Abstract
Recombination events associated with sexual replication in pathogens may generate new strains with altered virulence. Histoplasma capsulatum is a mating-competent, pathogenic fungus with two described phenotypic mating types, + and −. The mating (MAT) locus of H. capsulatum was identified to facilitate molecular studies of mating in this organism. Through syntenic analysis of the H. capsulatum genomic sequence databases, a MAT1-1 idiomorph region was identified in H. capsulatum strains G217B and WU24, and a MAT1-2 idiomorph region was identified in the strain G186AR. A mating type-specific PCR assay was developed, and two clinical isolates of opposite genotypic mating type, UH1 and VA1, were identified. A known − mating type strain, T-3-1 (ATCC 22635), was demonstrated to be of MAT1-2 genotypic mating type. The clinical isolates UH1 and VA1 were found to be mating compatible and also displayed mating-type-dependent regulation of the MAT transcription factors in response to extracts predicted to contain mating pheromones. These studies support a role for the identified MAT1 locus in determining mating type in H. capsulatum.
The mating process has the potential to play a role in the virulence of human pathogens. Recombination between two strains can result in a new strain with increased virulence. This has been documented in the parasite Toxoplasma gondii (12, 25) and also may have occurred in the fungus Cryptococcus gattii during the Vancouver Island outbreak (9). Populations of the pathogenic fungus Histoplasma capsulatum have been shown to recombine in nature (5, 13); however, little is known about mating on a molecular level in this organism.
H. capsulatum is a dimorphic ascomycete found worldwide. Inhalation of the conidia may result in pulmonary disease and, in some cases, severe disseminated disease and death (27). H. capsulatum is a heterothallic organism with two mating types, + and −, defined by phenotype (14). Mating between freshly isolated + and − strains occurs in the mycelial phase and has been demonstrated in the laboratory (14); however, strains lose the ability to mate with continuous culture (14, 15, 18). Strains of each mating type are not equally represented among clinical isolates of H. capsulatum. In two separate studies, − mating type strains were dominant among the clinical samples tested (17, 18). In contrast, strains isolated from the soil represented both mating types equally (18). It was also shown that the mating type disequilibrium occurs among strains isolated from patients with the pulmonary form of the disease but not the severe disseminated form (17). It is unknown whether these observations can be attributed to virulence differences between strains of opposite mating type.
While an organism's mating type can be determined phenotypically, it can also be determined by the genes present at the mating (MAT) locus (6). In filamentous ascomycetes, the MAT locus is a region of low sequence similarity between two organisms of opposite mating type, with each organism containing a different idiomorph of the MAT locus (6, 26). The MAT locus usually contains genes for one or more transcription factors with structural motifs such as α1 domains, high-mobility-group (HMG) DNA binding domains, amphipathic α-helices, and metallothioneins (6, 26). In a filamentous ascomycte, the first identified MAT idiomorph region encoding an α1 domain transcription factor is often designated MAT1-1, while the first identified MAT idiomorph region encoding an HMG DNA binding domain is often designated MAT1-2 (26).
The aim of this study was to identify the MAT1 locus in H. capsulatum and correlate the previous phenotypic mating type designations with genotypic mating type designations. Mating loci have been identified in other filamentous ascomycete fungi, including Neurospora crassa (11, 23), Aspergillus nidulans (8), and Aspergillus fumigatus (21). Using characteristics of these known MAT loci, predicted MAT1-1 and MAT1-2 idiomorph regions were identified in the genome sequences of three different strains of H. capsulatum. Genotypic mating types were assigned to strains based on the structure of the MAT1 idiomorph regions, and a known − mating type tester strain was shown to be of MAT1-2 genotype. Mating compatibility was confirmed between H. capsulatum strains of opposite genotypic mating type. Additionally, message levels of predicted MAT1 locus transcription factors were differentially regulated, depending on the genotypic mating type, in response to pheromone extracts. Taken together, these studies support a role for the predicted MAT1 locus in determining mating type in H. capsulatum.
MATERIALS AND METHODS
Strains and conditions.
H. capsulatum strains G217B (ATCC 26032; a kind gift from George Deepe, University of Cincinnati, Cincinnati, OH), G186A (ATCC 26029; a kind gift from William Goldman, Washington University, St. Louis, MO), G184A (ATCC 26027; from William Goldman), T-3-1 mating type − (ATCC 22635), UH1 (a clinical isolate obtained from a transplant patient with disseminated histoplasmosis), and VA1 (a clinical isolate obtained from a human immunodeficiency virus/AIDS patient with disseminated histoplasmosis) were grown in Histoplasma macrophage medium (HMM) broth or on HMM agarose plates. H. capsulatum strains WU8 (G186A ura5Δ32) and WU15 (G217B ura5Δ42), gifts from William Goldman and Russ Osguthorpe, were grown in HMM broth supplemented with 0.2 mg/ml uracil (Sigma-Aldrich, St. Louis, MO).
