Abstract
Stenotrophomonas maltophilia is a multiple-antibiotic-resistant opportunistic pathogen that is being isolated with increasing frequency from patients with health-care-associated infections and especially from patients with cystic fibrosis (CF). While clinicians feel compelled to treat infections involving this organism, its potential for virulence is not well established. We evaluated the immunostimulatory properties and overall virulence of clinical isolates of S. maltophilia using the well-characterized opportunistic pathogen Pseudomonas aeruginosa PAO1 as a control. The properties of CF isolates were examined specifically to see if they have a common phenotype. The immunostimulatory properties of S. maltophilia were studied in vitro by stimulating airway epithelial and macrophage cell lines. A neonatal mouse model of pneumonia was used to determine the rates of pneumonia, bacteremia, and mortality, as well as the inflammatory response elicited by S. maltophilia infection. Respiratory and nonrespiratory S. maltophilia isolates were highly immunostimulatory and elicited significant interleukin-8 expression by airway epithelial cells, as well as tumor necrosis factor alpha (TNF-α) expression by macrophages. TNF-α signaling appears to be important in the pathogenesis of S. maltophilia infection as less than 20% of TNFR1 null mice (compared with 100% of wild-type mice) developed pneumonia and bacteremia following intranasal inoculation. The S. maltophilia isolates were weakly invasive, and low-level bacteremia with no mortality was observed. Despite the lack of invasiveness of S. maltophilia, the immunostimulatory properties of this organism and its induction of TNF-α expression specifically indicate that it is likely to contribute significantly to airway inflammation.
There has been a notable increase in the prevalence of Stenotrophomonas maltophilia isolated from clinical specimens over the past several years, as documented by the SENTRY Antimicrobial Surveillance Program (18). This organism is often isolated as a nosocomial pathogen in hospitalized patients (7), as well as in cystic fibrosis (CF) (12), burn (36), human immunodeficiency-infected, and other immunosuppressed patients (2, 15). Although rarely associated with septic shock, S. maltophilia commonly causes persistent bacteremia and is frequently associated with respiratory tract and catheter-related infections. An analysis of 139 isolates from 105 non-CF patients established that S. maltophilia was a cause of infection in the central nervous system, bone, bloodstream, and urinary tract, as well as the respiratory tract (37). Many case reports have demonstrated the potential of S. maltophilia to cause invasive infection as an opportunistic pathogen in immunocompromised patients (24) or when it is inadvertently introduced into a normally sterile site (20).
S. maltophilia has been isolated from 10% of CF patients in the United States (Cystic Fibrosis Foundation registry data) (14) and from up to 25% of CF patients in Europe (12, 33). Epidemiological studies have suggested that, unlike Burkholderia cenocepacia complex and Pseudomonas aeruginosa infections, the presence of S. maltophilia in CF patients is not associated with a worse clinical outcome (14, 34). However, the contribution of this organism to chronic airway inflammation and its ability to persist within biofilms in vivo have not been well studied. Many CF clinicians feel compelled to treat S. maltophilia, a difficult task considering its innate resistance to β-lactam and aminoglycoside antibiotics and rapid development of resistance to fluoroquinolones. When S. maltophilia is isolated from normally sterile sites, eradication is similarly challenging.
S. maltophilia is of considerable general interest, as a PubMed search for 2006 yielded 165 articles covering diverse aspects of S. maltophilia biology, such as mechanisms of antimicrobial resistance, rapid identification, and descriptions of clinical illnesses. A prototypic strain has recently been sequenced, and annotation of the genome is in progress (www.sanger.ac.uk/Projects/S_maltophilia/). One recent clinical study of 89 S. maltophilia respiratory isolates indicated that the vast majority of these organisms were colonizers and not associated with a significant respiratory infection (26). The molecular mechanisms responsible for the virulence or lack of virulence of S. maltophilia have not been fully characterized. Although S. maltophilia has the high G+C content (63 to 70%) of the pseudomonads, it lacks the prodigious metabolic capabilities of these organisms. S. maltophilia strains are obligate aerobes, and most, but not all, strains require methionine or cysteine for growth (2). As might be expected for a respiratory pathogen, the organisms can form biofilms (5). Like P. aeruginosa, S. maltophilia expresses a homologue of algC, the gene encoding phosphoglucomutase, a key enzyme in the synthesis of extracellular polysaccharides (22). S. maltophilia expresses flagella, is motile (3), produces an extracellular protease (39), and synthesizes diverse lipopolysaccharide (LPS) structures with at least 31 different O antigens (40). While a single study has suggested that S. maltophilia LPS is less immunogenic than the LPS of Escherichia coli (41), the contribution of LPS to S. maltophilia virulence has not been well characterized. It is not clear if S. maltophilia isolates from CF patients have unique properties, as is the case for P. aeruginosa isolates.
Faced with an increasing number of infections with S. maltophilia and limited data regarding the potential of this organism for virulence, we surveyed selected properties of 24 S. maltophilia clinical isolates obtained from the Columbia University Medical Center. We examined strains from diverse clinical settings, including CF and non-CF respiratory specimens, as well as nonrespiratory (blood, skin, and soft tissue) specimens, and evaluated their immunogenic potential in established in vitro and in vivo assay systems by comparing them to the well-characterized laboratory strain P. aeruginosa PAO1.
