Abstract
We identified seven novel variants of streptococcal pyrogenic exotoxin G (SPEGG), a superantigen, in Streptococcus dysgalactiae subsp. dysgalactiae or equisimilis isolates from clinical cases of infection in humans and animals. Phylogenetic analysis of the SPEGG variants indicated two clades in the dendrogram: one composed of variants derived from the bacteria isolated from the humans and the other composed of variants from the bacteria isolated from the animals. Bovine peripheral blood mononuclear cells (PBMCs) were stimulated effectively by recombinant SPEGGs (rSPEGGs) expressed in Escherichia coli, while human PBMCs were not stimulated well by any of the rSPEGGs tested. SPEGGs selectively stimulated bovine T cells bearing Vβ1,10 and Vβ4. Bovine serum showed reactivity to the rSPEGG proteins. These results indicated that SPEGGs have properties as superantigens, and it is likely that SPEGGs play a pathogenic role in animals.
Bacterial superantigens (SAGs) bind simultaneously to major histocompatibility complex (MHC) class II molecules on antigen-presenting cells and T-cell receptor (TCR) molecules, and binding leads to the stimulation of large numbers of T cells in a TCR β-chain Vβ-selective manner. Streptococcus pyogenes (group A Streptococcus [GAS]), Staphylococcus aureus, Yersinia pseudotuberculosis, and Mycoplasma arthritidis are known as SAG producers (1, 18, 37). Overactivation of T cells by SAGs has been implicated in the pathogenesis of infectious diseases, such as toxic shock syndrome (TSS) and neonatal TSS-like exanthematous disease, as well as systemic Yersinia pseudotuberculosis infection.
It has been proposed that allelic variation in human leukocyte class II antigens affects the severity of invasive streptococcal infections, including streptococcal TSS (STSS) by regulating cytokine responses to streptococcal SAGs (17). However, we found that GAS isolated from STSS cases produced smaller amounts of SAGs than did GAS isolated from non-STSS cases (21). Most SAGs have highly conserved secondary and tertiary structures despite minimal amino acid sequence homology. Some SAGs, such as SMEZ (28), staphylococcal enterotoxin C (SEC), SPE-A (16, 22), SPE-G and SSA (30), and Y. pseudotuberculosis-derived mitogen show allelic variations, which are characterized by single- or multiple-amino-acid replacement.
In addition to GAS, serological group C and G streptococci possess genes that encode molecules similar to SAGs. For example, S. dysgalactiae subsp. dysgalactiae produces S. dysgalactiae-derived mitogen (SDM) (20). S. equi, the cause of equine strangles, produces S. equi pyrogenic exotoxin H (SePE-H), SePE-I, SPE-LSe, and SPE-M (3, 29). Recently, there have been a number of clinical case reports of STSS caused by S. dysgalactiae subsp. equisimilis, though S. dysgalactiae strains are generally pathogenic in animals (4, 8, 12, 15, 19, 25, 26, 38). In addition to SDM, S. dysgalactiae harbors a gene encoding a protein similar to SPE-G, which has been designated in different ways, such as spegg or speGdys (7, 11, 31). Here, we use “spegg” for the gene. Hashikawa et al. analyzed the prevalence of SAGs in 12 clinical isolates of S. dysgalactiae from STSS cases by PCR and found that only spegg was detected in 7 isolates, with none of the other superantigen genes being detected in any of the strains (11). Brandt et al. analyzed the mitogenic activity of S. dysgalactiae isolates carrying spegg and found no mitogenic activity in culture supernatants (7). However, there have been no previous studies of the biological properties of the spegg gene products. In this study, we analyzed the prevalence of spegg in S. dysgalactiae isolates from humans and animals and analyzed their biological activities using recombinant proteins encoded by the spegg genes. We also performed molecular modeling analysis to examine the point mutations found in SPEGG variants.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All S. dysgalactiae isolates used in this study were isolated from human subjects or animals. Individual strains were stored in brain heart infusion (BHI) broth (Difco, Franklin Lakes, NJ) containing 7% dimethyl sulfoxide at −80°C until use. They were cultured overnight in BHI broth at 37°C in a humidified 5% CO2 incubator as described previously (21). For expression of the six-His (His6)-tagged proteins, Escherichia coli M15(pREP4) (QIAGEN, Tokyo, Japan) was used for transformation with expression constructs derived from pQE30 (QIAGEN).
Preparation of genomic DNA and emm typing.
Chromosomal DNA was prepared from the S. dysgalactiae isolates as described previously (20). Briefly, bacteria grown in BHI broth overnight were collected and lysed by serial treatment with mutanolysin (Sigma-Aldrich, Tokyo, Japan), lysozyme, freeze-thaw cycling, and proteinase K (Sigma-Aldrich). S. dysgalactiae isolates were subjected to emm typing as described previously (http://www.cdc.gov/ncidod/biotech/strep/M-ProteinGene_typing.htm) (21). Briefly, genomic DNA from S. dysgalactiae was used as the template for PCR using specific primers A (5′-TATTAGCTTAGAAAATTAA-3′) and B (5′-GCAAGTTCTTCAGCTTGTTT-3′). The resulting PCR fragments were sequenced with a Genetic Analyzer 310 system (Applied Biosystems, Tokyo, Japan).
PFGE.
Large restriction fragment profiles of all of the isolates were obtained by SmaI digestion followed by pulsed-field gel electrophoresis (PFGE) as described previously (21). Briefly, plugs prepared from the isolates were treated sequentially with achromopeptidase, RNase, lysozyme, mutanolysin, sodium deoxycholate, sodium laurylsarcosine, Brij-58, EDTA, and proteinase K. After digestion with SmaI, the plugs were electrophoresed in a Genepath contour-clamped homogeneous electric field apparatus (Bio-Rad Labs, Tokyo, Japan) and the patterns were analyzed with Molecular Analyst Fingerprinting Plus software, version 1.6 (Bio-Rad Laboratories, Inc., Hercules, CA).
Amino acid sequence alignment and dendrogram preparation.
The spegg genes from S. dysgalactiae genomic DNA in Table 1 were amplified using several combinations of the primers listed in Table 2 and ExTaq (Takara, Shiga, Japan), because the sequences of spegg including the proximal region were different in several clinical isolates. The PCR products were sequenced directly after purification with a QIAquick PCR purification kit (QIAGEN). The SPEGG amino acid sequences were aligned with those of known bacterial SAGs, and a dendrogram was constructed using the search and analysis service based on Clustal W at DDBJ (http://www.ddbj.nig.ac.jp). The dendrogram was drawn using the TreeView program (27).