Yeast-phase organisms were grown at 37°C in liquid media on an orbital shaker or on plates at 37°C under 5% CO2 in a humidified incubator. Mycelial-phase organisms were grown in liquid culture at 25°C on an orbital shaker in HMM or yeast extract Medium (YEM) (14), with uracil supplementation as appropriate. Mating assays on solid media were performed at 25°C on Alphacel yeast extract medium (A-YEM) agarose plates (14).
Analysis of fungal mating loci.
The H. capsulatum genome sequence is available at the websites of the Washington University Genome Sequencing Center (http://genome.wustl.edu/genome_group_index.cgi) (strains G217B and G186AR) and the Broad Institute (http://www.broad.mit.edu/annotation/fgi) (strain WU24). Using BLASTX or TBLASTN, H. capsulatum MAT1-1-1 and MAT1-2-1 proteins and genes were compared to the fungal genomes at the Broad Institute or the database of the National Center for Biotechnology Information.
MAT locus PCR.
DNA was isolated from selected H. capsulatum strains grown in liquid culture by using the MasterPure yeast DNA kit (Epicentre Inc., Madison, WI). PCR was performed using sense primers MAT1-1S (5′-CGTGGTTAGTTACGGAGGCA-3′) and MAT1-2S (5′-ACACAGTAGCCCAACCTCTC-3′) and antisense primers MAT1-1AS (5′-TGAGGATGCGAGTGATGGGA-3′) and MAT1-2AS (5′-TCGACAATCCCATCCAATACCG-3′), designed based on the MAT1-1 and MAT1-2 idiomorph sequences. PCR was performed under standard conditions with a 60°C annealing temperature by using JumpStart Taq polymerase (Sigma-Aldrich). Products were analyzed by agarose gel electrophoresis.
Mating compatibility test.
The organisms to be tested were streaked onto HMM plates and grown at 25°C until mycelial growth was observed. A plug of each organism was transferred onto A-YEM agarose (14). Plugs were placed approximately 8 mm apart. Organisms were grown at 25°C for 3 weeks and then examined microscopically. Mycelial organisms were lifted from the plates by using clear Petri-Seal tape. Organisms on the tape were mounted on slides by using lactophenol mounting media and covered with a coverslip. Slides were observed using a Nikon E600 microscope equipped with a Spot RT slider camera.
Isolation of a and α pheromones.
Putative pheromone-containing suspensions were isolated using a protocol described by Strazdis and MacKay (24). Briefly, hydrophobic polystyrene resin Amberlite XAD2 was prepared by washing in water, followed by either 1:3 methylene chloride-methanol (for α factor isolation) or 1:3 dichloropropane-n-propanol (for a factor isolation), and stored in propanol. Prior to use, the resin was washed in excess water and sterilized by autoclaving in water. Fifty milliliters of the appropriate resin preparation was added to a 500-ml coculture of H. capsulatum strains WU8 and WU15. The culture was grown for 1 to 2 weeks in uracil-supplemented YEM at 25°C. The resin was then removed from the culture and washed extensively with water, and bound proteins were eluted from the beads by incubation in propanol (a factor) or 40% methanol (α factor) at 40°C for 2 h. The alcohol was evaporated by vacuum centrifugation, and the remaining protein was reconstituted in 100 μl dimethyl sulfoxide (DMSO). Eluted proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on 4 to 20% acrylamide gels in a Tris-Tricine buffer system and observed following silver staining.
Quantitative real-time PCR (qRT-PCR) of mating-related gene expression.