MATERIALS AND METHODS
Bacterial strains.
Twenty-four nonclonal clinical isolates of S. maltophilia were obtained from different patients over a 3-month period at the Columbia University Medical Center and the CF Referral Center in New York, NY. S. maltophilia was isolated from the respiratory tracts of CF patients (CF isolates) (n = 10) and non-CF patients (non-CF isolates) (n = 7), as well as from patients with blood, skin, and soft tissue infections (n = 7). Bacteria were isolated, identified as S. maltophilia by biochemical characteristics and antibiotic resistance analysis, and grown in Luria-Bertani (LB) broth, and aliquots were frozen in LB-glycerol at −80°C. For each experiment bacteria were grown from frozen stocks on LB agar. P. aeruginosa PAO1 and a lasl rhll mutant (26) were used as controls.
Cell culture and reagents.
1HAEo− (human airway epithelial) and 16HBE (human bronchoepithelial) cells were grown as described previously (6). RAW cells were grown in RPMI medium with 10% fetal calf serum (Invitrogen). Unless indicated otherwise, reagents were purchased from Sigma. All media used were supplemented with 100 U/ml penicillin, 100 μg/ml streptomycin, 50 μg/ml gentamicin, and 4 μg/ml amphotericin B.
Motility.
The motility of S. maltophilia was determined by examining its ability to diffuse in soft agar plates. PAO1 and a Fla− mutant (10) were used as positive and negative controls.
Biofilm assay.
Bacteria were grown overnight with agitation, the optical densities at 600 nm were standardized to 2, and the cultures were diluted 1:100 in LB broth. Aliquots (100 μl) were added to a 96-well plate, which was incubated for 18 h at 37°C (25). Growth was monitored by determining the optical density at 600 nm, and after two or three washes with water, crystal violet was added for 15 min, which was followed by three rinses with water and then addition of 95% ethanol. The material was then transferred to a fresh 96-well plate, and the absorbance at 540 nm was determined. Each sample was tested in triplicate.
IL-8 and TNF-α detection.
The level of interleukin-8 (IL-8) was determined by an enzyme-linked immunosorbent assay (ELISA) (R&D Systems) following exposure of 1HAEo− cells to 108 CFU of bacteria (29). The level of tumor necrosis factor alpha (TNF-α) was determined by an ELISA (DuoSet; R&D Systems) following exposure of RAW cells to 500 ng/ml of lipid A for 4 h. The cell viability was >75%, as assessed by using trypan blue. Each data point was determined in sextuplicate, and the data were normalized to the protein content.
LPS purification and lipid A isolation.
Large-scale LPS preparations were extracted using a hot phenol-water extraction method (38). LPS was treated with RNase A, DNase I, and proteinase K (11) and then extracted to remove contaminating proteins. Small-scale LPS preparations were isolated as described previously (9). Lipid A was isolated after hydrolysis in 1% sodium dodecyl sulfate at pH 4.5 (1). Samples were resuspended in 500 μl of water, frozen, and lyophilized. For RAW cell stimulation samples were standardized by weight.
Mass spectrometry.
Negative-ion matrix-assisted laser desorption ionization—time of flight (MALDI-TOF) experiments were performed as described previously, with the following modifications (9). Lyophilized lipid A was dissolved with 10 μl of a 5-chloro-2-mercaptobenzothiazole MALDI matrix in chloroform-methanol (1:1, vol/vol) and then applied (1 μl) onto a sample plate. All MALDI-TOF experiments were performed using a Bruker Autoflex II MALDI-TOF mass spectrometer (MS) (Bruker Daltonics Inc., Billerica, MA). Each spectrum was an average of 300 shots. ES tuning mixture (Agilent, Palo Alto, CA) was used to calibrate the MALDI-TOF MS.
Mouse model of infection.
Groups of 6 to 10 7- to 10-day-old C57BL/6 or C57BL/6-TnfrsflatmlImx (TNFR1 null; Jackson Laboratories) mice were intranasally inoculated with 108 CFU of bacteria in 10 μl of phosphate-buffered saline. Sixteen hours later the rates of pneumonia (defined as recovery of >103 CFU per lung), bacteremia (measured by determining the presence of bacteria in the spleen), and mortality were determined (35). For neutrophil detection, lung cell suspensions were analyzed for double expression of CD45 and Ly6C by flow cytometry (13). For lung TNF-α mRNA quantification, real-time PCR was performed using primers 5′-ATGAGCACAGAAAGCATGATC-3′ and 5′-TACAGGCTTGTCACTCGAATT-3′. Actin was used as a control for standardization. The studies were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee.
Serum sensitivity assay.
Bacteria were grown to an optical density of 0.5, washed in Hanks' balanced salt solution, and incubated with 60% serum, 60% heat-inactivated serum, or Hanks' balanced salt solution alone at 37°C with agitation for 90 min, and then they were plated on LB medium.
Phagocytosis assay.
Phagocytosis by RAW cells was determined by incubating cells with 108 CFU of bacteria for 30 min. Cells were washed, treated with 600 μg/ml of gentamicin (which killed all the S. maltophilia strains used) for 1 h, washed again, trypsinized, and plated on LB medium.
Invasion assay and measurement of transepithelial resistance.
