Abstract
Peptidoglycan muropeptides, potent proinflammatory components, are amidated in Staphylococcus aureus for unknown reasons. To study whether this modification may modulate proinflammatory capacity, cytokine induction by isogenic S. aureus strains with different amidation levels and by synthetic amidated/nonamidated muramyldipeptides was evaluated. However, amidation did not significantly affect cytokine induction. This finding contributes to defining peptidoglycan receptor specificities and indicates that further rationales for muropeptide amidation have to be considered.
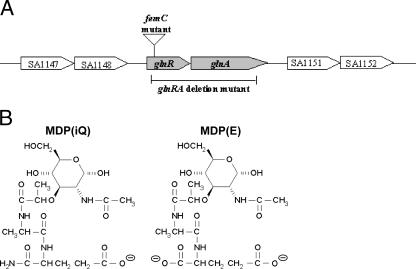
Staphylococcus aureus is a frequent constituent of human nasal microflora and a major cause of severe endogenous infections (11). The staphylococcal cell wall determines several key aspects of the infection process, such as adherence (11, 28), immune recognition (13), and resistance to host defenses (21, 22). While many adhesive surface proteins have been investigated (12), the structure, function, and variability of nonproteinaceous polymers, such as peptidoglycan (PG) or teichoic acids, have remained parts of a neglected field of research. S. aureus is known to modify the canonical PG structure by O acetylation of the glycan strand and by amidation of the α-carboxyl group of the d-glutamate (d-Glu) residue in muropeptides, resulting in the formation of d-isoglutamine (16, 29). While O acetylation confers lysozyme resistance (3), the primary role of d-Glu amidation has remained unclear. Nevertheless, the latter modification is known to affect the level of methicillin resistance (4, 17) and to contribute to vancomycin susceptibility in S. aureus (8). The amino group for d-Glu amidation in S. aureus muropeptides is most probably derived from free glutamine (17, 23). However, the responsible transamidase enzyme has not yet been identified. Gustafson et al. described a femC mutant with 48% decreased muropeptide amidation (17, 26). The femC mutant has a disrupted glnR gene encoding the regulator of the glutamine synthetase (Fig. 1A), which results in reduced glutamine synthetase activity. As a consequence of lower amounts of the amino group donor, muropeptide amidation is reduced in this mutant (17).
FIG. 1.
The S. aureus glutamine synthetase operon, with the insertion site of Tn551 in the femC mutant and the deleted region in the glnRA mutant (A), and structures of amidated MDP(iQ) and nonamidated MDP(E) (B).
PG has potent proinflammatory properties (5). It is sensed by the human innate immune system via NOD1 or NOD2 proteins (15, 18). An additional role of the TLR2 receptor in PG sensing has been described previously (10) but remains a matter of debate (27). The minimal structure required for NOD1 recognition is a dipeptide consisting of d-Glu and meso-diaminopimelate (mesoDAP), which is mainly produced by gram-negative bacteria (14, 15). d-Glu amidation (leading to d-isoglutamine) strongly impairs PG recognition by NOD1 (7), and the amidation of mesoDAP completely abrogates the ability of NOD1 to detect PG (15). NOD2 has a different substrate specificity as it requires a muramyldipeptide (MDP) composed of N-acetylmuramic acid, l-alanine, and d-Glu (Fig. 1B) (19, 27). S. aureus PG is recognized by NOD2 but not by NOD1 since it contains l-lysine instead of meso-DAP (7, 15). However, the relevance of d-Glu amidation for the efficiency of NOD2 recognition has remained unclear. MDP amidation has a profound impact on the physicochemical properties of the molecule since it removes the negative charge from the d-Glu α-carboxyl group (Fig. 1B). Accordingly, the net charge of MDP changes from −2 to −1 at physiological pH as a result of d-Glu amidation. In an attempt to study whether this structural difference has an impact on proinflammatory activity, we evaluated the tumor necrosis factor alpha (TNF-α)-stimulating capacities of S. aureus strains with different levels of PG amidation and of synthetic MDPs with and without amidation.