H. capsulatum strains VA1 and UH1, growing at 25°C on HMM agar, were used to start liquid cultures in HMM. Liquid cultures were grown to saturation at 25°C with shaking. Organisms were then transferred to YEM and grown at 25°C overnight with shaking. Five microliters of α or a pheromone-containing suspensions, extracted from H. capsulatum culture as described above, or DMSO was added to the cultures in YEM. RNA was extracted after 30 min by using the MasterPure yeast RNA kit (Epicentre). cDNA synthesis, primed from antisense primers specific to MAT1-1-1, MAT1-2-1, and GAPDH, was performed using Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA). The gene-specific antisense primers were MAT1-1-1 (5′-GTAATACGACTCACTATAGGGTTGTCCATGCTCACAGCCAATT-3′), MAT1-2-1 (5′-GTAATACGACTCACTATAGGGGGTGATGCCGGCGATACAAAAT-3′), and GAPDH (5′-GTAATACGACTCACTATAGGGGACAGCCTTGCCATTGACGGTC-3′).
cDNA samples were diluted 1:10 in water, and qRT-PCR was performed using SYBR green PCR master mix (Applied Biosystems, Foster City, CA) in the Applied Biosystems 7500 real-time PCR system (Applied Biosystems Inc.). qRT-PCR was performed in triplicate for each cDNA sample. The primer pairs for each qRT-PCR were designed such that one primer in each pair spanned an intron, preventing amplification from genomic DNA. The specificity of each primer pair for cDNA was confirmed by the lack of amplification, as evidenced by the lack of SYBR green incorporation and verified by analysis with agarose gel electrophoresis, using genomic DNA as the template. The primers and sequences used were as follows: MAT1-1-1-S, 5′-TTCGTTCATAGCCTTCAGAAGCTTC-3′; MAT1-1-1-AS, 5′-GGCCAGCATGACTGTCACGAAT-3′; MAT1-2-1-S, 5′-AAAATCAAAGACCGCTTGAGCGCA-3′; MAT1-2-1-AS, 5′-AACAACGGCAGCATCGACAATCCC-3′; GAPDH-S, 5′-ATTGGGCGTATTGTCTTCC-3′; and GAPDH-AS, 5′-TTGAGCATGTAGGCAGCATA-3′. The annealing temperatures for MAT1-1-1, MAT1-2-1, and GAPDH primers were 57°C, 62°C, and 60°C, respectively. cDNA for a relative standard curve was generated using a pool of RNA produced by pooling an aliquot of RNA from each sample collected. The standard cDNA was diluted 1:10, and a standard curve was generated using four samples, serially diluted 1:4 from the original 1:10 dilution. Relative expression levels of genes of interest were calculated using the standard curve method for relative quantification (2). Outliers were removed using Grubb's test for detecting outliers, and RNA levels under the different conditions were compared using analysis of variance (GraphPad Instat; GraphPad, San Diego, CA).
Nucleotide sequence accession numbers.
The sequences of the MAT1-1 and MAT1-2 loci have been deposited in GenBank and are available under the accession numbers EF433757 and EF433756, respectively.
RESULTS
Identification of MAT1 locus in H. capsulatum.
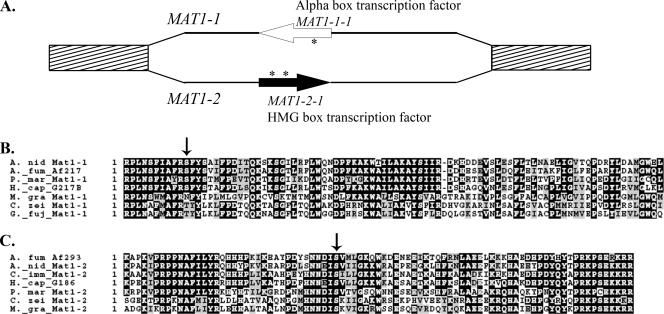
To identify the MAT1-1 idiomorph in H. capsulatum, a TBLASTN analysis was performed to compare the α1 region of a previously identified A. nidulans MAT1-1 peptide (8) with genome sequences of H. capsulatum strains G217B, G186AR, and WU24. HISTO_EA.Contig33 of strain G217B contained a region of 55% sequence identity and 71% similarity to the A. nidulans α1 region. Similarly, supercontig 1-1 of the WU24 genome sequence contained a region of 45% sequence identity and 63% similarity to the A. nidulans α1 region. No significant sequence similarity to the A. nidulans α1 region was noted in the G186AR genome sequence. The H. capsulatum α-box region was predicted to be located within a 1,256-bp gene, which was designated MAT1-1-1 (Fig. 1A) (26). The 1,206-bp MAT1-1-1 open reading frame, interrupted by one 50-bp intron, was predicted to encode a 401-amino-acid protein. The position of the intron was confirmed by sequencing and alignment of MAT1-1-1 PCR products generated from both DNA and cDNA (data not shown). The position of the intron was identical to that of introns found in other ascomycete α1 MAT transcription factors (Fig. 1B) (6).
