Abstract
Burkholderia cenocepacia, a member of the B. cepacia complex, is an opportunistic pathogen that causes serious infections in patients with cystic fibrosis. We identified a six-gene cluster in chromosome 1 encoding a two-component regulatory system (BCAL2831 and BCAL2830) and an HtrA protease (BCAL2829) hypothesized to play a role in the B. cenocepacia stress response. Reverse transcriptase PCR analysis of these six genes confirmed they are cotranscribed and comprise an operon. Genes in this operon, including htrA, were insertionally inactivated by recombination with a newly created suicide plasmid, pGPΩTp. Genetic analyses and complementation studies revealed that HtrABCAL2829 was required for growth of B. cenocepacia upon exposure to osmotic stress (NaCl or KCl) and thermal stress (44°C). In addition, replacement of the serine residue in the active site with alanine (S245A) and deletion of the HtrABCAL2829 PDZ domains demonstrated that these areas are required for protein function. HtrABCAL2829 also localizes to the periplasmic compartment, as shown by Western blot analysis and a colicin V reporter assay. Using the rat agar bead model of chronic lung infection, we also demonstrated that inactivation of the htrA gene is associated with a bacterial survival defect in vivo. Together, our data demonstrate that HtrABCAL2829 is a virulence factor in B. cenocepacia.
The Burkholderia cepacia complex (Bcc) comprises at least nine closely related bacterial species that are ubiquitous in the environment (6). Bcc bacteria are metabolically diverse, can degrade a variety of environmental pollutants (38), and also have plant-growth-promoting and antifungal properties (33, 38). However, Bcc bacteria are opportunistic pathogens that cause serious infections in immunocompromised individuals and in patients with cystic fibrosis (CF) (18).
Bcc infections in CF patients are complicated by the intrinsic resistance of the bacteria to most clinically relevant antimicrobial agents (18) and by their ability to be transmitted from person to person (17, 54). Bcc infections may also result in the “cepacia syndrome,” an often fatal necrotizing pneumonia (22). This severe outcome is rarely observed with other CF-related infections, distinguishing Bcc bacteria from other CF pathogens, such as Pseudomonas aeruginosa. B. cenocepacia is the most common Bcc species recovered from patients (45, 55) in most CF centers and is frequently associated with the most severe infections (32).
Since Bcc bacteria can survive in several different environmental niches, including humans, it is conceivable that these bacteria can readily adapt to changing environments. Adaptation to environmental stress in gram-negative bacteria is mediated in part by the extracytoplasmic stress response, which has been extensively characterized in Escherichia coli (43). Regulated expression of extracytoplasmic stress response genes in E. coli is mediated by the alternative sigma factor RpoE (σE) and the two-component regulatory systems BaeSR and CpxRA (9, 42, 47). Activation of the RpoE and Cpx regulons by accumulation of misfolded proteins in the outer membrane or in the periplasmic space (43) regulates the transcription of several genes encoding proteins that catalyze protein folding and degradation (10, 40).
One of these proteins is the periplasmic serine protease DegP, also known as HtrA (9, 10). Serine proteases of the HtrA family are highly conserved in bacteria, plants, and mammals (24). These proteases are defined by a conserved chymotrypsin-like protease domain and have at least one C-terminal PDZ domain (5). When misfolded proteins are not abundant, HtrA proteases generally function as chaperones (56). However, under conditions that cause misfolding of periplasmic proteins, HtrA functions as a protease (3, 56). The conserved PDZ domain, which promotes protein-protein interactions (51), is believed to play a key role in capturing substrates and regulating access of partially unfolded proteins to the catalytic domain (5, 24). In gram-negative and gram-positive bacteria, HtrA proteins are required for survival under environmental stress and for virulence (39, 58, 60). Analysis of the sequenced genome of B. cenocepacia J2315 (http://www.sanger.ac.uk/Projects/B_cenocepacia/) has revealed that this bacterium has five genes encoding predicted HtrA-like proteases, but the roles of these proteins in the physiology and pathogenesis of B. cenocepacia are unknown. In this study, we identified a six-gene operon encoding a putative two-component regulatory system and an HtrA-like serine protease (BCAL2829) in B. cenocepacia K56-2, which is clonally related to strain J2315 (31). Genetic analyses employing mutagenesis and complementation studies demonstrated that HtrA is required for growth under thermal and osmotic stress. Western blot analysis of periplasmic fractions and a colicin V secretion reporter assay also demonstrated that HtrABCAL2829 is exported to the periplasmic compartment. Finally, in vivo studies using the rat agar bead model of lung infection revealed that the htrABCAL2829 gene is required for B. cenocepacia survival during infection.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture media.
Bacterial strains and plasmids used in this study are listed in Table 1. Bacteria were routinely cultured in Luria broth (LB) (Difco) at 30 or 37°C with shaking. LB with an altered salt concentration was prepared with 1% (wt/vol) tryptone (Difco), 0.5% (wt/vol) yeast extract (Difco), and NaCl at final concentrations of 86 mM and 426 mM. For growth in the presence of an elevated KCl, NaH2PO4, or KH2PO4 concentration, LB was prepared with 86 mM NaCl and one of these salts at a concentration of 340 mM (final salt concentration, 426 mM). The pH of LB supplemented with NaH2PO4 or KH2PO4 was adjusted to 7.0. When required, E. coli cultures were supplemented with the following antibiotics (final concentrations): ampicillin, 100 μg/ml; tetracycline, 20 μg/ml; kanamycin, 40 μg/ml; trimethoprim, 50 μg/ml; chloramphenicol, 30 μg/ml; and gentamicin, 50 μg/ml. When required, B. cenocepacia cultures were supplemented with trimethoprim (100 μg/ml), tetracycline (100 μg/ml), chloramphenicol (100 μg/ml), and gentamicin (50 μg/ml).
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant characteristicsa | Source and/or reference |
|---|---|---|
| B. cenocepacia strains | ||
| K56-2 | ET12 clone related to J2315, CF clinical isolate | BCRRC; 31b |
| J2315 | ET12 clone, cbl+, CF clinical isolate | P. A. Sokol |
| RSF11 | K56-2, BCAM2160::pRF99, Tpr | This study |
| RSF12 | K56-2, BCAL2831::pRF103, Tpr | This study |
| RSF13 | K56-2, BCAL2829(htrA)::pRF109, Tpr | This study |
| RSF16 | K56-2, BCAL2826(coxG)::pRF115, Tpr | This study |
| RSF18 | K56-2, BCAL2828::pRF125, Tpr | This study |
| E. coli strains | ||
| DH5α | F− φ80dlacZΔM15 (ΔlacZYA-argF)U169 endA1 recA1 hsdR17(rK− mK+) supE44 thi-1 ΔgyrA96 relA1 | Laboratory stock |
| SY327 | araD Δ(lac pro) argE(Am) recA56 RifrnalA, λ pir | 35 |
| Plasmids | ||
| pGP704 | oriR6K, Apr, mob+ | 35 |
| pHP45ΩTet | oricolE1, ΩTetr cassette | 13 |
| pHP45ΩCm | oricolE1, ΩCmr cassette | 13 |
| pMLBAD | oripBBR1, Tpr, mob+, araC-PBAD | 28 |
| pUC18 | oricolE1, Apr, lacZ | 62 |
| pGPΩTp | oriR6K, ΩTpr cassette, mob+ | This study |
| pGPΩTet | oriR6K, ΩTetr cassette, mob+ | This study |
| pGPΩCm | oriR6K, ΩCmr cassette, mob+ | This study |
| pDA17 | oripBBR1, Tetr, mob+, Pdhfr, FLAG epitope | D. Aubert |
| pRK2013 | oricolE1, RK2 derivative, Kanr, mob+tra+ | 14 |
| pRF99 | pGPΩTp, 320-bp internal fragment from BCAM2160 | This study |
| pRF103 | pGPΩTp, 298-bp internal fragment from BCAL2831 | This study |
| pRF109 | pGPΩTp, 300-bp internal fragment from BCAL2829 | This study |
| pRF115 | pGPΩTp, 218-bp internal fragment from BCAL2826 | This study |
| pRF125 | pGPΩTp, 308-bp internal fragment from BCAL2828 | This study |
| pKMBAD | pMLBAD, Cmr | K. Maloney |
| pRF126 | pKMBAD, 3.3-kbp fragment containing BCAL2829, BCAL2828, BCAL2827, and BCAL2826 | This study |
| pRF127 | pRF126, BCAL2829::Kanr | This study |
| pRF128 | pRF126, BCAL2828::Kanr | This study |
| pRF129 | pRF126, BCAL2829/BCAL2828::Kanr | This study |
| pKD3 | Template plasmid, oriR6K, Kanr Apr | 11 |
| pKD46 | λ Red recombination system expression vector, ParaBAD, Apr | 11 |
| pColV | ColV expression vector, oripBBR1, Tpr, PrhaB, mob+ | D. Aubert |
| pRF130 | 210 bp encoding a 70-amino-acid N-terminal fragment of BCAL2829 | This study |
| pRF131 | 210 bp encoding a 70-amino-acid reverse N-terminal fragment of BCAL2829 | This study |
| pRF132 | BCAL2829, FLAG | This study |
| pRF134 | BCAL2829Δ PDZbp891-1485, FLAG | This study |
| pRF137 | BCAL2829 in pUC18 | This study |
| pRF138 | BCAL2829(S245A) | This study |
| pRF139 | BCAL2829(S245A), FLAG | This study |
Tpr, trimethoprim resistance; Kanr, kanamycin resistance; Tetr, tetracycline resistance; Cmr, chloramphenicol resistance; Apr, ampicillin resistance.