Proinflammatory capacities of S. aureus wild type and femC mutant.
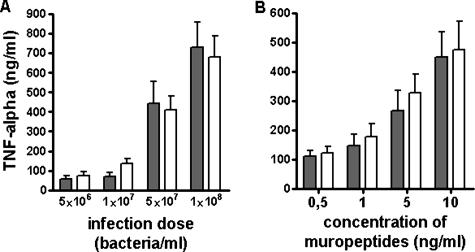
S. aureus NCTC8325 wild type and the isogenic femC mutant (17) were grown overnight in glutamine-deficient, lipopolysaccharide-free RPMI 1640 medium (Sigma), harvested, and washed twice in phosphate-buffered saline (PBS). The mutant grew slower than the wild type did, but both strains reached similar bacterial densities. After heat inactivation at 90°C for 15 min, bacteria were washed again with PBS and adjusted to 109 bacteria/ml by using a Neubauer chamber. Endotoxin contamination of these preparations was below the detection limit of 10 pg/ml of the Limulus amoebocyte lysate test (Cambrex). A total of 360 μl whole blood from healthy volunteers was incubated with 40 μl of PBS containing increasing numbers of bacteria at 37°C for 8 h. Subsequently, samples were centrifuged and TNF-α levels in the supernatant were determined by enzyme-linked immunosorbent assay (R&D Systems). Both bacterial strains stimulated TNF-α production in a dose-dependent fashion (Fig. 2A). However, there was no significant difference in TNF-α levels induced by wild-type or femC mutant bacteria, suggesting that the modulation of muropeptide structure by d-Glu amidation may not affect the inflammatory activity of S. aureus cells.
FIG. 2.
TNF-α induction by S. aureus strains (A) and synthetic MDPs (B). (A) Increasing numbers of S. aureus wild-type (gray bars) or femC mutant bacteria (white bars) were incubated with whole human blood, and TNF-α levels were determined. (B) MDP(iQ) (gray bars) or MDP(E) (white bars) was incubated with whole human blood, and TNF-α levels were determined. The means and standard errors of the means (error bars) of four independent experiments, run in duplicates, are shown. The small differences between the strains in panel A are not significant as calculated by Student's t test.
In an attempt to obtain a mutant strain that completely lacks d-Glu amidation and might cause a stronger phenotypic difference, we deleted the entire operon composed of glutamine synthetase (glnA) and a corresponding regulator gene (glnR) (Fig. 1A). However, the resulting mutant grew only upon supplementation with significant amounts of glutamine and did not have any advantage relative to the femC mutant (data not shown).
Proinflammatory capacities of synthetic MDPs with or without d-Glu amidation.
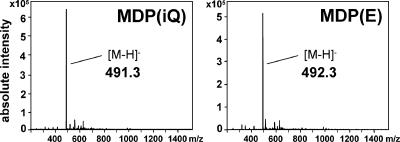
In order to study whether S. aureus MDPs with or without d-Glu amidation differ in their levels of proinflammatory activity when administered in the absence of other bacterial molecules, we chemically synthesized a nonamidated MDP with a d-Glu residue at position 2 of the peptide strand MDP(E), which is not commercially available. Synthesis of the acetylmuramyl-l-alanyl-d-glutamyl peptide was accomplished using Fmoc/tBu solid-phase peptide synthesis methodology (1) on acid-sensitive chlorotrityl chloride resin (2). Briefly, to 0.5 g 2-chlorotrityl chloride resin (0.6 mmol chloride/g resin) in 5 ml dichloromethane, 1 mmol of Fmoc-D-Glu(OBut)-OH and 2.5 mmol diisopropylethylamine (DIPEA) in 4 ml dichloromethane were added. After vigorous shaking for 60 min, the resin was filtered off, washed twice with dimethylformamide (DMF), and deactivated with a mixture of dichloromethane-methanol-DIPEA (80:15:5) for 15 min. After washing with DMF, the Fmoc group was deprotected with 5 ml of 20% piperidine in DMF for 5 min. After intensive washing with DMF, isopropanol, and diethyl ether, 1 mmol Fmoc-Ala-OH, 1 mmol diisoproyplcarbodiimide, and 1 mmol 1-hydroxybenzotriazole in 5 ml DMF were added and incubated for 60 min. Deprotection with piperidine and further coupling with N-acetylmuramic acid yielded the protected peptide. Removal of the side chain-protecting group and detachment from the resin were accomplished by treatment with trifluoroacetic acid-water-phenol (90:5:5). The peptide was precipitated by the addition of diethyl ether and lyophilized. The crude peptide was purified by preparative, reversed-phase, high-performance liquid chromatography on a Reprosil C8 column (150 × 10 mm) using a flat gradient from 100% A to 40% B in 40 min (A, 0.055% trifluoroacetic acid; B, 80% acetonitrile-0.05% trifluoroacetic acid) while monitoring absorbance at 214 nm. Appropriate fractions were lyophilized, and the identity of the product was confirmed by electrospray ionization mass spectrometry (ESI-MS). Our synthetic MDP(E) and the amidated MDP(iQ) purchased from Bachem were found to have the expected mass difference of 1 (Fig. 3).
