotenal and β-apo-4′-carotenal were kindly provided by BASF, Ludwigshafen, Germany. The substrates were purified using thin-layer chromatography (TLC) silica gel plates (Merck, Darmstadt, Germany), developed in petroleum benzene-diethylether-acetone (40:10:10, vol/vol/vol). Highly pure γ-carotene was obtained from Carotenature (Lupsingen, Switzerland). Torulene and neurosporaxanthin were purified from carotenoid extracts of Fusarium fujikuroi strains accumulating these substrates by preparative high-performance liquid chromatography (HPLC), as described below.
Enzyme assays.
To produce micelles, substrates were dried using a vacuum centrifuge, resuspended in 200 μl petroleum benzene, and then mixed with 150 μl of ethanolic detergent mixture consisting of 0.7% (vol/vol) Triton X-100 and 1.6% (vol/vol) Triton X-405. The mixture was dried again and resuspended in 110 μl of incubation buffer consisting of 200 mM HEPES-NaOH, pH 8.0, 2 mM TCEP [Tris(2-carboxy-ethyl)phosphine hydrochloride; obtained from Sigma-Aldrich (Deisenhofen, Germany)], 0.4 mM FeSO4, and 2 mg ml−1 catalase (Sigma-Aldrich, Deisenhofen, Germany). One hundred microliters of the prepared micelles was then used for an in vitro assay in a total volume of 200 μl containing CarX and the carotene substrate in final concentrations of 200 ng μl−1 and 20 μM, respectively. After an incubation of 2 h at 27°C, assays were stopped by adding 1 volume of acetone, extracted with petroleum benzene-diethylether (1:4, vol/vol), and subjected to HPLC analyses.
Photometric measurements.
Substrates were quantified spectrophotometrically at their individual λmax using extinction coefficients calculated from E1% (5, 13a). Protein concentration was determined using the Bio-Rad protein assay kit (Bio-Rad, CA).
HPLC analyses.
HPLC separations were performed through a C30 reversed-phase column (YMC Europe, Schermbeck, Germany) in a Waters system (Eschborn, Germany) equipped with a photodiode array detector (model 996). Cleavage products were analyzed using the solvent systems B, methanol (MeOH)-tert-butylmethyl ether-water (600:120:120, vol/vol/vol), and A, MeOH-tert-butylmethyl ether (500:500, vol/vol). The column was developed at a flow rate of 1 ml min−1, with a linear gradient from 100% B to 43% B within 45 min, then to 0% B within 1 min, and maintaining the final conditions for another 26 min at a flow rate of 2 ml min−1.
Torulene and neurosporaxanthin were purified from total carotenoid extracts of Fusarium fujikuroi using the solvent systems B, MeOH-tert-butylmethyl ether-water (120:4:40, vol/vol/vol), and A, MeOH-tert-butylmethyl ether (500:500, vol/vol). The column was developed at a flow rate of 1 ml min−1, with a linear gradient from 100% B to 50% B within 5 min, then to 0% B within 1 min, and maintaining the final conditions for another 26 min at a flow rate of 2 ml min−1.
GC-MS analyses.
The products obtained from β-CarX cells were prepurified by TLC using chloroform-washed RP-18 F245s plates (Merck, Darmstadt, Germany). The plates were developed in MeOH-water (100:1, vol/vol), and products were then scraped off and eluted with CHCl3, evaporated, and redissolved in acetone. Gas chromatography (GC)-mass spectrometry (MS) analyses were performed using a Finnigan Trace DSQ mass spectrometer coupled to a Trace GC gas chromatograph. Separations were carried out using a 30-m Zebron ZB5 column (5% phenyl-95% dimethylpolysilanoxane, 0.25-mm inner diameter, and 0.25-μm film thickness; Phenomenex, Aschaffenburg, Germany). For the identification of retinal, a temperature program was applied with an initial temperature of 100°C for 5 min, followed by a temperature ramp of 25°C min−1 to a final temperature of 320°C, which was maintained for an additional 5 min. A constant He carrier gas flow was maintained at 1 ml min−1 using a split flow of 1:30. The splitless time was 3 min, and the injector oven temperature was 220°C. Standard electron impact ionization was used at an ion source potential of 70 eV at 200°C. Identification of retinal was done by chromatographic comparison with the authentic references and by comparing the mass spectra with the NIST Mass Spectral Search Program version 2.0 (National Institute of Standards and Technology).
RESULTS
CarX converts β-carotene into retinal in vivo.