BCRRC, B. cepacia Research and Referral Repository for Canadian CF Clinics.
General molecular techniques.
DNA manipulations were performed as described previously (49). Restriction enzymes, T4 polynucleotide kinase, T4 DNA ligase (Roche Diagnostics), and mung bean nuclease (Amersham Pharmacia) were used as recommended by the manufacturers. E. coli DH5α cells were transformed by the calcium chloride protocol (7). E. coli SY327 cells were transformed by electroporation (12). Conjugation into B. cenocepacia K56-2 was accomplished by triparental mating (8) with E. coli strain DH5α carrying the helper plasmid pRK2013. DNA was amplified by PCR using the PTC-0200 or PTC-221 DNA engine (MJ Research) with either Taq DNA polymerase or Proof Start polymerase (QIAGEN). PCR mixtures used to amplify B. cenocepacia DNA were supplemented with QIAGEN Q solution according to the manufacturer's instructions. The DNA sequences of all primers used in this study and the specific PCR and reverse transcriptase PCR (RT-PCR) conditions are described in the supplemental material. DNA sequencing reactions were performed by the DNA Sequencing Facility at York University, Toronto, Ontario, Canada, and the Robarts Research Institute DNA Sequencing Facility at the University of Western Ontario, London, Ontario, Canada. The computer program BLAST was used to analyze the sequenced genome of B. cenocepacia strain J2315 (http://www.sanger.ac.uk/Projects/B_cenocepacia/).
Construction of the pGPΩ plasmids.
To rapidly inactivate B. cenocepacia genes in a targeted fashion, we constructed the suicide plasmids pGPΩTp, pGPΩTet, and pGPΩCm encoding resistance to trimethoprim, tetracycline, and chloramphenicol, respectively. These plasmids were derived from the pGP704 backbone, which carries the Pir protein-dependent R6K origin of replication (25). Insertion of any pGPΩ plasmid into a targeted gene results in a polar mutation due to the presence of the antibiotic resistance cassette flanked by omega (Ω) fragments (41). The details concerning plasmid construction and plasmid maps are provided in the supplemental material.
Mutagenesis of B. cenocepacia K56-2.
Insertional inactivation of B. cenocepacia K56-2 genes was performed using pGPΩTp. First, a PCR-amplified internal fragment from the target gene was cloned into pGPΩTp. Next, the resulting mutagenesis plasmid was conjugated into wild-type strain B. cenocepacia K56-2. Candidate mutants were identified by PCR and confirmed by Southern blot hybridization using the internal fragment labeled with digoxigenin as a probe. The BCAL2831, BCAL2829 (htrA), BCAL2828, BCAL2826, and BCAM2160 genes were targeted for mutagenesis and were inactivated using plasmids pRF103, pRF109, pRF115, pRF125, and pRF99, respectively. The corresponding mutant strains were designated RSF12, RSF13, RSF18, RSF16, and RSF11. Details concerning the construction of each mutant are provided in the supplemental material.
Bacterial growth.
Growth of B. cenocepacia was assessed by culture in LB alone or in LB with a modified salt content. Briefly, overnight cultures were used to inoculate fresh medium (LB or LB with excess salt; final volume, 5 ml) to obtain a starting optical density at 600 nm (OD600) of 0.005 (ca. 1.3 × 106 bacteria). Growth was monitored over time by determining the OD600 using a Beckman DU 530 spectrophotometer.
Thermal stress assay.
Single-cell suspensions with an OD600 of 0.1 were prepared from overnight cultures. Subsequently, the cell suspensions were serially diluted (100 to 10−4), and 10-μl portions of each dilution were dropped onto agar plates. The plates were then incubated at 30 and 44°C for 24 h and observed.
RT-PCRs.
RT-PCRs were performed as described previously (37) to investigate the transcriptional organization of htrA and neighboring genes. Total RNA was isolated using an RNeasy mini kit (QIAGEN) according to the manufacturers' protocol. Isolated RNA was treated with DNase (QIAGEN) for 30 min at 37°C and for 15 min at 75°C. To amplify the intergenic regions, reverse transcription reactions using RNA treated with RT and without RT (negative control) were performed. The reverse transcribed DNA was used as a template for PCR amplification, and genomic DNA served as a positive control. The primers used for each intergenic region are described in the supplemental material.
Complementation experiments.
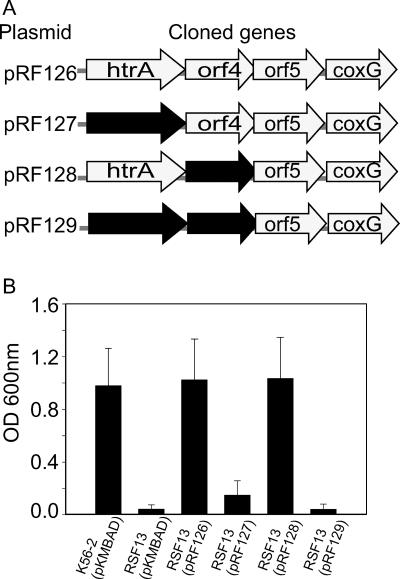
We constructed pRF126 to complement the salt-sensitive phenotype of mutant RSF13. This plasmid contained a 3.3-kbp fragment that included the four genes inactivated in RSF13, which was PCR amplified from B. cenocepacia K56-2 using primers 1801 and 1804, digested with KpnI and XbaI, and ligated into pKMBAD (Table 1). Derivatives of pRF126 with either BCAL2829 or BCAL2828 deleted and with both genes deleted were also constructed using the one-step PCR inactivation method, as described previously (11). These experiments resulted in generation of plasmids pRF127, pRF128, and pRF129 (Table 1), in which the appropriate targeted gene was replaced by a kanamycin resistance gene cassette (see Fig. 6A). Plasmids pRF126, pRF127, pRF128, and pRF129 were introduced into B. cenocepacia RSF13 by conjugation, and complementation was assessed by growth in LB containing excess salt.
FIG. 6.
Complementation of the B. cenocepacia RSF13 osmotic growth defect. (A) Genes cloned into complementation plasmids pRF126, pRF127, pRF128, and pRF129 for strain RSF13. The shaded arrows represent the genes (BCAL2831 to BCAL2826) and indicate the direction of transcription. The solid arrows represent genes that are replaced with a kanamycin resistance cassette. (B) Strains K56-2 and RSF13 carrying the control vector pKMBAD or derivatives of complementation plasmid pRF126 were cultured in LB with 426 mM NaCl at 37°C. The values are the means of three independent experiments in which each exconjugant was analyzed in triplicate. The error bars indicate the standard deviations.
