Abstract
Moraxella catarrhalis strains can express either a UspA2 protein or a UspA2H protein. The latter protein is encoded by a gene that possesses a homopolymeric nucleotide tract containing eight adenine (A) residues [i.e., a poly(A) tract] which is located near the 5′ end. A spontaneous UspA2H-negative variant of M. catarrhalis strain O46E, designated O46E.U2V, was found to have a uspA2H poly(A) tract that contained seven A residues. Northern blot analysis of total RNA from the O46E parent strain revealed a readily detectable uspA2H mRNA transcript, whereas little or no uspA2H transcript was detectable in total RNA from the UspA2H-negative variant O46E.U2V. The 5′ end of the uspA2H genes from both the O46E parent strain and the O46E.U2V variant were ligated to a promoterless lacZ gene to prepare translational fusions for use as reporter constructs. The level of β-galactosidase activity expressed by the fusion construct containing eight A residues in its poly(A) tract was 200-fold greater than that obtained with the construct that had seven A residues. Site-directed mutagenesis of the 5′ end of the uspA2H gene confirmed that translation was initiated at a GTG codon located 21 nucleotides (nt) upstream of the poly(A) tract. Primer extension analysis determined that the transcriptional start site of the uspA2H gene was located 291 nt upstream from the GTG translational start codon. This poly(A) tract was also found to be present in the uspA2H genes of other M. catarrhalis strains.
Once thought to be a harmless commensal inhabitant of the human upper respiratory tract, Moraxella catarrhalis is now acknowledged as being capable of causing disease in both the upper and lower respiratory tracts. In infants and very young children, this organism is an important cause of acute otitis media (19, 20, 32). In adults, M. catarrhalis can cause infectious exacerbations of chronic obstructive pulmonary disease and is likely responsible for 2 million to 4 million episodes of this type each year in this country (27). This gram-negative bacterium can also cause other diseases, including sinusitis and pneumonia and, very rarely, pericarditis, endocarditis, and meningitis (reviewed in reference 26).
Among the proteinaceous surface antigens of M. catarrhalis, there are at least two encoded by genes whose expression has been shown to be affected by changes in homopolymeric nucleotide tracts, also known as simple sequence repeats or microsatellites (5). The UspA1 adhesin protein of M. catarrhalis strain O35E is encoded by a gene that contains 10 consecutive guanine (G) residues [i.e., a poly(G) tract] in the 5′ untranslated region (UTR) (23). When the number of G residues in this poly(G) tract is reduced to nine, likely as a result of slipped-strand mispairing (14), the level of expression of both uspA1 mRNA and UspA1 protein is decreased significantly (23). The Hag protein (also known as MID), which is involved in hemagglutination, autoagglutination, attachment, and immunoglobulin D binding (11, 12, 18, 30), is encoded by a gene which has a poly(G) tract located just downstream from the translation initiation codon. According to at least three reports (25, 30; K. Sasaki, K., L. Myers, S. M. Loosmore, and M. H. Klein, 99th Gen. Meet. Am. Soc. Microbiol., abstr. B/D-306, 1999), when the number of G residues present in this poly(G) tract is not divisible by three, this results in premature termination of translation of hag mRNA. The occurrence of apparent slipped-strand mispairing affecting the poly(G) tract of the hag gene has been documented by at least two different laboratories (25; Sasaki et al., 99th Gen. Meet. Am. Soc. Microbiol.).
Virtually all strains of M. catarrhalis express either a UspA2 or a UspA2H surface protein (9, 22, 24). These two proteins have in common an epitope that binds a monoclonal antibody (MAb 17C7) that also binds the UspA1 adhesin protein (2, 22). On the basis of mutant analysis, UspA2 is directly involved in serum resistance (1, 3, 4). The UspA2H protein is a hybrid molecule with its N-terminal region being virtually identical to that of UspA1 proteins whereas its C-terminal region is very similar to that of the UspA2 protein (22). UspA2H proteins have previously been shown to function as adhesins (22), likely because of the presence of the UspA1-like domains. In addition, at least in M. catarrhalis strain O46E, insertional inactivation of the uspA2H gene converted the serum-resistant O46E wild-type strain to a serum-sensitive phenotype (22).
An apparently spontaneous UspA2H-negative variant of M. catarrhalis strain O46E was isolated during a study of biofilm formation by M. catarrhalis (31). In the present report, we show that this variant differed from its parent strain by the number of consecutive adenine (A) residues contained in a poly(A) tract located within the uspA2H open reading frame (ORF). By using recombinant DNA methods and reporter constructs, we found that this change in the poly(A) tract from eight to seven A residues resulted in lack of expression of the UspA2H protein and greatly reduced levels of uspA2H mRNA. We also report that the uspA2H genes of several other M. catarrhalis strains contain this same poly(A) tract.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The O46E strain of M. catarrhalis and its isogenic uspA2H mutant O46E.2 have been described previously (22). These and the other bacterial strains used in this study are described in Table 1. The O46E strain used as the parent strain in the majority of the experiments in this study was a spontaneous hag phase variant that had lost the ability to express the Hag protein (30) as a result of apparent slipped-strand mispairing in the poly(G) tract within the hag ORF (31). This strain also possessed nine G residues in the poly(G) tract of its uspA1 gene and consequently expressed a relatively low level of UspA1 (23, 31). M. catarrhalis strains were routinely propagated in brain heart infusion (BHI) broth (Difco/Becton Dickinson, Sparks, MD) at 37°C with aeration or on BHI agar plates at 37°C in an atmosphere of 95% air-5% CO2. Antibiotic supplementation for selection of M. catarrhalis mutants involved the use of spectinomycin (15 μg/ml) or kanamycin (15 μg/ml). Recombinant strains derived from Escherichia coli XL10-Gold (Stratagene, La Jolla, CA) were maintained on Luria-Bertani (LB) agar (35) supplemented with kanamycin (30 μg/ml) as appropriate. When necessary, X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) was incorporated into LB agar at a final concentration of 40 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Reference or source |
|---|---|---|
| M. catarrhalis | ||
| O46E | Wild-type disease isolate | 2 |
| O46E.2 | uspA2H mutant of O46E | 22 |
| O46E.U2V | UspA2H-negative variant of O46E | This study |
| O46E.U2VU1 | uspA1 mutant of O46E.U2V | This study |
| O46E.U1 | uspA1 mutant of O46E | This study |
| O46E.U2V-8A | O46E.U2V transformant with a poly(A) tract containing 8 A residues | This study |
| O46E.U2V-XXG | O46E.U2V transformant with a single G residue inserted 2 nt downstream from the poly(A) tract | This study |
| TTA37 | Wild-type disease isolate | 22 |
| V1166 | Wild-type isolate from a healthy child | Frederick Henderson |
| FR3227 | Wild-type disease isolate | Richard Wallace |
| ETSU-9 | Wild-type disease isolate | Steven Berk |
| E22 | Wild-type isolate | 7 |
| ATCC 25240 | Wild-type isolate | American Type Culture Collection |
| ATCC 43617 | Wild-type disease isolate | American Type Culture Collection |
| E. coli XL10-Gold | Host for cloning and expression | Stratagene |
| Plasmids | ||
| pAC7 | Used for construction of uspA2H-lacZ translational fusions | 37 |
| pWW105 | uspA2H-lacZ fusion with 8 A residues in the poly(A) tract | This study |
| pWW106 | uspA2H-lacZ fusion with 7 A residues in the poly(A) tract | This study |
| pWW105.1 | uspA2H-lacZ fusion derived from pWW105; first GTG codon changed to ACT | This study |
| pWW105.2 | uspA2H-lacZ fusion derived from pWW105; second GTG codon changed to ACT | This study |
| pWW105.3 | uspA2H-lacZ fusion derived from pWW105; third GTG codon changed to ACT | This study |
| pWW105.12 | uspA2H-lacZ fusion derived from pWW105.2; first and second GTG codons changed to ACT | This study |
| pWW105.13 | uspA2H-lacZ fusion derived from pWW105.3; first and third GTG codons changed to ACT | This study |
| pWW105.23 | uspA2H-lacZ fusion derived from pWW105.3; second and third GTG codons changed to ACT | This study |
| pWW105.123 | uspA2H-lacZ fusion derived from pWW105.13; all three GTG codons changed to ACT | This study |
| pWW105.M | uspA2H-lacZ fusion derived from pWW105; first ATG changed to ACT | This study |
| pWW106.M | uspA2H-lacZ fusion derived from pWW106; first ATG changed to ACT | This study |
Measurement of β-d-galactosidase activity.