FIG. 3.
Averaged negative ion mode ESI-MS of MDP(iQ) and MDP(E). Peaks with the expected masses for MDP(iQ) (491.3) or MDP(E) (492.3) are indicated.
MDP(E) and MDP(iQ) were dissolved in PBS. Again, the endotoxin contamination of the preparations was below the detection limit. Forty microliters of PBS containing increasing concentrations of these substances was incubated with 360 μl whole human blood at 37°C for 8 h to stimulate TNF-α production. TNF-α was quantified as described above. Both compounds stimulated TNF-α production in a dose-dependent manner. However, amidated and nonamidated MDP did not differ in their potency levels (Fig. 2B), which is in accordance with the equal proinflammatory activities of S. aureus wild type and the femC mutant.
Concluding comments.
The inflammation-eliciting capacity of S. aureus plays a crucial role in the host response to infections and contributes to life-threatening complications in septicemia (5, 13). However, the diversity, relative importance, and variability of proinflammatory staphylococcal molecules, such as peptidoglycan, lipopeptides, lipoteichoic acid, formylated peptides, and others have remained uncertain. While the relative importance of lipoproteins and formylated peptides has recently been confirmed using defined mutants lacking the corresponding classes of molecules (6, 9, 25), such a strategy is impossible for PG, which is essential for bacterial viability. However, bacterial mutants with altered PG structures might help to investigate the relative role of PG, along with the modulation of PG structure. This study demonstrates that the amidation of muropeptides does not influence the capacity of S. aureus or its MDP to stimulate the production of TNF-α, which contributes to our understanding of the specificity of PG-sensing host receptors. Why S. aureus amidates its muropeptides remains an open question. A possible reason may be the concomitant alteration of the net charge in the S. aureus cell envelope, which may play a role in the maintenance and turnover of the bacterial cell wall. Accordingly, muropeptide amidation has been implicated in PG cross-linking efficiency (20, 24). Identifying the muropeptide amidase gene would enable the construction of an S. aureus mutant lacking any muropeptide amidation without affecting the intracellular glutamine pool. Such an approach represents an important challenge for future studies.
Acknowledgments
We thank Gabriele Hornig and Katja Henseler for excellent technical assistance and Daniel Bächle and Alexander Beck (Panatecs, Tübingen) for ESI-MS analysis.
Our research is supported by grants from the German Research Foundation to A.P. and F.G. (FOR449, GRK685, and TR34) and to A.P. and H.K. (SFB685); the European Union (LSHM-CT-2004-512093) to A.P.; the German Ministry of Education and Research (NGFN2) to A.P. and F.G.; and the IZKF program of the Medical Faculty, University of Tübingen, to A.P.
Editor: J. N. Weiser
Footnotes
Published ahead of print on 29 January 2007.