TABLE 1.
Streptococcus dysgalactiae isolates used in this study
| Strain | Group | S. dysgalactiae subsp. | Hemolysis type | Origin | emm type | SPEGG | Accession no. | Gene name deposited in database |
|---|---|---|---|---|---|---|---|---|
| 125 | C | equisimilis | β | Human | stg653 | − | ||
| 152 | C | equisimilis | β | Human | − | − | ||
| 154 | C | equisimilis | β | Human | − | − | ||
| 164 | G | equisimilis | β | Human | stg485 | SPEGG2 | AB105080 | spegg4 |
| 162 | C | equisimilis | β | Human | − | − | ||
| 160 | G | equisimilis | β | Human | stg652 | − | ||
| 163 | G | equisimilis | β | Human | stg643 | − | ||
| 167 | C | equisimilis | β | Human | stL839 | SPEGG3 | AB105081 | spegg5 |
| 165 | G | equisimilis | β | Human | − | − | ||
| 168 | G | equisimilis | β | Human | NDa | SPEGG4 | AB105078 | spegg2 |
| 169 | G | equisimilis | β | Human | stg11 | SPEGG3 | ||
| 170 | G | equisimilis | β | Human | ND | SPEGG5 | AB105079 | spegg3 |
| 1586 | G | equisimilis | β | Human | stc36 | SPEGG5 | ||
| 1149 | G | equisimilis | β | Human | ND | + | ||
| 1317 | G | equisimilis | β | Human | ND | + | ||
| 1379 | G | equisimilis | β | Human | Stg4831 | + | ||
| 1434 | G | equisimilis | β | Human | Stg2028 | + | ||
| 1412 | G | equisimilis | β | Human | Stg485 | SPEGG2 | ||
| 8 | C | equisimilis | α | Cow | − | − | ||
| 9 | C | equisimilis | β | Cow | ND | SPEGG6 | AB105077 | spegg1 |
| 62 | C | equisimilis | β | Cow | stL2764 | − | ||
| 63 | C | equisimilis | β | Cow | − | − | ||
| 64α | C | equisimilis | β | Cow | − | − | ||
| 64β | C | equisimilis | β | Cow | stL2764 | − | ||
| 65 | C | dysgalactiae | β | Cow | stL2764 | − | ||
| 10 | C | dysgalactiae | α | Cow | − | − | ||
| 12 | C | dysgalactiae | α | Cow | − | − | ||
| SD-1 | C | dysgalactiae | β | Animal | − | SPEGG7 | AB105083 | spegg7 |
| SD-2 | C | dysgalactiae | β | Animal | stL2764 | − | ||
| SD-3 | C | dysgalactiae | β | Animal | − | SPEGG7 | ||
| SD-4 | C | dysgalactiae | β | Animal | − | − | ||
| SD-5 | C | dysgalactiae | β | Animal | − | − | ||
| SD-6 | C | dysgalactiae | β | Animal | stL2764 | SPEGG8 | AB105084 | spegg8 |
| SD-7 | C | dysgalactiae | β | Animal | stL2764 | SPEGG7 | ||
| 16008sα-8 | C | dysgalactiae | β | Animal | stL2764 | − | ||
| 16009sα-9 | C | dysgalactiae | β | Animal | stL2764 | − | ||
| SD-10 | C | dysgalactiae | β | Animal | − | − | ||
| SD-11 | C | dysgalactiae | α | Animal | − | − | ||
| 16021 | C | dysgalactiae | α | Animal | − | − | ||
| 124 | G | equisimilis | β | Animal | stg2028 | − | ||
| 151 | C | dysgalactiae | β | Animal | − | − |
ND, sequencing analysis was not done because of the presence of multiple amplicons.
TABLE 2.
Primers used in this study
| Primer name | Sequenceb |
|---|---|
| (direction)a | |
| A (forward) | TATTAGCTTAGAAAATTAA |
| B (reverse) | GCAAGTTCTTCAGCTTGTTT |
| G1 (forward) | TTAAGGATCCGATGAAATATTAAAAGATTTG |
| G2 (reverse) | CGCCGTCGACCTAGTGCGTTTTTAAGTAGAT |
| G3 (forward) | AAGCCCTTGCAAATGCATCA |
| G4 (forward) | CCTTTATGAACTTCCTCACT |
| G5 (reverse) | TGATTAACTCGACACCAATC |
| G6 (reverse) | AATCTACAGGCGAGCCATGA |
| G7 (forward) | TTAAGGATCCGATGAAATATCAAAAGATTTG |
| G8 (forward) | TTAAGGATCCGATGAAATATTAAATGATTTG |
| G9 (forward) | TTAAGGATCCGATGAAATATTAAAAGATTTG |
| G10 (reverse) | CGCCGTCGACCTAGTGTGTTTTTAAGTAGAT |
| G11 (reverse) | CGCCGTCGACCTAGTGCGTTTTTAAGTAGAT |
| G12 (reverse) | CGCCGTCGACCTAGTGTGTTTTTAAGTAGAC |
Primers A and B for emm typing were designed according to the previous description (http://www.cdc.gov/ncidod/biotech/strep/M-ProteinGene_typing.htm). Primers G1 to G12 were designed based on the sequence of accession no. AJ489606.
The sequences for recognition by the restriction enzymes are indicated in boldface.
Expression and purification of rSPEGG.
DNA fragments encoding the mature form of SPEGG, as speculated from the cleavage site of signal sequence in SPE-G (accession no. AAF60291), were amplified by PCR. Because there was sequence variation among spegg subtypes, two primers containing a BamHI or SalI site were chosen from the list of primers in Table 2. The fragments were amplified and cloned with a TOPO-TA cloning kit (Invitrogen) and then sequenced and subcloned in the corresponding restriction sites of pQE30 (QIAGEN). The resulting pQE30 derivatives were transformed into E. coli. Induction with isopropyl-thio-β-d-galactopyranoside resulted in production of His6-tagged recombinant SPEGGs (rSPEGGs). The His6-tagged proteins were purified by chromatography with chelating Sepharose 4B (Amersham Pharmacia Biotech) preloaded with Ni2+ according to the manufacturer's instructions.
Human and bovine PBMC proliferation assays and bovine T-cell repertoire analysis.