Bacteria (∼1 × 107 CFU) were added to the apical surface of polarized 16HBE cells on Transwell-Clear cell culture inserts (Corning-Costar) with 3-μm pores. After 4 h the number of organisms in the basolateral medium was determined. Transepithelial resistance was measured using Millicell-ERS (Millipore). Triplicate wells were used for both assays.
Statistical analysis.
Data obtained in the mouse experiments were analyzed using a Mann-Whitney nonparametric test. Categorical variable proportions were compared using Fisher's exact test.
RESULTS
S. maltophilia interactions with airway epithelial cells and macrophages.
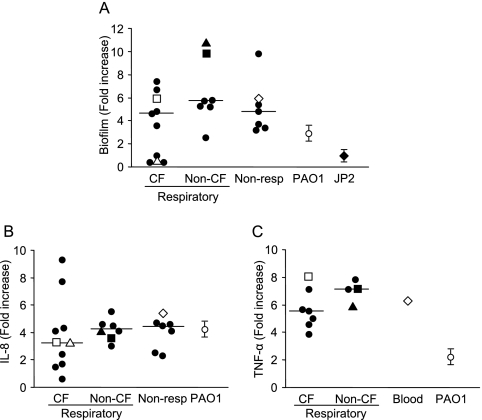
The virulence properties typically associated with airway pathogens include motility, the ability to form biofilms, and activation of chemokine expression (16, 30). The majority (80%) of the non-CF S. maltophilia isolates were as motile as P. aeruginosa PAO1, while only 30% of the CF strains were motile (data not shown). Whereas all S. maltophilia strains grew similarly in plastic, biofilm production was highly variable (Fig. 1A). Most CF isolates (6/10) formed biofilms that were appreciably more dense (increased staining with crystal violet) than a PAO1 biofilm. However, 4/10 CF strains did not produce a detectable biofilm and instead behaved like the negative control, JP2, a lasI rhlI mutant of PAO1 (27). All of the non-CF respiratory tract isolates, as well as the isolates from other clinical sites, synthesized at least as much biofilm as PAO1 synthesized, and the majority appeared to produce more extracellular material than PAO1 produced. We then assessed the ability of S. maltophilia to stimulate the expression of IL-8, the major polymorphonuclear leukocyte (PMN) chemokine produced by airway epithelial cells and a common marker of airway inflammation (Fig. 1B). The immunostimulatory capabilities of CF isolates were highly variable and more variable than the immunostimulatory capabilities of S. maltophilia strains from other sources. In a comparison with P. aeruginosa PAO1, some (4/10) of the CF strains were considerably less immunostimulatory, whereas the non-CF isolates did not differ from PAO1.
FIG. 1.
Biofilm production and chemokine-cytokine induction by S. maltophilia. (A) Amounts of biofilm produced by S. maltophilia isolates and P. aeruginosa (PAO1) expressed as fold increases compared to the biofilm produced by the PAO1 lasI rhl mutant (JP2). Each symbol represents an individual S. maltophilia isolate, and the median values are indicated by lines. □, CF1; ▵, CF2; ▪, N1; ▴, N2; ⋄, N3. (B) Levels of IL-8 determined by ELISA after stimulation of 1HAEo− cells for 1 h with S. maltophilia or P. aeruginosa PAO1. The IL-8 levels are expressed as fold increases compared to the IL-8 levels in unstimulated cells. Each symbol represents an individual S. maltophilia isolate, and the median values are indicated by lines. □, CF1; ▵, CF2; ▪, N1; ▴, N2; ⋄, N3. (C) Levels of TNF-α production determined by ELISA after stimulation of RAW cells for 4 h with 100 ng of lipid A from S. maltophilia or P. aeruginosa PAO1. The results are expressed as fold increases in TNF-α production compared to the production by unstimulated cells. Each symbol represents an individual S. maltophilia isolate, and the median values are indicated by lines. □, CF1; ▪, N1; ▴, N2; ⋄ N3.
In addition to airway epithelial cells, alveolar macrophages play an important role in the induction of inflammation by secreting TNF-α in response to bacterial stimulation. LPS is a potent inducer of TNF-α production through its lipid A moiety. The ability of the lipid A moiety of S. maltophilia LPS isolated from 12 clinical strains (7 CF isolates, 4 respiratory non-CF isolates, and 1 blood isolate) to induce TNF-α expression in RAW cells, a murine macrophage cell line, was tested (Fig. 1C). All of the S. maltophilia lipid A moieties were significantly more potent for stimulating TNF-α production by RAW cells than P. aeruginosa PAO1 lipid A was. The CF isolates exhibited a range of immunostimulation activities, but even the least stimulatory isolate elicited more TNF-α production than did PAO1. Similarly, the non-CF isolates induced six- to eightfold more cytokine production than PAO1 induced.
Properties of S. maltophilia lipid A.