The level of expression of β-d-galactosidase activity in recombinant E. coli cells was measured as described by Sambrook et al. (35) with the modification that a 0.1-ml portion of the broth-grown cells was mixed with 0.7 ml of Z buffer.
Western blot analysis.
Whole-cell lysates were prepared from bacterial cells grown on agar-solidified medium and were subjected to sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis as previously described (30). The primary antibody probes used for Western blot analysis were MAb 17C7, which is reactive with both the UspA1 and UspA2/UspA2H proteins of M. catarrhalis (2, 22), and MAb 24B5, which is specific for the UspA1 protein (22). Western blot analysis was performed with horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G (Jackson ImmunoResearch, West Grove, PA) or affinity-purified and radioiodinated goat anti-mouse immunoglobulin as the secondary antibody. With the former secondary antibody, chemiluminescent detection was accomplished with ECL Plus Western blotting detection reagents (Amersham Biosciences Corp., Piscataway, NJ).
PCR and nucleotide sequence analysis.
PCR was accomplished with ExTaq DNA polymerase (PanVera, Madison, WI) or Pfu DNA polymerase (Stratagene, La Jolla, CA) according to the manufacturer's instructions. The oligonucleotide primers used are listed in Table 2. Chromosomal DNA templates were prepared from M. catarrhalis cells with the Easy-DNA kit (Invitrogen, Carlsbad, CA). PCR products were sequenced with an ABI PRISM 377 DNA sequencer (Applied Biosystems, Foster City, CA) or by the McDermott Center for Human Growth and Development at the University of Texas Southwestern Medical Center. The MacVector analysis package (version 6.5; Oxford Molecular Group, Campbell, CA) was used for DNA sequence analysis.
TABLE 2.
Oligonucleotide primers used in this studya
| Primer | Sequence (5′-3′) |
|---|---|
| O46EU2F | GAGCATGCGGTTCGCTGTATTCCCCG |
| O46EU2B | GCGCATGCGCCAGCTTTATTTTATGCAGGG |
| U2HF4 | AAGATTTTAGGCGTTTTGCAG |
| WW72 | TCGGATCCATGCCCAATGCCCCAACAATCA |
| USPE | GAAGGAGGTGAAAGGTCTTG |
| USPL | CTTGCTGTTGTGCCAGCTC |
| U2CTR | CGGGATCCGCCAATAAAGCATCTGCGGATAC |
| WW56 | CATATAATACACTCCCTGATTAACAA |
| WW92 | CTGTATAGATACTCTGTTGGTA |
| WW102 | CAATTATTCAGGAGACTAAGACGAACAAAATTTATAAAGTGAA |
| WW103 | TTCACTTTATAAATTTTGTTCGTCTTAGTCTCCTGAATAATTG |
| WW104 | CTAAGGTGAACAAAATTTATAAAACGAAAAAAAATGCCGCAGGTCA |
| WW105 | TGACCTGCGGCATTTTTTTTCGTTTTATAAATTTTGTTCACCTTAG |
| WW106 | AAAATGCCGCAGGTCATTCGACGGCATGTTCTGAATTTGCCAAAG |
| WW107 | CTTTGGCAAATTCAGAACATGCCGTCGAATGACCTGCGGCATTTT |
| WW108 | CAATTATTCAGGAGACTAAGACGAACAAAATTTATAAAACGAA |
| WW109 | TTCGTTTTATAAATTTTGTTCGTCTTAGTCTCCTGAATAATTG |
| WW112 | GATTGTTGGGGCATTGGGCACGGATCCCGTCGTTTTACAAC |
| WW113 | GTTGTAAAACGACGGGATCCGTGCCCAATGCCCCAACAATC |
| WW126 | GACGAACAAAATTTATAAAACGAAAAAAAATGCCGCAGGTCATTC |
| WW127 | GAATGACCTGCGGCATTTTTTTTCGTTTTATAAATTTTGTTCGTC |
| WW261 | CGGTATTGGCTTGGTTACCT |
| WW263 | GGCGTTTTGCAGTTGCTACT |
| WW277 | GGTGAACAAAATTTATAAAGTGAAAAAAATGGCCGCAGGTCATTCGGTGGCATG |
| WW278 | CATGCCACCGAATGACCTGCGGCCATTTTTTTCACTTTATAAATTTTGTTCACC |
Bases used for mutagenesis are underlined.
Plasmid preparation and purification of DNA fragments.
Plasmid DNA was purified from E. coli XL10-Gold cells with the QIAprep Miniprep system (QIAGEN, Valencia, CA) as described by the manufacturer. DNA fragments generated by PCR or by restriction digestion were purified with the QIAquick gel extraction kit (QIAGEN).
Construction of uspA2H-lacZ translational fusions.
A 531-bp DNA fragment containing the putative promoter region of the uspA2H gene and the extreme 5′ end of the uspA2H ORF was amplified by PCR with primers U2HF4 and WW72 (Table 2; WW72 contains a BamHI site) and Pfu polymerase together with genomic DNA isolated from either O46E or O46E.U2V as the template. The PCR products were digested with BamHI and then cloned into pAC7 (44) which had been digested with both SmaI and BamHI, with E. coli XL10-Gold as the host strain. The resultant uspA2H-lacZ translational fusions derived from O46E and O46E.U2V were designated pWW105 and pWW106, respectively.
Site-directed mutagenesis.
Mutagenesis of putative translation initiation codons in the uspA2H gene fragment in pWW105 and pWW106 was accomplished with the QuickChange XL site-specific mutagenesis kit (Stratagene) according to the manufacturer's instructions. The oligonucleotide primers used for site-directed mutagenesis are listed in Table 2.
Construction of M. catarrhalis strains with altered uspA2H genes.
Two different methods were used to correct the frameshift in the uspA2H ORF in the O46E.U2V variant. To convert the poly(A) tract from seven A residues to eight A residues, a DNA fragment containing the 5′ UTR and approximately 500 bp of the 5′ end of the uspA2H ORF was amplified by PCR with the primer pair WW263-WW261 and with M. catarrhalis O46E chromosomal DNA as the template. This amplicon was gel purified, sequenced to confirm the presence of the poly(A) tract containing eight A residues, and used to transform the O46E.U2V variant by the plate transformation method (31). Approximately 30 of the resultant colonies were inoculated into tubes containing 0.6 ml of BHI broth, grown overnight, and screened for the ability to autoagglutinate rapidly (as evidence of production of the UspA2H protein). One of the rapidly autoagglutinating strains, designated O46E.U2V-8A, was shown to have a uspA2H poly(A) tract containing eight A residues and was selected for further analysis.
To insert a G residue 2 nt downstream from the poly(A) tract in the uspA2H ORF in O46E.U2V, a DNA fragment containing the 5′ UTR and a second fragment containing about 500 bp of the 5′ end of the uspA2H ORF were PCR amplified with the primer pairs WW263-WW278 and WW277-WW261, respectively, in two separate reactions with O46E.U2V chromosomal DNA as the template. These two amplicons were gel purified and then used in an overlapping extension PCR (16) with the primer pair WW263-WW261. The final amplicon was sequence confirmed and used in a plate transformation reaction with M. catarrhalis O46E.U2V as described above. Putative transformants were screened for rapid autoagglutination, and one of these, designated O46E.U2V-XXG, was shown to have a G residue inserted 2 nt downstream from the poly(A) tract in the uspA2H ORF. A G residue was used here to minimize the number of amino acid changes resulting from this downstream insertion.