FIG. 1.
MAT1 locus in H. capsulatum. (A) Representation of the MAT1-1 and MAT1-2 idiomorph regions, as identified in strains G217B and G186AR, respectively. Hatched boxes indicate regions with 96 to 98% sequence identity between the two strains. Black lines indicate the MAT1 idiomorph regions, with only 34% sequence identity. Predicted open reading frames are represented by arrows, white for MAT1-1-1 and black for MAT1-2-1, with introns designated by asterisks. (B and C) Alignment of H. capsulatum α1 (B) and HMG (C) regions with MAT1-1 and MAT1-2 protein sequences in other ascomycete fungi. The positions of conserved introns are noted by arrows. The organism abbreviations are as follows: A. nid, Aspergillus nidulans; A. fum, Aspergillus fumigatus; P. mar, Penicillium marneffei; H. cap, Histoplasma capsulatum; C. zei, Cercospora zeina; G. fuj, Gibberella fujikuroi; C. imm, Coccidioides immitis; M. gra, Mycosphaerella graminicola.
Since strain G186AR showed no sequence similarity to the A. nidulans α1 region, the G186AR genome was searched for a MAT1-2 idiomorph. When two ascomycetes of opposite mating type are compared, the MAT idiomorph regions generally contain little sequence similarity between the two but are flanked by regions of homology between the two (1, 6). Based on this, a BLASTN analysis was performed to compare the MAT1-1-1 gene sequence, and 10 kb of G217B sequence upstream and downstream of the gene, to the G186AR genome sequence. A region of approximately 5 kb with only 33.9% sequence identity was identified, flanked by regions with 95.8% and 96.4% sequence identity, respectively. In addition to sequence disparity, the regions of dissimilarity varied in length, containing 5,702 bp in the G217B strain and 4,997 bp in the G186AR strain, respectively. The MAT1-1 idiomorph contained only the one putative open reading frame described above. The 4,997-bp MAT1-2 idiomorph in G186AR was located in contig 30-9 and also contained a single predicted open reading frame. The open reading frame was designated MAT1-2-1 (26), as it was predicted to encode a 348-amino-acid protein with an HMG DNA binding domain (Fig. 1A). The MAT1-2-1 open reading frame was interrupted by two introns, which were confirmed in the same manner as the MAT1-1-1 intron (data not shown). Both introns were in the same relative position as introns found in other ascomycete MAT HMG transcription factors (Fig. 1C) (6, 22). Based on this information, the mating type of strain G186AR was designated MAT1-2, while the mating type of strains G217B and WU24 was designated MAT1-1 (26).
MAT locus PCR.
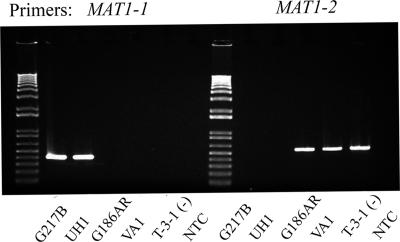
PCR primers were designed to amplify either the MAT1-1 or the MAT1-2 region specifically. Amplification of the MAT1-1 region would result in a 440-bp product, while amplification of the MAT1-2 region would result in a 528-bp product. This MAT locus PCR assay was used to determine the genotypic mating type of a known − mating type tester strain, T-3-1, to correlate genotypic and phenotypic mating type. The phenotypic − mating type strain was shown to be of the MAT1-2 genotypic mating type (Fig. 2). The assay was also used to determine the genotypic mating type of two clinical isolates, UH1 and VA1. UH1 and VA1 were shown to be of opposite mating type, MAT1-1 and MAT1-2, respectively (Fig. 2). An additional laboratory strain in common use, G184A, was similarly analyzed and shown to be of the MAT1-2 mating type (data not shown).
FIG. 2.
Results of MAT1 locus PCR assay. PCR was performed using primers specific to either the MAT1-1 or the MAT1-2 idiomorph region. Both primer pairs were used to test DNA isolated from various H. capsulatum strains. Lane 1, molecular weight marker; lanes 2 to 6, H. capsulatum strains as indicated under each lane; lane 7, “no template” control (NTC).
Mating compatibility test.