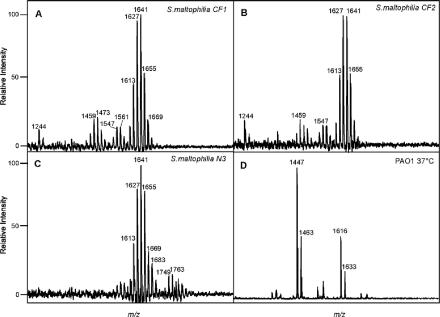
To better understand the immunogenicity of the S. maltophilia strains, particularly compared with the immunogenicity of PAO1, the lipid A moieties of 12 strains were analyzed by MS. MS analysis of S. maltophilia lipid A indicated that there was a high degree of overall heterogeneity in the ion species for all isolates, potentially due to increased fatty acid variability. The results of a detailed analysis of two randomly selected respiratory isolates (CF1 and CF2) and one randomly selected blood isolate (N3) are shown in Fig. 2. A dominant [M-H]− ion cluster at a mass-to-charge ratio (m/z) between 1613 and 1670 was observed for all S. maltophilia isolates (Fig. 2A to C). MALDI-TOF MS for all three isolates revealed a heterogeneous mixture of species (m/z 1613, 1627, 1641, 1655, 1669, and 1683) that suggests that fatty acids that differed by one carbon (Δm/z, 14) were added to the lipid A structure. Additionally, lipid A isolated from blood isolate N3 had an additional phosphate group (m/z 80) at m/z 1749 and 1763 (Fig. 2C). Compared to the increased heterogeneity observed in lipid A preparations isolated from the individual S. maltophilia clinical isolates, the lipid A from the laboratory-adapted wild-type P. aeruginosa isolate, PAO1, exhibited markedly less heterogeneity, with penta- and hexa-acylated ion species at m/z 1447 and 1616, respectively (Fig. 2D).
FIG. 2.
Characterization of S. maltophilia and P. aeruginosa lipid A. Isolated lipid A preparations from CF isolates CF1 (A) and CF2 (B), blood isolate N3 (C), and P. aeruginosa (D) were characterized by using negative-ion matrix-assisted laser desorption ionization—time of flight mass spectrometry.
S. maltophilia does not invade across epithelial monolayers.
After initial colonization of the airways, bacterial pathogens must cross the epithelial barrier to cause bacteremia. Five S. maltophilia strains (two CF isolates, two respiratory non-CF isolates, and one blood isolate) were randomly selected, and their invasiveness was determined in vitro by comparing their abilities to cross intact airway epithelial cell monolayers with tight junctions. S. maltophilia caused a reduction in the potential difference across the membrane (from 3,000 Ω to 1,500 Ω). This level of disruption of the tight junctions enabled at most 1 × 102 to 3 × 102 CFU of the S. maltophilia strains to cross the epithelial barrier, values which are 2 orders of magnitude lower than the number of PAO1 cells that were able to invade. P. aeruginosa PAO1 destroyed the integrity of the tight junctions by the end of the 4-h incubation. This reduced the resistance across the cells to that of the transwell membrane alone (200 Ω), which enabled >104 CFU to get across the monolayer (Table 1).
TABLE 1.
Invasion through polarized 16HBE airway epithelial cells
| Species | Isolate | Inoculum | No. of invading bacteria (CFU) |
|---|---|---|---|
| P. aeruginosa | PAO1 | 0.34 × 107 | >104 |
| S. maltophilia | CF1 | 1.4 × 107 | 278 |
| CF2 | 1.0 × 107 | 78 | |
| N1 | 1.8 × 107 | 186 | |
| N2 | 1.2 × 107 | 32 | |
| N3 | 1.4 × 107 | 236 |
S. maltophilia virulence in a mouse model of respiratory tract infection.
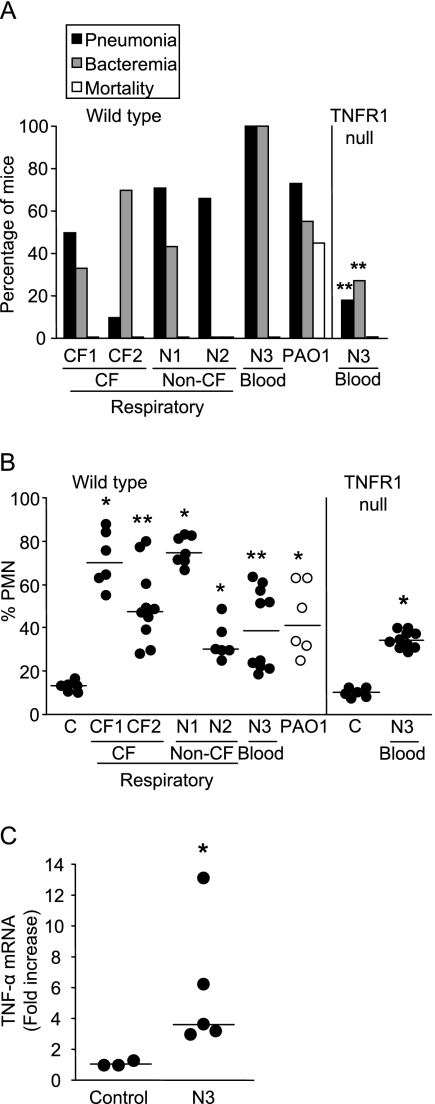
The net effects of the immunostimulatory and invasive properties of five S. maltophilia isolates were tested in a mouse model of infection and compared directly to the effects of P. aeruginosa PAO1 (Fig. 3A). The majority of the S. maltophilia isolates caused pneumonia in more than 50% of the mice inoculated. The importance of biofilm production in the ability to colonize and cause airway infection has been demonstrated for P. aeruginosa (32). Similarly, isolate CF2, a CF isolate, caused pneumonia in only 10% of the mice, which correlated with its inability to form a biofilm (Fig. 1A). The other CF and non-CF isolates all formed dense biofilms (Fig. 1A), which likely contributed to the development of pneumonia (Fig. 3A). Although a relatively high percentage of mice developed bacteremia, the bacterial counts in the spleens were in general very low (Table 2), in contrast to the results obtained with PAO1, which was recovered at a density of >103 CFU. The low levels of bacteremia did not lead to mortality, nor did any of the mice appear to be moribund at 16 h postinoculation.