Serum bactericidal activity, autoagglutination, and attachment assays.
The resistance of M. catarrhalis strains to complement-mediated killing in 10% (vol/vol) normal human serum was measured as previously described (3). The tendency of M. catarrhalis strains to autoagglutinate in phosphate-buffered saline was measured as previously described (30), except that the starting suspension of bacteria was adjusted to yield a reading of approximately 300 Klett units (approximately 2 × 109 CFU/ml) as determined by the use of a Klett-Summerson colorimeter (Klett Manufacturing Company, New York, NY). The ability of M. catarrhalis strains to attach to Chang human conjunctival epithelial cells (ATCC CCL-20.2) in vitro was measured as previously described (42).
Northern blot analysis.
Growth from a fresh overnight BHI agar plate culture of M. catarrhalis was used to inoculate 20 ml of BHI broth, and this culture was allowed to grow until the culture reached a density that yielded a reading of 130 Klett units. Total RNA was isolated from 10 ml of this culture with the RNeasy Midi kit (QIAGEN). An overnight culture of recombinant E. coli cells was used to inoculate 5 ml LB medium, and this culture was grown to the same density as the M. catarrhalis culture. Total RNA was isolated from 1 ml of these E. coli cultures with the RNeasy Mini kit (QIAGEN). A 5-μg portion of total RNA was resolved by electrophoresis in a 1% (wt/vol) agarose gel in 1× morpholinepropanesulfonic acid (MOPS) solution (Sigma) containing 6.6% (vol/vol) formaldehyde. This gel was then soaked in 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) twice for 20 min, and then the RNA was blotted onto a nylon membrane (Hybond N+; Amersham Pharmacia Biotech, Amersham, Buckinghamshire, United Kingdom) in 10× SSC. QuikHyb solution (Stratagene) was used for hybridization with either radiolabeled oligonucleotide probes overnight at 37°C or with digoxigenin-labeled, PCR-derived probes at 50°C. The latter probes were labeled with digoxigenin-dUTP by using DIG High Prime DNA Labeling and Detection Starter Kit II (Roche). The membrane was then washed in 2× SSC containing 0.1% (wt/vol) SDS twice at room temperature for 20 min and then in 0.2× SSC containing 0.1% (wt/vol) SDS twice. The latter two washes were performed at 42°C for radiolabeled oligonucleotide probes and at 50°C for digoxigenin-labeled, PCR-derived probes. The filter was exposed to X-ray film with or without digoxigenin processing, as appropriate.
Primer extension analysis.
Total RNA was prepared from recombinant E. coli cells as described above. For the primer extension reaction, a 5-μg portion of total RNA was incubated with 2 pmol of oligonucleotide primer WW56 or WW92 (Table 2) by using Invitrogen's SuperScript First-Strand Synthesis System in the presence of [α-32P]dCTP. The DNA sequencing ladder was prepared by using the same primer and pWW105 (Table 2) with the AmpliCycle Sequencing Kit (Applied Biosystems) and [α-32P]dCTP. The DNA sequencing ladder and the primer extension reaction products were resolved in a 6% (wt/vol) polyacrylamide sequencing gel (Long Ranger Gel Solutions, Cambrex, Rockland, ME) and then exposed to X-ray film.
Measurement of uspA2H mRNA stability.
Rifampin (150 μg/ml) was added to M. catarrhalis strains O46E and O46E.U2V in the mid-logarithmic phase of growth in BHI broth. RNA was isolated from cells obtained at time zero and at 1, 2, 5, and 10 min after rifampin addition as previously described (43) and was subjected to Northern blot analysis as described above. The intensity of the bands corresponding to full-length uspA2H transcripts in these X-ray films was quantified with the Image Station 440 System (Eastman Kodak Scientific Imaging Systems, Rochester, NY) and was analyzed with Kodak 1D Image Analysis Software. The half-life of the O46E uspA2H mRNA was determined as previously described (8).
Determination of the frequency of phase variation.
A broth culture of the M. catarrhalis O46E parent strain was serially diluted and plated on BHI agar. A total of 10,752 of the resultant colonies were inoculated into individual tubes containing 0.6 ml of BHI broth and incubated overnight with agitation. Cultures in which turbidity was readily apparent (i.e., in which autoagglutination did not occur) were streaked to obtain isolated colonies, which were then used to prepare genomic DNA for subsequent PCR amplification and nucleotide sequence analysis of the region of the uspA2H gene containing the poly(A) tract.
RESULTS
Identification of a UspA2H-negative variant of strain O46E.
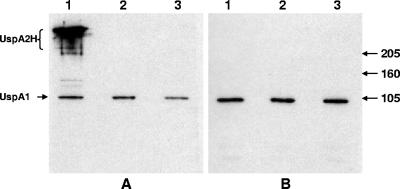
M. catarrhalis strain O46E (Fig. 1A, lane 1) has been shown to produce a UspA2H protein in place of a UspA2 protein (22). In a recent study, transposon-mutagenized cells of M. catarrhalis strain O46E were screened for the ability to form biofilms in vitro (M. M. Pearson and E. J. Hansen, data not shown). During routine Western blot-based screening, it was observed that the subculture of the O46E parent strain used for this experiment no longer expressed a UspA2H protein. This UspA2H-negative variant, designated O46E.U2V (Fig. 1A, lane 3), yielded a Western blot result with MAb 17C7 that was indistinguishable from that obtained with the isogenic uspA2H mutant O46E.2 (Fig. 1A, lane 2). All three of these strains still expressed the UspA1 protein (Fig. 1B, lanes 1 to 3), as detected by its reactivity with UspA1-specific MAb 24B5.
FIG. 1.
Expression of UspA2H by different strains of M. catarrhalis O46E. Whole-cell lysates prepared from the M. catarrhalis O46E parent strain (lane 1), the uspA2H mutant O46E.2 (lane 2), and the UspA2H-negative variant O46E.U2V (lane 3) were analyzed by Western blot analysis with MAbs 17C7 (A) and 24B5 (B) as the primary antibody probes. Molecular mass position markers (in kilodaltons) are shown to the right side of panel B.
Phenotype of the O46E.U2V strain.
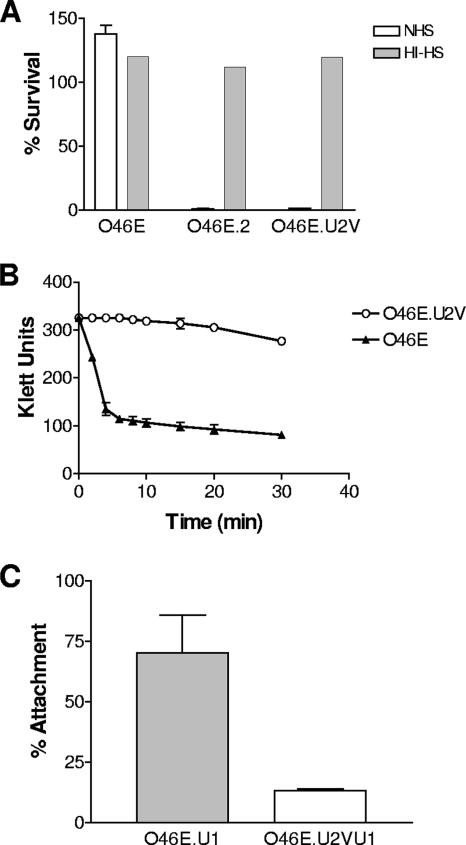
It was previously reported that M. catarrhalis strain O46E is resistant to killing by normal human serum and that inactivation of the uspA2H gene of this strain converted it to a serum-sensitive phenotype (22). To determine whether the UspA2H-negative variant O46E.U2V had a phenotype similar to that of the isogenic uspA2H mutant O46E.2, these two strains were compared to the O46E parent strain in a serum bactericidal activity assay. The O46E parent strain (Fig. 2A) was completely resistant to killing by 10% (vol/vol) normal human serum, whereas both the uspA2H mutant O46E.2 and the UspA2H-negative variant O46E.U2V (Fig. 2A) were readily killed. None of these strains were killed by heat-inactivated normal human serum (Fig. 2A).