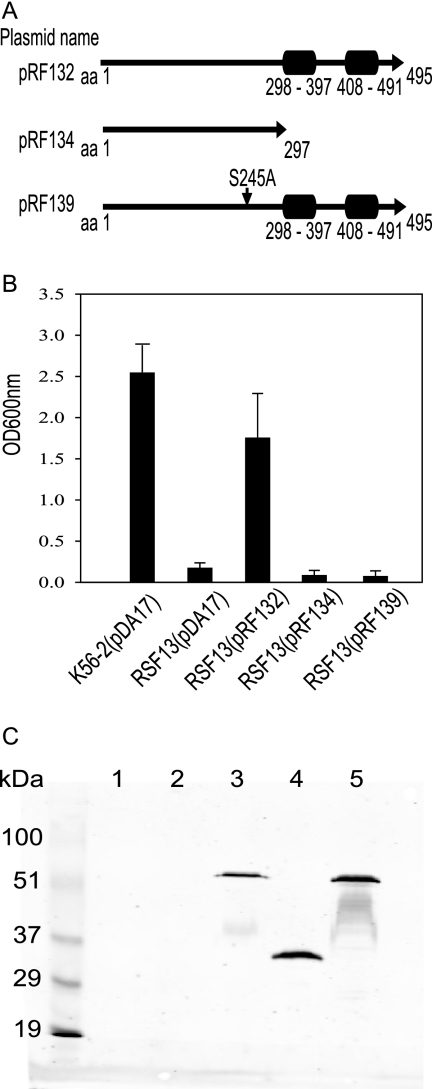
The complementation plasmids pRF132, pRF134, and pRF139 were created to assess the importance of the HtrABCAL2829 PDZ domains and the conserved active site serine residue (S245) in the complementation of RSF13. These plasmids included the full-length BCAL2829 gene (pRF132) or a truncated variant lacking both PDZ domains (pRF134). Each fragment was PCR amplified and cloned into pDA17 as an EcoRI and XbaI fragment and was fused with the FLAG epitope. To generate pRF139, which included BCAL2829 S245A, the BCAL2829 gene was excised from pRF132 and cloned into pUC18, resulting in pRF137. Site-directed mutagenesis of pRF137 using a QuikChange site-directed mutagenesis kit from Stratagene was performed as recommended by the supplier, resulting in pRF138. The single amino acid substitution, S245A, was confirmed by DNA sequencing. The resulting mutated BCAL2829 gene was excised from pRF138 and cloned into pDA17, resulting in pRF139 (see Fig. 8A). Each complementation plasmid was introduced into RSF13 by conjugation, and growth was analyzed.
FIG. 8.
Analysis of PDZ domain deletions and active site mutagenesis of HtrABCAL2829 for RSF13 complementation. (A) Schematic maps of the HtrABCAL2829 protein expressed from pRF132, pRF134, and pRF139. (B) Strains K56-2 and RSF13 carrying pDA17 or the complementing plasmid pRF132, pRF134, or pRF139 were cultured in LB with 426 mM NaCl at 37°C. The values are the means of three independent experiments in which each transformant was analyzed in triplicate. The error bars indicate standard deviations. (C) Anti-FLAG Western blot analysis of periplasmic fractions recovered from K56-2 and RSF13 carrying pDA17, pRF132, pRF134, or pRF139. Lane 1, B. cenocepacia K56-2(pDA17); lane 2, B. cenocepacia RSF13(pDA17); lane 3, B. cenocepacia RSF13(pRF132); lane 4, B. cenocepacia RSF13(pRF134); lane 5, B. cenocepacia RSF13(pRF139). The lane on the left contained molecular mass markers, and the sizes (in kDa) are indicated.
Preparation of periplasmic proteins and Western blot analysis.
Periplasmic proteins were isolated as described previously, with the following modifications (23). Bacterial cells were pelleted by centrifugation (9,000 × g for 10 min) and resuspended in 2.5 ml of lysis buffer (20% [wt/vol] sucrose, 30 mM Tris-HCl [pH 8.0], 1 mM EDTA [pH 8.0], 3.5 mg/ml lysozyme). Cell suspensions were incubated at 37°C for ∼3 h and visualized by light microscopy to confirm that spheroplasts were formed, and then they were centrifuged as described above. Supernatants containing periplasmic proteins were electrophoresed (15 μg/ml total protein) on 14% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels and transferred to nitrocellulose membranes for immunoblot analysis. The membranes were incubated with the FLAG M2 monoclonal antibody (Sigma), and Alexa Fluor 680 goat anti-mouse immunoglobulin G (Molecular Probes) was used as a secondary antibody. Detection was performed by infrared imaging, using an Odyssey infrared imager (LI-COR Biosciences).
Colicin V secretion reporter assay.
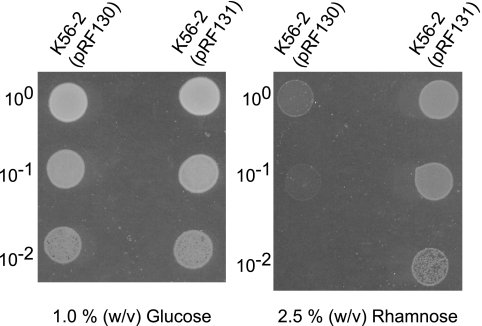
A colicin V secretion reporter system was used to determine whether HtrABCAL2829 is exported to the periplasm. Colicin V is cytotoxic only if it is present in the periplasm. The pColV expression plasmid encodes leaderless colicin V under control of the rhamnose-inducible promoter PrhaR, and its construction will be described elsewhere (D. Aubert and M. A. Valvano, unpublished). The 5′ coding region of BCAL2829 encoding the first 70 N-terminal amino acids of HtrABCAL2829, which contain a putative signal peptide sequence, was PCR amplified, digested with NdeI and SphI, and cloned into pColV in the forward and reverse orientations. The resulting plasmids, pRF130 and pRF131, encode in-frame HtrA-colicin V and revHtrA-colicin V protein fusions, respectively (Table 1). These reporter plasmids were introduced into B. cenocepacia K56-2 by conjugation, and bacteria carrying pRF130 or pRF131 were cultured overnight in LB supplemented with 1% (wt/vol) glucose. The overnight cultures were centrifuged, the cell pellets were resuspended in fresh LB without glucose, and the OD600 was adjusted to 0.05 (∼ 1.3 × 107 bacteria). The suspensions were serially diluted, and 10-μl aliquots were spotted onto LB agar containing 2.5% (wt/vol) rhamnose and LB agar containing 1% (wt/vol) glucose to induce and repress gene expression, respectively. The plates were incubated at 37°C for 20 to 24 h and observed.
Animal infections and CI.
Animal infection experiments were performed using the rat model of chronic respiratory infection described previously by Cash et al. (4). To determine the competitive index (CI), approximately equal numbers (5.0 × 105 CFU) of wild-type and mutant bacteria were coembedded in agar beads and used to inoculate each rat. The inoculation ratio of mutant to wild-type bacteria in the agar beads was determined by plating on B. cepacia selective agar with and without trimethoprim. On day 14 postinfection, the lungs of each infected rat were removed aseptically, homogenized, and plated as described previously (21). The CI was calculated by dividing the mean output ratio of mutant to wild-type bacteria by the mean input ratio of mutant to wild-type bacteria.
RESULTS
Identification and mutagenesis of two operons encoding candidate PmrA response regulators in B. cenocepacia K56-2.
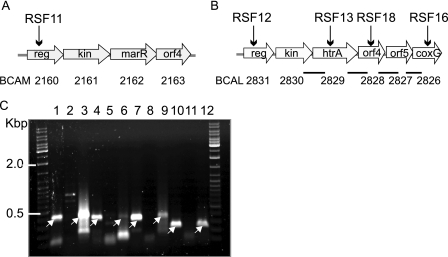
Resistance to antimicrobial peptides in P. aeruginosa and Salmonella enterica is controlled in part by the PmrA/PmrB two-component regulatory system (19, 34). In an attempt to identify similar regulators in Bcc species, we searched the sequenced genome of B. cenocepacia J2315 for genes encoding homologs of the PmrA response regulator. From several genes encoding putative response regulators, BCAM2160 and BCAL2831 were selected for further study as they encoded proteins that exhibited the highest levels of identity (48 and 53%, respectively) at the primary amino acid sequence level with PmrA of P. aeruginosa. BCAM2160 is the first gene of a predicted four-gene operon spanning bp 2409268 to 2411814 on chromosome 2 (Fig. 1A). The other genes in this putative operon encode a predicted sensor kinase (BCAM2161), a transcriptional regulator of the MarR family (BCAM2162), and a conserved uncharacterized bacterial protein (BCAM2163). The other gene encoding a protein similar to PmrA, BCAL2831, is the first gene of a putative six-gene operon spanning bp 3105477 to 3110139 on chromosome 1 (Fig. 1B). The downstream genes in this operon encode a sensor kinase (BCAL2830), a predicted HtrA (BCAL2829), a hypothetical protein (BCAL2828), a conserved protein with a domain having an unknown function (BCAL2827), and a putative membrane protein (BCAL2826) with a domain related to CoxG, an accessory protein of the carbon monoxide dehydrogenase complex (50). We confirmed by PCR analysis that these putative operons had the same gene organization in B. cenocepacia K56-2 (data not shown), which is clonally related to sequenced strain J2315 (31).
