Abstract
Enterococci are commensal organisms of the gastrointestinal (GI) tracts of a broad range of mammalian and insect hosts, but they are also leading causes of nosocomial infection. Little is known about the ecological role of enterococci in the GI tract consortia. To develop a tractable model for studying the roles of these organisms as commensals and pathogens, we characterized the Drosophila melanogaster microflora and examined the occurrence of enterococci in the gastrointestinal consortium of Drosophila. In a survey of laboratory-reared Drosophila and wild-captured flies, we found that Drosophila was naturally colonized by representatives of five bacterial phyla. Among these organisms were several species of enterococci, including Enterococcus faecalis, Enterococcus faecium, Enterococcus gallinaraum, and Enterococcus durans, as well as a previously detected but uncultured Enterococcus species. Drosophila could be cured of enterococcal carriage by antibiotic treatment and could be reassociated with laboratory strains. High-level colonization by a well-characterized strain expressing the enterococcal cytolysin was found to be detrimental to Drosophila compared to the effect of an isogenic, noncytolytic control. The anatomical distribution of enterococci in the Drosophila GI tract was determined by immunohistochemical staining of thin sections of naturally colonized and reassociated flies.
Enterococci are among the most common causes of hospital-acquired infections (24, 38). The enterococci that cause these infections are often resistant to multiple antibiotics and, with increasing frequency, to all antibiotics (38). It was recently proposed (30) that nosocomial enterococcal infections may be the result of a two-step process: asymptomatic colonization and amplification within the gastrointestinal (GI) tract of virulent, antibiotic-resistant enterococcal strains, followed by infection of extraintestinal sites, such as the bloodstream, urinary tract, or a surgical wound. However, little is known about the mechanisms used by enterococci to colonize the GI tracts of either healthy individuals or hospitalized patients. Studies of mammalian GI tract colonization and the transition to infection are exceedingly complex because of the number of variables involved. An estimated 500 taxa are represented among the bacterial species in the consortium in the human colon (23, 71, 72), and species representation in this consortium varies by age, diet, and genetics. This complexity is further compounded by the health status of the host and by antimicrobial therapy (72).
Enterococci are present naturally in the GI tracts of insects (8, 11, 49, 50, 70), reptiles (55), humans, and other mammals (1, 38, 55, 59). Enterococci are minority members of the human GI tract consortium, comprising up to 1% of the adult microflora (65). Enterococcus faecalis and Enterococcus faecium are the two species most commonly detected in the human bowel (27, 59, 72), while Enterococcus durans occurs in a small percentage of adults (72). Martin and Mundt (50) recognized the association of E. faecalis, E. faecium, and Enterococcus casseliflavus with a broad range of insect orders, including the Diptera. They found that E. faecalis, E. faecium, and E. casseliflavus were associated with 32, 22.4, and 43.5%, respectively, of wild insect isolates in 37 different taxa, but they did not include Drosophila in their analysis. Enterococci are auxotrophic for a number of vitamins, amino acids and other micronutrients (58). Nevertheless, they are remarkably resilient organisms that are capable of enduring a broad pH range, as well as hypotonic and hypertonic conditions, characteristics that likely contribute to their presence in insects, such as Drosophila, that possess diverticulated GI tracts consisting of acidic crops, highly alkaline midguts, and neutral to acidic hindguts (16, 21, 52).
Because enterococci have been reported to be commensal organisms of insects and because Drosophila is a genetically tractable model for studying host-pathogen interactions, it was of interest to characterize the microbial consortium of the fruit fly and to determine whether enterococci are a native component of that consortium. It was also of interest to determine whether the consortium could be manipulated to study the roles of enterococcal traits during colonization and infection.
MATERIALS AND METHODS
Stocks, strains, and growth conditions.
Laboratory-reared fly stocks were acquired from several geographically distinct facilities. Oregon R Bloomington and Canton S Bloomington stocks were obtained from the Drosophila Stock Center at Indiana University in Bloomington. Oregon R OU and Canton S OU stocks were obtained from James Thompson, Department of Zoology, University of Oklahoma in Norman. Oregon R UT stocks were donated by Glen Collier, Department of Biological Sciences, University of Tulsa, Tulsa, OK. Wild Drosophila melanogaster adults were captured during the summer of 2005 using fermenting banana traps (6) in two separate locations. The flies in the first group, designated OK, were collected in Oklahoma City, OK. The flies in the second group, designated MA, were collected in Cambridge, MA. All stocks were maintained on standard, sterilized cornmeal-molasses food (6) at 23°C, unless otherwise noted. Samples of newly acquired fly stocks were either surface decontaminated, homogenized, and plated immediately for quantification and identification of the commensal flora or frozen in 25% glycerol at −70°C for later use. Adult flies were confirmed to be D. melanogaster by physical examination of the genital structures (6).
Isogenic strains of E. faecalis (E. faecalis FA2-2/pAM714 [cytolytic] and FA2-2/pAM771 [noncytolytic]) (40) were used to reassociate Drosophila. These strains were routinely cultured in brain heart infusion (BHI) medium (Difco) at 37°C with antibiotic selection (50 μg/ml erythromycin).
Quantification of cultivable bacteria and Enterococcus in laboratory-reared and wild-captured fly stocks.
In order to increase the detection limit for identifying members of the Drosophila microbial consortium, pools of 10 flies each from the Oregon R Bloomington and Oregon R OU stocks and the wild-captured stocks from Massachusetts (MA) and Oklahoma (OK) were rinsed with 70% ethanol for surface decontamination, transferred to microcentrifuge tubes, and homogenized with disposable pellet pestles (Kontes, Vineland, NJ) in 500 μl sterile phosphate-buffered saline (PBS). Homogenates were serially diluted in PBS and spread plated on bile esculin azide agar (Difco, Sparks, MD) for enumeration of presumed enterococci and on BHI agar and Chromagar Orientation plates (BBL) for detection and differentiation of the cultivable flora. Plates were incubated at 37°C overnight under both aerobic and anaerobic conditions. Following incubation, colony morphological characteristics were recorded. Each colony type was then quantified to determine the number of CFU per fly.
Bacterial identification. (i) Culturable isolates.
For 16S rRNA gene sequence identification of culturable isolates, colonies representing each morphological type obtained on the three different media used were streaked for isolation on BHI agar and incubated overnight at 37°C under aerobic or anaerobic conditions. Following incubation, single colonies were suspended in 50 μl sterile water, and a colony PCR was performed using 16S rRNA gene eubacterial oligonucleotide primers 27F and 1492R (Integrated DNA Technologies, Coralville, IA) as previously described (45). Each 50-μl PCR mixture consisted of 1 μl of a bacterial colony suspension, 1 μl of a 10 μM forward primer 27F solution, 1 μl of a 10 μM reverse primer 1492R solution, 0.8 μl of a 10 mM deoxynucleotide triphosphate solution (Invitrogen, Carlsbad, CA), 5 μl of 10× Taq polymerase buffer (Promega, Madison, WI), and 1 μl of Taq DNA polymerase (5 U; Promega). The thermal cycling conditions included an initial 3-min denaturation step at 94°C, 30 cycles of 94°C for 30 s, 55°C for 90 s, and 72°C for 2.5 min, and a final 10-min extension at 72°C. All reactions were carried out in 0.2-ml reaction tubes in a Techne TC-412 thermocycler (Techne, Burlington, NJ). PCR products were confirmed by electrophoresis through a 1% agarose gel and were visualized by ethidium bromide staining.
(ii) Culture-independent identification.
For culture-independent identification of Drosophila commensal flora, pools of 10 flies were individually rinsed and homogenized as described above. Total DNA was then extracted from fly homogenates by a previously described method (3), modified as described by Broderick et al. (8). Briefly, fly homogenates were sonicated twice at 60 Hz for 30 s in a bench-top sonicator (PC3; L&R Ultrasonics, Kearny, NJ). To each homogenate, 60 μl of 10% sodium dodecyl sulfate (Sigma) and 3 μl of a 20-mg/ml proteinase K (Amresco, Solon, OH) solution were added and incubated for 90 min at 37°C. The homogenates were then pelleted by centrifugation for 30 s in a bench-top minicentrifuge (ISC Bioexpress, Kayesville, UT) to remove any chitinous debris (8). Following lysis and proteolytic digestion, 100 μl of 5 M NaCl and then 80 μl of 10% cetyltrimethylammonium bromide-0.7 M NaCl were added. The mixture was then incubated for 1 h at 65°C. DNA was extracted with equal volumes of chloroform-isoamyl alcohol (24:1, vol/vol) and phenol-chloroform-isoamyl alcohol (25:24:1, vol/vol/vol). DNA was precipitated with isopropanol and pelleted by centrifugation, and this was followed by resuspension in 100 μl of Tris-EDTA buffer. The DNA concentration and purity were determined by determining the absorbance at 260 nm/absorbance at 280 nm, and the preparations were stored at −20°C.