FIG. 2.
Phenotypic characteristics of the O46E parent strain and the UspA2H-negative variant O46E.U2V. (A) Resistance of M. catarrhalis strains to the bactericidal activity of normal human serum. Killing of the O46E parent strain, the uspA2H mutant O46E.2, and the UspA2H-negative variant O46E.U2V by 10% (vol/vol) normal human serum (NHS, open columns) and by heat-inactivated NHS (HI-HS, shaded columns) was measured as described in Materials and Methods. (B) Autoagglutination of the O46E parent strain (closed triangles) and the UspA2H-negative variant (open circles) after suspension of agar plate-grown bacteria in phosphate-buffered saline. (C) Attachment to Chang cells of uspA1 mutants derived from the O46E parent strain (O46E.U1, shaded bar) and the O46E.U2V variant (O46E.U2VU1, open bar). Results represent the mean from three independent experiments.
The O46E.U2V strain (Fig. 2B) was also shown to have a much slower rate of autoagglutination than the O46E parent strain (Fig. 2B). In addition, when the uspA1 gene of this variant was inactivated by insertion of a kan cartridge, the resultant mutant, O46E.U2VU1 (Fig. 2C), was found to have lost much of its ability to attach to Chang conjunctival epithelial cells in vitro. In contrast, the uspA1 mutant O46E.U1 (which is unable to express UspA1 but does express UspA2H) (Fig. 2C) readily attached to Chang cells. This latter result suggested that loss of expression of UspA2H in the O46E.U2V strain resulted in the loss of a functional Chang cell adhesin, as would have been predicted from the previously observed phenotype of a uspA1 uspA2H mutant of strain O46E (22). In addition, it was noted that the O46E.U1 organisms tended to adhere in clumps or clusters to the Chang cells (data not shown), a finding which suggested that the autoagglutination property of the UspA2H protein also contributed to the ability of this strain to interact with Chang cell monolayers.
Expression of uspA2H mRNA.
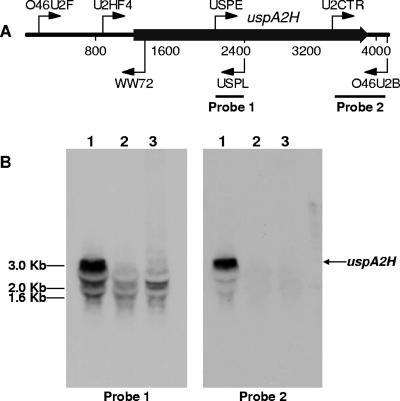
Nucleotide sequence analysis of the predicted uspA2H ORF in the O46E parent strain revealed the presence of a homopolymeric nucleotide tract that contained eight A residues [i.e., a poly(A) tract] near the 5′ end of this gene. In contrast, the same region of the uspA2H gene from the O46E.U2V variant contained only seven A residues. To determine whether this difference in the poly(A) tracts affected transcription of the uspA2H gene, total RNA was purified from both the O46E parent strain and the O46E.U2V variant, as well as from the isogenic uspA2H mutant O46E.2 and was subjected to Northern blot analysis. The two different uspA2H-derived probes used for Northern blot analysis bound to the middle or to the extreme 3′ end of the predicted uspA2H transcript (Fig. 3A). A 3-kb transcript that bound both of these probes was readily detectable in total RNA from the O46E parent strain (Fig. 3B, lane 1). In contrast, there was little or no detectable 3-kb transcript in the total RNA extracted from both the uspA2H mutant O46E.2 and the UspA2H-negative variant O46E.U2V (Fig. 3B, lanes 2 and 3, respectively) that hybridized to these probes.
FIG. 3.
Northern blot analysis of total RNA from M. catarrhalis O46E strains with probes for the uspA2H transcript. (A) Schematic diagram of the M. catarrhalis O46E uspA2H locus showing the locations of the primers used for nucleotide sequence analysis and for PCR-based amplification of probes for Northern blot analysis. Nucleotides are numbered in 800-nt increments. (B) Total RNA isolated from the M. catarrhalis O46E parent strain (lane 1), the uspA2H mutant O46E.2 (lane 2), and the UspA2H-negative variant O46E.U2V (lane 3) was analyzed by Northern blotting with probes 1 and 2. DNA molecular size markers are indicated on the left side of panel B.
Construction of a uspA2H-lacZ reporter construct.
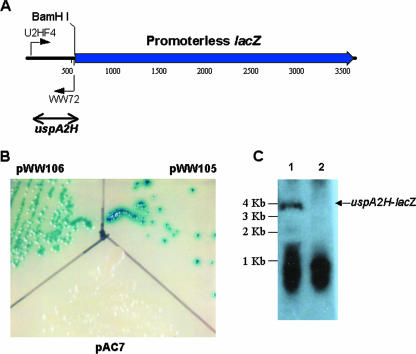
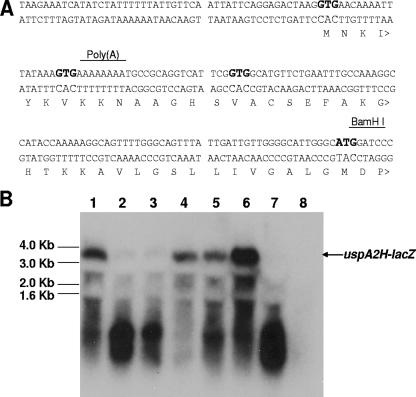
In order to determine more quantitatively the effect of changes in the poly(A) tract on expression of the uspA2H gene, the predicted 5′ end of the uspA2H gene from the O46E parent strain, including the poly(A) tract with eight A residues, was amplified by PCR and ligated into pAC7 (44). The resultant plasmid, pWW105 (Fig. 4A), contained a translational fusion between 524 nt from the 5′ end of the uspA2H gene (encompassing both the putative promoter region and the putative 5′ end of the ORF) and the promoterless lacZ gene in pAC7, such that the ATG codon located 95 nt downstream from the poly(A) tract became the ATG translation initiation codon of the lacZ gene. Similarly, the same region from the uspA2H gene in the UspA2H-negative variant O46E.U2V was amplified by PCR and ligated into pAC7 to yield pWW106. When these two recombinant strains were grown on LB agar containing X-Gal, the E. coli XL10-Gold strain containing pWW105 (Fig. 4B) produced very small colonies that were surrounded by a blue halo. In contrast, the colonies obtained with E. coli XL10-Gold(pWW106) (Fig. 4B) had a very light blue center and did not produce a colored halo. Northern blot analysis of total RNA from these two recombinant strains was performed with an oligonucleotide probe (WW72; Fig. 4A) specific for the uspA2H nucleotide sequence in the fusion construct. E. coli XL10-Gold(pWW105) had a readily detectable mRNA transcript (Fig. 4C, lane 1) of the size predicted (approximately 3.6 to 3.7 kb) for the uspA2H-lacZ fusion construct. In contrast, there was no detectable uspA2H-lacZ transcript in total RNA from E. coli XL10-Gold(pWW106) (Fig. 4C, lane 2).
FIG. 4.
Use of a translational fusion to study uspA2H gene expression. (A) Schematic diagram of the uspA2H-lacZ translational fusion protein construct. The putative uspA2H promoter region from both the O46E parent strain [with a poly(A) tract containing eight A residues] and the UspA2H-negative variant O46E.U2V [with a poly(A) tract containing seven A residues] was PCR amplified with primers U2HF4 and WW72 and cloned into plasmid pAC7 via SmaI and BamHI sites to yield pWW105 and pWW106, respectively. Nucleotides are numbered in 500-nt increments. (B) Expression of the UspA2H-LacZ fusion protein. E. coli XL10-Gold cells containing plasmid pAC7, pWW105, or pWW106 were grown on LB agar containing X-Gal at 37°C overnight and then photographed. (C) Northern blot analysis. Total RNA from E. coli XL10-Gold(pWW105) (lane 1) and E. coli XL10-Gold(pWW106) (lane 2) was analyzed by Northern blotting with the 32P-labeled oligonucleotide WW72 to detect uspA2H-lacZ mRNA. DNA molecular size markers are indicated on the left side of panel C.