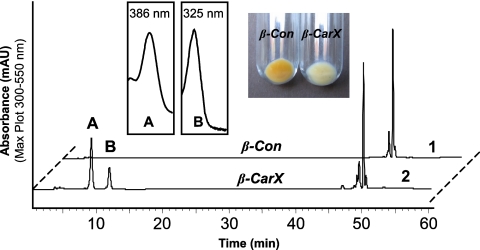
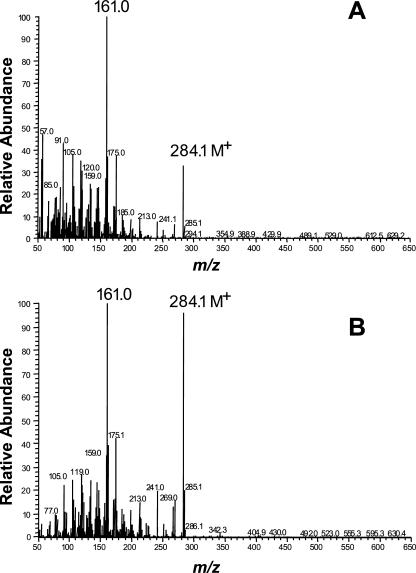
Sequence comparisons suggested that CarX is a member of the carotenoid oxygenase family (40). To investigate its ability to cleave carotenoids, we expressed CarX in lycopene- and β-carotene-accumulating E. coli cells using the pBAD/TOPO expression system (Invitrogen, Paisley, United Kingdom). While no enzymatic activity was detectable in the lycopene background, the expression of CarX in β-carotene-accumulating cells unveiled it as a β-carotene-cleaving enzyme. As shown in Fig. 1, the expression of CarX led to an obvious decolorization of the accumulating cells, indicating β-carotene cleavage. The subsequent HPLC analyses (Fig. 1) showed that CarX cleaved β-carotene into a compound resembling retinal in its UV-visible light spectrum and elution profile. In addition to the aldehyde, the extract contained two compounds likely to be the corresponding alcohol retinol and its corresponding ester. To prove the identity of the formed aldehyde, cell extracts were separated using TLC, and the corresponding product was purified and subjected to GC-MS analyses using retinal as a standard. The product of CarX and the retinal standard showed identical retention times and, as shown in Fig. 2, the mass spectrum was identical to published data (28) and to spectra in the NIST database, including the presence of the correct molecular ion of m/z = 284 as well as the typical m/z = 161 fragment, indicative of the loss of the ionone ring from the parent ion. These data demonstrate that CarX converts β-carotene into retinal in vivo.
FIG. 1.
In vivo test of CarX activity. CarX was expressed in β-carotene-accumulating E. coli cells (β-CarX) under the control of an arabinose-inducible promoter. The induction led to clear decolorization of β-CarX compared to the control cells (β-Con). The HPLC analysis revealed the formation of product (A) showing a retinal UV-visible light spectrum and elution behavior. In addition, variable amounts of retinyl ester (B) and retinol (not shown), coeluting with retinal in the used HPLC system, were produced.
FIG. 2.
GC-MS analysis of retinal produced in β-CarX cells. (A) The electron impact mass spectrum of compound (A) (Fig. 1) showed identity with the reference spectrum of retinal (B) and exhibited the expected molecular ion of m/z = 284 as well as the typical m/z = 161 fragment, indicative of loss of the β-ionone ring from the parent ion.
CarX synthesizes retinal from β-carotene, γ-carotene, and torulene in vitro.
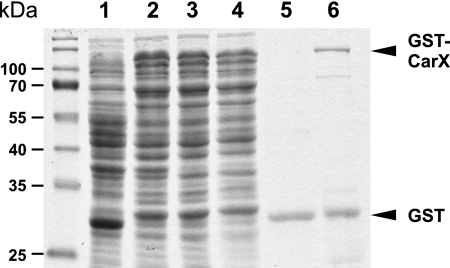
To investigate the enzyme activity in vitro, CarX was expressed and purified as a GST fusion protein (Fig. 3). The purified enzyme was then used for in vitro assays with β-carotene as a substrate. The HPLC analysis of the in vitro assays (Fig. 4) shows a clear conversion of β-carotene into retinal.
FIG. 3.
Coomassie-stained sodium dodecyl sulfate gel showing GST-CarX purification. Lanes: 1, total lysate of control cells expressing GST (control); 2, total lysate of cells expressing GST-CarX; 3, corresponds to sample 2 after removal of inclusion bodies; 4, nonbinding supernatant of sample 3; 5, 4.5-μg protein of the elution fraction of the control; 6, 4.5-μg protein of eluted GST-CarX.