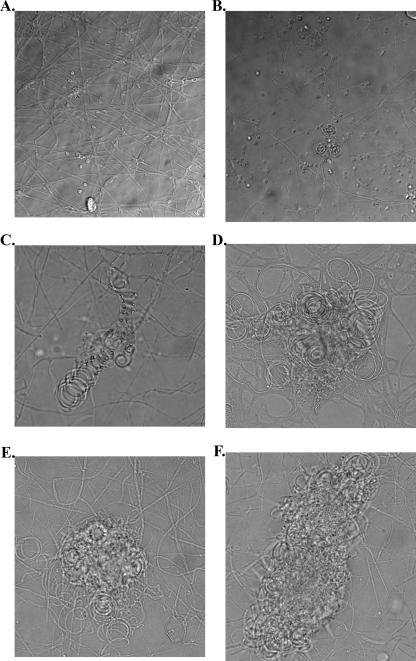
Phenotypic confirmation was sought for the assigned opposite genotypic mating types of clinical isolates UH1 and VA1. Mating compatibility assays were performed between UH1 + and VA1 − strains, with self-crosses of UH1 or VA1 as controls. After 10 to 14 days, a confrontation border appeared between the mycelial plugs of opposite mating type where the advancing hyphal colonies met (14). After an additional 7 days, the mycelia in this region were viewed microscopically. The formation of the ascocarp in H. capsulatum has been described previously (14), and several structures indicative of the mating process in this organism were observed. The structures observed included hyphal coiling and cleistothecia covered by short branching hyphae. These structures were not seen when strains were tested as self-crosses (Fig. 3).
FIG. 3.
Results of mating compatibility test between UH1 and VA1. Organisms were plated on A-YEM and grown for 3 weeks. (A) UH1 self-cross. (B to F) UH1 crossed with VA1. (B and C) Coiled hyphae, consistent with younger mating structures. (D to F) Coiled hyphae covered by short, branching hyphae, consistent with more mature mating structures.
Differential expression of MAT locus transcription factors.
Regulation of the expression of the MAT1-1 and MAT1-2 transcripts in H. capsulatum was examined under mating conditions to examine the association of MAT1 gene expression with mating. Mating pheromones have not been identified or isolated in H. capsulatum; however, based on the assumption that the pheromones produced by H. capsulatum resemble the peptide and lipopeptide pheromones of other ascomycetous fungi, procedures enriching for each class of pheromone produced by Saccharomyces cerevisiae were performed on mixed MAT1-1 and MAT1-2 mycelial cultures of H. capsulatum. When analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and examined by silver staining, the resulting suspensions contained only a few (∼5 to 10) bands, all with molecular masses below 20 kDa (data not shown).
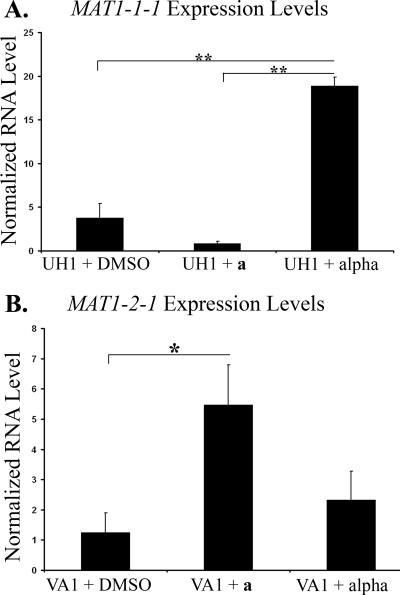
Clinical isolates of opposite genotypic mating type, UH1 (MAT1-1) and VA1 (MAT1-2), were stimulated either with each of the putative pheromone-containing extracts or with vehicle controls. The expression levels of the predicted MAT1 locus transcription factors were then measured by RT-PCR. Stimulation of UH1 with an extract predicted to contain the α pheromone resulted in a fivefold increase in MAT1-1-1 expression compared to stimulation with DMSO alone. This was not seen when UH1 was stimulated with an extract predicted to contain the a pheromone (Fig. 4). The opposite response was seen in VA1, the organism of MAT1-2 genotype. Stimulation of VA1 with an extract predicted to contain the a pheromone resulted in a 4.4-fold increase in MAT1-2-1 expression compared to stimulation with DMSO alone. Stimulation of VA1 with an extract predicted to contain the α pheromone caused no significant change in MAT1-2-1 expression compared to stimulation with DMSO (Fig. 4).
FIG. 4.
Expression levels of MAT1 transcription factors. H. capsulatum strains UH1 (A) and VA1 (B) were stimulated with DMSO, a pheromone extract, or α pheromone extract for 30 min. RNA levels of MAT1-1-1 (A) and MAT1-2-1 (B) were determined by qRT-PCR and normalized to RNA levels of GAPDH. *, P ≤ 0.05; **, P ≤ 0.01.