FIG. 1.
Genetic organization and analysis of the gene clusters encoding candidate PmrA response regulators in B. cenocepacia J2315 and K56-2. The positions of the genes and the direction of transcription are indicated by arrows. The vertical arrows indicate genes in which pGPΩTp was integrated, and the corresponding strain designation is indicated above each arrow. The BCAL and BCAM gene designations are based on a preliminary annotation of the B. cenocepacia J2315 genome (www.sanger.ac.uk/Projects/B_cenocepacia/). The abbreviated gene designations are as follows: reg, response regulator; kin, sensor kinase; orf, open reading frame. (A) Four-gene cluster located on chromosome 2 containing genes BCAM2160 to BCAM2163. (B) Six-gene operon located on chromosome 1 containing genes BCAL2831 to BCAL2826. The bars indicate the regions analyzed by RT-PCR as shown in panel C. (C) RT-PCR analysis of the intergenic regions for genes BCAL2831 to BCAL2826. Lanes 1 to 3, DNA with no RT and RT for BCAL2830 and BCAL2829; lanes 4 to 6, DNA with no RT and RT for BCAL2829 and BCAL2828; lanes 7 to 9, DNA with no RT and RT for BCAL2828 and BCAL2827; lanes 10 to 12, DNA with no RT and RT for BCAL2827 and BCAL2826. The arrows indicate bands at expected positions.
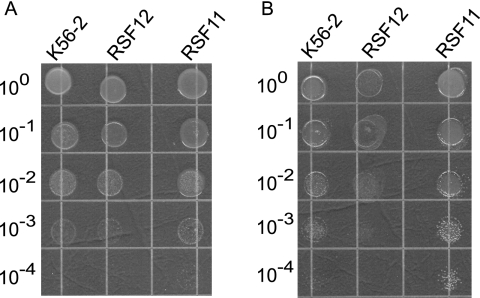
We constructed pGPΩTp (see Fig. S1A in the supplemental material) to assess the role of these regulators in antimicrobial peptides resistance. Derivatives of pGPΩTp carrying gene fragments from BCAM2160 and BCAL2831 were constructed, and the plasmids were integrated by homologous recombination into the response regulator genes. The appropriate insertion event was confirmed by PCR analysis and Southern blot hybridization (data not shown), resulting in B. cenocepacia strains RSF11 (BCAM2860::pRF99) and RSF12 (BCAL2831::pRF103).
Strains RSF11 and RSF12 were assessed to determine their sensitivity to 100 μg/ml polymyxin B. This concentration is threefold higher than the MIC for a polymyxin B-sensitive B. cenocepacia K56-2 mutant producing a truncated core lipopolysaccharide (30). RSF11, RSF12, and parental strain K56-2 were equally resistant to polymyxin B, while E. coli K-12 strain W3110, used as a negative control, was sensitive (data not shown). In some bacterial species, inactivation of PmrA is also associated with ferric iron toxicity (59). The RSF11 and RSF12 mutants did not show any growth defects in the presence of 800 mM FeCl3 compared to parental strain K56-2 (data not shown). Together, these observations suggest that the putative PmrA-like regulators encoded by BCAM2860 and BCAL2831 do not have the functional properties attributed to PmrA in S. enterica and P. aeruginosa.
Inactivation of the BCAL2831-BCAL2826 gene cluster affects growth under osmotic and thermal stress.
In B. cenocepacia RSF12, insertional inactivation of the response regulator gene BCAL2831 with pGPΩΤp should also inactivate the five downstream genes (BCAL2830 to BCAL2826) if they are part of an operon (Fig. 1B). The intergenic regions between BCAL2830, htrA (BCAL2829), BCAL2828, BCAL2827, and coxG (BCAL2826) were analyzed by RT-PCR, which resulted in amplification of the expected products from isolated RNA (Fig. 1B and C). The no-RT reaction used as a negative control for BCAL2829 and BCAL2828 produced a faint band at a slightly higher molecular weight that did not correspond to the size of the expected amplicon obtained in the DNA control or the RT reaction (Fig. 1C, lanes 4 to 6). The intergenic region between BCAL2831 and BCAL2830 was not analyzed as cotranscription of these genes was expected given their proximity to one another. These results demonstrated that the six genes in the BCAL2831-BCAL2826 cluster are indeed part of a single transcriptional unit.
Since BCAL2829 encodes a predicted serine protease of the HtrA family and its transcription should be affected by integration of pGPΩTp into BCAL2831 (Fig. 1B), we hypothesized that growth of RSF12 would be altered under stress conditions known to affect htrA null bacteria (39, 58, 60). However, RSF12 did not exhibit any growth defects when it was cultured in rich (brain heart infusion or LB) or minimal (M56) medium compared to the growth of parental strain K56-2 and the negative control strain RSF11 (Fig. 2A and data not shown). No differences were found when RSF12 was compared to RSF11 and the parental strain K56-2 for sensitivity to oxidative stress (H2O2 and paraquat), growth under acidic conditions (pH 4.8 and 6.2), detergent (8 μl of 10% [wt/vol] sodium dodecyl sulfate by disk diffusion), antibiotics in liquid culture (350 μg/ml puromycin, 10 μg/ml tetracycline, and 50 μg/ml chloramphenicol), indole (5 mM), or 50% (vol/vol) rat serum or for the ability to form biofilms (data not shown).
FIG. 2.
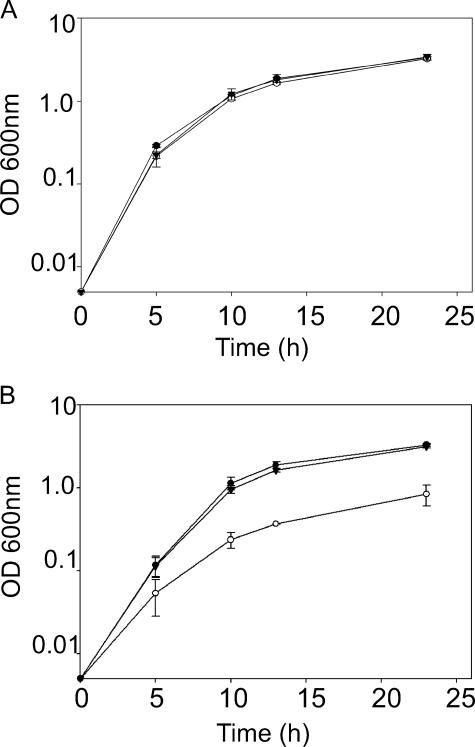
Growth of B. cenocepacia RSF12 in thermal stress conditions. B. cenocepacia strains were drop plated and cultured for 24 h at 30°C (A) or 44°C (B). The results are representative of at least three independent experiments.
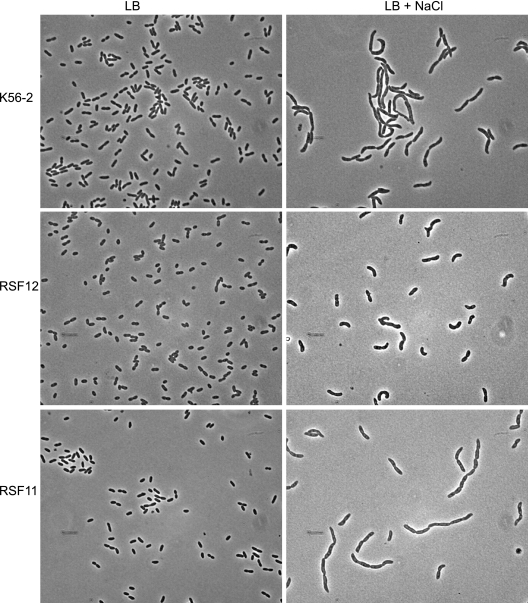
The RSF12 mutant did not exhibit a survival defect after incubation for 1 h at 42°C. However, a 10- to 100-fold reduction in growth was observed after incubation for 24 h at 44°C compared to the growth of K56-2 and RSF11 (Fig. 2B). The same strains grew equally well at 30 and 37°C (Fig. 2A and 3A). The RSF12 mutant also displayed a growth defect when it was cultured in the presence of 426 mM NaCl or KCl (Fig. 3B and Fig. 4) ; however, this defect was not apparent in standard LB (Fig. 3A). Similar growth impairment was observed in the presence of 426 mM salt when sodium phosphate and potassium phosphate were used (data not shown), which ruled out the possibility that Cl− ion toxicity is a cause of the salt-sensitive phenotype of RSF12. The RSF12 growth defect was also seen when the strain was cultured with 500 mM sucrose (data not shown). The growth defect of B. cenocepacia RSF12 in the presence of an elevated level of salt was also apparent on solid media, on which the mutant formed pinpoint colonies compared to the colonies of the control strains (data not shown). Exposure to an elevated level of NaCl also resulted in morphological differences between parental strain K56-2 and the RSF12 mutant. As determined by phase-contrast microscopy, cells of strains K56-2 and RSF11 became filamentous in the presence of a high salt concentration (Fig. 5), while RSF12 cells were much shorter (Fig. 5), resembling more closely the morphology of B. cenocepacia cultured in standard LB (Fig. 5). Together, the growth defect and the lack of cell filamentation suggest that RSF12 cells, in contrast to the cells of parental strain K56-2 and the control mutant RSF11, cannot adapt to thermal stress or high osmolarity, particularly excess NaCl or KCl.