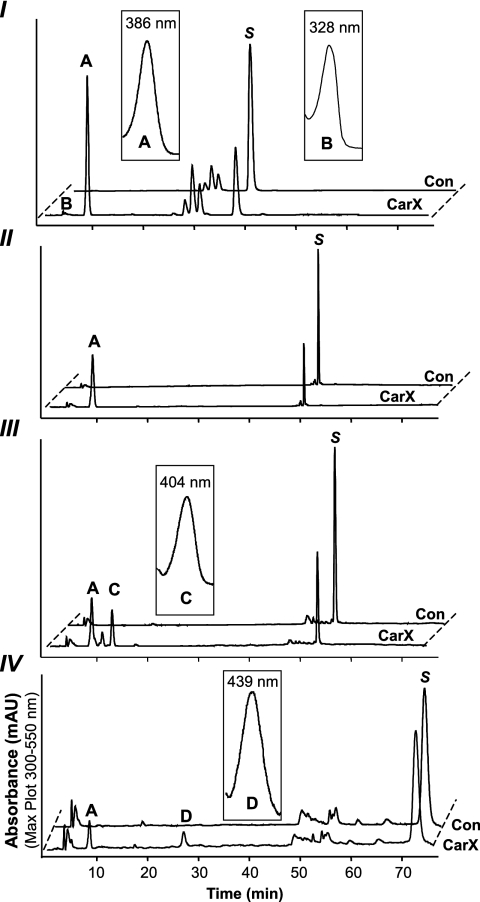
FIG. 4.
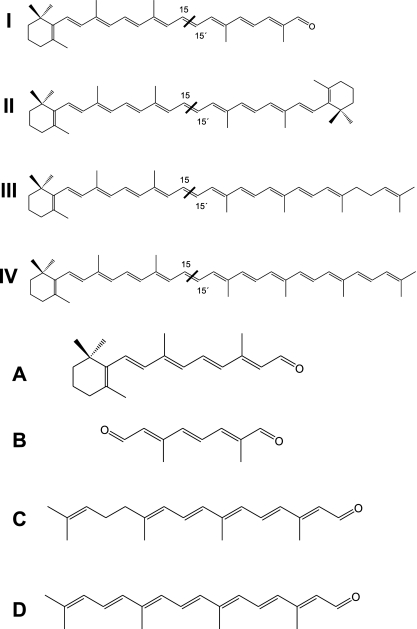
HPLC analyses of the in vitro assay products obtained from β-apo-8′-carotenal (I), β-carotene (II), γ-carotene (III), and torulene (IV) and the corresponding UV-visible light spectra (A to D). CarX formed retinal (A) from the four substrates (S). The conversion to retinal led also to the formation of apo-8′,15′-apo-carotene-dial (B) from β-apo-8′-carotenal, acycloretinal (C) from γ-carotene, and 3,4-didehydro-acycloretinal (D) from torulene. The structure of substrates and products is given in Fig. 5.
The expression of CarX in lycopene-accumulating E. coli cells did not lead to any detectable cleavage activity, indicating the need of a β-ionone ring in the carotenoid substrate. To determine whether monocyclic carotenes are also cleaved by CarX, in vitro assays were performed with γ-carotene and torulene (Fig. 4). As shown in the HPLC analysis (Fig. 4), CarX cleaved γ-carotene and torulene into retinal and the corresponding acyclic aldehydes acycloretinal and 3,4-didehydro-acycloretinal, respetively. The structures of applied substrates and obtained products are given in Fig. 5.
FIG. 5.
Structure of substrates and products of the in vitro assays. The enzyme converted β-apo-8′-carotenal (I) into retinal (A) and apo-8′,15′-apo-carotene-dial (B), β-carotene (II) into retinal (A), γ-carotene (III) into retinal (A) and acycloretinal (C), and torulene (IV) into retinal (A) and 3,4-didehydro-acycloretinal (D). As indicated, all substrates were cleaved at the 15-15′ double bond.
CarX converts the synthetic compound β-apo-8′-carotenal but not C35-apocarotenoids.
Recently, we reported that the cyanobacterial enzymes Synechocystis and Nostoc apocarotenoid oxygenase convert apocarotenoids into retinal and retinal-like compounds by cleaving substrates with different chain lengths at the central 15-15′ double bond (28, 30). To investigate the ability of CarX to produce retinal from apocarotenals, we performed in vitro assays using β-apo-8′-carotenal (C30) as a substrate. As shown in the HPLC analysis (Fig. 4), the incubation with the purified CarX enzyme resulted in the formation of two products which match those formed by the Synechocystis apocarotenoid oxygenase in their UV-visible light spectrum and chromatographic behavior (28). Thus, CarX converted the C30 substrate into retinal (C20) and the corresponding C10 compound apo-8′,15′-apo-carotene-dial (2,6-dimethyl-octa-2,4,6-trien-dial, C10O2H11). The structures of the substrate and the obtained products are given in Fig. 5.