Bovine and human peripheral blood mononuclear cells (PBMCs) were prepared as described previously (23, 36). Briefly, human or bovine PBMCs were obtained from healthy donors and separated by Ficoll-Conray density gradient centrifugation. T-cell-depleted bovine PBMCs were prepared by cell sorting of cells negative for CD2, CD4, and CD8 using an EPICS ALTRA cell sorter (Beckman-Coulter, Fullerton, CA). Purified PBMCs of human, bovine (1 × 105 cells/well), and bovine T-cell-depleted populations in 96-well plates were stimulated with rSPEGG2 to rSPEGG8 or SPE-G. Primary proliferative response was measured by [3H]thymidine uptake assay. Analysis of the Vβ repertories of bovine T cells stimulated by SPEGG was performed as described previously (23). Briefly, bovine PBMCs were incubated with recombinant proteins (1 μg/ml) at 105 cells/well for 3 days, and mRNAs were prepared using ISOGEN (Nippon Gene, Tokyo, Japan) from the induced T-cell blasts. cDNAs synthesized from the mRNA samples by avian myeloblastosis virus reverse transcriptase XL with random DNA hexamers (Takara, Shiga, Japan) were used as templates for PCR amplification of each Vβ fragment using Taq DNA polymerase (Sigma-Aldrich) with 26 5′ Vβ-specific primers and a 3′-Cβ-specific antisense primer.
Reactivity of bovine serum to the rSPEGG proteins.
To detect antibodies reactive to SPEGG in bovine serum, a conventional enyzyme-linked immunosorbent assay (ELISA) was performed. Briefly, each well in Maxisorp ELISA plates (Nunc, Rochester, NY) was filled with 50 μl of rSPEGG solution (1 μg/ml in 50 mM Na-CO3 [pH 9.0]) and incubated for 1 h at room temperature. After blocking the wells with SuperBlock (Pierce, Rockford, IL), the wells were incubated with 1:1,000-diluted bovine serum samples for 1 h at room temperature. After washing the wells, antibodies reactive with the rSPEGGs were detected with horseradish peroxidase-labeled anti-goat immunoglobulin G, which cross-reacts with bovine immunoglobulin G (Biosource Camarillo, CA).
Homology modeling.
Models of SPEGG based on SPEGG2 and SPEGG3 sequences were made based on the SPE-C structure (PDB entry 1AN8) using the program MODELLER (32).
Nucleotide sequence accession number.
The DNA sequences for spegg2 to spegg8 have been deposited in the DDBJ under the accession numbers listed in Table 1.
RESULTS
Distribution of spegg in S. dysgalactiae.
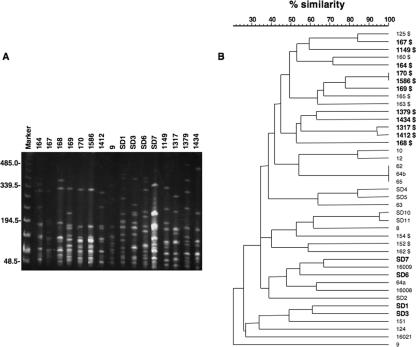
We screened 41 independent isolates of S. dysgalactiae subsp. dysgalactiae (GCS) or S. dysgalactiae subsp. equisimilis (GCS or GGS) from the humans or animals by PCR with spegg-specific primers (G1 and G2; Table 2). Sixteen of the 41 (39%) S. dysgalactiae isolates tested positive for spegg. Nineteen M/emm types were detected among the S. dysgalactiae isolates, although the emm gene was not amplified in 22 isolates with the primer pair used. The emm types of the S. dysgalactiae isolates carrying spegg genes were stg485, stL839, stg11, stc36, stg485, stL2764, stg4831, stg2028, and stg6, respectively. Several reports suggested that types of SPE in GAS strains correlate with their emm types (28). However, in the present study, the emm type did not correlate with the prevalence of spegg in S. dysgalactiae, in accordance with reports of S. dysgalactiae isolates from human subjects (11). PFGE analysis was performed with the isolates after SmaI digestion, and the PFGE profiles were found to vary among the isolates regardless of the presence of spegg (Fig. 1). PFGE profiles also varied among isolates carrying the same spegg variant, except for strains 170 and 1586, both of which carry spegg5. Although the prevalence of spegg was higher in the isolates from humans than in those from animals, spegg was not detected in some of the human isolates, such as 154, 152, and 162.
FIG. 1.
PFGE pattern of S. dysgalactiae isolates carrying spegg genes and a dendrogram constructed by computer-assisted comparison of PFGE patterns generated from all isolates used in this study. In the dendrogram, the isolates carrying spegg are shown in boldface. Isolates from humans are indicated with a dollar sign ($). The scale bar indicates percent similarity among the isolates.
Sequence variation of spegg.
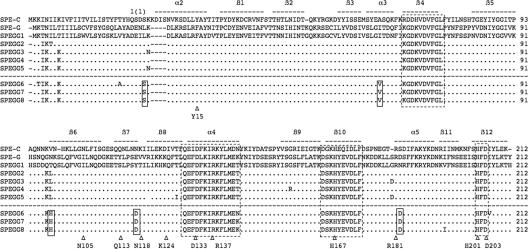
Although the presence of spegg in S. dysgalactiae has been reported (7, 11, 31), characterization of the SPEGG protein encoded by the spegg gene has not been performed. To investigate the possibility that spegg exhibits allelic variations, such as SPE-A and SMEZ, we sequenced 12 of the 16 spegg genes detected in this study (Table 1) and identified 7 different alleles based on the sequences within the open reading frames. None of the genes in the isolates examined in this study were identical to spegg1 reported previously (accession no. AJ294849). SPEGGs showed about 84% similarity to SPE-G. Forty-one mutations were detected at the DNA sequence level. Most of the changes were A/G (31.7%), T/C (19.5%), C/A (9.7%), and T/C (9.7%) transitions, while G/C translation occurred only at position 169. Eighteen of the variable positions resulted in amino acid changes, while the others were synonymous mutations. Interestingly, the positions of the SPEGG mutations in S. dysgalactiae isolates from humans were clearly different from those in isolates from animals, except for mutations at amino acid positions 4, 5, and 8 (IK.K) and 98 and 99 (KL or KH), which were found in both the human and animal isolates (Fig. 2). Because the mutations at amino acid positions 4, 5, and 8 are located in the putative signal peptide sequence, they do not seem to affect function when they are expressed.