FIG. 3.
Growth of B. cenocepacia RSF12 in high-salt conditions. B. cenocepacia K56-2 (•), RSF12 (○), and RSF11 (▴) were cultured at 37°C in LB containing either 86 mM NaCl (A) or 426 mM NaCl (B). The values are the means of three independent experiments in which each strain was analyzed in triplicate. The error bars indicate the standard deviations.
FIG. 4.
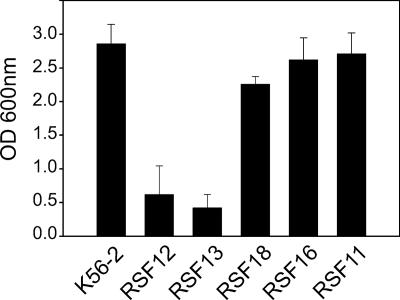
Growth of the B. cenocepacia BCAL2831-BCAL2830-BCAL2829-BCAL2828-BCAL2827-BCAL2826 operon mutants in the presence of excess salt. Parental strain K56-2 and insertional mutants were cultured at 37°C in LB with 426 mM NaCl. The end point OD600 was obtained after 24 h. The values are the means of three independent experiments. The error bars indicate the standard deviations.
FIG. 5.
Phase-contrast microscopy of mutant RSF12 and the B. cenocepacia control strains grown in the presence and absence of excess salt. Cells from B. cenocepacia K56-2, RSF12, and RSF11 cultures are shown. The bacteria were cultured in LB and LB with 426 mM NaCl. Magnification, ×800.
Osmotic and thermal growth defects are associated with inactivation of htrABCAL2829.
To determine which gene or genes were responsible for adaptation to high levels of salt, we mutagenized htrABCAL2829, BCAL2828, and BCAL2826 by insertional inactivation with pGPΩTp. These experiments produced mutant strains RSF13, RSF18, and RSF16, respectively. Strain K56-2 and mutants RSF12, RSF13, RSF18, RSF16, and RSF11 (as a negative control) were grown in the presence of excess NaCl. As expected, there were no growth differences among these strains when they were cultured in LB with the normal salt concentration (data not shown). In contrast, the growth of RSF12 and RSF13 was significantly reduced when these strains were cultured in media with a high concentration of salt (Fig. 4 and data not shown). Similar to RSF12, B. cenocepacia RSF13 exhibited the same morphology when it was cultured in high-salt conditions (data not shown). These data indicate that the salt-sensitive phenotype is due to inactivation of the htrABCAL2829 gene and not the downstream genes (BCAL2828 to BCAL2826).
To confirm the role of htrABCAL2829 in salt sensitivity, plasmid pRF126 was conjugated into the RSF13 mutant in an attempt to restore growth. This plasmid carries the htrA gene and the three other downstream genes (BCAL2828, BCAL2827, and BCAL2826) under control of the PBAD promoter. When strains were cultured in the presence of excess NaCl, the growth of RSF13(pRF126) was comparable to the growth of K56-2 containing the plasmid vector pKMBAD, while RSF13(pKMBAD) grew very poorly (Fig. 6B). Addition of arabinose was not required to observe complementation, likely due to the trace amounts of the sugar in the culture medium. Additional plasmids were constructed by replacing htrA, BCAL2828, and both htrA and BCAL2828 with a kanamycin resistance cassette, which resulted in plasmids pRF127, pRF128, and pRF129, respectively (Fig. 6A). Plasmid pRF128 restored growth of RSF13 in the presence of excess NaCl, while neither pRF127 nor pRF129 rescued the salt-sensitive phenotype of the mutant strain when htrABCAL2829 was inactivated (Fig. 6B).
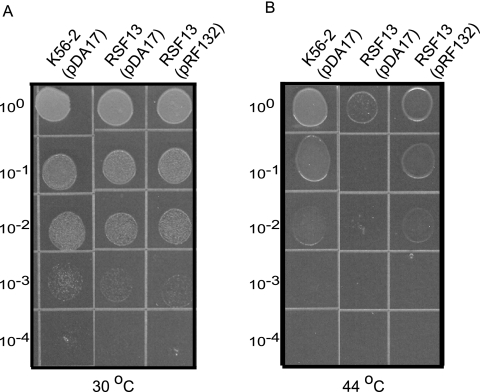
The requirement for the htrABCAL2829 gene for growth under thermal stress was also demonstrated by complementation of strain RSF13 with a plasmid that expressed only the htrA gene (Fig. 7B). In contrast, RSF13 carrying the control vector pDA17 exhibited a 100-fold reduction in growth at 44°C compared to the growth of K56-2(pDA17) or RSF13 carrying htrABCAL2829 complementation plasmid pRF132. Each strain grew equally well when it was cultured at 30°C (Fig. 7A). Together, these experiments conclusively demonstrated that the thermal and salt-sensitive growth defects of strain RSF13 are due to inactivation of the htrA gene. Moreover, complementation experiments using the plasmids could not restore growth of B. cenocepacia RSF12 under the same conditions, indicating that the two-component regulatory system also plays a role in adaptation to these stresses independent of HtrABCAL2829 (data not shown).
FIG. 7.
Complementation of the B. cenocepacia RSF13 temperature-dependent growth defect. B. cenocepacia K56-2 and RSF13 carrying either the control vector pDA17 or the complementation plasmid pRF132 were drop plated and cultured for 24 h at 30°C (A) or 44°C (B). The results are representative of at least three independent experiments.
Predicted active site serine residue and PDZ domains are required for HtrA function.
In E. coli, HtrA (DegP) functions as both a periplasmic chaperone and a serine protease (5, 51, 56). To characterize in part the mechanism of HtrABCAL2829 function in B. cenocepacia, we sought to determine whether the PDZ domains, the predicted active site serine residue, or both were needed to restore growth of the RSF13 mutant under osmotic stress conditions. We created plasmids expressing either the wild-type HtrABCAL2829 protein (pRF132), HtrABCAL2829 with both PDZ domains deleted (pRF134), or an HtrABCAL2829 mutant with the predicted active site serine residue (S245) changed to alanine (pRF139) (Fig. 8A). In each case, the protein expressed was C terminally tagged with the FLAG epitope. Introduction of pRF132 into RSF13 restored growth to approximately the same level as the growth of parental strain K56-2 carrying a vector control (Fig. 8B). Deletion of the PDZ domains or mutation of the active site serine residue abrogated complementation and resulted in a growth defect similar to that of RSF13 carrying only the vector control (Fig. 8B). In separate experiments expression of HtrABCAL2829 with deletion of only one PDZ domain also could not restore growth to RSF13 in high-salt conditions (data not shown). To verify that the lack of complementation was not due to poor protein expression, an anti-FLAG immunoblot analysis was performed with periplasmic protein extracts from RSF13 carrying pRF132, pRF134, and pRF139. The bands for full-length BCAL2829 protein and the PDZ deletion protein migrated at approximately 52 and 34 kDa, respectively (Fig. 8C, lanes 3 and 4). These molecular masses are in agreement with the predicted molecular masses of the wild-type protein and the protein with PDZ deleted. Neither parental strain K56-2 nor RSF13 expressed any periplasmic proteins that were detectable with the anti-FLAG monoclonal antibody, confirming that the polypeptides in Fig. 8, lanes 3, 4, and 5, contained the FLAG epitope (Fig. 8C, lanes 1 to 5). Together, these results indicate that BCAL2829 is a periplasmic protease that requires the active site serine residue and PDZ domains to promote growth of B. cenocepacia under high-salt conditions.