Based on the conversion of β-apo-8′-carotenal into retinal, we assumed that CarX may also cleave the acidic apocarotenoid neurosporaxanthin (C35), the major carotenoid in Fusarium, as well as its corresponding aldehyde β-apo-4′-carotenal. Therefore, we performed in vitro assays using these apocarotenoids as substrates. However, subsequent HPLC analyses revealed, in both cases, the formation of only traces of retinal (data not shown).
DISCUSSION
In this work, we show in vivo and in vitro that the fungal enzyme CarX is a carotenoid oxygenase which catalyzes the formation of retinal through central cleavage of β-carotene at the 15-15′ double bond. To determine whether monocyclic carotenes are also cleaved by CarX, in vitro assays were performed with γ-carotene and torulene (Fig. 4), two compounds also found in F. fujikuroi (35). In a control experiment, we also performed an in vitro assay with lycopene as a substrate. The subsequent analyses proved that lycopene does not represent a suitable substrate (data not shown). Similar results were reported for the human BCO I, which cleaved β-carotene but not lycopene (22). In contrast, the recombinant enzyme catalyzed the cleavage of γ-carotene and torulene into retinal and the corresponding acyclic aldehydes acycloretinal and 3,4-didehydro-acycloretinal. These data suggest that the occurrence of at least one β-ionone ring in the substrate is required for the cleavage reaction.
Carotenoids may be targets for destruction processes caused by oxidative stress and leading to the formation of apocarotenoids with different chain lengths. These unspecific cleavage products may represent substrates for carotenoid oxygenases which convert them into shorter apocarotenoids with a defined chain length. For instance, the cyanobacterial enzymes Synechocystis (28) and Nostoc apocarotenoid oxygenase (30) cleaved several apocarotenoids to retinal and retinal-like compounds. It was also shown that the A. thaliana enzyme AtCCD1, which cleaves all-trans-carotenoids at the 9-10 and 9′-10′ double bonds, also readily converts β-apo-8′-carotenal (C30) into β-ionone (C30) and a C17-dialdehyde (31). In light of these findings, we investigated the ability of CarX to cleave β-apo-8′-carotenal and observed a clear conversion of this substrate into retinal and apo-8′,15′-apo-carotene-dial. This suggests that the 15-15′ double bond is the target of the CarX-mediated cleavage, irrespective of the length of the substrate. Additionally, the formation of retinal from β-apo-8′-carotenal indicates that the distance to the β-ionone ring determines the cleavage site, as was shown for the apocarotenoid cleavage enzymes Synechocystis (28) and Nostoc apocarotenoid oxygenase (30).
The cleavage of β-apo-8′-carotenal indicated that the enzyme may also convert the natural C35 apocarotenoid neurosporaxanthin, accumulated in Fusarium, and its precursor β-apo-4′-carotenal. Surprisingly, in vitro assays with these substrates resulted in the formation of only traces of retinal. This obvious contradiction to the conversion of the C30 compound β-apo-8′-carotenal could be explained by interactions between the oxygene functional groups and the substrate-binding cavity. Such interactions may shift the 15-15′ cleavage site from the reaction center or impede the isomerization step, which is assumed to occur prior cleavage (20). The conversion of the shorter β-apo-8′-carotenal indicates that the occurrence of such interactions depends on the length of the substrate. Elucidation of the structure of CarX may provide an explanation for the different activities observed.
The discovery of carX as a fourth constituent of the car cluster, which encodes two carotenoid biosynthesis enzymes (CarRA and CarB) and an opsin-like protein (CarO), had led to the suggestion that CarX might be responsible for the formation of the end product, neurosporaxanthin (40), which is synthesized through the oxidative cleavage of torulene. However, targeted deletion of carX did not result in a loss of the capability of synthesizing neurosporaxanthin. On the contrary, the disruption of carX was accompanied by a duplication of the total carotenoid content under illumination and by an about 10-fold increase in the dark (40). This significant enhancement of the carotenoid amount suggests that the product formed by CarX is a signaling compound downregulating carotenoid biosynthesis. Accordingly, it was shown that the transcript levels of carRA and carO are increased in the carX mutant (40).