Eubacterial total 16S DNA in the extracts was amplified using primers 27F and 1492R described above. The PCR products were electrophoresed through 1% agarose, visualized by ethidium bromide staining, sliced from the gel, and extracted using a QIAquick gel extraction kit (QIAGEN, Valencia, CA). Purified PCR products were then ligated into the TOPO TA cloning plasmid pCR4-TOPO (Invitrogen, Carlsbad, CA) used according to the manufacturer's instructions. Clone libraries were created by transforming One Shot (Invitrogen), chemically competent Escherichia coli cells. Transformants were selected on LB agar containing 100 μg/ml ampicillin (Sigma) and 40 μg/ml 5-bromo-4-chloro-3- indolyl-β-d-galactopyranoside (X-Gal) (Sigma). Well-isolated white colonies were suspended in 50 μl distilled H2O. The cloned rRNA gene sequence was selectively PCR amplified using M13 forward and reverse primers (Promega) and the conditions described above. Amplified products from approximately 250 clones for each fly stock were then sequenced.
16S rRNA sequencing and analysis.
Sequencing reactions were performed by the DNA Sequencing Center for Vision Research at the Ocular Molecular Genetics Institute, Massachusetts Eye and Ear Infirmary, using the BigDye Terminator V3.1 cycle sequencing Ready Reaction mixture (Applied Biosystems, Foster City, CA). Prior to sequencing, all PCR products were treated with ExoSAP-It (Amersham Biosciences, Piscataway, NJ). PCR products were sequenced using universal eubacterial 16S rRNA primers 27F, 1492R, 704F, and 787R (45). All 16S rRNA sequences were compiled using the Codon Code Aligner software (Codon Code Corporation, Dedham, MA).
Phylogenetic analysis.
Approximately 250 16S rRNA gene sequences derived from each fly stock (a total of 1,000 sequences) were analyzed by individual fly of origin and collectively for all flies. Sequences were aligned with Clustal X (73) and were compared to the sequences in Ribosomal Database Project II (RDP-II) (18), using the Sequence Match function. Prior to phylogenetic analysis, each sequence was manually edited by examination of its sequencing chromatogram and tested as a possible chimera using Bellerophon (35) and Chimera Check v2.7 (part of the online analysis tool suite provided at the RDP-II website [http://wdcm.nig.ac.jp/RDP/cgis/chimera.cgi?su=SSU]) (18). All chimeric sequences and poor-quality sequences were excluded from further analysis. Single representatives of each phylotype from the combined libraries were then aligned using Clustal X. The resulting alignment was used to construct a neighbor-joining tree using the PAUP program (version 4; Sinauer Associates, Sunderland, MA) (http://paup.csit.fsu.edu/). Optimal PAUP input settings for tree construction were determined using Modeltest v3.7 (61). Trees were constructed using a general time-reversible model using base frequency and substitution values and gamma distribution shape parameters indicated by Modeltest. The tree topology was tested by bootstrap analysis with 1,000 iterations.
Phylotype determination.
Jukes-Cantor-corrected distance matrices (42) were constructed for individual sequence libraries, as well as for the combined library, using the DNAdist program in the PHYLIP v3.65 program suite, and they were analyzed using DOTUR (63) to identify phylotypes at multiple similarity cutoffs using the furthest-neighbor algorithm. Sequences with at least 97% similarity were assigned to operational taxonomic units (OTUs) as previously described (63).
Estimation of microbial diversity.
Collector's and rarefaction curves, richness estimates, and diversity indices were generated for individual libraries and for the combined data set with DOTUR, using a 97% similarity cutoff. Collector's and rarefaction curves were plotted using observed phylotype accumulation and the Chao1 estimator of species richness (13, 14). Chao1 estimates were generated after 1,000 randomizations without replacement. The percent coverage achieved by sequencing efforts was calculated by Good's method (31), using the formula (1 − n/N) × 100, where n is the number of singletons (phylotypes represented by a single clone) in a sample and N is the total number of sequences in the sample.
Diversity statistics were calculated using the Shannon and Simpson indices. The Shannon index of diversity (H′) was calculated using the formula H′ = −Σpi(ln pi), where pi is the abundance of phylotypes, calculated from n/nN (where n is the number of sequences in each OTU and N is the total number of sequences in the data set). The Simpson index of diversity (D) was calculated using the formula D = Σn(n − 1)/N(N − 1), and the results are reported below as the reciprocal (1/D). The Simpson index of evenness was calculated with the formula (1/D)/S, where S is the number of phylotypes in the data set.
Examination of shared phylotypes and similarity between fly stocks.
To determine the fraction of phylotypes shared by our Drosophila 16S rRNA gene libraries, the microbial consortia of Oregon R OU, Oregon R Bloomington, and wild-captured MA fly stocks were compared using SONS (64) (http://www.plantpath.wisc.edu/fac/joh/sons.html). The estimated phylotype richness within each library (determined using DOTUR as described above) was compared to the shared phylotypes in each possible combination of libraries at a level of similarity of 97%. The statistical estimates of the phylotypes determined to be shared by the three libraries were plotted by using a Venn diagram.
Antibiotic curing of enterococci from the Drosophila consortium.
Indigenous enterococci were assessed to determine their susceptibilities to erythromycin in vitro and were found to be uniformly sensitive to a concentration of 1 μg/ml or less. Therefore, 50 μg/ml erythromycin was added to freshly prepared, sterilized fly food. Oregon R Bloomington flies were then transferred to erythromycin-containing food for 24 h. Following removal of flies from the antibiotic-containing medium, repeated culture of fly homogenates revealed that detectable enterococci were not present during the analysis.
Stable colonization after removal from bacterium-infused food.
To examine the reassociation of Drosophila with E. faecalis, 1 × 109 cells of an E. faecalis strain, FA2-2, which was derived from a human clinical isolate (17) by selecting for spontaneous chromosomal mutations mediating resistance to rifampin and fusidic acid, were stirred into 50 ml of sterile fly food. For each experiment, 100 adult, erythromycin-treated Oregon R Bloomington flies were exposed to the E. faecalis-seeded food for 24 h at 30°C. The flies were then transferred to fresh, sterile food without antibiotics. Following the transfer to sterile food, 10 flies were homogenized at 24-h intervals for 7 days, and enterococci were enumerated as described above.
Cytolysin lethality in Drosophila.
To examine the effects of the enterococcal cytolysin on colonization of Drosophila, erythromycin-treated flies were placed on food containing either cytolytic strain FA2-2/pAM714 or the isogenic noncytolytic strain FA2-2/pAM771 (39). Plasmids pAM714 and pAM771 were derived from the E. faecalis cytolysin-encoding plasmid pAD1 by Tn917 insertional mutagenesis (39); Tn917 was inserted adjacent to the cytolysin operon in pAM714, leaving the operon intact, and this insertionally inactivated toxin precursor production in pAM771 (39). These plasmids are stably maintained in the E. faecalis host by an active partitioning system (77, 78). Flies treated with only erythromycin and then placed on sterile medium were included as controls. In these experiments, flies were examined every 24 h for 7 days, and the number of dead flies was recorded each day. The colonization rates for both the cytolytic and noncytolytic strains were determined in parallel by homogenization of 10 flies every 48 h, and the numbers of viable enterococci per fly were determined as described above using erythromycin-containing media. The number of viable enterococci per gram of fly food at each time was also determined for each experiment. The ability of erythromycin to cure flies of indigenous enterococci was verified for each experiment, and reassociation was verified after 24 h of feeding on Enterococcus-containing food to ensure that there was colonization of test populations. Killing curves were generated for each fly type-isogenic mutant strain pair. All experiments were conducted in triplicate.
Immunohistology.
Drosophila adults, either from an unperturbed laboratory-maintained wild-type stock or artificially colonized with E. faecalis FA2-2, were fixed in 37% paraformaldehyde in water and octane, using a method developed by Richard Carthew, Northwestern University, Chicago, IL (personal communication). Briefly, flies were anesthetized with ether (J. T. Baker) and transferred to a 2-ml microtube containing an octane fixation solution prepared by mixing 3.38 ml of 37% (wt/vol) paraformaldehyde prepared in water, 5 ml of octane (Sigma), and 0.5 ml of 1 M Na2PO4 (pH 6.8). Flies were fixed at room temperature for 20 min. Flies were then washed once with pure octane and then with PBS. Fixed and rinsed flies were suspended in a postfixation solution, which was prepared by mixing 430 μl of 37% paraformaldehyde in water, 400 μl of 1 M Na2PO4 (pH 6.8), and 3.17 ml of water, without octane, for 1 h. Fixed flies were rinsed twice with PBS, embedded in paraffin, and sectioned. Thin sections (10 μm) were mounted on charged glass slides (Colorfrost Plus; Fisher Scientific, Pittsburgh, PA) and stained using immunological reagents.
For immunohistochemical staining, antienterococcal polyclonal antibodies were produced by immunizing rabbits with heat-killed E. faecalis FA2-2. To determine whether this polyclonal antiserum exhibited any cross-species activity with other cultivable members of the Drosophila microflora, pre- and postimmune sera were compared by colony lift Western blotting and enzyme-linked immunosorbent assays. Colony lift Western blotting was performed as described previously (80), with modifications. Single colonies of native enterococci and other organisms cultured from Drosophila (including species of Serratia, Bacillus, Pseudomonas, and Staphylococcus from wild-captured and laboratory stocks, as well as E. faecalis FA2-2 as a control) were examined.