FIG. 2.
Multiple alignment of SPE-C, SPE-G, and SPEGG amino acid sequences. The amino acid sequences were aligned using Clustal W (35). The bars above the sequences indicate the secondary structure based on the crystal structure of SPE-C (PDB identification no. 1AN8). Highly conserved regions and the zinc-binding motif are boxed with dotted lines. SPEGG variants derived from isolates from animals (SPEGG6 to SPEGG8) are separated with a dotted line from those from humans, and the residues commonly mutated in SPEGG6 to SPEGG8 are indicated in boxes (also see Fig. 6). Corresponding residues of interest (see text for details) in the alignment are marked with Δ.
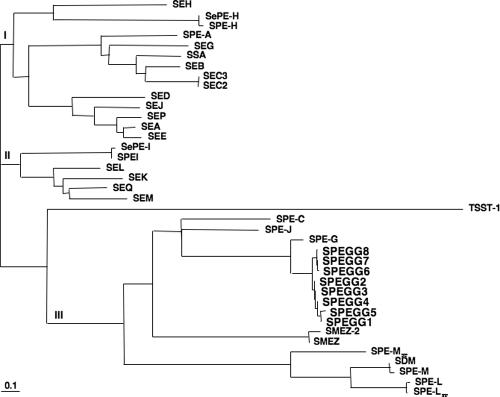
As expected from the sequence similarity, phylogenetic analysis of the SAGs indicated that SPEGGs were related to SPE-G (Fig. 3). There are three major clades in the phylogenetic tree: clades I and II are comprised of GAS, GCS, GGS, and staphylococcal SAGs, while clade III contains only streptococcal SAGs, including SPEGGs. In clade III, SPEGG variants are clearly divided into two subgroups: those in the animal isolates (SPEGG6 to SPEGG8 [SPEGG animal forms]) and those in the human isolates (SPEGG2 to SPEGG5 [SPEGG human forms]).
FIG. 3.
Phylogenetic tree of streptococcal and staphylococcal SAGs, including SPEGGs. This phylogenetic tree was constructed using Clustal W, and the GenBank accession numbers of the SAGs sequences are as follows: SEH, CAI77677; SePE-H, AAF72809; SPE-H, AAK33907; SPE-A, AAL97141; SEG, AAX11325; SSA, AAA65928; SEB, AAL04126; SEC3, AAA26624; SEC2, P34071; SED, P20723; SEJ, AAC78590; SEP, BAB43036; SEA, AAP37183; SEE, P12993; SePE-I, AAF72808; SPE-I, AAL31571; SEL, BAB58170; SEK, AAL04147; SEQ, AAL04146; SEM, AAG36925; TSST-1, BAB58173; SPE-C, AAA27017; SPE-J, AAZ50974; SPE-G, AAZ50801; SMEZ-2, AAD52087; SMEZ, CAD91900; SPE-L, BAC63752; SPE-Lse, CAH65000; SPE-Mse, CAH68555; SPE-M, AAL97849; and SDM, AB074529.
Comparison of SAG activities of rSPEGGs and SPE-G on human and bovine PBMCs.
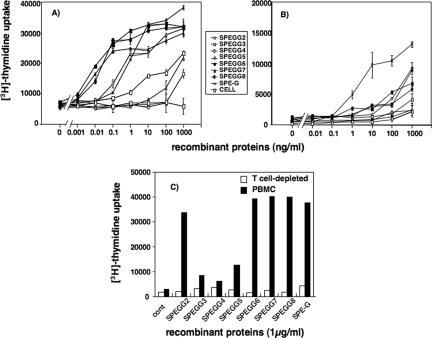
To determine whether SPEGG has SAG properties, we first analyzed the mitogenic activities of the culture supernatants from the S. dysgalactiae isolates. However, we did not detect any mitogenic activities in any of the isolates tested (data not shown), which may be due to the lack of expression of spegg under the bacterial culture conditions used. Therefore, all variants of SPEGGs were expressed in E. coli as recombinant proteins to analyze further their function as SAGs and to determine whether their function was affected by the mutations. rSPEGGs were used to stimulate human or bovine PBMCs, and [3H]thymidine uptake by the cells was measured to analyze their proliferation. Proliferation of bovine PBMCs was induced by the animal forms SPEGG6, SPEGG7, and SPEGG8 at a dose of 0.01 ng/ml, while induction of bovine PBMC proliferation by the human forms, SPEGG2 to SPEGG5, required concentrations of 0.1 ng/ml (Fig. 4). Among the human forms, only SPEGG2 showed higher activity than SPEGG3, SPEGG4, and SPEGG5 against bovine PBMCs. In contrast, concentrations of 100 ng/ml or more of the proteins were required to induce proliferation of human PBMCs. T-cell-depleted bovine PBMCs did not respond to any of the SPEGGs. These findings indicated that SPEGG animal forms can stimulate bovine PBMCs more effectively than the human forms and that stimulation of bovine PBMCs occurs in a T-cell-dependent manner.
FIG. 4.
Stimulation of human and bovine PBMCs with rSPEGGs. PBMCs were isolated from bovine (A) and human (B) blood samples and incubated with various concentrations of rSPEGGs. After 2 days, 0.1 μCi of [3H]thymidine was added, and cells were incubated for a further 18 h before being harvested and counted for the thymidine uptake with a beta counter. These experiments are representative of four independent experiments. In panel C, bovine PBMCs (solid bars) and T-cell-depleted bovine PBMCs (open bars) were stimulated with 1 μg/ml of SPEGGs and SPE-G for 2 days and the mitogenic response was measured as described in Materials and Methods. Serial dilution was performed without SPEGG and used as a negative control (marked as “cell”). cont, control.
Identification of TCR Vβ repertories of bovine T cells reactive to rSPEGGs.