The retinal-forming activity of CarX presented here provides a possible explanation for the effects observed in the ΔcarX mutant in the light. It can be speculated that CarX delivers the chromophore retinal to an opsin, which exerts a negative feedback on carotenoid biosynthesis via an unknown signaling cascade. This hypothesis does not contradict the previous finding that loss of function of the putative opsin gene carO from F. fujikuroi did not have a detectable impact on carotenoid biosynthesis (26). The absence of a carotenogenic phenotype in the carO− mutant could be explained by redundancy of opsins, since the genome of F. fujikuroi encodes at least two additional opsin-like proteins (A. F. Estrada, personal communication). This presumption is supported by the occurrence of three opsin-like proteins in the closely related Fusarium graminearum (Gibberella zae PH-1). Based on sequence homology, two of them (accession no. XP_383240 and XP_387730) can be considered closely homologous to CarO (accession no. CAD97459) and the opsin-like protein OpsA from F. fujikuroi (A. F. Estrada, personal communication), respectively, while a counterpart of the third (accession no. XP_381616) has not been identified so far. Interestingly, OpsA (accession no. XP_387730) is the closest homolog to NOP-1 from Neurospora (8), showing a similarity of 74% and identity of 54% over a stretch of 278 amino acids. The expression of nop1 in the yeast Pichia pastoris led to an active, retinal-binding opsin with absorption properties and a photochemical reaction cycle characteristic of the archaeal sensory rhodopsins (9). However, the Δnop-1 mutant did not show any phenotype with respect to light-regulated processes (8). Taken together, the up-regulation of carotenoid biosynthesis manifested by the ΔcarX mutant in the light could be due to the loss of function of another opsin because of the lack of retinal or might be, like the induction in the dark, opsin independent. In the latter case, it could be assumed that the CarX-mediated cleavage reactions sense the carotenoid content and thus implies that retinal or its derivatives are mediators for a negative feedback regulation of the pathway. Thewes et al. (40) reported that the supplementation of ΔcarX mutant with retinyl ester or retinal did not lead to rescue of the observed phenotype. However, our knowledge about the absorption and metabolism of these compounds in Fusarium is still limited. Therefore, the rescue experiments should be optimized, probably by using radiolabeled retinal. In addition, the effect of a third conceivable retinoid signaling molecule, retinoic acid, should be also investigated.
The previously observed ΔcarX phenotype and the data presented here indicate that the retinal-forming activity is solely catalyzed by CarX in F. fujikuroi. This conclusion coincides with the occurrence of only two carotenoid oxygenases in the closely related fungus Fusarium graminearum. The first (accession no. XP_383243) shows a similarity of about 84% to CarX, indicating its involvement in retinal formation; the second (accession no. XP_382801) is CarT (26a), the enzyme which catalyzes the initial cleavage of torulene which leads to the formation of neurosporaxanthin. Not surprisingly, BLAST analysis of the Neurospora genome reveals also two carotenoid oxygenases in this fungus (accession no. XP_961764.1 and XP_958452). The similarity of the carotenoid pathway of Neurospora with that of F. fujikuroi and the occurrence of opsins suggest equivalent roles for the Neurospora carotenoid oxygenases as CarX and CarT counterparts. The activities of the Neurospora enzymes are currently under investigation. The disruption of carX orthologues in other fungal species containing more than one opsin gene provides a powerful tool to unveil the functions of these light receptors.
Our results represent the first report on retinal biosynthesis in the fungal kingdom. This outstanding enzyme activity, needed for the production of vitamin A and retinoic acid in animals, was formerly found in very distant species, from eubacteria and archaea to higher eukaryotes. The occurrence of a retinal-producing enzyme also in a filamentous fungus provides valuable evidence on their ubiquity in all the major taxonomic groups and on their sequence conservation and opens the way for their identification in other fungi and lower eukaryotes.
Acknowledgments
We thank Peter Beyer for valuable discussions, Jorge Mayer for correcting the English version of the manuscript, and Hansgeorg Ernst for providing the synthetic apocarotenoids.
This work was supported by a grant to Peter Beyer by the Bill & Melinda Gates Foundation as part of the Grand Challenges in Global Health Initiative and Deutsche Forschungsgemeinschaft (DFG) grant AL892-1. A.P.-C. was supported by a short-term EMBO fellowship.
Footnotes
Published ahead of print on 9 February 2007.