REFERENCES
- 1.Baechle, D., A. Cansier, R. Fischer, J. Brandenburg, T. Burster, C. Driessen, and H. Kalbacher. 2005. Biotinylated fluorescent peptide substrates for the sensitive and specific determination of cathepsin D activity. J. Pept. Sci. 11:166-174. [DOI] [PubMed] [Google Scholar]
- 2.Barlos, K., O. Chatzi, D. Gatos, and G. Stavropoulos. 1991. 2-Chlorotrityl chloride resin. Studies on anchoring of Fmoc-amino acids and peptide cleavage. Int. J. Pept. Protein Res. 37:513-520. [PubMed] [Google Scholar]
- 3.Bera, A., S. Herbert, A. Jakob, W. Vollmer, and F. Götz. 2005. Why are pathogenic staphylococci so lysozyme resistant? The peptidoglycan O-acetyltransferase OatA is the major determinant for lysozyme resistance of Staphylococcus aureus. Mol. Microbiol. 55:778-787. [DOI] [PubMed] [Google Scholar]
- 4.Berger-Bächi, B., A. Strassle, J. E. Gustafson, and F. H. Kayser. 1992. Mapping and characterization of multiple chromosomal factors involved in methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 36:1367-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boneca, I. G. 2005. The role of peptidoglycan in pathogenesis. Curr. Opin. Microbiol. 8:46-53. [DOI] [PubMed] [Google Scholar]
- 6.Bubeck Wardenburg, J., W. A. Williams, and D. Missiakas. 2006. Host defenses against Staphylococcus aureus infection require recognition of bacterial lipoproteins. Proc. Natl. Acad. Sci. USA 103:13831-13836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chamaillard, M., M. Hashimoto, Y. Horie, J. Masumoto, S. Qiu, L. Saab, Y. Ogura, A. Kawasaki, K. Fukase, S. Kusumoto, M. A. Valvano, S. J. Foster, T. W. Mak, G. Nunez, and N. Inohara. 2003. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat. Immunol. 4:702-707. [DOI] [PubMed] [Google Scholar]
- 8.Cui, L., H. Murakami, K. Kuwahara-Arai, H. Hanaki, and K. Hiramatsu. 2000. Contribution of a thickened cell wall and its glutamine nonamidated component to the vancomycin resistance expressed by Staphylococcus aureus Mu50. Antimicrob. Agents Chemother. 44:2276-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dürr, M. C., S. A. Kristian, M. Otto, G. Matteoli, P. S. Margolis, J. Trias, K. P. Van Kessel, J. A. Van Strijp, E. Bohn, R. Landmann, and A. Peschel. 2006. Neutrophil chemotaxis by pathogen-associated molecular patterns—formylated peptides are crucial but not the sole neutrophil attractants produced by Staphylococcus aureus. Cell Microbiol. 8:207-217. [DOI] [PubMed] [Google Scholar]
- 10.Dziarski, R., and D. Gupta. 2005. Staphylococcus aureus peptidoglycan is a Toll-like receptor 2 activator: a reevaluation. Infect. Immun. 73:5212-5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foster, T. J. 2004. The Staphylococcus aureus “superbug.” J. Clin. Investig. 114:1693-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foster, T. J., and M. Hook. 1998. Surface protein adhesins of Staphylococcus aureus. Trends Microbiol. 6:484-488. [DOI] [PubMed] [Google Scholar]
- 13.Fournier, B., and D. J. Philpott. 2005. Recognition of Staphylococcus aureus by the innate immune system. Clin. Microbiol. Rev. 18:521-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Girardin, S. E., I. G. Boneca, L. A. Carneiro, A. Antignac, M. Jehanno, J. Viala, K. Tedin, M. K. Taha, A. Labigne, U. Zahringer, A. J. Coyle, P. S. DiStefano, J. Bertin, P. J. Sansonetti, and D. J. Philpott. 2003. Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science 300:1584-1587. [DOI] [PubMed] [Google Scholar]
- 15.Girardin, S. E., L. H. Travassos, M. Herve, D. Blanot, I. G. Boneca, D. J. Philpott, P. J. Sansonetti, and D. Mengin-Lecreulx. 2003. Peptidoglycan molecular requirements allowing detection by Nod1 and Nod2. J. Biol. Chem. 278:41702-41708. [DOI] [PubMed] [Google Scholar]
- 16.Götz, F., T. Bannerman, and K.-H. Schleifer. 2006. The genera Staphylococcus and Macrococcus, p. 45-75. In M. Dworkin (ed.), The prokaryotes: an evolving electronic resource for the microbiological community. Springer Verlag, New York, NY.