N-terminal leader of the HtrA protease is required for export to the periplasmic space.
The predicted amino acid sequence of HtrABCAL2829 revealed a cleavable N-terminal leader peptide (2). To confirm localization of HtrABCAL2829 to the periplasmic compartment, as predicted by Western blot analysis of periplasmic extracts, we used a rhamnose-inducible colicin V export reporter system. Colicin V kills bacterial cells by disrupting their membrane potential once the colicin gains access to the periplasmic face of the inner membrane (61). Plasmids expressing the first 70 amino acids of HtrABCAL2829 fused to leaderless colicin V in either the forward (pRF130) or reverse (pRF131) orientation were generated. B. cenocepacia K56-2 was transformed with these constructs, and cell viability was assessed in the presence of rhamnose and glucose. B. cenocepacia K56-2 carrying pRF131, which did not encode a functional leader fused to colicin V, grew in medium with rhamnose (Fig. 9). In contrast, K56-2 carrying pRF130, which should have encoded a functional HtrABCAL2829 leader peptide fused to colicin V, failed to grow in medium with rhamnose (Fig. 9). Conversely, B. cenocepacia K56-2 carrying either plasmid grew well in the presence of glucose when the promoter was repressed (Fig. 9). These data, together with the Western blot analysis results, demonstrated that HtrABCAL2829 has a functional leader peptide that directs the protein to the periplasmic compartment.
FIG. 9.
Expression of HtrABCAL2829/colicin V fusion proteins in B. cenocepacia K56-2. The indicated dilutions of cultures of strain K56-2 carrying plasmids encoding the N terminus of HtrA fused to colicin V in the forward (pRF130) and reverse (pRF131) orientations were spotted on LB agar plates containing either 1.0% (wt/vol) glucose or 2.5% (wt/vol) rhamnose. The results are representative of four independent experiments.
HtrABCAL2829 is required for survival in a rat model of chronic lung infection.
The in vivo role of the two operons inactivated in RSF12 and RSF11 was investigated using the rat agar bead model of lung infection (4) involving competition with parental strain K56-2. RSF12 was unable to compete with parental strain K56-2, as no mutant colonies were recovered after infection, whereas 4.7 × 104 ± 7.6 × 104 CFU/ml of K56-2 was recovered at this time (Table 2 and data not shown). In contrast, RSF11 could compete with parental strain K56-2, and the competitive index was 1.06, indicating that equal numbers of the two bacteria were present (Table 2). These data demonstrate that inactivation of BCAL2831 and the downstream genes compromises bacterial survival in vivo, while inactivation of BCAM2160 has no effect on survival. Additional infection experiments with mutants RSF13, RSF18, and RSF16 were performed to determine which of the genes inactivated in B. cenocepacia RSF12 were required for survival in the rat. Mutants RSF18 and RSF16 gave competitive indices of 1.08 and 1.04, respectively, indicating that these mutants could compete with K56-2 (Table 2). In contrast, 14 days after infection RSF13 could not be recovered from seven of nine rats in two independent experiments, demonstrating that the HtrABCAL2829 protein is important in the survival of B. cenocepacia in the rat model of chronic lung infection (Table 2). The RSF13 isolates that were recovered from two of the infected rats were analyzed by PCR, which demonstrated that the pGPΩTp plasmid remained integrated in the htrA gene. Interestingly, phenotypic analysis of these isolates showed that they had an intermediate ability to grow in high-salt media compared to the original RSF13 isolate. These observations indicate that the mutants recovered from the rats were in some way different from the original RSF13 mutant and may suggest that secondary mutations can compensate for the inactivation of htrABCAL2829 (data not shown).
TABLE 2.
Competition assays with the B. cenocepacia mutants
| B. cenocepacia strain | Locus of pGPΩTp integration | CIa |
|---|---|---|
| RSF12 | BCAL2831, response regulator | NR |
| RSF13 | BCAL2829, HtrA protease | NRb |
| RSF18 | BCAL2828, unknown function | 1.08 |
| RSF16 | BCAL2826, CoxG | 1.04 |
| RSF11 | BCAM2160, response regulator | 1.06 |
The CI was determined by dividing the mean output ratio of the mutant to the wild type by the mean input ratio of the mutant to the wild type. Six or seven rats were analyzed for each mutant. NR, not recovered.
For RSF13 two independent experiments were performed using a total of nine rats. Seven of the nine rats completely cleared RSF13; however, bacteria were recovered from two rats (see the text for details).
DISCUSSION
In this study, we identified a six-gene operon encoding a putative two-component regulatory system (BCAL2831 and BCAL2830) and a predicted HtrA-like protease (BCAL2829). Two-component regulatory systems in several bacteria regulate the transcription of HtrA proteases (9, 52, 57), and these proteases are required for growth under environmental stress conditions (39, 58, 60). Mutagenesis of the B. cenocepacia htrA gene was facilitated by the suicide plasmid pGPΩTp, which has been used successfully to inactivate several other B. cenocepacia genes in addition to those investigated here (26, 27, 30). We determined that several stresses known to adversely affect HtrA null bacteria do not cause toxicity or inhibit growth of a B. cenocepacia mutant lacking HtrABCAL2829. However, osmotic stress and prolonged heat stress significantly impaired the growth of the htrABCAL2829 null strain. Osmotic growth defects of htrA mutants have been reported for other bacteria, but these mutants generally exhibit sensitivity to several other stresses (2, 3, 39, 60), which was not observed here. Analysis of the sequenced genome of B. cenocepacia J2315 revealed that this bacterium contains five predicted HtrA-like proteases. Therefore, it is conceivable that other htrA alleles may be needed for adaptation to stress conditions for which HtrABCAL2829 does not appear to be required. We are currently investigating the functional role of these additional HtrA-like proteins.
Cell filamentation is observed in many bacteria under stress (29, 36, 46, 63), probably as a result of the normal stress response. In our experiments, control strains expressing a functional HtrABCAL2829 formed long filaments under salt stress conditions, consistent with previous observations of NaCl-stressed Salmonella and Listeria (16, 20). In contrast, the htrA mutant cells were short rods whose morphology resembled the morphology of unstressed bacteria, which suggests that the normal cellular response is in some way defective.
HtrA serine proteases are defined by the conserved catalytic triad comprised of histidine, aspartic acid, and serine residues and at least one C-terminal PDZ domain (5). The catalytic residues and the PDZ domains are essential for HtrA function in E. coli (51, 53). Expression of an HtrABCAL2829 site-directed mutant protein with an S245A substitution could not restore growth to B. cenocepacia RSF13 in high-salt conditions and is consistent with the notion that BCAL2829 does in fact code for an HtrA-like serine protease. Also, HtrABCAL2829 with PDZ domains deleted did not restore growth of RSF13 under osmotic stress, indicating that the predicted PDZ domains are required for function. Alternatively, the absence of PDZ domains may affect enzymatic activity by compromising HtrA integrity.
The E. coli DegP protein is in the periplasmic space, where it processes misfolded proteins (51). Our results from Western blot analysis of periplasmic fractions demonstrated that HtrABCAL2829 tagged with a FLAG epitope is located in the periplasmic space. Furthermore, we showed that the HtrABCAL2829 N-terminal signal peptide is sufficient to export colicin V to the periplasm. We concluded from these experiments that HtrABCAL2829 is a periplasmic HtrA-like serine protease that presumably functions in a manner analogous to the E. coli DegP protease activity.
In vivo infection experiments employing the rat agar bead model of chronic lung infection demonstrated that HtrABCAL2829 is required for survival of B. cenocepacia in infected animals. Temperature- and salt-sensitive phenotypes may suggest that htrABCAL2829 mutants are more sensitive to general stresses and therefore present with a survival defect in vivo. However, the htrA mutant is not sensitive to antimicrobial peptides, oxidative stress, and rat serum, all of which are stresses that bacteria encounter in vivo (1, 15, 48), making explanation of the survival defect more difficult at this time. It is probable that a combination of stresses encountered in vivo results in the killing of B. cenocepacia lacking HtrABCAL2829. However, our experiments have not excluded other immunological components of the lung that may kill the htrA mutant. In the phagosome lumen of the neutrophil the osmolarity approaches 500 mM (44), approximately the concentration of salt used here to inhibit the growth of the htrA mutant. It is tempting to speculate that B. cenocepacia may experience some form of osmotic stress in vivo, although this remains to be established.