Whole-cell enzyme-linked immunosorbent assays were also performed for each of the bacterial isolates as described previously (4) to quantify cross-reactivity. Polyclonal antiserum, serially diluted twofold from 1:800 to 1:1,638,400, was tested. Secondary goat anti-rabbit immunoglobulin G alkaline phosphatase conjugate (Sigma), diluted 1:500, was used for detection. p-Nitrophenyl phosphate hydrolysis was then measured spectrophotometrically at 405 nm, using a Genios 96-well plate reader (Tecan, Grödig, Austria).
As cross-reactivity was observed, antibodies were affinity purified by a method adapted from the method of Campbell et al. (12). Briefly, washed, gentamicin-killed enterococci were resuspended in 0.1 volume of antienterococcal antiserum and incubated for 1 h at 4°C with rocking. Antibody-bound cells were pelleted, washed with PBS, resuspended in an equal volume of 100 mM glycine elution buffer (pH 2.5), and incubated at 4°C for 20 min. Cells were then pelleted and discarded, and an equal volume of 1 M Tris (pH 8.0) was added to neutralize the supernatant.
For immunohistochemical staining, slides were deparaffinized, blocked by incubation with 1% bovine serum albumin (Sigma) and 10% normal horse serum (Vector Laboratories, Burlingame, CA), and incubated with antienterococcal antiserum or with preimmune serum as a specificity control. Antibody bound to enterococci was fluorescently labeled by incubation with a secondary, Alexaflour 594-labeled goat anti-rabbit antibody (Molecular Probes). Drosophila tissue was counterstained using Vectashield with 4′,6′-diamidino-2-phenylindole (DAPI) (Vector Laboratories). Immunohistochemically stained sections were then observed by fluorescence microscopy using a Nikon Eclipse E800 microscope (Nikon, El Segundo, CA), and images were obtained using a Spot 1.10 digital camera (Spot Diagnostic Instruments Inc., Sterling Heights, MI).
RESULTS
Phylogenetic analysis.
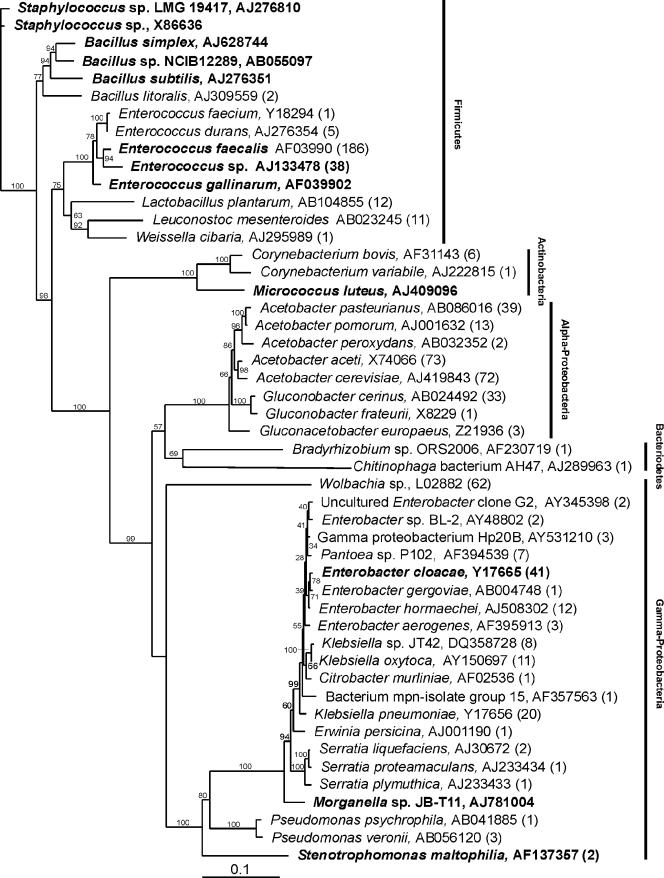
Figure 1 shows a phylogenetic tree consisting of 49 phylotypes identified in a combined 16S rRNA gene sequence library, which was made up of 686 clones from three Drosophila stocks (Oregon R OU, Oregon R Bloomington, and MA), as well as from cultured phylotypes isolated from fly homogenates. Sequences with at least 97% similarity were grouped into OTUs (phylotypes), as calculated with DOTUR. A fourth library, constructed from randomly selected, PCR-amplified eubacterial 16S rRNA genes isolated from the wild-captured OK fly stock, was found to consist of only two phylotypes, an unidentified Wolbachia species (239 clones, 95.6%) and an unidentified Enterococcus species (11 clones, 4.4%). By culturing, we identified nine other phylotypes that had unique colony morphologies on the media used. These phylotypes were identified as E. faecalis, an unidentified Enterococcus isolate, Bacillus subtilis, Bacillus cereus, Bacillus pumilus, Leuconostoc mesenteroides, Lactobacillus plantarum, Serratia marcescens, and Pseudomonas putida. However, because of the overwhelming infection of this fly line with Wolbachia, a pathogen of the Drosophila reproductive tract and other organ systems (15, 51), an analysis of the diversity of the flora of this fly line using the culture-independent approach was not possible.
FIG. 1.
Phylogenetic tree of the microbial flora identified in four separately reared Drosophila stocks. Phylotypes identified by alignment with the RDP-II database are followed by GenBank accession numbers for previously identified strains. The number of times that a clone of each phylotype occurred is indicated in parentheses. Cultured phylotypes are indicated by bold type, but because plating efficiencies were not determined, numbers are not indicated unless the phylotypes were also observed in the less biased rRNA gene census. The tree was constructed with PAUP by neighbor-joining analysis using a general time-reversible model. Bootstrap values calculated from 1,000 tree iterations are indicated at branch points. The scale bar indicates evolutionary distance (10 substitutions per 100 nucleotides).
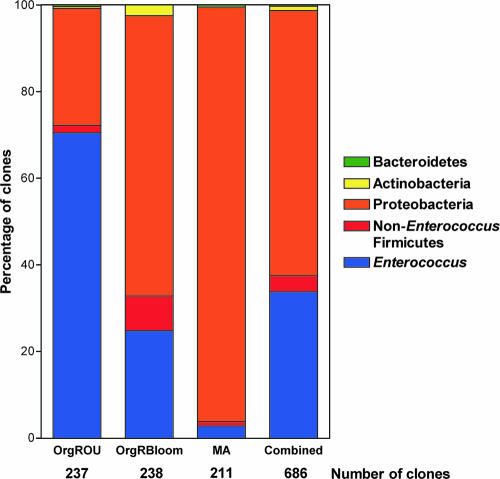
As shown in Fig. 1, the combined library was composed of sequences representing five bacterial phyla. Among these, we detected both low- and high-G+C-content bacterial species, all of which were either strict aerobes or facultatively anaerobic (28). Members of the Firmicutes (low-G+C-content gram-positive bacteria) accounted for 37.3% of the library and were represented by 256 clones and 14 phylotypes (Fig. 1 and 2). Enterococcus was the predominant Firmicutes phylotype observed, accounting for 90% of the phylum (230 clones, five phylotypes). Of the enterococci, E. faecalis was by far the most abundant (81%, 186 clones). An unidentified Enterococcus species was the next most abundant (16.5%, 38 clones). E. durans accounted for 2.2% (five clones) of the enterococcal phylotypes. E. faecium and Enterococcus gallinarum were found to account for less than 1% of the total enterococcal population. The remainder of the Firmicutes phylotypes observed constituted a small fraction of this phylum (Fig. 2). L. plantarum (12 clones) accounted for 4.7% of the total Firmicutes population, while L. mesenteroides (11 clones) accounted for 4.3%. Members of the genera Staphylococcus, Bacillus, and Weissella accounted for less than 1% each of the Firmicutes phylotypes.
FIG. 2.
Individual variation within phylotypes occurring in individual Drosophila stocks. Clones were grouped into bacterial phyla based on their positions in Fig. 1. Relative phylotype frequencies, including the percentages of Enterococcus clones, are shown for individual fly libraries and for the combined data set.
The Proteobacteria was the most abundant phylum (61%, 419 clones) (Fig. 2), and it was represented by members of the α- and γ-Proteobacteria. The α-Proteobacteria accounted for 32 56.3% of the Proteobacteria, with 236 clones and eight phylotypes. Acetobacter was the most abundant genus observed in this study, with 199 clones accounting for 29% of all phylotypes and 84.3% of the α-Proteobacteria. Among these, Acetobacter aceti (73 clones, 31%) and Acetobacter cerevisiae (72 clones, 30.5%) were the most abundant phylotypes. Other Acetobacter phylotypes observed included Acetobacter pasteurianus (39 clones, 16.5%), Acetobacter pomorum (13 clones, 5.5%), and Acetobacter peroxydans (2 clones, 0.9%). The remainder of the α-Proteobacteria were represented by Gluconobacter cerinus (33 clones, 14%), Gluconobacter frateruii (1 clone, 0.4%), and Gluconacetabacter europaeus (3 clones, 1.3%).