- 17.Gustafson, J., A. Strassle, H. Hachler, F. H. Kayser, and B. Berger-Bachi. 1994. The femC locus of Staphylococcus aureus required for methicillin resistance includes the glutamine synthetase operon. J. Bacteriol. 176:1460-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Inohara, N., M. Chamaillard, C. McDonald, and G. Nunez. 2005. NOD-LRR proteins: role in host-microbial interactions and inflammatory disease. Annu. Rev. Biochem. 74:355-383. [DOI] [PubMed] [Google Scholar]
- 19.Inohara, N., Y. Ogura, A. Fontalba, O. Gutierrez, F. Pons, J. Crespo, K. Fukase, S. Inamura, S. Kusumoto, M. Hashimoto, S. J. Foster, A. P. Moran, J. L. Fernandez-Luna, and G. Nunez. 2003. Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn's disease. J. Biol. Chem. 278:5509-5512. [DOI] [PubMed] [Google Scholar]
- 20.Komatsuzawa, H., K. Ohta, H. Labischinski, M. Sugai, and H. Suginaka. 1999. Characterization of fmtA, a gene that modulates the expression of methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 43:2121-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kraus, D., and A. Peschel. 2006. Molecular mechanisms of bacterial resistance to antimicrobial peptides. Curr. Top. Microbiol. Immunol. 306:231-250. [DOI] [PubMed] [Google Scholar]
- 22.Peschel, A., and H. G. Sahl. 2006. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat. Rev. Microbiol. 4:529-536. [DOI] [PubMed] [Google Scholar]
- 23.Siewert, G., and J. L. Strominger. 1968. Biosynthesis of the peptidoglycan of bacterial cell walls. XI. Formation of the isoglutamine amide group in the cell walls of Staphylococcus aureus. J. Biol. Chem. 243:783-790. [PubMed] [Google Scholar]
- 24.Sinha, R. K., and F. C. Neuhaus. 1991. Biosynthesis of peptidoglycan in Gaffkya homari: on the target(s) of benzylpenicillin. Antimicrob. Agents Chemother. 35:1753-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stoll, H., J. Dengjel, C. Nerz, and F. Götz. 2005. Staphylococcus aureus deficient in lipidation of prelipoproteins is attenuated in growth and immune activation. Infect. Immun. 73:2411-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strandén, A. M., M. Roos, and B. Berger-Bachi. 1996. Glutamine synthetase and heteroresistance in methicillin-resistant Staphylococcus aureus. Microb. Drug Resist. 2:201-207. [DOI] [PubMed] [Google Scholar]
- 27.Travassos, L. H., S. E. Girardin, D. J. Philpott, D. Blanot, M. A. Nahori, C. Werts, and I. G. Boneca. 2004. Toll-like receptor 2-dependent bacterial sensing does not occur via peptidoglycan recognition. EMBO Rep. 5:1000-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weidenmaier, C., J. F. Kokai-Kun, S. A. Kristian, T. Chanturyia, H. Kalbacher, M. Gross, G. Nicholson, B. Neumeister, J. J. Mond, and A. Peschel. 2004. Role of teichoic acids in Staphylococcus aureus nasal colonization, a major risk factor in nosocomial infections. Nat. Med. 10:243-245. [DOI] [PubMed] [Google Scholar]
- 29.Wilkinson, B. J. 1997. Biology, p. 1-38. In K. B. Crossley and G. L. Archer (ed.), The staphylococci in human disease. Churchill Livingstone, New York, NY.