Identification of the uspA2H translation initiation codon.
In a previous study (22), a putative translation initiation codon was predicted for the O46E uspA2H gene on the basis of similarities between the predicted signal peptide of the UspA2H protein and the confirmed signal peptide of the UspA1 protein (9). However, it was important to identify the true translation initiation codon for the uspA2H gene because there were several alternative translation initiation codons in relatively close proximity to the poly(A) tract. Specifically, there were two GTG codons immediately upstream of the poly(A) tract and one GTG codon between the poly(A) tract and a downstream ATG codon (Fig. 5A). To address this issue, site-directed mutagenesis was used to individually alter each of these four potential translation initiation codons in the uspA2H-lacZ fusion construct, resulting in four new constructs that were then tested for expression of β-galactosidase activity. Four additional constructs containing various combinations of these altered codons were also prepared.
FIG. 5.
Use of site-directed mutagenesis to identify the translation initiation codon of the O46E uspA2H ORF. (A) Nucleotide sequence of the distal portion of the uspA2H promoter region and the 5′ end of the uspA2H ORF. The four putative translation initiation sites (three GTG codons and one ATG codon) are indicated in bold. The BamHI site used for construction of the uspA2H-lacZ translational fusion in pAC7 is also indicated. (B) Northern blot analysis of uspA2H-lacZ transcription in E. coli XL10-Gold containing the following plasmids: lane 1, pWW105 [with a poly(A) tract containing eight A residues]; lane 2, pWW106 [with a poly(A) tract containing seven A residues]; lane 3, pWW105.1 with the first GTG changed to ACT; lane 4, pWW105.2 with the second GTG changed to ACT; lane 5, pWW105.3 with the third GTG changed to ACT; lane 6, pWW105.M with the ATG changed to ACT; lane 7, pWW106.M with the ATG changed to ACT; lane 8, pAC7 (negative control). The probe for Northern blot analysis was the uspA2H-specific, 32P-labeled oligonucleotide WW72 (Fig. 4A). DNA molecular size markers are indicated on the left side of panel B.
Conversion of the GTG codon located 21 nt upstream of the poly(A) tract to an ACT codon resulted in a drastic decrease in the level of β-galactosidase activity expressed by plasmid pWW105.1 in E. coli XL10-Gold (Table 3). In contrast, changing the other two GTG codons to ACT codons had little effect on the level of β-galactosidase activity expressed by pWW105.2 and pWW105.3 (Table 3). When the ATG codon located 94 nt downstream from the poly(A) tract was changed to ACT, this also had little effect on protein expression by pWW105.M (Table 3). When various combinations of the different GTG codons were altered, the only combinations which had severely reduced expression of β-galactosidase activity were those (pWW105.12, pWW105.13, and pWW105.123) in which the first GTG codon had been altered (Table 3). As expected, the presence of only seven A residues in the poly(A) tract of the uspA2H-lacZ fusion construct in pWW106 resulted in a level of β-galactosidase activity that was 2 orders of magnitude lower than that obtained with pWW105 (Table 3). It could be inferred from these protein expression data that the GTG codon located 21 nt upstream from the poly(A) tract was the primary translation initiation codon for the uspA2H ORF.
TABLE 3.
Expression of β-galactosidase activity by uspA2H-lacZ fusion constructs in E. coli XL10-Gold
| Plasmid | Primers/templatea | Miller unitsb | % Relative to pWW105 |
|---|---|---|---|
| pAC7 | 0 | 0 | |
| pWW105 | 36,959 ± 4,681 | 100 | |
| pWW106 | 201 ± 89 | 0.5 | |
| pWW105.1 | WW102-WW103/pWW105 | 214 ± 19 | 0.6 |
| pWW105.2 | WW104-WW105/pWW105 | 34,200 ± 7,389 | 92.5 |
| pWW105.3 | WW106-WW107/pWW105 | 33,643 ± 5,434 | 91.0 |
| pWW105.12 | WW108-WW109/pWW105.2 | 200 ± 12 | 0.5 |
| pWW105.13 | WW102-WW103/pWW105.3 | 184 ± 16 | 0.5 |
| pWW105.23 | WW104-WW105/pWW105.3 | 33,037 ± 5,871 | 89.3 |
| pWW105.123 | WW126-WW127/pWW105.13 | 130 ± 30 | 0.4 |
| pWW105.M | WW112-WW113/pWW105 | 31,110 ± 5,378 | 84.2 |
| pWW106.M | WW112-WW113/pWW106 | 110 ± 25 | 0.3 |
Oligonucleotide primers and template used for mutagenesis of selected codons.
Miller units were calculated with the following formula: 1,000 × A420/[time (minutes) × vol (milliliters) × A600]. This formula was modified from that described by Sambrook et al. (35). These values represent the mean ± the standard deviation.
Northern blot analysis of these same constructs (in E. coli XL10-Gold) revealed that, when protein expression was lost as a result of alteration of the GTG start codon located 21 nt 5′ from the poly(A) tract in pWW105.1 (Fig. 5B, lane 3), there was little or no detectable uspA2H-lacZ mRNA reactive with a uspA2H-specific probe. In contrast, transcripts of the appropriate size were detected in the total RNA from pWW105.2, pWW105.3, and pWW105.M (Fig. 5B, lanes 4, 5, and 6, respectively), all of which also expressed wild-type levels of β-galactosidase activity (Table 3). As expected, a 3.6- to 3.7-kb transcript was readily detectable when total RNA from E. coli(pWW105) (Fig. 5B, lane 1) was incubated with the uspA2H-specific probe and transcripts of the same size were either not detectable or barely detectable in total RNA from E. coli(pWW106) (Fig. 5B, lane 2). It should also be noted that, in contrast to all of the aforementioned samples in Fig. 5B, there was little or no material with an apparent size of less than 1.6 kb in the total RNA sample derived from pAC7 (Fig. 5B, lane 8) that bound the uspA2H-specific probe. It could be inferred from this result that the uspA2H-lacZ mRNA derived from the constructs that did not have detectable protein expression (e.g., pWW106) might have been degraded more rapidly than that from the constructs which had readily detectable β-galactosidase activity (e.g., pWW105).
Primer extension analysis of the uspA2H gene.
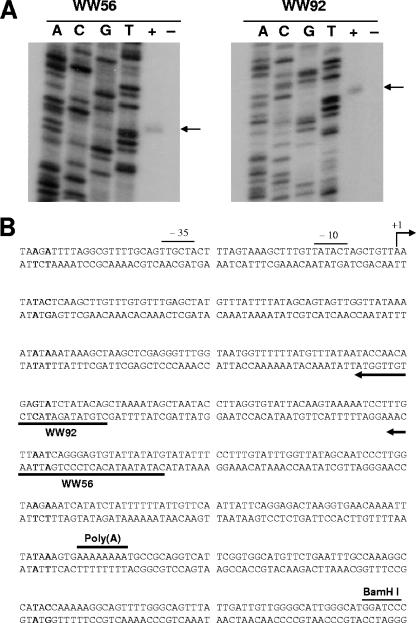
The transcriptional start of the uspA2H gene was determined by primer extension analysis with the uspA2H-lacZ fusion construct. When total RNA from E. coli(pWW105) was used in a primer extension analysis with oligonucleotide primer WW92 (Fig. 6B), a cDNA fragment of 136 nt was obtained (Fig. 6A, WW92 side, + lane). When this same primer was used for nucleotide sequence analysis, the transcriptional start site of uspA2H was mapped to an A residue (Fig. 6B) located 311 nt upstream from the poly(A) tract. This same A residue was again identified as the transcriptional start site with primer WW56 by primer extension analysis (Fig. 6A). These results indicate that the 5′ UTR of the O46E uspA2H gene is 291 nt in length. Nucleotide sequences with homology to the −10 and −35 consensus sequences for bacterial promoters were observed upstream of the transcriptional start site.