REFERENCES
- 1.Auldridge, M. E., D. R. McCarty, and H. J. Klee. 2006. Plant carotenoid cleavage oxygenases and their apocarotenoid products. Curr. Opin. Plant Biol. 9:315-321. [DOI] [PubMed] [Google Scholar]
- 2.Avalos, J., and E. Cerdà-Olmedo. 1987. Carotenoid mutants of Gibberella fujikuroi. Curr. Genet. 25:1837-1841. [Google Scholar]
- 3.Avalos, J., and E. L. Schrott. 1990. Photoinduction of carotenoid biosynthesis in Gibberella fujikuroi. FEMS Microbiol. lett. 66:295-298. [Google Scholar]
- 4.Avalos, J., and E. Cerdá-Olmedo. 2004. Fungal carotenoid production, p. 367-378. In D. Arora (ed.), Handbook of fungal biotechnology. Marcel Dekker, Inc., New York. NY.
- 5.Barua, A. B., and J. A. Olson. 2000. β-Carotene is converted primarily to retinoids in rats in vivo. J. Nutr. 130:1996-2001. [DOI] [PubMed] [Google Scholar]
- 6.Béjà, O., E. N. Spudich, J. L. Spudich, M. Leclerc, and E. F. DeLong. 2001. Proteorhodopsin phototrophy in the ocean. Nature 411:786-789. [DOI] [PubMed] [Google Scholar]
- 7.Bergo, V., E. N. Spudich, J. L. Spudich, and K. J. Rothschild. 2002. A Fourier transform infrared study of Neurospora rhodopsin: similarities with archaeal rhodopsins. Photochem. Photobiol. 76:341-349. [DOI] [PubMed] [Google Scholar]
- 8.Bieszke, J. A., E. L. Braun, L. E. Bean, S. Kang, D. O. Natvig, and K. A. Borkovich. 1999. The nop-1 gene of Neurospora crassa encodes a seven transmembrane helix retinal-binding protein homologous to archaeal rhodopsin-like photochemically reactive pigment. Proc. Natl. Acad. Sci. USA 96:8034-8039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bieszke, J. A., E. N. Spudich, K. L. Scott, K. A. Borkovich, and J. L. Spudich. 1999. A eukaryotic protein NOP-1, binds retinal to form an archaeal rhodopsin-like photochemically reactive pigment. Biochemistry 38:14138-14145. [DOI] [PubMed] [Google Scholar]
- 10.Booker, J., M. Auldridge, S. Wills, D. McCarty, H. Klee, and O. Leyser. 2004. MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Curr. Biol. 14:1232-1238. [DOI] [PubMed] [Google Scholar]
- 11.Bouvier, F., J. C. Isner, O. Dogbo, and B. Camara. 2005. Oxidative tailoring of carotenoids: a prospect towards novel functions in plants. Trends Plant Sci. 10:187-194. [DOI] [PubMed] [Google Scholar]
- 12.Bouvier, F., O. Dogbo, and B. Camara. 2003. Biosynthesis of the food and cosmetic plant pigment bixin (annatto). Science 300:2089-2091. [DOI] [PubMed] [Google Scholar]
- 13.Bouvier, F., C. Suire, J. Mutterer, and B. Camara. 2003. Oxidative remodeling of chromoplast carotenoids: identification of the carotenoid dioxygenase CsCCD and CsZCD genes involved in Crocus secondary metabolite biogenesis. Plant Cell 15:47-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13a.Davies, B. H. 1976. Carotenoids, p. 150-153. In T. W. Goodwin (ed.), Chemistry and biochemistry of plant pigments, 2nd ed., vol. 2. Academic Press, London, United Kingdom. [Google Scholar]
- 14.de la Torre, J. R., L. M. Christianson, O. Béjà, M. T. Suzuki, D. M. Karl, J. Heidelberg, and E. F. DeLong. 2003. Proteorhodopsin genes are distributed among divergent marine bacterial taxa. Proc. Natl. Acad. Sci. USA 100:12830-12835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giuliano, G., S. Al-Babili, and J. von Lintig. 2003. Carotenoid oxygenases: cleave it or leave it. Trends Plant Sci. 8:145-149. [DOI] [PubMed] [Google Scholar]
- 16.Hegemann, P. 1997. Vision in microalgae. Planta 203:265-274. [DOI] [PubMed] [Google Scholar]
- 17.Idnurm, A., and B. J. Howlett. 2001. Characterization of an opsin gene from the ascomycete Leptosphaeria maculans. Genome 44:167-171. [DOI] [PubMed] [Google Scholar]