The γ-Proteobacteria accounted for 43.6% (183 clones) of the Proteobacteria (26.7% of the total library) and was represented by 22 phylotypes. Of these, Wolbachia (62 clones, 34%) and Enterobacter (61 clones, 33.3%) were the most abundant phylotypes. All clones of the Wolbachia phylotype identified in this analysis originated in wild-captured MA flies. Presumably, a large proportion of these organisms occur in the reproductive tract and other organ systems of Drosophila. However, it is likely that some are also associated with the GI tract (15). Because the level of Wolbachia infection of the MA fly line did not preclude detection of other flora, we included this line in the analysis as a representative of Drosophila in the wild. Nevertheless, the relatively large number of Wolbachia clones obtained for this fly line likely skewed the representation of Proteobacteria in the consortium of the line. Enterobacter cloacae was the most abundant Enterobacter, with 41 clones (67.2% of the Enterobacter clones, accounting for 22.4% of the γ-Proteobacteria). The other Enterobacter phylotypes observed included a previously uncultured Enterobacter (two clones, 3.3% of the Enterobacter clones), Enterobacter sp. strain BL-2 (two clones, 3.3% of the Enterobacter clones), Enterobacter gergoviae (one clone, 1.6% of the Enterobacter clones), Enterobacter hormaechei (eight clones, 13.1% of the Enterobacter clones), and Enterobacter aerogenes (seven clones, 11.5% of the Enterobacter clones). Three Klebsiella phylotypes were observed. Klebsiella pneumoniae (20 clones) accounted for 11% of the γ-Proteobacteria sequences, Klebsiella sp. strain JT42 (8 clones) accounted for 4.4%, and Klebsiella oxytoca (11 clones) accounted for 6%. Pantoea sp. strain P102 (seven clones) accounted for 3.8% of the γ-proteobacterial sequences. The remainder of the γ-Proteobacteria clones, including those belonging to the Citrobacter, Erwinia, Serratia, Morganella, Pseudomonas, and Stenotrophomonas phylotypes, as well as an uncultured γ-Proteobacteria clone, each accounted for less than 1% of the γ-Proteobacteria.
Seven clones representing three phylotypes belonging to the Actinobacteria (1% of the combined sequences) were observed. Corynebacterium bovis (six clones), Corynebacterium variabile (one clone), and Micrococcus luteus (isolated by culture) were identified. Two phylotypes belonging to the Bacteriodetes (0.3% of the combined library) were observed and consisted of single clones belonging to the genera Bradyrhizobium and Chitinophaga.
Estimation of phylotype richness, diversity, and coverage.
Phylotype richness, diversity, evenness, and coverage were calculated for each Drosophila stock, as well as for the combined data set (Table 1). To determine the number of sequences necessary to obtain a robust estimate of microbial diversity in our Drosophila 16S rRNA gene libraries, nonrandomized species accumulation curves were constructed using DOTUR. Figure S1 in the supplemental material shows observed and Chao1-estimated nonrarefied accumulation curves for Oregon R OU (Fig. S1A), Oregon R Bloomington (Fig. S1B), and MA (Fig. S1C), which suggest that there is a simple eubacterial Drosophila consortium consisting of (at most) 25 phylotypes in any single fly line. Furthermore, the slope of each of the species accumulation curves provided a robust estimate of phylotype richness.
TABLE 1.
Drosophila microbial flora diversity and coverage estimates
| Fly line(s) | No. of clones | % Enterococcus clones | No. of observed phylotypes (culture independent) | Good's estimator of coverage (%) | No. of singletons | Chao1 estimator of species richness (no. of OTUs) | Shannon's index of diversity | Simpson's index of diversity (1/D) | Simpson's evenness |
|---|---|---|---|---|---|---|---|---|---|
| Oregon R OU | 237 | 70.5 | 9 | 98.3 | 4 | 11 | 0.9 | 1.8 | 0.2 |
| Oregon R Bloomington | 238 | 24.8 | 13 | 99.6 | 1 | 13 | 2.0 | 6.2 | 0.5 |
| Wild-captured MA | 211 | 2.8 | 23 | 97.6 | 5 | 25 | 2.3 | 6.8 | 0.3 |
| Combined | 686 | 33.8 | 37 | 99.3 | 5 | 38 | 2.7 | 9.8 | 0.3 |
To measure phylotype richness, a rarefaction analysis was performed for each 16S rRNA gene library. Figure 3A shows rarefaction curves for each library, as well as Chao1 estimates of species richness (Fig. 3B). Figure S2 in the supplemental material shows observed and Chao1-estimated rarefaction curves (Fig. S2A) and observed and Chao1-estimated species accumulation curves (Fig. S2B) for the combined data set. All rarefaction curves constructed in this study approached a plateau, indicating nearly complete coverage. This is in agreement with Good's estimate of coverage (Table 1), which indicated that there was 99.3% coverage for the combined library, providing evidence that the 16S rRNA gene sequences in the libraries represent the majority of bacterial phylotypes present in the Drosophila stocks analyzed in this study. A statistical analysis of library diversity and evenness was performed by conventional methods (66, 68) (Table 1). The Shannon and Simpson indices of diversity were highest for the wild-captured MA stock, followed by Oregon R Bloomington; the lowest values were the values for Oregon R OU.
FIG. 3.
Rarefaction analysis of 16S rRNA gene clone libraries. Observed (A) and Chao1 (B) rarefaction curves, plotted using a 97% similarity cutoff, were computed with DOTUR. The error bars in panel B indicate the 95% confidence intervals for Chao1 estimation of species richness after 1,000 randomizations.
Estimation of phylotypes shared by Drosophila microbial consortia.
To quantify the degree of overlap for the three Drosophila 16S rRNA gene libraries examined and to identify microbial phylotypes shared by them, we used the recently developed program SONS (64), which effectively extends the phylogenetic analysis performed using DOTUR (63) to allow comparisons between individual libraries. Figure 4 shows a Venn diagram illustrating the numbers of phylotypes shared by the three libraries with a 95% confidence interval. When data were examined with a 97% similarity cutoff, three phylotypes were shared by all three libraries. These phylotypes were A. aceti, A. pasteurianus, and E. faecalis.
FIG. 4.

Venn diagram describing a comparison of phylotype membership in Drosophila microbial consortia for three fly types. Oregon R OU (red), Oregon R Bloomington (blue), and wild-captured MA (yellow) 16S rRNA gene libraries were compared using SONS. The numbers in the circles indicate the estimated numbers of OTUs that are shared or are unique to each fly type. Below each Drosophila stock designation are the Chao1 estimate and 95% confidence interval for richness at a similarity level of 97%.
Native colonization of Drosophila by enterococci.
To obtain additional evidence that enterococci are an important part of the flora of Drosophila, three additional fly stocks consisting of both wild-captured and laboratory-maintained flies were directly assessed to determine carriage of culturable enterococci. All seven stocks (Oregon R Bloomington, Oregon R OU, OK, MA, Oregon R UT, Canton S Bloomington, and Canton S OU) yielded culturable Enterococcus on bile esculin azide agar. As shown in Table 2, the enterococci identified included E. faecalis, E. faecium, E. durans, and E. gallinarum, as well as enterococcal species yet to be named. E. faecalis was found in six of the seven stocks examined. To determine what fractions of the cultivable flora were enterococci for the four Drosophila lines studied further, the total cultivable flora was assessed by plating fly homogenates on BHI agar, followed by anaerobic and aerobic incubation. Drosophila flies were found to harbor 1.6 × 104 ± 5.8 × 103 cultivable CFU/fly, and the levels of cultivable enterococci present were 4.4 × 103 ± 2.9 × 103 CFU/fly.
TABLE 2.
Native enterococcal colonization of Drosophila
| Fly stock | Enterococcal species (≥98% identity) | Colonization of Drosophila
|
|
|---|---|---|---|
| Culture dependent | Culture independent | ||
| Wild-captured MA | E. faecalis | + | + |
| E. gallinarum | + | ||
| E. durans | + | ||
| Wild-captured OK | E. faecalis | + | |
| Enterococcus sp. | + | + | |
| Oregon R Bloomington | E. faecalis | + | + |
| Enterococcus sp. | + | + | |
| Oregon R OU | E. faecalis | + | + |
| E. faecium | + | ||
| Canton S OU | E. faecalis | + | NDa |
| Canton S Bloomington | E. faecalis | + | ND |
| E. durans | + | ND | |
| Oregon R UT | E. durans | + | ND |
ND, not determined.
Enterococcal reassociation for Drosophila.
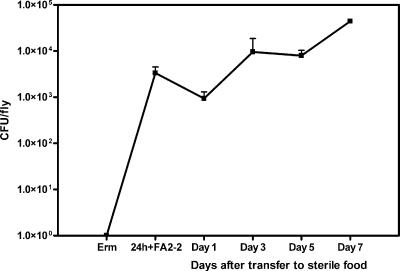
To determine whether Drosophila could be reassociated with known enterococcal strains and to determine whether a trait known to contribute significantly to enterococcal virulence (38) affected a fly when the trait was expressed by an ingested microbe, we conducted the following experiments. The levels of native enterococci in adult Oregon R Bloomington flies were reduced to levels below the detection limit by exposing the flies to food containing 50 μg/ml erythromycin for 24 h at room temperature. As shown in Fig. 5, flies were then transferred to antibiotic-free food that had been inoculated with E. faecalis strain FA2-2 (plasmid free) for 24 h, 30°C. Flies were then transferred to sterile food, and enterococcal carriage was quantified at 24-h intervals. After 24 h of exposure to bacterium-laden food, the level of enterococci was 3.3 × 103 CFU per fly (Fig. 5). Twenty-four hours after transfer of flies to sterile food, the level dropped slightly (9.4 × 102 CFU per fly). However, enterococcal colonization rebounded, and the level remained relatively stable during the 7-day experiment. Recovered enterococci were tested for chromosomal resistance to fusidic acid and rifampin, and the results verified that the bacteria were strain FA2-2 and did not represent resurgent colonization by native flora that had been present at levels below the detection limit.