FIG. 6.
Mapping of the transcriptional start site of the uspA2H-lacZ fusion construct by primer extension analysis. (A) A 5-μg portion of total RNA from E. coli XL10-Gold(pWW105) was used as the template, and oligonucleotide WW56 or WW92 was used as the primer for reverse transcription in the presence of [α-32P]dCTP. The DNA sequencing ladder was prepared by using each primer individually with pWW105 as the template. The arrows indicate the labeled cDNA from each primer extension reaction. Plus and minus signs indicate the presence or absence of reverse transcriptase, respectively. (B) Nucleotide sequence of the promoter region of the O46E uspA2H gene. The transcriptional start site (+1), the putative −10 and −35 regions, the oligonucleotide primers WW56 and WW92, the poly(A) tract, and the uspA2H-lacZ fusion site (BamHI) are indicated.
Restoration of the reading frame affects uspA2H mRNA levels.
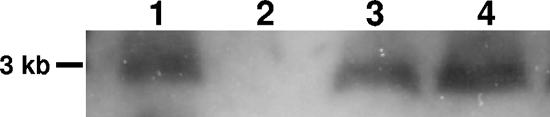
These data on the locations of the transcriptional and translational starts for the uspA2H gene allowed us to predict that the presence of the seven A residues in the poly(A) tract would result in the occurrence of a premature translation termination codon 74 nt downstream from the poly(A) tract. Therefore, the apparent lack of detectable uspA2H mRNA in the O46E.U2V variant could have been caused by destabilization of the transcript by this mutation. To address this possibility, we both corrected the original deletion mutation (i.e., seven A to eight A) and independently restored the reading frame by inserting a single G residue 2 nt downstream from the poly(A) tract in the uspA2H ORF in O46E.U2V. The two resultant M. catarrhalis strains, O46E.U2V-8A and O46E.U2V-XXG, respectively, expressed the UspA2H protein and were serum resistant like the O46E parent strain (data not shown). Both of these strains (Fig. 7, lanes 3 and 4) also expressed levels of uspA2H mRNA equivalent to that expressed by the O46E parent strain (Fig. 7, lane 1).
FIG. 7.
Restoration of the reading frame affects uspA2H mRNA levels. RNAs prepared from different M. catarrhalis strains were subjected to Northern blot analysis with probe 2 from Fig. 3. Lanes: 1, O46E parent strain; 2, uspA2H variant O46E.U2V; 3, O46E.U2V-8A; 4, O46E.U2V-XXG. A DNA molecular size marker is indicated on the left.
Determination of the half-life of uspA2H mRNA.
To determine the half-life of uspA2H transcripts, rifampin was added to M. catarrhalis cells in the logarithmic phase of growth and RNA was prepared from cells harvested at various time points. The uspA2H transcripts in the O46E parent strain were readily detectable at time zero but decreased gradually over a 10-min period (Fig. 8), with a calculated half-life of 2.87 ± 0.25 min. No uspA2H transcripts were detected in RNA prepared from O46E.U2V cells at any time point (Fig. 8), precluding determination of the half-life of this mRNA.
FIG. 8.
Determination of the half-life of uspA2H mRNA. Rifampin was added to cells of M. catarrhalis strains O46E and O46E.U2V in the logarithmic phase of growth. RNA prepared from samples of these cells harvested at five different time points was used in a Northern blot analysis with probe 2 from Fig. 3. Three independent experiments were performed to obtain densitometric measurements; the results of one of these experiments are depicted here. DNA molecular size markers are indicated on the left.
Frequency of phase variation.
The rapid autoagglutination phenotype associated with UspA2H expression was used to facilitate determination of the frequency of phase variation. A total of 10,752 colonies of the O46E parent strain were inoculated into individual broth tube cultures, which were then incubated overnight with agitation to identify phase variants (i.e., eight A to seven A) that could no longer express UspA2H and therefore did not autoagglutinate. By this method, we identified seven variants that had a poly(A) tract containing seven A residues, as confirmed by PCR and nucleotide sequence analysis. The calculated frequency of phase variation (eight A to seven A) was 6.5 × 10−4. It is assumed that the frequency of phase variation in the opposite direction (seven A to eight A) is lower than 10−4 because of the shorter poly(A) tract (10).
Presence of a poly(A) tract in other uspA2H genes and uspA1 genes.
We have identified at least six other M. catarrhalis strains (TTA37, E22, ETSU-9, V1166, ATCC 25240, and FR3227), in addition to strain O46E, that express a UspA2H protein, and the uspA2H genes of these six strains all possess a poly(A) tract near the 5′ end of the ORF that contains eight A residues (data not shown). In addition, it was noted previously that the amino acid sequence of the signal peptide of the UspA1 protein was virtually identical to the predicted signal peptides of the UspA2H proteins of strains O46E and TTA37 (22). Examination of the nucleotide sequences of the uspA1 genes present in these M. catarrhalis strains which express a UspA2H protein revealed the presence of a poly(A) tract containing eight A residues in nearly all of the uspA1 genes, located just downstream from the translation initiation codon (data not shown). In addition, the nucleotide sequences of most of the other uspA1 genes characterized in previous studies in this laboratory (2, 9) also contain this poly(A) tract.
DISCUSSION
There are at least three different types of intrachromosomal recombination that can effect genotypic variation in bacteria (see reference 34 for a review). These include RecA-dependent homologous recombination, site-specific recombination, and illegitimate recombination (34). Slipped-strand mispairing is one type of RecA-independent illegitimate recombination that occurs within the bacterial chromosome and can affect either transcription of the gene involved or translation of its mRNA. There are now many examples of slipped-strand mispairing involving homopolymeric or heteropolymeric nucleotide repeats that result in phase variation in bacteria (see references 14, 38, and 39 for reviews). Simple sequence repeats, including poly(A) tracts similar to the uspA2H gene feature that is the subject of the present study, have been reported to be more common in bacterial pathogens than in other bacteria (34), perhaps because of their involvement with contingency genes (5, 6, 38).
In M. catarrhalis, the UspA1 and Hag surface appendages (30) are known to undergo a phase variation that involves simple sequence repeats. More specifically, the poly(G) tracts in the uspA1 and hag genes have been reported to affect the expression of these two genes in different ways. In M. catarrhalis strain O35E, a decrease from 10 to 9 G residues in the poly(G) tract located in the 5′ UTR of the uspA1 gene causes a reduction in the level of uspA1 mRNA and the amount of UspA1 protein (23). In contrast, the poly(G) tract found within the hag ORF affects Hag expression by allowing frameshift mutations that cause premature termination of translation (25; Sasaki et al., 99th Gen. Meet. Am. Soc. Microbiol.). Both the UspA1 protein and the Hag protein function as adhesins, albeit for different human cell types (11, 15, 18, 22), and the ability of M. catarrhalis to alter the expression of these two different gene products via phase variation may affect the ability of this organism to colonize the mucosal surface of the nasopharynx or permit the selection of a population with a phenotypic trait that is advantageous for a particular environmental niche.
A change in the number of A residues in the uspA2H poly(A) tract of strain O46E had multiple phenotypic effects. The UspA2H-negative variant O46E.U2V lost its ability to resist killing by normal human serum, a trait that is most often associated with disease isolates of this organism (17, 40, 41). This UspA2H-negative variant also lost its ability to attach to Chang cells in vitro via this protein. What adhesin molecule is directly involved in the attachment of M. catarrhalis to the human nasopharyngeal mucosa in vivo is not known, but clearly this variant has lost one facet of its attachment capability. In addition, this variant also lacked the rapid-autoagglutination phenotype apparently caused by expression of the UspA2H protein in strain O46E. This resulted in the variant growing in suspension in broth cultures whereas the O46E parent strain rapidly fell out of suspension. This rapid-autoagglutination phenotype likely affected the ability of the O46E strain to attach to other M. catarrhalis bacteria and form microcolonies or otherwise interact with host cells. Interestingly, the lack of autoagglutination by the O46E.U2V variant is similar to that of a mutant or variant of M. catarrhalis strain 4223 that did not readily form aggregates and which was cleared from the lungs of mice more slowly than its wild-type parent strain (21). This same nonaggregating 4223 variant lacked or had altered expression of several antigens, including HMW-OMP (i.e., UspA2).