It is interesting that RSF13 was recovered from two infected rats, whereas in the seven other animals the same strain was completely cleared. Possibly these two rats had some inherent factor such that selective pressure in them resulted in a secondary mutation that compensated for the lack of HtrABCAL2829. The latter hypothesis is supported by the results of a PCR analysis of recovered isolates confirming that pGPΩTp was still integrated in the htrA gene and that the same isolates exhibited an intermediate growth defect in the presence of excess salt (R. S. Flannagan and M. A. Valvano, unpublished). Despite these few recovered isolates, the original RSF13 mutant could not persist in the environment of the rat lung, and this demonstrated that HtrABCAL2829 is required for survival in vivo. The RSF12 mutant may also not have competed with the parental strain in part due to inactivation of the htrABCAL2829 gene as a result of the polar effect of pGPΩTp. However, the inability to restore growth to B. cenocepacia RSF12 through expression of HtrABCAL2829 alone indicates that the two-component regulatory system comprised of BCAL2831 and BCAL2830 may regulate genes required for growth under stress conditions and for survival in vivo. The role of this regulatory system in the adaptation of B. cenocepacia to stress is currently being investigated.
In summary, we discovered an HtrA protease in B. cenocepacia that is required for bacterial survival in vivo and for growth under osmotic and long-term thermal stress. Our data indicate that HtrABCAL2829 functions as a periplasmic serine protease and that it presumably degrades misfolded proteins that arise when B. cenocepacia is subjected to stress. The presence of multiple genes encoding predicted HtrA-like proteases suggests that B. cenocepacia may be particularly well equipped to combat adverse conditions encountered in different environments, including mammalian hosts. Further investigation of these proteases and a better understanding of stress responses in B. cenocepacia should shed light on how this organism is able to persist in many different environments.
Supplementary Material
Acknowledgments
We thank the members of our laboratories for helpful discussions and J. Parkhill for providing access to the draft annotation of B. cenocepacia J2315. We specifically thank S. Loutet and T. Stephens for critically reading the manuscript.
This work was supported by a grant from the Canadian Institutes of Health Research and a special program grant in memory of Michael O'Reilly from the Canadian Cystic Fibrosis Foundation (to M.A.V.) and by a grant from the Canadian Cystic Fibrosis Foundation (to P.A.S.). D.A. was supported by a postdoctoral fellowship from the Canadian Institutes of Health Research and the Canadian Foundation for Infectious Disease. R.S.F. was supported by a graduate student fellowship from the Canadian Cystic Fibrosis Foundation. M.A.V. holds a Canada Research Chair in Infectious Diseases and Microbial Pathogenesis.
Editor: V. J. DiRita
Footnotes
Published ahead of print on 12 January 2007.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Babior, B. M. 2004. NADPH oxidase. Curr. Opin. Immunol. 16:42-47. [DOI] [PubMed] [Google Scholar]
- 2.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 3.Betton, J. M., N. Sassoon, M. Hofnung, and M. Laurent. 1998. Degradation versus aggregation of misfolded maltose-binding protein in the periplasm of Escherichia coli. J. Biol. Chem. 273:8897-8902. [DOI] [PubMed] [Google Scholar]
- 4.Cash, H. A., D. E. Woods, B. McCullough, W. G. Johanson, Jr., and J. A. Bass. 1979. A rat model of chronic respiratory infection with Pseudomonas aeruginosa. Am. Rev. Respir. Dis. 119:453-459. [DOI] [PubMed] [Google Scholar]
- 5.Clausen, T., C. Southan, and M. Ehrmann. 2002. The HtrA family of proteases: implications for protein composition and cell fate. Mol. Cell 10:443-455. [DOI] [PubMed] [Google Scholar]
- 6.Coenye, T., and P. Vandamme. 2003. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ. Microbiol. 5:719-729. [DOI] [PubMed] [Google Scholar]
- 7.Cohen, S. N., A. C. Chang, and L. Hsu. 1972. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc. Natl. Acad. Sci. USA 69:2110-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Craig, F. F., J. G. Coote, R. Parton, J. H. Freer, and N. J. Gilmour. 1989. A plasmid which can be transferred between Escherichia coli and Pasteurella haemolytica by electroporation and conjugation. J. Gen. Microbiol. 135:2885-2890. [DOI] [PubMed] [Google Scholar]
- 9.Danese, P. N., W. B. Snyder, C. L. Cosma, L. J. Davis, and T. J. Silhavy. 1995. The Cpx two-component signal transduction pathway of Escherichia coli regulates transcription of the gene specifying the stress-inducible periplasmic protease, DegP. Genes Dev. 9:387-398. [DOI] [PubMed] [Google Scholar]
- 10.Dartigalongue, C., D. Missiakas, and S. Raina. 2001. Characterization of the Escherichia coli sigma E regulon. J. Biol. Chem. 276:20866-20875. [DOI] [PubMed] [Google Scholar]
- 11.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16:6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fellay, R., J. Frey, and H. Krisch. 1987. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene 52:147-154. [DOI] [PubMed] [Google Scholar]
- 14.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ganz, T. 2003. Defensins: antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 3:710-720. [DOI] [PubMed] [Google Scholar]
- 16.Geng, T., K. P. Kim, R. Gomez, D. M. Sherman, R. Bashir, M. R. Ladisch, and A. K. Bhunia. 2003. Expression of cellular antigens of Listeria monocytogenes that react with monoclonal antibodies C11E9 and EM-7G1 under acid-, salt- or temperature-induced stress environments. J. Appl. Microbiol. 95:762-772. [DOI] [PubMed] [Google Scholar]
- 17.Govan, J. R., P. H. Brown, J. Maddison, C. J. Doherty, J. W. Nelson, M. Dodd, A. P. Greening, and A. K. Webb. 1993. Evidence for transmission of Pseudomonas cepacia by social contact in cystic fibrosis. Lancet 342:15-19. [DOI] [PubMed] [Google Scholar]
- 18.Govan, J. R., and V. Deretic. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60:539-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gunn, J. S., K. B. Lim, J. Krueger, K. Kim, L. Guo, M. Hackett, and S. I. Miller. 1998. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol. Microbiol. 27:1171-1182. [DOI] [PubMed] [Google Scholar]
- 20.Hazeleger, W. C., M. Dalvoorde, and R. R. Beumer. 2006. Fluorescence microscopy of NaCl-stressed, elongated Salmonella and Listeria cells reveals the presence of septa in filaments. Int. J. Food Microbiol. 112:288-290. [DOI] [PubMed] [Google Scholar]
- 21.Hunt, T. A., C. Kooi, P. A. Sokol, and M. A. Valvano. 2004. Identification of Burkholderia cenocepacia genes required for bacterial survival in vivo. Infect. Immun. 72:4010-4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Isles, A., I. Maclusky, M. Corey, R. Gold, C. Prober, P. Fleming, and H. Levison. 1984. Pseudomonas cepacia infection in cystic fibrosis: an emerging problem. J. Pediatr. 104:206-210. [DOI] [PubMed] [Google Scholar]
- 23.Kang, H. W., I. G. Wirawan, and M. Kojima. 1994. Cellular localization and functional analysis of the protein encoded by the chromosomal virulence gene (acvB) of Agrobacterium tumefaciens. Biosci. Biotechnol. Biochem. 58:2024-2032. [DOI] [PubMed] [Google Scholar]
- 24.Kim, D. Y., and K. K. Kim. 2005. Structure and function of HtrA family proteins, the key players in protein quality control. J Biochem. Mol. Biol. 38:266-274. [DOI] [PubMed] [Google Scholar]
- 25.Kolter, R., M. Inuzuka, and D. R. Helinski. 1978. Trans-complementation-dependent replication of a low molecular weight origin fragment from plasmid R6K. Cell 15:1199-1208. [DOI] [PubMed] [Google Scholar]
- 26.Lamothe, J., K. K. Huynh, S. Grinstein, and M. A. Valvano. Intracellular survival of Burkholderia cenocepacia in macrophages is associated with a delay in the maturation of bacteria-containing vacuoles. Cell Microbiol., in press. [DOI] [PubMed]
- 27.Lefebre, M. D., R. S. Flannagan, and M. A. Valvano. 2005. A minor catalase/peroxidase from Burkholderia cenocepacia is required for normal aconitase activity. Microbiology 151:1975-1985. [DOI] [PubMed] [Google Scholar]