DISCUSSION
Similarities between MAT loci of several ascomycete fungi have made the identification of MAT loci possible in organisms whose genomes have been sequenced. In this manner, a MAT1-1 region was identified in the homothallic fungus A. nidulans (8), and MAT1-1 and MAT1-2 idiomorphs were identified in A. fumigatus (21, 22). The predicted MAT1 locus, identified in H. capsulatum through the current study, shared common characteristics with MAT loci in other ascomycete fungi (6, 11, 21, 23, 26). The H. capsulatum MAT1-1 idiomorph contained a predicted transcription factor characterized by the presence of an α1 domain. Using standard nomenclature, this is a common feature of MAT1-1 idiomorphs found in ascomycete fungi (6, 26). Similarly, the MAT1-2 idiomorph contained a transcription factor characterized by an HMG domain, which is common among MAT1-2 idiomorph regions in ascomycete fungi (6, 26). Heterothallism, with strains containing either a MAT1-1 or a MAT1-2 idiomorph, appears to be common among the dimorphic ascomycetes. Using a similar genomic approach, MAT1-2 regions can be identified in Coccidioides immitis strains RS and RMSCC 2394 (10), while a MAT1-1 region can be identified in C. immitis strain 4538.4, using genome sequences available at the Broad Institute website (http://www.broad.mit.edu/annotation/fgi) (data not shown). MAT1-2 regions can also be identified in Coccidioides posadasii (10) and Blastomyces dermatitidis, using genome sequences available from The Institute for Genomic Research (http://www.tigr.org/tdb/fungal/index.shtml) and the Washington University Genome Sequencing Center (http://genome.wustl.edu/genome_group_index.cgi) websites, respectively (data not shown). Additionally, ascomycete α1 and HMG domains each contain introns at conserved positions within the domains (6), which were also identified in the sequences for the predicted MAT transcription factors in H. capsulatum.
H. capsulatum has a heterothallic mating system, and the mating morphology of this fungus has been described in great detail using strains of opposite phenotypic mating type (14). Through the present study, mating compatibility was confirmed morphologically in two organisms of opposite genotypic mating type. Asci and ascospores, the results of mating, were not sought in this study; however, many structures were observed that are associated with the formation of the ascocarp. This supports the genomic evidence that the predicted MAT1 locus is associated with mating type in H. capsulatum. This assertion is further supported by the mating type-specific regulation of the MAT transcription factors in response to pheromone extracts. As the pheromone extracts were not purified to homogeneity, it is feasible that other proteins, unrelated to the pheromone response in this organism, may have caused the changes in levels of MAT1-1-1 and MAT1-2-1 expression. If this were the case, however, one would have expected to see the α pheromone extract affect both UH1 and VA1, with the same holding true for the a pheromone extract. The mating type-specific regulation suggests that the effect was due to pheromones present in the semipurified extracts.
This study was carried out with the assumption that in H. capsulatum, organisms of opposite mating type produce different mating pheromones: a peptide α-type pheromone and a lipopeptide a-type pheromone. This is the case for S. cerevisiae (3, 4); however, this is not the case for the fungus Cryptococcus neoformans, which produces two lipid-modified pheromones (7, 20). The techniques used to purify the putative pheromones rely on the differences in structure between the two pheromone types, allowing differential purification of peptide and lipopeptide pheromones. The fact that up-regulation of MAT transcription factors in the present study occurred only in one mating type organism for each pheromone extract argues that the H. capsulatum coculture produced lipopeptide a-type and peptide α-type pheromones. This is further supported by the existence of predicted pheromone-processing machinery for both pheromone types as well as a predicted α-type pheromone sequence in the H. capsulatum genome (data not shown). The genes predicted to encode pheromone-processing proteins and the α-type pheromone are unlinked to the MAT1 locus or to each other.
Characterization of the MAT locus in H. capsulatum will allow a molecular comparison of + and − mating type organisms. Mating type disequilibrium exists among clinical isolates of this organism, with − mating type organisms dominating (17, 18). This disequilibrium was not seen in environmental isolates (18). While the mating type disequilibrium in clinical isolates may suggest differences in virulence between mating types, prior studies of H. capsulatum testing the virulence potentials of + and − mating type strains demonstrated no difference in a mouse model of disease (19). These findings may require reevaluation, as the studies were performed using unrelated strains of H. capsulatum and utilized an intravenous inoculation model of disease. Mating type disequilibrium is found among clinical isolates of C. neoformans, with organisms of the α mating type dominating (16). For the C. neoformans cases, this may be explained partly by the predominance of α mating type organisms in the environment (16). Characterization of the MAT locus in H. capsulatum will allow the possibility of constructing congenic mating type strains in this organism to reexamine questions regarding virulence and mating type. This will prove challenging, however, as few chromosomal markers are known for this organism and the rate of targeted homologous integration in this organism is low (27).