The amino terminus of the predicted mature UspA2H protein closely resembles that of UspA1 proteins (22), and the signal peptides are virtually identical in both molecules. In fact, the same poly(A) tract that is present at the 5′ end of the uspA2H ORF is also present at the 5′ end of the uspA1 ORF. However, the 5′ UTR of the uspA2H gene of strain O46E is very different from that of the uspA1 gene, lacking the poly(G) tract that is characteristic of all of the M. catarrhalis uspA1 genes analyzed to date (9). The observed change from eight to seven A residues in the uspA2H poly(A) tract of the O46E strain is most likely the result of slipped-strand mispairing (14, 38) and occurred at a frequency of 6.5 × 10−4. Efforts to determine the frequency of the change from seven to eight A residues were not successful, although there are reports of homopolymeric repeats containing as few as 6 nt in other bacteria being unstable and undergoing reversible loss or gain of nucleotides (13, 33). The presence of a poly(A) tract containing eight A residues near the 5′ end of the ORF of the uspA1 gene in several of the M. catarrhalis strains included in this study raises the possibility that expression of these particular UspA1 proteins could be subject to phase variation involving the deletion of an A residue from this poly(A) tract, resulting in a frameshift mutation that would cause premature termination of translation. In this event, these particular strains could experience phase-variable expression of their UspA1 protein by either of two different mechanisms, one involving the poly(G) tract in the 5′ UTR and the other involving the poly(A) tract within the ORF.
The lack of expression of the UspA2H protein by the O46E.U2V variant described in the present study was the result of a frameshift that occurred immediately downstream from the uspA2H translation initiation codon. This, in turn, resulted in the occurrence of a premature translational stop codon 74 nt downstream from the poly(A) tract. It is also apparent that this change in the poly(A) tract affected uspA2H mRNA levels, as measured by Northern blot analysis. It has been known for many years that the stability of some mRNA transcripts is negatively affected by early or premature translation termination (28). In the present study, both native M. catarrhalis uspA2H transcripts and uspA2H-lacZ fusion-derived transcripts were apparently more readily degraded when the number of A residues in the poly(A) tract was seven instead of eight. Moreover, when this frameshift in the O46E.U2V variant was corrected, either by converting the poly(A) tract from seven to eight A residues or by insertion of a G residue 2 nt downstream from the poly(A) tract, this restoration of the reading frame resulted in readily detectable levels of uspA2H transcripts. It could be inferred from these data that the frameshift-induced premature termination of translation and a possible reduction in the number of ribosomes on the uspA2H transcripts from O46E.U2V may have resulted in increased accessibility of these transcripts to endoribonucleolytic enzymes. Differences in RNA secondary structure were apparently not involved (data not shown). Whether the observed frameshift occurs in the host environment and whether the resultant altered phenotype would affect the colonization ability or virulence of M. catarrhalis remain to be determined.
Finally, in silico analysis of the available nucleotide sequence of the genome of M. catarrhalis ATCC 43617 (GenBank accession numbers AX067426 to AX067466) revealed the presence of homopolymeric nucleotide repeats containing eight or more nucleotides located just downstream from the beginning of at least nine different ORFs (data not shown), in a manner similar to that observed for the poly(A) tract near the start of the uspA2H ORF. These included the previously described poly(G) tract in the hag gene (25, 30; Sasaki et al., 99th Gen. Meet. Am. Soc. Microbiol.) and the poly(A) tract within the uspA1 ORF. In addition, at least 20 homopolymeric nucleotide repeats containing eight or more nucleotides were found to be located immediately upstream (within 100 nt) from ORFs (data not shown). Furthermore, it has been reported that several M. catarrhalis strains contained loci with the tetranucleotide repeat 5′-CAAC-3′ (29) and that this repeat was associated with phase-variable restriction-modification systems (36). Taken together, these data suggest that M. catarrhalis likely contains at least two dozen potentially phase-variable genes.
Acknowledgments
This study was supported by U.S. Public Health Service grant AI36344 to E.J.H. M.M.P. was supported by U.S. Public Health Service training grant 5-T32-AI007520.
We thank John Nelson, Steven Berk, Frederick Henderson, and Richard Wallace for supplying the isolates of M. catarrhalis used in this study.
Editor: V. J. DiRita
Footnotes
Published ahead of print on 12 January 2007.
REFERENCES
- 1.Aebi, C., E. R. Lafontaine, L. D. Cope, J. L. Latimer, S. R. Lumbley, G. H. McCracken, Jr., and E. J. Hansen. 1998. Phenotypic effect of isogenic uspA1 and uspA2 mutations on Moraxella catarrhalis O35E. Infect. Immun. 66:3113-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aebi, C., I. Maciver, J. L. Latimer, L. D. Cope, M. K. Stevens, S. E. Thomas, G. H. McCracken, Jr., and E. J. Hansen. 1997. A protective epitope of Moraxella catarrhalis is encoded by two different genes. Infect. Immun. 65:4367-4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Attia, A. S., E. R. Lafontaine, J. L. Latimer, C. Aebi, G. A. Syrogiannopoulos, and E. J. Hansen. 2005. The UspA2 protein of Moraxella catarrhalis is directly involved in the expression of serum resistance. Infect. Immun. 73:2400-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Attia, A. S., S. Ram, P. A. Rice, and E. J. Hansen. 2006. Binding of vitronectin by the Moraxella catarrhalis UspA2 protein interferes with late stages of the complement cascade. Infect. Immun. 74:1597-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bayliss, C. D., K. M. Dixon, and E. R. Moxon. 2004. Simple sequence repeats (microsatellites): mutational mechanisms and contributions to bacterial pathogenesis. A meeting review. FEMS Immunol. Med. Microbiol. 40:11-19. [DOI] [PubMed] [Google Scholar]
- 6.Bayliss, C. D., D. Field, and E. R. Moxon. 2001. The simple sequence contingency loci of Haemophilus influenzae and Neisseria meningitidis. J. Clin. Investig. 107:657-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beaulieu, D., M. Ouellette, M. G. Bergeron, and P. H. Roy. 1988. Characterization of a plasmid isolated from Branhamella catarrhalis and detection of plasmid sequences within the genome of a B. catarrhalis strain. Plasmid 20:158-162. [DOI] [PubMed] [Google Scholar]
- 8.Bechhofer, D. H., and W. Wang. 1998. Decay of ermC mRNA in a polynucleotide phosphorylase mutant of Bacillus subtilis. J. Bacteriol. 180:5968-5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cope, L. D., E. R. Lafontaine, C. A. Slaughter, C. A. Hasemann, Jr., C. Aebi, F. W. Henderson, and G. H. McCracken, Jr. 1999. Characterization of the Moraxella catarrhalis uspA1 and uspA2 genes and their encoded products. J. Bacteriol. 181:4026-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Bolle, X., C. D. Bayliss, D. Field, T. van de Ven, N. J. Saunders, D. W. Hood, and E. R. Moxon. 2002. The length of a tetranucleotide repeat tract in Haemophilus influenzae determines the phase variation rate of a gene with homology to type III DNA methyltransferases. Mol. Microbiol. 46:293. [DOI] [PubMed] [Google Scholar]