- 18.Iuchi, S., M. Kobayashi, T. Taji, M. Naramoto, M. Seki, T. Kato, S. Tabata, Y. Kakubari, K. Yamaguchi-Shinozaki, and K. Shinozaki. 2001. Regulation of drought tolearance by gene manipulation of 9-cis-epoxycarotenoids dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 27:325-333. [DOI] [PubMed] [Google Scholar]
- 19.Kiefer, C., S. Hessel, J. M. Lampert, K. Vogt, M. O. Lederer, D. E. Breithaupt, and J. von Lintig. 2001. Identification and characterization of a mammalian enzyme catalyzing the asymmetric oxidative cleavage of provitamin A. J. Biol. Chem. 276:14110-14116. [DOI] [PubMed] [Google Scholar]
- 20.Kloer, D. P., S. Ruch, S. Al-Babili, P. Beyer, and G. E. Schulz. 2005. The structure of a retinal-forming carotenoid oxygenase. Science 308:267-269. [DOI] [PubMed] [Google Scholar]
- 21.Leuenberger, M. G., C. Engeloch-Jarret, and W. Woggon. 2001. The reaction mechanism of the enzyme catalyzed central cleavage of β-carotene to retinal. Angew. Chem. 113:2684-2687. [DOI] [PubMed] [Google Scholar]
- 22.Lindqvist, A., and S. Andersson. 2002. Biochemical properties of purified recombinant human beta-carotene 15,15′-monooxygenase. J. Biol. Chem. 277:23942-23948. [DOI] [PubMed] [Google Scholar]
- 23.Linnemannstöns, P., M. M. Prado, R. Fernandez-Martin, B. Tudzynski, and J. Avalos. 2002. A carotenoid biosynthesis gene cluster in Fusarium fujikuroi: the genes carB and carRA. Mol. Genet. Genomics 267:593-602. [DOI] [PubMed] [Google Scholar]
- 24.Moise, A. R., J. von Lintig, and K. Palczewski. 2005. Related enzymes solve evolutionarily recurrent problems in the metabolism of carotenoids. Trends Plant Sci. 10:178-186. [DOI] [PubMed] [Google Scholar]
- 25.Naested, H., A. Holm, T. Jenkins, H. B. Nielsen, C. A. Harris, M. H. Beale, M. Andersen, A. Mant, H. Scheller, B. Camara, O. Mattsson, and J. Mundy. 2004. Arabidopsis VARIEGATED 3 encodes a chloroplast targeted, zinc-finger protein required for chloroplast and palisade cell development. J. Cell Sci. 117:4807-4818. [DOI] [PubMed] [Google Scholar]
- 26.Prado, M. M., A. Prado-Cabrero, R. Fernandez-Martin, and J. Avalos. 2004. A gene of the opsin family in the carotenoid gene cluster of Fusarium fujikuroi. Curr. Genet. 46:47-58. [DOI] [PubMed] [Google Scholar]
- 26a.Prado-Cabrero, A., A. F. Estrada, S. Al-Babili, and J. Avalos. Identification and biochemical characterization of a novel carotenoid oxygenase: elucidation of the cleavage step in the Fusarium carotenoid pathway. Mol. Microbiol., in press. [DOI] [PubMed]
- 27.Redmond, T. M., S. Gentleman, T. Duncan, S. Yu, B. Wiggert, E. Gantt, and F. X. Cunningham, Jr. 2001. Expression and substrate specificity of a mammalian β-carotene 15,15′-dioxygenase. J. Biol. Chem. 276:6560-6565. [DOI] [PubMed] [Google Scholar]
- 28.Ruch, S., P. Beyer, H. Ernst, and S. Al-Babili. 2005. Retinal biosynthesis in eubacteria: in vitro characterization of a novel carotenoid oxygenase from Synechocystis sp. PCC6803. Mol. Microbiol. 55:1015-1024. [DOI] [PubMed] [Google Scholar]
- 29.Sabehi, G., A. Loy, K. H. Jung, R. Partha, J. L. Spudich, T. Isaacson, J. Hirschberg, M. Wagner, and O. Béjà. 2005. New insights into metabolic properties of marine bacteria encoding proteorhodopsins. PLoS Biol. 3:e273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scherzinger, D., S. Ruch, D. P. Kloer, A. Wilde, and S. Al-Babili. 2006. Retinal is formed from apo-carotenoids in Nostoc sp. PCC7120: in vitro characterization of an apo-carotenoid oxygenase. Biochem. J. 398:361-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmidt, H., R. Kurtzer, W. Eisenreich, and W. Schwab. 2006. The carotenase AtCCD1 from Arabidopsis thaliana is a dioxygenase. J. Biol. Chem. 281:9845-9851. [DOI] [PubMed] [Google Scholar]