Characterization of the MAT locus in H. capsulatum will also allow a molecular characterization of the mating process in this organism. Not only will this provide a molecular means of comparing organisms of opposite mating type but this will allow a molecular comparison of mating-competent and non-mating-competent organisms. While it has been noted that, with time, H. capsulatum loses the ability to mate when grown in culture (14, 15, 18), the molecular mechanisms of this loss of mating competence have not been explored. This is an obstacle to the development of classical genetic systems in this organism. Laboratory strains G186AR and G217B did not generate a confrontation reaction or form mating structures in the mating compatibility assay described above (data not shown). However, cocultivated derivatives of these strains yielded the pheromone extracts used to stimulate MAT1 gene expression in a regulated manner, implying that the inability of laboratory strains to mate is not due to the loss of the ability to produce pheromone. Additional studies will attempt to determine the level of the defect resulting in the loss of mating competence.
In summary, genomic studies, mating compatibility tests, and the regulation of predicted MAT locus transcription factors in response to pheromone extracts together support the conclusion that the identified MAT1 locus in H. capsulatum plays a role in determining mating type in this organism. The results of these studies will be useful in answering questions regarding the virulence of this organism as well as in developing new tools to study the organism.
Acknowledgments
We thank William Goldman and Russ Osguthorpe for H. capsulatum strains, George Deepe for H. capsulatum strains, advice, and support, Judith Rhodes for advice and assistance, and Jeff Demland, Reiko Tanaka, Debbie Spaulding, Reta Gibbons, and Holly Allen for technical assistance.
Footnotes
Published ahead of print on 23 February 2007.
REFERENCES
- 1.Astell, C. R., L. Ahlstrom-Jonasson, M. Smith, K. Tatchell, K. A. Nasmyth, and B. D. Hall. 1981. The sequence of the DNAs coding for the mating-type loci of Saccharomyces cerevisiae. Cell 27:15-23. [DOI] [PubMed] [Google Scholar]
- 2.Bookout, A. L., C. L. Cummins, D. J. Mangelsdorf, J. M. Pesola, and M. F. Kramer. 2006. High-throughput real-time quantitative reverse transcription PCR, p. 15.8.1-28. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley and Sons, Hoboken, NJ. [DOI] [PubMed]
- 3.Bussey, H. 1988. Proteases and the processing of precursors to secreted proteins in yeast. Yeast 4:17-26. [DOI] [PubMed] [Google Scholar]
- 4.Caldwell, G. A., F. Naider, and J. M. Becker. 1995. Fungal lipopeptide mating pheromones: a model system for the study of protein prenylation. Microbiol. Rev. 59:406-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carter, D. A., A. Burt, J. W. Taylor, G. L. Koenig, and T. J. White. 1996. Clinical isolates of Histoplasma capsulatum from Indianapolis, Indiana, have a recombining population structure. J. Clin. Microbiol. 34:2577-2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coppin, E., R. Debuchy, S. Arnaise, and M. Picard. 1997. Mating types and sexual development in filamentous ascomycetes. Microbiol. Mol. Biol. Rev. 61:411-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davidson, R. C., T. D. Moore, A. R. Odom, and J. Heitman. 2000. Characterization of the MFα pheromone of the human fungal pathogen Cryptococcus neoformans. Mol. Microbiol. 38:1017-1026. [DOI] [PubMed] [Google Scholar]
- 8.Dyer P. S., M. Paoletti, and D. B. Archer. 2003. Genomics reveals sexual secrets of Aspergillus. Microbiology 149:2301-2303. [DOI] [PubMed] [Google Scholar]
- 9.Fraser, J. A., S. S. Giles, E. C. Wenink, S. G. Geunes-Boyer, J. R. Wright, S. Diezmann, A. Allen, J. E. Stajich, F. S. Dietrich, J. R. Perfect, and J. Heitman. 2005. Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature 437:1360-1364. [DOI] [PubMed] [Google Scholar]
- 10.Fraser, J. A., and J. Heitman. 2006. Sex, MAT, and the evolution of fungal virulence, p. 13-33. In J. Heitman, S. G. Filler, J. E. Edwards, Jr., and A. P. Mitchell (ed.), Molecular principles of fungal pathogenesis. ASM Press, Washington, DC.