- 11.Forsgren, A., M. Brant, M. Karamehmedovic, and K. Riesbeck. 2003. The immunoglobulin D-binding protein MID from Moraxella catarrhalis is also an adhesin. Infect. Immun. 71:3302-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forsgren, A., M. Brant, A. Mollenkvist, A. Muyombwe, H. Janson, N. Woin, and K. Riesbeck. 2001. Isolation and characterization of a novel IgD-binding protein from Moraxella catarrhalis. J. Immunol. 167:2112-2120. [DOI] [PubMed] [Google Scholar]
- 13.Hammerschmidt, S., A. Muller, H. Sillmann, M. Muhlenhoff, R. Borrow, A. Fox, J. van Putten, W. D. Zollinger, R. Gerardy-Schahn, and M. Frosch. 1996. Capsule phase variation in Neisseria meningitidis serogroup B by slipped-strand mispairing in the polysialyltransferase gene (siaD): correlation with bacterial invasion and the outbreak of meningococcal disease. Mol. Microbiol. 20:1211-1220. [DOI] [PubMed] [Google Scholar]
- 14.Henderson, I. R., P. Owen, and J. P. Nataro. 1999. Molecular switches—the ON and OFF of bacterial phase variation. Mol. Microbiol. 33:919-932. [DOI] [PubMed] [Google Scholar]
- 15.Hill, D. J., and M. Virji. 2003. A novel cell-binding mechanism of Moraxella catarrhalis ubiquitous surface protein UspA: specific targeting of the N-domain of carcinoembryonic antigen-related cell adhesion molecules by UspA1. Mol. Microbiol. 48:117-129. [DOI] [PubMed] [Google Scholar]
- 16.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 17.Hol, C., C. M. Verduin, E. van Dijke, J. Verhoef, and H. van Dijk. 1993. Complement resistance in Branhamella (Moraxella) catarrhalis. Lancet 341:1281. [DOI] [PubMed] [Google Scholar]
- 18.Holm, M. M., S. L. Vanlerberg, D. D. Sledjeski, and E. R. Lafontaine. 2003. The Hag protein of Moraxella catarrhalis strain O35E is associated with adherence to human lung and middle ear cells. Infect. Immun. 71:4977-4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karalus, R., and A. Campagnari. 2000. Moraxella catarrhalis: a review of an important human mucosal pathogen. Microbes Infect. 2:547-559. [DOI] [PubMed] [Google Scholar]
- 20.Klein, J. O. 1994. Otitis media. Clin. Infect. Dis. 19:823-833. [DOI] [PubMed] [Google Scholar]
- 21.Kyd, J. M., A. W. Cripps, and T. F. Murphy. 1998. Outer-membrane antigen expression by Moraxella (Branhamella) catarrhalis influences pulmonary clearance. J. Med. Microbiol. 47:159-168. [DOI] [PubMed] [Google Scholar]
- 22.Lafontaine, E. R., L. D. Cope, C. Aebi, J. L. Latimer, G. H. McCracken, Jr., and E. J. Hansen. 2000. The UspA1 protein and a second type of UspA2 protein mediate adherence of Moraxella catarrhalis to human epithelial cells in vitro. J. Bacteriol. 182:1364-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lafontaine, E. R., N. J. Wagner, and E. J. Hansen. 2001. Expression of the Moraxella catarrhalis UspA1 protein undergoes phase variation and is regulated at the transcriptional level. J. Bacteriol. 183:1540-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meier, P. S., R. Troller, I. N. Grivea, G. A. Syrogiannopoulos, and C. Aebi. 2002. The outer membrane proteins UspA1 and UspA2 of Moraxella catarrhalis are highly conserved in nasopharyngeal isolates from young children. Vaccine 20:1754-1760. [DOI] [PubMed] [Google Scholar]
- 25.Möllenkvist, A., T. Nordstrom, C. Hallden, J. J. Christensen, A. Forsgren, and K. Riesbeck. 2003. The Moraxella catarrhalis immunoglobulin D-binding protein MID has conserved sequences and is regulated by a mechanism corresponding to phase variation. J. Bacteriol. 185:2285-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murphy, T. F. 1996. Branhamella catarrhalis: epidemiology, surface antigenic structure, and immune response. Microbiol. Rev. 60:267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murphy, T. F., A. L. Brauer, B. J. Grant, and S. Sethi. 2005. Moraxella catarrhalis in chronic obstructive pulmonary disease: burden of disease and immune response. Am. J. Respir. Crit. Care Med. 172:195-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nilsson, G., J. G. Belasco, S. N. Cohen, and A. von Gabain. 1987. Effect of premature termination of translation on mRNA stability depends on the site of ribosome release. Proc. Natl. Acad. Sci. USA 84:4890-4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peak, I. R. A., M. P. Jennings, D. W. Hood, M. Bisercic, and E. R. Moxon. 1996. Tetrameric repeat units associated with virulence factor phase variation in Haemophilus also occur in Neisseria spp. and Moraxella catarrhalis. FEMS Microbiol. Lett. 137:109-114. [DOI] [PubMed] [Google Scholar]
- 30.Pearson, M. M., E. R. Lafontaine, N. J. Wagner, J. W. St. Geme III, and E. J. Hansen. 2002. A hag mutant of Moraxella catarrhalis strain O35E is deficient in hemagglutination, autoagglutination, and immunoglobulin D-binding activities. Infect. Immun. 70:4523-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pearson, M. M., C. A. Laurence, S. E. Guinn, and E. J. Hansen. 2006. Biofilm formation by Moraxella catarrhalis in vitro: roles of the UspA1 adhesin and the Hag hemagglutinin. Infect. Immun. 74:1588-1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pelton, S. I. 2005. Otitis media: re-evaluation of diagnosis and treatment in the era of antimicrobial resistance, pneumococcal conjugate vaccine, and evolving morbidity. Pediatr. Clin. N. Am. 52:711-728. [DOI] [PubMed] [Google Scholar]
- 33.Pericone, C. D., D. Bae, M. Shchepetov, T. McCool, and J. N. Weiser. 2002. Short-sequence tandem and nontandem DNA repeats and endogenous hydrogen peroxide production contribute to genetic instability of Streptococcus pneumoniae. J. Bacteriol. 184:4392-4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rocha, E. P. 2003. An appraisal of the potential for illegitimate recombination in bacterial genomes and its consequences: from duplications to genome reduction. Genome Res. 13:1123-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 36.Seib, K. L., I. R. A. Peak, and M. P. Jennings. 2002. Phase variable restriction-modification systems in Moraxella catarrhalis. FEMS Immunol. Med. Microbiol. 32:159-165. [DOI] [PubMed] [Google Scholar]
- 37.Shimotsu, H., and D. J. Henner. 1986. Construction of a single-copy integration vector and its use in analysis of regulation of the trp operon of Bacillus subtilis. Gene 43:85-94. [DOI] [PubMed] [Google Scholar]
- 38.van Belkum, A., S. Scherer, L. van Alphen, and H. Verbrugh. 1998. Short-sequence DNA repeats in prokaryotic genomes. Microbiol. Mol. Biol. Rev. 62:275-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van der Woude, M. W., and A. J. Bäumler. 2004. Phase and antigenic variation in bacteria. Clin. Microbiol. Rev. 17:581-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Verduin, C. M., C. Hol, A. Fleer, H. van Dijk, and A. Van Belkum. 2002. Moraxella catarrhalis: from emerging to established pathogen. Clin. Microbiol. Rev. 15:125-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verduin, C. M., M. Kools-Sijmons, J. van der Plas, J. Vlooswijk, M. Tromp, H. van Dijk, J. Banks, H. Verbrugh, and A. Van Belkum. 2000. Complement-resistant Moraxella catarrhalis forms a genetically distinct lineage within the species. FEMS Microbiol. Lett. 184:1-8. [DOI] [PubMed] [Google Scholar]
- 42.Wang, W., A. S. Attia, L. Liu, T. Rosche, N. J. Wagner, and E. J. Hansen. 2006. Development of a shuttle vector for Moraxella catarrhalis. Plasmid 55:50-57. [DOI] [PubMed] [Google Scholar]
- 43.Wang, W., and D. H. Bechhofer. 1997. Bacillus subtilis RNase III gene: cloning, function of the gene in Escherichia coli, and construction of Bacillus subtilis strains with altered rnc loci. J. Bacteriol. 179:7379-7385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weinrauch, Y., T. Msadek, F. Kunst, and D. Dubnau. 1991. Sequence and properties of comQ, a new competence regulatory gene of Bacillus subtilis. J. Bacteriol. 173:5685-5693. [DOI] [PMC free article] [PubMed] [Google Scholar]