As SPEGG showed T-cell-dependent PBMC stimulation, we next performed reverse transcription-PCR analyses to characterize the Vβ repertories of bovine T cells reactive to SPEGG (Table 3). The Vβ profiles of bovine PBMCs stimulated with rSPEGGs were compared with those stimulated with concanavalin A (ConA) and SPE-G (cow 1 in experiment 1), staphylococcal enterotoxin B (SEB) (cow 2 in experiment 2), or SPE-G (cow 3 in experiment 3). Blasts induced by stimulation with SPEGG2, SPEGG6, SPEGG7, SPEGG8, and SPE-G were significantly richer (two- to sevenfold) in Vβ1,10 and Vβ4 than those stimulated with ConA or SEB (experiment 1). Blasts induced by rSPEGG2, -3, and -6 were slightly richer (about twofold) in Vβ50 in cow 1 but not in cows 2 and 3. Blasts induced by rSPEGG3 and 5 were similar in Vβ50 to those induced by SPE-G in cow 1 but not in cows 2 and 3. These experimental variations might come from individual differences in the responses of T cells in cattle. There were more blasts with Vβ1,10 than with other Vβs, especially in experiments 2 and 3, suggesting that the T cells most reactive to SPEGGs were those with Vβ1,10. These results indicated that T-cell blasts induced by SPEGGs showed a bias in the Vβ repertoire, suggesting that SPEGGs act as SAGs on bovine T cells.
TABLE 3.
TCR Vβ repertories of bovine T cells stimulated with rSPEGG and SPE-G
| Bovine Vβ type | % of Vβ type induced when stimulated bya:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ConA | SEB | rSPEGG2b | rSPEGG3b,c | rSPEGG4c | rSPEGG5c | rSPEGG6 | rSPEGG7 | rSPEGG8 | SPE-G | |
| Expt 1 | ||||||||||
| 1,10 | 16 | 17 | 9 | 0 | 18 | 41 | 0 | 31 | 26 | |
| 4 | 15 | 13 | 8 | 0 | 15 | 40 | 0 | 32 | 24 | |
| 13 | 2 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | |
| 13C | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 18 | 13 | 13 | 0 | 0 | 5 | 3 | 0 | 0 | 2 | |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 27 | 13 | 19 | 26 | 0 | 29 | 3 | 0 | 16 | 24 | |
| 35,91 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 45 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 50 | 6 | 15 | 17 | 0 | 6 | 4 | 0 | 1 | 0 | |
| 82C | 3 | 6 | 3 | 0 | 2 | 2 | 0 | 2 | 0 | |
| 90 | 18 | 17 | 36 | 0 | 21 | 5 | 0 | 17 | 24 | |
| 93 | 12 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | |
| Expt 2 | ||||||||||
| 1,10 | 11 | 52 | 0 | 0 | 0 | 49 | 79 | 54 | ||
| 4 | 9 | 48 | 0 | 0 | 0 | 39 | 21 | 41 | ||
| 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| 13C | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| 18 | 10 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | ||
| 22 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| 27 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| 35,91 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| 45 | 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| 50 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| 82C | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | ||
| 90 | 40 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | ||
| 93 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Expt 3 | ||||||||||
| 1,10 | 0 | 52 | 0 | 0 | 100 | 85 | 100 | 39 | ||
| 4 | 0 | 16 | 0 | 0 | 0 | 12 | 0 | 27 | ||
| 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| 13C | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| 18 | 0 | 21 | 0 | 0 | 0 | 0 | 0 | 8 | ||
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| 27 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 14 | ||
| 35,91 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| 45 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| 50 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 9 | ||
| 82C | 0 | 6 | 0 | 0 | 0 | 1 | 0 | 0 | ||
| 90 | 0 | 5 | 0 | 0 | 0 | 2 | 0 | 2 | ||
| 93 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | ||
Bovine PBMCs were incubated with rSPEGGs (1 μg/ml) for 3 days. The numbers represent the percentage of each Vβ type, and significant responses in the rSPEGG variants are indicated in boldface.
Because of experimental variation, bovine blasts were not induced effectively by rSPEGG2 in experiment 3 and rSPEGG3 in experiment 2.
Induction of blasts was not as effective as for the other rSPEGG variants because of low mitogenic activity of the rSPEGG variants at 1 μg/ml against bovine PBMCs, although the entire experimental procedure was performed for those variants.
Recognition of rSPEGGs by bovine immune system.
There have been no reports of the expression of spegg in S. dysgalactiae isolates, and we could not detect any mitogenic activity in the culture supernatants of isolates carrying spegg. However, spegg may be expressed specifically in vivo. To address this possibility, we quantified antibodies reactive to SPEGG proteins in bovine sera (Fig. 5). Sera from 10 cows were incubated with rSPEGGs to detect the presence of antibodies against these proteins in ELISA. One of the serum samples reacted strongly to all SPEGGs, and the other three samples reacted with SPEGG3, indicating that a certain proportion of the cattle had been exposed to SPEGG.
FIG. 5.
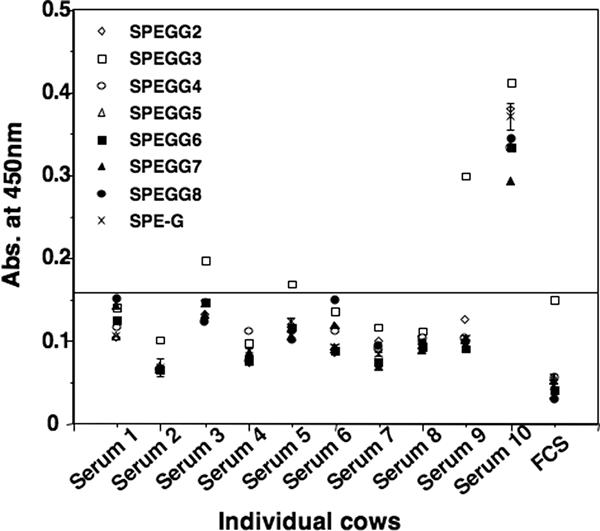
Reactivity of bovine serum to rSPEGGs. Serum samples obtained from 10 cows were diluted 1:1,000, and their reactivity to SPEGG was analyzed using ELISA. Fetal calf serum (FCS) was used as a negative control, and the threshold line is presented according to the absorbance (Abs.) obtained from FCS.
Structural comparison of SPEGG based on homology modeling.
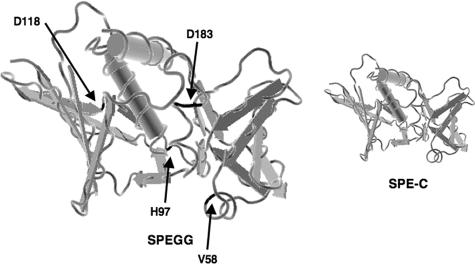
As the SPEGG variants showed marked differences with regard to mitogenic activity, we used a molecular modeling approach to analyze the influences of each mutation found in the SPEGG variants in comparison with the known tertiary structure of SPE-C (Fig. 6). The sequence identity was about 38%. The SPEGG model showed a very good match (0.28-Å root-mean-square deviation) with the SPE-C structure, although three residues of SPEGG did not have equivalent residues in SPE-C; these residues are different in the sequence alignment (Fig. 2) and in the model. These discrepancies are probably due to the difference in the methodologies of the alignment analysis and the modeling approach.