- 11.Glass, N. L., J. Grotelueschen, and R. L. Metzenberg. 1990. Neurospora crassa A mating-type region. Proc. Natl. Acad. Sci. USA 87:4912-4916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grigg, M. E., S. Bonnefoy, A. B. Hehl, Y. Suzuki, and J. C. Boothroyd. 2001. Success and virulence in Toxoplasma as the result of sexual recombination between two distinct ancestries. Science 294:161-165. [DOI] [PubMed] [Google Scholar]
- 13.Kasuga, T., J. W. Taylor, and T. J. White. 1999. Phylogenetic relationships of varieties and geographical groups of the human pathogenic fungus Histoplasma capsulatum Darling. J. Clin. Microbiol. 37:653-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwon-Chung, K. J. 1973. Studies on Emmonsiella capsulata. I. Heterothallism and development of the ascocarp. Mycologia 65:109-121. [PubMed] [Google Scholar]
- 15.Kwon-Chung, K. J. 1975. Perfect state (Emmonsiella capsulata) of the fungus causing large-form African histoplasmosis. Mycologia 67:980-990. [PubMed] [Google Scholar]
- 16.Kwon-Chung, K. J., and J. E. Bennett. 1978. Distribution of alpha and alpha mating types of Cryptococcus neoformans among natural and clinical isolates. Am. J. Epidemiol. 108:337-340. [DOI] [PubMed] [Google Scholar]
- 17.Kwon-Chung, K. J., M. S. Bartlett, and L. J. Wheat. 1984. Distribution of the two mating types among Histoplasma capsulatum isolates obtained from an urban histoplasmosis outbreak. Sabouraudia 22:155-157. [PubMed] [Google Scholar]
- 18.Kwon-Chung, K. J., R. J. Weeks, and H. W. Larsh. 1974. Studies on Emmonsiella capsulata (Histoplasma capsulatum). II. Distribution of the two mating types in 13 endemic states of the United States. Am. J. Epidemiol. 99:44-49. [DOI] [PubMed] [Google Scholar]
- 19.Kwon-Chung, K. J., and W. B. Hill. 1981. Virulence of the two mating types of Emmonsiella capsulata and the mating experiments with Emmonsiella capsulata var. duboisii, p. 48-56. In C. De Vroey and R. Vanbreuseghem (ed.), Sexuality and pathogenicity of fungi. Masson, Paris, France.
- 20.McClelland, C. M., J. Fu, G. L. Woodlee, T. S. Seymour, and B. L. Wickes. 2002. Isolation and characterization of the Cryptococcus neoformans MATa pheromone gene. Genetics 160:935-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paoletti, M., C. Rydholm, E. U. Schwier, M. J. Anderson, G. Szakacs, F. Lutzoni, J. P. Debeaupuis, J. P. Latge, D. W. Denning, and P. S. Dyer. 2005. Evidence for sexuality in the opportunistic fungal pathogen Aspergillus fumigatus. Curr. Biol. 15:1242-1248. [DOI] [PubMed] [Google Scholar]
- 22.Pöggeler, S. 2002. Genomic evidence for mating abilities in the asexual pathogen Aspergillus fumigatus. Curr. Genet. 42:153-160. [DOI] [PubMed] [Google Scholar]
- 23.Staben, C., and C. Yanofsky. 1990. Neurospora crassa a mating-type region. Proc. Natl. Acad. Sci. USA 87:4917-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strazdis, J. R., and V. L. MacKay. 1982. Reproducible and rapid methods for the isolation and assay of a-factor, a yeast mating hormone. J. Bacteriol. 151:1153-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su, C., D. Evans, R. H. Cole, J. C. Kissinger, J. W. Ajioka, and L. D. Sibley. 2003. Recent expansion of Toxoplasma through enhanced oral transmission. Science 299:414-416. [DOI] [PubMed] [Google Scholar]
- 26.Turgeon, B. G., and O. C. Yoder. 2000. Proposed nomenclature for mating type genes of filamentous ascomycetes. Fungal Genet. Biol. 31:1-5. [DOI] [PubMed] [Google Scholar]
- 27.Woods, J. P. 2002. Histoplasma capsulatum molecular genetics, pathogenesis, and responsiveness to its environment. Fungal Genet. Biol. 35:81-97. [DOI] [PubMed] [Google Scholar]