FIG. 3.
S. maltophilia lung infection. (A) Percentages of C57BL/6 mice (Wild type) and TNFR1 null mice that developed pneumonia or bacteremia or died. Two asterisks indicate that the P value is <0.01 for a comparison with wild-type mice inoculated with N3. (B) Percentages of PMNs in the total leukocytes in the lungs of wild-type and TNFR1 null mice. Each symbol represents an individual mouse, and the lines indicate the medians for the groups. One asterisk indicates that the P value is <0.05 and two asterisks indicate that the P value is <0.01 for a comparison with control mice inoculated with phosphate-buffered saline (C). (C) Lung TNF-α mRNA expression in TNFR1 null mice as determined by real-time PCR and standardized to actin. Each symbol represents an individual mouse, and the lines indicate the medians for the groups. The asterisk indicates that the P value is <0.05 for a comparison with control mice inoculated with phosphate-buffered saline.
TABLE 2.
S. maltophilia strains isolated from the spleens of infected mice
| Mice | Isolate | Total no. of CFU
|
|
|---|---|---|---|
| Median | Range | ||
| Wild type | CF1 | 5 | 4-6 |
| CF2 | 3 | 1-14 | |
| N1 | 2 | 2-11 | |
| N2 | 0 | ||
| N3 | 54a | 35-93 | |
| TNFR1 null | N3 | 2.5b | 1-100 |
P < 0.001, as determined by the Mann-Whitney test, for comparisons with the values for CF1, CF2, and N1.
P < 0.01, as determined by the Mann-Whitney test, for a comparison with the value for N3 in wild-type mice.
The nature of the immune response in the mouse lungs to each of the S. maltophilia isolates was evaluated by flow cytometry (Fig. 3B). The percentage of PMNs recruited to the lungs as a function of the total number of leukocytes was determined. Once again, there was a wide range of responses, but each of the isolates induced significant PMN recruitment and the number of PMNs was, in general, equivalent to or even greater than the number of PMNs elicited by the PAO1 control strain.
S. maltophilia virulence in a TNFR1 null mouse.
A major difference between the immune responses induced by the S. maltophilia strains and the immune responses elicited by P. aeruginosa PAO1 was the amount of TNF-α produced. To assess the importance of TNF-α signaling for clearance of S. maltophilia and for defense against invasive infection, we compared the responses of wild-type C57BL/6 and TNFR1 null mice to the most virulent isolate, isolate N3 (Fig. 3A and Table 2). N3 caused significantly less pneumonia and bacteremia in the TNFR1 null animals (100% in the wild type, compared to 20% pneumonia and 25% bacteremia in the TNFR1 null mice; P < 0.001 for both), and the bacterial counts in the spleens of bacteremic mice were significantly lower (P < 0.01). Similarly, TNFR1 null mice showed increased bacterial clearance during P. aeruginosa infection (31). The TNF-α expression in the lungs of TNFR1 null mice infected with S. maltophilia N3 was determined as a control (Fig. 3C). Whereas high levels of TNF-α were induced, the TNF-α could not contribute effectively to the pathogenesis of infection in the absence of TNFR1, the only TNF receptor expressed by airway epithelial cells. These results suggest that TNF-α-dependent signaling is a major cause of the pathology attributed to the organisms. Flow cytometry analysis of the cellular infiltrates in the wild-type and TNFR1 null mice demonstrated that the numbers of recruited PMNs were similar, indicating that chemokine signaling remained intact (Fig. 3B).
Phagocytosis and killing assays.
In addition to weak invasiveness, the low levels of bacteremia after intranasal infection with high levels of S. maltophilia may be attributed to specific systemic host clearance mechanisms. We compared the serum sensitivities and rates of phagocytosis and killing by RAW cells of the different isolates. While S. maltophilia isolates CF1, CF2, N1, and N2 were sensitive to serum, the N3 strain, like PAO1, was resistant. These results are consistent with the higher bacterial counts in spleens of mice infected with the N3 isolate (Table 2). However, all of the S. maltophilia strains tested were readily phagocytosed (range, 6 to 33%) even more efficiently than the PAO1 control (5%), indicating that control of bacterial replication in the blood prevents sepsis and mortality.
DISCUSSION
S. maltophilia has the properties expected of an opportunistic pathogen. These organisms have intrinsic antimicrobial resistance and cause infections that result in increased morbidity, but not usually in mortality, in patients with impaired host defenses. The clinical isolates evaluated in the present study were generally capable of biofilm formation and were highly immunostimulatory, features that are very important in the initial colonization of the airways and development of pneumonia. As might be predicted from the accumulating clinical reports, most of the isolates that we tested were not particularly virulent, and none caused death in a neonatal mouse model of respiratory tract infection using a high inoculum.