FIG. 6.
Molecular modeling of the SPEGG protein. SPEGG2 and -3 were modeled onto the crystal structures of SPE-C with MODELLER (32). Residues that differed between animal and human forms of SPEGG are shown in black.
Residues needed for intramolecular hydrogen bonds combining N and C domains in SPE-C are conserved in SPEGG, and their positions are structurally very similar to those in SPE-C (2). Salt bridge formation between Asp133 and Arg137 (SPE-C numbering) is also possible in SPEGG. Lys124 in SPE-C is superimposed on His of SPEGG, and this could be a difference between SPEGG and SPE-G because SPE-G has Lys, similar to SPE-C. SPE-C Lys124 forms a salt bridge with SPE-C Glu131. His is also a positively charged residue that can form salt bridges, but the distance between His and Glu can be longer in SPEGG than in SPE-C.
The zinc-binding residues of SPE-C (His167, His201, and Asp203 in SPE-C), which are necessary to provide a binding site for MHC class II in a zinc-dependent manner, are also conserved in SPEGG and SPE-G. MHC class II binding of SPE-G shows clear zinc dependency, and it seems that SPEGG could act in the same way. There is some variation between SPE-C and SPEGG sequences in the residues involved in MHC class II β-chain binding. SPE-C Asn105 (this Asn is conserved in other β-chain binding), which forms a hydrogen bond with MHC class II β-chain, is aligned and superimposed with Ile in SPEGG and SPE-G. Other residues involved in MHC class II binding are conserved, but neither SPEGG nor SPE-G has a residue similar to Gln113 of SPE-C, which is important for peptide binding.
On the other hand, there are clear differences between SPEGG and SPE-C in the residues involved in TCR binding, as judged from the SPE-C complex structures. The SPEGG residue equivalent to SPE-C Tyr15 (important for TCR binding) is replaced with Phe (also in SPE-G). Furthermore, in most cases, other SAGs have Asn in the same position and this residue is important for TCR binding. SPE-G and SPEGG must have some alternative way to bind TCR. Arg181 of SPE-C is another important residue for TCR binding, and both SPEGG and SPE-G have an equivalent Arg residue. Other residues of minor importance in TCR binding of SPE-C are dissimilar in SPEGG and SPE-G.
Taken together, the results of SPEGG modeling suggested strongly that there are some variations in MHC class II and TCR binding in comparison with SPE-C, but the sequence of SPEGG seems to fit the overall three-dimensional structure of SAG. The sequence variations among the SPEGGs probably have no direct effect on TCR or MHC class II binding because the variations are in regions peripheral to the putative binding sites for the MHC and TCR. Thus, the mechanism underlying the differences in mitogenic activities of SPEGG on bovine and human PBMCs and the differences in stimulation of bovine PBMCs between human and animal forms of SPEGG seem to be due to the differences in binding ability of human and bovine forms of SPEGG, which are affected indirectly by variations in the SPEGGs themselves.
DISCUSSION
In this study, we demonstrated that SPEGGs have mitogenic activity toward bovine PBMCs and also, to a lesser extent, toward human PBMCs. The mitogenic activity was dependent on the presence of the T-cell population in PBMCs, and SPEGG selectively activated bovine Vβ1,10- and Vβ4-positive T cells, strongly suggesting that SPEGGs act as SAGs.
Brandt et al. showed that S. dysgalactiae subsp. equisimilis clinical isolates from human invasive infections and from superficial infections lacked expression of spegg and had no mitogenic activity in their culture supernatants (7). As shown in this study, rSPEGG does not stimulate human PBMCs effectively. However, it stimulates bovine PBMCs effectively and a percentage of bovine sera contained antibodies reactive to SPEGG. These observations suggest that SPEGG has some pathogenic and/or physiological roles in cattle but not in humans. In addition, SPEGGs in animals seem to have been adapted to their environment, because all of the animal forms of SPEGG stimulated bovine PBMCs more effectively than the human forms. Igwe et al. reported that 10 of 20 S. dysgalactiae subsp. equisimilis isolates from the human invasive cases were positive for at least one SAG gene (13). Hashikawa et al. reported that 7 of 12 S. dysgalactiae subsp. equisimilis isolates from the human invasive cases were positive for spegg but none was positive for speA, speC, speH, speI, speJ, or speL (11). Brandt et al. reported that 6 of 46 S. dysgalactiae subsp. equisimilis isolates were positive for spegg but none was positive for speA, speC, speH, speI, speJ, speK, speL, speM, smeZ, or ssa (7). Due to the low prevalence and based on our analysis showing low mitogenic activity toward human PBMCs of rSPEGGs from bacteria isolated from the human STSS cases, SAGs of S. dysgalactiae seem to play very limited roles in the severity of invasive S. dysgalactiae subsp. equisimilis infection in humans.
Many SAGs have been shown to be located in the mobile genetic elements, such as defective prophages, active prophages, or bacteriophages, in the GAS genome (1, 14). These SAGs show sequence similarity to those of non-group A streptococci, suggesting that horizontal gene transfer has occurred between GAS and non-GAS. Such horizontal gene transfer may not be the case between SPEGG and SPE-G, which is a homologue of SPEGG in GAS. SPE-G is not located in any apparent mobile genetic elements in all of the GAS strains for which the genome sequences are available, whereas all of them have speG (5, 6, 9, 10, 24, 33, 34). Although Sachse et al. speculated that the region including spegg (speG) in GAS came from S. dysgalactiae, based on a comparison of the genomes of MGAS8232 and GAS SF370 (31), whole-genome data of the other GAS strains, such as MGAS10394 (accession no. NC_006086.1), which became available after their report, suggested that their speculation may not be true. Thus, the common ancestor of GAS and non-GAS may already have a common ancestor in SPEGGs and SPE-G instead of gene transfer among GAS and S. dysgalactiae. If this is true, SPEGG and SPE-G may be direct descendants of one of the ancestral SAGs of streptococci. Further genome-wide analyses of streptococci should provide the answers to these questions.