FIG. 5.
Colonization of Drosophila with E. faecalis FA2-2. One hundred adult Oregon R Bloomington flies were fed cornmeal-molasses food containing erythromycin (50 μg/ml) for 24 h, transferred to food inoculated with 2 ×107 CFU/ml E. faecalis FA2-2 for 24 h (24 h + FA2 - 2), and then transferred to sterile food. Enterococcal colonization was tracked for 7 days. The error bars indicate standard errors of the means for experiments conducted independently three times.
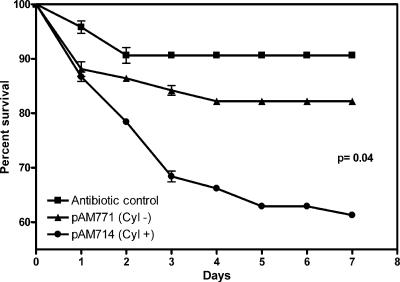
Since enterococcal cytolysin contributes to the severity of infection in all models tested so far (29, 38), this virulence factor was selected for testing in the Drosophila oral ingestion model. This cytolysin, in addition to being a toxin, is also a bacteriocin (67). Because we were able to reassociate Drosophila with known enterococcal strains, it was of interest to determine whether the cytolysin affected colonization of the fly through its bacteriocin properties or the health of the fly through its activity as a toxin. To determine the effect of the cytolysin on either colonization or health, we reassociated erythromycin-cured flies with FA2-2/pAM714, which produces active cytolysin, or FA2-2/pAM771, which produces the cytolysin activator protease but not the toxin subunits and is therefore noncytolytic (39). Both pAD1-derived plasmids were constructed previously by insertional mutagenesis using the transposon Tn917 (40) conveying erythromycin resistance and were previously shown to be stably maintained by a partitioning system (77, 78). Figure 6 shows the effect of ingestion of cytolysin toxin-expressing E. faecalis on the viability of Drosophila. The two groups responded in the same way to the initial 24-h exposure to Enterococcus, with 12% mortality. We observed a small decrease in the survival of flies colonized with the cytolysin-negative E. faecalis strain (17% killed by day 4) compared to the survival of an antibiotic-only control, which suggests that either perturbation of the flora followed by colonization with a human-derived E. faecalis strain or residual production of the cytolysin activator by the noncytolytic strain is deleterious to Drosophila. Lethality was significantly increased in flies colonized by the cytolysin toxin-producing strain, with 40% of the flies dying by the end of the week-long experiment (P = 0.04, as determined by Welch's corrected t test), compared to flies colonized by FA2-2/pAM771. These results were reproduced in an independently conducted second experiment.
FIG. 6.
Effect of cytolysin-producing E. faecalis on Drosophila survival. One hundred adult Oregon R Bloomington flies were associated with either E. faecalis FA2-2/pAM714 producing cytolysin or the isogenic cytolysin-negative control FA2-2/pAM771. The percentage of survival of colonized flies was determined for 1 week. Controls were placed on sterile food after elimination of native enterococci by erythromycin. The symbols indicate means of experiments conducted three separate times, and the error bars indicate standard errors of the means.
Figure 7 shows the relative rates of colonization of Drosophila by isogenic E. faecalis strains. The colonization rates with the cytolytic strain were consistently higher than those with the noncytolytic strain, but they were not statistically significantly different until the last day (P = 0.02). In order to determine whether the relative colonization rates observed in flies after transfer to sterile food were simply a reflection of the amount of enterococci deposited in the food throughout the experiment or whether flies remained stably colonized with levels below the levels of enterococci in the food, the relative numbers of cytolytic and noncytolyitic enterococci in the food were also determined. The levels of cytolytic and noncytolytic enterococci in the food were found to be similar at each time. However, the number of enterococcal CFU per gram of food was consistently approximately 1 order of magnitude higher than the number of CFU per colonized fly. While the relative colonization rates shown in Fig. 7 suggest that the cytolysin toxin provides Enterococcus with only a slight advantage during colonization of flies with antibiotic-perturbed consortia, the significant decrease in the survival of flies colonized with the cytolytic strain demonstrates that the toxin at these levels of colonization is deleterious to the host.
FIG. 7.
Colonization of Drosophila with cytolysin-producing E. faecalis FA2-2. One hundred adult Oregon R Bloomington flies were fed cornmeal-molasses food containing erythromycin (50 μg/ml) for 24 h, transferred to food inoculated with 2 × 107 CFU/ml E. faecalis FA2-2/pAM714 or FA2-2/pAM771 for 24 h (24 hr + bac), and then transferred to sterile food. Enterococcal colonization was tracked for 7 days. The error bars indicate standard errors of the means for experiments conducted independently three times.
Anatomical localization of naturally occurring enterococci.
Because Wolbachia had been identified by other workers as an organism that occurs in the reproductive tract and other organ systems of Drosophila, it was of interest to determine the anatomical localization of Enterococcus and to verify its presence in the GI tract. Enterococci were visualized by immunohistochemical staining of thin sections of intact Oregon R Bloomington adults, using affinity-purified antienterococcal antibodies (Fig. 8). Naturally occurring enterococci were observed by fluorescent microscopy, and immunostained Enterococcus-free flies were included as negative controls (Fig. 8C and F). In unperturbed adults, we observed enterococci in the foregut, midgut, and hindgut. In the foregut, enterococci were observed in the crop and crop stalk, which form an invagination off the esophagus, anterior to the stomadeal valve, but they were not observed in the esophagus (Fig. 8D). With the exception of the crop stalk, no enterococci were detected in any portion of GI tract anterior to the stomadeal valve, including the salivary glands, proboscis, or other mouthparts. In the midgut, enterococci were observed to be most abundant in the ventriculus (Fig. 8G). In the hindgut, enterococci were abundant in the rectum (Fig. 8B and H) and less abundant in the intestine (Fig. 8E) and anus (Fig. 8I). Enterococci were not detected in the Malpighian tubules or the reproductive organs.
FIG. 8.
Immunohistochemical staining of thin sections of Drosophila naturally colonized with Enterococcus. Ten-micrometer cross sections of Oregon R flies either naturally colonized with enterococci or hatched and raised with 50 μg/ml erythromycin (negative control) were stained with affinity-purified rabbit anti-Enterococcus antibodies and visualized with Alexafluor 594-conjugated goat anti-rabbit antibodies. (A) Schematic diagram of Drosophila GI tract. Red indicates portions of the GI tract naturally colonized by Enterococcus. Ph, pharynx (cibarium); Sd, salivary duct; Es, esophagus; Car, cardium; Vent, pro ventriculus; CrStlk, crop stalk; Cr, crop; Sg, salivary gland; Int, intestine; Mal, Malpighian tubule; Rect, rectum. (B) Hindgut section showing colonized rectum. Magnification, ×85. (C) Hindgut negative control. Magnification, ×85. (D) Esophagus and crop stalk of colonized fly. Magnification, ×850. (E) Intestine of colonized fly. Magnification, ×850. (F) Ventriculus of negative control fly. Magnification, ×850. (G) Ventriculus of colonized fly. Magnification, ×850. (H) Rectum of colonized fly. Magnification, ×850. (I) Anus of colonized fly. Magnification, ×850. The diagram (A) was adapted from the study by Hartenstein (32), with permission of the publisher.
DISCUSSION
The prevalence of Enterococcus in the microflora of insects was examined by Martin and Mundt (50). Using classic biochemical tests, as well as selective and differential plating techniques for identification of enterococcal species, these workers surveyed 37 insect phylotypes belonging to eight orders, including the Coleoptera, Diptera, Hemiptera, Homoptera, Hymenoptera, Isoptera, Lepidoptera, and Orthoptera. Enterococci were observed in approximately 53% of 403 insect homogenates examined, and the levels ranged from 103 to 3 × 107 bacteria/g. The largest faction (43.5%) of enterococcal isolates were identified as E. casseliflavus. E. faecalis (32%) and E. faecium (22.4%) were also prominent. Consistent with our observations with Drosophila, Martin and Mundt observed the presence of more than one enterococcal phylotype in a given insect phylotype. Interestingly, wide variation in the percentage of Enterococcus clones from fly stock to fly stock was observed (Table 1). While it is not known why the percentage of Enterococcus clones was lower in wild-caught Drosophila than in laboratory-reared strains, it was observed that the GI tract microbiota of wild-captured flies had a greater level of diversity (Table 1); therefore, enterococci accounted for a smaller percentage of the overall total for the more complex flora observed in wild flies. The higher level of diversity seen in wild flies may have been the result of consumption of a diet that was more varied than the diet of laboratory-reared Drosophila stocks passaged on artificial fly food, which may not support some members of the microbiota normally associated with wild Drosophila.