The properties of S. maltophilia strains isolated from patients with respiratory and nonrespiratory infections did not differ significantly. While CF isolates were heterogeneous, there were no marked differences between these isolates and other respiratory (non-CF) isolates that suggested a “CF phenotype,” analogous to the mucoid strains of P. aeruginosa. However, we found that only 30% of the CF isolates were motile, compared to 80% of the non-CF isolates. While motility is very important in the pathogenesis of pneumonia (10), one feature of P. aeruginosa adaptation to the CF lung is its loss of motility (20). Flagella are highly immunostimulatory (28) and can function as ligands for nonopsonic phagocytosis (21). Thus, it is thought that decreased expression of flagella may protect the bacteria from the host immune response. Loss of motility, likely due to attenuated expression of flagella, appears to be a common mechanism of adaptation to the CF airways for both P. aeruginosa and S. maltophilia.
All the S. maltophilia isolates tested were highly immunostimulatory. Overall, the S. maltophilia strains induced as much IL-8 expression as P. aeruginosa induced. IL-8 (or KC in mice) is a chemokine that recruits PMNs into the lungs. In a murine model of pneumonia, S. maltophilia and P. aeruginosa similarly induced significant PMN recruitment into the lungs. However, S. maltophilia induced substantially more TNF-α expression than the P. aeruginosa control induced. This TNF-α response may be associated with the high degree of lipid A heterogeneity detected in S. maltophilia (8, 23). TNF-α is a potent proinflammatory cytokine that induces neutrophil and macrophage activation. Airway inflammation, the hallmark of pneumonia, is required to clear the bacteria, but the accumulation of activated neutrophils and macrophages and their products is detrimental to normal lung function. TNF-α signaling contributes significantly to the pathophysiology of S. maltophilia pneumonia, as reflected by the minimal disease observed in the TNFR1 null mice. These findings are consistent with clinical studies that showed that there was deterioration of lung function after prolonged exposure to S. maltophilia in CF patients (19).
S. maltophilia strain N3, a blood isolate, was somewhat more virulent than the other strains in the mouse model of pneumonia. Mass spectrometry analysis of the lipid A of this strain, however, revealed larger peaks (m/z >1,700) that represented modifications not characterized yet. These modifications may explain why this isolate is more virulent, as it has been demonstrated that in other gram-negative bacteria modifications in lipid A play a role in increased virulence and immunostimulatory responses (23). Further studies that include analysis of the O antigen and LPS core modifications are required to confirm this hypothesis.
S. maltophilia binds to airway epithelial cells as efficiently as P. aeruginosa binds and it aggregates along the cell junctions (4), but it is poorly invasive. The sequenced S. maltophilia genome has few regions with low levels of homology (20 to 30%) to any of the P. aeruginosa type III secretion genes. Type III secretion systems mediate bacterial interactions with host cytoskeletal components in many gram-negative pathogens and, in P. aeruginosa, correlate highly with invasive infection (17). Thus, the potential lack of type III secretion genes in S. maltophilia may contribute to its limited invasive capabilities. Thus, in terms of invasive potential, S. maltophilia differs substantially from even the laboratory strain P. aeruginosa PAO1. Moreover, during pulmonary infection, the few organisms that cross the epithelial barrier are readily cleared, if not by lytic effects of serum, by phagocytosis, and they do not produce concentrations in the blood that are high enough to cause sepsis.
S. maltophilia, like P. aeruginosa, has the potential to contribute to the inflammatory process that compromises respiratory function in CF and in hospital-acquired pneumonias. Since few CF patients have an S. maltophilia infection without a concomitant P. aeruginosa infection (34), it is difficult to sort out the relative contribution of each organism to ongoing lung damage. However, our data suggest that targeting S. maltophilia with antimicrobial therapy and perhaps even anti-inflammatory therapy may decrease overall levels of inflammation that contribute to pathology.
Acknowledgments
This work was supported by the National Institutes of Health (grant RO1 DK36393 AI4793A1). V.J.W., M.I.G., and R.K.E. were supported by the U.S. Cystic Fibrosis Foundation.
Editor: J. N. Weiser
Footnotes
Published ahead of print on 12 January 2007.