Acknowledgments
The authors thank T. Fujino of the International Medical Center of Japan for excellent technical assistance.
This study was supported by a grant for research on emerging and reemerging infectious diseases (H12 Shinkou-27) from the Ministry of Health and Welfare, Japan. T. H. was supported by a High-Tech Research Center Project for Private Universities grant/matching-fund subsidy from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT), 2004-2008.
Editor: A. Camilli
Footnotes
Published ahead of print on 5 February 2007.
REFERENCES
- 1.Alouf, J. E., and M. Popoff. 2006. The comprehensive sourcebook of bacterial protein toxins, 3rd ed. Academic Press, San Diego, CA.
- 2.Arcus, V. L., T. Proft, J. A. Sigrell, H. M. Baker, J. D. Fraser, and E. N. Baker. 2000. Conservation and variation in superantigen structure and activity highlighted by the three-dimensional structures of two new superantigens from Streptococcus pyogenes. J. Mol. Biol. 299:157-168. [DOI] [PubMed] [Google Scholar]
- 3.Artiushin, S. C., J. F. Timoney, A. S. Sheoran, and S. K. Muthupalani. 2002. Characterization and immunogenicity of pyrogenic mitogens SePE-H and SePE-I of Streptococcus equi. Microb. Pathog. 32:71-85. [DOI] [PubMed] [Google Scholar]
- 4.Assimacopoulos, A. P., J. A. Stoehr, and P. M. Schlievert. 1997. Mitogenic factors from group G streptococci associated with scarlet fever and streptococcal toxic shock syndrome. Adv. Exp. Med. Biol. 418:109-114. [DOI] [PubMed] [Google Scholar]
- 5.Banks, D. J., S. F. Porcella, K. D. Barbian, S. B. Beres, L. E. Philips, J. M. Voyich, F. R. DeLeo, J. M. Martin, G. A. Somerville, and J. M. Musser. 2004. Progress toward characterization of the group A Streptococcus metagenome: complete genome sequence of a macrolide-resistant serotype M6 strain. J. Infect. Dis. 190:727-738. [DOI] [PubMed] [Google Scholar]
- 6.Beres, S. B., G. L. Sylva, K. D. Barbian, B. Lei, J. S. Hoff, N. D. Mammarella, M. Y. Liu, J. C. Smoot, S. F. Porcella, L. D. Parkins, D. S. Campbell, T. M. Smith, J. K. McCormick, D. Y. Leung, P. M. Schlievert, and J. M. Musser. 2002. Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc. Natl. Acad. Sci. USA 99:10078-10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brandt, C. M., K. G. Schweizer, R. Holland, R. Lutticken, and B. S. Freyaldenhoven. 2005. Lack of mitogenic activity of speG- and speG(dys)-positive Streptococcus dysgalactiae subspecies equisimilis isolates from patients with invasive infections. Int. J. Med. Microbiol. 295:539-546. [DOI] [PubMed] [Google Scholar]
- 8.Eskens, F. A., P. E. Verweij, J. F. Meis, and A. Soomers. 1995. Septic shock caused by group G beta-haemolytic streptococci as presenting symptom of acute myeloid leukaemia. Neth. J. Med. 46:153-155. [DOI] [PubMed] [Google Scholar]
- 9.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA. 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Green, N. M., S. Zhang, S. F. Porcella, M. J. Nagiec, K. D. Barbian, S. B. Beres, R. B. LeFebvre, and J. M. Musser. 2005. Genome sequence of a serotype M28 strain of group a streptococcus: potential new insights into puerperal sepsis and bacterial disease specificity. J. Infect. Dis. 192:760-770. [DOI] [PubMed] [Google Scholar]
- 11.Hashikawa, S., Y. Iinuma, M. Furushita, T. Ohkura, T. Nada, K. Torii, T. Hasegawa, and M. Ohta. 2004. Characterization of group C and G streptococcal strains that cause streptococcal toxic shock syndrome. J. Clin. Microbiol. 42:186-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirose, Y., K. Yagi, H. Honda, H. Shibuya, and E. Ozaki 1997. Toxic shock-like syndrome caused by non-group A beta-hemolytic streptococci. Arch. Intern. Med. 157:1891-1894. [PubMed] [Google Scholar]
- 13.Igwe, E. I., P. L. Shewmaker, R. R. Facklam, M. M. Farley, C. van Beneden, and B. Beall. 2003. Identification of superantigen genes speM, ssa, and smeZ in invasive strains of beta-hemolytic group C and G streptococci recovered from humans. FEMS Microbiol. Lett. 229:259-264. [DOI] [PubMed] [Google Scholar]
- 14.Ikebe, T., A. Wada, Y. Inagaki, K. Sugama, R. Suzuki, D. Tanaka, A. Tamaru, Y. Fujinaga, Y. Abe, Y. Shimizu, H. Watanabe, and the Working Group for Group A Streptococci in Japan. 2002. Dissemination of the phage-associated novel superantigen gene speL in recent invasive and noninvasive Streptococcus pyogenes M3/T3 isolates in Japan. Infect. Immun. 70:3227-3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keiser, P., and W. Campbell. 1992. ‘Toxic strep syndrome’ associated with group C Streptococcus. Arch. Intern. Med. 152:882-884. [DOI] [PubMed] [Google Scholar]
- 16.Kline, J. B., and C. M. Collins. 1996. Analysis of the superantigenic activity of mutant and allelic forms of streptococcal pyrogenic exotoxin A. Infect. Immun. 64:861-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kotb, M., A. Norrby-Teglund, A. McGeer, H. El-Sherbini, M. T. Dorak, A. Khurshid, K. Green, J. Peeples, J. Wade, G. Thomson, B. Schwartz, and D. E. Low. 2002. An immunogenetic and molecular basis for differences in outcomes of invasive group A streptococcal infections. Nat. Med. 8:1398-1404. [DOI] [PubMed] [Google Scholar]
- 18.Kotzin, B. L., D. Y. Leung, J. Kappler, and P. Marrack. 1993. Superantigens and their potential role in human disease. Adv. Immunol. 54:99-166. [DOI] [PubMed] [Google Scholar]
- 19.Kugi, M., H. Tojo, I. Haraga, T. Takata, K. Handa, and K. Tanaka. 1998. Toxic shock-like syndrome caused by group G Streptococcus. J. Infect. 37:308-309. [DOI] [PubMed] [Google Scholar]