- 28.Lefebre, M. D., and M. A. Valvano. 2002. Construction and evaluation of plasmid vectors optimized for constitutive and regulated gene expression in Burkholderia cepacia complex isolates. Appl. Environ. Microbiol. 68:5956-5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lemay, M. J., N. Rodrigue, C. Gariepy, and L. Saucier. 2000. Adaptation of Lactobacillus alimentarius to environmental stresses. Int. J. Food Microbiol. 55:249-253. [DOI] [PubMed] [Google Scholar]
- 30.Loutet, S. A., R. S. Flannagan, C. Kooi, P. A. Sokol, and M. A. Valvano. 2006. A complete lipopolysaccharide inner core oligosaccharide is required for resistance of Burkholderia cenocepacia to antimicrobial peptides and bacterial survival in vivo. J. Bacteriol. 188:2073-2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mahenthiralingam, E., T. Coenye, J. W. Chung, D. P. Speert, J. R. Govan, P. Taylor, and P. Vandamme. 2000. Diagnostically and experimentally useful panel of strains from the Burkholderia cepacia complex. J. Clin. Microbiol. 38:910-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahenthiralingam, E., P. Vandamme, M. E. Campbell, D. A. Henry, A. M. Gravelle, L. T. Wong, A. G. Davidson, P. G. Wilcox, B. Nakielna, and D. P. Speert. 2001. Infection with Burkholderia cepacia complex genomovars in patients with cystic fibrosis: virulent transmissible strains of genomovar III can replace Burkholderia multivorans. Clin. Infect. Dis. 33:1469-1475. [DOI] [PubMed] [Google Scholar]
- 33.McLoughlin, T. J., J. P. Quinn, A. Bettermann, and R. Bookland. 1992. Pseudomonas cepacia suppression of sunflower wilt fungus and role of antifungal compounds in controlling the disease. Appl. Environ. Microbiol. 58:1760-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McPhee, J. B., S. Lewenza, and R. E. Hancock. 2003. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol. Microbiol. 50:205-217. [DOI] [PubMed] [Google Scholar]
- 35.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mulvey, M. A., J. D. Schilling, and S. J. Hultgren. 2001. Establishment of a persistent Escherichia coli reservoir during the acute phase of a bladder infection. Infect. Immun. 69:4572-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ortega, X., T. A. Hunt, S. Loutet, A. D. Vinion-Dubiel, A. Datta, B. Choudhury, J. B. Goldberg, R. Carlson, and M. A. Valvano. 2005. Reconstitution of O-specific lipopolysaccharide expression in Burkholderia cenocepacia strain J2315, which is associated with transmissible infections in patients with cystic fibrosis. J. Bacteriol. 187:1324-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Sullivan, L. A., and E. Mahenthiralingam. 2005. Biotechnological potential within the genus Burkholderia. Lett. Appl. Microbiol. 41:8-11. [DOI] [PubMed] [Google Scholar]
- 39.Pedersen, L. L., M. Radulic, M. Doric, and Y. Abu Kwaik. 2001. HtrA homologue of Legionella pneumophila: an indispensable element for intracellular infection of mammalian but not protozoan cells. Infect. Immun. 69:2569-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pogliano, J., A. S. Lynch, D. Belin, E. C. Lin, and J. Beckwith. 1997. Regulation of Escherichia coli cell envelope proteins involved in protein folding and degradation by the Cpx two-component system. Genes Dev. 11:1169-1182. [DOI] [PubMed] [Google Scholar]
- 41.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 42.Raffa, R. G., and T. L. Raivio. 2002. A third envelope stress signal transduction pathway in Escherichia coli. Mol. Microbiol. 45:1599-1611. [DOI] [PubMed] [Google Scholar]
- 43.Raivio, T. L. 2005. Envelope stress responses and Gram-negative bacterial pathogenesis. Mol. Microbiol. 56:1119-1128. [DOI] [PubMed] [Google Scholar]
- 44.Reeves, E. P., H. Lu, H. L. Jacobs, C. G. Messina, S. Bolsover, G. Gabella, E. O. Potma, A. Warley, J. Roes, and A. W. Segal. 2002. Killing activity of neutrophils is mediated through activation of proteases by K+ flux. Nature 416:291-297. [DOI] [PubMed] [Google Scholar]
- 45.Reik, R., T. Spilker, and J. J. Lipuma. 2005. Distribution of Burkholderia cepacia complex species among isolates recovered from persons with or without cystic fibrosis. J. Clin. Microbiol. 43:2926-2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosenberger, C. M., R. L. Gallo, and B. B. Finlay. 2004. Interplay between antibacterial effectors: a macrophage antimicrobial peptide impairs intracellular Salmonella replication. Proc. Natl. Acad. Sci. USA 101:2422-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rouviere, P. E., A. De Las Penas, J. Mecsas, C. Z. Lu, K. E. Rudd, and C. A. Gross. 1995. rpoE, the gene encoding the second heat-shock sigma factor, sigma E, in Escherichia coli. EMBO J. 14:1032-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rus, H., C. Cudrici, and F. Niculescu. 2005. The role of the complement system in innate immunity. Immunol. Res. 33:103-112. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1990. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 50.Santiago, B., U. Schubel, C. Egelseer, and O. Meyer. 1999. Sequence analysis, characterization and CO-specific transcription of the cox gene cluster on the megaplasmid pHCG3 of Oligotropha carboxidovorans. Gene 236:115-124. [DOI] [PubMed] [Google Scholar]
- 51.Sassoon, N., J. P. Arie, and J. M. Betton. 1999. PDZ domains determine the native oligomeric structure of the DegP (HtrA) protease. Mol. Microbiol. 33:583-589. [DOI] [PubMed] [Google Scholar]
- 52.Sebert, M. E., L. M. Palmer, M. Rosenberg, and J. N. Weiser. 2002. Microarray-based identification of htrA, a Streptococcus pneumoniae gene that is regulated by the CiaRH two-component system and contributes to nasopharyngeal colonization. Infect. Immun. 70:4059-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Skorko-Glonek, J., A. Wawrzynow, K. Krzewski, K. Kurpierz, and B. Lipinska. 1995. Site-directed mutagenesis of the HtrA (DegP) serine protease, whose proteolytic activity is indispensable for Escherichia coli survival at elevated temperatures. Gene 163:47-52. [DOI] [PubMed] [Google Scholar]
- 54.Smith, D. L., L. B. Gumery, E. G. Smith, D. E. Stableforth, M. E. Kaufmann, and T. L. Pitt. 1993. Epidemic of Pseudomonas cepacia in an adult cystic fibrosis unit: evidence of person-to-person transmission. J. Clin. Microbiol. 31:3017-3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Speert, D. P., D. Henry, P. Vandamme, M. Corey, and E. Mahenthiralingam. 2002. Epidemiology of Burkholderia cepacia complex in patients with cystic fibrosis, Canada. Emerg. Infect. Dis. 8:181-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spiess, C., A. Beil, and M. Ehrmann. 1999. A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell 97:339-347. [DOI] [PubMed] [Google Scholar]
- 57.Stack, H. M., R. D. Sleator, M. Bowers, C. Hill, and C. G. Gahan. 2005. Role for HtrA in stress induction and virulence potential in Listeria monocytogenes. Appl. Environ. Microbiol. 71:4241-4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilson, R. L., L. L. Brown, D. Kirkwood-Watts, T. K. Warren, S. A. Lund, D. S. King, K. F. Jones, and D. E. Hruby. 2006. Listeria monocytogenes 10403S HtrA is necessary for resistance to cellular stress and virulence. Infect. Immun. 74:765-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wosten, M. M., L. F. Kox, S. Chamnongpol, F. C. Soncini, and E. A. Groisman. 2000. A signal transduction system that responds to extracellular iron. Cell 103:113-125. [DOI] [PubMed] [Google Scholar]
- 60.Yamamoto, T., T. Hanawa, S. Ogata, and S. Kamiya. 1996. Identification and characterization of the Yersinia enterocolitica gsrA gene, which protectively responds to intracellular stress induced by macrophage phagocytosis and to extracellular environmental stress. Infect. Immun. 64:2980-2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang, C. C., and J. Konisky. 1984. Colicin V-treated Escherichia coli does not generate membrane potential. J. Bacteriol. 158:757-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 63.Zaika, L. L., and J. S. Fanelli. 2003. Growth kinetics and cell morphology of Listeria monocytogenes Scott A as affected by temperature, NaCl, and EDTA. J. Food Prot. 66:1208-1215. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.