REFERENCES
- 1.Caroff, M., A. Tacken, and L. Szabo. 1988. Detergent-accelerated hydrolysis of bacterial endotoxins and determination of the anomeric configuration of the glycosyl phosphate present in the “isolated lipid A” fragment of the Bordetella pertussis endotoxin. Carbohydr Res. 175:273-282. [DOI] [PubMed] [Google Scholar]
- 2.Denton, M., and K. G. Kerr. 1998. Microbiological and clinical aspects of infection associated with Stenotrophomonas maltophilia. Clin. Microbiol. Rev. 11:57-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Oliveira-Garcia, D., M. Dall'Agnol, M. Rosales, A. C. Azzuz, M. B. Martinez, and J. A. Giron. 2002. Characterization of flagella produced by clinical strains of Stenotrophomonas maltophilia. Emerg. Infect. Dis. 8:918-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Vidipo, L. A., E. A. De Marques, E. Puchelle, and M. C. Plotkowski. 2001. Stenotrophomonas maltophilia interaction with human epithelial respiratory cells in vitro. Microbiol. Immunol. 45:563-569. [DOI] [PubMed] [Google Scholar]
- 5.Di Bonaventura, G., I. Spedicato, D. D'Antonio, I. Robuffo, and R. Piccolomini. 2004. Biofilm formation by Stenotrophomonas maltophilia: modulation by quinolones, trimethoprim-sulfamethoxazole, and ceftazidime. Antimicrob. Agents Chemother. 48:151-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DiMango, E., A. J. Ratner, R. Bryan, S. Tabibi, and A. Prince. 1998. Activation of NF-kappaB by adherent Pseudomonas aeruginosa in normal and cystic fibrosis respiratory epithelial cells. J. Clin. Investig. 101:2598-2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ensminger, S. A., R. S. Wright, L. M. Baddour, and B. Afessa. 2006. Suspected ventilator-associated pneumonia in cardiac patients admitted to the coronary care unit. Mayo Clin. Proc. 81:32-35. [DOI] [PubMed] [Google Scholar]
- 8.Ernst, R. K., A. M. Hajjar, J. H. Tsai, S. M. Moskowitz, C. B. Wilson, and S. I. Miller. 2003. Pseudomonas aeruginosa lipid A diversity and its recognition by Toll-like receptor 4. J. Endotoxin Res. 9:395-400. [DOI] [PubMed] [Google Scholar]
- 9.Ernst, R. K., E. C. Yi, L. Guo, K. B. Lim, J. L. Burns, M. Hackett, and S. I. Miller. 1999. Specific lipopolysaccharide found in cystic fibrosis airway Pseudomonas aeruginosa. Science 286:1561-1565. [DOI] [PubMed] [Google Scholar]
- 10.Feldman, M., R. Bryan, S. Rajan, L. Scheffler, S. Brunnert, H. Tang, and A. Prince. 1998. Role of flagella in pathogenesis of Pseudomonas aeruginosa pulmonary infection. Infect. Immun. 66:43-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer, W., H. U. Koch, and R. Haas. 1983. Improved preparation of lipoteichoic acids. Eur. J. Biochem. 133:523-530. [DOI] [PubMed] [Google Scholar]
- 12.Gibson, R. L., J. L. Burns, and B. W. Ramsey. 2003. Pathophysiology and management of pulmonary infections in cystic fibrosis. Am. J. Respir. Crit. Care Med. 168:918-951. [DOI] [PubMed] [Google Scholar]
- 13.Gomez, M. I., A. Lee, B. Reddy, A. Muir, G. Soong, A. Pitt, A. Cheung, and A. Prince. 2004. Staphylococcus aureus protein A induces airway epithelial inflammatory responses by activating TNFR1. Nat. Med. 10:842-848. [DOI] [PubMed] [Google Scholar]
- 14.Goss, C. H., K. Otto, M. L. Aitken, and G. D. Rubenfeld. 2002. Detecting Stenotrophomonas maltophilia does not reduce survival of patients with cystic fibrosis. Am. J. Respir. Crit. Care Med. 166:356-361. [DOI] [PubMed] [Google Scholar]
- 15.Graff, G. R., and J. L. Burns. 2002. Factors affecting the incidence of Stenotrophomonas maltophilia isolation in cystic fibrosis. Chest 121:1754-1760. [DOI] [PubMed] [Google Scholar]
- 16.Hall-Stoodley, L., J. W. Costerton, and P. Stoodley. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2:95-108. [DOI] [PubMed] [Google Scholar]
- 17.Hauser, A. R., E. Cobb, M. Bodi, D. Mariscal, J. Valles, J. N. Engel, and J. Rello. 2002. Type III protein secretion is associated with poor clinical outcomes in patients with ventilator-associated pneumonia caused by Pseudomonas aeruginosa. Crit. Care Med. 30:521-528. [DOI] [PubMed] [Google Scholar]
- 18.Jones, R. N., H. S. Sader, and M. L. Beach. 2003. Contemporary in vitro spectrum of activity summary for antimicrobial agents tested against 18569 strains of non-fermentative Gram-negative bacilli isolated in the SENTRY Antimicrobial Surveillance Program (1997-2001). Int. J. Antimicrob. Agents 22:551-556. [DOI] [PubMed] [Google Scholar]
- 19.Karpati, F., A. S. Malmborg, H. Alfredsson, L. Hjelte, and B. Strandvik. 1994. Bacterial colonisation with Xanthomonas maltophilia—a retrospective study in a cystic fibrosis patient population. Infection 22:258-263. [DOI] [PubMed] [Google Scholar]
- 20.Landrum, M. L., N. G. Conger, and M. A. Forgione. 2005. Trimethoprim-sulfamethoxazole in the treatment of Stenotrophomonas maltophilia osteomyelitis. Clin. Infect. Dis. 40:1551-1552. [DOI] [PubMed] [Google Scholar]