- 20.Miyoshi-Akiyama, T., J. Zhao, H. Kato, K. Kikuchi, K. Totsuka, Y. Kataoka, M. Katsumi, and T. Uchiyama. 2003. Streptococcus dysgalactiae-derived mitogen (SDM), a novel bacterial superantigen: characterization of its biological activity and predicted tertiary structure. Mol. Microbiol. 47:1589-1599. [DOI] [PubMed] [Google Scholar]
- 21.Miyoshi-Akiyama, T., J. Zhao, K. Kikuchi, H. Kato, R. Suzuki, M. Endoh, and T. Uchiyama. 2003. Quantitative and qualitative comparison of virulence traits, including murine lethality, among different M types of group A streptococci. J. Infect. Dis. 187:1876-1887. [DOI] [PubMed] [Google Scholar]
- 22.Musser, J. M., V. Kapur, S. Kanjilal, U. Shah, D. M. Musher, N. L. Barg, K. H. Johnston, P. M. Schlievert, J. Henrichsen, D. Gerlach, et al. 1993. Geographic and temporal distribution and molecular characterization of two highly pathogenic clones of Streptococcus pyogenes expressing allelic variants of pyrogenic exotoxin A (Scarlet fever toxin). J. Infect. Dis. 167:337-346. [DOI] [PubMed] [Google Scholar]
- 23.Mwaka, M., T. Kato, R. Hayashi, T. Abe, T. Miyoshi-Akiyama, T. Uchiyama, and K. Imanishi. 2003. Superantigenic stimulation of bovine T cells by Streptococcus dysgalactiae-derived mitogen (SDM). J. Tokyo Women's Med. Univ. 73:24-34. [Google Scholar]
- 24.Nakagawa, I., K. Kurokawa, A. Yamashita, M. Nakata, Y. Tomiyasu, N. Okahashi, S. Kawabata, K. Yamazaki, T. Shiba, T. Yasunaga, H. Hayashi, M. Hattori, and S. Hamada. 2003. Genome sequence of an M3 strain of Streptococcus pyogenes reveals a large-scale genomic rearrangement in invasive strains and new insights into phage evolution. Genome Res. 13:1042-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Natoli, S., C. Fimiani, N. Faglieri, L. Laurenzi, A. Calamaro, A. M. Frasca, and E. Arcuri. 1996. Toxic shock syndrome due to group C streptococci. A case report. Intensive Care Med. 22:985-989. [DOI] [PubMed] [Google Scholar]
- 26.Ojukwu, I. C., D. W. Newton, A. E. Luque, M. Y. Kotb, and M. Menegus. 2001. Invasive group C Streptococcus infection associated with rhabdomyolysis and disseminated intravascular coagulation in a previously healthy adult. Scand. J. Infect. Dis. 33:227-229. [DOI] [PubMed] [Google Scholar]
- 27.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 28.Proft, T., S. L. Moffatt, K. D. Weller, A. Paterson, D. Martin, and J. D. Fraser. 2000. The streptococcal superantigen SMEZ exhibits wide allelic variation, mosaic structure, and significant antigenic variation. J. Exp. Med. 191:1765-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Proft, T., P. D. Webb, V. Handley, and J. D. Fraser. 2003. Two novel superantigens found in both group A and group C Streptococcus. Infect. Immun. 71:1361-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reda, K. B., V. Kapur, D. Goela, J. G. Lamphear, J. M. Musser, and R. R. Rich. 1996. Phylogenetic distribution of streptococcal superantigen SSA allelic variants provides evidence for horizontal transfer of ssa within Streptococcus pyogenes. Infect. Immun. 64:1161-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sachse, S., P. Seidel, D. Gerlach, E. Gunther, J. Rodel, E. Straube, and K. H. Schmidt. 2002. Superantigen-like gene(s) in human pathogenic Streptococcus dysgalactiae, subsp equisimilis: genomic localisation of the gene encoding streptococcal pyrogenic exotoxin G (speG(dys)). FEMS Immunol. Med. Microbiol. 34:159-167. [DOI] [PubMed] [Google Scholar]
- 32.Sali, A., and T. L. Blundell. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234:779-815. [DOI] [PubMed] [Google Scholar]
- 33.Smoot, J. C., K. D. Barbian, J. J. Van Gompel, L. M. Smoot, M. S. Chaussee, G. L. Sylva, D. E. Sturdevant, S. M. Ricklefs, S. F. Porcella, L. D. Parkins, S. B. Beres, D. S. Campbell, T. M. Smith, Q. Zhang, V. Kapur, J. A. Daly, L. G. Veasy, and J. M. Musser. 2002. Genome sequence and comparative microarray analysis of serotype M18 group A Streptococcus strains associated with acute rheumatic fever outbreaks. Proc. Natl. Acad. Sci. USA 99:4668-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sumby, P., S. F. Porcella, A. G. Madrigal, K. D. Barbian, K. Virtaneva, S. M. Ricklefs, D. E. Sturdevant, M. R. Graham, J. Vuopio-Varkila, N. P. Hoe, and J. M. Musser. 2005. Evolutionary origin and emergence of a highly successful clone of serotype M1 group a Streptococcus involved multiple horizontal gene transfer events. J. Infect. Dis. 192:771-782. [DOI] [PubMed] [Google Scholar]
- 35.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uchiyama, T., Y. Kamagata, X. J. Yan, M. Kohno, M. Yoshioka, H. Fujikawa, H. Igarashi, M. Okubo, F. Awano, T. Saito-Taki, et al. 1987. Study of the biological activities of toxic shock syndrome toxin-1. II. Induction of the proliferative response and the interleukin 2 production by T cells from human peripheral blood mononuclear cells stimulated with the toxin. Clin. Exp. Immunol. 68:638-647. [PMC free article] [PubMed] [Google Scholar]
- 37.Uchiyama, T., X. J. Yan, K. Imanishi, and J. Yagi. 1994. Bacterial superantigens—mechanism of T cell activation by the superantigens and their role in the pathogenesis of infectious diseases. Microbiol. Immunol. 38:245-256. [DOI] [PubMed] [Google Scholar]
- 38.Wagner, J. G., P. M. Schlievert, A. P. Assimacopoulos, J. A. Stoehr, P. J. Carson, and K. Komadina. 1996. Acute group G streptococcal myositis associated with streptococcal toxic shock syndrome: case report and review. Clin. Infect. Dis. 23:1159-1161. [DOI] [PubMed] [Google Scholar]