- 32.Schwartz, S. H., X. Qin, and M. C. Loewen. 2004. The biochemical characterization of two carotenoid cleavage enzymes from Arabidopsis indicates that a carotenoid-derived compound inhibits lateral branching. J. Biol. Chem. 279:46940-46945. [DOI] [PubMed] [Google Scholar]
- 33.Schwartz, S. H., Q. Xiaoqiong, and J. A. Zeevart. 2001. Characterization of a novel carotenoid cleavage dioxygenase from plants. J. Biol. Chem. 276:25208-25211. [DOI] [PubMed] [Google Scholar]
- 34.Schwartz, S. H., B. C. Tan, D. A. Gage, J. A. Zeevart, and D. R. McCarty. 1997. Specific oxidative cleavage of carotenoids by VP14 of maize. Science 276:1872-1874. [DOI] [PubMed] [Google Scholar]
- 35.Sieiro, C., M. Poza, T. de Miguel, and T. G. Villa. 2003. Genetic basis of microbial carotenogenesis. Int. Microbiol. 6:11-16. [DOI] [PubMed] [Google Scholar]
- 36.Simkin, A. J., S. H. Schwartz, M. Auldridge, M. G. Taylor, and H. J. Klee. 2004. The tomato carotenoid cleavage dioxygenase 1 genes contribute to the formation of the flavor volatiles β-ionone, pseudoionone, and geranylacetone. Plant J. 40:882-892. [DOI] [PubMed] [Google Scholar]
- 37.Simkin, A. J., B. A. Underwood, M. Auldridge, H. M. Loucas, K. Shibuya, E. Schmelz, D. G. Clark, and H. J. Klee. 2004. Circadian regulation of the PhCCD1 carotenoid cleavage dioxygenase controls emission of beta-ionone, a fragrance volatile of petunia flowers. Plant Physiol. 136:3504-3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spudich, J. L., C. S. Yang, K. H. Jung, and E. N. Spudich. 2000. Retinylidene proteins: structures and functions from archaea to humans. Annu. Rev. Cell Dev. Biol. 16:365-392. [DOI] [PubMed] [Google Scholar]
- 39.Tan, B. C., L. M. Joseph, W. T. Deng, L. Liu, Q. B. Li, K. Cline, and D. R. McCarty. 2003. Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 35:44-56. [DOI] [PubMed] [Google Scholar]
- 40.Thewes, S., A. Prado-Cabrero, M. M. Prado, B. Tudzynski, and J. Avalos. 2005. Characterization of a gene in the car cluster of Fusarium fujikuroi that codes for a protein of the carotenoid oxygenase family. Mol. Genet. Genomics 274:217-228. [DOI] [PubMed] [Google Scholar]
- 41.Vogeley, L., O. A. Sineshchekov, V. D. Trivedi, J. Sasaki, J. L. Spudich, and H. Lueke. 2004. Anabaena sensory rhodopsin: a photochromic color sensor at 2.0 Å. Science 306:1390-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.von Lintig, J., and K. Vogt. 2000. Filling the gap in vitamin A research. J. Biol. Chem. 275:11915-11920. [DOI] [PubMed] [Google Scholar]
- 43.Waschuk, S. A., A. G. Bezerra, L. Shi, and L. S. Brown. 2005. Leptosphaeria rhodopsin: bacteriorhodopsin-like proton pump from a eukaryote. Proc. Natl. Acad. Sci. USA 102:6879-6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wyss, A., G. Wirtz, W. Woggon, R. Brugger, M. Wyss, A. Friedlein, H. Bachmann, and W. Hunziker. 2000. Cloning and expression of beta, beta-carotene 15,15′-dioxygenase. Biochem. Biophys. Res. Commun. 271:334-336. [DOI] [PubMed] [Google Scholar]
- 45.Xue, L., D. R. Gollapalli, P. Maiti, W. J. Jahng, and R. R. Rando. 2004. A palmitoylation switch mechanism in the regulation of the visual cycle. Cell 117:761-771. [DOI] [PubMed] [Google Scholar]
- 46.Yan, W., G. F. Jang, F. Haeseleer, N. Esumi, J. Chang, M. Kerrigan, M. Campochiaro, P. Campochiaro, K. Palczewski, and D. J. Zack. 2001. Cloning and characterization of a human beta, beta-carotene-15,15′-dioxygenase that is highly expressed in the retinal pigment epithelium. Genomics 72:193-202. [DOI] [PubMed] [Google Scholar]