- 21.Mahenthiralingam, E., and D. P. Speert. 1995. Nonopsonic phagocytosis of Pseudomonas aeruginosa by macrophages and polymorphonuclear leukocytes requires the presence of the bacterial flagellum. Infect. Immun. 63:4519-4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKay, G. A., D. E. Woods, K. L. MacDonald, and K. Poole. 2003. Role of phosphoglucomutase of Stenotrophomonas maltophilia in lipopolysaccharide biosynthesis, virulence, and antibiotic resistance. Infect. Immun. 71:3068-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller, S. I., R. K. Ernst, and M. W. Bader. 2005. LPS, TLR4 and infectious disease diversity. Nat. Rev. Microbiol. 3:36-46. [DOI] [PubMed] [Google Scholar]
- 24.Miyairi, I., J. A. Franklin, M. Andreansky, K. M. Knapp, and R. T. Hayden. 2005. Acute necrotizing ulcerative gingivitis and bacteremia caused by Stenotrophomonas maltophilia in an immunocompromised host. Pediatr. Infect. Dis. J. 24:181-183. [DOI] [PubMed] [Google Scholar]
- 25.O'Toole, G., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49-79. [DOI] [PubMed] [Google Scholar]
- 26.Pathmanathan, A., and G. W. Waterer. 2005. Significance of positive Stenotrophomonas maltophilia culture in acute respiratory tract infection. Eur. Respir. J. 25:911-914. [DOI] [PubMed] [Google Scholar]
- 27.Pearson, J. P., M. Feldman, B. H. Iglewski, and A. Prince. 2000. Pseudomonas aeruginosa cell-to-cell signaling is required for virulence in a model of acute pulmonary infection. Infect. Immun. 68:4331-4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prince, A. 2006. Flagellar activation of epithelial signaling. Am. J. Respir. Cell Mol. Biol. 34:548-551. [DOI] [PubMed] [Google Scholar]
- 29.Ratner, A. J., R. Bryan, A. Weber, S. Nguyen, D. Barnes, A. Pitt, S. Gelber, A. Cheung, and A. Prince. 2001. Cystic fibrosis pathogens activate Ca2+-dependent mitogen-activated protein kinase signaling pathways in airway epithelial cells. J. Biol. Chem. 276:19267-19275. [DOI] [PubMed] [Google Scholar]
- 30.Sadikot, R. T., T. S. Blackwell, J. W. Christman, and A. S. Prince. 2005. Pathogen-host interactions in Pseudomonas aeruginosa pneumonia. Am. J. Respir. Crit. Care Med. 171:1209-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skerrett, S. J., T. R. Martin, E. Y. Chi, J. J. Peschon, K. M. Mohler, and C. B. Wilson. 1999. Role of type 1 TNF receptor in lung inflammation after inhalation of endotoxin or Pseudomonas aeruginosa. Am. J. Physiol. Lung Cell. Mol. Physiol. 276:L715-L727. [DOI] [PubMed] [Google Scholar]
- 32.Soong, G., A. Muir, M. I. Gomez, J. Waks, B. Reddy, P. Planet, P. K. Singh, Y. Kaneko, M. C. Wolfgang, Y. S. Hsiao, L. Tong, and A. Prince. 2006. Bacterial neuraminidase facilitates mucosal infection by participating in biofilm production. J. Clin. Investig. 116:2297-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steinkamp, G., B. Wiedemann, E. Rietschel, A. Krahl, J. Gielen, H. Barmeier, and F. Ratjen. 2005. Prospective evaluation of emerging bacteria in cystic fibrosis. J. Cyst Fibros. 4:41-48. [DOI] [PubMed] [Google Scholar]
- 34.Talmaciu, I., L. Varlotta, J. Mortensen, and D. V. Schidlow. 2000. Risk factors for emergence of Stenotrophomonas maltophilia in cystic fibrosis. Pediatr. Pulmonol. 30:10-15. [DOI] [PubMed] [Google Scholar]
- 35.Tang, H. B., E. DiMango, R. Bryan, M. Gambello, B. H. Iglewski, J. B. Goldberg, and A. Prince. 1996. Contribution of specific Pseudomonas aeruginosa virulence factors to pathogenesis of pneumonia in a neonatal mouse model of infection. Infect. Immun. 64:37-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsai, W. P., C. L. Chen, W. C. Ko, and S. C. Pan. 2006. Stenotrophomonas maltophilia bacteremia in burn patients. Burns 32:155-158. [DOI] [PubMed] [Google Scholar]
- 37.Valdezate, S., A. Vindel, P. Martin-Davila, B. S. Del Saz, F. Baquero, and R. Canton. 2004. High genetic diversity among Stenotrophomonas maltophilia strains despite their originating at a single hospital. J. Clin. Microbiol. 42:693-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Westphal, O., K. Jann, and K. Himmelspach. 1983. Chemistry and immunochemistry of bacterial lipopolysaccharides as cell wall antigens and endotoxins. Prog. Allergy 33:9-39. [PubMed] [Google Scholar]
- 39.Windhorst, S., E. Frank, D. N. Georgieva, N. Genov, F. Buck, P. Borowski, and W. Weber. 2002. The major extracellular protease of the nosocomial pathogen Stenotrophomonas maltophilia: characterization of the protein and molecular cloning of the gene. J. Biol. Chem. 277:11042-11049. [DOI] [PubMed] [Google Scholar]
- 40.Winn, A. M., and S. G. Wilkinson. 2001. Structures of the O4 and O18 antigens of Stenotrophomonas maltophilia: a case of enantiomeric repeating units. Carbohydr. Res. 330:215-221. [DOI] [PubMed] [Google Scholar]
- 41.Zughaier, S. M., H. C. Ryley, and S. K. Jackson. 1999. Lipopolysaccharide (LPS) from Burkholderia cepacia is more active than LPS from Pseudomonas aeruginosa and Stenotrophomonas maltophilia in stimulating tumor necrosis factor alpha from human monocytes. Infect. Immun. 67:1505-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]