Analyses of the microbial flora of the gypsy moth, as well as termites, have been described. Broderick et al. (8) recently characterized the larval midgut flora of the gypsy moth, Lymantria dispar L. Their results indicate that this flora is a simple consortium composed of 23 phylotypes, and while the relative proportions of the various phylotypes found in the gypsy moth differ from the relative proportions in Drosophila, similar bacterial species were observed. The gypsy moth larval midgut flora is composed of similar bacterial phylotypes, including members of the α- and γ-Proteobacteria, Firmicutes, Actinobacteria, and Bacteriodetes. Notably, E. faecalis was a prominent member of the cultured gypsy moth midgut flora, and the levels were as high as 3.6 × 108 CFU/moth. Other phylotype representatives found in both the gypsy moth and Drosophila include species of Pseudomonas, Enterobacter, Pantoea, and Serratia. Members of the Firmicutes other than E. faecalis that were observed in both types of insects include species of Bacillus and Staphylococcus. As was the case in our study, the gypsy moth flora also lacked obligate anaerobic phylotypes, a fact that may be pertinent when the compositions of the various insect microbial consortia are compared to the composition of the human GI tract flora for the purposes of modeling colonization with clinically important human isolates.
The composition of the intestinal microflora of termites (Isoptera) was shown to be much more complex than the composition of the intestinal microflora of Drosophila; it consisted of obligate and facultative anaerobes, as well as microaerophilic phylotypes (34, 60, 74, 79). This complex microbiota, in addition to the termites' own cellulolytic enzymes (56, 81) and resident cellulolytic, flagellated protozoans (56, 62), plays an important role in the digestion of cellulose, a main component of the termite diet. In contrast to the Drosophila gut, not only is the enlarged, microbe-packed termite hindgut paunch large enough (diameter, ∼1 mm diameter [7]) to restrict the diffusion of oxygen into its innermost regions, but it also contains complex GI tract consortium members that act as an oxygen sink, which effectively consume residual oxygen at the organ's outer perimeter (10). The lower termite Reticulitermes speratus was found to harbor a much more complex microbial consortium consisting of 314 phylotypes belonging to five main bacterial phyla. These phyla included the Cytophaga-Flexibacter-Bacteroides group, Firmicutes, Proteobacteria, Spirochaeta, and a newly discovered termite-specific phylum, termite group I (60). The majority of clones in this study clustered in anaerobic phylotypes, including the genus Treponema, and the orders Clostridiales and Bacteroidales, all of which are unique to the termite.
Phylogenetic characterization of the GI tract flora of higher termites (Termes comis, Pericapritermes latignathus, Microcerotermes sp., and Seculitermes sp.) (74) produced results similar to the results obtained for the lower termite but revealed, in addition to obligate anaerobic phylotypes, the presence of numerous additional facultative anaerobes, including species of Enterococcus. These Enterococcus species included E. faecalis, E. casseliflavus, E. raffinosus, and E. hirae. Members of the genera Streptococcus, Lactobacillus, Lactococcus, and Bacillus were also observed. While this flora was similar to the Drosophila flora in terms of the composition of associated Firmicutes, notable differences were apparent, particularly in the complexity of the consortium found in the termites and the predominance of obligate anaerobes.
The Drosophila microbial consortium is considerably less complex than the human GI tract flora. Eckburg et al. (23) recently described a highly detailed analysis of the diversity of the human intestinal microbial flora. Using contemporary 16S rRNA gene sequencing techniques, these workers found 395 bacterial phylotypes, while further phylotype richness estimates based on their findings suggested that the number is more than 500 phylotypes (23). The most numerous human intestinal phylotypes analyzed belonged to the Firmicutes (301 phylotypes, 76% of the total), and the vast majority (95% of Firmicutes and 69% of all phylotypes identified [274 phylotypes]) belonged to the largely anaerobic or microaerophilic clostridia. While not specifically mentioned in the findings of Eckburg et al., enterococcal phylotypes fell into the relatively small fraction containing nonclostridial Firmicutes (0.2% of the total).
The Bacteriodetes was the next most abundant phylum in the human intestinal microbial consortium (48% of the total, 65 phylotypes). In contrast, Bacteriodetes phylotypes were relatively rare in Drosophila (0.3%, two phylotypes). Conversely, human proteobacterial phylotypes were relatively scarce (∼0.1% of the total), while the Drosophila microbial consortium was composed of approximately 61% proteobacterial phylotypes. As was the case with the Drosophila microbial consortium, the Actinobacteria also accounted for only a minor fraction of the human flora. Fusobacterium and Verrucomicrobium phylotypes (absent from Drosophila) were also rare.
Differences between human GI tract anatomy and physiology and Drosophila GI tract anatomy and physiology undoubtedly play a role in determining the composition and physical location of the respective microbial populations. Anatomically, the GI tract of Drosophila (6) has an overall organization that is comparable to that of humans. With the exception of the Drosophila crop, the two systems possess a single alimentary canal beginning at the esophagus, connecting to a ventriculus (stomach), extending to the intestine, and then proceeding to the rectum and terminating at the anus. In terms of digestive function, however, they are quite different. Drosophila possesses an acidic crop but maintains a highly alkaline ventriculus and a neutral to acidic hindgut (16, 21). In comparison, the human stomach is highly acidic, the intestine is neutral, and the colon is maintained at a pH gradient ranging from slightly acidic at the anterior end to neutral at the posterior end (22). As discussed above, another major difference between the two GI tracts is the absence of obligate anaerobic phylotypes in Drosophila, while the majority of human intestinal microbial phylotypes observed were obligate or microaerophilic anaerobes (23). The open circulatory system of the fly (6), together with the small diameter of the Drosophila alimentary canal, which even at its greatest diameter (225 μm [6]) does not exceed the diffusion limit of oxygen in tissue (140 μm [46]) when it is considered that diffusion can occur from all sides, may account for the lack of anoxic environments in the Drosophila GI tract.
As discussed above, we found that enterococci were most prominent throughout the midgut of Drosophila, localizing in the ventriculus, crop, and crop stalk, as well as in the hindgut, the intestine, and the rectum. Similarly, recent findings (33) suggest that enterococcal localization in the human GI tract is centered primarily in the small and large intestines, where enterococci are prominent members of the jejunal, ileal, cecal, and rectosigmoidal consortia. Enterococci have also been observed to a lesser extent in the stomach (5, 53) and, in contrast to Drosophila, in the oral cavity (69). The apparent absence of detectable enterococci in any alimentary canal structure proximal to the stomadeal valve in Drosophila may be the result of production of protective antimicrobial peptides or other digestive compounds by the labelar or salivary glands. Tzou et al. (75) recently demonstrated that tissue-specific inducible expression of antimicrobial peptides, such as defensin, which are known to have anti-gram-positive properties (47), occurs in the foregut, in the cardia, and in the anterior portion of the ventriculus but not in the more posterior regions of the GI tract.
Studies of enterococcal colonization and ecology in the GI tract using mammalian models (36, 37) have had limited success because of the complexity of the flora, animal-to-animal variation, and the possibility that lineages of enterococci may be host adapted (9). Recently, Caenorhabditis elegans was used to screen traits of enterococci for virulence in this model (29). Virulence properties known to be important in mammalian infection were verified in this model, but this model has limited value for studying host-commensal relationships in GI tract colonization, since C. elegans ingests bacteria as food. The extent to which C. elegans harbors a commensal flora is unknown.
While the anatomical localization of the Drosophila microbiota has not been determined in its entirety, we observed by using immunohistochemical staining of thin sections (Fig. 8) that enterococci were consistently located only within the confines of the GI tract, suggesting that they represented a consistent component of the Drosophila GI tract consortium, irrespective of whether flies were reared in the laboratory or captured from the wild. With the exception of the Oregon R UT laboratory stock, which was observed to be colonized with E. durans as the only detectable Enterococcus species, all fly stocks examined were colonized with E. faecalis. Wild-captured MA flies were observed to have the most diverse range of enterococcal species. Interestingly, we were able to isolate a previously unidentified Enterococcus strain from the OK and Oregon R Bloomington stocks, which was originally reported to be found in association with plants (54). In addition to finding enterococci natively associated with Drosophila, the use of a culture-independent approach allowed us to evaluate other members of the fruit fly bacterial consortium. While our assessment of the total Drosophila microbial consortium appears to be reasonably complete, it is possible that minor species, not detected by amplification and 16S rRNA gene sequence analysis, could be present. Nevertheless, the results presented here represent the first reported characterization of the total Drosophila microbial consortium that we are aware of.
Studies in which Drosophila was used to examine the pathogenesis of infection caused by oral ingestion of bacteria were described recently. Vodovar et al. (76) demonstrated that oral infection of Drosophila with the gram-negative γ-proteobacterium Pseudomonas entomophila resulted in 70% lethality in wild-type flies and induced a systemic immune response. Liehl et al. (48) showed that infection with this pathogen also induced a local immune response and demonstrated that inactivation of a secreted metalloprotease produced by P. entomophila attenuated its ability to persist and cause lethality. In the present study we found that enterococci are present in the GI tract consortia of humans and Drosophila. It was therefore of interest to determine whether a known enterococcal virulence trait also contributed to lethality in Drosophila. The enterococcal cytolysin was of particular interest for study in this model because of its dual role as both a bacteriocin and a toxin. Fly survival was significantly affected by colonization with the cytolytic strain, highlighting the utility of this model.
Drosophila has been used as a model organism in research for nearly a century (6). Its genome has recently been sequenced (2), it has been utilized as a nonmammalian model for studying some aspects of infectious diseases (19, 20, 26, 57), and it possesses signaling systems governing its innate immune response that are functionally conserved in mammals (25, 41, 43, 44). The fact that it is natively colonized with enterococci but has a simpler GI tract consortium suggests that it may be well suited for studies of the role of enterococci in a GI tract consortium and the mechanisms that underlie the ability to use the consortium as a springboard for causing disease.
Supplementary Material
Acknowledgments
This research was supported by NIH grants AI41108, AI054614, and 1RR020596-01A1.
We gratefully acknowledge James Thompson for his contribution of fly stocks and expertise, and we appreciate the assistance of Tracy Mincer, Bryan Jennings, and Peter Girguis with the construction of phylogenetic trees. Special thanks are extended to Pat Schloss for his expertise and assistance with phylogenetic analysis.
Editor: V. J. DiRita
Footnotes
Published ahead of print on 12 January 2007.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Aarestrup, F. M., P. Butaye, and W. Witte. 2002. Nonhuman reservoirs of enterococci, p. 55-99. In M. S. Gilmore (ed.), The enterococci: pathogenesis, molecular biology, and antibiotic resistance. ASM Press, Washington, DC.
- 2.Adams, M. D., et. al. 2000. The genome sequence of Drosophila melanogaster. Science 287:2185-2195. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M. 1994. Preparation of genomic DNA from bacteria, p. 2.4.1-2.4.5. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley and Sons, Inc., New York, NY.
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1994. Current protocols in molecular biology, p. 11.2.1-11.2.5. Greene Publishing Associates and Wiley-Interscience, New York, NY.
- 5.Bik, E. M., P. B. Eckburg, S. R. Gill, K. E. Nelson, E. A. Purdom, F. Francois, G. Perez-Perez, M. J. Blaser, and D. A. Relman. 2006. Molecular analysis of the bacterial microbiota in the human stomach. Proc. Natl. Acad. Sci. USA 103:732-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bodenstein, D., K. W. Cooper, G. F. Ferris, A. Miller, D. F. Poulson, and B. P. Sonnenblick. 1950. Biology of Drosophila. John Wiley & Sons, Inc., New York, NY.
- 7.Breznak, J. A., and H. S. Pankratz. 1977. In situ morphology of the gut microbiota of wood-eating termites [Reticulitermes flavipes (Kollar) and Coptotermes formosanus Shiraki]. Appl. Environ. Microbiol. 33:406-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broderick, N. A., K. F. Raffa, R. M. Goodman, and J. Handelsman. 2004. Census of the bacterial community of the gypsy moth larval midgut by using culturing and culture-independent methods. Appl. Environ. Microbiol. 70:293-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bruinsma, N., R. J. Willems, A. E. van Den Bogaard, M. Santen-Verheuvel, N. London, C. Driessen, and E. E. Stobberingh. 2002. Different levels of genetic homogeneity in vancomycin-resistant and -susceptible Enterococcus faecium isolates from different human and animal sources analyzed by amplified-fragment length polymorphism. Antimicrob. Agents Chemother. 46:2779-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brune, A., D. Emerson, and J. A. Breznak. 1995. The termite gut microflora as an oxygen sink: microelectrode determination of oxygen and pH gradients in guts of lower and higher termites. Appl. Environ. Microbiol. 61:2681-2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bucher, G. E. 1963. Survival of populations of Streptococus faecalis Andrewes and Horder in the gut of Galleria mellonella (Linnaeus) during metamorphosis, and transmission of the bacteria to the filial generation of the host. J. Insect Pathol. 5:336-343. [Google Scholar]
- 12.Campbell, D. H., E. Luescher, and L. S. Lerman. 1951. Immunologic adsorbents. I. Isolation of antibody by means of a cellulose-protein antigen. Proc. Natl. Acad. Sci. USA 37:575-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chao, A. 1984. Non-parametric estimation of the number of classes in a population. Scand. J. Stat. 11:265-270. [Google Scholar]
- 14.Chao, A., S. M. Lee, and S. L. Jeng. 1992. Estimating population size for capture-recapture data when capture probabilities vary by time and individual animal. Biometrics 48:201-216. [PubMed] [Google Scholar]
- 15.Clark, M. E., C. L. Anderson, J. Cande, and T. L. Karr. 2005. Widespread prevalence of Wolbachia in laboratory stocks and the implications for Drosophila research. Genetics 170:1667-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clark, T. M. 1999. Evolution and adaptive significance of larval midgut alkalinization in the insect superorder. J. Chem. Ecol. 25:1945-1960. [Google Scholar]
- 17.Clewell, D. B., P. K. Tomich, M. C. Gawron-Burke, A. E. Franke, Y. Yagi, and F. Y. An. 1982. Mapping of Streptococcus faecalis plasmids pAD1 and pAD2 and studies relating to transposition of Tn917. J. Bacteriol. 152:1220-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D'Argenio, D. A., L. A. Gallagher, C. A. Berg, and C. Manoil. 2001. Drosophila as a model host for Pseudomonas aeruginosa infection. J. Bacteriol. 183:1466-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dionne, M. S., N. Ghori, and D. S. Schneider. 2003. Drosophila melanogaster is a genetically tractable model host for Mycobacterium marinum. Infect. Immun. 71:3540-3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dow, J. A., and W. R. Harvey. 1988. Role of midgut electrogenic K+ pump potential difference in regulating lumen K+ and pH in larval lepidoptera. J. Exp. Biol. 140:455-463. [DOI] [PubMed] [Google Scholar]
- 22.Dunham, L. J., and A. Brunschwig. 1946. Calcium and potassium content of secretions from noncancerous and cancerous stomachs. Cancer Res. 6:54-56. [PubMed] [Google Scholar]
- 23.Eckburg, P. B., E. M. Bik, C. N. Bernstein, E. Purdom, L. Dethlefsen, M. Sargent, S. R. Gill, K. E. Nelson, and D. A. Relman. 2005. Diversity of the human intestinal microbial flora. Science 308:1635-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edwards, D. D. 2000. Enterococci attract attention of concerned microbiologists. ASM News 9:540-545. [Google Scholar]
- 25.Engstrom, Y. 1999. Induction and regulation of antimicrobial peptides in Drosophila. Dev. Comp. Immunol. 23:345-358. [DOI] [PubMed] [Google Scholar]
- 26.Erickson, D. L., J. L. Lines, E. C. Pesci, V. Venturi, and D. G. Storey. 2004. Pseudomonas aeruginosa relA contributes to virulence in Drosophila melanogaster. Infect. Immun. 72:5638-5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Finegold, S. M., V. L. Sutter, and G. E. Mathisen. 1983. Normal indigenous intestinal flora, p. 3-31. In D. J. Hentges (ed.), Human intestinal microflora in health and disease. Academic Press, New York, NY.
- 28.Garrity, G. M. (ed.) 2001. Bergey's manual of systematic bacteriology. Springer, New York, NY.
- 29.Garsin, D. A., C. D. Sifri, E. Mylonakis, X. Qin, K. V. Singh, B. E. Murray, S. B. Calderwood, and F. M. Ausubel. 2001. A simple model host for identifying Gram-positive virulence factors. Proc. Natl. Acad. Sci. USA 98:10892-10897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gilmore, M. S., and J. J. Ferretti. 2003. Microbiology. The thin line between gut commensal and pathogen. Science 299:1999-2002. [DOI] [PubMed] [Google Scholar]
- 31.Good, I. J. 1953. The population frequencies of species and the estimation of population parameters. Biometrika 40:237-264. [Google Scholar]
- 32.Hartenstein, V. 1993. Atlas of Drosophila development. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 33.Hayashi, H., R. Takahashi, T. Nishi, M. Sakamoto, and Y. Benno. 2005. Molecular analysis of jejunal, ileal, caecal and recto-sigmoidal human colonic microbiota using 16S rRNA gene libraries and terminal restriction fragment length polymorphism. J. Med. Microbiol. 54:1093-1101. [DOI] [PubMed] [Google Scholar]
- 34.Hongoh, Y., P. Deevong, T. Inoue, S. Moriya, S. Trakulnaleamsai, M. Ohkuma, C. Vongkaluang, N. Noparatnaraporn, and T. Kudo. 2005. Intra- and interspecific comparisons of bacterial diversity and community structure support coevolution of gut microbiota and termite host. Appl. Environ. Microbiol. 71:6590-6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huber, T., G. Faulkner, and P. Hugenholtz. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317-2319. [DOI] [PubMed] [Google Scholar]
- 36.Huycke, M. M., M. S. Gilmore, B. D. Jett, and J. L. Booth. 1992. Transfer of pheromone-inducible plasmids between Enterococcus faecalis in the Syrian hamster gastrointestinal tract. J. Infect. Dis. 166:1188-1191. [DOI] [PubMed] [Google Scholar]
- 37.Huycke, M. M., W. A. Joyce, and M. S. Gilmore. 1995. Enterococcus faecalis cytolysin without effect on the intestinal growth of susceptible enterococci in mice. J. Infect. Dis. 172:273-276. [DOI] [PubMed] [Google Scholar]
- 38.Huycke, M. M., D. F. Sahm, and M. S. Gilmore. 1998. Multiple-drug resistant enterococci: the nature of the problem and an agenda for the future. Emerg. Infect. Dis. 4:239-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ike, Y., and D. B. Clewell. 1984. Genetic analysis of the pAD1 pheromone response in Streptococcus faecalis, using transposon Tn917 as an insertional mutagen. J. Bacteriol. 158:777-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ike, Y., D. B. Clewell, R. A. Segarra, and M. S. Gilmore. 1990. Genetic analysis of the pAD1 hemolysin/bacteriocin determinant in Enterococcus faecalis: Tn917 insertional mutagenesis and cloning. J. Bacteriol. 172:155-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Imler, J. L., and J. A. Hoffmann. 2000. Signaling mechanisms in the antimicrobial host defense of Drosophila. Curr. Opin. Microbiol. 3:16-22. [DOI] [PubMed] [Google Scholar]
- 42.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism. Academic Press, New York, NY.
- 43.Khush, R. S., and B. Lemaitre. 2000. Genes that fight infection: what the Drosophila genome says about animal immunity. Trends Genet. 16:442-449. [DOI] [PubMed] [Google Scholar]
- 44.Kimbrell, D. A., and B. Beutler. 2001. The evolution and genetics of innate immunity. Nat. Rev. Genet. 2:256-267. [DOI] [PubMed] [Google Scholar]
- 45.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Inc., New York, NY.
- 46.Lanzen, J., R. D. Braun, B. Klitzman, D. Brizel, T. W. Secomb, and M. W. Dewhirst. 2006. Direct demonstration of instabilities in oxygen concentrations within the extravascular compartment of an experimental tumor. Cancer Res. 66:2219-2223. [DOI] [PubMed] [Google Scholar]
- 47.Lemaitre, B., J. M. Reichhart, and J. A. Hoffmann. 1997. Drosophila host defense: differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proc. Natl. Acad. Sci. USA 94:14614-14619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liehl, P., M. Blight, N. Vodovar, F. Boccard, and B. Lemaitre. 2006. Prevalence of local immune response against oral infection in a Drosophila/Pseudomonas infection model. PLoS Pathog. 2:e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Love, J. A., and G. D. Gill. 1965. Incidence of coliforms and enterococci in field populations of Stomoxys calcitrans (Linneaus). J. Invert. Pathol. 7:430-436. [Google Scholar]
- 50.Martin, J. D., and J. O. Mundt. 1972. Enterococci in insects. Appl. Microbiol. 24:575-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mateos, M., S. J. Castrezana, B. J. Nankivell, A. M. Estes, T. A. Markow, and N. A. Moran. 2006. Heritable endosymbionts of Drosophila. Genetics 174:363-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moffett, D. F., and A. Koch. 1992. Driving forces and pathways for H+ and K+ transport in insect midgut goblet cells. J. Exp. Biol. 172:403-415. [DOI] [PubMed] [Google Scholar]
- 53.Monstein, H. J., A. Tiveljung, C. H. Kraft, K. Borch, and J. Jonasson. 2000. Profiling of bacterial flora in gastric biopsies from patients with Helicobacter pylori-associated gastritis and histologically normal control individuals by temperature gradient gel electrophoresis and 16S rDNA sequence analysis. J. Med. Microbiol. 49:817-822. [DOI] [PubMed] [Google Scholar]
- 54.Muller, T., A. Ulrich, E. M. Ott, and M. Muller. 2001. Identification of plant-associated enterococci. J. Appl. Microbiol. 91:268-278. [DOI] [PubMed] [Google Scholar]
- 55.Mundt, J. O. 1963. Occurrence of enterococci in animals in a wild environment. Appl. Microbiol. 11:136-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakashima, K., H. Watanabe, H. Saitoh, G. Tokuda, and J. I. Azuma. 2002. Dual cellulose-digesting system of the wood-feeding termite, Coptotermes formosanus Shiraki. Insect Biochem. Mol. Biol. 32:777-784. [DOI] [PubMed] [Google Scholar]
- 57.Needham, A. J., M. Kibart, H. Crossley, P. W. Ingham, and S. J. Foster. 2004. Drosophila melanogaster as a model host for Staphylococcus aureus infection. Microbiology 150:2347-2355. [DOI] [PubMed] [Google Scholar]
- 58.Niven, C. F., and J. M. Sherman. 1944. Nutrition of the enterococci. J. Bacteriol. 47:335-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Noble, C. J. 1978. Carriage of group D streptococci in the human bowel. J. Clin. Pathol. 31:1182-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ohkuma, M., and T. Kudo. 1996. Phylogenetic diversity of the intestinal bacterial community in the termite Reticulitermes speratus. Appl. Environ. Microbiol. 62:461-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 62.Radek, R. 1999. Flagellagtes, bacteria, and fungi associated with termites: diversity and function in nutrition—a review. Ecotropica 5:183-196. [Google Scholar]
- 63.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schloss, P. D., and J. Handelsman. 2006. Introducing SONS, a tool that compares the membership of microbial communities. Appl. Environ. Microbiol. 72:6773-6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sghir, A., G. Gramet, A. Suau, V. Rochet, P. Pochart, and J. Dore. 2000. Quantification of bacterial groups within the human fecal flora by oligonucleotide probe hybridization. Appl. Environ. Microbiol. 66:2263-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shannon, C. E., and W. Weaver. 1949. The mathematical theory of communication. University of Illinois Press, Urbana.
- 67.Sherwood, N. P., B. E. Russell, A. R. Jay, and K. Bowman. 1949. Studies on streptococci. III. New antibiotic substances produced by beta hemolytic streptococci. J. Infect. Dis. 84:88-91. [DOI] [PubMed] [Google Scholar]
- 68.Simpson, E. H. 1949. Measurement of diversity. Nature 163:688. [Google Scholar]
- 69.Smyth, C. J., H. Matthews, M. K. Halpenny, H. Brandis, and G. Colman. 1987. Biotyping, serotyping and phage typing of Streptococcus faecalis isolated from dental plaque in the human mouth. J. Med. Microbiol. 23:45-54. [DOI] [PubMed] [Google Scholar]
- 70.Steinhaus, E. A. 1941. A study of the bacteria associated with thirty species of insects. J. Bacteriol. 42:757-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Suau, A., R. Bonnet, M. Sutren, J. J. Godon, G. R. Gibson, M. D. Collins, and J. Dore. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tannock, G. W., and G. Cook. 2002. Enterococci as members of the intestinal microflora of humans, p. 101-132. In Gilmore M.S. (ed.), The enterococci: pathogenesis, molecular biology, and antibiotic resistance. ASM Press, Washington, DC.
- 73.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thongaram, T., Y. Hongoh, S. Kosono, M. Ohkuma, S. Trakulnaleamsai, N. Noparatnaraporn, and T. Kudo. 2005. Comparison of bacterial communities in the alkaline gut segment among various species of higher termites. Extremophiles 9:229-238. [DOI] [PubMed] [Google Scholar]
- 75.Tzou, P., S. Ohresser, D. Ferrandon, M. Capovilla, J. M. Reichhart, B. Lemaitre, J. A. Hoffmann, and J. L. Imler. 2000. Tissue-specific inducible expression of antimicrobial peptide genes in Drosophila surface epithelia. Immunity 13:737-748. [DOI] [PubMed] [Google Scholar]
- 76.Vodovar, N., M. Vinals, P. Liehl, A. Basset, J. Degrouard, P. Spellman, F. Boccard, and B. Lemaitre. 2005. Drosophila host defense after oral infection by an entomopathogenic Pseudomonas species. Proc. Natl. Acad. Sci. USA 102:11414-11419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Weaver, K. E. 1995. Enterococcus faecalis plasmid pAD1 replication and maintenance. Dev. Biol. Stand. 85:89-98. [PubMed] [Google Scholar]
- 78.Weaver, K. E., and D. J. Tritle. 1994. Identification and characterization of an Enterococcus faecalis plasmid pAD1-encoded stability determinant which produces two small RNA molecules necessary for its function. Plasmid 32:168-181. [DOI] [PubMed] [Google Scholar]
- 79.Wenzel, M., I. Schonig, M. Berchtold, P. Kampfer, and H. Konig. 2002. Aerobic and facultatively anaerobic cellulolytic bacteria from the gut of the termite Zootermopsis angusticollis. J. Appl. Microbiol. 92:32-40. [DOI] [PubMed] [Google Scholar]
- 80.Xu, Y., L. Jiang, B. E. Murray, and G. M. Weinstock. 1997. Enterococcus faecalis antigens in human infections. Infect. Immun. 65:4207-4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yokoe, Y. 1964. Cellulase activity in the termite, Leucotermes speratus, with new evidence in support of a cellulase produced by the termite itself. Sci. Pap. Coll. Gen. Educ. Univ. Tokyo 14:115-120. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.